Abstract
Xenoturbellida and Acoelomorpha are marine worms with contentious ancestry. Both were originally associated with the flatworms (Platyhelminthes), but molecular data haverevised their phylogenetic positions, generally linking Xenoturbellida to the deuterostomes1,2 and positioning the Acoelomorpha as the most basally branching bilaterian group(s)3–6. Recent phylogenomic data suggested that Xenoturbellida and Acoelomorpha are sister taxa and together constitute an early branch of Bilateria7. Here we assemble three independent data sets—mitochondrial genes, a phylogenomic data set of 38,330 amino-acid positions and new microRNA (miRNA) complements—and show that the position of Acoelomorpha is strongly affected by a long-branch attraction (LBA) artefact. When we minimize LBA we find consistent support for a position of both acoelomorphs and Xenoturbella within the deuterostomes. The most likely phylogeny links Xenoturbella and Acoelomorpha in a clade we call Xenacoelomorpha. The Xenacoelomorpha is the sister group of the Ambulacraria (hemichordates and echinoderms). We show that analyses of miRNA complements8 have been affected by character loss in the acoels and that both groups possess one miRNA and the gene Rsb66 otherwise specific to deuterostomes. In addition, Xenoturbella shares one miRNA with the ambulacrarians, and two with the acoels. This phylogeny makes sense of the shared characteristics of Xenoturbellida and Acoelomorpha, such as ciliary ultrastructure and diffuse nervous system, and implies the loss of various deuterostome characters in the Xenacoelomorpha including coelomic cavities, through gut and gill slits.
In contrast to previous results1,2,4,9,10 (Fig. 1a), two recent phylogenomic studies have suggested a sister group relationship between Acoelomorpha and Xenoturbella. These studies disagree over where this clade might be placed, either at the base of Bilateria7 (Fig. 1b) or with the deuterostomes11 (Fig. 1c). The acoelomorph genes studied, however, show extremely high rates of sequence evolution. This bias could result in susceptibility to the LBA artefact: a systematic error that may be compounded by the short internal branches around the origin of the Bilateria12. Overcoming this potential artefact requires the analysis of large molecular data sets comprising many species and using a complex model of sequence evolution designed to reduce the impact of systematic errors12.
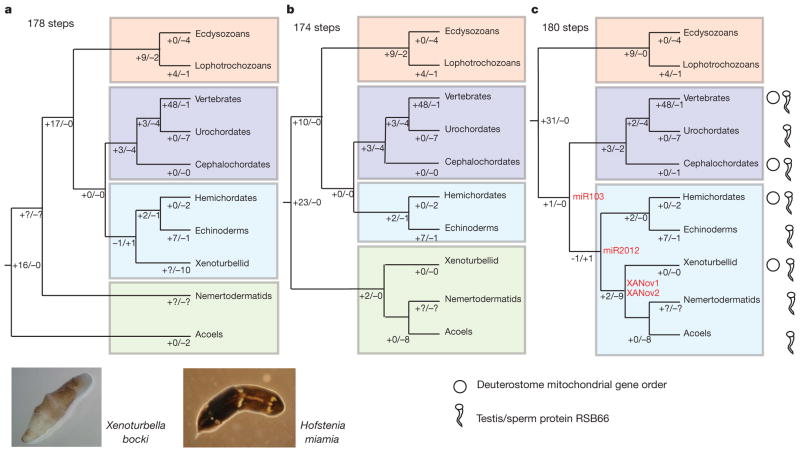
Figure 1. Alternative phylogenetic positions of Acoela, Nemertodermatida and Xenoturbellida with implied evolution of different characters.
a, Tree based on refs 6 and 9 for positions of nemertodermatids and acoels, and refs 1 and 2 for position of Xenoturbella. b, Tree based on analyses of ref. 7. c, Tree based on the results from this paper. Protein RSB66 and deuterostome mitochondrial gene order are also indicated. miRNAs representing possible synapomorphies of Deuterostomia, Xenambulacraria and Xenacoelomorpha are shown in red. The minimum number of total steps to explain miRNA distribution is shown above trees. Losses and gains of miRNAs are shown on each branch. Complete trees are shown in Supplementary Figs 10–14. Bottom left, X. bocki and H. miamia (photographs by M.J.T. and A.W.).
