Short abstract
Adhesive dynamics (AD) is a method for simulating the dynamic response of biological systems in response to force. Biological bonds are mechanical entities that exert force under strain, and applying forces to biological bonds modulates their rate of dissociation. Since small numbers of events usually control biological interactions, we developed a simple method for sampling probability distributions for the formation or failure of individual bonds. This method allows a simple coupling between force and strain and kinetics, while capturing the stochastic response of biological systems. Biological bonds are dynamically reconfigured in response to applied mechanical stresses, and a detailed spatio-temporal map of molecules and the forces they exert emerges from AD. The shape or motion of materials bearing the molecules is easily calculated from a mechanical energy balance provided the rheology of the material is known. AD was originally used to simulate the dynamics of adhesion of leukocytes under flow, but new advances have allowed the method to be extended to many other applications, including but not limited to the binding of viruses to surface, the clustering of adhesion molecules driven by stiff substrates, and the effect of cell-cell interaction on cell capture and rolling dynamics. The technique has also been applied to applications outside of biology. A particular exciting recent development is the combination of signaling with AD (so-called integrated signaling adhesive dynamics, or ISAD), which allows facile integration of signaling networks with mechanical models of cell adhesion and motility. Potential opportunities in applying AD are summarized.
Keywords: adhesion, integrins, inside-out signaling, Bell model, chemokine, HIV
1. Introduction
The first paper on adhesive dynamics (AD) was published 20 years ago [1]. Since then, AD has become widely used for a wide spectrum of problems in biological mechanics, as well as for applications outside of biology. The purpose of this review is to introduce AD, explain several demonstrated uses of AD, and to discuss new and emerging applications of the method to problems both within and outside biology.
Adhesive dynamics was first used to understand the dynamics of leukocyte adhesion under flow. Starting in the late 1980s, key experiments from the McIntire and Springer laboratories showed that when leukocytes were under flow, they displayed a wide variety of dynamic phenomena, including a curious dynamic state called rolling, in which receptor-ligand bonds formed and broke between the cell and substrate at a coordinated rate [2,3]. Our laboratory had tried to get other cell types to roll via antibody-antigen interactions [4] with limited success, and it led us to consider ways of mapping the rheology of adhesion molecules to the dynamics of adhesion itself.
The groundwork for AD had been laid by three key previous papers. The first was the classic paper by Bell [5], who was the first to suggest that the kinetic rate of failure of adhesion bonds should be modulated by force, F,
| (1) |
where kf is the observed off rate, is the unstressed off rate (in the absence of force), γ is the length to the transition state, and kbT the thermal energy [5]. We refer to γ as the reactive compliance, as it sets the sensitivity of a bond to dissociation to a force; the larger the reactive compliance, the more sensitive the molecule. The Bell model only allows for cases where force accelerates dissociation, but it is now known that exceptions exist [6]. The second paper was a simple model of cell capture under shear flow that Doug Lauffenburger and I wrote, in which we could determine whether adhesion was controlled by kinetics (when the reaction rates were too slow) or by mechanics (when the reaction rates were fast) [7]. Finally, Micah Dembo and I, with colleagues, developed a tape peeling model of the dynamics of adhesion, in which the rate of peeling under an applied tension could be calculated from the density, strength and mechano-chemical response of the adhesion molecules [8]. This was the first paper to introduce the concept of “slip” and “catch” bonding, in which forces could accelerate or inhibit the dissociation of bonds. This paper suggested that membrane peeling was due to a small amount of bond slippage, in which the bond failure rate has to increase with force, but not too fast to prevent continued adhesion with the substrate.
2. Methodology
When I constructed AD, it was from a simple desire to understand how various modes of bond failure would lead to the dynamics of adhesion. I also wanted a model in which the mechanical energy balance was satisfied rigorously, without simplifying assumptions. Also, the dynamics of rolling showed significant saltation [3], which I inferred were due to small numbers of adhesive bonds between the cell and substrate. This required a method that had some stochasticity.
Adhesive dynamics is a simulation, in which the current configuration is used to calculate the likelihood of future events. The method is Eulerian, in that current information is used to calculate the configuration of the system at the end of the next time step. AD has four principle elements: a rendering of each adhesion molecule, a mechanical energy balance, a chemical reaction scheme, and a prescription for how the particle and all its attached parts will advance for every time step. We keep track of the end point position of each molecule, which was made possible by computing power that easily allowed tracking tens of thousands of receptors. The mechanical energy balance solves F = m a for a rigid spherical particle. However, because the particle Reynolds number is low, we ignored inertia, and the velocity emerged from a balance of forces
| (2) |
from which the velocity can be determined since it is incorporated in the drag. Note, the force balance includes the torque on the particle, and both the velocity and the angular velocity are determined in all directions. The mobility matrix, which includes the drag in all directions, including cross terms, was assembled from known solutions of the motion of a hard sphere near a wall in viscous shear flow [1]. From this relationship, the instantaneous velocity is known from the sum of applied forces. The adhesive bonds were modeled as springs, so calculating the net adhesive force acted along the bond axis in proportion to strain was straightforward. The nonspecific forces are body forces which include gravity, van der Waals attraction and steric stabilization forces that prevent the particle from getting too close to the wall [1].
In AD, the connection point between each adhesion molecule and the cell body is known. Generally, kinetics are coupled to either the strain on the molecules, or the force. Dembo and co-workers posited a relationship between dissociation and strain which involved the unstressed forward and reverse rates, the spring constant, σ, and a transition state spring constant, σts [8]; this formulation was used in the earliest versions of AD. Once the kinetics are known, calculating the formation and breakage of bonds during each time step of the simulation is done by sampling random numbers and comparing to calculated probabilities of events. These probabilities are,
| (3) |
where Pf is the probability of formation in the time step Δt corresponding to the forward reaction rate, kf, which depends on the distance between unbound ligands and receptors; and Pr is the probability of failure of a bond in the time step, which results from kr, which depends on the molecule's length or force. Therefore, there is natural coupling between mechanics and bond failure; this feature of AD has been widely adopted into many biomechanical simulations [9]. This feature of the algorithm is so obvious (at least now) and so compelling that it is often subconsciously incorporated into simulations of force driven bond failure. Each unbound and bound receptor pair is interrogated in each time step to determine whether bonds will form or break, but the reconfiguration of the system is enacted at the end of the time step.
Correspondingly, the velocity, as calculated from Eq. (2), determines the kinematics of the particle, and all the attached receptors, during the time step. The particle, and the positions of all receptors, are repositioned according to the velocity. In the original version of AD, the particle exhibited rigid body motion, but in later versions of AD, deformation of the membrane or cell must be taken into account to accurately reposition the receptors.
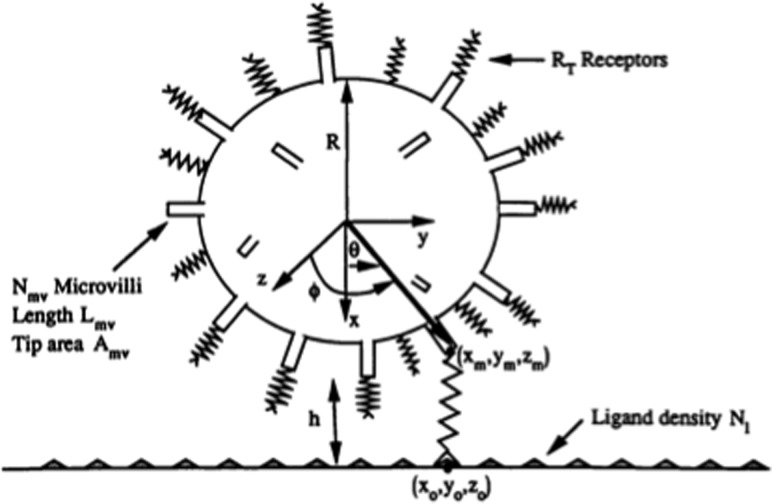
In the original version of AD, the cell was equipped with rigid microvilli of a number and topology that was suggested by electron micrographs of cells [10] (see Fig. 1). Several adhesion molecules, such as P-selectin glycoprotein ligand-1 (PSGL-1) and L-selectin are both known to cluster on the tips of microvilli [10,11], and given that microvilli lengths are much longer than the receptor length, only the receptors at the tips of microvilli are relevant for the early stages of leukocyte rolling. We assumed that the hydrodynamic resistance of the microvilli resembled that of a Brinkman bed [12] so the leukocyte behaved hydrodynamically as if it had an increased radius equal to that of the cell body plus the microvilli length.
