Abstract
Prions are self-propagating infectious protein isoforms. A growing number of prions have been identified in yeast, each resulting from the conversion of soluble proteins into an insoluble amyloid form. These yeast prions have served as a powerful model system for studying the causes and consequences of prion aggregation. Remarkably, a number of human proteins containing prion-like domains, defined as domains with compositional similarity to yeast prion domains, have recently been linked to various human degenerative diseases, including amyotrophic lateral sclerosis. This suggests that the lessons learned from yeast prions may help in understanding these human diseases. In this review, we examine what has been learned about the amino acid sequence basis for prion aggregation in yeast, and how this information has been used to develop methods to predict aggregation propensity. We then discuss how this information is being applied to understand human disease, and the challenges involved in applying yeast prediction methods to higher organisms.
Keywords: Yeast, Prion, Amyloid, Prion-like domains, Amyotrophic lateral sclerosis
Introduction
Prions result from the conversion of soluble proteins to an insoluble aggregated form. Typically, these aggregates are assembled into organized amyloid fibers with cross β-sheet structure and are capable of acting as self-propagating infectious agents sans nucleic acid [1]. Though this structural characterization of prion aggregates is generally accepted, the specific features that drive prion nucleation, aggregation, and propagation have proven more difficult to determine. Further complicating matters, prions can be subdivided into unique classes with fundamentally different features driving prion formation.
Several naturally occurring prion-forming proteins have been identified in Saccharomyces cerevisiae, including Mot3, Rnq1, Swi1, Cyc8, Sfp1, Mod5, Ure2, Sup35, and Nup100 [2–10]. All of these yeast prion proteins, with the exception of Mod5, contain prion-forming domains (PFDs) rich in glutamine and asparagine (Q/N). However, many other amyloid- and prion-forming proteins are not Q/N-rich, so this feature is not required for either amyloid formation or prion activity.
The presence of such a large number of proteins that can act as prions in yeast is somewhat enigmatic. It is clear that for some proteins, amyloid or prion formation can serve beneficial functions, acting as regulatory or structural elements [11]. However, the role of prions in normal yeast physiology is less clear. Some argue that prions may be advantageous to yeast under particular conditions, allowing them to act as means of survival and adaptation in fluctuating environments [12–14]. Others maintain that since yeast prions are relatively rare in wild strains despite their ability to form spontaneously and spread, these prions likely do not confer a selective advantage [15–18]. Rather, yeast PFDs may have evolved for reasons unrelated to prion formation.
The presence of prions in humans is currently thought to be limited to a single prion-forming protein, PrP, which is responsible for Creutzfeldt–Jakob disease (CJD), fatal familial insomnia (FFI), Gerstmann–Sträussler–Scheinker syndrome (GSS), and kuru [19]. Although many diseases involve protein misfolding, prions are distinguished from other protein-misfolding diseases based on transmissibility of the misfolded form. These PrP-associated prion diseases often have long incubation times; however, upon onset of initial symptoms, these diseases generally progress rapidly. Symptoms include neurodegeneration and progressive dementia, as well as other disease-specific pathologies. Currently, all of the human prion diseases are incurable and invariably fatal.
Although PrP is the only known human prion protein, many other human proteins contain “prion-like domains” (PrLDs), defined as regions with high compositional similarity to yeast PFDs [20, 21], and Q/N-rich proteins are overrepresented among certain eukaryotic genomes, including the human genome [22, 23]. A few of these PrLD-containing proteins have recently been linked to various neurodegenerative disorders in humans, suggesting that lessons learned from yeast prions may be applicable to non-prion neurodegenerative diseases, and that there may be additional beneficial or disease-associated prion-like domains in humans yet to be identified.
Although some progress has been made in predicting prion-like activity in yeast and in humans, it is clear that current prion prediction methods harbor significant limitations. Therefore, in this review we will discuss a brief history of yeast prion discovery and characterization, the systematic attempts to identify and predict prion behavior, ways that these predictions may be adapted and applied to human prion-like neurodegenerative disorders, and the challenges ahead in developing better prediction methods.
Yeast prion discovery and characterization
The first two yeast prions discovered, [PSI +] and [URE3], were initially identified in genetic screens as non-chromosomal genetic elements with non-Mendelian inheritance [24, 25]. However, the basis for this non-Mendelian inheritance was not initially known. Decades later, [PSI +] and [URE3] were proposed to be the prion forms of Sup35 and Ure2, respectively, based on their unusual genetic properties [9]. Subsequent careful analyses of these and other yeast prions have revealed a series of common sequence characteristics that have allowed for more targeted searches for new prion proteins.
Ure2 and Sup35 have similar domain layouts, with an N-terminal PFD that is responsible for prion activity but dispensable for the major cellular function of the prion protein (Table 1), and a C-terminal functional domain [26–30]. Sup35 contains an additional highly charged middle domain, termed “M”, which is not required for either prion formation or the normal cellular function of Sup35, but which helps to stabilize [PSI +] [31]. Sup35 and Ure2 are modular in nature, meaning the PFDs can be transferred to unrelated proteins and still support prion formation [32, 33].
Table 1.
Amyloid-based prions from Saccharomyces cerevisiae
Both the Ure2 and Sup35 PFDs are Q/N-rich and intrinsically disordered [34, 35]. Scrambling the primary sequence of the Ure2 and Sup35 PFDs does not does not eliminate the ability to form prions, indicating that amino acid composition, not primary sequence, is predominantly responsible for prion activity [36, 37]. The Sup35 and Ure2 PFDs share a number of compositional features, including an under-representation of charged and highly hydrophobic residues relative to the yeast proteome, and an over-representation of polar amino acids and glycine (Table 2).
Table 2.
Percent amino acid composition of yeast PFDs and human disease-associated PrLDs
| Gln/Asn | Ser | Gly | Tyr | Chargeda | Hydrophobicb | |
|---|---|---|---|---|---|---|
| Yeast PFDsc | ||||||
| Ure2 | 48.3 | 11.2 | 5.6 | 0 | 11.2 | 15.7 |
| Rnq1 | 43.1 | 15.4 | 16.7 | 5.9 | 2.4 | 8.3 |
| Sup35 | 45.6 | 3.5 | 16.7 | 17.5 | 4.4 | 3.5 |
| PFD averaged | 45.7 | 10.0 | 13.0 | 7.8 | 6.0 | 9.2 |
| Yeast genome | 10.0 | 9.0 | 5.0 | 3.4 | 24.0 | 28.3 |
| Human PrLDse | ||||||
| TDP-43 | 21.8 | 15.9 | 26.8 | 0.7 | 3.5 | 15.9 |
| FUS | 21.5 | 22.8 | 28.3 | 12.2 | 3.8 | 0.8 |
| TAF15 | 27.7 | 22.4 | 15.1 | 15.1 | 10.6 | 1.3 |
| EWSR1 | 18.9 | 15.4 | 9.6 | 13.6 | 3.6 | 4.0 |
| hnRNPA2B1 | 12.7 | 9.6 | 45.2 | 10.8 | 9.5 | 6.4 |
| hnRNPA1 | 12.3 | 16.0 | 42.2 | 8.0 | 9.7 | 8.1 |
| TIA1 | 31.6 | 4.2 | 15.8 | 9.5 | 2.1 | 12.6 |
| PrLD average | 20.9 | 15.2 | 26.1 | 10.0 | 6.1 | 7.0 |
| Human genome | 8.3 | 8.1 | 6.6 | 2.8 | 22.9 | 26.5 |
| PrPf | 12.8 | 5.7 | 9.9 | 7.8 | 20.6 | 19.9 |
aCharged residues include D, E, K, R
bHydrophobic residues include F, I, L, M, V
cPFDs are as defined in Table 1
dAverage of the Ure2, Sup35, and Rnq1 PFDs
eDisease-associated PrLDs found in RRM-containing proteins, as defined by the Alberti algorithm [21]
fAmino acids 90–230, which constitute the protease-resistant core of prion aggregates
Many of these same general features are also found in the other Q/N-rich PFDs. All of the known Q/N-rich PFDs are predicted to be intrinsically disordered. Each has relatively few charged and highly hydrophobic residues (for a detailed review of yeast PFD composition, see Du, 2011 [38]). Consequently, a number of the subsequent prions to be discovered were identified based on compositional similarity to known prions [39]. Other compositional biases, including biases towards serine, tyrosine, and glycine, are only seen in a subset of PFDs [38].
Minimum PFD length requirements vary
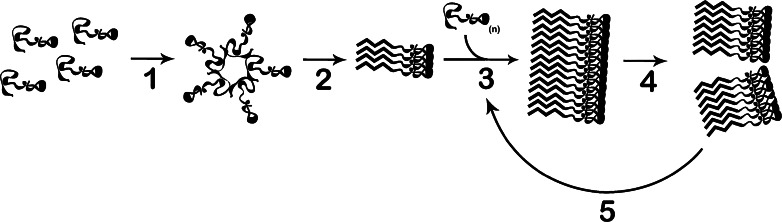
Although Sup35 and Ure2 contain clearly defined PFDs, determining the exact sequence features within these PFDs that are required for prion activity has proven more challenging. One challenge is that prion activity involves a series of discrete steps that may have distinct sequence requirements (Fig. 1). Prion proteins must be able to form prion aggregates (Fig. 1, steps 1 and 2). These prion aggregates must then be able to recruit additional soluble protein and convert it to the prion form (Fig. 1, Step 3). Finally, prion aggregates must be fragmented to generate new independently segregating aggregates (seeds) to offset dilution by cell division (Fig. 1, Step 4).