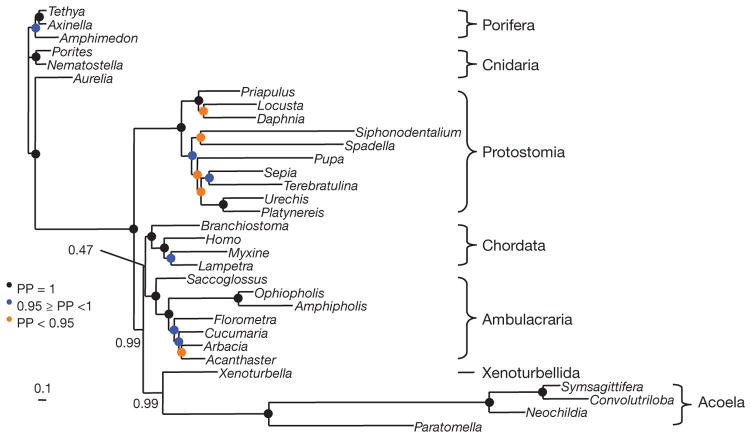
We assembled a largely complete set of mitochondrial protein sequences from four acoels using expressed sequence tag (EST) databases. Better-fitting models of molecular evolution are expected to be less sensitive to systematic errors, and cross validation13 shows that the CAT model with a general time reversible (GTR) exchange rate matrix and gamma correction (CAT + GTR + Γ) fits best, followed by GTR + Γ then CAT + Γ. In the phylogeny inferred with the best model, acoels are the sister-group of Xenoturbella (posterior probability (PP) of 0.99) within deuterostomes (PP = 0.99) (Fig. 2). However, the relationship between chordates, ambulacrarians and the Xenoturbella/acoel group is unresolved (PP = 0.47). A notable feature of Fig. 2 is the extremely fast evolutionary rate of acoels, which are nevertheless grouped with the slow-evolving Xenoturbella.
Figure 2. Animal phylogeny based on mitochondrial proteins reconstructed using the CAT + GTR + Γ model under a Bayesian analysis.
Xenoturbella and the four acoel species are sister taxa (PP = 0.99). This clade is grouped with the deuterostomes (PP = 0.99), but is excluded from within the clade with weak support (PP = 0.47). Cross-validation demonstrates that the GTR + Γ model has a better fit than the CAT + Γ model, albeit without statistical significance (ΔlnL = 20 ± 24), and that the CAT + GTR + Γ model has a significantly better fit than the GTR + Γ model (ΔlnL = 96 ± 21). Using less fit models (GTR + Γ and CAT + Γ), the support for association with the deuterostomes decreases (Supplementary Fig. 1). Scale bar, substitutions per position.
We exaggerated the effect of LBA by using the less-fit models (Supplementary Fig. 1). Using GTR + Γ we recover acoels + Xenoturbella (PP = 1.0) as basal deuterostomes (PP = 0.99). Only using the least fit model (CAT + Γ) do we find that the acoels are located as basal bilaterians (PP = 0.65). The fast evolutionary rate of acoels is therefore likely to be responsible for their early emergence revealed in previous studies14. Interestingly, the acoel Symsagittifera roscoffensis does not possess a protostometype mitochondrial NAD5 gene15, finally ruling out a close relationship between acoels and rhabditophoran platyhelminthes3 (Supplementary Fig. 2). Despite the limited size and heterogeneous evolutionary rates of mitochondrial genomes, these analyses provide evidence for grouping acoels and Xenoturbella together with deuterostomes.
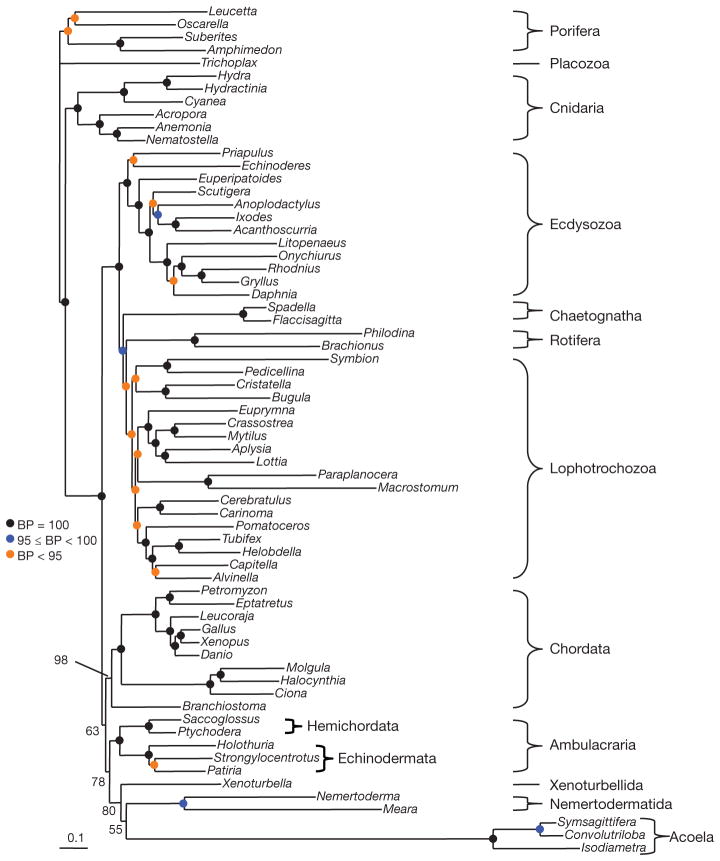
We also constructed a large alignment from EST and genome data (66 species, 197 genes, 38,330 positions, 30% missing data) including all major animal phyla represented by slowly evolving species (Supplementary Tables 1 and 2). For this new data set, CAT + Γ has a better fit than GTR + Γ (CAT + GTR + Γ was not investigated as the computation is too time-consuming)13. The phylogeny inferred under CAT + Γ (Fig. 3) recovers all expected clades (Bilateria, Ecdysozoa, Lophotrochozoa, etc.) with high support (generally a bootstrap support (BS) of 100%). Acoels are seen to be very fast evolving and are the sister group of the nemertodermatids (BS = 55%). As in the mitochondrial analyses, the acoelomorph clade is sister to the slow-evolving Xenoturbella (BS = 80%). Xenoturbella plus Acoelomorpha are sister to Ambulacraria (BS = 78%) within deuterostomes.
