Significance
Connectivity within the brain’s resting-state default mode network (DMN) has been shown to be compromised in multiple genetically complex/heritable neuropsychiatric disorders. Uncovering the source of such alterations will help in developing targeted treatments for these disorders. To our knowledge, this study is the first attempt to do so by using a multivariate data-driven fusion approach. We report five major DMN subnodes, all of which were found to be hypo-connected in probands with psychotic illnesses. Further, we found an overrepresentation of genes in major relevant pathways such as NMDA potentiation, PKA/immune response signalling, synaptogenesis, and axon guidance that influenced altered DMN connectivity in psychoses. The study thus identifies several putative genes and pathways related to an important biological marker known to be compromised in psychosis.
Keywords: genetics, BSNIP, architecture, molecular
Abstract
The brain’s default mode network (DMN) is highly heritable and is compromised in a variety of psychiatric disorders. However, genetic control over the DMN in schizophrenia (SZ) and psychotic bipolar disorder (PBP) is largely unknown. Study subjects (n = 1,305) underwent a resting-state functional MRI scan and were analyzed by a two-stage approach. The initial analysis used independent component analysis (ICA) in 324 healthy controls, 296 SZ probands, 300 PBP probands, 179 unaffected first-degree relatives of SZ probands (SZREL), and 206 unaffected first-degree relatives of PBP probands to identify DMNs and to test their biomarker and/or endophenotype status. A subset of controls and probands (n = 549) then was subjected to a parallel ICA (para-ICA) to identify imaging–genetic relationships. ICA identified three DMNs. Hypo-connectivity was observed in both patient groups in all DMNs. Similar patterns observed in SZREL were restricted to only one network. DMN connectivity also correlated with several symptom measures. Para-ICA identified five sub-DMNs that were significantly associated with five different genetic networks. Several top-ranking SNPs across these networks belonged to previously identified, well-known psychosis/mood disorder genes. Global enrichment analyses revealed processes including NMDA-related long-term potentiation, PKA, immune response signaling, axon guidance, and synaptogenesis that significantly influenced DMN modulation in psychoses. In summary, we observed both unique and shared impairments in functional connectivity across the SZ and PBP cohorts; these impairments were selectively familial only for SZREL. Genes regulating specific neurodevelopment/transmission processes primarily mediated DMN disconnectivity. The study thus identifies biological pathways related to a widely researched quantitative trait that might suggest novel, targeted drug treatments for these diseases.
Schizophrenia (SZ) and psychotic bipolar disorder (PBP) are common, serious, heritable, genetically complex illnesses, sharing multiple characteristics, including risk genes and abnormalities in cognition, neural function, and brain structure (1–4). However, despite recent advances, their underlying biological mechanisms are largely undetermined and may be shared across the two diagnostic groups. Recent large-scale analyses have used various statistical informatics strategies to dissect these biological underpinnings better (5, 6). A recent study using a pathway-enrichment strategy showed that genes involved in neuronal cell adhesion, synaptic formation, and cell signaling are overrepresented in SZ and bipolar disorder (BP) (6). Another study using an informatics-based approach identified several cohesive genetic networks related to axon guidance, neuronal cell mobility, and synaptic functioning as key players in schizophrenia (5).
Although risk for psychotic illnesses is driven in small part by highly penetrant, often private mutations such as copy number variants, substantial risk also is likely conferred by multiple genes of small effect sizes interacting together (7). According to the “common disease common variant” (CDCV) model, one would expect both common and unique quantitative/heritable traits associated with the above syndromes, regulated by these underlying genes, to provide a good starting point for understanding the etiology of SZ and BP. Because definitions of psychiatric diseases are based on clinical phenomenology and lack biological validity (2, 8) a recent strategy has been to use intermediate phenotypes (9, 10) to elucidate quantitative/mechanistic aspects of the underlying disease processes, thereby reducing phenotypic heterogeneity and increasing association power (9, 11, 12). Various properties of intrinsic networks derived from resting-state functional MRI (RS-fMRI) connectivity are promising putative endophenotypes (4, 13). A core component within the resting state is the default mode network (DMN), comprising posterior cingulate cortex (PCC), retrosplenial cortex/precuneus, medial prefrontal cortex (MPFC), medial and lateral parietal cortex, inferior/middle temporal gyri, and parts of cerebellum and basal ganglia that is thought to characterize basal neural activity (14). Connectivity within the DMN is compromised in multiple mental disorders including SZ and PBP (4, 15–19), although reports are inconsistent as to whether hypo- vs. hyper-connectivity (13, 16, 17, 19–21) within this circuit is related to risk of psychosis. Ongür et al. (17) reported reduced MPFC resting-state DMN connectivity in both SZ and BP. Abnormal recruitment involved parietal cortex in BP and frontopolar cortex/basal ganglia in SZ, indicating a dysfunctional core resting-state network with some shared and some unique features in these disorders. Importantly, several studies report DMN dysfunction in siblings who are at increased genetic risk for these disorders (13, 22). DMN connectivity is strongly heritable, making it a promising endophenotype candidate (23).
A challenge in imaging–genetic analyses is correcting for multiple univariate statistical tests, making it difficult to observe weak effects across multiple variables, as presumed in the CDCV model (10). To overcome this problem and to identify aggregate effects, there has been a recent shift toward using multivariate techniques (12, 24–26) such as parallel-independent component analysis (para-ICA), an approach previously validated in psychiatric disorders including SZ and Alzheimer’s disease, yielding robust results with practical sample sizes (12, 24, 25, 27). Given the substantial overlap in the clinical, neurophysiologic, genetic, and molecular characteristics of SZ and BP, we first sought to clarify similarities and differences between SZ and PBP using DMN connectivity as a quantitative disease marker. We next wanted to test whether trait measures found to be abnormal in probands were transmitted to their unaffected relatives and to quantify heritability. Our third goal was to identify the genes and the underlying molecular/biological mechanisms associated with such SZ and PBP intermediate phenotypes to illuminate the etiology of these disease processes.
In line with prior evidence, we hypothesized that (i) we would observe strongly altered (both increased and decreased) connectivity within the DMN in SZ and PBP probands, specifically in regions including precuneus, PCC, and medial prefrontal cortex; (ii) we would observe similar traits in their unaffected first-degree relatives, exhibiting significant heritability; and (iii) variation in DMN connectivity would be characterized by sets of genes that primarily govern axon guidance, neuronal ion channels, neuronal/neurotransmitter signaling, synaptic transmission, and/or cellular signaling and that may have been identified previously in association with risk for neurodevelopmental disorders, including SZ and/or BP.
Results
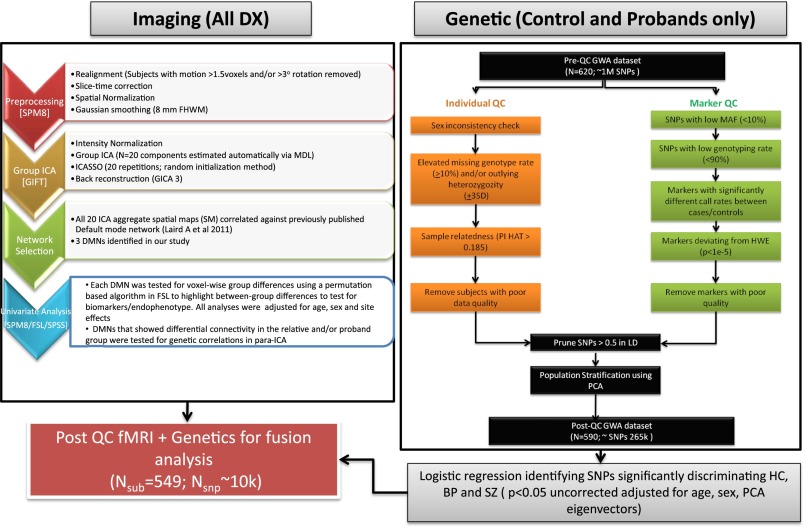
Group differences were noted for age, sex, and site in the overall sample (Table 1). There were no significant between-group differences on average translational and rotational motion parameters estimated during the realignment process. General quality-control parameters used for both the imaging and genetic data are shown in Fig. 1.
Table 1.