Fig. 1.
Idealized rendering of a leukocyte in the original version of adhesive dynamics, where the leukocyte was modeled as a hard sphere with adhesive springs. The net motion of the cell comes from a balance of forces, easily derived from tracking the endpoints of each adhesion molecule. Figure reproduced from Ref. [1] with permission from Elsevier.
Because the method is Eulerian, all decisions about the behavior of the system at the end of the time step (t + Δt) are made at the current time, t. Eulerian methods are inherently unstable for large time-steps. Generally, the time step should be set to be smaller than the fastest event. Large time-steps lead to instabilities that can be either subtle or apparent, and numerical checks with smaller time-steps should be performed to make sure results are invariant. Also, one should be careful with the generation of random numbers; identical zeros can cause bonds to form and break in unlikely places, leading to further instabilities. Also, because AD is a simulation, it is a computer experiment, and like all experiments, it needs to be conducted some number of times to obtain a statistically meaningful result.
3. Relating Bond Mechanics to Dynamics of Adhesion
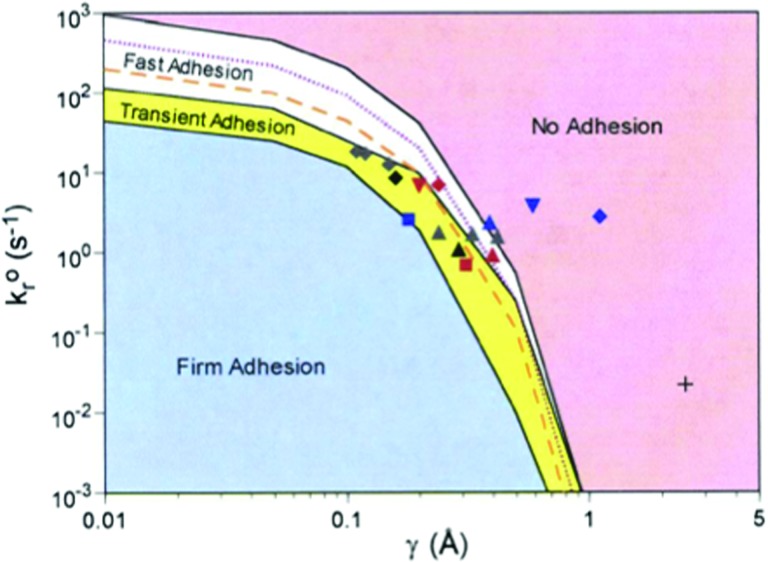
Using the Bell model to describe bond failure, we performed a series of calculation that related Bell model parameters ( and γ) to the dynamics of adhesion. The conclusions of both Dembo and co-workers [8] and Hammer and Apte [1] suggested that the mechanical response to bond failure controlled the dynamics of adhesion, but the relationship had not been systematically mapped. We therefore, performed simulations varying kro and γ, and characterized the results by the rolling velocity, with zero representing firm adhesion, small velocities (much less than the hydrodynamic velocity) representing rolling, and large velocities representing no adhesion. This simple binning of the dynamic states greatly oversimplifies the dynamics of adhesion, yet is useful to ascertain broad relationships between bond mechanics and dynamics. The main result, illustrated in Fig. 2, which was produced with all other parameters constant (such as on-rate and shear rate), illustrates a very small area in kro − γ gives rise to rolling [13]. Larger values of kro and γ make adhesion weaker, and small values of kro and γ support firm arrest. The symbols in the kro − γ envelope are independent measurements of Bell-model parameters for chemistries that are known to support rolling (for example, see Ref. [14]). Most of these measurements were taken by measuring the pause-times of leukocytes on sparsely-coated molecular surfaces under flow [14]. The mapping of these parameters, obtained from independent measurements, to the appropriate region of the phase diagram that corresponds to rolling gives great confidence in the ability of AD to simulate leukocyte adhesion.
Fig. 2.
The state diagram for leukocyte adhesion, reprinted from Ref. [13] with permission from the National Academy of Sciences. The dynamic states of leukocyte adhesion are calculated as a function of the unstressed off rate (kro) and reactive compliance (γ). Different dynamic states of adhesion are realized, including firm adhesion, transient or rolling adhesion, and no adhesion. Symbols represent independent measurements of kro- γ for molecules known to support rolling, consistent with the model predictions.
An insight that comes from the diagram in Fig. 2 is that every horizontal line (every constant value of kro) is a locus of constant chemical affinity, kf/kro, because the entire diagram is calculated at the same value of kf. It is clear that every dynamic state can be observed at one chemical affinity, illustrating that chemical affinity does not control the dynamics of adhesion. The dynamics of adhesion are controlled by other features of the molecules—their unstressed off rate and their mechano-chemical response.
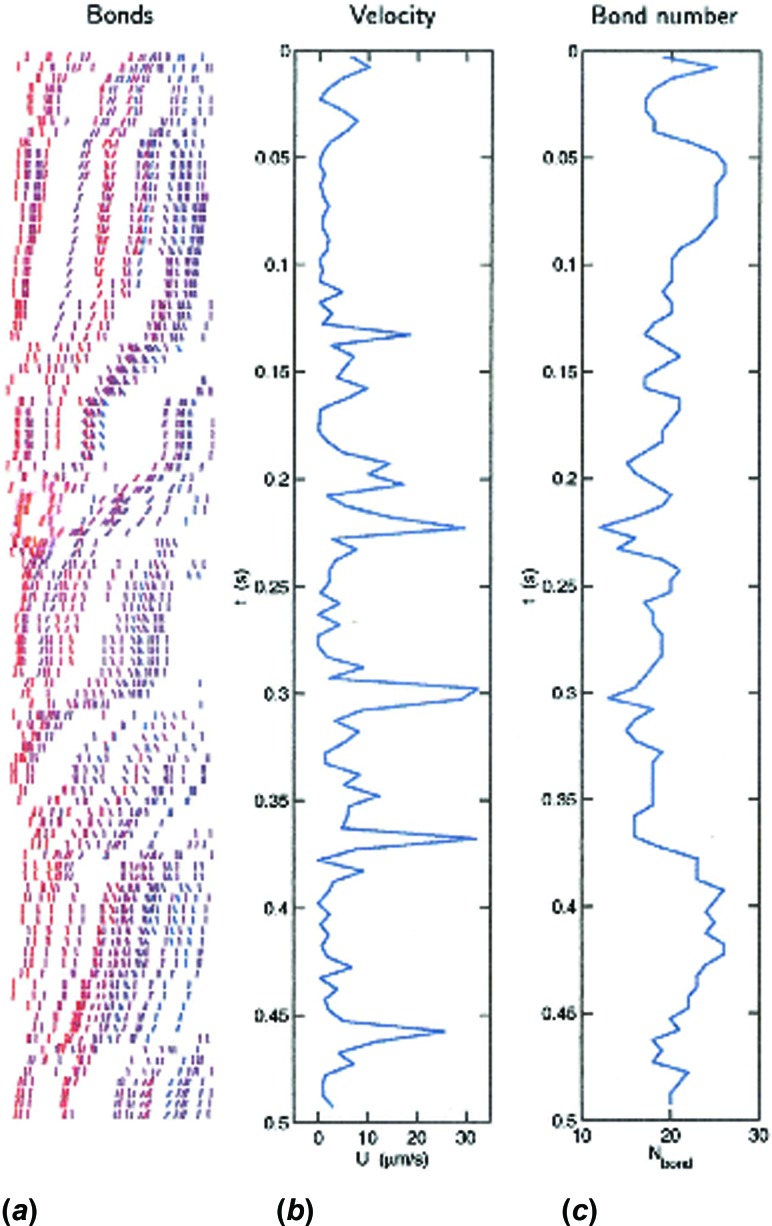
An illustration of the spatio-temporal dynamics during bond formation and breakage, published by King and Hammer [15], is shown in Fig. 3 [16]. This calculation was done for a hard sphere rolling over a ligand-coated substrate in Couette flow. The approximation of a hard sphere was appropriate because we had developed “cell-free” rolling in which the dynamics of rolling was recreated by decorating hard spheres with a selectin ligand, such as the tetra-saccharide sialyl-Lewisx [17]; we have made rigorous comparisons between the dynamics of bead rolling and the results of simulations [18] to verify that the model was correct. The left panel shows a spatio-temporal map of bonds in the contact zone between a bead and surface, in the reference frame of the bead, with each line segment indicating a bond, and red bonds under strain. As the particle moves to the right, bonds are convected to the back end of the contact zone to break. Therefore, rolling involves the coordinated formation of bonds at the front and breakage at the back. A spatio-temporal map of bond location and stress is a feature of every AD simulation. The right-most vertical panel shows how the total number of bonds fluctuates during the rolling motion of a cell over a surface. Clearly, the number of bonds fluctuates around an average, owing to the fact that small numbers of bonds control rolling. Correspondingly, minima in the number of bonds lead to maxima in the rolling velocity. This fluctuation explains the process of saltation, in which cells can roll with sudden leaps in velocity [19].