Figure 3. Phylogeny of 66 animal species based on EST sequences.
Analysis of 197 genes, 38,330 unambiguously aligned positions, 30% missing data. Tree was reconstructed using the CAT + Γ model under a Bayesian analysis. Major accepted metazoan clades (for example, Lophotrochozoa, Ecdysozoa, Protostomia) are supported. Acoela and Nemertodermatida are sister groups (Acoelomorpha). Xenoturbella and Acoelomorpha are sister groups (phylum Xenacoelomorpha). Xenacoelomorpha is the sister taxon of Ambulacraria (Xenambulacraria) within the deuterostomes. Level of bootstrap support is indicated. Similar support is obtained when jack-knifing 50% of the genes (Supplementary Fig. 9). Scale bar, substitutions per position.
Although the analysis of ESTs is congruent with the mitochondrial genome result, our topology differs from the recent phylogenomic analysis of Hejnol et al.7 (Fig. 1b). To test the possibility that the fast evolutionary rate of Acoelomorpha might have an effect on phylogenetic inference due to LBA, we pruned our data set to 37 species and compared alternative models (including CAT + GTR + Γ) and different taxon sampling schemes aimed at lessening or exaggerating a potential LBA artefact. The backbone topology inferred with the CAT + Γ model is unchanged when the number of taxa is reduced (Supplementary Fig. 3a and Fig. 3).
Cross-validation demonstrates that the site-heterogeneous CAT + GTR + Γ model has a significantly better fit than the CAT + Γ model (ΔlnL = 490 ± 48), which itself is significantly better than the GTR + Γ model (ΔlnL = 3,195 ± 127). Regardless of the species sampling, the best available model (CAT + GTR + Γ) locates Xenoturbella, Acoela and Nemertodermatida within deuterostomes. The fast-evolving Nemertodermatida are consistently found as a sister-group to Ambulacraria, and the very fast-evolving Acoela are either grouped with Nemertodermatida and Xenoturbella or basal to deuterostomes (Supplementary Fig. 4a–f). The sub-optimal site-heterogeneous CAT + Γ model yields results that are more difficult to interpret: for example, deuterostomes are paraphyletic when Xenoturbella is absent (Supplementary Fig. 3b, d), but in no case are acoelomorphs basal to Bilateria (Supplementary Fig. 3). The least-fit site-homogeneous GTR + Γ model, in contrast, leads to variable positions of the members of Acoelomorpha, depending on whether slow- or fast-evolving representatives are included (Supplementary Fig. 5), reflecting the expected behaviour of a method sensitive to LBA artefacts. Interestingly, even with the least-fit GTR + Γ model, Xenoturbella plus Acoelomorpha are monophyletic and sister to Chordata + Ambulacraria (Supplementary Fig. 5a). When the very-fast-evolving Acoela are removed, even GTR + Γ recovers the monophyly of Xenoturbella, Nemertodermatida and Ambulacraria, showing that the very-fast-evolving Acoela is the only group that is difficult to locate and that its placement requires the use of complex models to avoid artefactual results.
We propose that the basal emergence of Xenoturbella plus Acoelomorpha observed by Hejnol et al. is the result of an LBA artefact stemming from the use of a sub-optimal site-homogeneous model7. Computational constraints prevented the analysis of the original data set (94 species and 270,580 positions, with 84% missing data) with the site-heterogeneous CAT + Γ model; we therefore assembled an alignment of 145 genes (with the same gene coverage of the pivotal Xenoturbella and Acoelomorpha as used by Hejnol et al.) for the same 94 species (24,633 positions, and 30% missing data). The resulting phylogeny (Supplementary Fig. 6) is very similar to our results, with Nemertodermatida the sister-group to Xenoturbella (PP = 1.0), this clade being sister to Ambulacraria (PP = 0.98); the very fast-evolving acoels are included in deuterostomes (PP = 0.89), but with an unstable position (PP = 0.61 at the base of deuterostomes). Given that CAT + Γ has a better fit on this data set than the site-homogeneous model previously used, this suggests that the topology of the analysis of Hejnol et al.7 was affected by an LBA artefact.
