While translational regulation is widely thought to contribute significantly to cellular differentiation programs, there are few genome-wide studies approaching this question. Here, the authors use ribosome profiling and mRNA-seq to assess gene-specific translation activity genome-wide in response to glucose starvation in yeast. Interestingly, a minority of transcripts comprise the majority of translating mRNA in both rapidly dividing and glucose-starved cells, though the identities of the translationally enriched mRNAs are distinct. Additionally, the authors identify features of the translational response that are specifically mediated by protein kinase A (PKA). These studies nicely reveal growth-state specialization of the “translatome.”
Keywords: adaptation to stress, glucose starvation, PKA signaling, translatome specialization, yeast invasive growth
Abstract
Cellular differentiation is driven by coordinately regulated changes in gene expression. Recent discoveries suggest that translation contributes as much as transcription to regulating protein abundance, but the role of translational regulation in cellular differentiation is largely unexplored. Here we investigate translational reprogramming in yeast during cellular adaptation to the absence of glucose, a stimulus that induces invasive filamentous differentiation. Using ribosome footprint profiling and RNA sequencing to assay gene-specific translation activity genome-wide, we show that prolonged glucose withdrawal is accompanied by gene-specific changes in translational efficiency that significantly affect expression of the majority of genes. Notably, transcripts from a small minority (<5%) of genes make up the majority of translating mRNA in both rapidly dividing and starved differentiating cells, and the identities of these highly translated messages are almost nonoverlapping between conditions. Furthermore, these two groups of messages are subject to condition-dependent translational privilege. Thus the “housekeeping” process of translation does not stay constant during cellular differentiation but is highly adapted to different growth conditions. By comparing glucose starvation to growth-attenuating stresses that do not induce invasive filamentation, we distinguish a glucose-specific translational response mediated through signaling by protein kinase A (PKA). Together, these findings reveal a high degree of growth-state specialization of the translatome and identify PKA as an important regulator of gene-specific translation activity.
INTRODUCTION
Free-living microbes must adapt to the availability of specific nutrients in their environments. Remarkably, the yeast Saccharomyces cerevisiae can adjust its growth and division rate over a 10-fold range in response to nutrient levels (Brauer et al. 2008). In addition to regulating growth rate, depletion of specific nutrients induces adaptive cellular differentiation programs including sporulation, filamentation, and quiescence. In haploid yeast, glucose starvation induces invasive growth, a developmental program in which yeast cells form filamentous networks that invade the growth medium. This is thought to function as a foraging response allowing the yeast colony to “search” for glucose (Broach 2012; Cullen and Sprague 2012). Invasive filamentous differentiation involves changes in cell polarity, actin cytoskeletal organization, cell surface properties, and cell-cycle progression. Two conserved signaling pathways control glucose-regulated filamentation–the Ras/PKA pathway and a mitogen-activated protein kinase (MAPK) pathway (for review, see Cullen and Sprague 2012). Recent genome-scale gene deletion studies have identified hundreds of genes that are specifically required for invasive filamentous growth but are dispensable for rapid growth in the presence of glucose (Chin et al. 2012; Ryan et al. 2012), highlighting the profound differences between these two growth states.
In addition to controlling filamentous differentiation, glucose availability regulates sweeping changes in gene expression. The Ras/PKA pathway mediates most of the glucose-induced transcriptional changes, with additional regulation by the AMP-activated protein kinase (AMPK) Snf1, the Hap2/3/4/5 transcription complex, and the Rgt network (Zaman et al. 2009; Broach 2012). The mRNA levels of ∼40% of yeast genes change ≥2-fold in response to glucose addition or withdrawal (Brauer et al. 2005; Zaman et al. 2009; Arribere et al. 2011). Thus, the availability of glucose substantially affects the pool of cellular mRNA.
The acute response to glucose withdrawal also profoundly affects cellular translation activity. Within 1 min, polysomes collapse and protein synthesis rates plummet (Ashe et al. 2000). We, and others, have recently shown that some genes escape translational repression during the early stages of glucose withdrawal (Arribere et al. 2011; Castelli et al. 2011), demonstrating that the translation activity of individual genes can be differentially regulated in glucose-starved cells. It is not known how long-term adaptation to the absence of glucose affects translation globally or for individual genes.
Here we have investigated translational regulation in starved differentiating yeast using ribosome footprint profiling to determine gene-specific translation activity genome-wide (Ingolia et al. 2009). We show that cellular adaptation to the absence of glucose is accompanied by widespread and functionally coherent translational changes, which could not be detected by bulk polysome profiling approaches (Arribere et al. 2011; Castelli et al. 2011). Specifically, transcripts from growth-promoting genes, including ribosomal proteins (RPs) and ribosome biogenesis (RiBi) factors, are very efficiently translated in rapidly dividing cells and preferentially translationally repressed under differentiation-promoting starvation conditions. In contrast, transcripts encoding mitochondrial proteins (mito mRNAs) are specifically translationally activated in starved cells. As a class, genes required for invasive growth are significantly translationally up-regulated in response to glucose withdrawal, consistent with a positive role for translational regulation in cellular differentiation. Using a candidate gene approach, we identify the conserved cAMP-dependent protein kinase (PKA) as a major regulator of these gene-specific glucose-responsive translation changes. Together, our results identify mechanisms for pervasive translational changes in response to glucose availability and reveal a surprising degree of growth state-dependent specialization of the process of translation.
RESULTS
Global translational reprogramming in response to glucose starvation
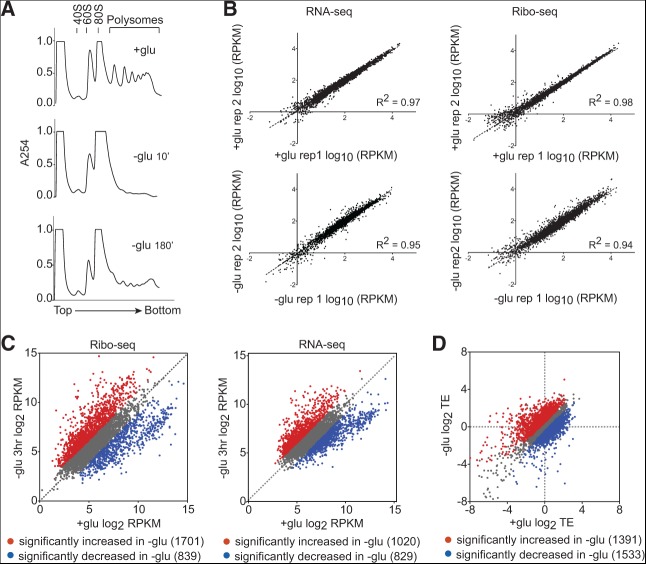
To examine translational changes following glucose withdrawal, we performed ribosome footprint profiling (Ribo-seq), which consists of isolating and sequencing ribosome-protected mRNA fragments and gives a precise measure of gene-specific translation activity genome-wide (Ingolia et al. 2009). Wild-type Sigma 1278b cells were grown to mid-log phase in rich media containing 2% glucose and then transferred to rich media base lacking glucose. Immediately following glucose withdrawal, polysomes collapsed (Fig. 1A), as previously described (Ashe et al. 2000). Polysomes gradually recovered to a reduced level by 3 h, at which point cells had resumed growth at a reduced rate (Fig. 1A; data not shown). Ribo-seq and RNA-seq were highly reproducible (R2 = 0.94–0.98) between replicates (Fig. 1B) and revealed profound changes in the transcriptome and translatome of starved cells. mRNA abundance was significantly affected for 32% of genes, and ribosome footprints were significantly affected for 45% of genes (P < 0.05, based on analysis of replicates using counting statistics) (Fig. 1C). Gene expression changes were correlated with changes previously observed 2 h after glucose withdrawal using microarrays to total mRNA and polysomal mRNA (R2= 0.64 and 0.65 for RNA-seq and Ribo-seq, respectively), although Ribo-seq revealed larger fold changes than those previously detected by bulk polysome profiling (Arribere et al. 2011).