Fig. 3.
A simulation of a single particle rolling in Couette flow on a P-selectin surface, reproduced from Ref. [15] with permission of Elsevier. Panel (a) shows the distribution of bonds in the reference frame of the cell, which is moving from left to right. Bonds are line segments that are color coded, where red indicates bonds that are under strain. Bond are convected from the back to the front of the contact zone. Panel (c) shows the total number of bonds as a function of time, which fluctuates. The number of bonds needed to support rolling is very small. Panel (b) shows the rolling velocity, which fluctuates and is anti-correlated with the bond number.
Single molecule physicists have measured the failure rate of adhesion molecules as a function of the applied force. Insightfully, Evans used Kramer's rate theory to argue that the rate at which bonds are pulled could influence the rate of failure, and that bonds with two energy minima would be sensitive to changes in the rate of applied force, since this rate could affect the relative influence of each minima [20]. In some circumstances, this shifting of minima could make the bonds appear like catch bonds. It had been suspected that such apparent catch bond behavior could be responsible for biological effects such as the “shear threshold effect,” first observed by Springer's laboratory, in which adhesion under flow appears to increase, then decrease with shear rate [21]. An alternative explanation for the shear threshold effect would be that the on-rate for bond formation increases with shear rate [22], leading to an apparent increase in adhesion, before increasing shear caused bond failure. The relative contribution of these two competing effects—increased bond formation and shifting bond minima—were tested in AD simulations in two papers from our laboratory. We conclusively showed that the shear threshold emerges from the shifting minima in bond failure, owing to the emergence of minima with lower dissociation rates at higher shear rates, and not increasing bond formation rates at higher shear [23]. Further calculations by Beste and Hammer [6], showed when the shear threshold should emerge, and for precisely what conditions, and which molecules. For instance, a key prediction is that the conditions for which the shear threshold will emerge for P- and L-selectin are different, and these results are consistent with experiment [6].
4. Conditions for Firm Arrest
Leukocyte rolling is a precondition for firm arrest, but other adhesion molecules must participate in the adhesion cascade to give rise to firm binding. This is clear from the state diagram, which illustrates that selectins are equipped to support rolling, but not firm adhesion. In leukocytes, integrins are responsible for firm adhesion, either β 2-integrins which binds to ICAM-1, or that switch conformation in response to signals, or the β 1-integrin VLA-4 which supports rolling and then, through conformational change, takes on a conformation required for firm adhesion. Springer has performed work to identify the conformational states of integrins and integrin sub-domains, such as the I-domain, that are responsible for activation and firm adhesion [24].
The main question we addressed with AD was, what combination of integrins and selectins, and in what concentrations, are required for firm adhesion? One can expect the interaction between the two adhesion molecules to be complex, made more complicated by the switch in conformation and hence mechanical state of the β 2-integrin. But at a more basic level, one can ask a simple question: given a certain number of mechanically stronger integrins, and a certain number of selectins, what is the expected outcome of an encounter with a surface? Furthermore, can there be a synergistic relationship between the two molecules, such that the presence of one enhances the adhesive effect of the other. For example, for a given number of activated integrins, will the level of firm arrest be the same or greater if there are additional selectin interactions? We hypothesized, and later showed in cell free systems, that combinations of selectins and integrins could enhance firm adhesion synergistically [25].
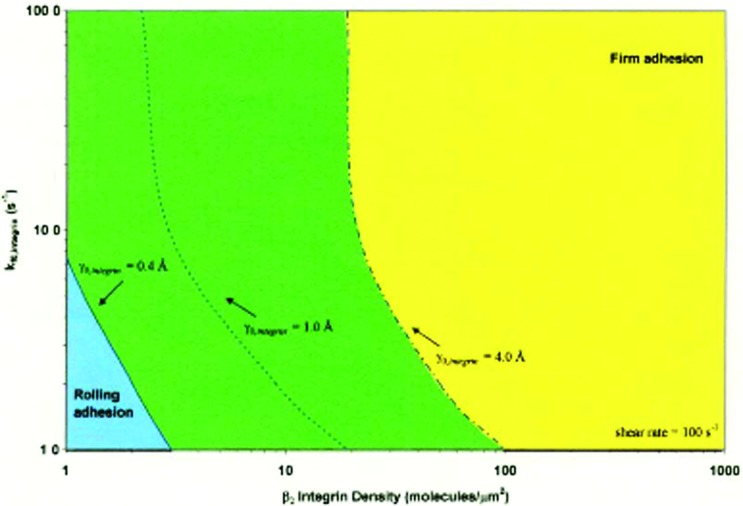
The goal of these calculations is to understand the requirements for firm adhesion. With a given level of selectins, which by themselves support rolling, how many additional interactions are necessary to give rise to firm adhesion? Since AD can straightforwardly be extended to multiple adhesion receptors, this calculation was a natural extension of the method which illustrates its power. The result is a Leukocyte Activation State Diagram, illustrated in Fig. 4, where one can assess how many integrins need to be activated, to what mechanical strength, on rate, and density, to secure firm binding, given a number of selectin interactions [26]. The change in mechanical strength is illustrated by a leftward shift in the dotted line, corresponding to lower values of the reactive compliance, which captures a larger region in firm binding. Thus, the diagram illustrates how much of a change in mechanical strength, or how many molecules must change mechanical strength, or how quickly they must react, to give firm arrest.
Fig. 4.
The leukocyte activation diagram, calculated in Bhatia and Hammer [26]. This diagram calculates the state of adhesion as a function of the properties of integrin receptors, including the on rate, the density, and the strength of integrins that are necessary to stop a leukocyte. Any combination of these parameters to the right of the dotted line indicates a cell will adhere firmly if the reactive compliance is below the number indicated on the line, or above the integrin density or greater than the on rate associated with the line. Integrin activation is tantamount to increasing integrin density (such as increased expression of Mac-1), increasing the integrin on-rate (moving up on this diagram) or moving the dotted line to the left, which captures a larger region of the diagram in firm adhesion. Reprinted from [26] with permission of the National Academy of Sciences.
5. Integrated Signaling Adhesive Dynamics (ISAD)
A reasonable question is how these integrins become activated through signaling pathways. There are several hypotheses that have been advanced for the onset of firm adhesion [27]. One is that chemokine receptors engage chemokines presented on endothelium during leukocyte rolling, leading to an outside-in signal that activates internal pathways, in turn activating integrins. The other is that engagement of integrins in passive, low strength states lead to an outside-signal through integrins that primes the internal signaling machinery, putting the integrins on a hair trigger. This priming may be enough to activate integrins by itself, or may need the assistance of a small amount of local chemokine.
The elucidation of the connection between signaling and adhesion requires some sort of integrated model. The model is integrated because it combines a mechanical model of adhesion, such as AD, with some description of the signaling pathways in a cell. Over the course of several years, we developed a sequence of such models, each of which incorporated unique architectural features of the signaling cascade. In all of these models, the signaling ultimately leads to changes in adhesion through a chemical reaction that convert an adhesion receptor between different biomechanical states. These activated integrins bind with mechanical strength to cognate ligands, increasing the probability of firm adhesion, as predicted by the state diagrams described above. A number of biochemical analyses have identified the key molecular players that bind to integrin receptor cytoplasmic tails, inducing conversion to mechanically strong states, including RapL, talin, and kindlin [28]. This class of simulation is called integrated signaling adhesive dynamics (ISAD).
ISAD was used to simulate the progressive rolling and firm adhesion of cells on chemokine coated substrates that bear both integrin and selectin ligands. We developed models that incorporated both deterministic and stochastic signaling networks, with increasing degree of detail for the signaling networks, from coarse graining to complete articulation of each element of the relevant biochemical pathways [29–31]. We focus on the most recent versions of these which employs the next subvolume method to simulate each biochemical pathway within the cell in each reaction volume [32]. Philosophically, the reason why the articulation of each biochemical step in the pathway is valuable is that this allows facile testing of models using the tools of molecular biology, including siRNA knock down and transfection. When signaling is coarse grained, the coupling between individual mutation and the signaling can be unclear.