Although our two data sets are consistent with a deuterostome affinity for Acoelomorpha and Xenoturbella (see also Supplementary Figs 7–9), the paucity of bilaterian miRNAs in one acoel, Symsagittifera roscoffensis, has supported the idea that the acoels are a basal clade relative to other Bilateria8. To examine this conclusion we have constructed and sequenced libraries of small RNAs from a second acoel, Hofstenia miamia and from Xenoturbella bocki. From the Hofstenia library we found ten miRNAs that were not detected in the S. roscoffensis library (Supplementary Table 3). From Xenoturbella we detected reads from all ten of these miRNAs, as well as eight additional miRNAs found in Bilateria. Xenoturbella has all but ten of the miRNAs typically found in bilaterian genomes16.
The most parsimonious tree derived from an analysis of miRNA data places the two species of acoels and Xenoturbella as three independent branches basal to the Bilateria (Symsagittifera (Hofstenia (Xenoturbella, Bilateria))) (Supplementary Figs 10 and 11). This result rather implausibly implies non-monophyly of Acoela. Alternative interpretations of these data assuming monophyly of acoels and based either on the results of refs 1, 2, 6, 9, 14 (Supplementary Fig. 12), or the tree of Hejnol et al. (Supplementary Fig. 13) or on our tree (Supplementary Fig. 14) imply large-scale losses of miRNAs, in particular from Symsagittifera. Numerous miRNAs must have been lost from at least some Acoels, suggesting that their absence cannot be considered a credible contra-indication of deuterostome affinity, fitting a picture of miRNA evolution occurring through continuous addition and mosaic loss16. Locating Acoelomorpha and Xenoturbella inside Deuterostomia, yet outside Chordata or Ambulacraria, means almost all possible losses are of bilaterian level characters—there is only a single known deuterostome specific miRNA—and this is exactly what we observe.
Limited additional support for our tree comes from this unique deuterostomian miRNA (miR-103/107/2013), which we find in both acoels and in Xenoturbella (Fig. 1 and Supplementary Fig. 14). Xenoturbella, at least, possesses a second miRNA, miR-2012 (Fig. 1 and Supplementary Fig. 15), previously found only in Ambulacraria. We suggest that miR-103/107/2013 is a plausible synapomorphy of Xenoturbella, Acoelomorpha, Ambulacraria and Chordata and that miR-2012 is a likely synapomorphy of Xenambulacraria. Furthermore, we find that Xenoturbella and acoels share two miRNAs found in no other animals: a novel miRNA family (XANov-1) and a paralogue of miR-92 (XANov-2) (Supplementary Fig. 15). Finally, we have detected an additional gene, coding for the sperm protein RSB66, uniquely in the genomes of Ambulacraria, Chordata, Acoelomorpha and Xenoturbella (Fig. 1 and Supplementary Fig. 16). The Rsb66 genes of acoelomorphs and Xenoturbella share a small and rather variable insertion relative to Chordata and Ambulacraria; this, in addition to their two novel miRNAs and their eight shared miRNA losses, gives further support to the idea that they are sister taxa (Fig. 1 and Supplementary Fig. 14).
Difficult phylogenetic questions such as that addressed here must ultimately be solved by the congruent patterns emerging from what, inevitably, are not highly supported results. Our three independent data sources indicate a sister group relationship between the acoelomorphs and Xenoturbella17–19 within the deuterostomes; we propose the name Xenacoelomorpha for this clade, noting that a deuterostome affinity for both Xenoturbella and Acoelomorpha has been previously suggested based on morphological considerations20–22. The Xenacoelomorpha are excluded from the deuterostome phyla of Hemichordata, Echinodermata and Chordata and hence constitute an independent fourth phylum of deuterostomes. Our results suggest that characters shared by the Xenacoelomorpha are likely to be synapomorphies inherited from a common ancestor19,23.
Our findings also indicate that the Acoelomorpha are not early branches on the stem leading to the Bilateria. This phylogenetic relationship, first reported over a decade ago6, had led to the acoelomorphs being interpreted as modern representatives of a lineage intermediate between the diploblasts and the Bilateria24,25, a position that made sense of the paucity of HOX genes and miRNAs found in their genomes. The supposed presence of a small, simple, directly developing ancestor of the Bilateria with a weakly centralized nervous system and blind gut also led to the assumption—not supported by our findings—that these characteristics were present in Precambrian bilaterians26.
