Abstract
Evolutionary relationships among the eight major lineages of Mollusca have remained unresolved despite their diversity and importance. Previous investigations of molluscan phylogeny, based primarily on nuclear ribosomal gene sequences1–3 or morphological data4, have been unsuccessful at elucidating these relationships. Recently, phylogenomic studies using dozens to hundreds of genes have greatly improved our understanding of deep animal relationships5. However, limited genomic resources spanning molluscan diversity has prevented use of a phylogenomic approach. Here we use transcriptome and genome data from all major lineages (except Monoplacophora) and recover a well-supported topology for Mollusca. Our results strongly support the Aculifera hypothesis placing Polyplacophora (chitons) in a clade with a monophyletic Aplacophora (worm-like molluscs). Additionally, within Conchifera, a sister-taxon relationship between Gastropoda and Bivalvia is supported. This grouping has received little consideration and contains most (>95%) molluscan species. Thus we propose the node-based name Pleistomollusca. In light of these results, we examined the evolution of morphological characters and found support for advanced cephalization and shells as possibly having multiple origins within Mollusca.
With over 100,000 described extant species in eight major lineages, Mollusca is the second most speciose animal phylum6. Manymolluscs are economically important as food and producers of pearls and shells whereas others cause economic damage as pests, biofoulers and invasive species. Molluscs are also biomedically important as models for the study of brain organization, learning and memory as well as vectors of parasites. Although shelled molluscs have one of the best fossil records of any animal group, evolutionary relationships among major molluscan lineages have been elusive.
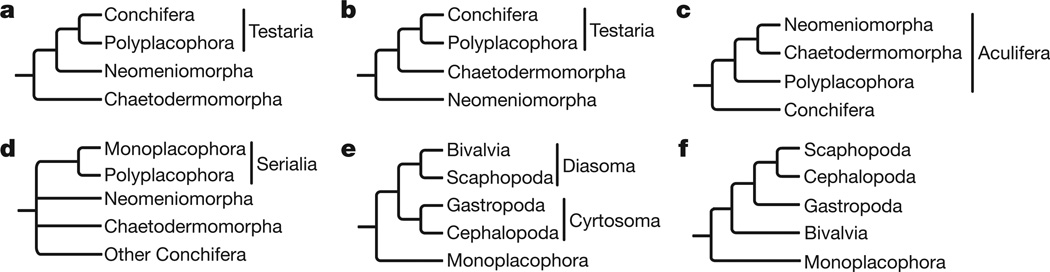
Morphological disparity among the major lineages of Mollusca has prompted numerous conflicting phylogenetic hypotheses (Fig. 1). The vermiform Chaetodermomorpha (also known as Caudofoveata) and Neomeniomorpha (also known as Solenogastres) traditionally have been considered to represent the plesiomorphic state of Mollusca because of their ‘simple’ internal morphology and lack of shells7.Whether these two lineages constitute a monophyletic group, Aplacophora8, or a paraphyletic grade4,9 has been widely debated. Some workers have considered the presence of sclerites a synapomorphy for a clade Aculifera, uniting Polyplacophora (chitons; which have both sclerites and shells) and Aplacophora. In contrast, Polyplacophora has alternatively been placed with Conchifera (Bivalvia, Cephalopoda, Gastropoda, Monoplacophora and Scaphopoda) in a clade called Testaria uniting the shelled molluscs4. Morphology has been interpreted to divide Conchifera into a gastropod/cephalopod clade (Cyrtosoma) and a bivalve/scaphopod clade (Diasoma)6.Unfortunately, because of varying interpretations of features as derived or plesiomorphic, a lack of clear synapomorphies, and often unclear character homology, the ability of morphology to resolve such deep phylogenetic events is limited.
Figure 1. Leading hypotheses of molluscan phylogeny.
a, Adenopoda hypothesis placing Chaetodermomorpha basal. b, Hepagastralia hypothesis placing Neomeniomorpha basal. c, Aculifera hypothesis placing Aplacophora sister to Polyplacophora. d, Serialia hypothesis allying Polyplacophora and Monoplacophora. e, Diasoma and Cyrtosoma hypotheses allying bivalves to scaphopods and gastropods to cephalopods, respectively. f, Unnamed hypothesis, allying scaphopods and cephalopods.
Molecular investigations of molluscan phylogeny have relied primarily on nuclear ribosomal gene sequences (18S and 28S)1–3,10, and have also offered little resolution. Maximum likelihood (ML) analyses of 18S, 28S or both1 recovered most major lineages monophyletic, but support at deeper nodes was generally weak. Subsequent analyses of a combined data set (18S, 28S, 16S, cytochrome c oxidase I and histone H3)2 yielded similar results, namely that bivalves were not monophyletic and support values at most deep nodes were low. Expanding on this study, further work supported a sister-taxon relationship between chitons and monoplacophorans (Serialia) but support at other deep nodes was generally low3. Moreover, Mollusca was not recovered monophyletic (a result significantly supported by Approximately Unbiased, AU, tests; Supplementary Table 1) possibly due to contaminated neomenioid sequences10.
Morphological and traditional molecular phylogenetic approaches have failed to robustly reconstruct mollusc phylogeny. Notably, several recent phylogenomic studies (for example, refs 5 and 11) have significantly advanced our understanding of metazoan evolution by using sequences derived from genome and transcriptome data. With this approach, numerous orthologous protein-coding genes can be identified and employed in phylogeny reconstruction. Many of these genes are constitutively expressed and can be easily recovered from even limited expressed sequence tag (EST) surveys. Additionally, these genes are usually informative for inferring higher-level phylogeny because of their conserved nature due to their functional importance.