The numerical strategy of this technique is similar to that employed in the Gillespie method [33], where all possible dynamic changes are placed in an event queue from which the most rapid events are pulled. In this case, time-steps in the AD part of the simulation, in which the mechanical steps of adhesion are considered, are compared to time-steps for the biochemical reactions in the signaling pathway, and the most rapid event is taken from the event queue.
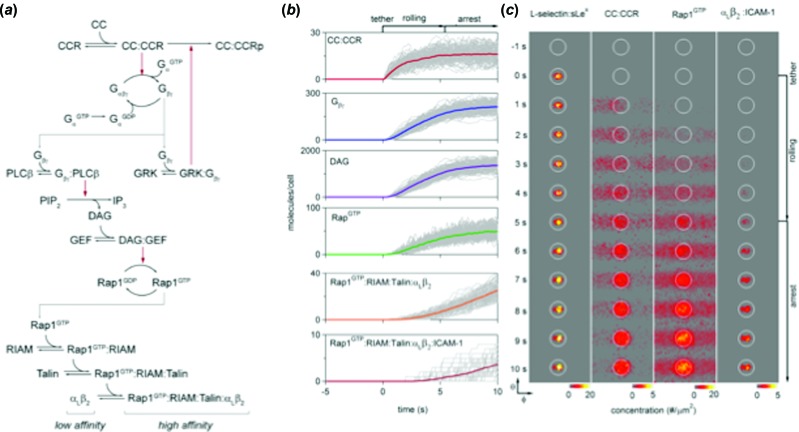
A characteristic simulated pathway and spatio-temporal result of simulation is shown in Fig. 5. Panel (a) illustrates the full signaling network that was simulated using the reaction-diffusion models. Panel (b) represents the temporal emergence of different molecules in a simulation of leukocyte stopping under flow on a substrate bearing selectin/ICAM-1/chemokine, starting with ligation of selectin to ligand, and progressing to down stream effectors, such as RapL, and finally activated integrins. Panel (c) shows the spatio-temporal emergence of each molecule under a projection of the contact zone during a trajectory during which the cell stops. Adhesion molecules are restricted to be within the contact zone but cytoplasmic signaling molecules diffuse away from the contact zone.
Fig. 5.
Integrated adhesive dynamics (ISAD) for the simulation of T-cell adhesion under flow. Panel (a) shows the reaction pathways that are simulated as a part of ISAD, starting with the chemokine and ending at the activation of the integrin into a high strength state. Panel (b) shows the progressive accumulation of each important molecule, starting from the chemokine and culminating in the ligation of Rap1 at the integrin. Panel (c) shows the spatio-temporal dynamics of molecular engagement from adhesion molecules to signaling molecules in the vicinity of the contact zone (the circle) during the progressive activation of the cell. Image taken from Ref. [32] with permission of the American Chemical Society.
These simulations were used to determine the effects of multiple chemokines on cell adhesion. Chemokine diversity is a major regulator of leukocyte activation, and chemokines can ligate multiple chemokine receptors. We used ISAD to explore the role that chemokines play in leukocyte stopping [32]. First, one question is whether chemokines drive leukocyte activation through repeated binding of receptors, or by the occupancy of receptors? Chemokine receptor turnover would be given as kf[CC] + kr, whereas total chemokine ligation would be given [CC]/([CC] + KD). We showed that cell stopping is correlated with total chemokine receptor occupancy. The simulations also provided insight into the effect that multiple chemokine receptors can have on cell stopping. If two chemokines bind the same receptor and operate through a single chemokine, the results are additive; the net effect on cell stopping is as if the two chemokines were added. However, if each chemokine acts through two separate receptors, and each of these receptors act through the same chemokine, the arrest dynamics suggest a synergistic interaction, where the effect of the two chemokines is greater than the sum of their concentrations [32]. The results of these simulations also present a hypothesis for how chemokines control cell recruitment, in that chemokines-chemokine receptor interactions need to be sufficiently weak, either in number or affinity, to give rise to selective recruitment of leukocytes.
6. Role of Cell Mechanics in Leukocyte Adhesion
Since bonds are under mechanical load in cell adhesion under flow, cell deformation should have a quantitative effect on the number and type of adhesion receptors that are needed for adhesion. Fundamentally, the dynamics of adhesion are controlled primarily by the mechano-chemical properties of adhesion receptors, as shown previously, so mechanics can modulate the dynamics, not define it. Yet, precise quantitative relationships between cell adhesion, numbers of receptors and cell mechanics require more accurate descriptors of cell shape.
There are two types of cell deformation—local and global. Local deformation includes deformation of microvilli, which are known to display viscoelastic deformation [34]. Global deformation includes calculation of the changes in shape of the cell. Side views of cells during rolling under high shear conclusively show that cells adopt a tear drop shape, which leads to increasing the contact zone and promoting adhesion [35].
Caputo and Hammer addressed the role of deformable, cylindrical microvilli in the dynamics of cell adhesion when the remainder of the cell body was rigid [36]. The simulations included microvilli that followed a classical Voigt model for viscoelastic deformation. Previously, Shao and Hochmuth made elegant measurements of the elasticity and viscosity of microvilli on human neutrophils [34]. We performed numerous simulations with different values of the spring constant and viscosity of the microvilli, and showed that minima in velocity for neutrophils rolling on selectins occurred at precisely the measured materials properties of the microvilli [36].
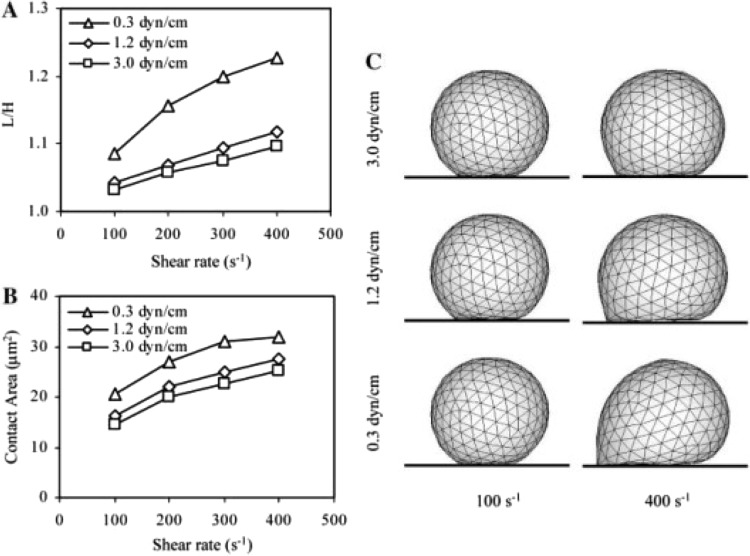
Several laboratories have addressed the ambitious undertaking of including all cell deformation in models of cell rolling. Jadhav and coworkers used an immersed boundary element method to calculate changes in cell shape under hydrodynamic stress and correspondingly calculate changes in cell shape for an elastic shell which surrounds and is surrounded by a viscous fluid [35]. Briefly, the technique requires placement of nodes into the boundary outlining the periphery of the cell, and then recalculates the Navier-Stokes equations for the displaced nodes. As illustrated in Fig. 6, these calculations illustrate that the contact area increases and the rolling velocity decreases when the cells are deformable. Khismatullin and Truskey performed a similar calculation that appeared a bit later, using a volume-of-fluid method for dealing with a more complex cell rheology that was more similar to neutrophil rheology [37].
Fig. 6.
Simulation by Jadhav and coworkers [35] on the effect of macroscopic deformation on the progressive rolling and firm adhesion of a cell, modeled with an elastic shell of varying stiffness. The calculations account for the deformation, spreading, increased contact zone and decreased rolling velocity for more deformable cells in higher shear fields. Reprinted from [35] with the permission of the authors and Elsevier.