FIGURE 1.
Glucose withdrawal caused widespread translational changes. (A) Polysome profiles of cells grown in rich media with or without 2% glucose for the indicated times. (B) Ribo-seq (right) and RNA-seq measurements (left) were reproducible. (C) Ribo-seq and RNA-seq libraries were prepared from cells starved for 3 h. Average ribosome densities (reads per kilobase per million reads) for 4576 genes that passed a minimum read cutoff were calculated from two biological replicates. Genes that were significantly (P < 0.05) increased (red) or decreased (blue) were identified by analysis of replicates using counting statistics as described (Larsson et al. 2011). (D) Translational efficiencies (TEs) in glucose-fed cells vary widely from those in glucose-starved cells. Average TEs for 5447 genes that passed a minimum read cutoff were calculated from two biological replicates. Genes that were significantly (FDR < 1%) increased (red) or decreased (blue) were identified by analysis of propagated RNA-seq and Ribo-seq variances (Materials and Methods).
Combining Ribo-seq with measurement of total transcript abundance by RNA-seq can reveal changes in the translational efficiency (TE) for individual genes. Adaptation to glucose withdrawal was accompanied by pervasive translational regulation: 54% of all genes showed significant differences in TE between glucose-grown and glucose-starved cells (FDR < 1%, based on propagation of RNA-seq and Ribo-seq variances) (Fig. 1D). Of particular note were 1391 genes with significantly increased translation activity in starved cells, which were not identified by bulk polysome profiling (Arribere et al. 2011). This difference could be explained by the fact that Ribo-seq can distinguish between mRNAs with different numbers of ribosomes per mRNA instead of counting all polysomal mRNAs equally.
Both microarray and sequencing-based methods can measure fold changes for individual genes between conditions, but only RNA-seq and Ribo-seq allow accurate quantitation of the contributions of individual genes to the total and translating mRNA pools. We exploited this characteristic of sequencing-based gene expression data to determine how different are the translatomes of two cellular growth states—rapidly dividing and starved differentiating. The translated mRNA pools were strikingly specialized in both growth conditions, as most ribosome-protected fragments came from a small subset of genes (Gini coefficients of 0.85 and 0.79) (Fig. 2A). After correction for differences in ORF length, a mere 111 genes were sufficient to account for 50% of translated mRNA in rapidly dividing cells, and 184 genes comprised the top 50% in glucose-starved cells. Remarkably, there were only 12 genes in common between the two conditions (Fig. 2B; Supplemental Table 1). The identities of these genes were reproducible between replicates (Fig. 2C). Based on these results, we infer that the majority of translation initiation events occurred on different mRNAs in glucose-starved compared to rapidly dividing cells. This conclusion has profound implications for the roles of general translation factors (Discussion). The short gene lists comprising the top 50% of translating mRNAs were dominated by two functional categories, one per condition. In glucose, cytoplasmic ribosomal proteins accounted for 80/111 genes (72%, P = 2.7 × 10−112). In cells adapting to glucose withdrawal, 119/184 of the most highly translated genes encode proteins that localize to mitochondria (65% by GO component analysis, P = 2.7 × 10−49). Thus, the majority of the translational machinery was dedicated to synthesizing a different small set of proteins, which depended on the growth state of the cells.
FIGURE 2.
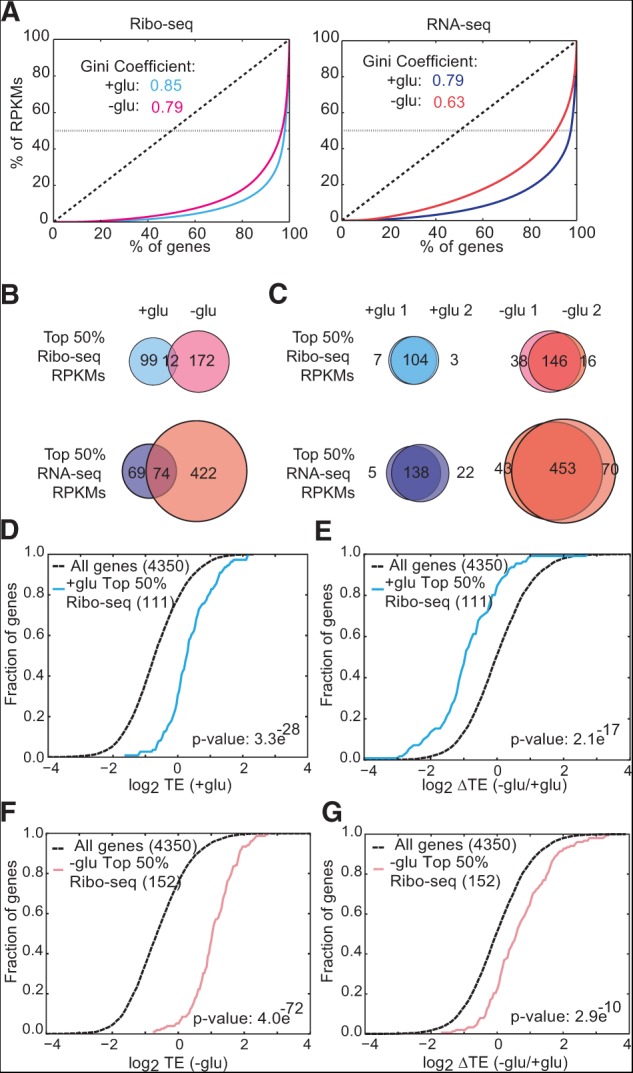
Gene-specific translation efficiencies are adapted to growth conditions. (A) Gini coefficient plots of yeast genes versus Ribo-seq (left) and RNA-seq (right) reads per kilobase per million mapped reads (RPKMs) represent the heavily skewed gene expression profiles in different growth conditions. The dotted horizontal lines represent the top 50% of RPKMs from each library. (B) The translated mRNA pool differed substantially depending on growth conditions (±glucose). Venn diagrams represent the genes that make up the top 50% of RPKMs from each library. (C) The identities of the most highly expressed and translated mRNAs were reproducible between biological replicates. Venn diagrams represent the genes that make up the top 50% of RPKMs from each library. (D) Cumulative distribution (CDF) plot for translational efficiency (Ribo-seq RPKM divided by RNA-seq RPKM) in +glu of the top 50% of genes by Ribo-seq RPKMs in +glu. (E) CDF plot for changes in TE upon glucose withdrawal of the top 50% of genes by Ribo-seq RPKMs in +glu. (F) CDF plot for TE in −glu of the top 50% of genes by Ribo-seq RPKMs in −glu. (G) CDF plot for changes in TE upon glucose withdrawal of the top 50% of genes by Ribo-seq RPKMs in −glu. Individual replicates were used to identify the top 50% genes. TEs are the average of two biological replicates. Significance was determined by the Kolmogorov-Smirnov test.