Here, we used such a phylogenomic approach to investigate evolutionary relationships among the major lineages of Mollusca. High-throughput transcriptome data were collected from 18 operational taxonomic units (OTUs; Supplementary Table 2), and augmented with publicly available ESTs and genomes (Supplementary Table 3). To increase data set completeness, data from closely related species were combined in eleven cases, resulting in a total of 42 mollusc OTUs. Every major lineage of Mollusca was represented in the data set by at least two distantly related species, except for monoplacophorans that live in deep marine habitats and could not be procured in adequate condition for transcriptome analyses. For sequence processing and orthology determination, a bioinformatic pipeline was developed that builds upon previous studies (see Methods and Supplementary Fig. 2). This pipeline identified 308 orthologous genes suitable for concatenation and phylogenetic analyses (Supplementary Table 4), totalling 84,614 amino acid positions.
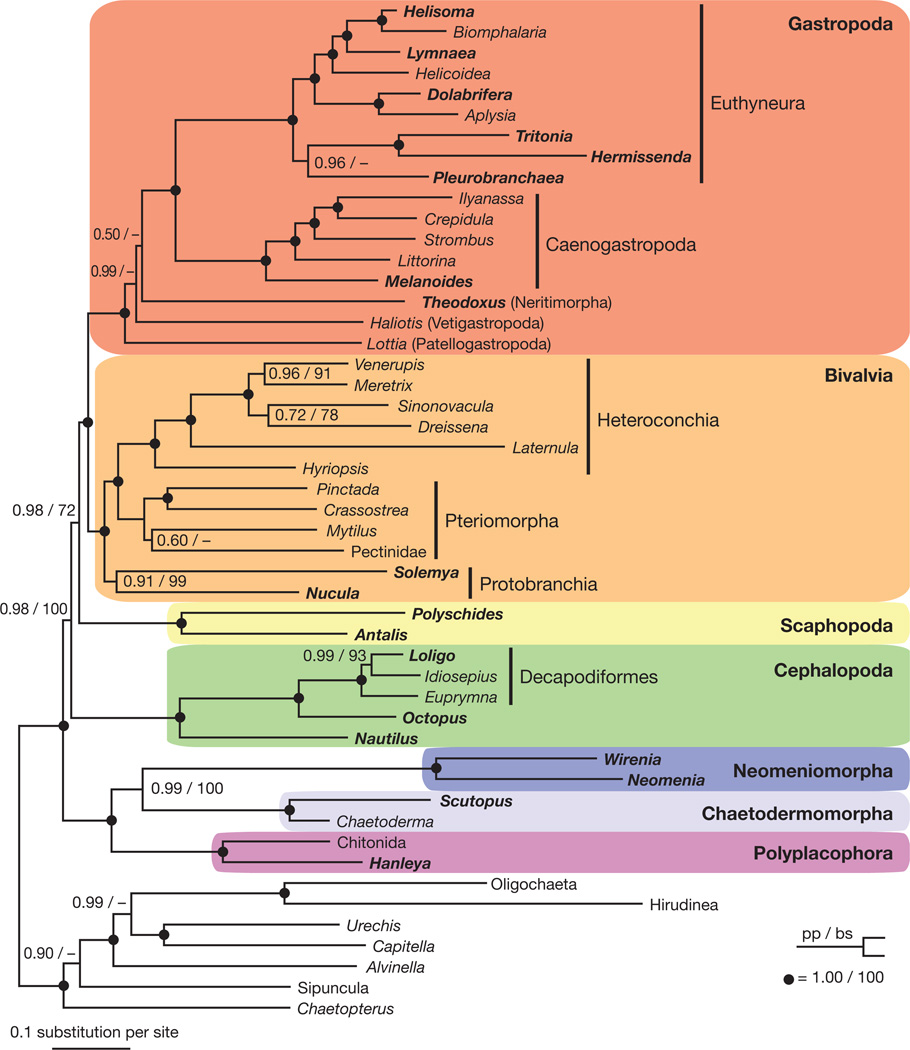
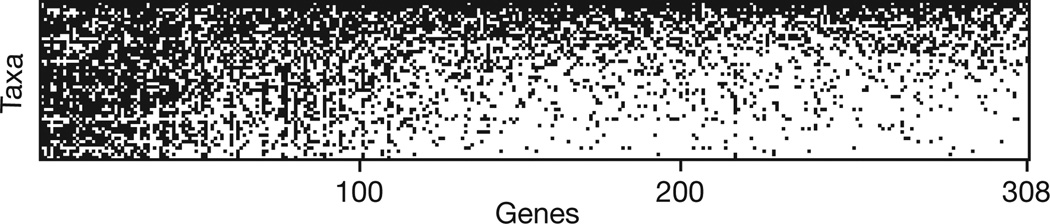
To determine the appropriate outgroup to Mollusca, preliminary analyses including a broad range of lophotrochozoans and the cnidarian Nematostella were conducted. Nematostella was included to verify that neomenioid data did not contain cnidarian contamination (see Methods). Maximum likelihood (ML) analyses using the best-fitting model for each gene strongly supported Annelida as the sister taxon of Mollusca (bootstrap support, bs = 100, Supplementary Fig. 3), whereas Bayesian inference (BI) placed Entoprocta + Cycliophora sister to Mollusca with poor support (posterior probability, pp = 0.62, Supplementary Fig. 4). Relationships among major lineages of Mollusca were consistent between analyses with multiple outgroups (Supplementary Figs 3–4) or with only Annelida as outgroup (Fig. 2 and Supplementary Fig. 5; additional information on outgroup selection in Supplementary Results). On the basis of these results, Annelida was selected as outgroup for all other analyses to reduce computational complexity and potential homoplasy from distant or fast-evolving out-groups. This final data matrix including all 308 genes (Fig. 3) had an average percentage of genes sampled per taxon of 41% and an overall matrix completeness of 25.6%, comparable to other major phylogenomic data sets (for example, ref 11).ML and BI analyses of this matrix yielded nearly identical topologies within Mollusca, except for relationships among basal gastropods and placements of the sea slug Pleurobranchaea and the bivalve Mytilus (Fig. 2 and Supplementary Fig. 5). High leaf stability scores for all OTUs (Supplementary Table 3) and strong support for most nodes suggest all OTUs were represented by sufficient data to be reliably placed. Remarkably, branch lengths were relatively uniform; cephalopods did not show long branches as previously reported in analyses of 18S and 28S1–3,10.