7. Multicellular Adhesive Dynamics
A natural extension of AD would extend it to the interaction among multiple cells. The first steps were taken by including pairwise cell-cell interactions, in which the two cells could interact both adhesively and hydrodynamically. Adhesive dynamics was first extended to multiple cells by Michael R. King while he was a postdoctoral associate in my laboratory, who developed multiparticle adhesive dynamics (or MAD). In these calculations, the particles were rigid, and the hydrodynamics were resolved using a boundary integral method [15,16,38]. We performed a series of calculations that included a recreation of training of leukocytes, when leukocytes are rolling on substrates and they align behind each other without directly interacting molecularly [15]. The mechanism of training is that the stochastic fluctuation of cell motion in the direction normal to the flow ultimately leads to a preference for the cell to follow the streamline of the neighbor immediately preceding; fluctuations in motion that place a rolling particle in the wake of another rolling particle are favorable, and this effect continues until the particles are aligned [15]. Another calculation that was performed is to understand how already adherent cells can help capture other cells from a free stream [39]. This effect would have obvious physiological effects on cell recruitment during inflammation, where it is important that many cells marginate to the blood vessel wall in numbers for a robust cellular response. At first glance, it might appear that the ability to capture a cell downstream violates the reversibility of low Reynolds number Stokes flow, but in this case, the fluctuation in cell motion, due to bond fluctuation, breaks reversibility and allows incoming cells to cross streamlines. This crossing need be only tens of nanometers for the cells to come in molecular contact with the substrate, and as a result the capture zone could extend many cell diameters downstream of the already adherent particle [39].
Finally, we used MAD to perform a calculation on the effect of passing cells on the dynamics of adhesion that were already adherent. In these calculations, we did not allow any specific interaction between cells [38]. Reflexively, one might assume that collisions would lead to greater fluctuations in motion of already adherent rolling cells, but in actuality, the opposite is true. Passing “collisions” with already rolling cells increases their adhesion; the collisions lead to decreased fluctuations in the rolling velocity, because collisions press the already adherent cell close in contact with the substrate, increasing bond formation [38]. This effect was predicted by simulations and confirmed by experiments.
The increase in computational power makes it possible to increase volume fraction (hematocrit) of particles. The hematocrit of whole blood is 40–45 (volume fraction – 0.4–0.45), and researchers are progressively increasing volume fraction to approach a realistic simulation of whole blood, at least in a small vessels. As a note added in proof, Freund and coworkers found that at a hematocrit of 25%, collisions of red cells with a passing, firmly adherent leukocyte increase the normal force (in addition to increasing the force acting on the adherent particle in the direction of flow) [40]. I have no doubt that in the next decade we will see a fully realistic simulation of cell adhesion under flow in a blood vessel at the hematocrit of blood.
8. Beyond Leukocytes
Although AD was originally developed for leukocyte adhesion under flow, there is no a priori reason its use should be restricted to leukocyte adhesion. Recent developments have shown how AD can be used for other blood cells, such as platelets [41–45], sickle cells and infected erythrocytes [46–50], viruses [51,52] and bio-adhesive interfaces [53].
9. Blood Cell Adhesion
The King laboratory applied AD to dynamics of platelet adhesion, and platelet-surface interactions [41–44]. This version of AD is called platelet adhesive dynamics, or “PAD.” The complication in this problem is the hydrodynamics of motion of a platelet, which was modeled as a rigid oblate spheroid. The hydrodynamic are solved with a completed double layer-boundary integral equation method (CBL-BIEM). Because an oblate spheroid has two axes, the dynamics of motion is complicated by the orientation of these axes relative to the axes of extension and vorticity. The problem is further complicated when there are two platelets near the wall, and their principle axes are orientated arbitrarily relative to each other. As a result, the dynamics of platelet motion near a wall in shear flow are fascinating, and given to sudden changes of orientation, flipping, orbits, and other interesting dynamic phenomena [42]. The model was used to understand the role of von Willebrand factor in controlling platelet aggregation in flow via the GP-Ibα receptor [43], comparing between experiments and theory.
The long-range application for simulations such as these would be the formation of thrombi on damaged blood vessel walls. As a significant first step toward this process, PAD, was used to simulate adhesion and tethering oblate spheroids via GP-Ibα onto the damaged vessel wall followed by simulation of collisions between a flowing platelet with a downstream adherent platelet [54]. The simulations illustrated parts of platelets that would be responsible for platelet-platelet adhesive interactions. The simulations show that the platelet's spheroid shape plays a unique role in promoting both platelet-surface and platelet-platelet interactions.
Another application of AD is to the adhesion of blood cells infected with malaria. Fedosov and coworkers performed ambitious simulations of the adhesion and flipping of an infected red cell over a surface using AD, modeling the red cell as a network with 500 nodes with an appropriate rheology shown to faithfully recreate the material properties of the erythrocyte membrane [46]. The malarial parasite was included in the cell as a rigid body, and its shape, relative to the shape of the erythrocyte, induces higher variances in cell motion, and pronounced flipping of the red cell as it tethers to the surface [48]. Simulations of the average velocity of rolling motion agree with that observed for infected red cells. Additionally, the Karniadakis laboratory applied AD to the effects of sickle cell disease, and the effect sickle cells can have on the occlusion of blood flow in small vessels. These simulations involved the simulation of sickle cell adhesion to a wall with a simulation of the flow of blood cells in the vessel at a hematocrit approaching that of blood using a particle dissipation method. The simulation address the conditions of sickle cell shape, adhesiveness and elasticity that can cause the occlusion of a small blood vessel, resulting in a vasal occlusion crisis [55].
10. Brownian Adhesive Dynamics (BRAD)
Of course, AD can be used for the adhesion of particles of different sizes, provided the appropriate body forces are added to the simulations. For nano-particles or viruses, fluctuations in viral motion, caused by Brownian forces, can be included in the simulations. This version of AD is called brownian adhesive dynamics, or BRAD. The motion of the particles comes from solution of the Langevin equation with a fluctuating adhesive force added to the thermal forces to drive particle motion.
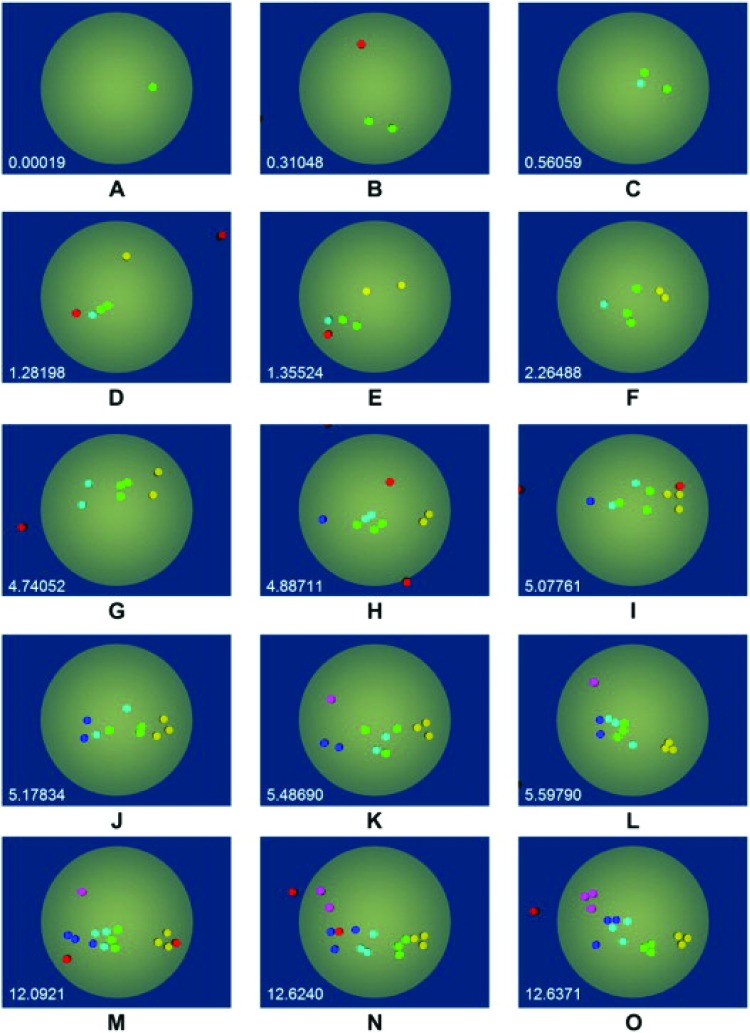
We used the method to simulate viral docking, with specific interest in the docking of HIV to T-cell surfaces [51,52]. The calculations were intended to make adjustments to the estimates of viral docking calculated by Hlavacek and Perelson, based on simple models for the binding of multivalent particles to surfaces that involved steric hinderance factors [56]. We reasoned that features such as receptor overlap and radius of curvature would affect the quantity of viral attachment proteins that could bind to the cell surface. Indeed, BRAD simulations show that while viral binding is rapid, the number of engaged VAPs is less than previously thought, due to geometric surface constraints [51,52]. However, we also used BRAD to address a conundrum in the dynamics of HIV binding, which is why the time scale for fusion of HIV at the cell surface is so long. It is known that gp-120, the glycoprotein for biding the T-cell receptors (TCR), is present as a trimer, attached to a single gp41 stalk. Each of the arms of the triskelion can be engaged to a single TCR. We used AD to explore the dynamics of the reorganization of the linked arms of gp120-TCR using AD, where we modeled the reorganization of TCRs in the T-cell membrane due to fluid drag [57]. The hypothesis was that the delay in time for entry is due to the delay for forming organized clusters of TCR that efficiently promote fusion. Illustrated in Fig. 7, we simulate the temporal reorganization of receptors in the T-cell interface, where the color-coding indicates TCR which are engaged to the same triskelion. After gp120-TCR bonds are formed, the combined actions of TCR diffusion and reconfiguration due to unbinding unfavorable configurations and reformation of favorable configurations lead to the formation of tight clusters of TCRs into distinct trimers over a period of minutes [57]. We hypothesize this delay accounts for the delay in HIV entry during fusion.