Demographic characteristics of the overall sample (n = 1,305) used for primary ICA analysis
| Demographic characteristic | Controls (n = 324) | SZ probands (n = 296) | PBP probands (n = 300) | SZREL (n = 179) | BPREL (n = 206) | Statistic | P value | |||||
| N | % | N | % | N | % | N | % | N | % | χ2 | ||
| Sex | ||||||||||||
| Male | 180 | 55.5 | 199 | 0.6 | 112 | 0.4 | 53 | 0.3 | 74 | 0.4 | 101.1 | <<0.001 |
| Female | 144 | 44.4 | 97 | 0.3 | 188 | 0.63 | 126 | 0.7 | 132 | 0.6 | ||
| Ethnicity | ||||||||||||
| Non-Hispanic | 298 | 91.9 | 271 | 91.6 | 270 | 90.0 | 159 | 88.8 | 187 | 90.7 | 1.8 | NS |
| Hispanic | 26 | 8.1 | 25 | 8.4 | 30 | 10.0 | 20 | 11.2 | 19 | 9.3 | ||
| Site | ||||||||||||
| Hartford | 126 | 38.8 | 106 | 35.8 | 89 | 29.6 | 76 | 42.4 | 88 | 42.7 | 67.0 | <<0.001 |
| Baltimore | 58 | 17.9 | 92 | 31.1 | 47 | 15.6 | 45 | 25.2 | 28 | 13.6 | ||
| Chicago | 55 | 16.9 | 41 | 13.8 | 81 | 27.0 | 22 | 12.4 | 47 | 22.8 | ||
| Detroit | 26 | 8.0 | 19 | 6.4 | 21 | 7.00 | 9 | 5.0 | 7 | 3.4 | ||
| Dallas | 59 | 18.2 | 38 | 12.8 | 62 | 20.7 | 27 | 15.1 | 36 | 17.5 | ||
Fig. 1.
Overall workflow of the study describing the steps in processing both the imaging and genetic data leading to para-ICA analysis.
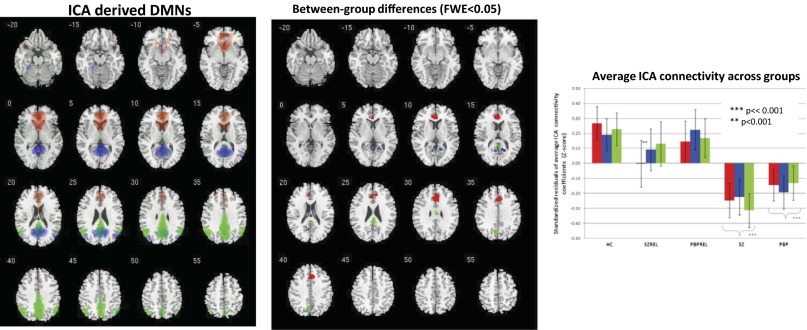
Using a spatial correlation approach (SI Materials and Methods), we identified three data-driven global (parent) DMNs based on their relatively high correlation values (Pearson r >0.4) compared with other intrinsic networks from the current study (Fig. S1). All three networks were highly stable, as assessed using ICASSO (http://research.ics.aalto.fi/ica/icasso), with a stability quotient >0.95. The identified global (parent) DMNs (Fig. 2, Left) were (i) anterior DMN (a-DMN; Red Network): MPFC–anterior cingulate (ACC)–caudate; (ii) inferior–posterior DMN (ip-DMN; Blue Network): PCC–inferior parietal lobule (IPL)–middle temporal gyrus–cuneus/pre-cuneus; and (iii) superior posterior DMN (sp-DMN; Green Network): cuneus/pre-cuneus–IPL–cingulate.
Fig. 2.
(Left) Spatial topology of ICA-derived DMNs identified in the study thresholded at Z > 2. (Center) Between-group voxelwise differences within DMNs derived from Randomize-TFCE (FWE <0.05). (Right) Bar graphs show average connectivity values across groups in clusters identified in the center panel as being significantly different among groups.
Conventional Analysis of DMNs.
No voxels showed a significant diagnosis × site interaction when thresholded at familywise error (FWE) <0.05; k = 10, whole-brain corrected. Initial omnibus F-tests (P < 0.05 FWE corrected) (Fig. 2, Center) generated using the Randomize threshold-free cluster enhancement (TFCE) option in FSL (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/Randomise) revealed all three DMNs to have significantly less connectivity in both proband groups. No region with greater connectivity was identified. Clusters in MPFC (x, y, z = 3, 31, 34; F = 4.5), cingulate (x, y, z = −3, 25, 9; F = 7.1), and ACC (x, y, z = −9, 35, 9; F = 8.4) were most affected in probands in the a-DMN. Similarly, PCC (x, y, z = 12, −52, 14; F = 8.9) and cuneus (x, y, z = −12, −61, 9; F = 6.9) showed lower connectivity in probands within the ip-DMN. Cingulate (x, y, z = 0, −28, 26; F = 5.7) and precuneus (x, y, z = −9, −59, 34; F = 12.6) were most disrupted within the sp-DMN. The sp-DMN also significantly differentiated SZ from PBP, with SZ showing a more pronounced hypo-connectivity within this network. Post hoc t tests in SPSS evaluating average regional differences in relatives in the above abnormal clusters showed significant hypo-connectivity within the a-DMN only in relatives of the SZ probands (SZREL) (t = 2.8; P < 0.005) but not in the relatives of the PBP probands (PBPREL) within this component. See Fig. 2, Right for all three DMNs networks and standardized regional connectivity coefficients across all groups in significant regions.
Heritablity.
Heritability scores (h2) of the above DMNs calculated in Sequential Oligogenic Linkage Analysis Routines (SOLAR) (28) showed that ip-DMN connectivity was significantly but modestly heritable (h2 = 0.18; P = 0.03) and a-DMN connectivity was trend-heritable (h2 = 0.14; P = 0.07).
Correlation Analyses.
Connectivity across all three global (parent) DMNs showed a significant positive correlation with the Social Functioning Scale (SFS) measure (P < 0.05, Bonferroni corrected), suggesting poorer social functioning was associated with diminished connectivity within DMNs. Sp-DMN connectivity correlated negatively with negative symptom scores on the Positive and Negative Symptom Scale (PANSS), and significant negative correlations were observed between positive symptom scores on the PANSS and DMN connectivity across both a-DMN and ip-DMN (although these correlations did not survive correction for multiple comparisons). These results suggest that higher clinical symptomology is associated with lower connectivity within the above networks. Because of the exploratory nature of these correlations, Table S1 shows both Bonferroni-corrected and uncorrected P values.
Para-ICA.
Before conducting para-ICA, we confirmed that the genetic sample had a low genomic inflation factor (∼1) using a logistic regression (adjusted for age, sex, site, and PCA factors) in PLINK (http://pngu.mgh.harvard.edu/∼purcell/plink/) tested separately for both proband groups to verify the absence of stratification bias.
a-DMN (Red Network).
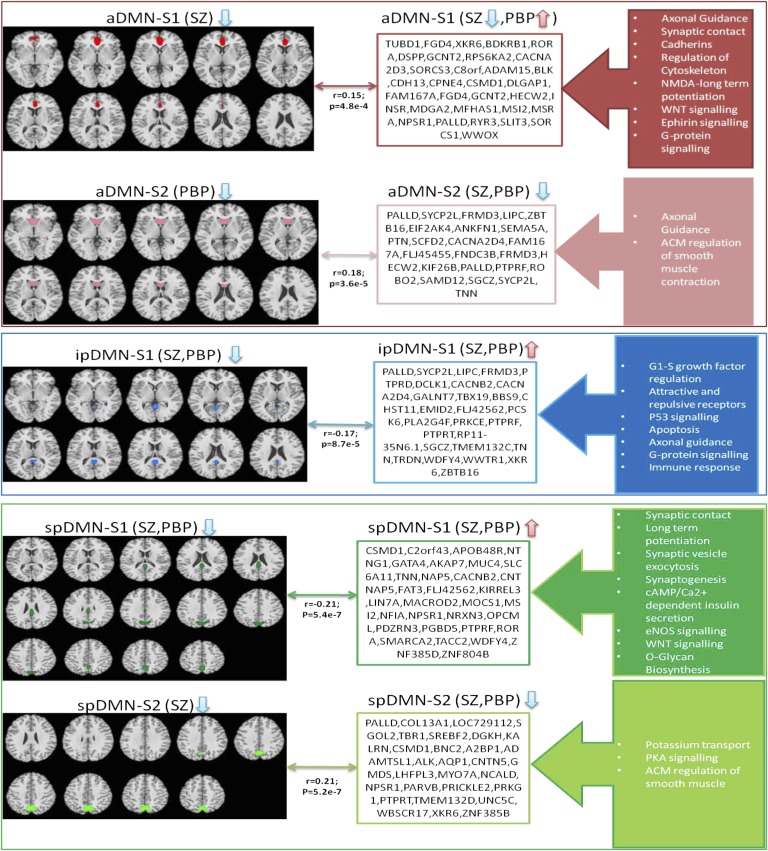
Para-ICA identified two significant positively correlated phenotype–genotype relationships for the a-DMN. Loading coefficients summarizing the strength of network modulation across subjects were derived for both imaging and genetic features. The phenotypic subnetworks encompassed the ACC/MPFC (aDMN-S1) and caudate (aDMN-S2). The aDMN-S1 network was positively linked to a genetic cluster (r = 0.15; P = 4.8E-4) containing 454 SNPs in 386 genes (each having varying weights toward the cluster’s overall signal). Significant functional ontologies representing this gene cluster included (but are not limited to) neurophysiologic processes (NMDA-dependent postsynaptic long-term potentiation, netrin-1 regulation of axon guidance), development [endothelin receptor type A/B (EDNRA/B), Wnt signaling], cell adhesion (cadherins, synaptic contact), and regulation of cytoskeleton rearrangement. Similarly, the aDMN-S2 network was linked positively to a genetic network (r = 0.18; P = 3.6E-5) whose top biological enrichments pertained (but were not limited) to cell adhesion (synaptic contact, attractive/repulsive receptors), neurogenesis, neuron differentiation, and smooth muscle contraction. Supplementary analyses on loading coefficients revealed that connectivity within the aDMN-S1 network was decreased significantly only in SZ. Functional connectivity within the aDMN-S2 network showed a strong trend of reduced connectivity among the PBP probands and was not affected in SZ probands.
ip-DMN (Blue Network).
