Abstract
Indolethylamine-N-methyltransferase (INMT) is a Class 1 transmethylation enzyme known for its production of N,N-dimethyltryptamine (DMT), a hallucinogen with affinity for various serotonergic, adrenergic, histaminergic, dopaminergic, and sigma-1 receptors. DMT is produced via the action of INMT on the endogenous substrates tryptamine and S-adenosyl-l-methionine (SAM). The biological, biochemical, and selective small molecule regulation of INMT enzyme activity remain largely unknown. Kinetic mechanisms for inhibition of rabbit lung INMT (rabINMT) by the product, DMT, and by a new novel tryptamine derivative were determined. After Michaelis–Menten and Lineweaver–Burk analyses had been applied to study inhibition, DMT was found to be a mixed competitive and noncompetitive inhibitor when measured against tryptamine. The novel tryptamine derivative, N-[2-(1H-indol-3-yl)ethyl]-N′,N′-dimethylpropane-1,3-diamine (propyl dimethyl amino tryptamine or PDAT), was shown to inhibit rabINMT by a pure noncompetitive mechanism when measured against tryptamine with a Ki of 84 μM. No inhibition by PDAT was observed at 2 mM when it was tested against structurally similar Class 1 methyltransferases, such as human phenylethanolamine-N-methyltransferase (hPNMT) and human nicotinamide-N-methyltransferase (hNNMT), indicating selectivity for INMT. The demonstration of noncompetitive mechanisms for INMT inhibition implies the presence of an inhibitory allosteric site. In silico analyses using the computer modeling software Autodock and the rabINMT sequence threaded onto the human INMT (hINMT) structure (Protein Data Bank entry 2A14) identified an N-terminal helix–loop–helix non-active site binding region of the enzyme. The energies for binding of DMT and PDAT to this region of rabINMT, as determined by Autodock, were −6.34 and −7.58 kcal/mol, respectively. Assessment of the allosteric control of INMT may illuminate new biochemical pathway(s) underlying the biology of INMT.
Indolethylamine-N-methyltransferase (INMT), as a transmethylation enzyme, transfers one or more methyl groups from S-adenosyl-l-methionine (SAM) to appropriate amino group acceptors on indole-containing compounds. The activity of this enzyme was first reported in rabbit lung homogenate by J. Axelrod,1 and rabbit and human INMT were eventually cloned and sequenced by Thompson and collegues in the late 1990s.2,3 INMT is a member of a large family of N-methyltransferases that can methylate a variety of small molecule acceptors such as tryptamine,2 serotonin, and other endogenous indole-containing compounds.4−7 This enzyme is widely distributed in mammalian tissues, including the lungs, adrenal gland, thyroid, placenta, heart, pancreas, lymph nodes,2−6 retina, pineal gland, and spinal cord ventral horn motoneurons.8,9 Tryptamine, the substrate normally associated with INMT, is derived from the in vivo decarboxylation of tryptophan. Transmethylation produces N-methyltryptamine (MMT) and N,N-dimethyltryptamine (DMT). DMT is found in trace amounts in humans and other animals.6,10−17 For a thorough review of the synthesis of DMT, its regulation, and its metabolism, see ref (7). Recently, Barker and colleagues, using microdialysis techniques, detected DMT in real time in the pineal glands of living rats.18
The precise role of INMT in biological systems is currently unresolved. However, several observations indicate the pleiotropic importance of the INMT gene. For example, expression of the INMT gene has been shown to be downregulated in prostate cancer19 and lung cancer,20 implying a role for INMT in inhibiting tumor progression. Further, successful implantation of the embryo in the mouse appears to be supported by the presence of the INMT/TEMT gene (together with several other genes).21 INMT enzyme activity is elevated in the rabbit fetal lung.22 Refer further to a recent review of the possible peripheral [in addition to the central nervous system (CNS)] role(s) of INMT/DMT in mammalian systems involving the sigma-1 receptor by Frecska and colleagues.23
Increased INMT activity had been suggested to be involved in schizophrenia and stress-related psychoses in humans, and this served as the basis for early work on the proposed utility of inhibiting in vivo INMT enzyme activity to treat schizophrenia.24−26 Further, the INMT product DMT, when ingested, produces a psychological state characterized by colorful visual illusions, altered time and space perceptions, and changes in body image.27 Because of these known psychoactive effects of DMT, the proposition that INMT activity and DMT production may be involved in producing exceptional mental states may be worth investigating.
The psychoactive effects of DMT are mediated through various mechanisms, including binding to and activating serotonin receptors,28−30 exhibiting substratelike behavior at serotonin and vesicular uptake transporters,31,32 and inhibiting monoamine oxidase enzymes.31−34 The sigma-1 receptor is the latest identified receptor target for DMT, where it binds at low micromolar concentrations, inhibits voltage-activated sodium ion channels via sigma-1 receptor interactions at higher concentrations, and induces a hypermobility response in wild-type mice that is abolished in sigma-1 receptor knockout mice.35 INMT has been shown to colocalize with the sigma-1 receptors in primate spinal cord motoneurons containing unique synapses called C-terminals9 and may be involved in future therapeutic strategies for the treatment of amyotrophic lateral sclerosis (ALS).36,37 Whether INMT colocalizes with sigma-1 receptors in other neural tissue remains unknown.
Product inhibition of INMT by S-adenosyl-l-homocysteine (SAH) has been demonstrated. SAH’s inhibitory mechanism has been reported to be competitive against SAM and noncompetitive against N-methylserotonin.22 Little is known regarding the relationship between the structure and activity of INMT and synthetic or endogenous small molecule regulators or their kinetic mechanisms. Thompson et al.3 identified several compounds, including the β-carboline, norharmane, that when compared to tryptamine, served as relatively effective substrates for rabbit INMT. Several inhibitors of rabINMT were identified by Thompson et al.,3 but the kinetic mechanism(s) was not reported. The in vivo activity of INMT appears to be inhibited by uncharacterized dialyzable endogenous compounds.38,39 A competitive inhibition mechanism of 1,8-diaminooctane and 1,7-diaminoheptane of rabbit lung INMT when measured against tryptamine has been reported.40 Here, we report the mechanism of inhibition of rabINMT by DMT and a novel derivative of tryptamine, N-[2-(1H-indol-3-yl)ethyl]-N′,N′-dimethylpropane-1,3-diamine (PDAT). Inhibition by PDAT was compared to that by N-[2-(1H-indol-3-yl)ethyl]propane-1,3-diamine (PAT), which is lacking the two methyl groups on the N-propyl amino moiety.
Experimental Procedures
Materials
All chemicals were purchased from Aldrich Chemical Co. (Milwaukee, WI) and utilized without further purification unless stated otherwise. [14C]-S-Adenosyl-l-methionine ([14C]SAM) was purchased from PerkinElmer Life Sciences (Wellesley, MA). Frozen rabbit lungs were purchased from Pel-Freez (Rogers, AR). Histidine-tagged human indolethylamine-N-methyltransferase was obtained from the Structural Genomics Consortium (University of Toronto, Toronto, ON). Recombinant human phenylethanolamine-N-methyltransferase (hPNMT) and nicotinamide-N-methyltransferase (hNNMT) were purchased from Creative BioMart (Shirley, NY). All reagents for the hPNMT assay were prepared in 50 mM Tris-HCl buffer (pH 8.5) unless stated otherwise, excluding PDAT, which was dissolved in dimethyl sulfoxide (DMSO). All reagents for the hNNMT assay were prepared in 50 mM Tris-HCl buffer (pH 7.2).