Fig. 7.
A simulation of the organization of TCR-gp120 bonds during the docking of HIV to the cell surface. Color coding indicates TCR that are bound to the same virus triskelion. The simulations indicate that after 12 s, the disorganized binding has minimized energy to lead to the formation of several well organized trimers. Figure reproduced from Ref. [57] with the permission of Elsevier.
11. Bio-Adhesive Interfaces
The principles of AD are not limited to the adhesion of a particle to a surface, but rather can be used for all biological processes that involve the mechanics of assembly via biological adhesion molecules that bear stress. In some sense, AD becomes an agent-based simulation, in which simple design rules for the formation and breakage of bonds govern how the assembly forms, breaks, and reconfigures. As such, the class of algorithms involving AD is probably much more widely applicable than has been previously realized.
Two recent examples include the clustering of adhesion receptors due to the mechanical resistance from the substrate or the glycocalyx [53], and the clutch model for the extension of filopodia [58].
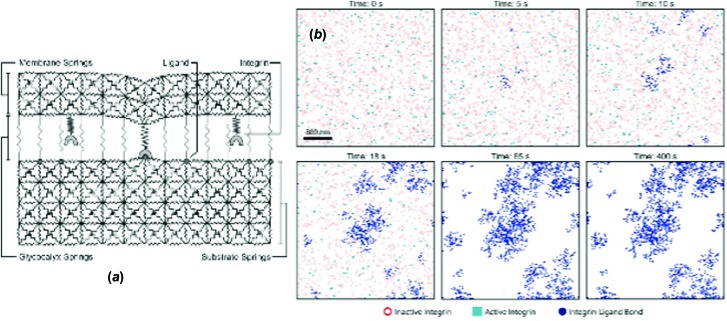
We developed a model of integrin receptor dynamics and clustering that was designed to address a number of seemingly unrelated observations. One observation is that cells, when placed on stiffer substrates, exert large traction forces and become invasive and motile [59]. A second observation is that cancer cells are known to have aberrant profiles of receptor glycosylation [60], and that steric stabilization is known to give rise to receptor clustering [61]. We developed a model, illustrated in Fig. 8, in which springlike receptors, embedded in an elastic membrane, bind to elastic substrates, among the resistance of springlike repellers. The two resistances—the glycocalyx and the elastic substrate—present an energetic penalty to the formation of bonds. The system responds by clustering the receptors, which minimizes the areas that have to be deformed or the areas in which the glycocalyx must be compressed. Increasing resistances, such as increasing substrate elasticity, leads to increasing receptor clustering and focal adhesion assembly, which in turn leads to increased signal transduction [53]. This mechanical effect may provide a simple explanation for the biochemical effects on cells due to changes in substrate elasticity [62].
Fig. 8.
AD simulation of the clustering of integrin receptors against elastic substrate with a glycocalyx. Panel (a) shows a schematic of the simulation, and panel (b) shows the progressive accumulation of receptor clusters on the adhesive interface, driven by resistance from both the glycocalyx and the stiffness of the substrate. Image taken from Ref. [53].
The principles of AD can be used to understand the dynamic interaction of extensible structures on adhesive substrates. Chan and Odde [9] developed a simple model for the extension of a filopod on a compliant substrate, in which integrins could dissociate and rebind to a substrate as a function of stress, using an AD like algorithm (using Eq. (3) to simulate the formation and breakage of adhesion bonds). The models predicted two regimes of filopodial extension, depending on the stiffness of the substrate: a frictional slippage on high stiffnesses and an oscillatory extension on softer substrates [9]. These calculations illustrate that AD can be used to calculate the formation and breakage of bonds, and the formation and dynamic reassembly of cells interacting with substrates under mechanical stiffness.
12. Concluding Remarks
Following the initial developments of AD, in which leukocyte adhesion under flow was faithfully recreated, substantial new uses for AD have emerged. Within leukocyte adhesion, the main new development, which should propel future research, is the integration of signaling with AD, to understand how intracellular processes can affect cell adhesion. Extension to different cell types, and to physiologically hematocrits, should allow the simulation of realistic dynamics in blood vessels, particularly in states of disease. The dynamics of viral binding may be simulated, although multiscale models of the behavior of viral attachment proteins, with a bit more sophistication than adhesive springs, may be necessary to bring these models to fruition. And finally, AD may be used to understand complex interfacial phenomena, such as the formation of cell clusters and filopodia, and with realistic, agent based models of receptor-cytoskeletal assembly, and the integration of signaling networks with interfacial mechanics, may be able to model the assembly of surface structures such as focal adhesions [63,64] and podosomes [65]. In the long term, AD may also be used for dynamic stochastic regulation of any stochastic cell process, including gene regulation, enzymatic reactions, and channel opening.
Acknowledgment
We are grateful to the NIH through support from Grants HL18208 and AI082292.
References
- [1]. Hammer, D. A. , and Apte, S. M. , 1992, “Simulation of Cell Rolling and Adhesion on Surfaces in Shear Flow: General Results and Analysis of Selectin-Mediated Neutrophil Adhesion,” Biophys. J., 63, pp. 35–57. 10.1016/S0006-3495(92)81577-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Lawrence, M. B. , Berg, E. L. , Butcher, E. C. , and Springer, T. A. , 1995, “Rolling of Lymphocytes and Neutrophils on Peripheral Node Addressing and Subsequent Arrest on ICAM-1 in Shear Flow,” Eur. J. Immunol., 25, pp. 1025–1031. 10.1002/eji.1830250425 [DOI] [PubMed] [Google Scholar]
- [3]. Lawrence, M. B. , McIntire, L. V. , and Eskin, S. G. , 1987, “Effect of Flow on Polymorphonuclear Leukocyte/Endothelial Cell Adhesion,” Blood, 70(5), pp. 1284–1290. [PubMed] [Google Scholar]
- [4]. Tempelman, L. A. , and Hammer, D. A. , 1994, “Receptor-Mediated Binding of IgE-Sensitized Rat Basophilic Leukemia Cells to Antigen-Coated Substrates Under Hydrodynamic Flow,” Biophys. J., 66, pp. 1231–1243. 10.1016/S0006-3495(94)80907-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Bell, G. I. , 1978, “Models for the Specific Adhesion of Cells to Cells,” Science, 200, pp. 618–627. 10.1126/science.347575 [DOI] [PubMed] [Google Scholar]
- [6]. Beste, M. T. , and Hammer, D. A. , 2008, “Selectin Catch-Slip Kinetics Encode Shear Threshold Adhesive Behavior of Rolling Leukocytes,” Proc. Natl. Acad. Sci. U.S.A., 105(52), pp. 20716–20721. 10.1073/pnas.0808213105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Hammer, D. A. , and Lauffenburger, D. A. , 1987, “A Dynamic Model for Receptor-Mediated Cell Adhesion to Surfaces,” Biophys. J., 52, pp. 475–487. 10.1016/S0006-3495(87)83236-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Dembo, M. , Torney, D. C. , Saxman, K. , and Hammer, D. A. , 1988, “The Reaction-Limited Kinetics of Membrane-to-Surface Adhesion and Detachment,” Proc. R. Soc. London, Ser. B, 234, pp. 55–83. 10.1098/rspb.1988.0038 [DOI] [PubMed] [Google Scholar]
- [9]. Chan, C. E. , and Odde, D. J. , 2008, “Traction Dynamics of Filopodia on Compliant Substrates,” Science, 322(5908), pp. 1687–1691. 10.1126/science.1163595 [DOI] [PubMed] [Google Scholar]