Next we asked whether condition-dependent differences in translational efficiency (TE) were consistent with growth state optimization of cellular translation activity. Elongation rates were assumed to be constant between genes, as suggested by recent analysis (Ingolia et al. 2011). Gene-specific TEs were computed by normalizing ribosome footprint density to total mRNA abundance. The most highly translated mRNAs in glucose-grown cells showed elevated TE compared to all genes in the presence of glucose (P = 3.3 × 10−28) (Fig. 2D), and this group was significantly more translationally repressed following glucose withdrawal, i.e., had a greater reduction in the average number of ribosomes per transcript (P = 2.1 × 10−17) (Fig. 2E). On the other hand, the most highly translated mRNAs from glucose-starved cells had moderate TEs in the presence of glucose (data not shown) and very high TEs following glucose withdrawal (P = 4.0 × 10−72) (Fig. 2F) due to a significant increase in TE (P = 2.9 × 10−10) (Fig. 2G). Fewer genes were required to account for the top 50% of Ribo-seq reads compared to RNA-seq reads (Fig. 2B), which may be partly explained by translational privileging of highly expressed genes. Together, these results show that glucose starvation induced pronounced gene-specific translational changes. These TE changes are consistent with a retooling of the translation machinery to better translate a dramatically altered mRNA substrate pool.
Translational control of starvation adaptation genes
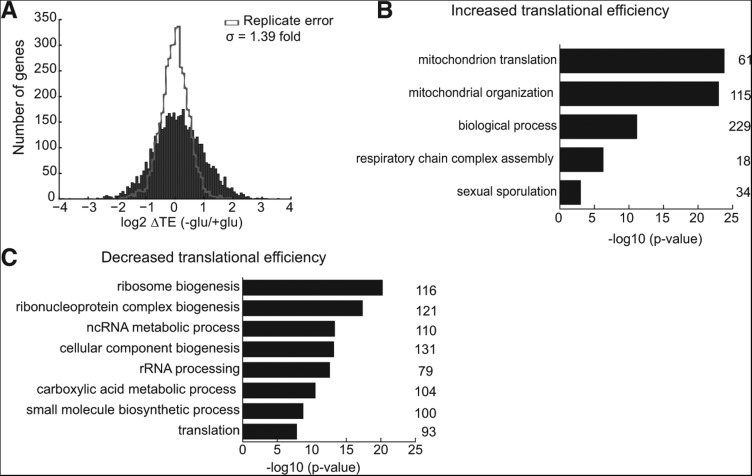
Next we examined whether translational regulation disproportionately impacted genes with important functions in cellular adaptation to starvation. Global gene deletion analysis has identified the genes required for starvation-induced invasive growth (430) and adhesion to agar (329) (Chin et al. 2012; Ryan et al. 2012). These sets of genes were preferentially translationally up-regulated in glucose-starved cells compared to all genes (P = 2.1 × 10−4, 1.9 × 10−8). Globally, glucose withdrawal resulted in significant (FDR < 1%) changes in TE for 54% of yeast genes and ≥2-fold changes in TE for 29% of yeast genes (Figs. 1D, 3A). Notably, of the 870 genes that were substantially translationally activated (▵TE ≥ +2-fold) in glucose-starved cells, mRNAs encoding mitochondrial proteins (mito mRNAs) formed the dominant category (Fig. 3B). Moreover, as a group, nuclear-encoded mito mRNAs were significantly better translated compared to all genes in glucose-starved cells (P = 3 × 10−33). Mitochondrial respiratory function is critical for yeast growth and filamentation in the absence of glucose (Broach 2012), and nuclear-encoded mitochondrial protein genes are substantially overrepresented among the genes reported to be required for invasive growth (159/430, P = 2.6 × 10−29) and adhesion (152/329, P = 6.1 × 10−38). Thus, translational regulation enhances synthesis of proteins that are required for the adaptive cellular response to glucose starvation.
FIGURE 3.
RP and mito mRNAs differentially translated upon glucose withdrawal. (A) Distribution of changes in TE in glucose-starved cells. (B) GO category enrichment of genes whose TE increases ≥2-fold (FDR < 1%) in glucose starvation. The number of genes in each category is indicated to the right of the graph. (C) GO category enrichment of genes whose TE decreases ≥2-fold (FDR < 1%) in glucose starvation. The number of genes in each category is indicated to the right of the graph.
On the other hand, genes that are important for rapid growth in the presence of glucose were selectively translationally repressed during glucose starvation. Genes with TE reduced ≥2-fold fell into a few main GO categories: ribosome biogenesis (16% of translationally repressed genes), cytoplasmic translation (13%), metabolic processes (69%), and glucose catabolism (3%). Furthermore, most genes within these functional categories showed significant reductions in TE following glucose withdrawal. Thus, widespread translational regulation leads to enhanced expression of specific genes under the cellular conditions where the gene products are required for optimal growth.
The translational and transcriptional changes caused by glucose starvation were largely coherent—ribosomal protein genes were expressed at lower levels and also translated less efficiently, while mitochondrial protein genes were both transcriptionally up-regulated and translationally privileged in starved cells. It was recently argued, based on mathematical principles, that using RNA-seq values to normalize Ribo-seq data may not be sufficient to eliminate the contribution of transcriptional changes to TE changes quantified by taking log2 ratios of Ribo-seq RPKMs (reads per kilobase per million mapped reads) to RNA-seq RPKMs. An alternative approach was proposed, which analyzes partial variance and uses a variance shrinkage method for improving statistical inference, in a minimum of three biological replicates to assess changes in gene-specific translational efficiency (Larsson et al. 2011). We therefore performed a third independent experiment and analyzed gene-specific translation changes caused by glucose withdrawal using the ANOTA R package. This alternative analysis revealed similarly significant translational regulation of RP and mito mRNAs in glucose-starved cells (P = 5.3 × 10−20 and 1.9 × 10−12, respectively), thus ruling out the trivial explanation of inadequate normalization of ribosome footprints to total RNA levels as an explanation for these TE changes.