Figure 2. Relationships among major lineages of Mollusca based on 308 genes.
Bayesian inference topology shown with ML bootstrap support values (bs) >50 and posterior probabilities (pp) >0.50 are listed at each node. Filled circles represent nodes with bs = 100 and pp = 1.00. Taxa from which new data were collected are shown in bold.
Figure 3. Data matrix coverage.
Genes are ordered along the x-axis from left to right from best sampled to worst sampled. Taxa are ordered along the y-axis from top to bottom from most genes sampled to fewest genes sampled. Black squares represent a sampled gene fragment and white squares represent a missing gene fragment.
All major lineages of Mollusca were monophyletic with strong support (bs = 100%, pp = 1.00). Importantly, there was strong support at all deep nodes, although the node placing Scaphopoda received moderate support in ML (bs = 72%) but strong support in BI (pp = 0.98). A clade including Aplacophora and Polyplacophora was unequivocally supported (bs = 100%, pp = 1.00) and placed sister to Conchifera, consistent with the Aculifera hypothesis. Moreover, we found strong support (bs = 100%, pp = 0.99) for a sister relationship between Neomeniomorpha and Chaetodermomorpha, supporting the Aplacophora hypothesis but contrary to previous molecular1–3,10 and morphological4 studies. To evaluate alternatives to the Aculifera and Aplacophora hypotheses, we used AU tests (Supplementary Table 5). These tests rejected the Testaria hypothesis, which allies chitons with the other shelled molluscs (P < 0.02) and placement of either aplacophoran taxon as sister to all other molluscs (both P < 0.01). Aculiferan monophyly supports interpretation of the Palaeozoic taxon ‘Helminthochiton’ thraivensis as possessing features intermediate between chitons and aplacophorans12, and interpretation of dorsal, serially arranged calcareous structures as a possible aculiferan synapomorphy13. Specifically, the chaetoderm Chaetoderma14 and some, but not all, neomenioids15 possess dorsal, serially repeated sclerite-secreting regions during development. Notably, chiton valves are not thought to be homologous to aculiferan sclerites16, although certain genes involved in patterning these structures may be. Our results highlight a need for developmental gene expression studies of aculiferans to address this issue.
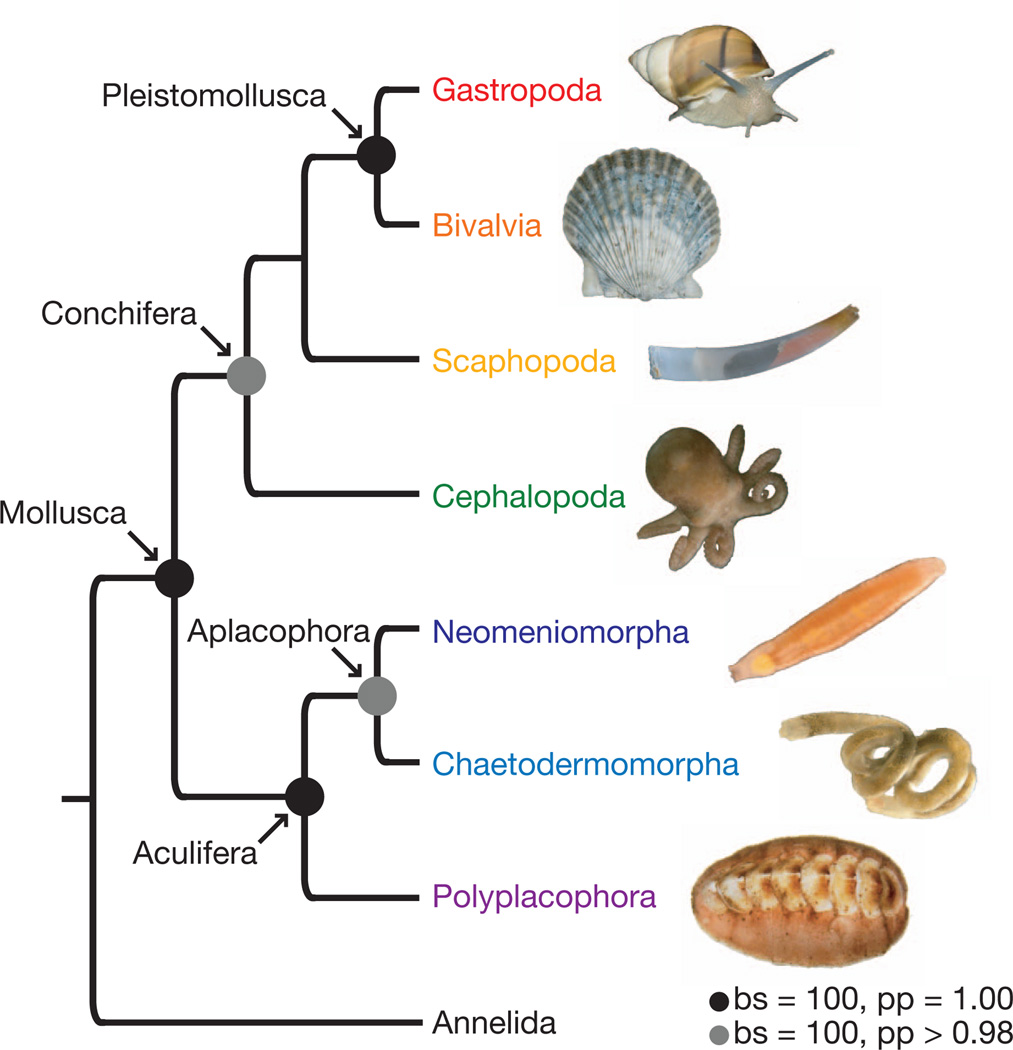
Within a monophyletic Conchifera (bs = 100%, pp = 0.98), Gastropoda and Bivalvia were supported as derived sister taxa (bs = 100%, pp = 1.0). Traditionally, a sister relationship between gastropods and bivalves, which relates the two most speciose lineages of molluscs, has received little consideration. However, this relationship has been recovered in molecular studies with relatively limited taxon sampling across Mollusca5,17. Similarities between the veliger larvae of gastropods and lamellibranch bivalves have been long recognized. Most notably, both possess larval retractor muscles and a velum muscle ring18. Another potential synapomorphy is loss of the anterior ciliary rootlet in locomotory cilia of gastropods and bivalves19. Because of strong support for a gastropod/bivalve clade in most analyses and the implications of this hypothesis for understanding molluscan evolution, we propose the node-based name Pleistomollusca, which includes the last common ancestor of Gastropoda and Bivalvia and all descendents (Fig. 4). Etymology of this name (pleistos from Greek for ‘most’) recognizes the incredible species diversity of this clade of molluscs which we conservatively estimate to contain >95% of described mollusc species.
Figure 4. Deep molluscan phylogeny as inferred in the present study.
Black circles represent nodes with bs = 100 and pp = 1.00. Gray circles represent nodes with bs = 100 and pp ≥ 0.98. The actual specimens of Polyschides and Hanleya used in this study are shown. Photos are not to scale. A full-page version of this figure is presented in Supplementary Fig. 1.