Fig. 1.
Basic steps in prion formation and propagation. Soluble proteins interact to form non-amyloid oligomers (step 1). These aggregates undergo a structural conversion to form amyloidogenic oligomers (step 2). The amyloidogenic aggregates recruit additional soluble proteins to form amyloid fibrils and to grow these fibrils (step 3). Fragmentation of these fibrils (step 4) creates new fiber ends for growth, while also creating new independently segregating aggregates to offset dilution by cell division
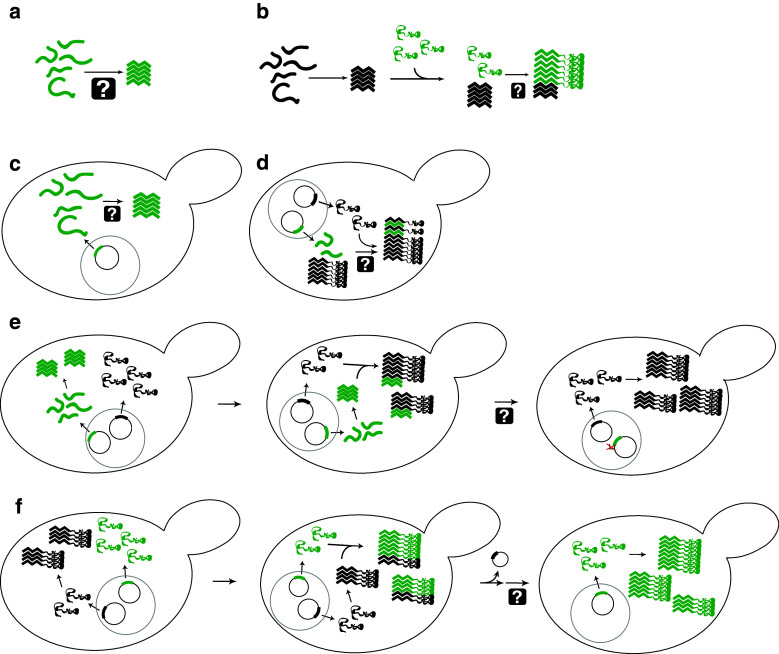
A wide variety of in vitro (Fig. 2a, b) and in vivo (Fig. 2c–f) assays have been developed to define the sequence elements required for prion activity, but many of these assays only test a subset of the steps in prion formation and propagation, and subtle differences in experimental set-up can lead to very different outcomes. Consequently, attempts to define these sequence requirements have yielded seemingly contradictory results, with some experiments suggesting that very short segments are responsible for driving prion formation, and other experiments indicating that larger regions are required for prion activity.
Fig. 2.
Assays to monitor prion-like activity and to define the regions of PFDs responsible for various aspects of prion activity. a In vitro aggregation. Protein fragments are incubated, generally with shaking, and aggregation is monitored using various techniques including Congo red binding, thioflavin T fluorescence, or pelleting assays. b Seeded in vitro aggregation. Preformed aggregates (green) are mixed with soluble protein (black) to test the ability of fragments to seed aggregation, or to test the ability of mutants to add onto preformed aggregates. c De novo aggregation in vivo. PFD fragments are transiently over-expressed. Aggregation is monitored either by fusing the fragments to GFP to observe foci formation, or through biochemical methods such as semi-denaturing detergent agarose gel electrophoresis (SDD–AGE). d Decoration of aggregates. PFD fragments are expressed in prion-positive cells to determine whether the fragments are capable of adding to preexisting aggregates. e Induction assays. PFD fragments (green) are transiently overexpressed in the presence of the full-length prion protein (black) to determine whether these fragments are sufficient to seed aggregation of the full-length protein. Aggregation is generally assayed by monitoring loss of function of the full-length protein. f Prion propagation assays. A prion-positive cell in which the chromosomal copy of the prion gene is deleted, but that carries a maintainer plasmid expressing the wild-type prion protein (black), is transformed with a plasmid expressing a prion protein mutant (green). The prion phenotype is assayed after selection for loss of the maintainer plasmid to determine if the mutant is capable of maintaining the prion
A widely used method to identify key nucleating segments within PFDs is to test the ability of mutated proteins to incorporate into wild-type prion aggregates either in vivo (Fig. 2d) or in vitro (Fig. 2b). Various single point mutations are sufficient to substantially reduce incorporation into wild-type Sup35 aggregates, both in vivo and in vitro [26, 40]. Many of these mutations cluster in a small 19-amino-acid segment of the Sup35 PFD (amino acids 8–26), suggesting a critical role for this segment. This segment also appears critical for mediating the [PSI +] prion species barrier between S. cerevisiae and C. albicans. Insertion of amino acids 8–26 from S. cerevisiae Sup35 into C. albicans Sup35 was sufficient to allow for efficient cross-seeding between S. cerevisiae and C. albicans Sup35 [41]. Other studies similarly indicate that short segments can play an important role in mediating the species barrier [42].
However, these short segments are not sufficient for prion activity. The Sup35 PFD contains two subdomains: an extreme N-terminal nucleation domain (amino acids 1–39) and an oligopeptide repeat domain (ORD; amino acids 40–114), which consists of five and a half copies of an imperfect nine-amino-acid sequence. The nucleation domain and the first repeat (amino acids 1–49) are required for incorporation into pre-existing aggregates. A slightly longer fragment (amino acids 1–64, which includes the first two repeats) is required for de novo aggregation (Fig. 2c) or induction of prion formation by full-length Sup35 (Fig. 2e; [43]). Furthermore, the ORD is necessary for efficient prion propagation (Fig. 2f); deletion of some or all of the repeats destabilizes or eliminates [PSI +] [43–45].
These experiments demonstrate that while short segments may act as mediators of prion aggregation, larger PFD segments are required for full prion activity, and different regions of a PFD are important for different aspects of this prion activity. Other studies further argue against the importance of short sequence motifs. The relative insensitivity of PFDs to scrambling suggests either that short sequence motifs are not important for prion activity or that the sequence requirements for any such motifs are sufficiently flexible that they are likely to be generated by random chance within scrambled PFDs. In addition, deletion analysis of one of the scrambled versions of Ure2 showed that while progressively larger truncations resulted in gradually decreasing prion-forming ability, no single segment within the PFD was absolutely required for prion activity [37]. Together, these results suggest that length and composition of PFDs are more important than any particular primary sequence element.
Curiously, much smaller segments are sufficient for in vitro aggregation (Fig. 2a). Six- and seven-amino-acid segments from Sup35 can form amyloid aggregates in vitro [46]. Likewise, eight-residue peptides from Ure2 form amyloid fibrils in vitro [47]. Peptide arrays of 20-amino acid fragments from Sup35 revealed multiple fragments spanning amino acids 9–39 that efficiently nucleate aggregation of the Sup35 PFD [48]. The basis for this dramatic difference in length requirements for in vitro versus in vivo aggregation is unclear.
Although some similar results have been seen for other PFDs, each has its own variations. For many of the prion proteins, the minimal prion domain has not been rigorously mapped, making it difficult to draw broad conclusions. For Ure2, amino acids 1–65 are sufficient to maintain [URE3] [28]. A smaller 42-amino-acid segment (amino acids 1,20–65) is capable of inducing prion formation by full-length Ure2 [37], but this fragment has not been tested for prion maintenance, so the exact minimum requirements for prion maintenance are unclear.
Intriguingly, amino acids 1–37 of Swi1 are sufficient for in vivo aggregation, induction, and transmission of the [SWI +] prion [49]; this fragment is notably shorter than other minimal PFDs. By contrast, Rnq1, which forms the [PIN +] (also known as [RNQ +]) has a much larger and more complex PFD. The PFD spans residues 153–405, and contains four Q/N-rich segments [50]. Three of these are capable of supporting amyloid aggregation in vitro [51]. Deletion of any one of the Q/N-rich segments does not result in loss of [PIN +] in vivo, indicating that Rnq1 contains multiple distinct prion determinants. Indeed, either the second or fourth Q/N-rich segment (spanning amino acids 218–263 and 337–405, respectively) is sufficient to maintain a very weak form of [PIN +] when fused to the non-Q/N-rich N-terminal domain (amino acids 1–132).
Collectively, analysis of these PFDs creates a series of challenges that must be accounted for in building effective prion prediction methods. Specifically, while short stretches appear to act as key nucleating elements, much longer segments are required for in vivo prion activity for each of the characterized PFDs. Furthermore, for most proteins, the exact boundaries for prion activity are not rigidly defined, as progressive PFD truncations frequently result in progressively diminishing prion activity. Finally, while PFD length seems to be a key factor in determining prion activity, the exact length requirements vary substantially between proteins.