Finally, the deuterostome affinity of the Xenacoelomorpha implies that they have lost characters present in the common ancestor of deuterostomes. This ancestor must have possessed panbilaterian apomorphies (for example, through-gutandprotonephridia) as well as the homologous attributes of Ambulacraria and Chordata (deuterostomy, enterocoely, gill slits and endostylar tissue27). Although it is clear that certain of these characters have been lost in the living Xenacoelomorpha, we predict that more deuterostome characters will be discovered in the morphology, embryology or genomes of members of the Xenacoelomorpha.
METHODS
Phylogenetic data assembly
Mitochondrial data
Metazoan mitochondrial protein coding genes were downloaded from OGRE (http://drake.physics.mcmaster.ca/ogre/). To assemble acoel partial genomes we used TBLASTN against EST collections from Convolutriba longifissura, Neochildia fusca and Symsagittifera roscoffensis from the Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/) at the National Center for Biotechnology Information. We used Priapulus caudatus mitochondrial proteins as a BLAST query. Open reading frames of positive hits were identified by aligning ESTs to the homologous protein sequence from P. caudatus using Genewise31. Multiple positives from a given species were then assembled into a contig using CAP3 (ref. 32). Nucleotides from each gene were aligned using TranslatorX33 with the appropriate genetic code and using ClustalW34 for the amino-acid alignment. Phylogenetic analyses were performed on the amino-acid translations.
Phylogenomic data
Alignments from Philippe et al.28 and Dunn et al.35 were updated with new sequences from GenBank (http://www.ncbi.nlm.nih.gov/sites/entrez?db=protein), dbEST (http://www.ncbi.nlm.nih.gov/dbEST/) and the Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/) at the National Center for Biotechnology Information. Single gene alignments were assembled using new features of the program Ed from the MUST software package36. Ambiguously aligned regions were detected and removed with Gblocks37 (b2 = 75%, b3 = 5, b4 = 5, b5 = half); this automated selection was slightly refined by eye using Net (also from MUST).
Concatenations of single gene alignments into supermatrices were performed with SCaFoS38. When multiple orthologous sequences were available for a particular operational taxonomic unit, SCaFoS helped to select the slowest-evolving sequence as determined from ML distances computed under a WAG + F model with TREE-PUZZLE39. To minimize the amount of missing data, SCaFOS was allowed to create chimaerical operational taxonomic units by merging partial sequences from closely related species (Supplementary Table 1) when full-length sequences were not available. The amount of missing data per gene was limited to 25 species out of 66. Information about the names of the 197 selected genes, their size and the distribution of missing data are available in Supplementary Table 2.
To produce a data set that was tractable for the most time-consuming CAT+ GTR + Γ model we reduced the number of taxa from 66 to 37. Our strategy for selecting taxa for elimination was as follows.
We first discarded seven species because of their incompleteness (that is, species with fewer than 16,000 amino acids); acoelomorphs, except the very incomplete Convolutriloba longifissura (5,458 amino acids), were exempt from this cull for obvious reasons.
Then we removed the most incomplete species within well-established clades. For example, within sponges, Suberites 23,000 amino acids versus 37,000 in Amphimedon; within urochordates, Halocynthia 24,000 amino acids versus 36,000 in Molgula and 37,000 in Ciona; within chelicerates, Anoplodactylus and Acanthoscurria 18,000 and 26,000 amino acids versus 37,000 in Ixodes.
This reduction of less complete taxa was balanced by the need to maintain a homogeneous taxon sampling (that is, about three species per major phylum Porifera, Cnidaria, Arthropoda, Mollusca, Annelida, Vertebrata). This strategy, while making the data set of a size that permits the use of the best (and most time-consuming) models, also allowed us to reduce the proportion of missing data from 30% (66 species) to 22% (37 species).
Another data set was assembled from the set of genes with the taxon sampling of Hejnol et al.7. In that case, only genes for which sequences were available for at least 55 species out of 94 were retained, yielding a set of 145 genes (24,632 unambiguously aligned positions, 30% missing data).
Phylogenetic inference
For mitochondrial and phylogenomic data sets, PhyloBayes analyses were performed with the CAT + Γ4 mixture model. This accounts for across-site heterogeneities in the amino-acid replacement process29. This model is implemented in an MCMC framework by the program PhyloBayes version 3.2 (ref. 40). Two independent runs were performed with a total length of 10,000 cycles (250 topological moves per cycle) with the same operators as in Lartillot et al.41, saved every ten cycles, for most data sets; however, for the super-matrix of 94 species, 20,000 cycles were necessary. The first 5,000 points were discarded as burn-in for all the data sets (expect for the mitochondrial alignments where a burn-in of 1,000 was sufficient), and the posterior consensus was computed on the 500 (900 for mitochondrial alignments) remaining trees.