After adjusting for covariates and multiple comparisons, we found that one significant relationship remained for this network. The phenotype feature (ip-DMN-S1) primarily comprised PCC and was associated (r = −0.17; P < 8.7E-5) with a genetic cluster enriched for G protein signaling, immune response, signal transduction, apoptosis, cell adhesion, inflammation, and neuron differentiation. Post hoc tests showed diminished fMRI connectivity in both SZ and PBP probands.
sp-DMN (Green Network).
Para-ICA revealed two significant fMRI–gene relationships. The first subnetwork, sp-DMN-S1, contained PCC/precuneus and was associated (r = −0.21; P = 5.34E-7) with a genetic network in which ontologies including neurophysiologic processes (NMDA long-term potentiation, regulation of axonal guidance), cAMP/Ca2+-dependent insulin secretion, synaptogenesis, nervous system regulation/development, and regulation of excitatory postsynaptic potential were overrepresented. The second subnetwork, sp-DMN-S2 (r = 0.21; P = 5.2E-7). mainly consisted of precuneus, whose connectivity was associated with proteins responsible for PKA signaling, K+ transport, axonal guidance, neuronal generation/development, and cell differentiation. Supplementary analysis of fMRI loading coefficients revealed reduced connectivity in both SZ and PBP within sp-DMN-S1 and selectively for SZ in sp-DMN-S2.
Brief descriptions of the top 10 ranked genes (based on Z-score weighting) from each component along with their functional significance and properties related to BP and SZ are listed in Table 2. Overall para-ICA results are shown in Fig. 3; Table S2 lists detailed functional enrichment properties for each significant gene network/cluster. Fig. S2 summarizes the identified functional enrichments across all DMN subnetworks.
Table 2.
Summary information on top 10 genes from each para-ICA genetic component
| Default-mode subnetworks | Gene | SNP | Location | Para-ICA weighted score | Curated function/annotation | BP, SZ, or other CNS association at GWAS level* | Allen Human Brain Atlas expression† |
| a-DMNS1 | TUBD1 | rs1292045 | 17q23.1 | 1.00 | Microtubule movement and protein polymerization | A, B, C | |
| FGD4 | rs10844258 | 12p11.21 | 0.92 | Actin cytoskeleton and cell shape | IV (30) | D, B, A | |
| XKR6 | rs10101292 | 8p23.1 | 0.92 | Unknown | C | ||
| BDKRB‡ | rs12050217 | 14q31.1 | 0.82 | Inflammation/pain, Ca2+ signaling and actin complement | I, III, VIII (31) | D/Para-D, C, A | |
| RORA | rs2433025 | 15q22.2 | 0.82 | Regulates transcription from RNA polymerase II circadian rhythm | XIV,III, VI, V (32, 33) | B, C | |
| DSPP | rs13131929 | 4q21.3 | 0.82 | Extracellular matrix development and Ca2+ ion binding | D, C, K, B, C, R, F, A1, M, L, O | ||
| GCNT2 | rs17576994 | 6p24.2 | 0.82 | Glycosphingolipid biosynthesis | VII (34) | K, C, M | |
| RPS6KA2 | rs6456121 | 6q27 | 0.80 | Cell growth/differentiation. Neurotrophic and MAPK/mTOR signaling. | K, R, B, A, M, S | ||
| CACNA2D3‡ | rs9849795 | 3p21.1 | 0.79 | Ca2+ current density/channel kinetics, neurotransmission and synaptogenesis | U, R, D, B, N | ||
| SORCS3 | rs790661 | 10q23-q25 | 0.79 | Neuropeptide receptor activity | VI (35) | K, U, M, D, THA, W§, L, SI | |
| a-DMNS2 | PALLD | rs10022002 | 4q 32.3 | 1.00 | Actin cytoskeleton and cell shape | K, R§, D, B, A,S, L | |
| SYCP2L | rs1225741 | 6p24.2 | 0.84 | Unknown | K, THA, E, OG, C, F | ||
| FRMD3 | rs4014024 | 9q21.32 | 0.71 | Unknown | K, D, B, M, C | ||
| LIPC | rs4774302 | 15q21-q23 | 0.67 | HDL metabolic enzyme | VI, XIII (36) | K, D, C, FO, A, F, B, Motor | |
| ZBTB16 | rs495248 | 11q23.1 | 0.65 | Myeloid maturation, tissue development/maintenance | E/Pre-E, C, F, Y, G, A2, A1, A3, H, B2, I, J, C1 | ||
| EIF2AK4 | rs16970137 | 15q15.1 | 0.64 | Translational initiation | K, D, B, A, A1, G, L | ||
| ANKFN1 | rs7209891 | 17q22 | 0.62 | Unknown | K, M, B | ||
| SEMA5A | rs3846574 | 5p15.31 | 0.62 | Axonal guidance during neural development | V, IX (37, 38) | K, D, A, B, M, C, O | |
| PTN | rs13245911 | 7q33 | 0.61 | Neurite outgrowth, anti-apoptotic signaling and cell proliferation | K, SI, C, N, B, A1, G, M, S, C1 | ||
| SCFD2 | rs301088 | 4q12 | 0.61 | Protein transport | K, C, B, M, SN, T, D | ||
| ip-DMN | PALLD | rs6836618 | 4q 32.3 | 1.00 | Actin cytoskeleton and cell shape | K, R, D, B, A,S, L | |
| SYCP2L | rs1225741 | 6p24.2 | 0.63 | Unknown | K, THA, E§, OG, C§, F | ||
| LIPC | rs4774302 | 15q21-q23 | 0.55 | HDL metabolism | VI (39) | K, D, C§, FO, A, F, B, Motor | |
| FRMD3 | rs4014024 | 9q21.32 | 0.54 | Unknown | K, D, B, M, C | ||
| PTPRD | rs1543083 | 9p23-p24.3 | 0.52 | Cell/neurite growth, differentiation and neuronal axon guidance | XI, XII (40, 41) | N§, E§, A, B, D, C§, O | |
| DCLK1 | rs9546331 | 13q13.3 | 0.51 | Neuronal migration, retrograde transport, neuronal apoptosis and neurogenesis | G, A1, A2, A3, K, D, Pre-E, C2, C1, N§, P, Y, Z, F, B2, B1, Motor, Q | ||
| CACNB2 | rs1277738 | 10p12.33 | 0.51 | Ca2+ channel functioning | Cross-disorder PGC (29) | P, G, A1, A2, A3, Y, Q, N§, J, D, C§ | |
| CACNA2D4‡ | rs4765847 | 12p13.33 | 0.50 | Ca2+ current density and Ca2+ channel kinetics | I, VI, XII (21, 42) | K, B, R, A, S, C§, D, M, | |
| GALNT7 | rs10213525 | 4p31.1 | 0.49 | Protein O-linked glycosylation | C§, B | ||
| TBX19 | rs4656579 | 1q24.2 | 0.48 | Developmental processes | K, D, N§, B, M, A, L, O, C§ | ||
| DAAM2 | rs2504789 | 6p21.2 | 0.46 | Actin cytoskeleton/Cellular organization. WNT signaling. | C§, B, A, M, N§ | ||
| sp-DMNS1 | CSMD1 | rs2930353 | 8p23.2 | 1.00 | Complement control | II, XII (43, 44) | K, N§, G, Q, B2, I, M,Z, U, C1, D |
| C2orf43 | rs667529 | 2p24.1 | 0.98 | Unknown | K, C, B, A1, A, M, R, V, D | ||
| APOB48R | rs40831 | 16p11.2 | 0.96 | Lipid transport, steroid/cholesterol metabolism | |||
| NTNG1 | rs6677537 | 1p13.3 | 0.94 | Neurite outgrowth of axons and dendrites | II, IX, V (45, 46) | K, C, B, A1, M, O, E§, I, D | |
| GATA4 | rs6983129 | 8p23.1-p22 | 0.92 | Transcription factors in neurogenesis | K, C, B, A1, M, SN, O, L, R, D | ||
| AKAP7 | rs6942184 | 6q23.2 | 0.90 | Excitatory synaptic plasticity | K, C, N§, B, X, G, M, SN, O, I, E§, C1§, L, R, D | ||
| MUC4 | rs2259292 (MISSENSE) | 3q29 | 0.89 | Cell proliferation and differentiation. Interacts with ERBB2 to regulate apoptosis. | K, C, B, A2, A, M, SN, O, D | ||
| SLC6A11 | rs971930 | 3p25.3 | 0.89 | GABAnergic signaling and transmission | C, B, CP, M, SN, O | ||
| TNN | rs6701127 | 1q23-q24 | 0.88 | Neurite outgrowth and cell migration | K, C, B, A1, M, Q, F, D | ||
| NAP5‡ | rs6430390 | 2q21.2 | 0.88 | Unknown | II, I (47, 48) | ||
| sp-DMNS2 | PALLD | rs10022002 | 4q 32.3 | 1.00 | Actin cytoskeleton and cell shape | K, R, D, B, A,S, L | |
| COL13A1 | rs2637229 | 10q22 | 0.80 | Cell-matrix, collagen production and cell-cell adhesion | C, B, A1, I, E§, F | ||
| LOC729112 | rs7690087 | 0.80 | Unknown | Unknown | |||
| SGOL2 | rs842938 | 2q33.1 | 0.79 | Gametogenesis | K, C, B, A, M, SN, O, T, N, D | ||
| TBR1 | rs10175058 | 2q24 | 0.76 | Critical role in normal brain development | K, C, N, A1, Motor, M, Q, E§, I, H, B1, U§, F, Z, D | ||
| SREBF2 | rs9607850 | 22q13 | 0.75 | Lipid homeostasis | K, C, N, B, G, M, Q, Pre-E§, A2, Z, D | ||
| DGKH | rs943390 | 13q14.11 | 0.74 | Cell growth and signal transduction. Inhibits PKC signaling | I (49) | K, C, N, B, | |
| KALRN | rs1708320 | 3q21.2 | 0.74 | Neuronal shape, growth and plasticity | K, D, N, E§, P, A, R, A2, I, Q, B2, S/Y§ | ||
| CSMD1 | rs7006552 | 8p23.2 | 0.73 | Unknown | II, XII (43, 44) | K, N, G, Q, B2, I, Z, U§, C1§, D | |
| LOC100133332 | rs2612781 | UNK | 0.73 | Unknown | Unknown |
Information provided was manually curated from PubMed, gene cards, and gene association databases. All SNPs in the table were intronic except otherwise noted.