Surrounding regions exert subtle effects on prion activity
Although PFDs are generally thought of as functionally independent domains, prion activity does appear to be somewhat context dependent. For example, one common assay for prion activity is carried out by replacing part or all of the PFD of Sup35 with a suspected PFD fragment from another protein and testing for loss of Sup35 activity. Although this method has helped identify new prions and candidate prion proteins in yeast [2, 7], the PFDs from two known yeast prions, Cyc8 and Mot3, show no prion activity when fused to the Sup35 C-terminus [2]. Conversely, the suspected PFD of another yeast protein, New1, shows prion activity when fused to Sup35 [52], but full-length New1 has not been shown to exhibit prion activity.
Mutations outside of PFDs can also substantially affect prion activity. For Ure2, deletion of an eight amino acid segment from the middle of the functional domain increases prion induction by about 100-fold [28]. For Sup35, select mutations or deletions within the C-terminal domain of wild-type Sup35 result in minor changes in prion formation efficiency [28, 53, 54]. Likewise, mutations in the M domain can affect the efficiency of [PSI +] propagation, potentially by affecting chaperone interactions [31, 55].
Regions outside core PFDs could influence prion activity by a variety of mechanisms. First, such regions could actively stabilize amyloid fibrils. For example, while the M domain of Sup35 is not required for amyloid aggregation, solid state NMR suggests that it may participate in cross-β-sheet interactions within Sup35 fibers [56]. Second, the non-prion domains could affect accessibility of the PFD; for example, the non-prion domains could directly bind to the PFD and reduce the PFD’s structural flexibility. Finally, non-prion domains could affect interactions with factors, such as chaperones, that influence amyloid aggregation.
Yeast chaperone Hsp104 mediates prion maintenance
Prion propagation in yeast requires fragmentation of prion aggregates to offset dilution by cell division (Fig. 1, Step 4; [57]). This fragmentation is carried out by the chaperone protein Hsp104, along with co-chaperones Hsp70 and Hsp40 ([58–61]; reviewed in [62, 63]). Deletion of HSP104 results in loss of the [PSI +] or [URE3] prions [58, 64]. Additionally, overexpression of Hsp104 results in loss of [PSI +] [65]. Prion loss due to Hsp104 overexpression has not been observed for any of the other yeast prions, although all of the known yeast prions, with the exception of [ISP +] [6], appear to be Hsp104-dependent [8, 10, 55].
Currently, it is unclear exactly how Hsp104 recognizes and fragments prion fibers. A long-standing model suggests that Hsp104, a hexameric member of the ATPases associated with diverse cellular activities (AAA) superfamily, recognizes amyloid fibrils, threads a single protomer through its central pore, and delivers it to associated chaperones for refolding [66]. If the extracted protomer was embedded within the fibril, the fibril would be fragmented, producing two daughter seeds. Recent in vitro evidence [67] supports a more subtle model of Hsp104-dependent amyloid fibril fragmentation in which, rather than threading the entire protomer through the pore, a single β-strand is only partially released from the fibril [68]. Initially, this fibril would still be intact but unstable. The freed β-strand could then either re-insert into the fibril thereby re-stabilizing it, remain free and cause the fibril to fragment spontaneously as a result of the instability, or integrate into another amyloid fibril, resulting in a large, stable aggregate.
Sup35 has been widely used as a model for studying the role of Hsp104 in prion propagation. Deletion of one or more of the 5½ repeats in the ORD results in both an unstable [PSI +] state and a larger average aggregate size in the remaining [PSI +] cells. This suggests a possible role for the ORD in Hsp104-dependent fibril recognition or cleavage. The ORD could either directly bind Hsp104 or alter the conformation of the prion fibrils to allow Hsp104 access [43, 45]. However, while the ability of the ORD to support [PSI +] propagation is dependent on the composition of this region, it is largely primary sequence independent [37, 69]. This observation argues against a primary sequence-dependent Hsp104 recognition site in the ORD, although it is possible that Hsp104 simply has highly flexible requirements for binding. More recent experiments suggest direct binding of Hsp104 to a short segment within the Sup35 M domain [55, 68]. Peptide arrays were used to identify a lysine-rich 20-amino-acid segment (amino acids 129–148) that binds Hsp104 in vitro; deletion of this segment reduces Hsp104 binding and ATPase activity, weakens the [PSI +] phenotype, and makes [PSI +] resistant to curing by Hsp104 over-expression [55, 68]. Although these results strongly suggest that this region acts as an Hsp104 binding site, the exact sequence requirements for this recognition have yet to be defined.
Predicting prion propensity in yeast
Attempts at prion prediction
Many algorithms have been generated to predict aggregation propensity, each using a unique set of parameters. Examples include BETASCAN [70], its more recent relative STITCHER [71], Zyggregator [72], Zipper DB [73], Tango [74], SALSA [75], PASTA [76], and Waltz [77]. Although many of these algorithms have successfully predicted some amyloid proteins, none have demonstrated the ability to predict either the aggregation activity or prion activity of Q/N-rich proteins [78]. The failure to predict prion activity is not surprising, as these algorithms are specifically designed to predict aggregation, and therefore do not account for the other steps in prion activity (Fig. 1). However, the inability to predict aggregation activity of Q/N-rich domains, as measured by both in vivo GFP fusion assays (Fig. 2c) and in vitro amyloid aggregation assays (Fig. 2a), suggests that there may be differences in the sequence requirements for aggregation between Q/N-rich and non-Q/N-rich proteins. Most amyloid prediction algorithms are designed to identify short, highly amyloidogenic peptide segments that seem to characterize the majority of non-Q/N-rich amyloid domains. However, it appears that yeast PFDs are characterized by relatively long stretches of disorder-promoting, moderately aggregation-prone amino acids, rather than short stretches of high amyloid propensity [79, 80].
Therefore, while short segments may be sufficient for aggregation either in isolation or in the context of non-Q/N-rich domains, their presence is not sufficient for aggregation activity in the context of Q/N-rich domains. For example, the structure-based algorithm ZipperDB uses a 6-amino-acid window size to identify aggregation-prone segments; sequences are threaded into a known NNQQNY amyloid-forming hexapeptide crystal structure and the energetic fit is determined [73, 81]. Remarkably, insertion of a single aggregation-prone 6-amino-acid segment into an exposed loop in RNAse A is sufficient to cause amyloid formation [82]. However, the same does not seem to be true for Q/N-rich proteins. Because Q/N-rich segments tend to be intrinsically disordered, the RNAse A result would seem to suggest that Q/N-rich regions containing ZipperDB-positive segments should form amyloid aggregates. Instead, ZipperDB-positive segments are found in many Q/N-rich domains that show little or no detectable amyloid aggregation activity, and the presence of ZipperDB-positive segments shows little correlation with amyloid aggregation propensity for Q/N-rich domains [2, 78].
Similar results are seen for Waltz, another prediction algorithm that uses a 6-amino-acid window size [77]. Maurer-Stroh et al. analyzed over 200 hexapeptide sequences for cross β-sheet structure formation, and used these results to generate a position-specific matrix to predict amyloid propensity. Waltz-positive amyloid stretches do appear to be more common in Q/N-rich proteins that show prion activity; in one analysis of 36 Q/N-rich proteins (half that show prion-like activity, and half that are unable to support either prion or amyloid formation), Waltz-positive segments were found in 89 % of the prion-like proteins, but only 50 % of the non-prion proteins [2, 78]. However, subsequent analysis suggests that this modest success is due predominantly to the compositional aspects of Waltz, not due to the position-specific components of the matrix. Amino acid composition is an inherent characteristic of any primary sequence motif. Therefore, any method focused on primary sequence inevitably runs the risk of misattributing compositional effects to primary sequence. In the Waltz scoring matrix, certain amino acids tend to be favored across most or all positions, so prions may tend to have more Waltz-positive sequences simply because they have more of these favored residues. A simple method to determine whether a primary-sequence-dependent algorithm like Waltz is truly identifying primary sequence patterns (rather than simply acting as an imperfect surrogate for assessing composition) is to make the algorithm blind to the original primary sequence of a test set of proteins by scrambling the sequences in silico and re-analyzing them with the algorithm. After scrambling, the prion sequences still had substantially more Waltz-positive segments than the non-prion sequences, suggesting that Waltz is detecting compositional differences between the prion and non-prion set [78].
Collectively, these results argue that algorithms that are built based on in vitro analysis of short fragments may have little ability to predict aggregation propensity of Q/N-rich proteins. However, the insensitivity of yeast PFDs to scrambling [36, 37] indicates a possible alternative prediction approach. Specifically, the dominant role of composition suggests that compositional similarity to known prions could be used to predict prion activity.
Curiously, this does not seem to be the case. Alberti et al. [2] used a Hidden Markov Model to identify the 100 yeast protein domains with greatest compositional similarity to known yeast PFDs. All candidates were tested in four prion-like activity assays. A remarkable number of proteins (18 out of 100) showed prion-like activity in all four assays, suggesting that compositional similarity does reasonably well at separating potential prion candidates from the bulk yeast proteome [2]. However, there was little correlation between the degree of compositional similarity to the known yeast PFDs and observed prion-like activity [80]. Other composition-based searches yield similar results: they successfully identify prion candidates but cannot identify the actual prion-forming proteins among those candidates [7, 22, 23].