For the alignment of 66 species and 38,330 amino-acid positions, we applied a standard, time-consuming, bootstrap procedure42: 100 pseudo-replicates were generated with SEQBOOT43; each data set was analysed with PhyloBayes, trees were collected after the initial burn-in period and a consensus tree was computed by PhyloBayes; finally, a consensus tree was inferred from these 100 consensus trees using CONSENSE to compute the bootstrap support values for each node. To test the robustness of our results to gene sampling, we also performed a jack-knife analysis of genes. We randomly sampled 50% of our 197 genes and for each of the ten replicates a PhyloBayes analysis was done using the CAT + Γ4 model. The consensus trees obtained from all the post-burn-in trees (Supplementary Fig. 9) is identical to the tree based on the complete gene sample (Fig. 3), and jack-knife supports are very similar to bootstrap supports.
For the mitochondrial alignment (32 species and 2,118 positions) and a reduced EST-based alignment (37 species, 38,330 positions, 22% missing data), we also used the site-homogeneous GTR + Γ4 and the time-consuming site-heterogeneous CAT + GTR + Γ4. We first performed statistical comparisons of the CAT + GTR + Γ4 model, the CAT + Γ4 model and the GTR + Γ4 model using cross-validation tests as described in ref. 41. Ten replicates were run: 9/10 of the positions randomly drawn from the alignment were used as the learning set and the remaining 1/10 as the test set. For the GTR + Γ4 model, MCMC were run for 1,100 cycles, 100 being discarded as burn-in. For the CAT + Γ4 and CAT + GTR + Γ4 models, MCMC were run for 1,600 (2,100) cycles with the mitochondrial (EST) alignment, 600 (1,100) being discarded as burn-in. Other matrix based models—WAG, JTT or LG, which are generally used—are special cases of the GTR model; for large data sets, the amount of data available is generally sufficient to learn the 190 free parameters of the GTR model40; we verified by cross-validation that the GTR + Γ4 model had a better fit to our data set than the WAG + Γ4 model (ΔlnL = 1219 ± 60). We therefore only used the GTR + Γ4 model as the best site-homogeneous model in phylogenetic inference.
For these two smaller data sets (mitochondrial alignment of 32 species and 2,118 positions, and an EST-based alignment of 37 species and 38,330 positions), phylogenetic inference with the CAT + GTR + Γ4 and GTR + Γ4 models was performed with PhyloBayes 3.2 as for the CAT + Γ4 model. For the GTR + Γ4 model, the tree was also inferred with RAxML 7.0.4.1 (ref. 44), with 100 rapid bootstrap replicates, for the EST alignment.
For the six samples of the 37 species data set, we inferred the trees with the GTR + Γ4 model using RAxML and PhyloBayes. We expect minor, and even no, differences between the ML and Bayesian inference because the same model is used (priors are known to have an effect on Bayesian analysis, but it should be very small given the large size of the data set).
In five of these taxon samples, the RAxML and PhyloBayes topologies are identical, only the position of Trichoplax varies in some cases, but bootstrap supports for the placement of Trichoplax are around 50%. In one taxon sample (that excluding nemertodermatids), various chains of PhyloBayes failed to converge towards the same topology, differing only by the position of Acoela (which corresponds to bootstrap support close to 30%); interestingly, all the topologies found by PhyloBayes were also found among the bootstrap replicates of RAxML (corresponding to bootstrap support between 10 and 20%). This indicates that the various topologies have very similar likelihoods and that PhyloBayes is unable to switch readily among these various local minima, at least in a reasonable time (that is, several months of computation).
Compositional heterogeneity
The amino-acid composition of the 66 species EST-based data set was visualized by assembling a 20 × 66 matrix containing the frequency of each amino acid per species using the program Net from the MUST package36. This matrix was then displayed as a two-dimensional plot in a principal component analysis, as implemented in the R package. Supplementary Fig. 7 demonstrates that the amino-acid compositions of Xenoturbella, Nemertodermatida and Acoela are not similar and therefore that their monophyly is not likely to be due to a compositional artefact.
To verify that amino-acid compositional heterogeneity does not bias our inference, we cannot use the time-heterogeneous CAT-BP45 because of the intractable computational burden. Instead, we used the Dayhoff coding46 in which the amino acids are recoded according to the six classes defined by M. Dayhoff. The recoded alignments were analysed with PhyloBayes 3.2 using the CAT + Γ4 model, under the same conditions as previously. The resulting phylogeny (Supplementary Fig. 8) is almost identical to the tree of Fig. 3; some minor rearrangements within Porifera, Lophotrochozoa, Vertebrata and Xenacoelomorpha correspond to poorly supported groups. Our inference thus does not seem to be biased by compositional heterogeneity.
The mitochondrial analysis is complicated to an unknown extent by the existence of multiple variants of the genetic code within deuterostomes. The resulting compositional biases may impede correct reconstruction of relationships within the deuterostomes1. Different mitochondrial genetic codes are found in vertebrates, urochordates, cephalochordates, echinoderms and hemichordates, and the observation that the acoelomorph and Xenoturbella code (invertebrate code) differs from all of these makes the deuterostome affinity observed in our analyses conservative.