Key codes: I, bipolar; II, schizophrenia; III, major depression; IV, Charcot Marie Tooth; V, autism; VI, Alzheimer’s disease; VII, cardiovascular CNS; VIII, substance abuse; IX, Parkinson’s disease; X, dyslexia; XI, epilepsy; XII, stroke; XIII, cognitive performance; XIV, posttraumatic stress disorder.
Allen Atlas Brain Expression: A, globus pallidus; B, thalamus; C, cerebellar cortex; D, hippocampus; E, cuneus; F, Herschl gyrus; G, orbitofrontal cortex; H, occipito-temporal gyrus; I, lingual gyrus; J, supramarginal gyrus; K, amygdala; L, nucleus accumbens; M, hypothalamus; N, cingulate; O, ventral tegmental area; P, fusiform gyrus; Q, insula; R, caudate; S, putamen; T, locus ceruleus; U, angular gyrus; V, temporal pole; W, anterior cingulate; X, superior rostral gyrus; Y/Z, middle/superior temporal gyrus; A1/A2/A3, inferior/middle/superior frontal gyrus; B1/B2, inferior/superior occipital gyrus; C1/C2, superior/inferior parietal lobule.
Gene appears in the top 100 genes associated with SZ and/or BP in the mega-analysis published by the Psychiatric Genomic Consortium (28).
Expression regions overlapping with functional subnetwork.
Fig. 3.
The five default mode subnetworks and their associated genetic components as identified by para-ICA. Each genetic component is represented by the top 10 genes plus genes that feature with more than three SNPs within each network. Also identified are select significantly enriched ontology term(s) (FDR < 0.05) within each genetic cluster. Arrows pointing up and down indicate whether the loading coefficient for that particular feature (fMRI or gene) was significantly higher or lower for probands than for controls.
Pooled Enrichment Analysis.
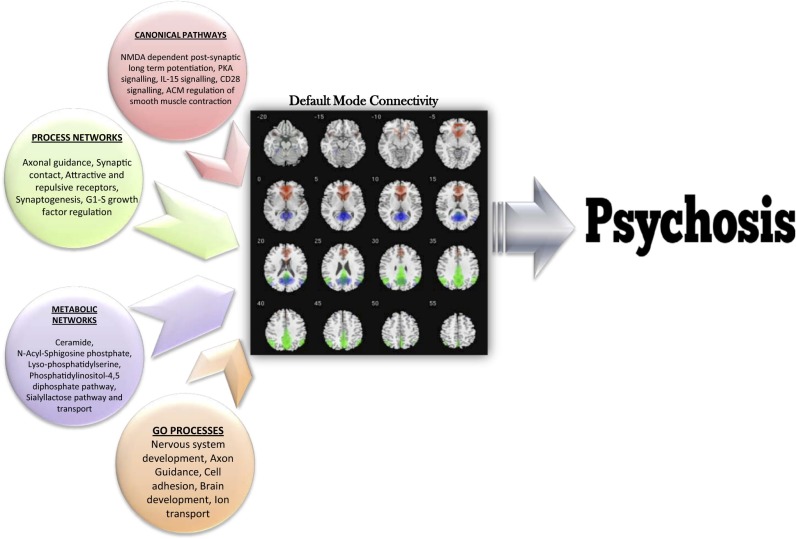
The top five significantly [false discovery rate (FDR) <0.05] overrepresented pathways, processes, and metabolic networks from the pooled enrichment analysis are shown in Fig. 4.
Fig. 4.
Significant ontology terms derived from a pooled analysis representing genes identified in our study in a variety of processes/pathways/networks whose overrepresentation suggests that they mediate the risk of psychosis via default mode connectivity in probands.
Discussion
DMN is the most widely examined intrinsic network, because of its unique characteristics and its role in indexing more fundamental aspects of brain function than those evoked by cognitive demands. DMN abnormalities have been associated with multiple mental disorders (50). The current study was one of the largest to date to quantify DMN connectivity across SZ and PBP probands and their relatives. In addition, this was the first study, to our knowledge, to assess genetic/biological associations of this vital network(s) in the context of psychoses.
Conventional Voxelwise Analysis of DMN.
As is consistent with the global DMN comprising multiple subsystems, each exhibiting varying degrees of differential connectivity (51, 52), the current data-driven analysis identified three separate DMNs. As predicted, both proband groups exhibited reduced global ICA connectivity in regions such as MPFC, ACC, PCC, cuneus, and precuneus, as is consistent with disconnectivity hypotheses in these disorders and previously published results using similar connectivity methods (15, 17, 18). However, contrary to previous reports showing DMN hyper-connectivity among probands (13, 53), we saw no such effects. Our sample was larger than that in prior reports showing hyper-connectivity, had different demographic and possibly different medication characteristics, and used a relatively new statistical thresholding technique designed to minimize potential confounds accompanying conventional techniques and a data-driven ICA approach, all of which might have contributed to these different findings. We also found that one of the three DMNs showed a statistical between-group difference in DMN connectivity between the SZ and PBP probands, suggesting the differential sensitivity of DMN nodes in capturing both unique and common functional traits across these two major psychiatric disorders. A recent study by Khadka and colleagues (19) that used a small subset of the current study sample found comparable results, wherein the p-DMN showed reduced connectivity in both the SZ and PBP probands but no effect across unaffected relatives. In contrast to the current study, however, Khadka et al. noted no differences in the a-DMN and similarly no correlations with behavioral indices across the DMNs, most likely because of the smaller sample size. Exploring the potential utility of these DMNs as endophenotypes, we observed that altered connectivity was detected only in SZRELs and was specific to a-DMN connectivity. As further validation, this network also showed a strong trend for heritability in the current sample, even though the study design was not optimally designed to capture dense family pedigrees. Abnormal connectivity patterns for other networks were not detected in relatives. However, despite the normal fMRI patterns in relatives, the ip-DMN was significantly, albeit modestly, heritable. Therefore our study may have been underpowered to detect abnormal patterns in relatives for this particular fMRI network.
Para-ICA.
A primary goal was to identify the genetic architecture and the underlying biological/molecular mechanisms associated with DMN disruptions in psychoses, using multivariate para-ICA that captures maximal association between variations among high-dimensional feature sets across subjects (here, DMNs and dosage-coded SNPs) (see Fig. S3). The major advantage of this approach in the context of imaging–genetics is its power to leverage differential multivariate patterns with minimal loss of statistical power resulting from necessary multiple comparison corrections.