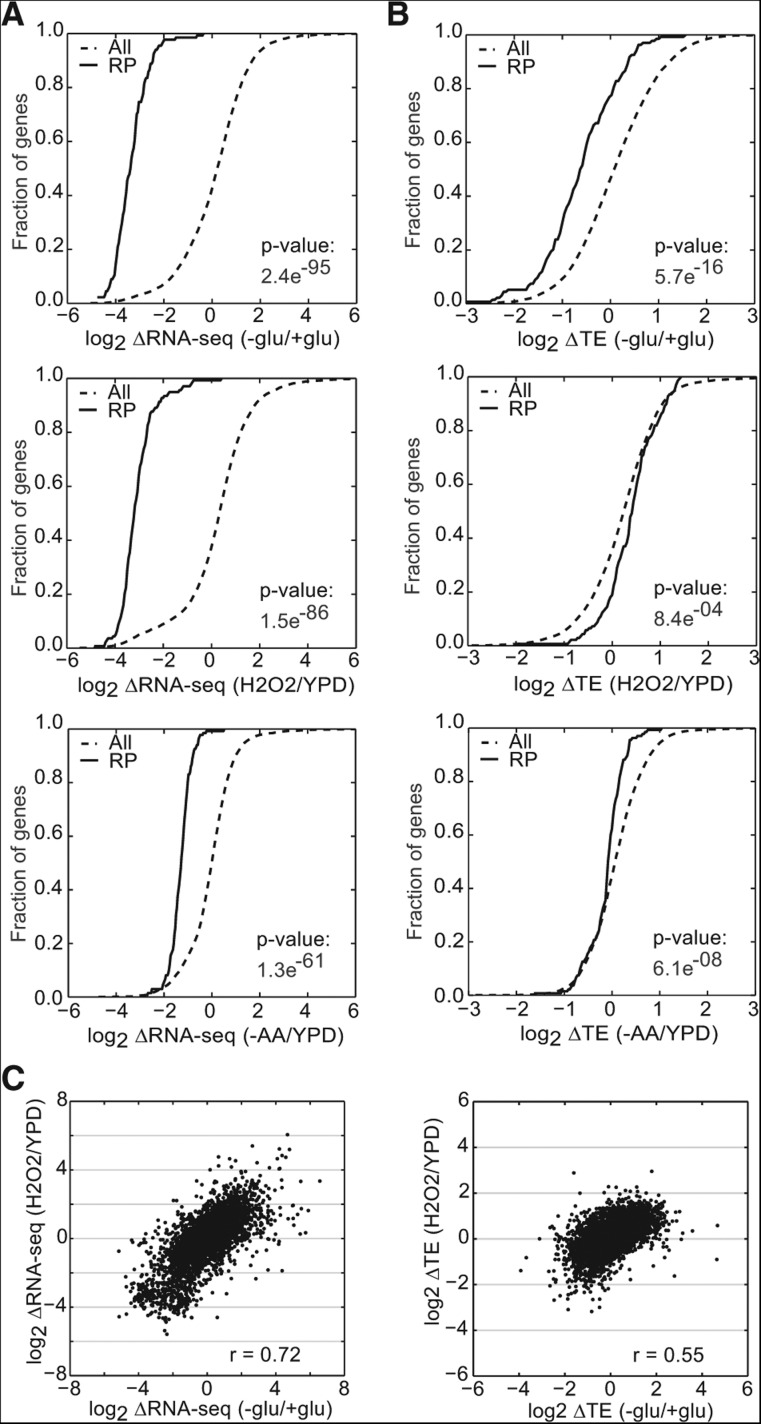
Next, we considered the possibility that increased and decreased competition by mRNAs for limiting translation initiation factors could be responsible for corresponding changes in mRNA abundance and translational efficiency for RP and mito mRNAs following glucose starvation. According to this model, the translational efficiency of RP mRNAs decreases as their relative abundance decreases, while transcriptionally up-regulated mRNAs such as the mito mRNAs become more competitive under conditions where they comprise a much larger portion of the total mRNA pool. To test this competition hypothesis, we compared changes in total mRNA abundance and TE for RP mRNAs in other conditions where the RP genes are transcriptionally down-regulated. Yeast Ribo-seq and RNA-seq data were available from four relevant stress conditions: glucose starvation (this study), amino acid starvation (Ingolia et al. 2009), nutrient deprivation during meiosis (Brar et al. 2012), and oxidative stress induced by hydrogen peroxide (H2O2) (Gerashchenko et al. 2012). Treatment with H2O2 and induction of meiosis caused reductions in RP mRNA levels that were well correlated with and comparable in magnitude to glucose starvation, yet H2O2 treatment did not lead to translational repression of RP mRNAs (Fig. 4A–C; data not shown). Amino acid starvation caused more moderate reductions in RP mRNA levels than either glucose starvation or H2O2 treatment and did not lead to consistent reductions in TE (Fig. 4A,C). Thus, decreased mRNA abundance is not sufficient to cause decreased translational efficiency of RP mRNAs.
FIGURE 4.
Translational regulation of RP genes is specific to individual stresses. (A,B) Translational and transcriptional changes of RP genes can be unlinked. CDF plots for changes in RNA-seq (A) and TE (B) of ribosomal protein genes (RP genes) upon glucose withdrawal, oxidative stress (H2O2) (Gerashchenko et al. 2012), and amino acid starvation (−AA) (Ingolia et al. 2009). (C) Comparison of changes in RNA-seq (left) and TE (right) between oxidative stress and glucose starvation. TEs were computed only for genes with sufficient reads in both RNA-seq and Ribo-seq libraries from +glu and −glu samples. RNA-seq and TE changes are the averages of two biological replicates. Significance was determined by the Kolmogorov-Smirnov test.
Strong transcriptional up-regulation of mito mRNAs was specific to glucose starvation. Thus, to determine whether increased mRNA abundance is generally associated with increased TE, we examined a group of genes that are induced in response to a variety of stresses as part of the environmental stress response (ESR) (Gasch et al. 2000). The ESR consists of regulated changes in ∼900 genes—down-regulation of the transcription of growth promoting genes and induction of stress-responsive genes—which are believed to allow cells to adapt to suboptimal environments. Stress response genes were induced in all four stresses (Fig. 5A). However, the presence and magnitudes of TE changes varied significantly between individual stresses and did not simply parallel changes in transcriptional activity (Fig. 5B). Taken together, these results show that changing the relative abundance of an mRNA does not lead to changes in its TE. Thus, translational regulatory mechanisms beyond simple competition for limiting factors are at work in glucose-starved cells.
FIGURE 5.
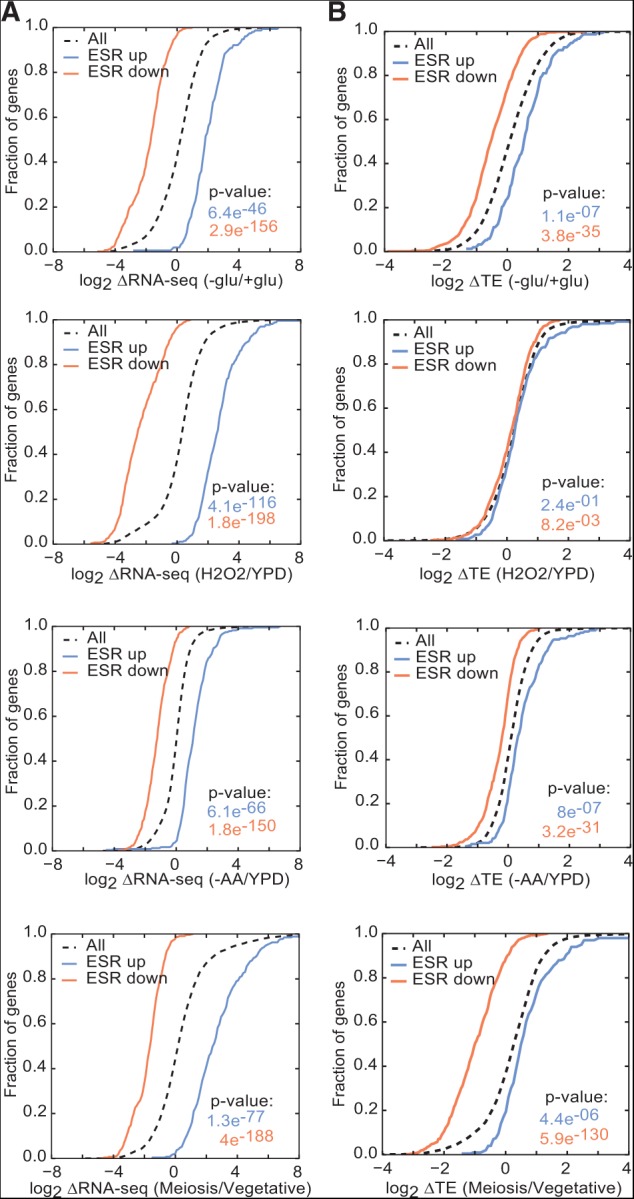
Translational control mechanisms are stress-specific. (A,B) Translational and transcriptional changes of environmental stress response (ESR) genes can be unlinked. CDF plots for changes in RNA-seq (A) and TE (B) of ESR genes transcriptionally induced (ESR up) or repressed (ESR down) in a variety of stresses including glucose starvation, amino acid starvation (−AA) (Ingolia et al. 2009), oxidative stress (H2O2) (Gerashchenko et al. 2012), and nutrient stress during the meiotic program (Meiosis) (Brar et al. 2012). TEs were computed only for genes with sufficient reads in both RNA-seq and Ribo-seq libraries from +glu and −glu samples. RNA-seq and TE changes are the averages of two biological replicates. Significance was determined by the Kolmogorov-Smirnov test.