Sister to Pleistomollusca is Scaphopoda (albeit with moderate support in ML; bs = 72%, pp = 0.98) and Cephalopoda represents the sister taxon of all other conchiferan lineages sampled. Despite strong support values for a gastropod/bivalve clade, AU tests failed to reject Scaphopoda as sister to any other conchiferan lineage (P > 0.5). Given the limited sampling for Scaphopoda, additional data may help solidify its position. Nonetheless, all results presented here clearly refute the traditional view of a sister relationship between gastropods and cephalopods (Cyrtosoma; P < 0.01). Features thought to be diagnostic of this clade include a well-developed, free head with cerebrally innervated eyes and a nervous system with visceral loop inwards of the dorsoventral musculature6. However, these characters must be reinterpreted as either symplesiomorphies lost in scaphopods and bivalves, or convergences. Notably, the high degree of cephalization in gastropods and cephalopods has recently been suggested to have evolved independently20.
The phylogenomic approach used here also holds promise for resolving relationships within major lineages. For example, although their phylogenyhas been widely debated, our broadly sampled caenogastropod subtree was strongly supported throughout (bs = 100, pp = 1.0) and consistent with previous morphological analysis21. We also recovered opisthobranchs paraphyletic with respect to Pulmonata, agreeing with recent morphological and molecular studies22. Additionally, our analyses confirm bivalve monophyly with deposit-feeding protobranchs sister to filter-feeding lamellibranchs.
To assess robustness of the reconstructed topology further, we examined the influences of matrix completeness, gene inclusion and substitution models on phylogenetic reconstruction (Supplementary Table 6). Analyses of the 200 and 100 best-sampled genes (Supplementary Figs 6 and 7) recovered the same branching order and relative level of support among major lineages as the full data set. For gene inclusion, matrices of only non-ribosomal (Supplementary Fig. 8) and only ribosomal protein genes (Supplementary Fig. 9) were analysed to address issues of different gene classes (for example, ribosomal proteins) biasing phylogenetic signal5. Support values for deep nodes inferred from non-ribosomal protein genes were generally weak and Aplacophora, Polyplacophora and Bivalvia were not recovered monophyletic. In contrast, analysis of only ribosomal protein genes recovered all major lineages monophyletic with strong support in BI but moderate support for most deep nodes in ML (see also ref. 17). Although ribosomal protein and non-ribosomal protein genes seem to be contributing different amounts of phylogenetic signal, support for most nodes was greater when all gene classes were included, in accordance with previous phylogenomic studies5,11. We also performed an analysis based on very conservative orthology determination using only the 243 genes for which our method and InParanoid identified the same Lottia sequence as orthologous to the primer taxon (Drosophila) sequence (see Methods). Branching order (Supplementary Fig. 10) was identical to the tree based on all 308 genes (Fig. 2). Our ML analyses differ from other phylogenomic studies by using gene-specific amino acid substitution models rather than a single model across the entire matrix. Thus, for comparative reasons, we also ran single-model ML analyses using the WAG + CAT + F model (Supplementary Fig. 11) and the LG + CAT + F model (Supplementary Fig. 12). These analyses yielded the same relationships as the ML analysis using the best-fitting model for each gene (Supplementary Fig. 5) with similar overall support in all three analyses. We also assessed the effect of model selection by performing a BI analysis using the CAT-GTR model on the data set of the 100 best-sampled genes (Supplementary Fig. 7); this model is too computationally intensive for the full 308 gene data set. Except for the placement of Pleurobranchaea, this analysis yielded the samebranching order as the analysis using the CAT model (Fig. 2) with similar support values. Finally, even an approximately ML analysis (Supplementary Fig. 13), which is less computationally intensive, yielded the same relationships among major lineages as the fully parameterized ML analysis.
