Abstract
The functional redundancy of the three mammalian Golgi-localized, γ-ear–containing, ADP-ribosylation factor-binding proteins (GGAs) was addressed in a previous study. Using insertional mutagenesis, we found that Gga1 or Gga3 homozygous knockout mice were for the most part normal, whereas mice homozygous for two different Gga2 gene-trap alleles exhibited either embryonic or neonatal lethality in the C57BL/6 background, depending on the source of the vector utilized (Byg vs. Tigm, respectively). We now show that the Byg strain harbors a disrupted Gga2 allele that is hypomorphic, indicating that the Byg lethality is attributable to a mechanism independent of GGA2. This is in contrast to the Tigm Gga2 allele, which is a true knockout and establishes a role for GGA2 during the neonatal period. Placement of the Tigm Gga2 allele into the C57BL6/Ola129Sv mixed background results in a lower incidence of neonatal lethality, showing the importance of genetic background in determining the requirement for GGA2 during this period. The Gga2−/− mice that survive have reduced body weight at birth and this runted phenotype is maintained through adulthood.
Keywords: GGA2, gene-trap, hypomorphic allele, neonatal lethality, genetic background
The Golgi-localized, γ-ear–containing, Arf-binding proteins (GGAs) are a family of monomeric clathrin adaptor proteins that function in receptor trafficking between the trans-Golgi network (TGN) and endosomes (Bonifacino 2004; Braulke and Bonifacino 2009). There are three homologous GGAs in mammalian species termed GGA1, GGA2, and GGA3. These proteins have identical domain organizations with an N-terminal VHS (VPS-27, Hrs, and STAM) domain followed by a GAT (GGA and Tom1) domain, a connecting hinge segment, and a C-terminal GAE domain that is homologous to the ear domain of γ-adaptin (Braulke and Bonifacino 2009). The VHS and GAT domains of the three human GGAs are conserved with 60–75% identity (Boman et al. 2000), but the similarity between each GGA becomes much less pronounced across the entire molecule within a given species. However, the individual GGAs display approximately 90% identity between the human and mouse counterparts (Govero et al. 2012).
We and others have noted a number of similarities between GGA1 and GGA3 that are not shared by GGA2. For instance, human GGA1 and GGA3, but not GGA2, are phosphorylated in vivo and subject to autoinhibition mediated by binding of internal acidic cluster-dileucine motifs present in the hinge to the ligand binding site on the VHS domain (Doray et al. 2002; McKay and Kahn 2004). In addition, the GAT domains of human GGA1 and GGA3, but not GGA2, bind ubiquitin and ubiquitinated proteins (Shiba et al. 2004; Yogosawa et al. 2006), and GGA1 and GGA3, but not GGA2, are depleted in the brains of patients with Alzheimer disease (Walker et al. 2012). Also, GGA2 has a shorter half-life than GGA1 and GGA3 (Hirst et al. 2007). Finally, only GGA2 is detectable in isolated HeLa cell clathrin-coated vesicles (CCVs) by either Western blotting or mass spectrometry and is dependent on AP-1 for incorporation into CCVs (Hirst et al. 2009; Hirst et al. 2012). Taken together, these findings suggest that GGA1 and GGA3 may perform more overlapping functions in vivo than does GGA2, and/or that GGA2 may perform some unique role not mediated by GGA1 and GGA3.
Consistent with this idea, we previously showed that loss of either GGA1 or GGA3 is well-tolerated in mice, indicating that the remaining members are able to compensate for the loss (Govero et al. 2012). However, gene-trap disruption of the Gga2 gene resulted in either embryonic lethality with the BayGenomics (Byg) gene-trap or neonatal lethality with the Texas Institute for Genomic Medicine (Tigm) gene-trap (Govero et al. 2012). We initially considered whether this difference in phenotype was attributable to differences in genetic backgrounds, because the Byg strain was generated in the C57BL6/Ola129Sv mixed genetic background, while the Tigm strain was in the C57BL/6NJ background. However, backcrossing of the Byg strain into the C57BL/6J background still resulted in embryonic lethality, leaving the basis for this difference in outcome unclear at the time and the question of a role for GGA2 in embryonic development unresolved (Govero et al. 2012).
Subsequently, on review of the Western blot data of brain tissue from wild-type (wt)and heterozygous (het) progeny of the Gga2+/− crosses corresponding to the Byg strain, we observed that the het mice frequently expressed GGA2 protein at a level similar to wt, indicating that the Byg Gga2 gene-trap allele is likely to be a hypomorphic allele. To verify this, we have generated compound het mice by crossing Byg and Tigm Gga2+/− mice in the C57BL/6J and C57BL/6NJ backgrounds, respectively. This has allowed us to unequivocally show that the embryonic lethality associated with the Byg Gga2 gene-trap allele is not attributable loss of GGA2 because this allele is clearly hypomorphic. In addition, we introduced the Tigm Gga2 gene-trap allele into the C57BL6/Ola129Sv mixed genetic background and demonstrated that the severity of the neonatal lethality associated with this allele is strongly influenced by the genetic background of the mice in which the allele occurs. These studies show that the Gga2−/− mice have a runted phenotype.