It should be noted that it is difficult to evaluate exactly how good the Alberti et al. algorithm is at identifying prion candidates. The number of prions in yeast is not known; no one has tested what fraction of randomly selected protein fragments would show prion-like activity in these four assays, so there is no benchmark against which to judge the observation that 18 out of 100 tested fragments had clear prion-like activity. A related concern is that all four assays involve removal of the predicted PFDs from their native context, which may artificially inflate the number of fragments showing prion activity. Tartaglia et al. [83] have eloquently argued that evolutionary selection tends to reduce the aggregation propensity of proteins to just below the threshold for aggregation in their normal biological environment; consequently, even minor changes in sequence, expression level or environment may cause aggregation. Therefore, while the work of Alberti et al. provides strong data about the intrinsic aggregation propensity of each candidate PFD (and thus provides an incredibly powerful dataset for testing any prediction algorithm), it is possible that many domains showing prion-like activity in these assays will not form prions in their native context.
However, it is unlikely that this issue fully explains the large number of proteins showing prion-like activity in the Alberti et al. assays. Of the 100 proteins tested, the 50 with highest compositional similarity to known prions on average had significantly higher prion-like activity than the next 50 [78]; this suggests that the algorithm has some ability to enrich for likely prion candidates. But, among the top 50 proteins, there was actually a small, statistically insignificant inverse correlation between compositional similarity to known prions and prion-like activity, suggesting that the algorithm has no ability to distinguish among the top candidates. The simplest explanation for this apparent contradiction is that the sequence features that most clearly distinguish Q/N-rich PFDs from the rest of the proteome are not necessarily the same features that would be most effective at distinguishing Q/N-rich PFDs from non-prion-forming Q/N-rich domains.
A related issue is that compositional similarity analyses implicitly assume that all deviations from the known PFDs will decrease prion-forming capacity. In reality, prion formation is an exceedingly rare event, so it is unlikely that PFDs are optimized for maximum prion propensity. Therefore, it is possible that some compositional changes may increase prion propensity. More accurate prion prediction requires an understanding of how deviations from the compositions of known PFDs will affect prion activity.
Determining the prion propensity of each amino acid would provide a means to predict exactly how compositional changes will affect prion propensity. In a preliminary attempt to determine these prion propensities, a segment from a scrambled version of Sup35 was replaced with a random sequence, thereby generating a library of mutants [80]. By comparing the frequency of occurrence of each amino acid in the initial library to the frequency of the amino acid among the subset of mutants that maintained the ability to form prions, a prion propensity score was developed for each amino acid. In general, hydrophobic and aromatic amino acids were found to be strongly prion-promoting, polar amino acids were relatively neutral, and charged residues and prolines were strongly prion-inhibiting.
These prion propensity scores were then used to generate a prediction algorithm, called prion aggregation prediction algorithm (PAPA); http://combi.cs.colostate.edu/supplements/papa/ [78, 80]. PAPA uses a 41-amino acid sliding window, calculating the prion propensity of each window by averaging the prion propensity scores for each amino acid within the window. In addition to calculating prion propensity, PAPA uses the FoldIndex algorithm [84] to predict ordered and disordered regions within the protein. A key feature of yeast PFDs is that they are intrinsically disordered [34, 35]. Therefore, PAPA scores the overall prion propensity of each protein by identifying the 41 consecutive 41-amino acid windows that have both the highest predicted prion propensity and a negative fold index score (i.e., they are predicted to be disordered). The window size was chosen based on the observation that approximately 40 amino acids seem to be required for aggregation of most yeast PFDs, yet the flanking sequences can also affect aggregation propensity.
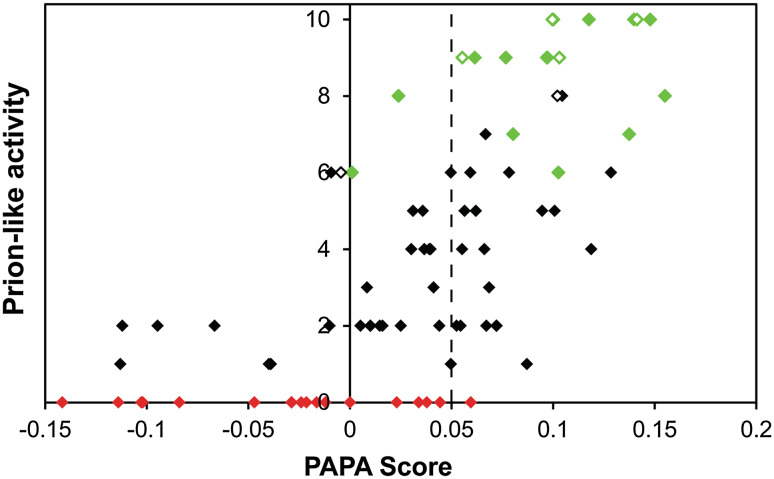
Strikingly, a strong correlation was seen between PAPA scores and observed prion propensity. The scores for the 100 domains tested by Alberti et al. ranged from approximately −0.13 to 0.15. A cutoff of 0.05 was most effective at discriminating between proteins with and without prion-like activity (Fig. 3). Of the 18 proteins that showed no prion-like activity in any of the Alberti et al. assays (Fig. 3, red diamonds), 17 scored below 0.05. By contrast, of the 18 proteins that showed prion-like activity in all four assays (Fig. 3, green diamonds), 16 scored above 0.05. Additionally, of the 37 proteins that scored above 0.05, 36 showed prion-like activity in at least one assay. However, this cut-off is not absolute. Among proteins that scored between 0.00 and 0.05, many showed at least some prion-like activity. Thus, these scores may be more accurately viewed as a gradient. In general, Q/N-rich proteins scoring below 0.00 are likely to have little or no prion activity; proteins scoring between 0.00 and 0.05 may have some prion activity; proteins scoring from 0.05 to 0.10 are likely to have some prion-like activity; and proteins scoring greater than 0.10 are likely to have strong prion activity. Further supporting the utility of PAPA, the algorithm was subsequently used to design completely synthetic Q/N-rich PFDs; when these domains were substituted in place of the Sup35 PFD, they were able to support prion activity [78].
Fig. 3.
PAPA predictions for yeast prion-like proteins. Alberti et al. identified the 100 proteins with greatest compositional similarity to known yeast prions. Each was tested in four assays for prion-like activity, and given a prion-activity score from 0 to 10 based on these results. PAPA was then used to predict the prion-like activity of each protein. Domains that were not testable in one or more assays are excluded. Proteins that showed prion-like activity in all four assays are indicated in green. Proteins that failed to show prion-like activity in any assay are indicated in red. Known PFDs are indicated with open diamonds. The PAPA cutoff shown to most effectively discriminate between proteins with and without prion-like activity (0.05) is indicated with a dotted line. However, this is not an absolute cut-off; between 0 and 0.05, most proteins show at least some prion-like activity. Updated and adapted from [80]
Surprisingly, there is little correlation between the frequency of occurrence of each amino acid among yeast PFDs and the amino acid’s PAPA score. As expected, charged residues and prolines have low prion propensity according to PAPA, consistent with their relative rarity in yeast PFDs [22, 80]. Unexpectedly, Q/N residues scored relatively neutral despite their prevalence in yeast PFDs, while hydrophobic residues, which are rare in yeast PFDs [22], scored as highly prion promoting. The importance of intrinsic disorder likely explains this apparent contradiction, and offers a simple theory to explain the compositional make-up of yeast PFDs [80]. The disordered nature of yeast PFDs makes the individual residues more accessible for prion formation. Q and N are likely common in yeast PFDs because they nicely balance prion propensity and disorder propensity. Most disorder-promoting residues are strongly aggregation-inhibiting. By contrast, Q/N residues promote intrinsic disorder while also providing a slight positive contribution to prion formation. In this context, very few hydrophobic residues are needed to drive aggregation. Additional hydrophobic residues would likely either make proteins excessively aggregation-prone, or create aggregates that are too stable, and thus not easily fragmented.
This theory also helps to reconcile other apparent contradictions. The proposed importance of short stretches for nucleating prion formation [40, 42, 48] seems to conflict with the insensitivity of PFDs to scrambling [36, 37]. However, if PFDs contain relatively few strongly prion-promoting amino acids, then the distribution of these amino acids will naturally create pockets of strong nucleating potential. Scrambling will simply redistribute these key amino acids, again creating nucleating sites wherever prion-promoting amino acids cluster. Indeed, the region spanning amino acids 8–26 of Sup35, which is thought to act as a critical nucleating site [40, 42, 48], contains two strongly prion-promoting amino acids (both tyrosine), and contains the longest stretch in the Sup35 PFD without any strongly prion-inhibiting amino acids. Thus, the presence of nucleating stretches can be rationalized based entirely on composition.
This may also explain why algorithms such as Waltz and ZipperDB show some ability to correctly identify key nucleating sites, but are less effective at distinguishing between proteins with and without prion-like activity. Consistent with the findings of Toombs et al. [80], aromatic and hydrophobic residues tend to score high at most positions in Waltz, while charged residues tend to score low at most positions. Therefore, although algorithms such as Waltz incorporate an additional layer of primary sequence, they may be acting predominantly as a screen for local amino acid composition.