Translation changes are due to altered efficiency of initiation and mRNA sequestration
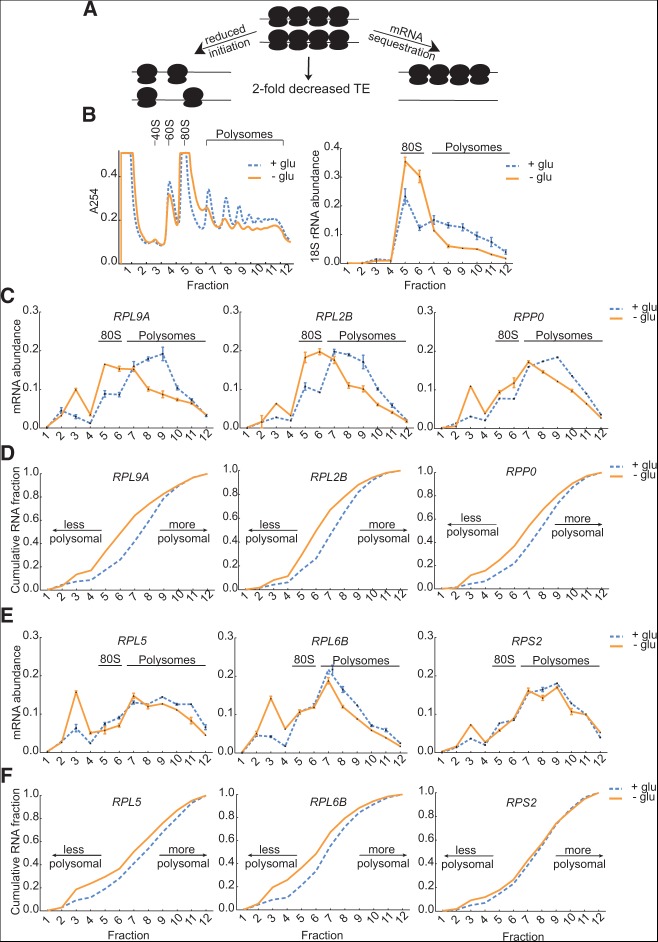
Reduced TE can arise by two distinct mechanisms that affect ribosome recruitment: an increase in the size of the nontranslating pool of mRNA (e.g., by mRNA sequestration), or a decrease in the average number of ribosomes associated with each transcript due to reduced efficiency of initiation. These mechanisms are indistinguishable by Ribo-seq, as either leads to a decrease in the number of ribosome footprints recovered per mRNA molecule (Fig. 6A). To determine the mechanism underlying reduced TE of RP mRNAs, we performed polysome profiling followed by qRT-PCR for representative RP genes. Relative mRNA abundance per fraction from polysome profiles before or after glucose withdrawal was quantified by normalization to a dope-in control firefly luciferase RNA (FLUC). The distribution of 18S rRNA after normalization was determined for reference (Fig. 6B). In the presence of glucose, almost all RP mRNA was associated with ribosome-containing fractions (5–12). Two modes of translational repression were observed 3 h after glucose withdrawal. For RPL9A, RPL2B, and RPP0, there was a modest increase in ribosome-free mRNA (fractions 1–4), but most mRNA (74%–93%) remained associated with one or more ribosomes. Although still ribosome-associated, these RP mRNAs shifted toward smaller polysomes in glucose-starved cells (Fig. 6C,D). On the other hand, RPL5, RPL6B, and RPS2 showed a pronounced accumulation of mRNA in the nontranslating fractions relative to the polysome fractions (Fig. 6E,F). Thus the translational repression of RP mRNAs in cells adapting to prolonged glucose starvation proceeds by two mechanisms—a decrease in the frequency of initiation and an increase in the size of the nontranslating pool.
FIGURE 6.
Two modes of translational regulation of RP mRNAs in glucose-starved cells. (A) Model for reduced TE by mRNA sequestration (right) or reduced frequency of translation initiation (left). (B) Polysome profiles of cells grown in rich media with or without 2% glucose for 3 h (left panel). qPCR analysis of 18S rRNA in polysome gradient fractions (right panel). (C) Translating RP mRNAs (RPL9A, RPL2B, RPP0) associated with fewer ribosomes after 3 h of glucose starvation. Relative mRNA abundance per fraction from polysome profiles before or after glucose withdrawal was quantified by qPCR normalized to dope-in control (FLUC) RNA. Error bars indicate standard deviation for technical replicates (n = 3). This is a representative of two replicate experiments. (D) CDF plots of mRNA abundance from fractions in C. (E) RP mRNAs (RPL5, RPL6B, RPS2) accumulated in ribosome-free fractions after 3 h of glucose starvation. Relative mRNA abundance per fraction from polysome profiles before or after glucose withdrawal was quantified by qPCR normalized to dope-in control (FLUC) RNA. Error bars indicate standard deviation for technical replicates (n = 3). This is a representative of two replicate experiments. (F) CDF plots of mRNA abundance from fractions in E.