Materials and Methods
All protocols involving the use of animals were in compliance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals and approved by the Animal Studies Committee in the Division of Comparative Medicine at Washington University School of Medicine in St. Louis (Protocol #20130010). Mice were housed in a barrier facility maintained under standards meeting federal, state, and local guidelines and under the supervision of licensed veterinarians.
Generation of Gga knockout mice
The Byg (cell-line ID SYA176) and Tigm (cell-line ID IST10483E10) Gga2+/− gene-trap mice in the C57BL/6J and C57BL/6NJ backgrounds, respectively, have been described (Govero et al. 2012). Primer sequences used for genotyping of mice and distinguishing between the two mutant Gga2 alleles, PCR conditions, and the sizes of all PCR products have been presented (Govero et al. 2012). The screen used in this study to identify the compound hets is illustrated in Supporting Information, Figure S1.
Harvesting mouse tissues and Western blot analyses
Adult mice were killed by CO2 inhalation whereas newborns were killed by decapitation. For serum preparation, blood was collected in a microfuge tube immediately after decapitation of newborns, allowed to sit at room temperature for 30 min, and centrifuged at 2000×g for 10 min to separate the clotted components. Brain tissue was removed and cellular extracts were prepared exactly as described (Govero et al. 2012). All protein samples were diluted to 5 mg/ml before being boiled in SDS sample buffer for gel loading. Proteins were resolved by SDS-PAGE, transferred to either nitrocellulose or PVDF membrane, and probed with the following antibodies: anti-GGA2 (GGA2 E-3; Santa Cruz); anti-GAPDH (Sigma); anti-CI-MPR (Zhu et al. 2001); anti-SorLA (BD Transduction Laboratories); and anti-Sortilin (Sigma),.
Serum IGF-II measurement by ELISA
Serum IGF-II levels of newborn mice were measured using a modification of the ELISA procedure that has been described by Mason et al. (2011). All reactions were performed in 96-well tissue culture plates. Briefly, purified monoclonal rat anti-mouse IGF-II antibody (R&D) was coated on the plate [2.5 μg in a final volume of 200 μl phosphate-buffered saline (PBS) per well] overnight on a shaker at 4°. The next day, the wells were washed three times with wash buffer (PBS with 0.05% Tween-20), followed by three washes with SuperBlock T20 blocking buffer (Thermo Scientific).
For serum preparation, 100 μl acid/ethanol reagent (12.5% 2N HCl, 87.5% ethanol) was added to 15 μl serum sample and incubated at room temperature for 30 min, followed by centrifugation at 10,000×g for 10 min. All the supernatant was removed and neutralized with 120 μl neutralization buffer (4× PBS containing 0.25% BSA and 500 ng/ml IGF-I). Standards were prepared by diluting recombinant mouse IGF-II (R&D) in dilution buffer (PBS with 0.1% Tween-20 and 0.25% BSA). Standards ranged in concentration from 2000 pg/ml to 125 pg/ml. A blank control (500 ng/ml IGF-I in dilution buffer) was also included for subtraction. Standard, control, or samples (100 μl/well) were combined with 50 ng (100 μl) biotinylated goat anti-mouse IGF-II antibody (R&D) and incubated for 3 hr on a shaker at room temperature. The wells were washed three times with wash buffer, after which 200 μl of a 1:200 dilution of streptavidin-HRP conjugate (R&D) in dilution buffer was added to each well and incubated for 45 min at room temperature. The wells were then washed four times with wash buffer and 200 μl of 3,3′,5,5′-Tetramethylbenzidine in buffered hydrogen peroxide (Sigma) was added to each well and incubated at room temperature for 30 min to allow the color to develop. Then, 50 μl of 1 M H2SO4 was added per well to stop the reaction and produce a yellow color, and the absorbance was read at 450 nm.
