Abstract
The base excision repair (BER) pathway is a conserved DNA repair system required to maintain genomic integrity and prevent mutagenesis in all eukaryotic cells. Nevertheless, how BER operates in vivo (i.e. in the context of chromatin) is poorly understood. We have investigated the role of an essential ATP-dependent chromatin remodeling (ACR) complex RSC (Remodels the Structure of Chromatin) in BER of intact yeast cells. We show that depletion of STH1, the ATPase subunit of RSC, causes enhanced sensitivity to the DNA alkylating agent methyl methanesulfonate (MMS) and results in a substantial inhibition of BER, at the GAL1 locus and in the genome overall. Consistent with this observation, the DNA in chromatin is less accessible to micrococcal nuclease digestion in the absence of RSC. Quantitative PCR results indicate that repair deficiency in STH1 depleted cells is not due to changes in the expression of BER genes. Collectively, our data indicates the RSC complex promotes efficient BER in chromatin. These results provide, for the first time, a link between ATP-dependent chromatin remodeling and BER in living cells.
Keywords: SWI/SNF, nucleosome, alkylated DNA damage, MMS
1. INTRODUCTION
DNA of all living cells is constantly damaged by exogenous and endogenous factors such as radiation, alkylating agents and reactive oxygen species [1]. DNA lesions pose a significant threat to genomic stability and need to be repaired quickly and efficiently throughout the genome, to prevent mutagenesis and maintain vital cellular functions. Base excision repair (BER) is a front-line defence system, which repairs damaged (or modified) DNA bases, a basic (AP) sites and DNA single strand breaks (SSBs). BER involves concerted action of enzymes, which sequentially process damaged bases and restore DNA to its unmodified state. This repair pathway and its individual components are important for genomic integrity and maintenance of the epigenetic states in chromatin [2, 3]. Deficiency in BER is tightly linked to aging and human diseases such as neurodegeneration, developmental abnormalities and several types of cancers [4–6].
DNA alkylation is one of the major DNA lesions induced by a variety of DNA damaging agents. Alkylating agents such as methyl methanesulfonate (MMS), methylate DNA bases by adding a methyl group to nucleophilic sites. The predominant forms of MMS-induced DNA damage are the N-methylpurines (NMPs) N7-methylguanine and N3-methyladenine, which cause base mispairing and replication blocks [7]. In Saccharomyces cerevisiae, the NMPs are mainly repaired by BER. The repair is initiated by the DNA glycosylase, MAG1, which recognizes and removes methylated bases, leaving AP (apurinic/apyrimidinic) sites. These sites are then cleaved by either (a) endonucleases APN1 or APN2, yielding single strand DNA (ssDNA) breaks with 5′-deoxyribosephosphate (5′-dRP) ends, or (b) AP lyases, such as NTG1 or NTG2, yielding ssDNA breaks with 3′-dRP ends. The 5′-dRP ends are excised by DNA endonuclease RAD27, while 3′-dRP ends are removed by either the 3-phosphodiesterase activity of APN1 and APN2, or by flap endonuclease RAD1-RAD10. The gap is filled by polymerase delta (δ) or epsilon (ε) and repair is completed by DNA ligase CDC9 [8–10].
The BER pathway has been reconstituted in vitro with purified proteins and naked, damaged DNA substrates [11]. In this manner, the components and major steps of BER are well characterized in both yeast and mammalian systems [12]. However, it remains unclear how BER operates in vivo, in a chromatin landscape, and whether chromatin remodeling plays a role in facilitating BER.
Eukaryotic DNA is packaged with histone proteins into nucleosomes [13]. Nucleosomes are further packaged with linker histones and other architectural proteins into higher-order chromatin structures and ultimately form mitotic chromosomes [14]. In the unmodified state, nucleosomal DNA is highly inaccessible to DNA binding proteins, such as repair factors. It has been well established that chromatin at the nucleosome level has an intrinsic ability to inhibit the efficiency of several DNA repair pathways, including BER, as the access of DNA repair proteins is limited and the activity of repair enzymes is severely compromised [15, 16]. DNA repair in compact, higher-order chromatin structures, such as mitotic chromosomes, faces greater challenges, with increased chromatin compaction rendering the DNA virtually inaccessible. Nevertheless, BER appears to operate efficiently in highly condensed chromatin. For example, Hildrestrand et al. found that human glycosylase NEIL1 is localized in centrosomes and mitotic chromosomes during mitosis [17]. In addition, Okano et al. demonstrated translocation of BER repair factors XRCC1 and DNA ligase IIIα from centrosomes to mitotic chromosomes following DNA damage [18].