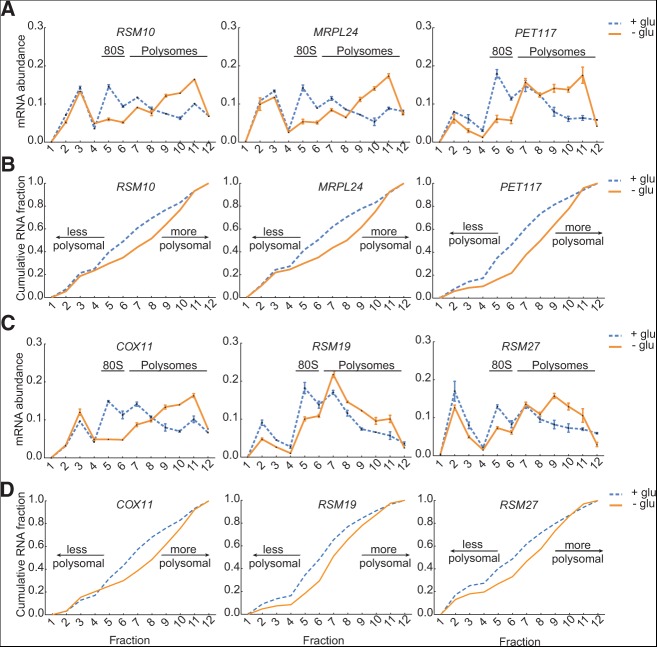
In contrast, the increase in TE of mito mRNAs in glucose-depleted cells was almost entirely due to increased polysome size, rather than an increased fraction of actively translating mRNA. Polysome profiling by qPCR revealed that representative mito mRNAs had relatively large amounts of free mRNA (13%–26% of total mRNA) in glucose-grown cells, consistent with previous genome-wide assessments, which observed low ribosome occupancy for mito mRNAs under these conditions (Arava et al. 2003; Arribere et al. 2011). However, in glucose-starved cells, this ribosome-free pool changed very little, whereas the ribosome-associated mito mRNAs shifted from light to heavier polysomes (Fig. 7A–D). This constancy of the ribosome-free pool explains why translational up-regulation of mito mRNAs following glucose withdrawal was not detected previously (Arribere et al. 2011). Therefore, mito mRNAs changed TE primarily due to altered translation initiation rather than altered partitioning between translating and nontranslating pools.
FIGURE 7.
Increased polysome size of mito mRNAs in glucose-starved cells. (A) Mitochondrial protein mRNAs (RSM10, MRPL24, PET117) associated with heavier polysomes after 3 h of glucose starvation. mRNA abundance per fraction was determined as in Figure 5. This is a representative of two replicate experiments. (B) CDF plots of mRNA abundance from fractions in A. (C) Mitochondrial protein mRNAs (COX11, RSM19, RSM27) associated with heavier polysomes after 3 h of glucose starvation. mRNA abundance per fraction was determined as in Figure 5. This is a representative of two replicate experiments. (D) CDF plots of mRNA abundance from fractions in C.
Inhibition of PKA signaling partially recapitulated translational effects of glucose withdrawal
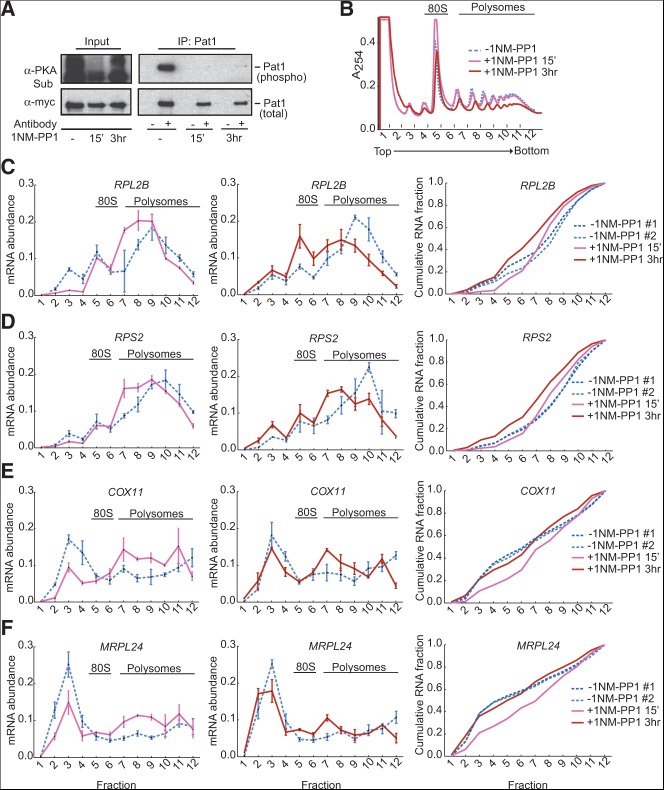
What could be responsible for widespread changes in gene-specific translational efficiencies specifically in response to glucose availability? Signaling through the Ras/PKA pathway is required for the majority of glucose-responsive transcriptional changes, and Ras/PKA activation is sufficient to recapitulate ∼90% of glucose-induced changes in mRNA abundance (Zaman et al. 2009). Additionally, glucose starvation has been shown to inhibit PKA activity (Broach 2012). We therefore investigated whether the Ras/PKA pathway also regulates translation of glucose-responsive mRNAs. PKA kinase activity was monitored by following phosphorylation of Pat1, which is phosphorylated by PKA on Ser-456 and Ser-457 (Ramachandran et al. 2011). To separate the effects of inhibiting PKA from the glucose starvation response, we exploited an analog-sensitive strain (pka-as) in which all three genes encoding the catalytic subunit of PKA (TPK1, TPK2, and TPK3) were mutated to render them sensitive to the ATP-analog 1NM-PP1. This inhibitor inactivates kinases whose active sites have been altered to accommodate binding of a bulky side group and has no effect on the growth of cells lacking the sensitizing mutation(s) (Bishop et al. 2001). Treatment of pka-as cells with 1NM-PP1 inhibited PKA activity (Fig. 8A) and caused a small reduction in bulk translation activity within 15 min and a larger reduction after 3 h based on a reduction in polysomes (Fig. 8B). Thus, inhibition of PKA was sufficient to decrease translation even in the presence of glucose.
FIGURE 8.
PKA activity promoted efficient translation of RP mRNAs and inhibited translation of mito mRNAs. (A) PKA-dependent phosphorylation of Pat1 was lost in an analog-sensitive PKA strain (tpk1-as tpk2-as tpk3-as) following treatment with 1NM-PP1 for 15 min and 3 h. A myc-tagged version of the PKA substrate Pat1 was immunoprecipitated, and the relative level of phosphorylation was determined by Western blotting with an anti-PKA substrate antibody (α-PKA Sub). Phosphorylated Pat1 in the IP lanes does not necessarily align with any of the bands in the input lane because there are hundreds of PKA substrates in the cell (Ptacek et al. 2005) that may be contributing to the signal in the input panel. 1NM-PP1 is stable in yeast over the time scale of this experiment, thus the slight increase in reactivity to the anti-PKA sub antibody observed in the input lanes after 3 h +1NM-PP1 is unlikely to be due to reactivation of PKA. It may be due to increased protein kinase C (PKC)-dependent phosphorylation when PKA is inhibited, because PKC recognizes a closely related sequence, (R/K) (R/K) X (S*) (Hydrophobic) (R/K). (B) Inhibiting PKA reduced bulk translation activity in the presence of glucose. Polysome profiles were analyzed after treatment with 1NM-PP1 for 15 min and 3 h. Representative traces from three independent replicates are shown. (C,D) Inhibiting PKA for 15 min or for 3 h decreased translation of RP mRNAs (RPL2B [C] and RPS2 [D]) in the presence of glucose. Polysome analysis of mRNAs (left and center) was performed as in Figure 5. Combined CDF plots of mRNA abundance from fractions including both time points of 1NM-PP1 inhibition are shown on the right. (E,F) Inhibiting PKA in the presence of glucose for 15 min increased translation of mito mRNAs (COX11 [left panel, E] and MRPL24 [left panel, F]). However, after 3 h of PKA inhibition, translation of mito mRNAs was similar to uninhibited glucose-replete conditions (center panels, E and F). Relative mRNA abundance per fraction was determined as in Figure 5. Combined CDF plots of mRNA abundance from fractions including both time points of 1NM-PP1 inhibition are shown on the right.