A primary goal of resolving molluscan phylogeny is to improve our understanding of their early evolutionary history. Perhaps more than any other animal group, understanding of molluscan early evolution has been constrained by the notion of a generalized bauplan or ‘archetype’ which is still propagated by some invertebrate zoology textbooks. Arguably, such a viewpoint has hindered our ability to consider how individual characters have evolved within Mollusca. Using a modified version of a morphological character matrix4, we performed ancestral state reconstruction using maximum parsimony and a simplified topology based on our results (Fig. 4) to infer ancestral states for 60 characters across Mollusca (Supplementary Table 7). Even though monoplacophoran transcriptome data were unavailable herein, we were able to evaluate how placement of Monoplacophora influences our understanding of early molluscan evolution. Ancestral state reconstruction of most characters for the last common ancestor of Mollusca was unaffected by the placement of monoplacophorans. We considered three possibilities: (1) Monoplacophora basal within Conchifera, (2) sister to Polyplacophora, and (3) absent from the analysis. In all three cases, only 6 out of 60 characters were influenced (Table 1). For example, ancestral state reconstruction for shell(s) secreted by a shell gland and periostracum changed between absent (Monoplacophora basal conchiferan) and equivocal (Monoplacophora sister to Polyplacophora, or not considered).
Table 1.
Ancestral states affected by placement of Monoplacophora
| Inferred plesiomorphic state of Mollusca | |||
|---|---|---|---|
| Character | Monoplacophora not considered | Monoplacophora basal in Conchifera | Monoplacophora sister to Polyplacophora |
| Shell by shell gland | Absent | Absent | Equivocal |
| Periostracum | Absent | Absent | Equivocal |
| Position of mantle cavity | Equivocal | Circumpedal | Equivocal |
| Number of D-V muscles | Equivocal | Eight or more | Equivocal |
| Pedal ganglia | Equivocal | Absent | Equivocal |
| Cerebral (pretrochal) eyes | Equivocal | Absent | Equivocal |
Only six of 60 characters were affected by the placement of Monoplacophora. See Supplementary Table 7 for additional characters and coding for all characters.
Results of these ancestral state reconstructions shed light on the early evolution of Mollusca. Odontogriphus, a Middle Cambrian form proposed to be a stem-group mollusc, showed character states consistent with our reconstructions (ventral muscular foot, dorsal cuticular mantle, mantle cavity containing ctenidia or gills, and regionalized gut)23. However, whereas Odontogriphus and Wiwaxia (another Middle Cambrian putative stem-group mollusc) apparently had a narrow, distichous (bipartite, aplacophoran-like) radula23,24, ancestral state reconstruction indicates that the plesiomorphic state of the radula was broad and rasping with multiple teeth per row attached to a flexible radular membrane supported by muscular and cartilage-like bolsters as in chitons and most conchiferans.
The origin and evolution of molluscan epidermal hardparts (shells and sclerites) is another contentious issue. Although aculiferan sclerites, chiton valves and conchiferan shells are all calcareous secretions of the mantle, developmental and structural differences indicate that these structures are not homologous16. Sclerites are only present in aculiferans, and shells secreted by a shell gland are only present in conchiferans. Moreover, fossil taxa do not help clarify the plesiomorphic state of the molluscan scleritome as Odontogriphus lacked both sclerites and shells23, Wiwaxia had uncalcified, chitinous sclerites, and other putative stemgroup molluscs had calcareous sclerites and/or shells7. Therefore, organization of the ancestral scleritome, if present, remains ambiguous.
In summary, our robustly supported evolutionary framework for Mollusca consists of two major clades: Aculifera, which includes a monophyletic Aplacophora sister to Polyplacophora, and Conchifera (as sampled), including a gastropod/bivalve clade we term Pleiostomollusca. Neomeniomorpha was not placed as the basal-most molluscan lineage as previously suggested nor is the Testaria hypothesis supported. Thus, several aplacophoran features commonly argued to be molluscan plesiomorphies (for example, non-muscular foot, organization of midgut, primarily distichous radula without subradular membrane) are reinterpreted as aplacophoran synapomorphies, whereas others are reinterpreted as neomenioid apomorphies (for example, prepedal cirri, pericalymma-type larva). Within Conchifera, our results show that gastropods are sister to bivalves (not cephalopods), a result that has important implications for molluscan model systems. Also, possible independent evolution of highly cephalized morphologies in gastropods and cephalopods suggests additional work addressing neural features across conchiferans is needed20.