Emerging evidence suggest that ATP-dependent chromatin remodeling (ACR) complexes play important roles in the rearrangement of nucleosomes at DNA damage sites, checkpoint signalling, recruitment and assembly of DNA repair machinery [19, 20]. Chromatin and nucleosome remodeling mechanisms involving ACR and histone modifications have been implicated in DNA repair pathways such as double strand break (DSB) repair and nucleotide excision repair (NER), which repair strand breaks and DNA helix distorting lesions, respectively. Repair of DSBs in chromatin, and NER, rely on the “access-repair-restore” (ARR) paradigm [21], and a direct link between chromatin remodeling and ARR is well established in vivo [22, 23]. In contrast, the role of chromatin remodeling and the involvement of ACR complexes in facilitating BER in chromatin are not yet well understood.
The yeast RSC complex belongs to the highly conserved SWI/SNF subfamily of ATP-dependent chromatin remodeling complexes. RSC is an essential and abundant (~2000 complexes/cell) remodeler, required for cell viability and cell cycle progression [24, 25]. The complex contains a central DNA- dependent ATPase subunit (STH1) and 17 additional accessory subunits [26]. The STH1 subunit is homologous to known human tumour suppressor proteins hBRM and BRG1, which are the catalytic subunits of hSWI/SNF. The remodeling activity of RSC can make nucleosomal DNA more accessible in an ATP-dependent manner. RSC is able to bind to the nucleosome core particle, translocate along DNA and pull the nucleosomal DNA off of the histone octamer surface, producing DNA loops of a broad size range (20–1200bp, average ~100bp) [27]. RSC can reposition, evict or restructure nucleosomes and the STH1 subunit alone is sufficient for the remodeling activity [28]. RSC complex is important for establishing and maintaining specific chromatin structures, including nucleosome positions and occupancy, both genome-wide and at specific loci in S. cerevisiae [29, 30]. The chromatin remodeling activity of RSC is crucial for regulation of nuclear processes such as DNA replication, transcription, and repair [31–33], and several studies have now demonstrated a direct role for RSC in DSB repair in vivo [34–37].
Recently, in vitro studies have demonstrated a direct role of RSC in facilitating BER in di-nucleosomal templates [38]. In the present study, we show that RSC complexes play a role in the repair of NMPs by BER in yeast, indicating that RSC activity promotes BER in chromatin of living cells. Indeed, RSC depleted cells are more sensitive to MMS treatment, and are deficient in repair of methylated DNA bases. Moreover, the global chromatin structure is less accessible to micrococcal nuclease (MNase) in RSC-depleted cells. These results highlight a novel role for RSC in gaining access to themethylated bases in chromatin during BER in intact yeast cells.
2. MATERIAL AND METHODS
2.1. Conditional depletion of STH1 protein from the yeast cells
Two alternative conditional STH1 knockdown systems (‘Tet-off’ and ‘degron’) were used in these studies. For the Tet-off system, yeast strains WT (URA3::CMV-tTA MATa his3-1 leu2-0 met15-0) and Tet-STH1 (pSTH1::kanR-tet07-TATA URA3::CMV-tTA MATa his3-1 leu2-0 met15-0) were obtained from Open Biosystems. In order to deplete STH1 from these cells, both WT and Tet-STH1 strains were first grown in 5 ml YPD liquid media at 30° C until log phase (OD600: 0.5–1). Cultures were diluted into larger volumes (50–100 ml) of fresh YPD supplemented with doxocycline (10 μg/ml), and incubated for an additional 20 hours. The rationale for 20 hour-long incubation with doxocycline was based on the predicted stability of STH1 protein [39]. As confirmed by qPCR and western blot analyses, this protocol yielded substantial depletion of STH1 protein (Fig. S2B, S3). For synchronization of both cultures at the G2/M phase of the cell cycle, the strains were grown on YPD supplemented with doxocycline (10 μg/ml) for approximately 17 hours, followed by addition of nocodazole (15 μg/ml) and grown for an additional 3 hours.