- [10]. Bruehl, R. E. , Springer, T. A. , and Bainton, D. F. , 1996, “Quantitation of L-Selectin Distribution on Human Leukocyte Microvilli by Immunogold Labeling and Electron Microscopy,” J. Histochem. Cytochem., 44(8), pp. 835–844. 10.1177/44.8.8756756 [DOI] [PubMed] [Google Scholar]
- [11]. Picker, L. J. , Warnock, R. A. , Burns, A. R. , Doerschuk, C. M. , Berg, E. L. , and Butcher, E. C. , 1991, “The Neutrophil Selectin LECAM-1 Presents Carbohydrate Ligands to the Vascular Selectins ELAM-1 and GMP-140,” Cell, 66(5), pp. 921–933. 10.1016/0092-8674(91)90438-5 [DOI] [PubMed] [Google Scholar]
- [12]. Bussell, S. J. , Koch, D. L. , and Hammer, D. A. , 1995, “The Effect of Hydrodynamic Interactions on the Diffusion of Integral Membrane Proteins. Diffusion in Plasma Membranes,” Biophys. J., 68, pp. 1836–1849. 10.1016/S0006-3495(95)80360-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Chang, K. C. , Tees, D. F. J. , and Hammer, D. A. , 2000, “The State Diagram for Cell Adhesion Under Flow: Leukocyte Rolling and Firm Adhesion,” Proc. Natl. Acad. Sci. U.S.A., 97(21), pp. 11262–11267. 10.1073/pnas.200240897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Alon, R. , Hammer, D. A. , and Springer, T. A. , 1995, “Lifetime of the P-selectin: Carbohydrate Bond and its Response to Tensile Force in Hydrodynamic Flow,” Nature, 374, pp. 539–542. 10.1038/374539a0 [DOI] [PubMed] [Google Scholar]
- [15]. King, M. R. , and Hammer, D. A. , 2001, “Multiparticle Adhesive Dynamics. Interactions Between Stably Rolling Cells,” Biophys. J., 81(2), pp. 799–813. 10.1016/S0006-3495(01)75742-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. King, M. R. , Kim, M. B. , Sarelius, I. H. , and Hammer, D. A. , 2003, “Hydrodynamic Interactions Between Rolling Leukocytes in vivo,” Microcirculation, 10, pp. 401–409. [DOI] [PubMed] [Google Scholar]
- [17]. Brunk, D. K. , and Hammer, D. A. , 1997, “Quantifying Rolling Adhesion With a Cell-Free Assay: E-Selectin and its Carbohydrate Ligands,” Biophys. J., 72(6), pp. 2820–2833. 10.1016/S0006-3495(97)78924-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Chang, K. C. , and Hammer, D. A. , 2000, “Adhesive Dynamics Simulations of Sialyl-Lewis(x)/E-Selectin- Mediated Rolling in a Cell-Free System,” Biophys. J., 79(4), pp. 1891–1902. 10.1016/S0006-3495(00)76439-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Lipowsky, H. , Riedel, H. D. , and Shi, G. S. , 1991, “in vivo Mechanical Properties of Leukocytes During Adhesion to Venular Endothelium,” Biorheology, 28(1–2), pp. 53–64. [DOI] [PubMed] [Google Scholar]
- [20]. Evans, E. , Leung, A. , Hammer, D. , and Simon, S. , 2001, “Chemically Distinct Transition States Govern Rapid Dissociation of Single L-Selectin Bonds Under Force,” Proc. Natl. Acad. Sci. U.S.A., 98(7), pp. 3784–3789. 10.1073/pnas.061324998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Finger, E. B. , Puri, K. D. , Alon, R. , Lawrence, M. B. , von Andrian, U. , and Springer, T. A. , 1996, “Adhesion Through L-Selectin Requires a Threshold Hydrodynamic Shear,” Nature, 379, pp. 266–269. 10.1038/379266a0 [DOI] [PubMed] [Google Scholar]
- [22]. Chang, K.-C. , and Hammer, D. A. , 1999, “The Forward Rate of Binding of Surface-Tethered Reactants: Effect of Relative Motion Between Two Surfaces,” Biophys. J., 76, pp. 1280–1292. 10.1016/S0006-3495(99)77291-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Caputo, K. E. , Lee, D. , King, M. R. , and Hammer, D. A. , 2007, “Adhesive Dynamics Simulations of the Shear Threshold Effect for Leukocytes,” Biophys. J., 92(3), pp. 787–797. 10.1529/biophysj.106.082321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Lu, C. , Shimaoka, M. , Ferzly, M. , Oxvig, C. , Takagi, J. , and Springer, T. A. , “An Isolated, Surface-Expressed I-Domain of the Integrin AlphaLbeta2 is Sufficient for Strong Adhesive Function When Locked in the Open Conformation With a Disulfide Bond,” Proc. Natl. Acad. Sci. U.S.A., 98, pp. 2387–2392. 10.1073/pnas.041606398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Eniola, A. O. , Willcox, J. , and Hammer, D. A. , 2003, “Interplay Between Rolling and Firm Adhesion Elucidated With a Cell-Free System Engineered With Two Distinct Receptor-Ligand Pairs,” Biophys. J., 85, pp. 2720–2731. 10.1016/S0006-3495(03)74695-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Bhatia, S. K. , King, M. R. , and Hammer, D. A. , 2003, “The State Diagram for Cell Adhesion Mediated by Two Receptors,” Biophys. J., 84, pp. 2671–2690. 10.1016/S0006-3495(03)75073-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Alon, R. , and Ley, K. , 2008, “Cells on the Run: Shear-Regulated Integrin Activation in Leukocyte Rolling and Arrest on Endothelial Cells,” Curr. Opin. Cell Biol., 20, pp. 1–8. 10.1016/j.ceb.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Morrison, V. L. , Macpherson, M. T. , Savinko, T. , San Lek, H. , Prescott, A. , and Fagerholm, S. C. , 2013, “The Beta2 Integrin-kindlin-3 Interaction is Essential for T-cell Homing but Dispensable for T-Cell Activation in vivo,” Blood, 122(8), pp. 1428–1436. 10.1182/blood-2013-02-484998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. Krasik, E. F. , Caputo, K. E. , and Hammer, D. A. , 2008, “Adhesive Dynamics Simulations of Neutrophil Arrest With Stochastic Activation,” Biophys. J., 95(4), pp. 1716–1728. 10.1529/biophysj.107.119677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Krasik, E. F. , Yee, K. Y. , and Hammer, D. A. , 2006, “Adhesive Dynamics Simulation of Neutrophil Arrest With Deterministic Activation,” Biophys. J., 91, pp. 1145–1155. 10.1529/biophysj.105.070706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Caputo, K. E. , and Hammer, D. A. , 2008, “Adhesive Dynamics Simulation of G-protein-mediated Chemokine-activated Neutrophil Adhesion,” Biophys. J. (submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Beste, M. T. , Lee, D. , King, M. R. , Koretzky, G. A. , and Hammer, D. A. , 2012, “An Integrated Stochastic Model of ‘Inside-Out’ Integrin Activation and Selective T-lymphocyte Recruitment,” Langmuir, 28(4), pp. 2225–2237. 10.1021/la203803e [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Gillespie, D. T. , 1975, “Exact Method for Numerically Simulating Stochastic Coalescence Process in a Cloud,” J. Atmos. Sci., 32(10), pp. 1977–1989. [DOI] [Google Scholar]
- [34]. Shao, J.-Y. , Ting-Beall, H. P. , and Hochmuth, R. M. , 1998, “Static and Dynamic Lengths of Neutrophil Microvilli,” Proc. Natl. Acad. Sci., 95, pp. 6797–6802. 10.1073/pnas.95.12.6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Jadhav, S. , Eggleton, C. D. , and Konstantopoulos, K. , 2005, “A 3-D Computational Model Predicts That Cell Deformation Affects Selectin-Mediated Leukocyte Rolling,” Biophys. J., 88(1), pp. 96–104. 10.1529/biophysj.104.051029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Caputo, K. E. , and Hammer, D. A. , 2005, “Effect of Microvillus Deformability on Leukocyte Adhesion Explored Using Adhesive Dynamics Simulations,” Biophys. J., 89, pp. 187–200. 10.1529/biophysj.104.054171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Khismatullin, D. B. , and Truskey, G. A. , 2005, “Three-Dimensional Numerical Simulation of Receptor-Mediated Leukocyte Adhesion to Surfaces: Effects of Cell Deformability and Viscoelasticity,” Phys. Fluids, 17, p. 031505. 10.1063/1.1862635 [DOI] [Google Scholar]