Next we examined gene-specific translational responses to PKA inhibition to determine whether inactivating PKA was sufficient to recapitulate the selective translational effects of glucose withdrawal. Treatment of rapidly dividing pka-as cells with 1NM-PP1 for 15 min caused a shift of RP mRNAs (RPL2B, RPS2) toward smaller polysomes, indicating reduced translation initiation despite the presence of glucose. RP mRNA translation was further inhibited after 3 h of incubation with the PKA inhibitor (Fig. 8C,D). In contrast, inhibiting PKA for 15 min increased translation activity of mito mRNAs (COX11, MRPL24) in the presence of glucose (left panels in Fig. 8E,F), but unlike RP mRNAs, translation changes of mito mRNAs were attenuated after 3 h of PKA inhibition in the presence of glucose (Fig. 8E,F). Taken together, these results show that PKA activity is necessary for highly efficient translation of RP mRNAs even when glucose is available. In contrast, glucose availability eventually trumps PKA inactivity for translational repression of the mito mRNAs, but it takes time for cells to adapt to the atypical situation of having high glucose and low PKA activity. Thus, the mechanisms for PKA-mediated RP mRNA translational activation and mito mRNA translational repression are distinct and not merely two sides of the same coin (see Discussion).
DISCUSSION
Glucose starvation induces filamentous differentiation accompanied by pervasive transcriptional changes that affect >40% of the genome. Here we showed that glucose starvation also causes widespread translational reprogramming of the transcriptome. Genes encoding mitochondrial proteins, which were transcriptionally up-regulated in response to glucose withdrawal, were also translationally activated in glucose-starved cells. Conversely, genes encoding cytoplasmic ribosomal proteins were translated less efficiently in addition to being transcribed at lower levels in the absence of glucose. These concerted changes in gene-specific TEs contributed to a profound growth state-dependent specialization of translation. In fact, the majority of translating ribosomes were engaged with mRNAs from <5% of genes in each condition, and translatomes were substantially less diverse than transcriptomes, particularly in glucose-starved cells.
Not only were translatomes highly specialized in both growth conditions, the identities of the most highly translated messages were almost nonoverlapping between conditions. This observation has profound implications for studying translation and for understanding the impact of translation factors and RNA-binding proteins on gene expression. According to our data, a hypothetical factor that was absolutely required for translation of 95% of genes might cause only a twofold change in bulk translation, depending on which 95% of genes it affected. Thus, the general importance of such a factor could easily be overlooked by relying on bulk assays. Conversely, a factor that causes bulk translation defects when mutated or depleted in a particular condition, and which has therefore been considered as a “general” translation factor, may actually play a specialized role in promoting translation of relatively few highly translated mRNAs. Because the identities of the ∼5% of genes that comprise the majority of translation activity differ between cellular growth conditions, the factor requirements for bulk translation could be strongly condition-dependent. It will be important for future work to assess the gene-specific effects of translation factors and to examine their roles under different growth conditions.
We have shown that the mechanisms of translational repression of RP mRNAs and translational activation of mito mRNAs are downstream of PKA signaling. Increased translation of mito mRNAs in glucose starvation or following inhibition of PKA is not simply an indirect consequence of reduced RP mRNA translation (e.g., by reduced competition for limiting initiation factors), because the two translational responses could be uncoupled. After 3 h of PKA inhibition with 1NM-PP1 in the presence of glucose, RP mRNA translation was still repressed, while mito mRNA translation had returned to the lower level typical of these transcripts in glucose-grown cells. Thus, separate PKA-dependent mechanisms regulate the efficiency of translation initiation on RP and mito mRNAs. Differential activity of PKA isoforms is a potential source of stress specificity in the gene-specific effects of PKA on translation. The catalytic subunit of PKA is encoded by three separate genes in S. cerevisiae (TPK1, TPK2, and TPK3). In our experiments, all three PKA catalytic subunits were inhibited with 1NM-PP1. Given that the Tpk isoforms have distinct substrate preferences (Ptacek et al. 2005) and subcellular localizations and show isoform-specific changes in localization upon glucose withdrawal (Tudisca et al. 2010), it will be important to determine which isoform(s) mediate translational activation of RP mRNAs and translational repression of mito mRNAs. Additionally, other glucose-responsive signaling pathways may also contribute to gene-specific translational control. Indeed, the minor differences in polysomal distributions of RP mRNAs between glucose-starved and PKA-inhibited cells offer some support for this possibility. It will be important for future studies to measure the relative contributions of each of these signaling networks toward starvation-induced translational reprogramming.
Our comparative analysis of different stress conditions suggests that only a small fraction of the genome is subject to similar translational regulation in all stresses, which contrasts with much greater similarity between stresses at the transcriptional level. Thus, control mechanisms that regulate gene translation in one condition may not necessarily be utilized in a different growth or stress condition. Because the functions of most yeast translation factors and cytoplasmic RNA-binding proteins have been characterized primarily, if not exclusively, in cells growing in glucose, the factors responsible for translational activation of mito mRNAs and other starvation-induced transcripts are likely to be relatively uncharacterized. Intriguingly, more than 70 annotated yeast RNA binding proteins changed expression ≥2-fold in starved cells, suggesting that many aspects of post-transcriptional gene regulation may be altered to adapt to the transformation of the mRNA pool by glucose starvation. It will be interesting to see whether cellular differentiation in multicellular eukaryotes is associated with a similarly high degree of translatome specialization. If so, this could illuminate the basis for the surprisingly tissue-specific effects of some translation factors and regulators.
MATERIALS AND METHODS
Yeast strains and culture conditions
Ribo-seq was performed on WT yeast strains in the Sigma 1278b background (YWG025) (see Table 1). Yeast cultures were grown in YPAD (1% yeast extract, 2% peptone, 0.01% adenine hemisulfate, 2% dextrose) at 30°C in baffled flasks with vigorous shaking. Glucose-starved cultures were prepared from YPAD cultures grown to OD600 = 1.0–1.1, harvested by centrifugation for 5 min at 12,000g, resuspended in prewarmed YPA medium lacking glucose and returned to shaking at 30°C for various times. The pka-as strain (a gift from James Broach) was grown in synthetic SD medium with 2% dextrose to an OD600 = 0.4. PKA activity was inhibited with 20 μM final concentration 1NM-PP1 (EMD Millipore) for various times.
TABLE 1.
Yeast strains
Ribo-seq and RNA-seq
Ribo-seq and RNA-seq were performed essentially as described (Ingolia et al. 2009; Brar et al. 2012) with minor modifications. Yeast cultures were harvested by centrifugation. Cells were lysed in PLB (20 mM HEPES-KOH, pH 7.4, 2 mM Mg(OAc)2, 100 mM KOAC, 1% Triton-X 100, 0.1 mg/mL CHX, 3 mM DTT) by vortexing with glass beads. RNA-seq libraries were prepared from total RNA purified from cell extract by the hot phenol method.