Results
Generation and analysis of Gga2−/− compound heterozygotes
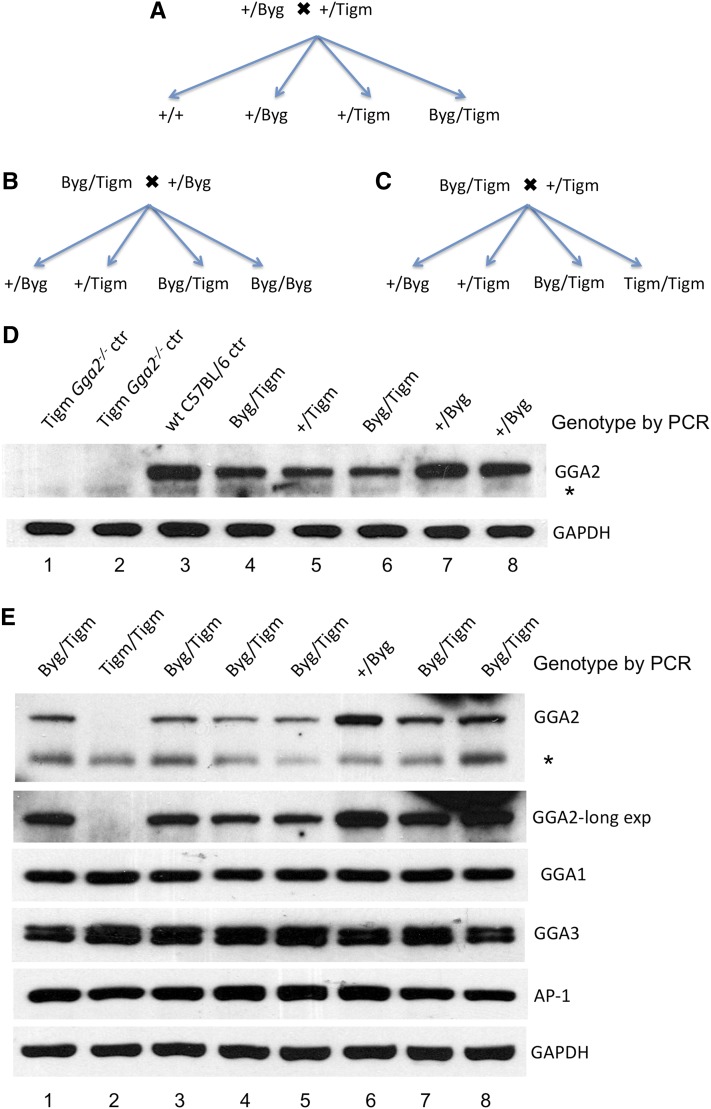
We initially crossed Byg Gga2+/− mice in the C57BL/6J genetic background with Tigm Gga2+/− mice in the C57BL/6NJ background using the mating scheme illustrated in Figure 1A. To facilitate the description of the compound heterozygotes, +/Byg and +/Tigm refer to mice that have one wild-type (wt) (+) and one Gga2 gene-trap allele (either the Byg or the Tigm Gga2 gene-trap allele, respectively), whereas Byg/Tigm are compound hets that harbor both the Byg and Tigm Gga2 gene-trap alleles. Byg/Byg and Tigm/Tigm, however, refer to homozygous mice carrying two copies of either the Byg or the Tigm Gga2 gene-trap alleles, respectively. Although the gene-trap cassette is inserted into intron 1 of the Gga2 gene in both strains, the position of insertion was different (Figure S1), as was the nature of the vectors utilized. Details for genotyping that distinguished between the two alleles and allowed for identification of compound hets (Byg/Tigm) are provided in Figure S1. As expected if the Byg allele is hypomorphic, all the compound hets that were born were viable and normal in all respects even though these mice are Gga2−/− at the DNA level. We next mated the Byg/Tigm compound hets with either Byg Gga2+/− mice (+/Byg)( Figure 1B) or Tigm Gga2+/− (+/Tigm) (Figure 1C). The results of the genotyping and the Western blot data of the progeny (day 1 pups of a representative litter) from the former mating scheme are shown in Figure 1D. As controls, wt or Tigm Gga2−/− (Tigm/Tigm) samples were included. Strikingly, in this litter the two Byg Gga2+/− pups (Figure 1D +/Byg, lanes 7 and 8) had higher brain expression of GGA2 than the Tigm Gga2+/− pup (+/Tigm, lane 5) similar to wt (compare lane 3 with lanes 7 and 8). The two compound hets (Byg/Tigm, lanes 4 and 6), however, although genetically Gga2−/−, had similar expression as the Tigm Gga2+/− pup (+/Tigm, lane 5).
Figure 1.
Generation and immunoblot analysis of Byg/Tigm compound hets. (A–C) Mating schemes used to generate compound heterozygous mice harboring both the Byg and Tigm mutant Gga2 alleles. The Byg strain was backcrossed for at least eight generations into the C57BL/6 genetic background before being crossed with the Tigm strain, also in the C57BL/6 background. (D and E) Genotyping results and Western blot analysis of brain tissue from day 1 pups of representative litters resulting from the mating schemes shown in (B and C), respectively. Twenty-five μg of protein extract for each sample was subjected to SDS-PAGE and immunoblot analysis of GGA1, GGA2, GGA3, AP-1, and GAPDH (5 μg of lysate) as a control. *A non-specific band. Use of a 4–12% Bris-Tris gradient gel (E) resulted in better separation of the GGA2 band from the non-specific band compared to a 10% Tris-Glycine gel (D).