METHODS
Overview
Data and analyses were conducted in four basic steps: (1) RNA was extracted from mollusc species, cDNA was prepared and then sequenced; (2) EST data were processed with a bioinformatics pipline incorporating EST2Uni25 and HaMStR26; (3) trees were reconstructed with RAxML 7.27 (ref. 27) and Phylobayes 2.3 (ref. 28). (4) Additional measures, including leaf stability with Phyutility29 and Approximately Unbiased (AU) tests30 were used to assess robustness of the results. Molecular techniques. Complementary DNA was prepared using standard protocols and sequenced using either 454 GS-FLX or Titanium. Sanger EST libraries generated for Scutopus and Wirenia were also included in this study. See Supplementary Methods for detailed laboratory methods.
Sequence processing
RawESTs were processed and assembled using the EST2uni pipeline25. This software removes low-quality regions with lucy31, removes vector sequences with lucy and SeqClean (http://compbio.dfci.harvard.edu/tgi/software), masks low complexity regions with RepeatMasker (http://www.repeatmasker.org), and assembles contigs with CAP3 (ref. 32). Data on sequence quality were used by CAP3 when available. Unigenes were translated with ESTScan33 and sequences shorter than 100 amino acids were deleted. Manual BLAST searches of samples of unigenes for vector sequences as well as examination of contig assembly diagrams generated by EST2uni indicated that these programs performed well at removing vector and low-quality sequences and assembling contigs, respectively.
To reduce the amount of missing data per taxon, sequences from two or more closely related taxa were combined to create the following 11 chimaerical OTUs: Chitonida, Crassostrea, Dreissena, Haliotis, Helicoidea, Loligo, Mytilus, Pectinidae, Pedicellina, Sipuncula and Venerupis.
Orthology assignment and data set assembly
OG identification used HaMStR local 7 (ref. 26), which uses profile hidden Markov models (pHMMs) generated from completely sequenced reference taxa in the In Paranoid database34. Translated unigenes were searched against the 1,032 single-copy OGs of HaMStR’s ‘model organism’ pHMMs derived from Homo, Ciona, Drosophila, Caenorhabditis and Saccharomyces. Translated unigenes matching an OG’s pHMM were then compared to the proteome of Drosophila using BLASTP. If the Drosophila protein contributing to the pHMM was the best BLASTP hit, the unigene was then placed in that OG.
If one of the first or last 20 characters of an amino acid sequence was an X (corresponding to a codon with an ambiguity, gap, or missing data), all characters between the X and that end of the sequence were deleted and treated as missing data. This step was important as ends of singletons were occasionally, but obviously, mistranslated. Each OG was aligned with MAFFT35 using the default alignment strategy. Aligned OGs were then manually inspected and subjected to trimming or deleting of partially mistranslated sequences, screening for paralogues, and combining incomplete sequences from the same OTU into one, more complete consensus sequence. These alignments were then trimmed with Aliscore and Alicut36 to remove regions with ambiguous alignment or little to no phylogenetic signal. Lastly, any alignments less than 25 amino acids in length were discarded.
Maximum likelihood (ML) trees were inferred for each OG using RaxML 7.2.7 (ref. 27) using the best-fitting amino acid substitution model as determined using the RAxML amino acid substitution model selection Perl script. OGs with strongly supported deep nodes suggesting the inclusion of paralogs were edited to delete obviously paralogous sequences or discarded. To reduce missing data in the final matrices, only OGs with sequences from at least ten molluscs were retained for analysis.
If an OG still possessed more than one sequence from one or more OTUs (inparalogues), the sequence with the shortest average pairwise distance to all others was retained. Pairwise distances were calculated using a gamma distribution with four rate categories as implemented in SCaFoS37. If two or more sequences from the same taxon were >10% divergent, all sequences from that taxon were discarded from that OG. To visualize the amount of data sampled for each taxon, a gene sampling diagram (Fig. 3) was created using MARE (http://mare.zfmk.de).
Contamination screening
Neomenioids have been reported to harbour nucleic acid contamination from their prey38. Given this, specimens of Wirenia argentea (which feed on cnidarians) were starved for 2 months before RNA extraction. Gut content analysis of Neomenia sp. confirmed that this undescribed Antarctic species (see Supplementary Results) also feeds on cnidarians. Therefore, Neomenia unigenes were compared to predicted transcripts of Lottia and Nematostella using TBLASTX and sequences with a lower E-value for Nematostella than Lottia (that is, sequences more similar to a sequence in the proteome of Nematostella than Lottia) were discarded. ML trees for each gene were manually evaluated and any remaining cnidarian contamination in the neomenioid data sets was removed by deleting sequences which either formed a clade with Nematostella or were part of a polytomy that included Nematostella. Finally, Nematostella was included in analyses with broad outgroup sampling (Supplementary Figs 3 and 4) to demonstrate that there is no obvious attraction between it and either neomenioid.