Future challenges in yeast prion prediction
Although great strides have been made in predicting prion activity, much remains to be understood. The Alberti algorithm and PAPA have potentially complementary strengths and weaknesses (Table 3). The Alberti algorithm was very successful at identifying prion candidates from the S. cerevisiae genome, but could not accurately predict which of the candidates would demonstrate prion activity [2, 80]. Conversely, PAPA was able to accurately predict prion activity within the Alberti et al. candidate dataset, and could even be used to build synthetic PFDs, but it is unclear whether PAPA may itself be used to identify prion candidates from whole genomes [78]. A unified prediction method could improve prion prediction, but designing such an algorithm requires overcoming a number of current challenges.
Table 3.
Strengths and limitations of PAPA and the Alberti algorithm for prion prediction
| Strengths | Limitations |
|---|---|
| Alberti et al. [2] | |
| Built based on analysis of multiple PFDs | Ineffective at ranking the highest scoring candidates |
| Can identify prion candidates from proteomes | Ability to predict the effects of point mutations is unclear |
| High fraction of candidates show prion-like activity | Does not consider primary sequence effects |
| Does not consider effects of regions outside of the PFD or interactions with heterologous proteins | |
| PAPA | |
| Reasonably effective at ranking candidate PrLDs | Built based on mutagenesis of a small region of a single protein |
| Sufficient for de novo design of Q/N-rich PFDs | Validated only on Q/N-rich proteins |
| Uses experimentally derived prion propensity values for each amino acid, allowing for prediction of the effects of amino acid substitutions | Effectiveness for genomic searches is unclear |
| Does not consider primary sequence effects, other than proline spacing | |
| Does not consider effects of regions outside of the PFD or interactions with heterologous proteins |
One major challenge is the lack of good datasets on which to train and test potential algorithms. For example, the Alberti algorithm was trained on the four prion proteins that were known at the time: Sup35, Ure2, Rnq1, and the prion candidate New1 [2]. This small training set may have limited the algorithm’s prediction accuracy. One of the greatest contributions of the work of Alberti et al. is that it provides a large, rigorously tested dataset; importantly, it includes domains that compositionally resembled yeast PFDs, but that show no prion activity. Recently, Espinosa Angarica et al. [85] took advantage of this to develop a new prediction algorithm. They used the full set of prion-like proteins from the Alberti et al. dataset to develop a probabilistic representation of Q/N-rich PFDs. This algorithm was reasonably effective both at discriminating between proteins with and without prion activity from among the Alberti et al. dataset and at picking known PFDs out of larger datasets. The algorithm identified 20,540 predicted PFDs in 1,536 organisms, but none of these new candidates have yet been tested for prion activity.
Although the growing list of known PFDs provides a broader dataset for training potential algorithms, this dataset may contain its own biases. The majority of known yeast prions or prion-like domains were identified either because of their sequence similarity to Sup35 and Ure2 or due to their ability, when overexpressed, to support [PSI +] formation in a [pin −] strain [39]. Therefore, they may not provide a representative sample of all yeast prion proteins, and any algorithm that uses this set of proteins as a training set runs the risk of being too narrowly focused.
In theory, one advantage of PAPA is that, because it actually scores the prion propensity of each amino acid rather than simply looking for compositional similarity, it is not constrained by any biases present in the current set of known prions. However, PAPA faces its own challenges in scoring proteins whose composition deviates from that of Sup35. PAPA’s prion propensity scores for each amino acid are only estimates, based on a random sampling of prion and non-prion isolates from a library of scrambled Sup35 mutants; therefore, each prion propensity score carries large confidence intervals, which creates errors in PAPA’s predictions. The further a protein’s composition deviates from that of Sup35, the more these errors will likely compound. Additionally, PAPA assumes a linear relationship between the frequency of occurrence of a given amino acid and prion propensity. This assumption almost certainly is an over-simplification; some amino acids may have non-linear relationships with prion propensity or show a threshold effect. For example, within Sup35, insertion or deletion of a single Q/N residue generally has little effect on prion activity; however, it is possible that proteins with lower concentrations of Q/N residues might be more sensitive to changes in Q/N content. The more a protein’s composition deviates from that of Sup35, the higher probability that this sort of non-linear relationship will affect prediction accuracy.
While both PAPA and the Alberti algorithm focus on composition, there may be minor primary sequence elements that these algorithms do not account for. Scrambled versions of Sup35 and Ure2 form and maintain prions with different efficiencies, indicating slight primary sequence effects [36, 37]. Elucidating these subtle primary sequence features is difficult, because most experimental methods affect both primary sequence and composition. For example, deletion experiments remove specific primary sequence elements but concomitantly result in a disproportionate loss of particular amino acids, thereby altering composition. This could result in misattribution of compositional effects to primary sequence elements.
PAPA does include in its prediction method one commonly recognized primary sequence feature governing prion formation. Since proline is a known β-sheet breaker [86], the distribution of prolines can greatly affect prion formation [2, 80]. A cluster of prolines would be expected to disrupt β-strand formation at just a single location, while these same prolines dispersed across a sequence would result in multiple disruptions of the β-sheet structure. Accordingly, PAPA classifies any set of two or more prolines, separated by no more than one amino acid each, as a single proline. This is only one of potentially many subtle primary sequence features. However, analysis of the libraries used to build PAPA has not revealed any other clear primary sequence biases. There is neither clear co-variance between particular amino acids nor positional biases of individual amino acids. But, because of the limited library sizes, subtle effects could easily have been missed.
Another major challenge is that two recent studies suggest that the distinct steps required for prion activity (Fig. 1) may have distinct compositional requirements. As previously discussed, the nucleation domain of Sup35 (amino acids 1–39) is thought to be primarily responsible for the initial nucleating events in prion formation and for fiber growth (Fig. 1, steps 1–3), while the ORD is thought to be primarily responsible for chaperone-dependent prion maintenance (Fig. 1, step 4). While the activity of the ORD is primary-sequence independent, when the composition of the ORD is changed to match that of the nucleation domain, it is no longer able to support prion maintenance [69]; this suggests that prion formation and prion maintenance have distinct compositional requirements. Further complicating matters, recent evidence suggests that individual amino acids can have differential effects on the discrete steps in prion formation (Fig. 1, steps 1–3; [87]). Specifically, prion proteins are thought to first associate with each other to form soluble oligomers (Fig. 1, step 1), and these oligomers then undergo a structural conversion to form ordered amyloid fibrils (Fig. 1, steps 2–3). Interestingly, glutamines seem to promote the formation of soluble non-amyloid oligomers, while asparagines seem to promote the formation of mature amyloid fibrils [87]. Thus, an accurate prediction algorithm needs to consider not only the overall prion propensity of each amino acid, but also the effect of each amino acid on each step in prion formation and propagation.
Prion-like domains in disease
Many protein aggregation-based diseases, like the prion diseases, involve self-templating structural conversions. But, since most protein aggregates are not infectious, the prion diseases have historically been viewed as fundamentally distinct from other aggregation-based disorders. However, recent developments have begun to blur this distinction between prion and non-prion aggregation diseases [88–90].
There is growing evidence that various proteins implicated in many neurodegenerative disorders show prion-like behavior. Notable examples of these proteins include α-synuclein; amyloid precursor protein (APP) and tau; and Huntingtin. These proteins are implicated in Parkinson’s disease, Alzheimer’s disease, and Huntington’s disease, respectively [89]. Although the clinical manifestations of these disorders vary, the prion-like behavior of the implicated proteins is roughly the same. Presumably, an initial misfolding event results in aggregation. The aggregates can then induce other proteins to similarly misfold and aggregate. This aggregation is thought to originate in a single epicenter and spread to neighboring tissues by an unknown mechanism, imposing aggregation commitment on nearby cells [91]. Despite this remarkable similarity to prion propagation, the hesitation in classifying these neurodegenerative proteins as bona fide prions arises from the lack of evidence of transmission between individuals [89, 92].
Additionally, a number of proteins containing domains with similar amino acid composition to yeast PFDs have recently been linked to degenerative diseases (Table 4), further highlighting the connection between infectious and non-infectious protein aggregation diseases. Interestingly, many of these disease-associated PrLD-containing proteins also have RNA recognition motifs (RRMs) that generally do not overlap with the predicted PrLD’s. This suggests that disruptions in RNA homeostasis via prion-like aggregation may represent a common mechanism of degenerative disease. The Alberti algorithm predicts 246 out of 21,873 genes in the human genome to have sequences encoding PrLDs [20, 21]. This group contains a relatively high proportion of RNA-binding proteins—nearly 12 % of predicted PrLD-containing proteins also contain at least one RRM. Furthermore, 20 % of the top 60 proteins predicted to contain PrLDs also contained at least one RRM, raising the possibility that more of these proteins could eventually be linked to disease.
Table 4.