We then examined brain extracts from another representative litter (Figure 1E) representing the mating scheme shown in Figure 1C. The large number of compound hets in this litter provided a good opportunity to monitor the GGA2 protein level in these mice. Immunoblot analysis of brain tissue extracts (Figure 1E) showed that the Tigm Gga2−/− pup in this litter (Tigm/Tigm, lane 2) had no detectable GGA2, even on long exposure of the blot, whereas the Byg Gga2+/− pup (+/Byg, lane 6) had the highest expression. Of note, GGA2 was detected in every compound het (Byg/Tigm, lanes 1, 3, 4, 5, 7, and 8), although there was substantial variability in the expression level (compare lanes 4 and 5 to 7 and 8.). It has been reported that a gene-trap inserted in intron 1 of the Prep1 gene gave rise to a hypomorphic mutation that also resulted in variable expression of the Prep1 protein and an embryonic lethal penetrance that was dependent on the degree of leakiness of the gene-trap cassette (Ferretti et al. 2006). GGA1, GGA3, and AP-1, in contrast, did not display this variability between the different genotypes. Analysis of brain, heart, and lung tissue extracts from other litters also showed variation in the expression of GGA2 among the compound hets and Byg Gga2+/− (+/Byg) pups (Figure S2), indicating that the Byg allele must be producing wt message in addition to the chimeric Gga2-βgal transcript. The presence of this chimeric mRNA in Byg Gga2+/− mice was demonstrated previously by reverse-transcriptase polymerase chain reaction (RT-PCR) (Govero et al. 2012). Long exposure of the GGA2 blots also revealed that an occasional Tigm Gga2−/− pup expresses very low levels of GGA2 that could at times be detected (Figure S2, long exposure of brain and lung GGA2) but, for the most part, even if there was low level expression, it was below the detection limit (Figure 1E and Figure 2C long exposure of GGA2). Western blot analysis of varying amounts of wt brain lysate run alongside a Gga2−/− brain lysate where some expression was observed, indicating that this low level expression is in the range of 1–2.5% relative to wt (Figure S3). Importantly, progeny corresponding to every possible genotype were obtained from the crosses shown in Figure 1, A–C, with the exception of Byg Gga2−/− mice (Byg/Byg) (Figure 1B), highlighting the fact that the embryonic lethality associated with the Byg allele in the homozygous state persists in the C57BL/6NJ background.
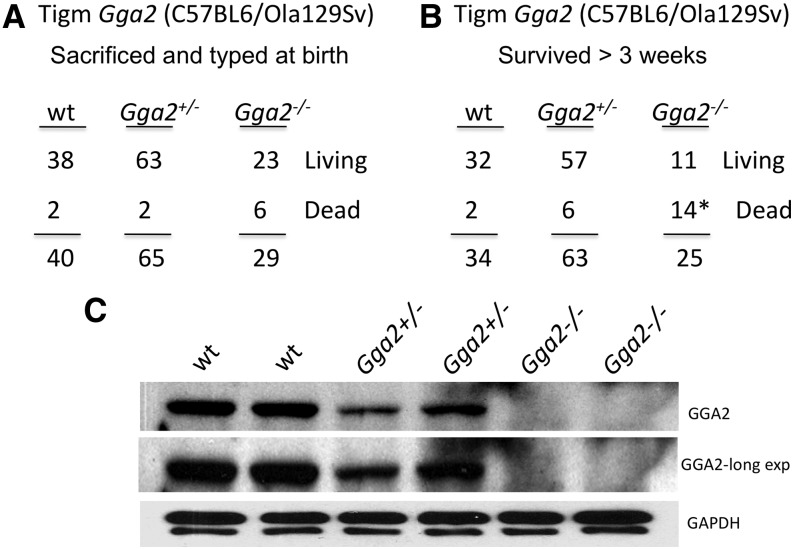
Figure 2.
Influence of genetic background on severity of neonatal lethality associated with loss of GGA2. (A and B) The progeny of Tigm Gga2+/− crosses in the C57BL6/Ola129Sv mixed genetic background were genotyped by PCR within 24 hr after birth (A), or after 1 wk (B), and includes all dead pups that were recovered. The six deaths at birth of the Gga2−/− mice were not significant compared to the two wt deaths at birth, whereas the 14 deaths (*) that occurred neonatally were highly significant, as determined by the Fisher exact test (P < 0.0001). (C) Western blot analysis of brain tissue from representative day 1 pups resulting from Tigm Gga2+/− crosses in the mixed genetic background. Twenty-five μg of protein extract for each sample was subjected to SDS-PAGE and immunoblot analysis of GGA2 and GAPDH (5 μg of lysate) as a control.
These results confirm that the embryonic lethality of the Byg Gga2−/− mice is not attributable to the absence of GGA2. In view of this, we decided not to pursue this Byg strain further.