Chemistry
Synthesis of N,N-Dimethyltryptamine (DMT)
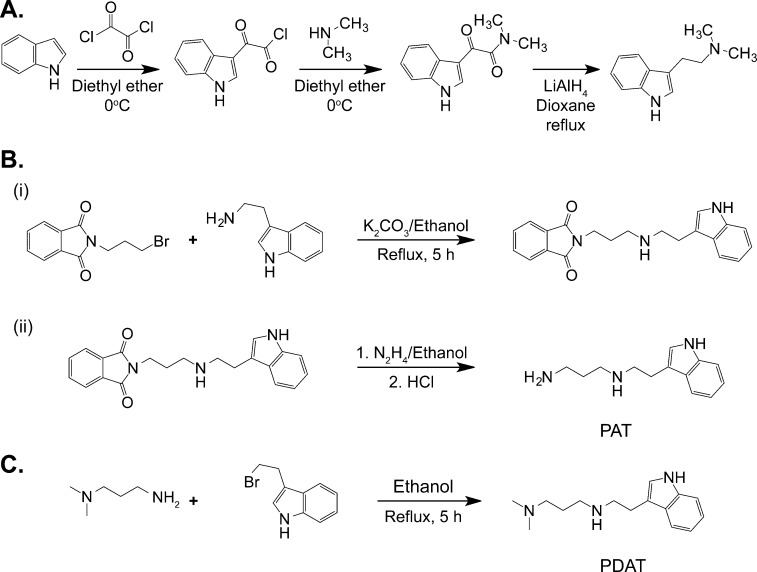
DMT was synthesized by the method of Speeter and Anthony41 with minor modifications. Briefly, indole, dissolved in ice-cold diethyl ether, was reacted with a solution containing 2 equiv of oxalyl chloride in diethyl ether followed by a 20 wt %/vol solution of dimethylamine in diethyl ether until the pH reached 9–10, while maintaining the reaction mixture at 0 °C with an ice bath. The resulting 3-indoleglyoxyl-N,N-dimethylamide was reduced to DMT with lithium aluminum hydride (LAH) in refluxing dioxane essentially as described previously.42 After workup and solvent removal, DMT free base was obtained. The DMT was purified by several recrystallizations from hot heptane followed by air drying. The product migrated as a single spot on silica gel thin layer plates (4:1:1 n-BuOH/AcOH/H2O mobile phase) and comigrated with authentic DMT. The melting point for DMT was determined to be 64–65 °C and is consistent with previously reported data.431H nuclear magnetic resonance (NMR) was consistent with the assigned structure.
Syntheses of N-[2-(1H-Indol-3-yl)ethyl]propane-1,3-diamine (PAT)
This compound was prepared as the nonmethylated derivative of PDAT to assess the possible importance of the N,N-dimethylation of the aminopropyl side chain. The synthesis occurred in two steps according to the schemes outlined in panels A and B of Figure 1. In reaction A, N-(3-bromopropyl)phthalimide (1 mmol, 0.268 g) and 5 equiv of tryptamine (5 mmol, 0.8 g) were refluxed together in ethanol and 2 mmol of potassium carbonate (0.27 g) for 5 h. The solid was filtered off, and the solvent was evaporated under reduced pressure using a rotary evaporator. Then the reaction was quenched with water and the mixture extracted three times with 5 mL of ethyl acetate. The combined extracts were dried in MgSO4 and evaporated under reduced pressure. The product was purified by column chromatography using silica gel and a 4:6 n-hexane/EtOAc mixture, giving a 90% yield. The product was subsequently taken to the next step without further characterization.
Figure 1.
Synthetic schemes of DMT and inhibitors of INMT: (A) DMT, (B) N-[2-(1H-indol-3-yl)ethyl]propane-1,3-diamine (PAT), and (C) N-[2-(1H-indol-3-yl)ethyl]-N′,N′-dimethylpropane-1,3-diamine (PDAT).
The phthalimide tryptamine product from reaction A was subsequently reduced with hydrazine in a Gabriel synthesis as shown in Figure 1B. Briefly, 1 mmol (0.349 g) of phthalimide tryptamine and 2 mmol (0.064 g) of hydrazine were refluxed together in ethanol for 1.5 h. The mixture was subsequently cooled to room temperature, and a 5-fold molar ratio of concentrated HCl was added dropwise. The mixture was refluxed for an additional 4 h and the reaction quenched with H2O. Workup procedures for this compound were performed in a manner similar to those described for reaction A. The product was purified by column chromatography using silica gel and a 9:1 toluene/diethylamine solvent system, giving an 82% yield.
Synthesis of N-[2-(1H-Indol-3-yl)ethyl]-N′,N′-dimethylpropane-1,3-diamine (PDAT)
This compound was prepared according to the synthetic scheme shown in Figure 1C. The reaction was conducted using a straightforward alkylation using 1 mmol (0.224 g) of 3-(2-bromoethyl)indole and 5 mmol (0.52 g) of 3-(dimethylamino)-1-propylamine. The reaction mixture was refluxed in ethanol for 5 h and the reaction quenched with H2O. Workup procedures for this compound were performed in a manner similar to that of the procedures described for reaction A. The product was purified by column chromatography using silica gel and a 9:1 toluene/diethylamine solvent system, giving an 80% yield.
Characterizations of PAT and PDAT
Yields refer to isolated products after column chromatography, and products were characterized by 1H NMR. 1H NMR spectra were recorded at 300 MHz in CDCl3 relative to TMS (0.00 ppm).
PAT: 1H NMR δ 10.1 (NH, 1 H), 7.60–7.47 (m, 5 H), 5.11 (d, 2 H), 2.63–2.57 (m, 6 H), 2.3 (d, 1 H), 1.84 (m, 2 H); 13C NMR (75 MHz, CDCl3) δ 136.2, 127.41, 123.0, 119.7, 118.6, 113.4, 111.6, 49.8, 46.9, 39.3, 31.725.3; EMS [MH+] for C13H19N3 calcd 218.1579, found 218.2892.
PDAT: 1H NMR δ 10.2 (d, 1 H), 7.60–7.47 (m, 2 H), 2.78 (m, 2 H), 2.37–2.46 (m, 3 H), 2.26 (d, 7 H), 1.62 (m, 2 H); 13C NMR (75 MHz, CDCl3) δ 136.8, 127.0, 123.6, 119.7, 118.6, 111.2, 54.9, 49.6, 46.5, 45.7, 26.7, 25.1; EMS [MH+] for C15H23N3 calcd 246.1892, found 246.2958.
Preparation of Rabbit Lung Homogenates
Rabbit lung homogenates were prepared according to the method described by Thompson and Weinshilboum.3 Briefly, one pair of frozen rabbit lungs was thawed in 50 mM Tris-HCl (pH 7.3), minced with a scissors, and homogenized using four bursts of 10 s each with a brinkman polytron (American Laboratory Trading Inc., East Lyme, CT) on setting 6 on ice. Homogenates were centrifuged at 15000g for 15 min at 4 °C, and the supernatant from this first low-speed centrifugation was subjected to a second centrifugation step at 100000g for 60 min at 4 °C. The supernatant from the second high-speed spin was aliquoted and frozen at −80 °C until it was used in the INMT enzymatic assay.
Rabbit Lung INMT Assays
Rabbit lung INMT assays were modified from those described by Thompson et al.3 A final incubation volume of 100 μL in 15 mL capped tubes contained ice-cold tryptamine solutions in Tris-HCl (pH 8.5) at final concentrations of 0.1, 0.3, 0.6, 0.8, and 1.0 mM, 250 μg/mL bovine serum albumin (BSA), and 35.5 μM [14C]-S-adenosyl-l-methionine ([14C]SAM) (specific radioactivity varied between 28.15 and 56.3 μCi/μmol). DMSO solutions of PDAT and DMT (when tested as inhibitors) were diluted 100-fold to a final concentration of 0.1 mM, and the same concentration of DMSO alone was added to the noninhibitor controls. The assay was initiated by the addition of 10 μL of rabbit lung supernatant (rabINMT) in Tris-HCl (pH 8.5) (stock protein concentrations between 10 and 25 mg/mL). The final volume was adjusted to 100 μL with Tris-HCl (pH 8.5). The reaction was allowed to proceed at 32 °C in a gently rotating water bath for 45 min. The assay was terminated by the addition of 0.6 mL of an ice-cold 0.5 M potassium borate solution (pH 10). Tubes without tryptamine were used as background controls. The [14C]-N-monomethyltryptamine (MMT) and DMT reaction products were extracted by addition of 5 mL of a 3% isoamyl alcohol/toluene mixture. Tubes were vortexed three times for 5 s each and then centrifuged at 1620g for 2–3 min. Following centrifugation, 3.5 mL of the top organic layer was assessed for the [14C]methylated tryptamine levels using a Beckman LS 6500 scintillation counter. The presence of authentic [14C]MMT and [14C]DMT, which comprised >95% of the reaction products, was confirmed by silica gel thin layer chromatography using a 12:5:3 n-butanol/water/acetic acid mixture followed by autoradiography (Figure S1 of the Supporting Information). Each radioactive spot was confirmed by comigration with nonradioactive MMT (Rf = 0.48) and DMT (Rf = 0.39). Assays were performed in duplicate, and each experiment was repeated. The results reported are averages of quadruplicate counts from each duplicate sample.