For the degron system, WT control (MATa ura3-52 tryp1Δ63 his3Δ200 leu2::PET56 ubr1Δ::HIS3 sth1Δ::pCUP1-sth1td::URA3) and STH1degron strains (MATa ura3-52 tryp1Δ63 his3Δ200 leu2::PET56 ubr1Δ::pGAL1-UBR1::HIS3 sth1Δ::pCUP1-sth1td::URA3) were obtained from Dr. Bradley Cairns, Huntsman Cancer Institute, Univ. of UT. Strains were grown as described by Parnell et al. [40]. Briefly, both WT and STH1degron strains were grown overnight at room temperature in 10ml starter cultures in YP media containing 2% raffinose. For experiments, 50 ml cultures were started and grown overnight at room temperature to early log phase (OD600:0.4–0.6). Galactose (2% final concentration) was then added and cultures were incubated at room temperature for 1 hour to induce UBR1, which encodes an E3 ubiquitin ligase. The temperature was then shifted to 37°C for 2 hrs to induce STH1 protein degradation.
2.2. Treatment of cells with MMS
The Tet-STH1 mutant cells were depleted of STH1, and both WT mutant cells were synchronized at G2/M as described above. MMS was added to liquid cultures at a final concentration of 0.2%v/vand cultures were incubated for 10 min at 30° C, in an orbital shaker. Cells were then harvested by centrifugation, washed with ice-cold phosphate buffered saline (PBS), and re-suspended in a pre-warmed YPD media supplemented with doxocycline and nocodazole. Cultures were incubated with continuous shaking and cell aliquots were collected after different repair times.
The STH1 degron and isogenic WT strains were grown at room temperature, as described above. Subsequently, 0.2% MMS (final concentration) was added to both cultures and they were incubated at room temperature for 10 minutes. Cells were washed in ice-cold PBS, re-suspended in pre-warmed media (YP + 2% raffinose supplemented with 2% galactose) and allowed to repair at 37°C for 3 hours. Aliquots for the damage-repair time courses were collected and processed as described below, and genomic DNA was extracted as described previously [41].
2.3. Metabolic competence assay
The metabolic competence of cells was assessed as described previously [42].
2.4. Global genomic BER repair analysis
Cells were treated with MMS as described above for BER analysis. BER of MMS-induced NMPs in yeast cells was performed as described previously [43]. Briefly, genomic DNA, isolated from cells incubated for different repair times, was treated with AAG glycosylase and APE1 endonuclease, to cleave the DNA specifically at NMPs. These damage-specific single strand breaks, were resolved on alkaline agarose gels and stained with SYBR gold (Invitrogen). To examine global BER, the number-average length of genomic DNA (± AAG/APE treatment) was determined and used to calculate the average number of ssDNA/kb, as described previously [44].
2.5. Quantification of gene expression using qPCR
The Tet-STH1 and WT strains were depleted of STH1 and synchronized at G2/M as described above. The protocol for RNA extraction was adopted from Schmitt et al. 1990 [45], with some modifications. For total RNA preparation, cells were washed in water, re-suspended in 400 μl of TES buffer (10 mM Tris pH 7.5, 10 mM EDTA, 0.5% SDS) and 400 μl PCI (phenol: chloroform: isoamyl alcohol), and vortexed vigorously for 4 min. Samples were then incubated at 65° C for 30 min, placed on ice for 5 min, and centrifuged at top speed and 4° C for 5 min. The supernatant was recovered and two additional PCI extractions were performed. RNA was precipitated with 1 ml of cold ethanol/ammonium acetate and centrifuged at 4° C for 15 min. RNA pellets were washed with 70% ethanol, air dried, resuspended in 40 μl water, and stored at −80° C. Total RNA was treated with DNAase (TURBO DNA–free™ kit, Life Technologies) to remove contaminating DNA, and converted to cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Transcript levels were quantitated using the comparative CT threshold cycle (ΔΔCT) method and SYBR green sequence detection on a 7500 Fast Real-Time PCR system (Applied Biosystems). Relative quantitation, via the (ΔΔCT) method, is expressed as a difference in target gene expression with respect to an internal control (actin mRNA or 18s rRNA) in different samples. Each cDNA sample was assayed in duplicate and RNAs were obtained from two separate biological samples.