Influence of genetic background on neonatal lethality of Tigm Gga2−/− gene-trap mice
We previously presented data showing that the Tigm Gga2−/− phenotype in the C57BL/6NJ background is neonatal lethal, exhibiting more than 95% mortality rate with one single survivor that lived beyond 3 wk (Govero et al. 2012). The surviving Gga2−/− mouse was a female that was 25% smaller than its littermates at 4 wk of age but had a normal lifespan. Genotyping of an additional 65 offspring from Tigm Gga2+/− intercrosses that survived beyond 2 wk identified only one Gga2−/− mouse that had survived up to that point. This surviving mouse was a male that weighed 50% less than its littermates and had to be killed at 3 wk due to marked cachexia. Histological examination did not reveal any pathologic changes in the internal organs (not shown), similar to the situation with the newborn Gga2 knockouts (Govero et al. 2012). These results confirm our previous finding that Gga2−/− mice display a neonatal lethal phenotype in the C57BL/6NJ background.
To analyze the influence of the genetic background on the consequence of the loss of GGA2, the Tigm Gga2 allele was introduced into the mixed background by mating Tigm Gga2+/− females in the C57BL/6NJ background with a wt male in the C57BL6/Ola129Sv mixed background. All the resulting N1 Gga2+/− mice were subsequently mated. Initially, all progeny of the Gga2+/− crosses were collected soon after birth and genotyped, including pups that died during or immediately after birth. Figure 2A shows that Gga2−/− mice in the mixed genetic background are born in near accordance with the expected Mendelian ratio, as was the case in the C57BL/6NJ background (Govero et al. 2012). Of note, the Gga2−/− mice exhibited increased mortality compared to wt and hets at this time point, although the difference was not statistically significant based on our sample size. In a separate experiment, 122 offspring were genotyped by PCR 1 wk after birth, along with the dead pups recovered during this period (Figure 2B). An additional three mice were recorded as being born but were missing and presumably cannibalized, so they could not be genotyped. Of the 25 Gga2−/− offspring that were born within this group, 14 (56%) died within the first week, with almost all dying within the first 48 hr. This is in stark contrast to the mortality rate of wt and Gga2+/− mice in the same background (6% and 10%, respectively). The result is also significantly different from the finding in the C57BL/6NJ background, where the lethality rate is more than 95% in the Gga2−/− homozygous state (Govero et al. 2012). Of the surviving Tigm Gga2−/− mice in the C57BL6/Ola129Sv mixed background, none have died beyond the 3-wk period, with the oldest mouse now 9 months old, and both males and females are fertile. Because 11 of 25 Tigm Gga2−/− mice in the mixed background survived beyond 3 wk, it was conceivable that these mice might express more GGA2 than do the Tigm Gga2−/− mice in the C57BL/6NJ background. However, Western