hPNMT Assay
The activity of hPNMT was measured following the method of Gee et al.44 Briefly, phenylethanolamine (PEA) at 2.48 mM and [14C]SAM (20.95 μM) were prepared in the absence (2% DMSO) or presence of PDAT (2 μM) in a final volume of 100 μL in 50 mM Tris-HCl (pH 8.5). The reaction was initiated via the addition of 5 μL of hPNMT (0.025 μg/μL) and incubated for 90 min at 32 °C. The reaction was quenched with 0.6 mL of 0.5 M potassium borate (pH 10.0); the product, [14C]-N-methyl-PEA, was extracted with 5 mL of 3% isoamyl alcohol in toluene, and the organic layers were separated by centrifugation for 2 min at 1620g in a swinging bucket centrifuge. The organic layer (approxiamtely 3.5 mL) was combined with 6 mL of a scintillation solution (Ultima Gold, PerkinElmer Life Sciences) and counted. The assay for hINMT was performed similarly except tryptamine (8 mM) and hINMT (0.14 μg/μL) were used.
hNNMT Assay
The hNNMT assay was performed in a manner similar to that used for hPNMT with modifications following Rini et al.45 Nicotinamide (10 mM) and 5 μL of [14C]SAM (20.95 μM) were prepared in 50 mM Tris-HCl (pH 7.2) in a final volume of 100 μL in the absence (2% DMSO) or presence of PDAT (2 μM). As a positive control, a similar reaction mixture was prepared in the presence of 1-N-methylnicotinamide (1 mM). The reaction was initiated by adding 5 μL of hNNMT (0.025 μg/μL). After 90 min at 32 °C, each reaction was quenched with 0.6 mL of 0.5 M potassium borate and 100 μL of 1.3 M 1-heptanesulfonic acid to neutralize the charge on the product 1-N-methylnicotinamide.45 The product was extracted with 5 mL of 60% isoamyl alcohol in toluene. The organic and aqueous layers were separated by centrifugation at 1620g for 3.5 min, after which 3.5 mL of the organic layer was extracted, combined with 6 mL of a scintillation solution (Ultima Gold, PerkinElmer Life Sciences), and counted.
The Ki for PDAT was determined from the Km and Vmax values by use of the equation Vmax (+inhibitor) = Vmax (−inhibitor)/(1 + [inhibitor]/Ki) to be ∼84 μM.
In Silico Modeling
The rabINMT sequence was aligned with the human INMT (hINMT) sequence using ClustalW. The human and rabbit INMT are 90% identical in amino acid sequence (Figure S2 of the Supporting Information). This aligned rabINMT sequence was threaded onto the hINMT structure [Protein Data Bank (PDB) entry 2A14] by sequential mutation of residues using the ‘mutate’ option in the biopolymer module of the molecular modeling program Sybyl (Tripos Corp., St. Louis, MO). The fits were examined with molecular graphics and the side chains torsioned to accommodate any clashes with the surrounding residues. To remove residual clashes, when fully threaded, the resultant model was energy-minimized using the Tripos force field. The allosteric loop (residues 25–34) was examined for conformational similarity to the hINMT and its suitability for docking.
All dockings were performed with Autodock4 (Scripps Institute) with a box size adequate to include the allosteric loop and the active site. The active site in hINMT has no ligand (other than SAH) and is an apo conformation that excludes ligands as part of the docking process. All the docking of ligands occurs in the allosteric loop. All minimizations and dockings included the SAH.
Results
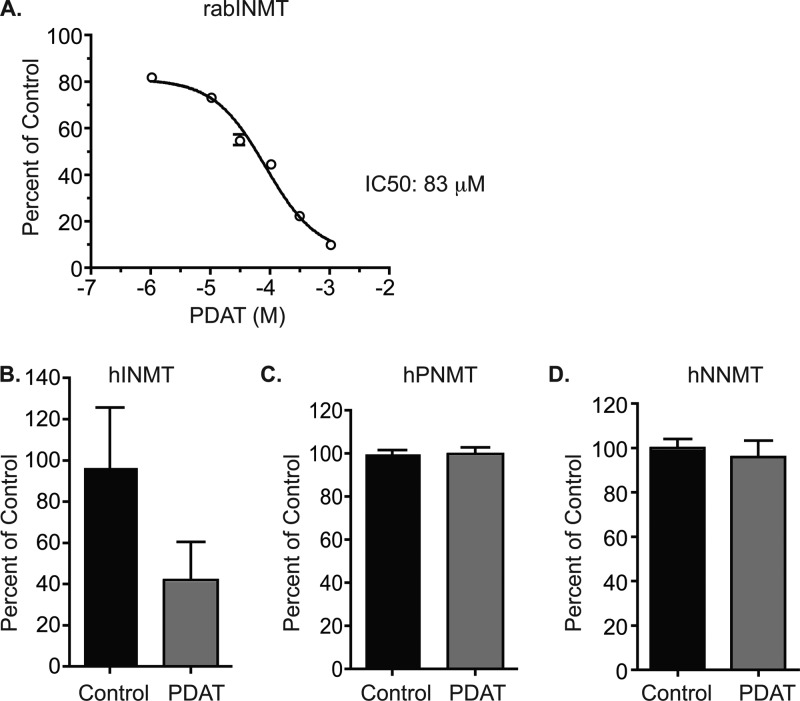
The lead compound, PDAT, showed inhibition of INMT in rabbit lung homogenates (rabINMT) with a half-maximal inhibitory concentration (IC50) of 83 μM (Figure 2A). PAT was decidedly less potent than PDAT for inhibition of rabINMT, showing an IC50 of approximately 1 mM (data not shown). These data demonstrate the importance of N,N-dimethylation of the propylamino moiety for effective INMT inhibition. PDAT inhibition of pure human INMT (hINMT) was also observed with an estimated IC50 of 1 mM (data not shown).
Figure 2.
Selectivity profile of PDAT for N-methyltransferases. (A) Inhibition of rabINMT by PDAT. Data are presented as a percentage of the control after background subtraction, and the absence of PDAT was normalized as 100%. (B) Inhibition of hINMT by PDAT. hINMT assays were conducted with tryptamine as a substrate and [14C]SAM as the methyl donor, in the absence or presence of 2 mM PDAT. (C) Lack of inhibition of human recombinant PNMT by PDAT. PNMT assays were conducted like hINMT assays but with phenylethanolamine (PEA) as a substrate. (D) Lack of PDAT inhibition of hNNMT. hNNMT assays were conducted as described for hINMT assays but using nicotinamide as a substrate.
The affinities of PDAT for two structurally related N-methyltransferases, human hPNMT and hNNMT, were assessed. PDAT did not inhibit either enzyme at a concentration of 2 mM, supporting its selectivity for INMT (Figure 2B). Inhibitors for both PNMT and NNMT,46−48 when measured against their respective substrates, phenylethanolamine and nicotinamide, respectively, showed complete inhibition of these enzymes (data not shown).