Data analysis
Sequencing reads were processed as described (Ingolia et al. 2009) using Python scripts developed in-house. To determine differential expression of genes, raw sequencing read counts were analyzed using the edgeR package (Robinson et al. 2010). Genes for which there were at least 128 read counts across both replicate libraries in each condition were included. The first eight codons from each gene were excluded from the gene expression calculation in all libraries. The translation efficiency of a gene was computed as the ratio of the Ribo-seq read density on the gene to the RNA-seq read density. To be included in the TE calculation, we required that genes pass a minimum read cutoff of at least 128 counts over both replicates of the Ribo-seq and RNA-seq libraries. To determine significant TE changes, raw footprint and total counts for each gene (excluding the first eight codons) were first processed by DESeq (Anders and Huber 2010). TEs were computed and variances determined by standard error propagation (Lee and Forthofer 2006). The resulting means and variances were used to compute an unpaired Z statistic, which was used to compute a two-tailed P-value by integration of the standard normal distribution. P-values were adjusted for multiple testing by the method of Benjamini and Hochberg (Benjamini and Hochberg 1995). An alternative metric for computing translation changes using the ANOTA R package (Larsson et al. 2011) gave similar results. Plots were made using GraphPad Prism version 5.0b for Mac OSX and the Matplotlib Python package (Hunter 2007).
Extract preparation, polysome gradient fractionation, and RNA isolation
Yeast polysomes were prepared as described (Arribere et al. 2011). In brief, cycloheximide was added to cell cultures to a final concentration of 0.1 mg/mL for 2 min before harvesting cells by centrifugation (5 min at 4,000g). Cells were lysed by vortexing with glass beads in cold PLB. The crude extracts were clarified (20 min at 14,700g), and 20 OD260 units from the resulting supernatants were applied to linear 10%–50% sucrose gradients in PLB and spun for 2.75 h at 35,000 rpm in a Beckman SW41 rotor. Fractions were collected on a BioComp gradient master, and 20 ng of control firefly luciferase RNA was added to each fraction before further processing.
RNA was purified from polysomal gradient samples by extraction with 25:24:1 acidic phenol:chloroform: isoamyl alcohol and followed by ethanol precipitation.
Quantitative RNA analysis
mRNA abundance per polysome fraction was determined by qRT-PCR. To control for differences in RNA recovery, RP and mito mRNA signals were normalized to synthetic poly(A)-tailed firefly luciferase control mRNA or synthetic rluB mRNA from Escherichia coli. Reverse transcription was performed using SuperScript III (Invitrogen) or Avian Myeloblastosis Virus Reverse Trancriptase (AMV-RT; Promega) using a mix of oligo-dT and random hexamer primers. Quantitative PCR was performed with SYBR Fast reagents (Kapa Biosystems) according to the manufacturer's instructions using a Roche Lightcycler 480. Gene-specific primer sequences are available upon request.
PKA activity assay
PKA activity was determined by monitoring the phosphorylation state of PKA substrate Pat1 as described (Ramachandran et al. 2011). Briefly, equal amounts of log-phase yeast cultures were harvested and resuspended in 5% TCA for ≥30 min to precipitate all cellular proteins. TCA-treated cells were washed first in 50 mM Tris-Cl, pH 7.5, then in acetone; and dried in a speed vac. Cell extracts for immunoprecipitation were prepared in breakage buffer (50 mM Tris-Cl, pH 7.5, 1 mM EDTA, 15 mM PNPP, 60 mM β-glycerophosphate, 5 mM DTT, 100 µg/mL aprotonin, 1 µg/mL pepstatin, 10 µg/µL leupeptin, 1 mM PMSF, and one tablet of Roche complete protease inhibitor cocktail [EDTA-free] per 50 mL buffer) by agitation with glass beads on a Fastprep-24 (MP Biomedicals) at maximum speed for 3 × 45 sec or on a multivortexer for 15 min at 3000 rpm. Myc-tagged Pat1 was immunoprecipitated from the clarified lysates with anti-myc monoclonal antibody (Sigma). Immunoprecipitates were collected on Protein A Mag Sepharose beads (GE Healthcare) and washed six times with washing buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% Igepal, 60 mM β-glycerophosphate, 0.1 mM sodium orthovanadate, 15 mM p-nitrophenylphosphate, 100 µg/mL aprotinin, 20 µg/mL leupeptin, 1 mM DTT, and one tablet of Roche complete protease inhibitor cocktail [EDTA-free] per 25 mL buffer). The level of PKA phosphorylation was assessed by Western blotting with an anti-PKA substrate antibody (Cell Signaling) that recognizes phospho-Ser/Thr peptides of the sequence RRXS*/T*.
DATA DEPOSITION
Sequencing data were deposited in the NCBI Gene Expression Omnibus (GEO) with accession number GSE51532.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank H. Madhani, G. Fink, and J. Broach for yeast strains; P. Herman for plasmids; J. Weissman for advice on ribosome profiling; MIT BioMicro Center for assistance with sequencing; and D. Bartel, C. Burge, Q. Justman, and members of the Gilbert laboratory for helpful discussions. This work was supported by the NIH R01 (GM094303).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.044552.114.
REFERENCES
- Anders S, Huber W 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci 100: 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Doudna JA, Gilbert WV 2011. Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol Cell 44: 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe MP, De Long SK, Sachs AB 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell 11: 833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Bishop AC, Buzko O, Shokat KM 2001. Magic bullets for protein kinases. Trends Cell Biol 11: 167–172 [DOI] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Saldanha AJ, Dolinski K, Botstein D 2005. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol Biol Cell 16: 2503–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D 2008. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell 19: 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR 2012. Nutritional control of growth and development in yeast. Genetics 192: 73–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli LM, Lui J, Campbell SG, Rowe W, Zeef LAH, Holmes LEA, Hoyle NP, Bone J, Selley JN, Sims PFG, et al. 2011. Glucose depletion inhibits translation initiation via eIF4A loss and subsequent 48S preinitiation complex accumulation, while the pentose phosphate pathway is coordinately up-regulated. Mol Biol Cell 22: 3379–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin BL, Ryan O, Lewitter F, Boone C, Fink GR 2012. Genetic variation in Saccharomyces cerevisiae: circuit diversification in a signal transduction network. Genetics 192: 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF 2012. The regulation of filamentous growth in yeast. Genetics 190: 23–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Lobanov AV, Gladyshev VN 2012. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci 109: 17394–17399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng 9: 90–95 [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, Sonenberg N, Nadon R 2011. anota: analysis of differential translation in genome-wide studies. Bioinformatics 27: 1440–1441 [DOI] [PubMed] [Google Scholar]
- Lee ES, Forthofer RN 2006. Analyzing complex survey data In Series: Quantitative applications in the social sciences (ed. Liao TF), Vol. 71 SAGE Publications, Thousand Oaks, CA [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. 2005. Global analysis of protein phosphorylation in yeast. Nature 438: 679–684 [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Shah KH, Herman PK 2011. The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol Cell 43: 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, Lin Z-Y, Cox MJ, Vizeacoumar F, Cheung D, et al. 2012. Global gene deletion analysis exploring yeast filamentous growth. Science 337: 1353–1356 [DOI] [PubMed] [Google Scholar]
- Tudisca V, Recouvreux V, Moreno S, Boy-Marcotte E, Jacquet M, Portela P 2010. Differential localization to cytoplasm, nucleus or P-bodies of yeast PKA subunits under different growth conditions. Eur J Cell Biol 89: 339–348 [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR 2009. Glucose regulates transcription in yeast through a network of signaling pathways. Mol Syst Biol 5: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]