blot analysis of brain tissue extracts prepared from day 1 pups in the mixed background probed for GGA2 showed no detectable protein in the Gga2−/− mice in most instances (representative pups shown in Figure 2C), although occasionally the same low-level expression was observed as with the Tigm Gga2−/− mice in the C57BL/6NJ background. Taken together, these results indicate that the partial rescue of the neonatal lethality in the mixed background is not due to increased expression of GGA2, but rather it is the genetic background of the mice that plays an important role in determining viability in the absence or near-complete loss of GGA2.
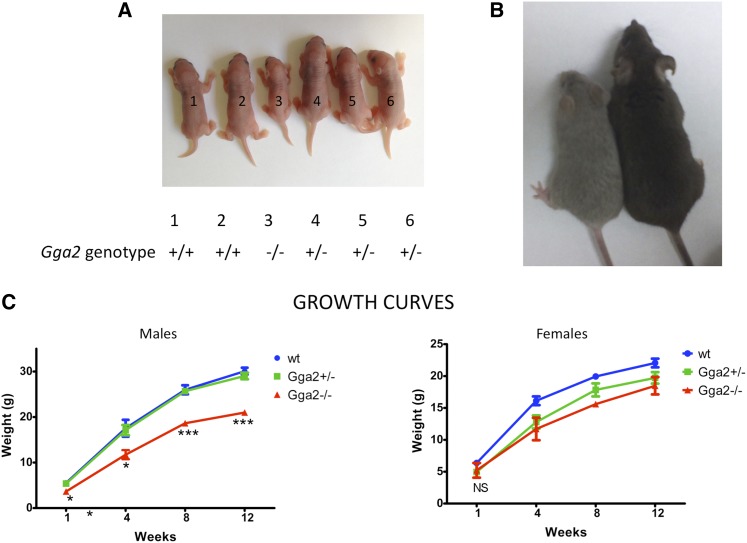
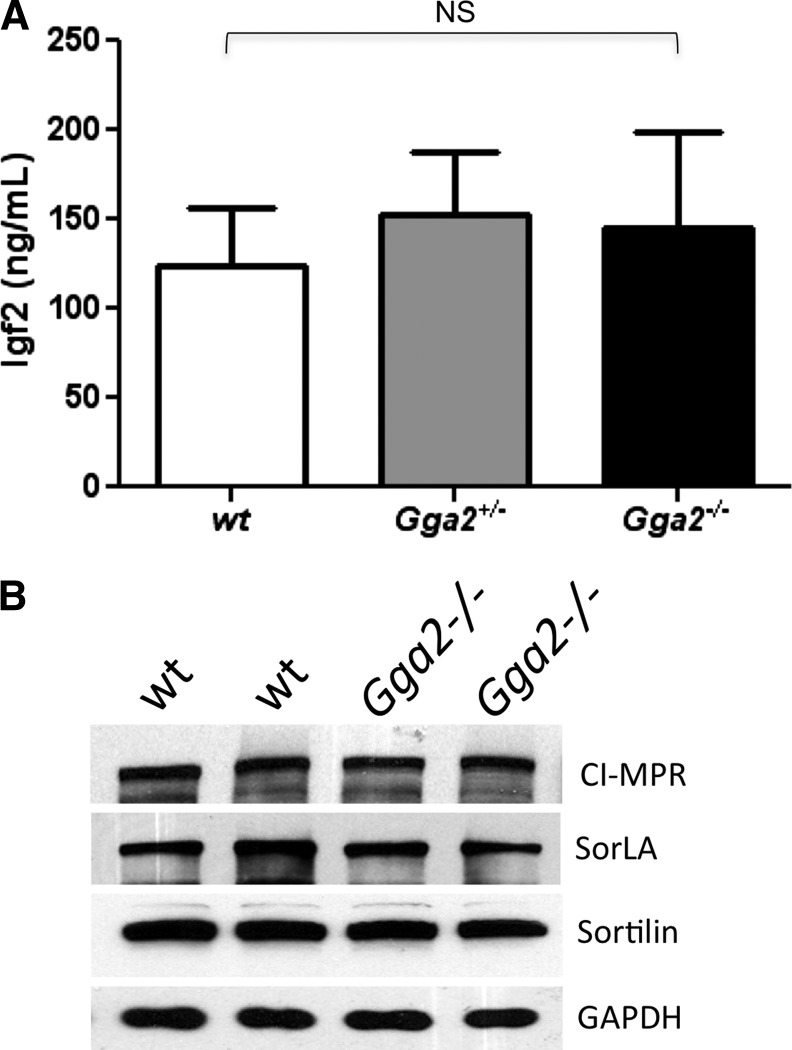
One consistent phenotype that was observed with the Gga2−/− mice in the mixed genetic background was that both males and females were significantly smaller than their siblings (Figure 3, A–C). Because the GGAs are known to be involved in the trafficking of the cation-independent mannose 6-phosphate receptor (CI-MPR) (Nielsen et al. 2001; Puertollano et al. 2001; Takatsu et al. 2001; Zhu et al. 2001), which clears insulin-like growth factor-II (IGF-II) from blood (Morgan et al. 1987), we considered the possibility that loss of GGA2 might result in increased levels of the CI-MPR at the plasma membrane, which, in turn, might lower the blood IGF-II level and inhibit growth. To investigate this possibility, serum IGF-II levels of day 1 pups were measured by enzyme- linked immunosorbent assay (ELISA). The levels were similar in all three genotypes, indicating that lack of circulating IGF-II is not the cause of the reduced body weight of Gga2−/− mice (Figure 4A). We also found that the expression level of the CI-MPR, SorLA, and Sortilin are unaltered in brain tissue of Gga2−/− mice (Figure 4B), excluding major differences in the turnover of these receptors in the absence of GGA2. Moreover, plasma levels of four different lysosomal acid hydrolases were assayed and found to be similar between wt and Gga2−/− mice (not shown), indicating that acid hydrolase sorting is normal in the absence of GGA2.
Figure 3.
Growth retardation of Gga2−/− mice. (A) Picture of day 1 pups from a single litter of a Tigm Gga2+/− cross in the mixed genetic background shows that the homozygous knockout has a markedly reduced body size. (B) The male Gga2−/− mouse (left mouse) in the mixed genetic background is much smaller than its female wt littermate at 6 wk of age but is otherwise healthy. (C) Weights were obtained from week 1 to week 12 and plotted according to gender and genotype. Results shown are mean ± SEM for each time point. Males: wt, n = 7; Gga2+/−, n = 11; Gga2−/−, n = 6. Females: wt, n = 3; Gga2+/−, n = 2; Gga2−/−, n = 2. P values for pairwise comparisons of mean weights between wt and Gga2−/− male mice are as follows: *P < 0.05, **P < 0.005, and ***P < 0.0005. NS, not significant. Statistical analysis was not performed with the females due to the small sample size.