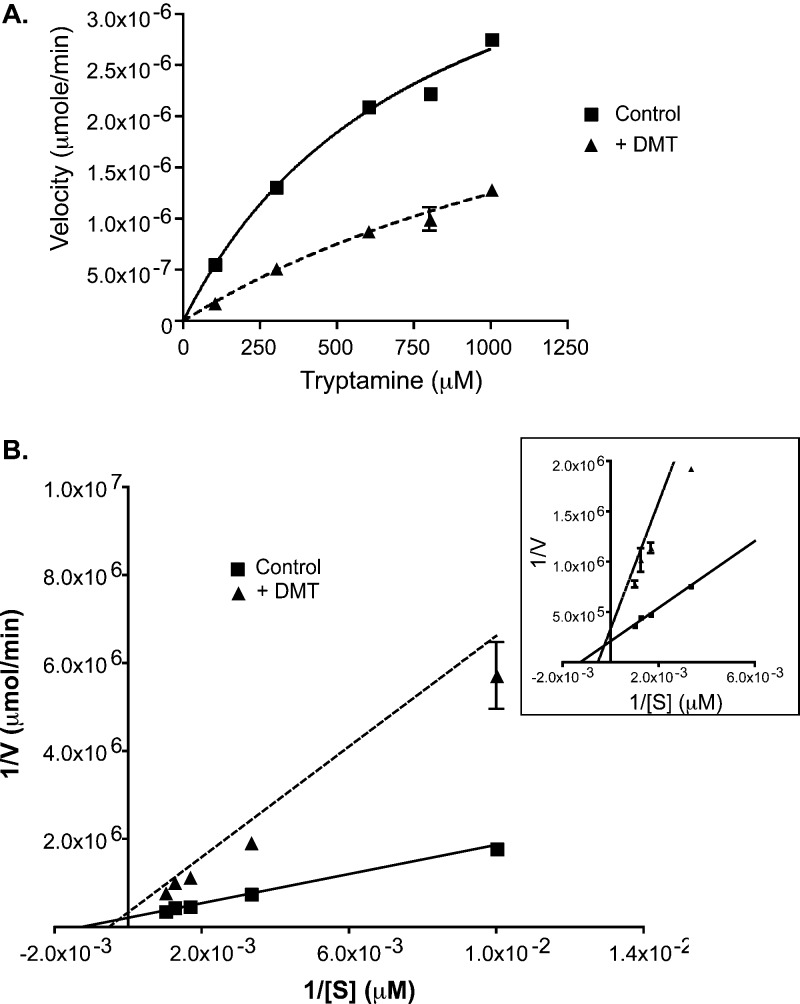
DMT is a known inhibitor of INMT,3 but its kinetic mechanism of inhibition has not been elucidated. DMT showed a mixed noncompetitive mechanism when measured against its substrate, tryptamine, as assessed by Michaelis–Menten and Lineweaver–Burk analysis. The rabINMT Vmax was reduced from 4.63 ± 0.14 μmol/min for the control to 2.75 ± 0.85 μmol/min in the presence of DMT, and the Km for tryptamine was increased from 852 ± 61 to 1618 ± 275 μM (Figure 3, Figure S3 of the Supporting Information, and Table 1).
Figure 3.
Mechanism of binding of DMT to rabINMT. (A) Representative Michaelis–Menten plot for DMT in the absence or presence of cold DMT (100 μM). Concentrations of tryptamine varied from 100 to 1000 μM. (B) Lineweaver–Burk transformation of the data presented in panel A. Velocity (V) was calculated by determining the amount of [14C]DMT formed over the time course of 60 min, and [S] represents the tryptamine concentration. The inset shows a close-up of panel B showing the intersections of the two lines. In the absence of an inhibitor, the Km for DMT is 852.2 ± 61.35 μM, and the Km for DMT is 1618 ± 275.5 μM in the presence of cold DMT. The Km values were determined using the Michaelis–Menten analyses from GraphPad Prism, and the standard error of the mean was calculated from two separate experiments performed in duplicate.
Table 1.
| Km (μM) | Vmax (μmol/min) | |
|---|---|---|
| control | 852 ± 61 | 4.63 ± 0.14 |
| 100 μM DMT | 1618 ± 275 | 2.75 ± 0.85 |
| control | 499 ± 68 | 2.47 ± 1.22 |
| 100 μM PDAT | 523 ± 85 | 1.13 ± 0.37 |
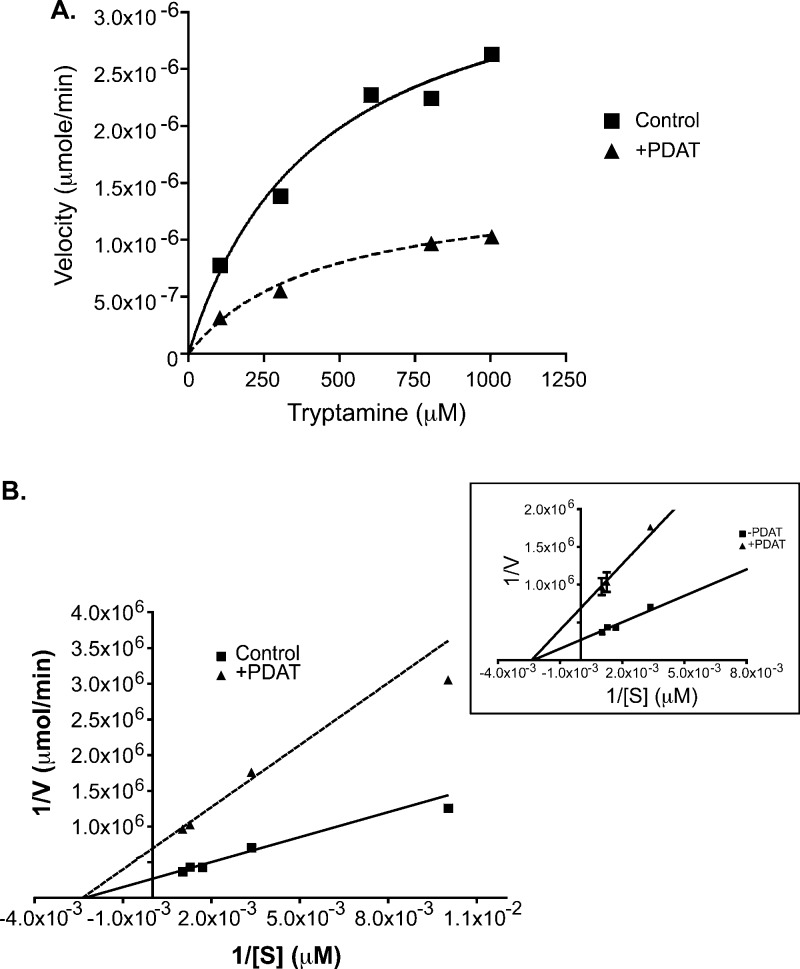
PDAT inhibition of rabINMT showed a pure noncompetitive inhibition mechanism when measured against the variable substrate, tryptamine. Vmax was reduced from 2.47 ± 1.22 μmol/min for the control to 1.13 ± 0.37 μmol/min in the presence of PDAT, but the Km for tryptamine remained largely unaltered (from 499 ± 68 to 523 ± 85 μM) (Figure 4, Figure S4 of the Supporting Information, and Table 1). These data are consistent with an INMT allosteric inhibitory mechanism for DMT and DMT-like molecules, such as PDAT.
Figure 4.
Mechanism of binding of PDAT to rabINMT. (A) Representative Michaelis–Menten plot for DMT in the absence or presence of 100 μM PDAT. Concentrations of tryptamine varied from 100 to 1000 μM. (B) Lineweaver–Burk transformations of the data presented in panel A. Velocity (V) was calculated by determining the amount of [14C]DMT formed over the time course of 60 min, and [S] represents the tryptamine concentration. The inset shows a close-up of panel B showing the intersections of the two lines. In the absence of inhibitor, the Km for DMT is 499.6 ± 68.2 μM, and the Km for DMT is 523.4 ± 85.4 μM in the presence of 100 μM PDAT. The Km values were determined using the Michaelis–Menten analyses from GraphPad Prism, and the standard error of the mean was calculated from two separate experiments performed in duplicate.