Unlike univariate studies [e.g., genome-wide association studies (GWAS)], para-ICA illuminates clusters of interacting SNPs, each with a weighted contribution toward a quantitative trait and readily interpreted in the context of illness-associated molecular/biological pathways. Thus, the current discussion focuses on two important aspects of the genetic information that enable better understanding of biological underpinnings of these complex data: (i) the functionality of genes with top rankings and/or multiple hits within each network and (ii) pathway/biological functional enrichments of the gene clusters.
a-DMN (Red Network).
Two positive genotype–phenotype relationships were identified for the a-DMN, indicating that lower neural network connectivity was associated with a lower genetic load contributed by the underlying genes. The two fMRI subnetworks within the a-DMN showed preferential functional disconnectivity among disorders. The a-DMN-S1 subnetwork primarily containing dorsal-MPFC/ACC was impaired in only SZ probands, as is consistent with prior reports examining other measures of RS-DMN connectivity (16, 17, 54). Several genes had relatively high loading coefficients and/or multiple occurrences within the genetic network (e.g., CDH13 from the cadherin family, which had nine SNPs). Cadherins mediate cell adhesion and intracellular signaling, playing a pivotal role in the development of neural circuitry and maturation of synaptic function. Cadherins, including CDH13, are implicated in multiple neuropsychiatric disorders (55). Other cadherins were CDH12, previously implicated in both SZ and BP, and CDH4, 5, associated with cognitive impairment (55). Multiple SNPs in the well-known complement-control CSMD1 gene also were found in this network. This gene is involved in activity of immunity-related pathways and is associated with SZ risk (56) and memory deficits in SZ (57). MSRA, implicated in protection against oxidative stress and protein maintenance, was shown recently to be a candidate SZ risk gene (58). RORA was another gene that showed a notable presence within the component and that has been implicated in a variety of neuropsychiatric disorders, including major depression, Alzheimer’s disease, and autism (32, 33, 59). Being a pivotal nuclear receptor for the survival and differentiation of Purkinje neuronal cells (60), it also regulates aromatase, which is reduced significantly in the frontal cortex of autistic subjects (59).
The second subnetwork, a-DMN-S2, containing mostly caudate, showed a strong trend of aberrance only in the PBP probands. Caudate connectivity is frequently implicated in major affective disorders (61, 62). Major gene candidates represented by multiple SNPs identified in this network included pallidin (PALLD), which encodes for a cytoskeletal protein that controls cell shape, adhesion, and contraction/migration. Multiple cytoskeletal regulatory proteins, including DISC1, Dysbindin, and NRG1, already have been identified as major risk genes for SZ/PBP (63). The association of PALLD with DMN connectivity in SZ/BP further supports the notion that mutations affecting cytoskeleton regulators play a major role in the pathology of cortical function and neuronal migration in neuropsychiatric disorders. CACNA2D4 is part of the CACN gene family responsible for encoding voltage-dependent calcium channels; its deletion was implicated recently in the risk for late-onset BP (21); another study showed it to interact significantly with another calcium subunit gene, RYR2, in BP (42). These genes are closely related to CACNA1A (whose direct association was not found in the current study), one of the genes more strongly associated with the risk of BP (64).
ip-DMN (Blue Network).
A single linked feature set was found for the ip-DMN. The phenotype network primarily involved PCC, showing reduced connectivity in both proband groups. PCC is a key DMN hub, shown repeatedly to be aberrant in both SZ (65, 66) and affective disorders, including BP (67). Similar to the discussion above, genes in this network, including CACNB2 and CACNA2D4 that regulate calcium voltage channels and PALLD, associated with the actin-cytoskeleton organization, featured multiple significant SNPs. Multiple SNPs also occurred in ZBTB16, a glucocorticoid-response gene whose deletion has been previously linked to mental retardation (68) and more recently implicated as a potential SZ candidate gene in a study using a pathway-based bioinformatics approach (69). Many of the top-ranked genes in this network, including COL4A2, HOMER2, DAAM2, ROBO2, and RGNEF, indirectly or directly regulate structural synaptic elements, thus providing further supporting evidence that intra- and extracellular structural elements might be dysregulated, leading to weaker synaptic stability, in the brains of SZ patients (70).
sp-DMN (Green Network).
The first subcomponent from the sp-DMN, sp-DMNS1, contained mostly superior PCC–pre-cuneus and had diminished connectivity in both the SZ and PBP probands, albeit to a larger degree in the latter. The top 10 candidates from the correlated genetic network included CSMD1, NTNG1 (Netrin), NAP5, and SLC6A11 (GAT-3), all previously implicated by GWAS as genes conferring risk for SZ and/or BP (46, 47, 56). CSMD1 also has been shown to be associated with cognitive deficits in SZ (57) and, along with CACNA1C, is an important miR-137 target, indicating its direct involvement in neurogenesis/maturation in SZ/BP (71). In addition, several genes from the glutamate-GABA system (GRIA1, GABRA3, and GBRB3) were associated with dysfunctional brain connectivity, highlighting the importance of balance among these neurotransmitters in maintaining normal cortical circuit connectivity. This finding is consistent with recent evidence showing that regional glutamate/GABA concentrations play a strong role in governing DMN functional connectivity (72).
The second subnetwork, sp-DMNS, containing precuneus was disconnected significantly only in the SZ probands, and the corresponding genetic network was enriched with ion transport, voltage-gated regulatory, and intracellular signaling genes, all processes associated with neural transmission, whose dysfunction might lead to abnormal brain function (73, 74). Some notable top-ranked genes associated with sp-DMNS2 connectivity, such as DGKH, whose differential effects were shown recently to manifest as a failure to disengage DMN during a verbal fluency task in subjects at high familial risk of BP, were candidates (75). Another top-ranked gene, SREBF2, a lipid homeostasis regulator, has been shown previously to be associated with SZ (76). Neuronal plasticity/synaptic regulators such as KALRN also were among the heavily weighted genes. More importantly KALRN has been reported to interact with prime SZ candidate genes such as NRG1 and ERB4 to promote dendrite growth (77).
Juxtaposing enriched functional categories across all DMN subnetworks revealed neurodevelopment, cell adhesion, and smooth muscle contraction (primarily calcium/voltage regulatory genes) as processes associated with DMN connectivity in at least three of the five subnetworks identified (Fig. S1). Interestingly, even though analysis initially was blind to group membership, all genetic subclusters identified via para-ICA differentiated both proband groups from controls; however, they seemed to mediate disease risk in SZ and BP selectively via DMN connectivity (i.e., some DMN sub-nodes were aberrant in one disease group but not the other, despite the associated genetic cluster showing significant differences across both disorders).
Global Pooled Functional Pathway/Process Enrichment.
Pooling all significant risk genes identified in the current study across all networks allowed us to dissect the overall biological/molecular underpinnings of DMN connectivity that is suggested to mediate disease risk in psychosis. We identified several relevant and plausible disease-related mechanisms, some of which have been implicated for decades in the pathology of both SZ and PBP, including candidate genes in the serotonin, glutamate, and dopamine systems (7, 24, 25, 78, 79). NMDA-related postsynaptic long-term potentiation was the top enriched pathway. We also found the l-glutamate transport system to be a significantly enriched molecular network, although not among the top 10. The results of our study reported above suggest underlying glutamatergic involvement with abnormal DMN connectivity. This finding is supported by recent fMRI–spectroscopy fusion data showing that glutamate levels are related to RS activity (80). Other top pathways were PKA signaling, involving phospholipid-dependent enzymes whose activity is directly dependent on cellular cAMP levels. This system is particularly important, because all dopamine receptors are coupled to the cAMP system (81), providing a secondary link to the dopaminergic pathophysiology. Our results also highlight key developmental pathways, including immune system-mediated neuronal responses via IL-15 and CD28 pathways. CD28 gene polymorphisms confer increased risk of SZ with affective symptoms (82). Interleukin family genes (IL3RA, IL-18, IL-12, IL-10, and IL1A/B/RA), representing a group of cytokines expressed in white blood cells, may play an important role in SZ (83, 84). Although “regulation of smooth muscle contraction” was a top enriched pathway, upon examining the encompassed genes, they were primarily Ca2+ channel/voltage regulation genes (including CACNA1C, CACNB2, and CACNA2D4), their strong interactors such as RYR2/3, and second-messenger molecules such as PKC and PLC-β1 that are crucial in smooth muscle contraction but also act as important cellular signaling molecules whose functions are linked strongly to psychiatric disorders (21, 29, 42).
Specific, highly enriched network processes included neurodevelopmental, transport, and cellular signaling networks comprising axonal guidance (developmental) and cell adhesion (synaptic contact, attractive repulsive receptors). These results are consistent with recent findings from a large-scale study using molecular pathway analysis identifying neuronal cell adhesion and membrane scaffolding proteins as primary contributors to SZ and BP susceptibility (6). Another recent computational neuroscience study reported cohesive gene networks related to axon guidance, neuronal cell mobility, synaptic function, and chromosomal remodeling as potential causative pathways in SZ (5). Our finding also bolsters the neurodevelopment hypothesis of SZ and affective disorders (85, 86).