The non-monophyly of Cnidaria observed in the mitochondrial tree is likely to be incorrect. This problem is particularly difficult because of the extreme rate heterogeneity in the tree (the distance between Porifera and Anthozoa is smaller than the distances within Echinodermata). This heterogeneity is coupled with a change in the properties of the evolutionary process47. Importantly, we do not see any tree reconstruction artefact that would erroneously cluster the fast-evolving acoels with the slow-evolving Xenoturbella (whereas the fast rate of Aurelia easily explains its incorrect position by an attraction with the very-long-branched Bilateria). As a result, it is reasonable to attribute the position of Acoela to genuine phylogenetic signal rather than to non-phylogenetic signal.
miRNA data collection
Specimens of Xenoturbella bocki were collected as previously described2 and were starved for 5 months to avoid contamination by their food. Specimens of H. miamia were extracted from algae and leaf litter collected among mangroves at Walsingham Pond, Bermuda. The worms were starved for 2 weeks before miRNA extraction. Small RNA libraries were constructed and sequences analysed as described elsewhere30. miRNA presequences were also recovered from Xenoturbella genomic DNA traces by BLAST searches.
Supplementary Material
Acknowledgments
We thank N. Lartillot for reading the manuscript, W. Sterrer for helping collect material, and E. Sperling for help with small RNA library construction. H.P. is funded by Canada Research Chairs, Natural Sciences and Engineering Research Council and Réseau Québécois de Calcul de Haute Performance for computational resources: more than 220,000 central processing unit (CPU)–hours were used producing at least 7 tonnes of CO2 excluding grey energy. R.R.C. and M.J.T. were part-funded by the Biotechnology and Biological Sciences Research Council SYNTAX scheme. K.J.P. is supported by the National Science Foundation and NASA Ames. R.R.C. was also supported by a Wellcome Trust core award, grant number 075491/Z/04. A.J.P. was supported by the Max-Planck Society for the Advancement of Sciences e.V. A.W. was funded by Inez Johanssons Stiftelse and Stiftelsen Lars Hiertas Minne.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions H.P. and M.J.T. conceived and designed the study. M.J.T. assembled mitochondrial data. H.P. and H.B. assembled EST data and performed phylogenetic analyses of ESTs and mitochondrial genomes. M.J.T., L.L.M. and R.R.C. performed preliminary phylogenomic analyses. H.N. collected Xenoturbella for genomic and miRNA data. A.W. collected Hofstenia. K.J.P. and A.W. produced Xenoturbella and Hofstenia miRNA libraries. K.J.P. assembled and analysed the miRNA matrix. M.J.T., R.R.C. and A.J.P. produced Xenoturbella genomic data. M.J.T. drafted the paper with H.P. and K.J.P. All authors commented on the manuscript.
Author Information MicroRNA sequences are deposited in www.mirbase.org and can be found in the Supplementary Information. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature).
References
- 1.Bourlat S, et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- 2.Bourlat S, Nielsen C, Lockyer A, Littlewood DTJ, Telford MJ. Xenoturbella is a deuterostome that eats molluscs. Nature. 2003;424:925–928. doi: 10.1038/nature01851. [DOI] [PubMed] [Google Scholar]
- 3.Egger B, et al. To be or not to be a flatworm: the acoel controversy. PLoS ONE. 2009;4:e5502. doi: 10.1371/journal.pone.0005502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Telford MJ, Lockyer AE, Cartwright-Finch C, Littlewood DTJ. Combined large and small subunit ribosomal RNA phylogenies support a basal position of the acoelomorph flatworms. Proc R Soc Lond B. 2003;270:1077–1083. doi: 10.1098/rspb.2003.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sempere LF, Cole CN, McPeek MA, Peterson KJ. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zool B. 2006;306:575–588. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz Trillo I, Riutort M, Littlewood DTJ, Herniou EA, Baguñà J. Acoel flatworms: earliest extant bilaterian metazoans, not members of Platyhelminthes. Science. 1999;283:1919–1923. doi: 10.1126/science.283.5409.1919. [DOI] [PubMed] [Google Scholar]
- 7.Hejnol A, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc R Soc B. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sempere LF, Martinez P, Cole C, Baguñà J, Peterson KJ. Phylogenetic distribution of microRNAs supports the basal position of acoel flatworms and the polyphyly of Platyhelminthes. Evol Dev. 2007;9:409–415. doi: 10.1111/j.1525-142X.2007.00180.x. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz-Trillo I, et al. A phylogenetic analysis of myosin heavy chain type II sequences corroborates that Acoela and Nemertodermatida are basal bilaterians. Proc Natl Acad Sci USA. 2002;99:11246–11251. doi: 10.1073/pnas.172390199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourlat SJ, Rota-Stabelli O, Lanfear R, Telford MJ. The mitochondrial genome structure of Xenoturbella bocki (phylum Xenoturbellida) is ancestral within the deuterostomes. BMC Evol Biol. 2009;9:107. doi: 10.1186/1471-2148-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philippe H, Brinkmann H, Martinez P, Riutort M, Baguñà J. Acoelflatwormsare not platyhelminthes: evidence from phylogenomics. PLoS ONE. 2007;2:e717. doi: 10.1371/journal.pone.0000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez-Ezpeleta N, et al. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst Biol. 2007;56:389–399. doi: 10.1080/10635150701397643. [DOI] [PubMed] [Google Scholar]