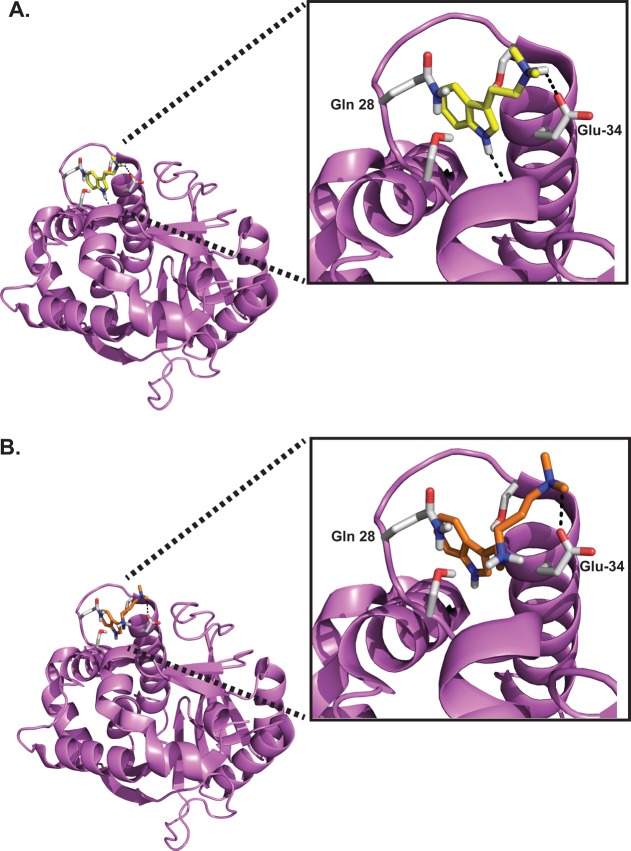
Because the crystal structure of SAH-bound hINMT has been reported,49 we assessed the potential docking site of DMT and PDAT for hINMT using an in silico approach (Figure 5A,B). Both molecules docked to the N-terminal helix–loop–helix region of INMT with binding energies of −6.34 kcal/mol for DMT and −7.58 kcal/mol for PDAT. There are multiple hydrogen bonds that may contribute to binding of both DMT and PDAT to this loop region. As shown in Figure 5, the indole nitrogen of DMT forms two hydrogen bonds with aspartic acid 28 with the terminal nitrogen hydrogen bonding with glutamic acid 34. Unlike DMT, PDAT is predicted to form four hydrogen bonds in which the middle nitrogen forms two hydrogen bonds with aspartic acid 28, and the indole nitrogen bonds with tyramine 24. The terminal nitrogen of PDAT bonds to glutamic acid 34. This mechanism of binding may contribute to the energy of PDAT being lower (or the affinity higher) than that of DMT.
Figure 5.
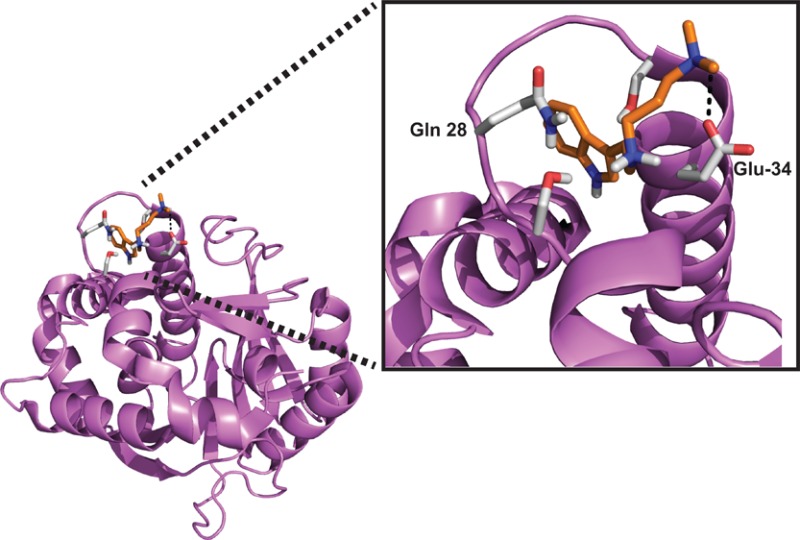
In silico docking of DMT and PDAT on the hINMT (PDB entry 2A14). The crystal structure of hINMT was determined with SAM bound. (A) Structure of hINMT docked with DMT. The optimal DMT docking to hINMT indicates a terminal nitrogen hydrogen bond to Glu-34, with the indole nitrogen hydrogen bonded to the loop Asp-28 carboxylate. The free energy of binding of the displayed fit is −7.06 kcal/mol. (B) Structure of hINMT docked with PDAT. Stabilization of PDAT with the protein is predicted to be mediated by the hydrogen bonding interactions between the terminal nitrogen and Glu-34, between the middle nitrogen and Asp-28 of the protein, and with the indole nitrogen hydrogen bonded to the backbone of the Tyr-24 carbonyl of helix Leu-16–Tyr-24. The free energy of binding of the displayed fit is −8.24 kcal/mol.
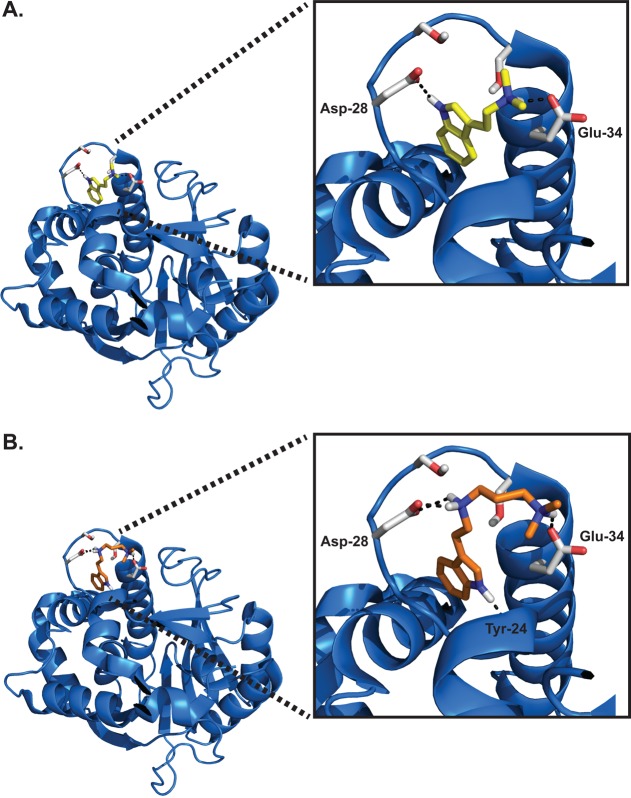
Additionally, we threaded the sequence of rabINMT onto the sequence of hINMT (Figure 6A,B). When DMT and PDAT were assessed in silico for their interactions with rabINMT using Autodock, the highest-affinity interactions of DMT and PDAT were found to occur exclusively in the N-terminal helix–loop–helix region like the docking of DMT and PDAT to hINMT. The proposed allosteric N-terminal helix–loop–helix region is not within the active site of the enzyme as assessed by the position of SAH in the cocrystal structure of hINMT (PDB entry 2A14). In addition to the hydrogen bonding interactions between these compounds and INMT, however, rabINMT contains a glutamine at position 28 that is likely to form π-stacking interactions with the indole ring of both DMT and PDAT to contribute to the binding of both compounds for rabINMT. The N-terminal helix–loop–helix sequences (amino acids 24–34) are aligned and identified in Figure S2 of the Supporting Information.
Figure 6.
In silico docking of DMT and PDAT on the threaded structure of rabINMT. The threaded structure of rabINMT was modeled after that of hINMT (PDB entry 2A14). (A) Threaded structure of rabINMT docked with DMT. Optimal DMT docking in threaded the rabINMT model uses a terminal hydrogen bond to Glu-34, with the indole nitrogen hydrogen bonded to the helix backbone. The free energy of binding is −6.34 kcal/mol. (B) Structure of rabINMT docked with PDAT. Optimal PDAT docking in the threaded rabINMT also uses a hydrogen bond from the terminal nitrogen to Glu-24, but the middle nitrogen is placed at the negative polar end of the Leu-16–Tyr-24 helix. The docking free energy of binding is calculated to be −7.58 kcal/mol. A π–π stacking interaction may occur between the indole rings of PDAT and DMT with Gln-28.