Metabolic pathway enrichments pointed to ceramides and the N-acyl-sphingosine pathways as the top related networks associated with DMN connectivity in psychoses. There is growing evidence that sphingolipid metabolism may be involved in the pathology of neuropsychiatric disorders (87–89). Ceramides (composed of sphingosines and fatty acids) regulate cellular differentiation, proliferation, and apoptosis. Recent research implicates abnormal sphingolipid metabolism through alterations in peripheral ceramide levels in first-episode SZ (90), and abnormal plasma ceramide levels are reported in affective disorders, mild cognitive impairment, and memory disturbances (87, 91).
Taken together, our results are highly consistent with processes identified in recent studies using bioinformatics-based network approaches studying gene–disease relationships directly (5, 6). However, the crucial distinctions between our study and the other approaches include our investigation across the psychosis diagnostic spectrum and the inclusion of a quantitative intermediate phenotype shown to mediate disease risk. Having measurable phenotypes and knowing their molecular underpinnings in a transdiagnostic, multivariate-context suggest novel strategies for drug targeting and customization in these serious disorders.
Limitations.
First, the current study was limited to the DMN because of the scope and complexity of the data. We intend to follow up by performing similar analyses on other non-DMN intrinsic resting networks we identified. Second, because of design limitations (generally having only one first-degree relative per proband), we were unable to capture a dense kinship structure required for the optimal estimation of heritability, and therefore we likely underestimated true heritability. Third, future studies should build on our results by further exploring genetic associations among siblings in addition to probands. Fourth, even though we found several known risk genes as part of para-ICA, some of these are yet to be replicated. Finally, our study included patients taking psychoactive medications that may bias our results. We tried to address this issue by testing for functional differences in relatives who, by definition, carry higher genetic risk for the disease but are not on antipsychotic medications. Two recent studies (54, 90) noted decreased DMN connectivity within MPFC regions in drug-naive SZ patients, and their findings are consistent with our data. One of these studies (90) showed that bilateral prefrontal cortex, parietal, caudate, and superior temporal regions had increased regional connectivity 6 wk after antipsychotic treatment. None of these regions (except the caudate) was affected differentially in probands vs. relatives in the current study, suggesting that regions reported in the current study might have minimal medication interactions. However, no similar studies have been conducted in treatment-naive BP patients.
Conclusion
Overall this study reconfirms the role of the DMN as a strong biomarker for SZ and PBP in a large sample. We also show that selective nodes within the DMN are differentially affected and are modestly heritable among probands and unaffected relatives. Using a multivariate technique, we were able to successfully identify several known large-scale GWAS risk genes and also discover additional genes acting in synchrony that could be prioritized usefully in future studies. Most importantly we were able to dissect the underlying biological/molecular pathways and processes that might mediate genetic risk of psychosis via a valuable, noninvasive imaging marker.
Materials and Methods
Study Sample.
Subjects were recruited as part of the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) Consortium study that used identical diagnostic and recruitment approaches at multiple sites (Baltimore, Chicago, Dallas, Detroit, and Hartford, CT). Detailed information on the whole study sample is provided elsewhere (91).
Initial analyses were restricted to quality-controlled RS-fMRI (Fig. 1) comprising 1,305 subjects; (324 healthy controls; 296 in the SZ proband group; 300 in the PBP proband group; 179 SZREL; and 206 PBPREL), a subset of which was used later for imaging–genetics fusion analyses. Detailed demographic descriptions are given in Table 1, clinical characteristics are given in Table 3, and medication information is provided in Table S3. Because the validity of diagnostic status of schizoaffective disorder (SZA) is controversial (92), we decided not to analyze SZA probands and relatives as separate groups. In the current study SZA probands were classified either as either PBP (schizoaffective manic probands) or SZ (schizoaffective depressed probands), as suggested elsewhere (8). As a secondary benefit, this classification allowed us to retain more subjects, providing greater statistical power. To maximize information and limit cost, only a random subset of the controls and probands was genotyped, comprising 549 subjects (190 SZ patients, 189 PBP patients, and 170 healthy controls) who survived extensive quality control of both fMRI and genetic data.
Table 3.
Clinical characteristics of the overall sample (n = 1,305) used for primary ICA analysis
| Clinical characteristic | Controls (n = 324) | SZ probands (n = 296) | PBP probands (n = 300) | SZREL (n = 179) | BPREL (n = 206) | Statistic | P value | |||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | F | ||
| Age, y | 35.2 | 13.4 | 34.9 | 12.2 | 36.7 | 12.6 | 43.8 | 15.8 | 39.8 | 16.1 | 15.8 | <<0.001 |
| Clinical scores | ||||||||||||
| YMRS | 0.3 | 1.1 | 5.1 | 5.7 | 6.4 | 6.3 | 0.3 | 1.4 | 0.3 | 1.4 | 212.2 | <<0.001 |
| MADRS | 0.3 | 1.5 | 8.7 | 8.3 | 11.6 | 9.8 | 0.8 | 2.8 | 0.7 | 3.0 | 160.1 | <<0.001 |
| SFS | 154.0 | 19.3 | 99.1 | 52.52 | 102.6 | 56.6 | 149.1 | 19.3 | 149.6 | 22.6 | 92.6 | <<0.001 |
| PANSS_POS | N/A | N/A | 16.3 | 5.3 | 15.1 | 5.5 | N/A | N/A | N/A | N/A | N/A | N/A |
| PANSS_NEG | N/A | N/A | 15.8 | 5.5 | 13.1 | 4.6 | N/A | N/A | N/A | N/A | N/A | N/A |
| SBS | N/A | N/A | 7.6 | 1.3 | 2.5 | 1.9 | N/A | N/A | N/A | N/A | N/A | N/A |
For medication information on subjects please refer to Table S3. MADRS, Montgomery-Asberg Depression Rating Scale; PANSS: Positive and Negative Symptom Scale; SBS: Schizo-Bipolar Scale; SFS: Social Functioning Scale; YMRS, Young Mania Rating Scale.
Data Acquisition and Preprocessing.
Fig. 1, Table S4 and SI Materials and Methods provide additional information on data acquisition and preprocessing.
Primary Data Analysis.
We used a two-step analytic approach in the current study. First we analyzed all the fMRI data from all 1,305 subjects in our study using a permutation-based technique to identify DMNs and to determine if any DMNs were valid biomarkers and/or endophenotypes. Then we used a subset (n = 549) who were genotyped to conduct our imaging–genetics analysis using para-ICA to determine multilocus associations with different DMN connectivity traits as a quantitative marker.
Detailed information on these analyses is given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. David Glahn for his helpful comments, discussions, and suggestions for improving the manuscript. We would also like to thank Zheng Wang, for her timely analytic help towards addressing some of the reviewers comments. This study was funded by National Institute of Mental Health Grants MH077851, MH078113, MH077945, MH077852, MH077862, and 2R44 MH075481.
Footnotes
Conflict of interest statement: J.A.S. receives support from Janssen, Takeda, Bristol-Myers Squibb, Roche, and Lilly. G.P. has been a consultant for Bristol-Myers Squibb. G.R. is the president and A.W. is a former employee of Genomas Inc. M.S.K. has received support from Sunovion.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313093111/-/DCSupplemental.