- [38]. King, M. R. , Rodgers, S. D. , and Hammer, D. A. , 2001, “Hydrodynamic Collisions Suppress Fluctuations in the Rolling Velocity of Adhesive Blood Cells,” Langmuir, 17(14), pp. 4139–4143. 10.1021/la010234b [DOI] [Google Scholar]
- [39]. King, M. R. , and Hammer, D. A. , 2001, “Multiparticle Adhesive Dynamics: Hydrodynamic Recruitment of Rolling Leukocytes,” Proc. Natl. Acad. Sci. U.S.A, 98(26), pp. 14919–14924. 10.1073/pnas.261272498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Isfahani, A. H. , and Freund, J. B. , 2012, “Forces on a Wall-Bound Leukocyte in a Small Vessel Due to Red Cells in the Blood Stream,” Biophys. J., 103(7), pp. 1604–1615. 10.1016/j.bpj.2012.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Mody, N. A. , and King, M. R. , 2007, “Influence of Brownian Motion on Blood Platelet Flow Behavior and Adhesive Dynamics Near a Planar Wall,” Langmuir, 23(11), pp. 6321–6328. 10.1021/la0701475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Mody, N. A. , and King, M. R. , 2008, “Platelet Adhesive Dynamics. Part I: Characterization of Platelet Hydrodynamic Collisions and Wall Effects,” Biophys. J., 95(5), pp. 2539–2555. 10.1529/biophysj.107.127670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43]. Mody, N. A. , and King, M. R. , 2008, “Platelet Adhesive Dynamics. Part II: High Shear-Induced Transient Aggregation via GPIbalpha-vWF-GPIbalpha Bridging,” Biophys. J., 95(5), pp. 2556–2574. 10.1529/biophysj.107.128520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Mody, N. A. , Lomakin, O. , Doggett, T. A. , Diacovo, T. G. , and King, M. R. , 2005, “Mechanics of Transient Platelet Adhesion to von Willebrand Factor Under Flow,” Biophys. J., 88(2), pp. 1432–1443. 10.1529/biophysj.104.047001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. Wang, W. , Mody, N. A. , and King, M. R. , 2013, “Multiscale Model of Platelet Translocation and Collision,” J. Comput. Phys., 244, pp. 223–235. 10.1016/j.jcp.2012.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Fedosov, D. A. , Caswell, B. , and Karniadakis, G. E. , 2010, “A Multiscale Red Blood Cell Model With Accurate Mechanics, Rheology, and Dynamics,” Biophys. J., 98(10), pp. 2215–2225. 10.1016/j.bpj.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Fedosov, D. A. , Caswell, B. , and Karniadakis, G. E. , 2011, “Wall Shear Stress-Based Model for Adhesive Dynamics of Red Blood Cells in Malaria,” Biophys. J., 100(9), pp. 2084–2093. 10.1016/j.bpj.2011.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48]. Fedosov, D. A. , Caswell, B. , Suresh, S. , and Karniadakis, G. E. , 2011, “Quantifying the Biophysical Characteristics of Plasmodium-Falciparum-Parasitized Red Blood Cells in Microcirculation,” Proc. Natl. Acad. Sci. U.S.A., 108(1), pp. 35–39. 10.1073/pnas.1009492108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. Fedosov, D. A. , Lei, H. , Caswell, B. , Suresh, S. , and Karniadakis, G. E. , 2011, “Multiscale Modeling of Red Blood Cell Mechanics and Blood Flow in Malaria,” PLoS Comput. Biol., 7(12), p. e1002270 10.1371/journal.pcbi.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. Pan, W. , Fedosov, D. A. , Caswell, B. , and Karniadakis, G. E. , 2011, “Predicting Dynamics and Rheology of Blood Flow: A Comparative Study of Multiscale and Low-Dimensional Models of Red Blood Cells,” Microvasc. Res., 82(2), pp. 163–170. 10.1016/j.mvr.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. English, T. J. , and Hammer, D. A. , 2005, “The Effect of Cellular Receptor Diffusion on Receptor-Mediated Viral Binding Using Brownian Adhesive Dynamics (BRAD) Simulations,” Biophys. J., 88(3), pp. 1666–1675. 10.1529/biophysj.104.047043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. English, T. J. , and Hammer, D. A. , 2004, “Brownian Adhesive Dynamics (BRAD) for Simulating the Receptor-Mediated Binding of Viruses,” Biophys. J., 86(6), pp. 3359–3372. 10.1529/biophysj.103.027813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Paszek, M. , Boettiger, D. , Weaver, V. M. , and Hammer, D. A. , 2009, “Integrin Clustering is Driven by Mechanical Resistance From the Glycocalyx and the Substrate,” PLoS Comput. Biol., 5(12), p. e1000604. 10.1371/journal.pcbi.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Wang, W. , Mody, N. A. , and King, M. K. , 2013, “Multiscale Model of Platelet Translocation and Collision,” J. Comput. Phys., 244, pp. 223–235. 10.1016/j.jcp.2012.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55]. Lei, H. , and Karniadakis, G. E. , 2013, “Probing Vasoocclusion Phenomena in Sickle Cell Anemia Via Mesoscopic Simulations,” Proc. Natl. Acad. Sci. U S A, 110(28), pp. 11326–11330. 10.1073/pnas.1221297110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56]. Hlavacek, W. S. , Posner, R. G. , and Perelson, A. S. , 1999, “Steric Effects on Multivalent Ligand-Receptor Binding: Exclusion of Ligand Sites by Bound Cell Surface Receptors,” Biophys. J., 76(6), pp. 3031–3043. 10.1016/S0006-3495(99)77456-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Trister, A. D. , and Hammer, D. A. , 2008, “Role of gp120 Trimerization on HIV Binding Elucidated With Brownian Adhesive Dynamics,” Biophys. J., 95(1), pp. 40–53. 10.1529/biophysj.107.118430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58]. Chan, C. E. , and Odde, D. J. , 2008, “Traction Dynamics of Filopodia on Compliant Substrates,” Science, 322(5908), pp. 1687–1691. 10.1126/science.1163595 [DOI] [PubMed] [Google Scholar]
- [59]. Paszek, M. , Zahir, N. , Johnson, K. R. , Lakins, J. N. , Rozenberg, G. I. , Gefen, A. , Reinhart-King, C. A. , Margulies, S. S. , Dembo, M. , Boettinger, D. , Hammer, D. A. , and Weaver, V. , 2005, “Tensional Homeostasis and the Malignant Phenotype,” Cancer Cell, 8, pp. 241–254. 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- [60]. Timpe, L. C. , Yen, R. , Haste, N. V. , Litsakos-Cheung, C. , Yen, T. Y. , and Macher, B. A. , 2013, “Systemic Alteration of Cell-Surface and Secreted Glycoprotein Expression in Malignant Breast Cancer Cell Lines,” Glycobiology, 11, pp. 1240–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61]. Ward, M. D. , and Hammer, D. A. , 1993, “Morphology of Cell-substratum Adhesion: Influence of Receptor Heterogeneity and Nonspecific Forces,” Cell Biophys., 20, pp. 177–222. [DOI] [PubMed] [Google Scholar]
- [62]. Wang, Y.-K. , and Chen, C. S. , 2013, “Cell Adhesion and Mechanical Stimulation in the Regulation of Mesenchymal Stem Cell Differentiation,” Journal of Cellular and Molecular Medicine, 27(7), pp. 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63]. Zhao, T. , Li, Y. , and Dinner, A. R. , 2009, “How Focal Adhesion Size Depends on Integrin Affinity,” Langmuir, 25(3), pp. 1540–1546. 10.1021/la8026804 [DOI] [PubMed] [Google Scholar]
- [64]. Yang, M. T. , Sniadecki, N. J. , and Chen, C. S. , 2007, “Geometric Considerations of Micro- to Nanoscale Elastomeric Post Arrays to Study Cellular Traction Forces,” Adv. Mater., 19(20), pp. 3119–3123. 10.1002/adma.200701956 [DOI] [Google Scholar]
- [65]. Gimona, M. R. , Buccione, S. A. , Courtneidge, and Linder, S. , 2008, “Assembly and Biological Role of Podosomes and Invadopodia,” Curr. Opin. Cell Biol., 20(2), pp. 235–241. 10.1016/j.ceb.2008.01.005 [DOI] [PubMed] [Google Scholar]