Phylogenetic analyses
Phylogenetic analyses were conducted using ML in RAxML 7.2.7 (ref. 27) and BI in PhyloBayes 2.3 (ref. 28) on the Alabama Supercomputer Authority Dense Memory Cluster (http://www.asc.edu/). For ML analyses, the best fitting amino acid substitution model for each gene was determined using the RAxML model selection Perl script. This script tests the fit of each available model of amino acid substitution by optimizing model parameters and branch lengths on a JTT start tree for each OG. Additionally, for comparative purposes, ML analyses using one model for the entire matrix were performed using the WAG + CAT + F and LG + CAT + F models in RAxML (Supplementary Figs 11 and 12) and an approximately ML analysis was performed using the JTT + CAT model in FastTree 2.1 (ref. 39, Supplementary Fig. 13). Topological robustness (that is, nodal support) for all ML analyses was assessed with 100 replicates of nonparametric bootstrapping. Stabilities of OTUs among the boot-strapped trees were calculated using the leaf stability index in Phyutility29. Competing hypotheses of mollusc phylogeny were evaluated using the AU test30 with the best-fitting model for each partition. For all BI analyses, the CAT model was used to account for site-specific rate heterogeneity28. Unless otherwise noted, all BI analyses were conducted with five parallel chains run for 15,000 cycles each, with the first 5,000 trees discarded as burn-in. A 50% majority rule consensus tree was computed from the remaining 10,000 trees from each chain. Topological robustness was assessed using posterior probabilities. Maxdiff values below 0.3 indicated that all chains in a run had converged.
Ancestral state reconstruction
Ancestral character state reconstruction was performed using an updated and modified version of the morphological matrix from ref. 4 in Mesquite 2.74 (http://mesquiteproject.org/) using maximum parsimony as the reconstruction method.
Supplementary Material
Acknowledgements
We thank W. Jones and K. T. Fielman for help with cDNA library preparation, R. M. Jennings, N. Mikkelsen, and the crews of the RV Håkon Mosby, RV Hans Brattstrom and RV Laurence M. Gould for assistance collecting aplacophorans, and J. C. Havird, P. J. Krug, S. C. Kempf, D. R. Lindberg, M. V. Matz, L. R. Page and T. H. Struck for discussions. D. Speiser kindly shared the photo of Argopecten. F. W. Goetz, A. Gracey and M. L. Blaxter kindly provided sequence quality data for Dreissena rostriformis, Mytilus californianus and Lumbricus rubellus, respectively. We thank A. Di Cosmo, P. Burbach, V. Rehder, W. Wright and R. Gillette for providing samples of Octopus, Loligo, Helisoma, Dolabrifera and Pleurobranchaea as well as sharing some sequencing cost for these species. We also thank D. Young and the Alabama Supercomputer Authority for access to computational resources. The genomes of Capitella teleta, Helobdella robusta, Lottia gigantea and Nematostella vectensis were produced by the US Department of Energy Joint Genome Institute in collaboration with the user community. This work was supported by National Science Foundation (NSF) grants (0744649 and 0821622) to K.M.H., National Institute of Health (NIH) grants (1RO1NS06076, 1R01GM097502, R21 RR025699, R21DA030118) and the McKnight Brain Research Foundation to L.L.M., the Deep Metazoan Phylogeny (DMP) program of the German Science Foundation (Li 998/9-1) to B.L., and The University of Bergen (Norway) free researcher initiated project grant to C.T. (project no. 226270). This work represents contributions 82 and 4 to the Auburn University (AU) Marine Biology Program and Molette Biology Laboratory for Environmental and Climate Change Studies, respectively.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions K.M.H., C.T., B.L., C.S. and K.M.K. conceived and designed this study. K.M.H., L.L.M., B.L. and C.T. supervised cDNA preparation and sequencing. L.L.M., A.B.K., K.M.K., J.T.C. and A.M. prepared and sequenced cDNA. K.M.K., J.T.C., S.R.S. and M.R.C. developed the bioinformatics pipeline. K.M.K. performed phylogenetic and ancestral state reconstruction analyses. K.M.K. and J.T.C. prepared the figures. C.S., C.T. and K.M.K. modified the morphological character matrix. A.B.K., K.M.K. and A.M. submitted sequences to GenBank. All authors contributed in preparing the Letter.
Capillary sequence data are available from the NCBI EST database (http://www.ncbi.nlm.nih.gov/projects/dbEST) under accession numbers JG454968.1–JG456874.1 and 454 sequence data are available from the NCBI SRA database (http://www.ncbi.nlm.nih.gov/sra) accession number SRA030407.1. Matrices and trees from this study are available from TreeBASE (http://www.treebase.org) accession number S11762.
The authors declare no competing financial interests.