References
- 1.Ellison-Wright I, Bullmore E. Anatomy of bipolar disorder and schizophrenia: A meta-analysis. Schizophr Res. 2010;117(1):1–12. doi: 10.1016/j.schres.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 2.Keshavan MS, et al. A dimensional approach to the psychosis spectrum between bipolar disorder and schizophrenia: The Schizo-Bipolar Scale. Schizophr Res. 2011;133(1-3):250–254. doi: 10.1016/j.schres.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lichtenstein P, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: A population-based study. Lancet. 2009;373(9659):234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meda SA, et al. Differences in resting-state functional magnetic resonance imaging functional network connectivity between schizophrenia and psychotic bipolar probands and their unaffected first-degree relatives. Biol Psychiatry. 2012;71(10):881–889. doi: 10.1016/j.biopsych.2012.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilman SR, et al. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci. 2012;15(12):1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Dushlaine C, et al. International Schizophrenia Consortium Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2011;16(3):286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, et al. Multifaceted genomic risk for brain function in schizophrenia. Neuroimage. 2012;61(4):866–875. doi: 10.1016/j.neuroimage.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keshavan MS, Clementz BA, Pearlson GD, Sweeney JA, Tamminga CA. Reimagining psychoses: An agnostic approach to diagnosis. Schizophr Res. 2013;146(1-3):10–16. doi: 10.1016/j.schres.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 9.Allen AJ, Griss ME, Folley BS, Hawkins KA, Pearlson GD. Endophenotypes in schizophrenia: A selective review. Schizophr Res. 2009;109(1-3):24–37. doi: 10.1016/j.schres.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. Am J Psychiatry. 2003;160(4):636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 11.Derks EM, Vorstman JA, Ripke S, Kahn RS, Ophoff RA. Schizophrenia Psychiatric Genomic Consortium Investigation of the genetic association between quantitative measures of psychosis and schizophrenia: A polygenic risk score analysis. PLoS ONE. 2012;7(6):e37852. doi: 10.1371/journal.pone.0037852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meda SA, et al. A large scale multivariate parallel ICA method reveals novel imaging-genetic relationships for Alzheimer’s disease in the ADNI cohort. Neuroimage. 2012;60(3):1608–1621. doi: 10.1016/j.neuroimage.2011.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitfield-Gabrieli S, et al. Hyperactivity and hyperconnectivity of the default network in schizophrenia and in first-degree relatives of persons with schizophrenia. Proc Natl Acad Sci USA. 2009;106(4):1279–1284. doi: 10.1073/pnas.0809141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raichle ME, Snyder AZ. A default mode of brain function: A brief history of an evolving idea. Neuroimage. 2007;37(4):1083–1090, discussion 1097–1099. doi: 10.1016/j.neuroimage.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Calhoun VD, et al. Exploring the psychosis functional connectome: Aberrant intrinsic networks in schizophrenia and bipolar disorder. Front Psychiatry. 2011;2:75. doi: 10.3389/fpsyt.2011.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mingoia G, et al. Default mode network activity in schizophrenia studied at resting state using probabilistic ICA. Schizophr Res. 2012;138(2-3):143–149. doi: 10.1016/j.schres.2012.01.036. [DOI] [PubMed] [Google Scholar]
- 17.Ongür D, et al. Default mode network abnormalities in bipolar disorder and schizophrenia. Psychiatry Res. 2010;183(1):59–68. doi: 10.1016/j.pscychresns.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Repovs G, Csernansky JG, Barch DM. Brain network connectivity in individuals with schizophrenia and their siblings. Biol Psychiatry. 2011;69(10):967–973. doi: 10.1016/j.biopsych.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khadka S, et al. Is aberrant functional connectivity a psychosis endophenotype? A resting state functional magnetic resonance imaging study. Biol Psychiatry. 2013;74(6):458–466. doi: 10.1016/j.biopsych.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazza M, et al. Dysfunctional neural networks associated with impaired social interactions in early psychosis: An ICA analysis. Brain Imaging Behav. 2013;7(3):248–259. doi: 10.1007/s11682-013-9223-6. [DOI] [PubMed] [Google Scholar]
- 21.Van Den Bossche MJ, et al. Identification of a CACNA2D4 deletion in late onset bipolar disorder patients and implications for the involvement of voltage-dependent calcium channels in psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(4):465–475. doi: 10.1002/ajmg.b.32053. [DOI] [PubMed] [Google Scholar]
- 22.van Buuren M, Vink M, Kahn RS. Default-mode network dysfunction and self-referential processing in healthy siblings of schizophrenia patients. Schizophr Res. 2012;142(1-3):237–243. doi: 10.1016/j.schres.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 23.Glahn DC, et al. Genetic control over the resting brain. Proc Natl Acad Sci USA. 2010;107(3):1223–1228. doi: 10.1073/pnas.0909969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, et al. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum Brain Mapp. 2009;30(1):241–255. doi: 10.1002/hbm.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meda SA, et al. A pilot multivariate parallel ICA study to investigate differential linkage between neural networks and genetic profiles in schizophrenia. Neuroimage. 2010;53(3):1007–1015. doi: 10.1016/j.neuroimage.2009.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tura E, Turner JA, Fallon JH, Kennedy JL, Potkin SG. Multivariate analyses suggest genetic impacts on neurocircuitry in schizophrenia. Neuroreport. 2008;19(6):603–607. doi: 10.1097/WNR.0b013e3282fa6d8d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jamadar S, et al. Genetic influences of cortical gray matter in language-related regions in healthy controls and schizophrenia. Schizophr Res. 2011;129(2-3):141–148. doi: 10.1016/j.schres.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smoller JW, et al. Cross-Disorder Group of the Psychiatric Genomics Consortium Genetic Risk Outcome of Psychosis (GROUP) Consortium Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet. 2013;381(9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boubaker C, et al. A novel mutation in FGD4/FRABIN causes Charcot Marie Tooth Disease Type 4H in patients from a consanguineous Tunisian family. Ann Hum Genet. 2013 doi: 10.1111/ahg.12017. [DOI] [PubMed] [Google Scholar]
- 31.Gratacòs M, et al. Psychiatric Genetics Network Group Identification of new putative susceptibility genes for several psychiatric disorders by association analysis of regulatory and non-synonymous SNPs of 306 genes involved in neurotransmission and neurodevelopment. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(6):808–816. doi: 10.1002/ajmg.b.30902. [DOI] [PubMed] [Google Scholar]
- 32.Furney SJ, et al. Alzheimer’s Disease Neuroimaging Initiative AddNeuroMed Consortium Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Mol Psychiatry. 2011;16(11):1130–1138. doi: 10.1038/mp.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terracciano A, et al. Genome-wide association scan of trait depression. Biol Psychiatry. 2010;68(9):811–817. doi: 10.1016/j.biopsych.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seshadri S, et al. Genetic correlates of brain aging on MRI and cognitive test measures: A genome-wide association and linkage analysis in the Framingham Study. BMC Med Genet. 2007;8(Suppl 1):S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grupe A, et al. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet. 2006;78(1):78–88. doi: 10.1086/498851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cirulli ET, et al. Common genetic variation and performance on standardized cognitive tests. Eur J Hum Genet. 2010;18(7):815–820. doi: 10.1038/ejhg.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maraganore DM, et al. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77(5):685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Y, Quinn JF, Weiss LA. An eQTL mapping approach reveals that rare variants in the SEMA5A regulatory network impact autism risk. Hum Mol Genet. 2013;22(14):2960–2972. doi: 10.1093/hmg/ddt150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds CA, et al. Analysis of lipid pathway genes indicates association of sequence variation near SREBF1/TOM1L2/ATPAF2 with dementia risk. Hum Mol Genet. 2010;19(10):2068–2078. doi: 10.1093/hmg/ddq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasperaviciūte D, et al. Common genetic variation and susceptibility to partial epilepsies: A genome-wide association study. Brain. 2010;133(Pt 7):2136–2147. doi: 10.1093/brain/awq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mälarstig A, et al. Genetic Analysis of Idiopathic Thrombophilia (GAIT) and Precocious Coronary Artery Disease (PROCARDIS) consortia Identification of ZNF366 and PTPRD as novel determinants of plasma homocysteine in a family-based genome-wide association study. Blood. 2009;114(7):1417–1422. doi: 10.1182/blood-2009-04-215269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prabhu S, Pe’er I. Ultrafast genome-wide scan for SNP-SNP interactions in common complex disease. Genome Res. 2012;22(11):2230–2240. doi: 10.1101/gr.137885.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SH, et al. Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ) International Schizophrenia Consortium (ISC) Molecular Genetics of Schizophrenia Collaboration (MGS) Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44(3):247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luke MM, et al. Polymorphisms and noncardioembolic stroke in three case-control studies. Cerebrovasc Dis. 2012;33(1):80–85. doi: 10.1159/000333444. [DOI] [PubMed] [Google Scholar]
- 45.Fukasawa M, et al. Case-control association study of human netrin G1 gene in Japanese schizophrenia. J Med Dent Sci. 2004;51(2):121–128. [PubMed] [Google Scholar]
- 46.Zhu Y, et al. Positive association between NTNG1 and schizophrenia in Chinese Han population. J Genet. 2011;90(3):499–502. doi: 10.1007/s12041-011-0112-8. [DOI] [PubMed] [Google Scholar]
- 47.Wang KS, Liu XF, Aragam N. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res. 2010;124(1-3):192–199. doi: 10.1016/j.schres.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Smith EN, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009;14(8):755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baum AE, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13(2):197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Broyd SJ, et al. Default-mode brain dysfunction in mental disorders: A systematic review. Neurosci Biobehav Rev. 2009;33(3):279–296. doi: 10.1016/j.neubiorev.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 51.Allen EA, et al. A baseline for the multivariate comparison of resting-state networks. Front Syst Neurosci. 2011;5:2. doi: 10.3389/fnsys.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skudlarski P, et al. Brain connectivity is not only lower but different in schizophrenia: A combined anatomical and functional approach. Biol Psychiatry. 2010;68(1):61–69. doi: 10.1016/j.biopsych.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karbasforoushan H, Woodward ND. Resting-state networks in schizophrenia. Curr Top Med Chem. 2012;12(21):2404–2414. doi: 10.2174/156802612805289863. [DOI] [PubMed] [Google Scholar]
- 54.He Z, et al. Aberrant intrinsic brain activity and cognitive deficit in first-episode treatment-naive patients with schizophrenia. Psychol Med. 2013;43(4):769–780. doi: 10.1017/S0033291712001638. [DOI] [PubMed] [Google Scholar]
- 55.Redies C, Hertel N, Hübner CA. Cadherins and neuropsychiatric disorders. Brain Res. 2012;1470:130–144. doi: 10.1016/j.brainres.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 56.Håvik B, et al. The complement control-related genes CSMD1 and CSMD2 associate to schizophrenia. Biol Psychiatry. 2011;70(1):35–42. doi: 10.1016/j.biopsych.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 57.Xiang B, et al. 2012. [Genome-wide association study with memory measures as a quantitative trait locus for schizophrenia]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 29(3):255–259. Chinese.