Figure 4.
Analysis of serum IGF-II and brain IGF-II receptor (CI-MPR) levels. (A) Serum IGF-II levels of newborn mice were determined by ELISA as described in Materials and Methods. The serum obtained from each pup was sufficient to perform the assay in duplicate. Results are mean ± SD, n = 16 per group. (B) Western blot analysis of brain tissue from two wt and two Tigm Gga2−/− mice in the mixed background. Twenty-five μg protein extract for each sample was subjected to SDS-PAGE and immunoblot analysis of CI-MPR, SorLA, and Sortilin. GAPDH (5 μg of lysate) was included as a loading control. NS, not significant as determined by Student t test.
Discussion
The data presented here extend and clarify our previous report concerning the consequences of Gga2 gene-trap disruption (Govero et al. 2012). We reported that the Byg Gga2−/− mice in both the mixed and C57BL/6J genetic backgrounds exhibit early embryonic lethality, whereas the Tigm Gga2−/− mice in the C57BL/6NJ background display neonatal lethality. The current experiments unequivocally demonstrate that the early embryonic lethality of the Byg Gga2−/− mice is not due the absence of GGA2 because the Byg Gga2 gene-trap allele is clearly hypomorphic. Because the Gga1 and Gga3 genes were also disrupted using the same vector as in the Byg Gga2 strain and the corresponding gene-trap homozygotes are complete knockouts (Govero et al. 2012), this result was unexpected. However, similar phenomena have been reported for a number of other trapped genes, including the iron regulatory protein (IRP)-1 and IRP-2 genes where the pre-mRNAs transcribed from identical gene-trap cassettes are processed very differently within the two different gene contexts, with the IRP1 mRNA being undetectable while the IRP2 mRNA was unchanged (Galy et al. 2004). In the case of the Byg Gga2 gene-trap allele, this alternate splicing around the gene-trap cassette in many instances gives rise to near-normal levels of GGA2, showing that the early embryonic lethality is the consequence of another factor that has yet to be identified. Based on the finding with the Tigm Gga2−/− mice, we conclude that GGA2 is important for viability of neonates but appears to be dispensable once the mice are past 3 wk of age. In this regard, the genetic background has a major impact in terms of the tolerance to the loss of GGA2, with more than 95% lethality occurring in the C57BL/6NJ background (Govero et al. 2012) compared with 56% in the C57BL6/Ola129Sv mixed genetic background. This difference is not due to increased expression of GGA2 in the mixed background. A similar phenotype has been reported for Nedd4-2−/− mice, in which the targeted homozygous nulls in the C57BL/6J background exhibit almost complete neonatal lethality, which was partially rescued by crossing into the same mixed genetic background as our mice (Boase et al. 2011). In addition, a number of other gene knockouts in mice have been shown to present considerable variation in the penetrance of lethality, depending on the genetic background in which the knockouts were made (Doetschman 2009).
In our previous study, it was shown that single knockouts of Gga1 and Gga3 in the mixed genetic background are, for the most part, phenotypically normal (Govero et al. 2012). In addition, we reported that Gga1−/− mice in the C57BL/6J background are viable. We have now backcrossed Gga3−/− mice into the C57BL/6J background as well and have found no obvious difference between the two genetic backgrounds with either knockout strain, indicating that loss of GGA1 or GGA3 is well-tolerated in both strains, in contrast to the loss of GGA2. We have shown that even though brain expression of the three GGAs was detected at all stages of development, GGA2 expression was highest at embryonic day 9 (day E9) through the end of week 1, and then declined substantially reaching the lowest level in the adult mouse brain (Govero et al. 2012). Brain expression levels of GGA1 and GGA3, however, remained consistently high from day E9 through the adult stage (Govero et al. 2012). These findings are in concordance with some non-redundant and vital function(s) mediated by GGA2 early in the life of a mouse. This is not to imply that GGA1 and GGA3 together are completely dispensable for survival. Our previous study demonstrated that GGA1/GGA3 double knockout mice show 58% neonatal lethality in the mixed genetic background, similar to the 56% lethality due to loss of GGA2 in the same background. What are the overlapping compared to distinct physiological roles served by GGA1/GGA3 compared to GGA2? This is an important question that warrants future investigation.
Supplementary Material
Acknowledgments
We thank members of the Kornfeld laboratory for their valuable comments regarding the manuscript and Sarah Lesorogol for performing the lysosomal enzyme assays. We also thank Junxiang Wang at USC-Davis for her input in performing the IGF-II ELISA. We are very grateful to Rachel Idol for help with preparation of the figures and statistical analysis. This work was supported by the National Institutes of Health grant CA-008759-44 (to S.K.) and partial funding from the Alzheimer's Disease Research Center at Washington University School of Medicine in St. Louis (NIH P50-AG05681).