RNA-binding proteins containing PrLDs that have been linked to degenerative disease
| Protein | Alberti algorithm | PAPA analysis | Disease | ||
|---|---|---|---|---|---|
| PrLD Ranka | PrLD amino acids | PAPA score | Highest scoring segment (isoform)b | ||
| TDP-43 | 43 | 277–414 | 0.042 | 339–414 | ALS, FTLD |
| FUS | 12 | 1–237 | 0.109 | 21–121 (2) | ALS, FTLD |
| TAF15 | 22 | 1–152 | 0.127 | 12–92 (2) | ALS, FTLD |
| EWSR1 | 25 | 1–280 | 0.057 | 194–274 (1) | ALS, FTLD |
| hnRNPA2B1 | 32 | 197–353 | 0.043c | 241–321 (A2) | IBMPFD |
| hnRNPA1 | 38 | 186–372 | 0.093d | 257–337 (b) | ALS, IBMPFD |
| TIA1 | 53 | 292–386 | 0.131 | 269–349 (1) | Welander distal myopathy |
aPrLD rank among the human genome [21]
bSome of the proteins have multiple isoforms that differ either in their PAPA scores or the exact location of the highest scoring region. Shown are the amino acid positions for the highest scoring segments from the highest scoring isoform, with the isoform indicated in parentheses
cThe PAPA score for the disease-associated mutant is 0.088
d PAPA score for Isoform B, the highest scoring isoform. hnRNPA1 has two isoforms with substantially different PAPA scores. The IBMPFD-associated mutation increases the PAPA score of Isoform B to 0.125. Isoform A scores 0.041, but the IBMPFD-associated mutation increases the PAPA score to 0.087
RNA-binding proteins with prion-like domains in human disease
TDP-43 was the first PrLD-containing RRM protein to be associated with a degenerative disease. It was identified as a major component of aggregates in patients with either amyotrophic lateral sclerosis (ALS) or some forms of frontotemporal lobar degeneration (FTLD) [93]. TDP-43 contains two RRM’s and a C-terminal PrLD. Normally, TDP-43 is primarily located in the nucleus, but in patients with ALS, it is found in cytoplasmic inclusions [93]. Interestingly, TDP-43 has since been found in inclusions in patients with a variety of other neurodegenerative diseases, including Alzheimer’s and Parkinson’s diseases [94]. A variety of evidence implicates the PrLD in disease. Overexpression of TDP-43 in yeast, C. elegans, or Drosophila results in TDP-43 aggregation and toxicity, and the PrLD is required for this aggregation and toxicity [95–97]. Furthermore, at least 44 mutations in TDP-43 have been identified in patients with ALS or FTLD; of these, 41 reside in the PrLD [98]. However, regions outside the PrLD also affect aggregation and toxicity. For example, the C-terminal domain is necessary, but not sufficient, for aggregation and toxicity in yeast [96]. The presence of at least one RRM is also required for toxicity in yeast. Additionally, some of the ALS-associated mutations do not accelerate aggregation in vitro or cause toxicity in yeast. Together, these results suggest a more complex mechanism of toxicity [99].
FUS was the second RRM-containing protein to be linked to neurodegenerative disease. It contains an N-terminal PrLD, a single RRM, and two C-terminal “RGG” domains (multiple gly–gly motifs interspersed with arginine and aromatic residues), one of which barely misses the defined PrLD cutoff according to the Alberti algorithm [21, 100]. Mutations in FUS cause familial ALS [101, 102] and aggregation of FUS has been linked to both ALS and FTLD [98]. FUS is normally a predominantly nuclear protein, but these mutations cause FUS to form cytoplasmic aggregates. FUS has since been found in cytoplasmic inclusions in patients with other neurodegenerative diseases, including Huntington’s Disease [94]. FUS is also highly aggregation-prone in vitro and causes toxicity when expressed in yeast [100]. The PrLD is necessary, but not sufficient for this aggregation and toxicity [100], and the RNA-binding ability of FUS seems to be critical for toxicity in yeast and Drosophila models [103]. The ALS-associated mutations in FUS seem to cluster in two regions: the N-terminal PrLD and a short C-terminal segment containing a predicted nuclear localization signal [98].
Two other RNA-binding proteins that ranked highly in prion prediction analyses, TAF-15 and EWSR-1, have also been linked to sporadic ALS and FTLD. Again, each protein contains a predicted N-terminal PrLD, as well as a single RRM and two RGG domains [21]. For both TAF-15 and EWSR1, mutations have been found in a small number of ALS patients that do not appear in control subjects [20, 104]. These mutations all occur outside of the PrLD, clustering in and around the RGG domains. Both proteins are inherently aggregation-prone in vitro, and the disease-associated mutations accelerate aggregation in each case [20, 104].
Additionally, point mutations in either hnRNPA2B1 or hnRNPA1 have been shown to cause familial inclusion body myopathy with Paget’s disease of bone, frontotemporal dementia, and ALS (IBMPFD/ALS) [105]. In both cases, the causative mutation is a single aspartic acid to valine substitution within the PrLD. Additional mutations were identified in patients with both familial and sporadic forms of ALS. In normal muscle cells, hnRNPA2B1 or hnRNPA1 are predominantly nuclear; however, in patients carrying the mutations, the proteins form large cytoplasmic inclusions. In vitro, the wild-type proteins are intrinsically aggregation-prone, and the disease-causing mutations accelerate this aggregation. In Drosophila and mouse models, expression of the mutant proteins leads to formation of large cytoplasmic inclusions and severe muscle degeneration. Furthermore, when the core PrLD from mutant hnRNPA1 and hnRNPA2 are substituted in place of the nucleation domain of Sup35, they can support prion formation in yeast, while the wild-type PrLDs cannot.
Mutations in another prion-like protein, TIA1, have recently been shown to cause Welander distal myopathy, a dominant adult-onset disorder characterized by progressive distal limb weakness [106]. Patient muscle biopsies showed TIA1 and TDP-43 staining adjacent to intracellular inclusions.
In all, of the 20 RNA-binding proteins that are scored highest by the Alberti algorithm, 10 have been linked to degenerative disease [107]. An obvious question is why so many of these RNA-binding proteins have maintained aggregation-prone PrLDs if aggregation of these domains is associated with neurodegenerative disease. A growing body of evidence suggests that the PrLDs may play a functional role, such as recruiting RNA-binding proteins to P bodies or stress granules under cellular stress [107, 108]. TIA-1 plays a critical role in stress granule formation, and the TIA-1 PrLD is required for stress granule formation [109]. Remarkably, the Sup35 PFD can substitute for the TIA-1 PrLD in supporting stress granule assembly, linking prion-like aggregation to stress granule formation [109]. Many of the disease-associated PrLD-containing proteins are recruited to stress granules, and mutations enhance this recruitment [105, 108]. This suggests a model in which these RNA-binding proteins form reversible stress granule aggregates, but where mutations or changes in the cellular environment, such as prolonged stress, can lead to excessive and pathogenic stress granule formation [108, 110].
Consistent with this theory, mutations that disrupt the turnover of RNA–protein aggregates have also been linked to degenerative disease. Specifically, VCP/p97 is a well-characterized AAA ATPase that is involved in disassembling protein complexes containing ubiquitinated proteins [111]. Mutations in VCP have been shown to cause both IBMPFD [112] and ALS [113]. These mutations result in the formation of cytoplasmic inclusions containing TDP-43 and other stress granule markers [110]. Additionally, over-expression of these mutants inhibits stress granule clearance by autophagy [114]. Collectively, these results suggest that these diseases result from impairment of the normal dynamics of RNA granule assembly, disassembly, and clearance.
Predicting disease-associated proteins: successes and future challenges
The various disease-associated mutations in human PrLDs demonstrate both the strengths and weaknesses of existing prediction algorithms, and highlight some of the challenges in predicting human PrLDs.
hnRNPA1 and hnRNPA2 offer the best examples of proteins that are accurately predicted by current algorithms. Wild-type hnRNPA2 scores below PAPA’s 0.05 threshold for high aggregation propensity, but the disease-associated mutation pushes the PAPA score past the aggregation threshold ([99]; Table 4). The same is true for the most highly expressed isoform of hnRNPA1 (Isoform A; [105])—the wild-type protein scores below 0.05, but both of the mutations linked to familial forms of IBMPFD or ALS are predicted by PAPA to push the proteins beyond the threshold of aggregation. hnRNPA1 has another isoform (Isoform B) that surpasses the PAPA threshold even in the wild-type state, but in which the mutations are predicted to further enhance the aggregation activity. ZipperDB also correctly predicts the effects of each mutation, predicting that they should create strong steric zipper motifs. By contrast, while the Alberti algorithm identifies PrLDs in both proteins, it predicts that the mutations will have little effect on prion propensity. These mutations may offer a good example of the limitations of using algorithms based on compositional similarity to known PFDs to predict the effects of mutations. The two mutations linked to IBMPFD both involve substitution of an aspartic acid with a valine. Because both residues are extremely rare in yeast PFDs, algorithms based on compositional similarity will score this as a relatively neutral substitution. However, aspartic acid and valine are likely rare in yeast PFDs for opposite reasons—aspartic acid because it strongly inhibits prion formation, and valine because it too strongly promotes prion formation, creating a strong selective pressure against its inclusion in PFDs.
The Alberti algorithm correctly predicts PrLDs in each of the other disease-associated RNA-binding proteins (Table 4). Likewise, PAPA scores EWSR1, FUS, TAF15, and TIA-1 above the predicted threshold for aggregation. TDP-43 scores just below the 0.05 threshold, within a range that is generally associated with some aggregation activity (Table 4; Fig. 3). The fact that both the Alberti algorithm and PAPA score the wild-type proteins as prion-like or aggregation-prone could be considered accurate, since the wild-type proteins each appear to be aggregation-prone [20]. However, this also highlights a key limitation of these algorithms—while both PAPA and the Alberti algorithm have shown success at identifying candidate disease-associated proteins, at this point none of the existing algorithms can consistently predict the exact effects of mutations on either aggregation propensity or pathogenicity.