- 58.Walss-Bass C, et al. Methionine sulfoxide reductase: A novel schizophrenia candidate gene. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(2):219–225. doi: 10.1002/ajmg.b.30791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarachana T, Xu M, Wu RC, Hu VW. Sex hormones in autism: Androgens and estrogens differentially and reciprocally regulate RORA, a novel candidate gene for autism. PLoS ONE. 2011;6(2):e17116. doi: 10.1371/journal.pone.0017116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boukhtouche F, et al. RORalpha, a pivotal nuclear receptor for Purkinje neuron survival and differentiation: From development to ageing. Cerebellum. 2006;5(2):97–104. doi: 10.1080/14734220600750184. [DOI] [PubMed] [Google Scholar]
- 61.Bluhm R, et al. Resting state default-mode network connectivity in early depression using a seed region-of-interest analysis: Decreased connectivity with caudate nucleus. Psychiatry Clin Neurosci. 2009;63(6):754–761. doi: 10.1111/j.1440-1819.2009.02030.x. [DOI] [PubMed] [Google Scholar]
- 62.Ma C, et al. Resting-state functional connectivity bias of middle temporal gyrus and caudate with altered gray matter volume in major depression. PLoS ONE. 2012;7(9):e45263. doi: 10.1371/journal.pone.0045263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sawa A, Snyder SH. Schizophrenia: Neural mechanisms for novel therapies. Mol Med. 2003;9(1-2):3–9. [PMC free article] [PubMed] [Google Scholar]
- 64.Sklar P, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13(6):558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alonso-Solís A, et al. Altered default network resting state functional connectivity in patients with a first episode of psychosis. Schizophr Res. 2012;139(1-3):13–18. doi: 10.1016/j.schres.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woodward ND, Rogers B, Heckers S. Functional resting-state networks are differentially affected in schizophrenia. Schizophr Res. 2011;130(1-3):86–93. doi: 10.1016/j.schres.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu CH, et al. Abnormal baseline brain activity in bipolar depression: A resting state functional magnetic resonance imaging study. Psychiatry Res. 2012;203(2-3):175–179. doi: 10.1016/j.pscychresns.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 68.Fischer S, et al. Biallelic loss of function of the promyelocytic leukaemia zinc finger (PLZF) gene causes severe skeletal defects and genital hypoplasia. J Med Genet. 2008;45(11):731–737. doi: 10.1136/jmg.2008.059451. [DOI] [PubMed] [Google Scholar]
- 69.Sun J, et al. Schizophrenia gene networks and pathways and their applications for novel candidate gene selection. PLoS ONE. 2010;5(6):e11351. doi: 10.1371/journal.pone.0011351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmitt A, et al. Structural synaptic elements are differentially regulated in superior temporal cortex of schizophrenia patients. Eur Arch Psychiatry Clin Neurosci. 2012;262(7):565–577. doi: 10.1007/s00406-012-0306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry. 2013;18(1):11–12. doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- 72.Kapogiannis D, Reiter DA, Willette AA, Mattson MP. Posteromedial cortex glutamate and GABA predict intrinsic functional connectivity of the default mode network. Neuroimage. 2013;64:112–119. doi: 10.1016/j.neuroimage.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Askland K, Read C, O’Connell C, Moore JH. Ion channels and schizophrenia: A gene set-based analytic approach to GWAS data for biological hypothesis testing. Hum Genet. 2012;131(3):373–391. doi: 10.1007/s00439-011-1082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smolin B, Karry R, Gal-Ben-Ari S, Ben-Shachar D. Differential expression of genes encoding neuronal ion-channel subunits in major depression, bipolar disorder and schizophrenia: Implications for pathophysiology. Int J Neuropsychopharmacol. 2012;15(7):869–882. doi: 10.1017/S1461145711001428. [DOI] [PubMed] [Google Scholar]
- 75.Whalley HC, et al. Effect of variation in diacylglycerol kinase η (DGKH) gene on brain function in a cohort at familial risk of bipolar disorder. Neuropsychopharmacology. 2012;37(4):919–928. doi: 10.1038/npp.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le Hellard S, et al. Polymorphisms in SREBF1 and SREBF2, two antipsychotic-activated transcription factors controlling cellular lipogenesis, are associated with schizophrenia in German and Scandinavian samples. Mol Psychiatry. 2010;15(5):463–472. doi: 10.1038/mp.2008.110. [DOI] [PubMed] [Google Scholar]
- 77.Cahill ME, et al. Control of interneuron dendritic growth through NRG1/erbB4-mediated kalirin-7 disinhibition. Mol Psychiatry. 2012;17(1):1, 99–107. doi: 10.1038/mp.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bertolino A, et al. Additive effects of genetic variation in dopamine regulating genes on working memory cortical activity in human brain. J Neurosci. 2006;26(15):3918–3922. doi: 10.1523/JNEUROSCI.4975-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7(10):818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- 80.Enzi B, et al. Glutamate modulates resting state activity in the perigenual anterior cingulate cortex - a combined fMRI-MRS study. Neuroscience. 2012;227:102–109. doi: 10.1016/j.neuroscience.2012.09.039. [DOI] [PubMed] [Google Scholar]
- 81.Muly C. Signal transduction abnormalities in schizophrenia: The cAMP system. Psychopharmacol Bull. 2002;36(4):92–105. [PubMed] [Google Scholar]
- 82.Frydecka D, et al. The role of genetic variations of immune system regulatory molecules CD28 and CTLA-4 in schizophrenia. Psychiatry Res. 2013;208(2):197–198. doi: 10.1016/j.psychres.2012.11.035. [DOI] [PubMed] [Google Scholar]
- 83.Ozbey U, Tug E, Namli M. Interleukin-10 gene promoter polymorphism in patients with schizophrenia in a region of East Turkey. World J Biol Psychiatry. 2009;10(4 Pt 2):461–468. doi: 10.1080/15622970802626580. [DOI] [PubMed] [Google Scholar]
- 84.Xu M, He L. Convergent evidence shows a positive association of interleukin-1 gene complex locus with susceptibility to schizophrenia in the Caucasian population. Schizophr Res. 2010;120(1-3):131–142. doi: 10.1016/j.schres.2010.02.1031. [DOI] [PubMed] [Google Scholar]
- 85.Bassett AS, Chow EW, O’Neill S, Brzustowicz LM. Genetic insights into the neurodevelopmental hypothesis of schizophrenia. Schizophr Bull. 2001;27(3):417–430. doi: 10.1093/oxfordjournals.schbul.a006884. [DOI] [PubMed] [Google Scholar]
- 86.Guilmatre A, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66(9):947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gracia-Garcia P, et al. Elevated plasma ceramides in depression. J Neuropsychiatry Clin Neurosci. 2011;23(2):215–218. doi: 10.1176/appi.neuropsych.23.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smesny S, et al. Skin ceramide alterations in first-episode schizophrenia indicate abnormal sphingolipid metabolism. Schizophr Bull. 2013;39(4):933–941. doi: 10.1093/schbul/sbs058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mielke MM, et al. Serum ceramides increase the risk of Alzheimer disease: The Women’s Health and Aging Study II. Neurology. 2012;79(7):633–641. doi: 10.1212/WNL.0b013e318264e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lui S, et al. Short-term effects of antipsychotic treatment on cerebral function in drug-naive first-episode schizophrenia revealed by “resting state” functional magnetic resonance imaging. Arch Gen Psychiatry. 2010;67(8):783–792. doi: 10.1001/archgenpsychiatry.2010.84. [DOI] [PubMed] [Google Scholar]
- 91.Tamminga CA, et al. Clinical phenotypes of psychosis in the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) Am J Psychiatry. 2013;170(11):1263–1274. doi: 10.1176/appi.ajp.2013.12101339. [DOI] [PubMed] [Google Scholar]
- 92.Jablensky A. Prototypes, syndromes and dimensions of psychopathology: An open agenda for research. World Psychiatry. 2012;11(1):22–23. doi: 10.1016/j.wpsyc.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.