Discussion
INMT is a member of a large class of N-methyltransferases that utilize SAM as a methyl donor. The enzyme INMT transfers methyl groups from SAM to the nitrogen of substrates containing indolyl alkyl amino groups with the resultant formation of SAH.1,3,51 One product of INMT N-methylation of tryptamine is DMT. In light of the recent discovery that DMT activates the sigma-1 receptor,35 we sought to find DMT-like small molecule regulators of INMT to study the crosstalk between this metabolic pathway and that of the sigma-1 receptor, as well as to better understand the biology of INMT. Two N-propylamino derivatives of tryptamine, PAT and PDAT (panels B and C of Figure 1, respectively) were synthesized, and their INMT regulatory properties were compared to those of DMT (Figure 1A).
The biological role of DMT remains largely unknown. There are suggestions that DMT may act as a neurotransmitter in humans and may be involved in psychosis, dreaming, near-death experiences, and spiritual exaltation.27,51−54 INMT/DMT/sigma-1 receptor mechanisms involving peripheral as well as CNS roles may also be important in mammalian biology.23 Recently, N-methylated derivatives of tryptamine, including N-monomethyl-, N,N-dimethyl-, and N,N,N-trimethyltryptamines, have been identified in the leaves and seeds of a wide variety of citrus plants, including Bergamot orange (Citrus bergamia)55 and other Citrus fruits.56 Presumably, these naturally occurring compounds are formed through INMT transmethylation of tryptamine, but this is yet to be established. DMT and related tryptamines also occur in the Illinois bundleflower (Desmanthus illinoensis, common in the United States), chacruna (Psychotria viridis, native to Central and South America), and trees of the Virola genus, native to South American rainforests. Archeological evidence indicates that South American native cultures have used these plants for shamanistic rituals for at least 3000 years.57
The work reported in this paper on the inhibition of rabbit lung INMT by DMT and by a novel synthetic tryptamine derivative, PDAT (Figure 1C), enhances our knowledge of the mechanisms of INMT regulation. Both compounds showed noncompetitive inhibition kinetics when measured against tryptamine at saturating concentrations of SAM. DMT showed mixed competitive and noncompetitive kinetics (Figure 3), consistent with DMT binding to INMT alone and to the INMT–tryptamine complex with different Ki values (see Table 1 and Figure S5A of the Supporting Information). PDAT, on the other hand, showed pure noncompetitive kinetics (Figure 4) consistent with an equal affinity for INMT alone and for the INMT–tryptamine complex (see Figure S5B of the Supporting Information). The Ki for PDAT was determined to be 84 μM (Figure 2A). The presence of the dimethyl substitution on the propylamino side chain was found to be important for the affinity of PDAT for INMT, because the nonmethylated propylamino tryptamine derivative, PAT (Figure 1B), was significantly weaker in INMT inhibition (data not shown). PDAT was shown to be a selective inhibitor of INMT because 2 mM PDAT showed no significant inhibition of NNMT or PNMT (Figure 2B–D).
These data support an allosteric inhibitory mechanism for DMT and PDAT for rabbit INMT. An in silico analysis of binding of DMT and PDAT to both hINMT and rabINMT (Figures 5 and 6) identified the N-terminal helix–loop–helix region of INMT as a possible location for a proposed allosteric inhibitory site. The volumes of the loop regions of PNMT and NNMT are substantially reduced because of an extra partial turn of the N-terminal helix as it enters the loop,58 consistent with the observation that PDAT did not inhibit these N-methyltransferases because it likely cannot bind.
There were several reports of developing inhibitors of INMT in the 1970s and 1980s. These compounds include derivatives of SAM22 that are broad-based inhibitors of SAM-dependent methyltransferases. These compounds are less selective for specific N-methyltransferases such as INMT. Cyclic amidine inhibitors of INMT were also previously developed.24−26 These compounds were tested for inhibition of both human and rabbit INMT, but their kinetic mechanisms, with the exception of a compound identified as DBN (2,3,4,6,7,8-hexahydropyrrolo[1,2-a]pyrimidine),24 were not thoroughly characterized. Our work with DMT thus provides the first kinetic analysis of a mixed competitive–noncompetitive mechanism of product inhibition of INMT. Further, characterization of the pure noncompetitive inhibition by PDAT suggests PDAT as a lead compound for the development of effective and selective allosteric regulators of INMT. In the broad family of methyltransferases, unique allosteric mechanisms of regulation may be common. As examples, a small molecule allosteric inhibition of a protein N-methyltransferase has been recently reported,59 and the type 1 protein arginine methyltransferase 3 (PRMT3) asymmetrically dimethylates one of the guanidine nitrogens of arginine on 40S ribosomal protein S2, resulting in stabilization of the protein.60 Siarheyeva et al.59 show that a unique phenethyl thiadiazolyl urea compound is an allosteric inhibitor of PRMT3 by binding at the nonactive site interface of a PRMT3 dimer. Other methyltransferases appear to be allosterically activated by interaction with their protein partners.61 For example, the DNA methyltransferase, Dnmt 1, preferentially methylates hemimethylated CpG sites on DNA after DNA replication,62,63 and Bashtrykov et al.61 report that the ubiquitin-like PHD and RING finger domains protein 1 (Uhrf1) allosterically activates Dnmt 1 by unblocking an inhibitory domain of Dnmt 1, thus allowing better access of the CpG substrate to the active site of Dnmt 1. In a similar manner, because the sigma-1 receptor colocalizes with INMT in primate motoneuron cell bodies,9 a regulatory allosteric interaction may occur between these two important signaling proteins, as well.
In summary, PDAT selectively inhibited hINMT in comparison to the structurally similar N-methyltransferases, PNMT and NNMT. DMT and PDAT were found to inhibit INMT activity, and the kinetic mechanisms for inhibition were determined to be mixed competitive and noncompetitive and pure noncompetitive, respectively. In silico docking analyses supported the experimentally derived kinetic data through the identification of a putative allosteric inhibitory binding site in the N-terminal helix–loop–helix region of the computer-threaded structure of rabINMT using hINMT as the template. Pharmacological approaches to regulating INMT enzymatic activity are highly desirable for assessing the biological importance of INMT and its possible role in cancer, autoimmunity, normal and aberrant mental states, and neurodegeneration.
Acknowledgments
We thank Dr. Masoud Vedadi from the Structural Genomics Consortium at the University of Toronto for providing the human INMT for our work.
Glossary
Abbreviations
- DMT
N,N-dimethyltryptamine
- hINMT
human indolethylamine-N-methyltransferase
- hNNMT
human nicotinamide-N-methyltransferase
- hPNMT
human phenethanolamine-N-methyltransferase
- PAT
propyl amino tryptamine or N-[2-(1H-indol-3-yl)ethyl]propane-1,3-diamine
- PDAT
propyl dimethyl amino tryptamine or N-[2-(1H-indol-3-yl)ethyl]-N′,N′-dimethylpropane-1,3-diamine
- rabINMT
rabbit indolethylamine-N-methyltransferase
- SAH
S-adenosyl-l-homocysteine
- SAM
S-adenosyl-l-methionine.
Supporting Information Available
Representative thin layer chromatography showing the end products of rabINMT methylation reactions (Figure S1), sequence alignment of human and rabbit INMT (Figure S2), mechanism of binding of DMT to rabINMT (Figure S3), mechanism of binding of PDAT to rabINMT (Figure S4), and chematic representation of the kinetic mechanisms of binding of DMT and PDAT (Figure S5). This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
U.B.C. and S.K.V. contributed equally to this work.