There are a number of possible reasons for this failing. First, aggregation propensity and toxicity are not always coupled, as evidenced by the observed mutations that cause disease without a detectable change in aggregation propensity [99]. These diseases appear to broadly result from disruptions in normal RNA homeostasis, so aggregation may simply be one of many causes of such disruption.
Second, these algorithms are designed to identify aggregation-prone protein fragments; however, in a cellular context, aggregation-prone fragments may be prevented from aggregating due to factors such as protein–protein interactions, interactions with other domains in the protein, protein modifications, or cellular localization. This is particularly true for the RNA-binding proteins, which appear to form regulated aggregates [110]. Disruption of any of these regulatory mechanisms could increase the aggregation propensity of the protein without changing the intrinsic aggregation propensity of the PrLD.
Third, differences between yeast PFDs and the human disease-associated PrLDs may limit the prediction accuracy of both the Alberti algorithm and PAPA. Both algorithms were designed and validated on proteins within a relatively narrow range of compositions. For example, 96 of the 100 candidate PFDs tested by Alberti et al. had greater than 24 % Q/N-content. This is significant because, although the yeast PFDs and human PrLDs share many compositional features including an under-representation of charged residues relative to their respective proteomes (Table 2), they differ significantly in other ways. The eight Q/N-rich yeast PFDs range from 28.6 to 46.8 % Q/N content, while among the human disease-associated RRM proteins, only the TAF15 and TIA1 PrLDs have greater than 22 % Q/N content. Thus, while Q and N are overrepresented among both yeast PFDs and human PrLDs, they are far less overrepresentated among the human PrLDs. Conversely, serine and glycine are more overrepresented among the human PrLDs than among the yeast PFDs (Table 2). Because the prediction accuracy of any algorithm is likely to decrease the further a protein’s composition deviates from that of the algorithm’s training set, these differences may limit the prediction accuracy of yeast-derived algorithms for human PrLDs. Therefore, these algorithms may need to be optimized for human PrLDs.
Finally, differences in cellular environment between yeast and human cells may impose distinct compositional requirements for prion-like activity. For example, Hsp104 is required for propagation of almost all yeast prions, and amino acid composition can affect the efficiency of Hsp104-dependent fiber fragmentation [115]. Therefore, some of the compositional biases seen in yeast prions may be due to specific requirements for Hsp104-dependent fragmentation. However, humans do not possess an Hsp104-homologue (although an Hsp110 in humans appears to be able to perform a subset of Hsp104 activities [116]), so human proteins may have very different compositional requirements for propagation. Additionally, the reason Hsp104 is required for most yeast prions is that prion aggregates need to be fragmented to create new independently segregating seeds to offset dilution by cell division; because mammalian neuronal cells typically do not divide rapidly, the levels of fiber fragmentation required for aggregate propagation are likely very different. The mechanism of spread of these neurodegenerative prion-like proteins to neighboring tissues may also differ from the mechanisms of propagation in yeast. Each of these differences may affect the specific compositional requirements for prion-like activity in yeast versus humans.
Conclusions
Significant progress has been made in defining the sequence features that drive yeast prion formation and in predicting the prion propensity of PrLDs. However, perfecting prion prediction will require overcoming a number of challenges. Translating results from yeast PFDs into methods to predict aggregation and toxicity of human PrLDs creates additional challenges due to the differences between the two systems. Collectively, these issues highlight the need for additional research to unveil the fundamental features of prion formation and propagation, as well as how prion-like activity relates to disease. As our knowledge of these fundamental features grows, application of this knowledge to prion prediction will lead to more accurate prediction methods and identification of new prions or prion-like proteins, potentially resulting in additional targets for treating human neurodegenerative disorders.
Acknowledgments
This work was supported by a National Institutes of Health grant (GM105991) and Muscular Dystrophy Association grant (255893) to E.D.R.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137(1):146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106(2):171–182. doi: 10.1016/S0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 4.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae . Nat Genet. 2008;40(4):460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11(3):344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, Volkov K, Mironova L. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc Natl Acad Sci USA. 2010;107(23):10573–10577. doi: 10.1073/pnas.1005949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5(1):163–172. doi: 10.1016/S1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336(6079):355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- 9.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae . Science. 1994;264(5158):566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 10.Halfmann R, Wright J, Alberti S, Lindquist S, Rexach M. Prion formation by a yeast GLFG nucleoporin. Prion. 2012;6(4):391–399. doi: 10.4161/pri.20199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid–from bacteria to humans. Trends Biochem Sci. 2007;32(5):217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 1999;18(7):1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407(6803):477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 14.Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6(11):e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bateman DA, Wickner RB. [PSI+] Prion transmission barriers protect Saccharomyces cerevisiae from infection: intraspecies ‘species barriers’. Genetics. 2012;190(2):569–579. doi: 10.1534/genetics.111.136655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly AC, Wickner RB. Saccharomyces cerevisiae: a sexy yeast with a prion problem. Prion. 2013;7(3):215–220. doi: 10.4161/pri.24845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci USA. 2005;102(30):10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wickner RB, Edskes HK, Bateman D, Kelly AC, Gorkovskiy A. The yeast prions [PSI+] and [URE3] are molecular degenerative diseases. Prion. 2011;5(4):258–262. doi: 10.4161/pri.17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couthouis J, Hart MP, Shorter J, Dejesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, Epstein J, Chiang A, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Solski JA, Williams KL, Mojsilovic-Petrovic J, Ingre C, Boylan K, Graff-Radford NR, Dickson DW, Clay-Falcone D, Elman L, McCluskey L, Greene R, Kalb RG, Lee VM, Trojanowski JQ, Ludolph A, Robberecht W, Andersen PM, Nicholson GA, Blair IP, King OD, Bonini NM, Van Deerlin V, Rademakers R, Mourelatos Z, Gitler AD. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci USA. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4(6):R40. doi: 10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci USA. 2000;97(22):11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox BS. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;26:211–232. doi: 10.1038/hdy.1971.28. [DOI] [PubMed] [Google Scholar]
- 25.Lacroute F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol. 1971;106(2):519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doel SM, McCready SJ, Nierras CR, Cox BS. The dominant PNM2- mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics. 1994;137(3):659–670. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kushnirov VV, Ter-Avanesyan MD, Telckov MV, Surguchov AP, Smirnov VN, Inge-Vechtomov SG. Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae . Gene. 1988;66(1):45–54. doi: 10.1016/0378-1119(88)90223-5. [DOI] [PubMed] [Google Scholar]
- 28.Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270(5233):93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 29.Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae . Genetics. 1994;137(3):671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7(5):683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu JJ, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+] Proc Natl Acad Sci USA. 2002;99(Suppl 4):16446–16453. doi: 10.1073/pnas.252652099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci USA. 2002;99(8):5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287(5453):661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- 34.Pierce MM, Baxa U, Steven AC, Bax A, Wickner RB. Is the prion domain of soluble Ure2p unstructured? Biochemistry. 2005;44(1):321–328. doi: 10.1021/bi047964d. [DOI] [PubMed] [Google Scholar]
- 35.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289(5483):1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 36.Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Mol Cell Biol. 2004;24(16):7206–7213. doi: 10.1128/MCB.24.16.7206-7213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci USA. 2005;102(36):12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du Z. The complexity and implications of yeast prion domains. Prion. 2011;5(4):311–316. doi: 10.4161/pri.18304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maclea KS, Ross ED. Strategies for identifying new prions in yeast. Prion. 2011;5(4):263–268. doi: 10.4161/pri.17918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93(7):1241–1252. doi: 10.1016/S0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 41.Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100(2):277–288. doi: 10.1016/S0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 42.Chen B, Bruce KL, Newnam GP, Gyoneva S, Romanyuk AV, Chernoff YO. Genetic and epigenetic control of the efficiency and fidelity of cross-species prion transmission. Mol Microbiol. 2010;76(6):1483–1499. doi: 10.1111/j.1365-2958.2010.07177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol. 2004;2(4):E86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parham SN, Resende CG, Tuite MF. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J. 2001;20(9):2111–2119. doi: 10.1093/emboj/20.9.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shkundina IS, Kushnirov VV, Tuite MF, Ter-Avanesyan MD. The role of the N-terminal oligopeptide repeats of the yeast sup35 prion protein in propagation and transmission of prion variants. Genetics. 2006;172(2):827–835. doi: 10.1534/genetics.105.048660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435(7043):773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fei L, Perrett S. Disulfide bond formation significantly accelerates the assembly of Ure2p fibrils because of the proximity of a potential amyloid stretch. J Biol Chem. 2009;284(17):11134–11141. doi: 10.1074/jbc.M809673200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature. 2007;447(7144):556–561. doi: 10.1038/nature05848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crow ET, Du Z, Li L. A small, glutamine-free domain propagates the [SWI(+)] prion in budding yeast. Mol Cell Biol. 2011;31(16):3436–3444. doi: 10.1128/MCB.05338-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vitrenko YA, Pavon ME, Stone SI, Liebman SW. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr Genet. 2007;51(5):309–319. doi: 10.1007/s00294-007-0127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kadnar ML, Articov G, Derkatch IL. Distinct type of transmission barrier revealed by study of multiple prion determinants of Rnq1. PLoS. 2010;6(1):e1000824. doi: 10.1371/journal.pgen.1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106(2):183–194. doi: 10.1016/S0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 53.Kabani M, Cosnier B, Bousset L, Rousset JP, Melki R, Fabret C. A mutation within the C-terminal domain of Sup35p that affects [PSI(+)] prion propagation. Mol Microbiol. 2011;81(3):640–658. doi: 10.1111/j.1365-2958.2011.07719.x. [DOI] [PubMed] [Google Scholar]