References
- 1.Passamaneck YJ, Schander C, Halanych KM. Investigation of molluscan phylogeny using large-subunit and small-subunit nuclear rRNA sequences. Mol. Phylogenet. Evol. 2004;32:25–38. doi: 10.1016/j.ympev.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 2.Giribet G, et al. Evidence for a clade composed of molluscs with serially repeated structures: Monoplacophorans are related to chitons. Proc. Natl. Acad. Sci. USA. 2006;103:7723–7728. doi: 10.1073/pnas.0602578103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson NG, Rouse GW, Giribet G. Assessing the molluscan hypothesis Serialia (Monoplacophora+ Polyplacophora) using novel molecular data. Mol. Phylogenet. Evol. 2010;54:187–193. doi: 10.1016/j.ympev.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 4.Haszprunar G. Is the Aplacophora monophyletic? A cladistic point of view. Am. Malacol. Bull. 2000;15:115–130. [Google Scholar]
- 5.Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 6.Haszprunar G, Schander C, Halanych KM. In: Phylogeny and Evolution of the Mollusca. Ponder W, Lindberg DR, editors. Univ. of California Press; 2008. pp. 19–32. [Google Scholar]
- 7.Todt C, Okusu A, Schander C, Schwabe E. In: Phylogeny and evolution of the Mollusca. Ponder W, Lindberg DR, editors. Univ. of California Press; 2008. pp. 105–141. [Google Scholar]
- 8.Scheltema AH. Aplacophora as progenetic aculiferans and the coelomate origin of mollusks as the sister taxon of Sipuncula. Biol. Bull. 1993;184:57–78. doi: 10.2307/1542380. [DOI] [PubMed] [Google Scholar]
- 9.Salvini-Plawen L. On the phylogenetic significance of the aplacophoran Mollusca. Iberus. 2003;21:67–97. [Google Scholar]
- 10.Meyer A, Todt C, Mikkelson N, Lieb B. Fast evolving 18S rRNA sequences from Solenogastres (Mollusca) resist standard PCR amplification and give new insights into mollusk substitution rate heterogeneity. BMC Evol. Biol. 2010;10:70. doi: 10.1186/1471-2148-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Struck TH, et al. Phylogenomic analyses unravel annelid evolution. Nature. 2011;471:95–98. doi: 10.1038/nature09864. [DOI] [PubMed] [Google Scholar]
- 12.Sigwart JD, Sutton MD. Deep molluscan phylogeny: synthesis of palaeontological and neontological data. Proc. R. Soc. B. 2007;274:2413–2419. doi: 10.1098/rspb.2007.0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheltema AH, Ivanov DL. An aplacophoran postlarva with iterated dorsal groups of spicules and skeletal similarities to Paleozoic fossils. Invertebr. Biol. 2002;121:1–10. [Google Scholar]
- 14.Nielsen C, Haszprunar G, Ruthensteiner B, Wanninger A. Early development of the aplacophoran mollusc. Chaetoderma. Acta Zool. 2007;88:231–247. [Google Scholar]
- 15.Todt C, Wanninger A. Of tests, trochs, shells, and spicules: Development of the basal mollusk Wirenia argentea (Solenogastres) and its bearing on the evolution of trochozoan larval key features. Front. Zool. 2010;7:6. doi: 10.1186/1742-9994-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheltema AH, Schander C. Exoskeletons: tracing molluscan evolution. Venus. 2006;65:19–26. [Google Scholar]
- 17.Meyer A, Witek A, Lieb B. Selecting ribosomal protein genes for invertebrate phylogenetic inferences: how many genes to resolve the Mollusca? Method. Ecol. Evol. 2011;2:34–42. [Google Scholar]
- 18.Wanninger A, Haszprunar G. Muscle development in Antalis entalis (Mollusca, Scaphopoda) and its significance for scaphopod relationships. J. Morphol. 2002;254:53–64. doi: 10.1002/jmor.10004. [DOI] [PubMed] [Google Scholar]
- 19.Lundin K, Schander C, Todt C. Ultrastructure of epidermal cilia and ciliary rootlets in Scaphopoda. J. Molluscan Stud. 2008;75:69–73. [Google Scholar]
- 20.Moroz LL. On the independent origins of complex brains and neurons. Brain Behav. Evol. 2009;74:177–190. doi: 10.1159/000258665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simone LRL. PhD thesis. Univ. São Paulo; 2000. Filogenia das superfamílias de Caenogastropoda (Mollusca) com base em morfologia comparativa. [Google Scholar]
- 22.Jörger KM, et al. On the origin of Acochlidia and other enigmatic euthyneuran gastropods, with implications for the systematics of Heterobranchia. BMC Evol. Biol. 2010;10:323. doi: 10.1186/1471-2148-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caron JB, Scheltema A, Schander C, Rudkin D. A soft-bodied mollusc with radula from the Middle Cambrian Burgess Shale. Nature. 2006;442:159–163. doi: 10.1038/nature04894. [DOI] [PubMed] [Google Scholar]
- 24.Scheltema AH, Kerth K, Kuzirian AM. Original molluscan radula: comparisons among Aplacophora, Polyplacophora, Gastropoda, and the Cambrian fossil Wiwaxia corrugata. J. Morphol. 2003;257:219–245. doi: 10.1002/jmor.10121. [DOI] [PubMed] [Google Scholar]
- 25.Forment J, et al. EST2uni: an open, parallel tool for automated EST analysis and database creation, with a data mining web interface and microarray expression data integration. BMC Bioinformatics. 2008;9:5. doi: 10.1186/1471-2105-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebersberger I, Strauss S, Von Haeseler A. HaMStR: Profile hidden markov model based search for orthologs in ESTs. BMC Evol. Biol. 2009;9:157. doi: 10.1186/1471-2148-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixedmodels. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 28.Lartillot N, Philippe HA. Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 29.Smith SA, Dunn CW. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 2008;24:715–716. doi: 10.1093/bioinformatics/btm619. [DOI] [PubMed] [Google Scholar]
- 30.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
References
- 31.Chou HH, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- 32.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lottaz C, Iseli C, Jongeneel CV, Bucher P. Modeling sequencing errors by combining Hidden Markov models. Bioinformatics. 2003;19 doi: 10.1093/bioinformatics/btg1067. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misof B, Misof K. A Monte Carlo approach successfully identifies randomness in multiple sequence alignments: a more objective means of data exclusion. Syst. Biol. 2009 doi: 10.1093/sysbio/syp006. [DOI] [PubMed] [Google Scholar]
- 37.Roure B, Rodriguez-Ezpeleta N, Philippe H. SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol. Biol. 2007;7 doi: 10.1186/1471-2148-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okusu A, Giribet G. New18S rRNA sequences from neomenioid aplacophorans and the possible origin of persistent exogenous contamination. J. Molluscan Stud. 2003;69:385–387. [Google Scholar]
- 39.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.