This research was supported by National Institutes of Health Grant NS075820 (to A.E.R.), the PhRMA Foundation (to U.B.C.), and the Science Research Intern Program (Madison Metropolitan School District, 545 W. Dayton St., Madison, WI 53703).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Axelrod J. (1961) Enzymatic formation of psychotomimetic metabolites from normally occurring compounds. Science 134, 343. [DOI] [PubMed] [Google Scholar]
- Thompson M. A.; Moon E.; Kim U. J.; Xu J.; Siciliano M. J.; Weinshilboum R. M. (1999) Human indolethylamine N-methyltransferase: cDNA cloning and expression, gene cloning, and chromosomal localization. Genomics 61, 285–297. [DOI] [PubMed] [Google Scholar]
- Thompson M. A.; Weinshilboum R. M. (1998) Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization. J. Biol. Chem. 273, 34502–34510. [DOI] [PubMed] [Google Scholar]
- Mandell A. J.; Morgan M. (1971) Indole(ethyl)amine N-methyltransferase in human brain. Nat. New Biol. 230, 85–87. [DOI] [PubMed] [Google Scholar]
- Morgan M.; Mandell A. J. (1969) Indole(ethyl)amine N-methyltransferase in the brain. Science 165, 492–493. [DOI] [PubMed] [Google Scholar]
- Wyatt R. J.; Saavedra J. M.; Axelrod J. (1973) A dimethyltryptamine-forming enzyme in human blood. Am. J. Psychiatry 130, 754–760. [DOI] [PubMed] [Google Scholar]
- Barker S. A.; Monti J. A.; Christian S. T. (1981) N,N-Dimethyltryptamine: an endogenous hallucinogen. Int. Rev. Neurobiol. 22, 83–110. [DOI] [PubMed] [Google Scholar]
- Cozzi N. V., Mavlyutov T. A., Thompson M. A., and Ruoho A. E. (2011) Indolethylamine N-methyltransferase expression in primate nervous tissue. Society for Neuroscience Meeting Abstract 840.19.
- Mavlyutov T. A.; Epstein M. L.; Liu P.; Verbny Y. I.; Ziskind-Conhaim L.; Ruoho A. E. (2012) Development of the sigma-1 receptor in C-terminals of motoneurons and colocalization with the N,N′-dimethyltryptamine forming enzyme, indole-N-methyl transferase. Neuroscience 206, 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrist B.; Gershon S.; Sathananthan G.; Walker R. W.; Lopez-Ramos B.; Mandel L. R.; Vandenheuvel W. J. (1976) Dimethyltryptamine levels in blood of schizophrenic patients and control subjects. Psychopharmacology (Heidelberg, Ger.) 47, 29–32. [DOI] [PubMed] [Google Scholar]
- Barker S. A.; McIlhenny E. H.; Strassman R. (2012) A critical review of reports of endogenous psychedelic N,N-dimethyltryptamines in humans: 1955–2010. Drug Test. Anal. 4, 617–635. [DOI] [PubMed] [Google Scholar]
- Corbett L.; Christian S. T.; Morin R. D.; Benington F.; Smythies J. R. (1978) Hallucinogenic N-methylated indolealkylamines in the cerebrospinal fluid of psychiatric and control populations. Br. J. Psychiatry 132, 139–144. [DOI] [PubMed] [Google Scholar]
- Franzen F.; Gross H. (1965) Tryptamine, N,N-dimethyltryptamine, N,N-dimethyl-5-hydroxytryptamine and 5-methoxytryptamine in human blood and urine. Nature 206, 1052. [DOI] [PubMed] [Google Scholar]
- Oon M. C.; Murray R. M.; Rodnight R.; Murphy M. P.; Birley J. L. (1977) Factors affecting the urinary excretion of endogenously formed dimethyltryptamine in normal human subjects. Psychopharmacology (Heidelberg, Ger.) 54, 171–175. [DOI] [PubMed] [Google Scholar]
- Saavedra J. M.; Axelrod J. (1972) Psychotomimetic N-methylated tryptamines: Formation in brain in vivo and in vitro. Science 175, 1365–1366. [DOI] [PubMed] [Google Scholar]
- Smythies J. R.; Morin R. D.; Brown G. B. (1979) Identification of dimethyltryptamine and O-methylbufotenin in human cerebrospinal fluid by combined gas chromatography/mass spectrometry. Biol. Psychiatry 14, 549–556. [PubMed] [Google Scholar]
- Tanimukai H.; Ginther R.; Spaide J.; Bueno J. R.; Himwich H. E. (1970) Detection of psychotomimetic N,N-dimethylated indoleamines in the urine of four schizophrenic patients. Br. J. Psychiatry 117, 421–430. [DOI] [PubMed] [Google Scholar]
- Barker S. A.; Borjigin J.; Lomnicka I.; Strassman R. (2013) LC/MS/MS analysis of the endogenous dimethyltryptamine hallucinogens, their precursors, and major metabolites in rat pineal gland microdialysate. Biomed. Chromatogr. 27, 1690–1700. [DOI] [PubMed] [Google Scholar]
- Larkin S. E.; Holmes S.; Cree I. A.; Walker T.; Basketter V.; Bickers B.; Harris S.; Garbis S. D.; Townsend P. A.; Aukim-Hastie C. (2012) Identification of markers of prostate cancer progression using candidate gene expression. Br. J. Cancer 106, 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopantzev E. P.; Monastyrskaya G. S.; Vinogradova T. V.; Zinovyeva M. V.; Kostina M. B.; Filyukova O. B.; Tonevitsky A. G.; Sukhikh G. T.; Sverdlov E. D. (2008) Differences in gene expression levels between early and later stages of human lung development are opposite to those between normal lung tissue and non-small lung cell carcinoma. Lung Cancer 62, 23–34. [DOI] [PubMed] [Google Scholar]
- Nuno-Ayala M.; Guillen N.; Arnal C.; Lou-Bonafonte J. M.; de Martino A.; Garcia-de-Jalon J. A.; Gascon S.; Osaba L.; Osada J.; Navarro M. A. (2012) Cystathionine β-synthase deficiency causes infertility by impairing decidualization and gene expression networks in uterus implantation sites. Physiol. Genomics 44, 702–716. [DOI] [PubMed] [Google Scholar]
- Lin R. L.; Narasimhachari N.; Himwich H. E. (1973) Inhibition of indolethylamine-N-methyltransferase by S-adenosylhomocysteine. Biochem. Biophys. Res. Commun. 54, 751–759. [DOI] [PubMed] [Google Scholar]
- Frecska E.; Szabo A.; Winkelman M. J.; Luna L. E.; McKenna D. J. (2013) A possibly sigma-1 receptor mediated role of dimethyltryptamine in tissue protection, regeneration, and immunity. J. Neural Transm. 120, 1295–1303. [DOI] [PubMed] [Google Scholar]
- Mandel L. R. (1976) Inhibition of indoleamine-N-methyltransferase by 2,3,4,6,7,8-hexahydropyrrololo[1,2-a]pyrimidine. Biochem. Pharmacol. 25, 2251–2256. [DOI] [PubMed] [Google Scholar]
- Rokach J.; Girard Y.; Hamel P.; Reader G.; Rooney C. S.; Mandel L. R.; Cragoe E. J. Jr.; Zacchei A. G. (1980) Inhibitors of indoleethylamine N-methyltransferase. Derivatives of 3-methyl-2-thiazolidinimine. In vitro, in vivo, and metabolic studies. J. Med. Chem. 23, 773–780. [DOI] [PubMed] [Google Scholar]
- Rokach J.; Hamel P.; Hunter N. R.; Reader G.; Rooney C. S.; Anderson P. S.; Cragoe E. J. Jr.; Mandel L. R. (1979) Cyclic amidine inhibitors of indolamine N-methyltransferase. J. Med. Chem. 22, 237–247. [DOI] [PubMed] [Google Scholar]
- Strassman R. J. (2001) DMT: The Spirit Molecule: A Doctor’s Revolutionary Research into the Biology of Near-Death and Mystical Experiences, Park Street Press, South Paris, ME. [Google Scholar]
- Deliganis A. V.; Pierce P. A.; Peroutka S. J. (1991) Differential interactions of dimethyltryptamine (DMT) with 5-HT1A and 5-HT2 receptors. Biochem. Pharmacol. 41, 1739–1744. [DOI] [PubMed] [Google Scholar]
- Glennon R. A.; Liebowitz S. M.; Mack E. C. (1978) Serotonin receptor binding affinities of several hallucinogenic phenylalkylamine and N,N-dimethyltryptamine analogues. J. Med. Chem. 21, 822–825. [DOI] [PubMed] [Google Scholar]