- 54.Kochneva-Pervukhova NV, Poznyakovski AI, Smirnov VN, Ter-Avanesyan MD. C-terminal truncation of the Sup35 protein increases the frequency of de novo generation of a prion-based [PSI+] determinant in Saccharomyces cerevisiae . Curr Genet. 1998;34(2):146–151. doi: 10.1007/s002940050379. [DOI] [PubMed] [Google Scholar]
- 55.Helsen CW, Glover JR. Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104) J Biol Chem. 2012;287(1):542–556. doi: 10.1074/jbc.M111.302869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA. 2006;103(52):19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15(12):3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 58.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268(5212):880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 59.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94(1):73–82. doi: 10.1016/S0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 60.Schirmer EC, Lindquist S. Interactions of the chaperone Hsp104 with yeast Sup35 and mammalian PrP. Proc Natl Acad Sci USA. 1997;94(25):13932–13937. doi: 10.1073/pnas.94.25.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27(20):2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grimminger-Marquardt V, Lashuel HA. Structure and function of the molecular chaperone Hsp104 from yeast. Biopolymers. 2010;93(3):252–276. doi: 10.1002/bip.21301. [DOI] [PubMed] [Google Scholar]
- 63.Reidy M, Masison DC. Modulation and elimination of yeast prions by protein chaperones and co-chaperones. Prion. 2011;5(4):245–249. doi: 10.4161/pri.17749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20(23):8916–8922. doi: 10.1128/MCB.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volkov KV, Aksenova AY, Soom MJ, Osipov KV, Svitin AV, Kurischko C, Shkundina IS, Ter-Avanesyan MD, Inge-Vechtomov SG, Mironova LN. Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae . Genetics. 2002;160(1):25–36. doi: 10.1093/genetics/160.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279(28):29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 67.DeSantis ME, Shorter J. Hsp104 drives “protein-only” positive selection of Sup35 prion strains encoding strong [PSI(+)] Chem Biol. 2012;19(11):1400–1410. doi: 10.1016/j.chembiol.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Helsen CW, Glover JR. A new perspective on Hsp104-mediated propagation and curing of the yeast prion [PSI (+)] Prion. 2012;6(3):234–239. doi: 10.4161/pri.19913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toombs JA, Liss NM, Cobble KR, Ben-Musa Z, Ross ED. [PSI+] maintenance is dependent on the composition, not primary sequence, of the oligopeptide repeat domain. PLoS ONE. 2011;6(7):e21953. doi: 10.1371/journal.pone.0021953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bryan AW, Jr, Menke M, Cowen LJ, Lindquist SL, Berger B. BETASCAN: probable beta-amyloids identified by pairwise probabilistic analysis. PLoS Comput Biol. 2009;5(3):e1000333. doi: 10.1371/journal.pcbi.1000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bryan AW, Jr, O’Donnell CW, Menke M, Cowen LJ, Lindquist S, Berger B. STITCHER: dynamic assembly of likely amyloid and prion beta-structures from secondary structure predictions. Proteins. 2011 doi: 10.1002/prot.23203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tartaglia GG, Pawar AP, Campioni S, Dobson CM, Chiti F, Vendruscolo M. Prediction of aggregation-prone regions in structured proteins. J Mol Biol. 2008;380(2):425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 73.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA. 2010;107(8):3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22(10):1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 75.Zibaee S, Makin OS, Goedert M, Serpell LC. A simple algorithm locates beta-strands in the amyloid fibril core of alpha-synuclein, Abeta, and tau using the amino acid sequence alone. Protein Sci. 2007;16(5):906–918. doi: 10.1110/ps.062624507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trovato A, Seno F, Tosatto SC. The PASTA server for protein aggregation prediction. Protein Eng Des Sel. 2007;20(10):521–523. doi: 10.1093/protein/gzm042. [DOI] [PubMed] [Google Scholar]
- 77.Maurer-Stroh S, Debulpaep M, Kuemmerer N, Lopez de la Paz M, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L, Schymkowitz JW, Rousseau F. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods. 2010;7(3):237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 78.Toombs JA, Petri M, Paul KR, Kan GY, Ben-Hur A, Ross ED. De novo design of synthetic prion domains. Proc Natl Acad Sci USA. 2012;109(17):6519–6524. doi: 10.1073/pnas.1119366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Esteras-Chopo A, Serrano L, de la Paz ML. The amyloid stretch hypothesis: recruiting proteins toward the dark side. Proc Natl Acad Sci USA. 2005;102(46):16672–16677. doi: 10.1073/pnas.0505905102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol. 2010;30(1):319–332. doi: 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thompson MJ, Sievers SA, Karanicolas J, Ivanova MI, Baker D, Eisenberg D. The 3D profile method for identifying fibril-forming segments of proteins. Proc Natl Acad Sci USA. 2006;103(11):4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Teng PK, Eisenberg D. Short protein segments can drive a non-fibrillizing protein into the amyloid state. Protein Eng Des Sel. 2009;22(8):531–536. doi: 10.1093/protein/gzp037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M. Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Biochem Sci. 2007;32(5):204–206. doi: 10.1016/j.tibs.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 84.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21(16):3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 85.Espinosa Angarica V, Ventura S, Sancho J. Discovering putative prion sequences in complete proteomes using probabilistic representations of Q/N-rich domains. BMC Genom. 2013;14:316. doi: 10.1186/1471-2164-14-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li SC, Goto NK, Williams KA, Deber CM. Alpha-helical, but not beta-sheet, propensity of proline is determined by peptide environment. Proc Natl Acad Sci USA. 1996;93(13):6676–6681. doi: 10.1073/pnas.93.13.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Halfmann R, Alberti S, Krishnan R, Lyle N, O’Donnell CW, King OD, Berger B, Pappu RV, Lindquist S. Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol Cell. 2011;43(1):72–84. doi: 10.1016/j.molcel.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Costanzo M, Zurzolo C. The cell biology of prion-like spread of protein aggregates: mechanisms and implication in neurodegeneration. Biochem J. 2013;452(1):1–17. doi: 10.1042/BJ20121898. [DOI] [PubMed] [Google Scholar]
- 89.Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123(Pt 8):1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hall GF, Patuto BA. Is tau ready for admission to the prion club? Prion. 2012;6(3):223–233. doi: 10.4161/pri.19912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 2009;64(6):783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 92.Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, Fraser G, Stalder AK, Beibel M, Staufenbiel M, Jucker M, Goedert M, Tolnay M. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11(7):909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 94.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19(R1):R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ash PE, Zhang YJ, Roberts CM, Saldi T, Hutter H, Buratti E, Petrucelli L, Link CD. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum Mol Genet. 2010;19(16):3206–3218. doi: 10.1093/hmg/ddq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2008;105(17):6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Y, Ray P, Rao EJ, Shi C, Guo W, Chen X, Woodruff EA, III, Fushimi K, Wu JY. A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci USA. 2010;107(7):3169–3174. doi: 10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21(6):904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284(30):20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9(4):e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 102.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Daigle JG, Lanson NA, Jr, Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. 2013;22(6):1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, Kim CE, Frackelton EC, Solski JA, Williams KL, Clay-Falcone D, Elman L, McCluskey L, Greene R, Hakonarson H, Kalb RG, Lee VM, Trojanowski JQ, Nicholson GA, Blair IP, Bonini NM, Deerlin VM, Mourelatos Z, Shorter J, Gitler AD. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21(13):2899–2911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Klar J, Sobol M, Melberg A, Mabert K, Ameur A, Johansson AC, Feuk L, Entesarian M, Orlen H, Casar-Borota O, Dahl N. Welander distal myopathy caused by an ancient founder mutation in TIA1 associated with perturbed splicing. Hum Mutat. 2013;34(4):572–577. doi: 10.1002/humu.22282. [DOI] [PubMed] [Google Scholar]
- 107.Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201(3):361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15(12):5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154(4):727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamanaka K, Sasagawa Y, Ogura T. Recent advances in p97/VCP/Cdc48 cellular functions. Biochim Biophys Acta. 2012;1823(1):130–137. doi: 10.1016/j.bbamcr.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 112.Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36(4):377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 113.Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, Galassi G, Scholz SW, Taylor JP, Restagno G, Chio A, Traynor BJ. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68(5):857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153(7):1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Alexandrov AI, Polyanskaya AB, Serpionov GV, Ter-Avanesyan MD, Kushnirov VV. The effects of amino acid composition of glutamine-rich domains on amyloid formation and fragmentation. PLoS ONE. 2012;7(10):e46458. doi: 10.1371/journal.pone.0046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE. 2011;6(10):e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maddelein ML, Wickner RB. Two prion-inducing regions of Ure2p are nonoverlapping. Mol Cell Biol. 1999;19(6):4516–4524. doi: 10.1128/mcb.19.6.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]