Footnotes
Communicating editor: D. W. Threadgill
Literature Cited
- Boase N. A., Rychkov G. Y., Townley S. L., Dinudom A., Candi E., et al. , 2011. Respiratory distress and perinatal lethality in Nedd4–2-deficient mice. Nat Commun 2: 287 10.1038/ncomms1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman A. L., Zhang C., Zhu X., Kahn R. A., 2000. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell 11: 1241–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S., 2004. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 5: 23–32 [DOI] [PubMed] [Google Scholar]
- Braulke T., Bonifacino J. S., 2009. Sorting of lysosomal proteins. Biochim. Biophys. Acta 1793: 605–614 [DOI] [PubMed] [Google Scholar]
- Doetschman T., 2009. Influence of genetic background on genetically engineered mouse phenotypes. Methods Mol. Biol. 530: 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B., Bruns K., Ghosh P., Kornfeld S., 2002. Autoinhibition of the ligand- binding site of GGA1/3 VHS domains by an internal acidic cluster-dileucine motif. Proc. Natl. Acad. Sci. USA 99: 8072–8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E., Villaescusa J. C., Di Rosa P., Fernandez-Diaz L. C., Longobardi E., et al. , 2006. Hypomorphic mutation of the TALE gene Prep1 (pKnox1) causes a major reduction of Pbx and Meis proteins and a pleiotropic embryonic phenotype. Mol. Cell. Biol. 26: 5650–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy B., Ferring D., Benesova M., Benes V., Hentze M. W., 2004. Targeted mutagenesis of the murine IRP1 and IRP2 genes reveals context-dependent RNA processing differences in vivo. RNA 10: 1019–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govero J., Doray B., Bai H., Kornfeld S., 2012. 2012 Analysis of Gga knockout mice demonstrates a non-redundant role for mammalian GGA2 during development. PLoS ONE 7: e30184 10.1371/journal.pone.0030184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Seaman M. N., Buschow S. I., Robinson M. S., 2007. The role of cargo proteins in GGA recruitment. Traffic 8: 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Sahlender D. A., Choma M., Sinka R., M. E. Harbour ME et al, 2009. Spatial and functional relationship of GGAs and AP-1 in Drosophila and HeLa cells. Traffic 10: 1696–1710 [DOI] [PubMed] [Google Scholar]
- Hirst J., Borner G. H., Antrobus R., Peden A. A., Hodson N. A., et al. , 2012. Distinct and overlapping roles for AP-1 and GGAs revealed by the “knocksideways” system. Curr. Biol. 22: 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason E. J., Grell J. A., Wan J., Cohen P., Conover C. A., 2011. Insulin-like growth factor (IGF)-I and IGF-II contribute differentially to the phenotype of pregnancy associated plasma protein-A knock-out mice. Growth Horm. IGF Res. 21: 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay M. M., Kahn R. A., 2004. Multiple phosphorylation events regulate the subcellular localization of GGA1. Traffic 5: 102–116 [DOI] [PubMed] [Google Scholar]
- Morgan D. O., Edman J. C., Standring D. N., Fried V. A., Smith M. C., et al. , 1987. Insulin-like growth factor II receptor as a multifunctional binding protein. Nature 329: 301–307 [DOI] [PubMed] [Google Scholar]
- Nielsen M. S., Madsen P., Christensen E. I., Nykjaer A., Gliemann J., et al. , 2001. The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 20: 2180–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Aguilar R. C., Gorshkova I., Crouch R. J., Bonifacino J. S., 2001. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science 292: 1712–1716 [DOI] [PubMed] [Google Scholar]
- Shiba Y., Katoh Y., Shiba T., Yoshino K., Takatsu H., et al. , 2004. GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J. Biol. Chem. 279: 7105–7111 [DOI] [PubMed] [Google Scholar]
- Takatsu H., Katoh Y., Shiba Y., Nakayama K., 2001. Golgi-localizing, gamma-adaptin ear homology domain, ADP-ribosylation factor-binding (GGA) proteins interact with acidic dileucine sequences within the cytoplasmic domains of sorting receptors through their Vps27p/Hrs/STAM (VHS) domains. J. Biol. Chem. 276: 28541–28545 [DOI] [PubMed] [Google Scholar]
- Walker K. R., Kang E. L., Whalen M. J., Shen Y., Tesco G., 2012. Depletion of GGA1 and GGA3 mediates postinjury elevation of BACE1. J. Neurosci. 32: 10423–10437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogosawa S., Kawasaki M., Wakatsuki S., Kominami E., Shiba Y., 2006. Monoubiquitylation of GGA3 by hVPS18 regulates its ubiquitin-binding ability. Biochem. Biophys. Res. Commun. 350: 82–90 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Doray B., Poussu A., Lehto V. P., Kornfeld S., 2001. Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science 292: 1716–1718 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.