- McKenna D. J.; Repke D. B.; Lo L.; Peroutka S. J. (1990) Differential interactions of indolealkylamines with 5-hydroxytryptamine receptor subtypes. Neuropharmacology 29, 193–198. [DOI] [PubMed] [Google Scholar]
- Cozzi N. V.; Gopalakrishnan A.; Anderson L. L.; Feih J. T.; Shulgin A. T.; Daley P. F.; Ruoho A. E. (2009) Dimethyltryptamine and other hallucinogenic tryptamines exhibit substrate behavior at the serotonin uptake transporter and the vesicle monoamine transporter. J. Neural Transm. 116, 1591–1599. [DOI] [PubMed] [Google Scholar]
- Nagai F.; Nonaka R.; Satoh Hisashi Kamimura K. (2007) The effects of non-medically used psychoactive drugs on monoamine neurotransmission in rat brain. Eur. J. Pharmacol. 559, 132–137. [DOI] [PubMed] [Google Scholar]
- Reimann W.; Schneider F. (1993) The serotonin receptor agonist 5-methoxy-N,N-dimethyltryptamine facilitates noradrenaline release from rat spinal cord slices and inhibits monoamine oxidase activity. Gen. Pharmacol. 24, 449–453. [DOI] [PubMed] [Google Scholar]
- Smith T. E.; Weissbach H.; Udenfriend S. (1962) Studies on the mechanism of action of monoamine oxidase: Metabolism of N,N-dimethyltryptamine and N,N-dimethyltryptamine-N-oxide. Biochemistry 1, 137–143. [DOI] [PubMed] [Google Scholar]
- Fontanilla D.; Johannessen M.; Hajipour A. R.; Cozzi N. V.; Jackson M. B.; Ruoho A. E. (2009) The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science 323, 934–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Saif A.; Al-Mohanna F.; Bohlega S. (2011) A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann. Neurol. 70, 913–919. [DOI] [PubMed] [Google Scholar]
- Mavlyutov T. A.; Epstein M. L.; Verbny Y. I.; Huerta M. S.; Zaitoun I.; Ziskind-Conhaim L.; Ruoho A. E. (2013) Lack of sigma-1 receptor exacerbates ALS progression in mice. Neuroscience 240, 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. L.; Sargeant S.; Narasimhachari N. (1974) Indolethylamine-N-methyltransferase in developing rabbit lung. Dev. Psychobiol. 7, 475–481. [DOI] [PubMed] [Google Scholar]
- Marzullo G.; Rosengarten H.; Friedhoff A. J. (1977) A peptide-like inhibitor of N-methyltransferase in rabbit brain. Life Sci. 20, 775–783. [DOI] [PubMed] [Google Scholar]
- Porta R.; Camardella M.; Esposito C.; Della Pietra G. (1977) Inhibition of indolethylamine-N-methyltransferase by aliphatic diamines. Biochem. Biophys. Res. Commun. 77, 1196–1202. [DOI] [PubMed] [Google Scholar]
- Speeter M. E.; Anthony W. C. (1954) The action of oxalyl chloride on indoles: A new approach to tryptamines. J. Am. Chem. Soc. 76, 6208–6210. [Google Scholar]
- Brutcher F.; Vanderwerff W. (1958) Notes: Concerning a Preparation of Tryptamine. J. Org. Chem. 23, 146–147. [Google Scholar]
- Falkenberg G. (1972) The Crystal and Molecular Structure of (N,N)-Dimethyltryptamine. Acta Crystallogr. B28, 3075. [Google Scholar]
- Gee C. L.; Tyndall J. D.; Grunewald G. L.; Wu Q.; McLeish M. J.; Martin J. L. (2005) Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: Implications for catalysis. Biochemistry 44, 16875–16885. [DOI] [PubMed] [Google Scholar]
- Rini J.; Szumlanski C.; Guerciolini R.; Weinshilboum R. M. (1990) Human liver nicotinamide N-methyltransferase: Ion-pairing radiochemical assay, biochemical properties and individual variation. Clin. Chim. Acta 186, 359–374. [DOI] [PubMed] [Google Scholar]
- Grunewald G. L.; Romero F. A.; Seim M. R.; Criscione K. R.; Deupree J. D.; Spackman C. C.; Bylund D. B. (2005) Exploring the active site of phenylethanolamine N-methyltransferase with 3-hydroxyethyl- and 3-hydroxypropyl-7-substituted-1,2,3,4-tetrahydroisoquinolines. Bioorg. Med. Chem. Lett. 15, 1143–1147. [DOI] [PubMed] [Google Scholar]
- Grunewald G. L.; Seim M. R.; Regier R. C.; Criscione K. R. (2007) Exploring the active site of phenylethanolamine N-methyltransferase with 1,2,3,4-tetrahydrobenz[h]isoquinoline inhibitors. Bioorg. Med. Chem. 15, 1298–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton R. G.; Kaiser C.; Gessner G. (1976) Studies on adrenal phenylethanolamine N-methyltransferase (PNMT) with S K & F 64139, a selective inhibitor. J. Pharmacol. Exp. Ther. 197, 623–632. [PubMed] [Google Scholar]
- Wu H., Dong A., Zeng H., Loppnau P., Sundstrom M., Arrowsmith C. H., Edwards A. M., Bochkarev A., and Plotnikov A. N. (2005) The Crystal Structure of Human Indolethylamine N-methyltransferase in complex with SAH. Protein Data Bank entry 2A14. [Google Scholar]
- Wallach J. V. (2009) Endogenous hallucinogens as ligands of the trace amine receptors: A possible role in sensory perception. Med. Hypotheses 72, 91–94. [DOI] [PubMed] [Google Scholar]
- Callaway J. C. (1988) A proposed mechanism for the visions of dream sleep. Med. Hypotheses 26, 119–124. [DOI] [PubMed] [Google Scholar]
- Jacob M. S.; Presti D. E. (2005) Endogenous psychoactive tryptamines reconsidered: An anxiolytic role for dimethyltryptamine. Med. Hypotheses 64, 930–937. [DOI] [PubMed] [Google Scholar]
- Szara S. (2007) DMT at fifty. Neuropsychopharmacologia Hungarica 9, 201–205. [PubMed] [Google Scholar]
- Servillo L.; Giovane A.; Balestrieri M. L.; Cautela D.; Castaldo D. (2012) N-methylated tryptamine derivatives in Citrus genus plants: Identification of N,N,N-trimethyltryptamine in bergamot. J. Agric. Food Chem. 60, 9512–9518. [DOI] [PubMed] [Google Scholar]
- Servillo L.; Giovane A.; Balestrieri M. L.; Casale R.; Cautela D.; Castaldo D. (2013) Citrus genus plants contain N-methylated tryptamine derivatives and their 5-hydroxylated forms. J. Agric. Food Chem. 61, 5156–5162. [DOI] [PubMed] [Google Scholar]
- Pochettino M. L.; Cortella A. R.; Ruiz M. (1999) Hallucinogenic snuff from northwestern Argentina: Microscopical identification of Anandenanthera colubrina var. Cebil (fabaceae) is powdered archaelogical material. Econ. Bot. 53, 127–132. [Google Scholar]
- Martin J. L.; Begun J.; McLeish M. J.; Caine J. M.; Grunewald G. L. (2001) Getting the adrenaline going: Crystal structure of the adrenaline-synthesizing enzyme PNMT. Structure 9, 977–985. [DOI] [PubMed] [Google Scholar]
- Siarheyeva A.; Senisterra G.; Allali-Hassani A.; Dong A.; Dobrovetsky E.; Wasney G. A.; Chau I.; Marcellus R.; Hajian T.; Liu F.; Korboukh I.; Smil D.; Bolshan Y.; Min J.; Wu H.; Zeng H.; Loppnau P.; Poda G.; Griffin C.; Aman A.; Brown P. J.; Jin J.; Al-Awar R.; Arrowsmith C. H.; Schapira M.; Vedadi M. (2012) An allosteric inhibitor of protein arginine methyltransferase 3. Structure 20, 1425–1435. [DOI] [PubMed] [Google Scholar]
- Swierez R.; Person M. D.; Bedford M. T. (2005) Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J. 386, 85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashtrykov P.; Jankevicius G.; Jurkowska R. Z.; Ragozin S.; Jeltsch A. (2014) The Uhrf1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase Dnmt1 by an allosteric mechanism. J. Biol. Chem. 289, 4106–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann A.; Gowher H.; Jeltsch A. (2004) Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 61, 2571–2587. [DOI] [PubMed] [Google Scholar]
- Hermann A.; Goyal R.; Jeltsch A. (2004) The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 279, 48350–48359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.