- 13.Lartillot N, Philippe H. Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Phil Trans R Soc B. 2008;363:1463–1472. doi: 10.1098/rstb.2007.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz Trillo I, Riutort M, Fourcade HM, Baguña J, Boore J. Mitochondrial genome data support the basal position of Acoelomorpha and the polyphyly of the Platyhelminthes. Mol Phyl Evol. 2004;33:321–332. doi: 10.1016/j.ympev.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Papillon D, Perez Y, Caubit X, Le Parco Y. Identification of chaetognaths as protostomes is supported by the analysis of their mitochondrial genome. Mol Biol Evol. 2004;21:2122–2129. doi: 10.1093/molbev/msh229. [DOI] [PubMed] [Google Scholar]
- 16.Sperling EA, Peterson KJ. In: Animal Evolution Genomes, Fossils and Trees. Telford MJ, Littlewood DTJ, editors. Ch 15. Oxford Univ. Press; 2009. pp. 157–170. [Google Scholar]
- 17.Lundin K. Degenerating epidermal cells in Xenoturbella bocki (phylum uncertain), Nemertodermatida and Acoela(Platyhelminthes) Belg J Zool. 2001;131:153–157. [Google Scholar]
- 18.Westblad E. Xenoturbella bocki n.g, n.sp, a peculiar, primitive turbellarian type. Arkiv Zool. 1949;1:3–29. [Google Scholar]
- 19.Nielsen C. After all: Xenoturbella is an acoelomorph! Evol Dev. 2010;12:241–243. doi: 10.1111/j.1525-142X.2010.00408.x. [DOI] [PubMed] [Google Scholar]
- 20.Franzen A, Afzelius BA. The ciliated epidermis of Xenoturbella bocki (Platyhelminthes, Xenoturbellida) with some phylogenetic considerations. Zool Scr. 1987;16:9–17. [Google Scholar]
- 21.Pardos F. Fine structure and function of pharynx cilia in Glossobalanus minutus Kowalewsky (Entropneusta) Acta Zool. 1988;69:1–12. [Google Scholar]
- 22.Tyler S. In: Interrelationships of the Platyhelminthes. Littlewood DTJ, Bray RA, editors. Taylor & Francis; 2001. pp. 3–12. [Google Scholar]
- 23.Telford MJ. Xenoturbellida: the fourth deuterostome phylum and the diet of worms. Genesis. 2008;46:580–586. doi: 10.1002/dvg.20414. [DOI] [PubMed] [Google Scholar]
- 24.Baguña J, Martinez P, Paps J, Riutort M. Back in time: a new systematic proposal for the Bilateria. Proc R Soc B. 2008;363:1481–1491. doi: 10.1098/rstb.2007.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hejnol A, Martindale MQM. Acoel development supports a simple planula-like urbilaterian. Phil Trans R Soc B. 2008;363:1493–1501. doi: 10.1098/rstb.2007.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson KJ, McPeek MA, Evans DA. Tempo and mode of early animal evolution: inferences from rocks, Hox, and molecular clocks. Paleobiology. 2005;31:36–55. [Google Scholar]
- 27.Ruppert EE. Key characters uniting hemichordates and chordates: homologies or homoplasies? Can J Zool. 2005;83:8–23. [Google Scholar]
- 28.Philippe H, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 29.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 30.Wheeler B, et al. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- 31.Birney E, Clamp M, Durbin R. GeneWise and GenomeWise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang X, Madan A. CAP3: a DNA assembly programme. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 35.Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 36.Philippe H. MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 38.Roure B, Rodriguez-Ezpeleta N, Philippe H. SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol Biol. 2007;7 (Suppl. 1):S2. doi: 10.1186/1471-2148-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 40.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 41.Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007;7 (Suppl. 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 43.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.69. Department of Genome Sciences, Univ. Washington; Seattle: 2005. [Google Scholar]
- 44.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 45.Blanquart S, Lartillot N. A site- and time-heterogeneous model of amino acid replacement. Mol Biol Evol. 2008;25:842–858. doi: 10.1093/molbev/msn018. [DOI] [PubMed] [Google Scholar]
- 46.Hrdy I, et al. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- 47.Roure B, Philippe H. Site-specific time heterogeneity of the substitution process and its impact on phylogenetic inference. BMC Evol Biol. doi: 10.1186/1471-2148-11-17. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.