2.6. Analysis of NMP repair at the GAL1 locus
Yeast cells were treated with MMS, and genomic DNA was extracted, as described above. High resolution NMP repair was performed as described previously [46]. Briefly, the GAL1 fragment was released from the genomic DNA using DraI and BanII restriction enzymes, and the restricted DNA was cleaved at NMP sites by incubation with AAG and APE1 enzymes. The non-transcribed strand of theGAL1 gene was first annealed to biotinylated oligonucleotides, then attached to streptavid in magnetic beads (Life Technologies) and labelled using [α-32P]dATP (Perkin Elmer) and Sequenase™ (Affymetrix). The labelled fragments were eluted, resolved on sequencing gels, and exposed to Phosphor Imager screens (Molecular Dynamics). The band intensities were quantified using ImageQuant, version 5.2 (Molecular Dynamics), and peak deconvolution software [Peak-Fit version 4.12 (SPSS Inc.)] as described previously [46].
2.7. Micrococcal nuclease (MNase) digestions
Chromatin accessibility was analysed in the nuclei from Tet-STH1 and WT strains synchronized at G2/M as described above. The MNase accessibility was measured as previously described [47]. Briefly, 50 ml cultures of mid-log phase yeast cells (~ 1.0× 107 cells/ml) were pelleted, washed with PBS, suspended in yeast lytic enzyme YLE buffer containing10 mg/ml zymolaze (MP Biomedicals) in 1M sorbitol and 5 mM β-mercaptoetanol, and incubated at 30° C for 20 min. Spheroplasts were resuspended in digestion buffer (SDB, 1 M sorbitol, 50 mM NaCl, 10 mM Tris-HCl [pH 7.5], 5 mM MgCl2,1 mM CaCl2, 1 mM β-mercaptoetanol, 0.5 mM spermidine and 0.075% v/v IGEPAL CA-630), divided into 150 μl aliquots, and digested with varying concentrations of MNase (Affymetrix) at 37° C for 10 min. Reactions were stopped with 5% SDS and 250 mM EDTA, and treated with proteinase K for 2 h at 55° C. Digested DNA was purified and resolved on 1.2% native agarose gels.
Southern blotting was performed to analyze chromatin accessibility at the HML locus. DNA was separated on a1% native agarose gel, transferred to Hybond N+ membranes (GE Healthcare) and hybridised with radiolabeled gene specific probes generated from the HMLα-1 ORF by PCR amplification of nucleotides +1 to +518 and radiolabeled with [α-32P]dATP using a Random Priming Kit (Agilent, Stratagene).
3. RESULTS
3.1. Depletion of STH1 results in increased sensitivity to MMS
The remodeling function of RSC is dependent on its ATPase subunit, STH1 [28]. Studies with ACRs indicate that lack of the central ATPase subunit prevents assembly of the whole complex and eliminates its remodeling activity [48]. The STH1 gene encodes essential protein and complete deletion of the gene is lethal. Therefore, we used two different conditional STH1 knockdown systems, both ‘Tet-off’ and ‘STH1-degron’ cells. In Tet-off cells, conditional knockdown targets gene expression, leading to progressive depletion of STH1 mRNA during the 20-hour long depletion time course, while the degron system is based on rapid, 2-hour long, temperature-induced, protein degradation. For the Tet-off experiments, we used a conditional mutant Tet-STH1 strain, bearing a tetracycline-regulated element (Tet) in the promoter of the STH1 gene [49]. Growth of the Tet-STH1 strain in the absence of doxocycline (a stable homolog of tetracycline) results in the functional expression of STH1 and essentially a wild type phenotype. Addition of doxocycline to the growth media deactivates the Tet promoter and shuts down the expression of STH1, leading to depletion of the STH1 protein (Fig. S1, S3B). As determined in previous studies, the majority ofSTH1-depleted cells arrest at the G2/M phase of the cell cycle and show typical phenotypes, such as reduced growth rate and small colony size [37, 50, 51].
We have confirmed that the concentration of doxocycline used in our studies has little (or no) effect on growth of the WT strain, not containing the Tet element (Fig. S2 A), indicating that doxocycline, at the concentration used is not toxic.. Suppressed expression of the STH1 gene indicates that doxocycline significantly reduces transcription of STH1 only in the Tet-STH1 strain, as expected (Fig. S2B). Protein depletion in both Tet-off and degron strains has been verified by western blotting (Fig. S3A,B). Importantly, we have determined that STH1-depleted cells remain viable, as demonstrated by the metabolic competence assay (Fig. S2 C).
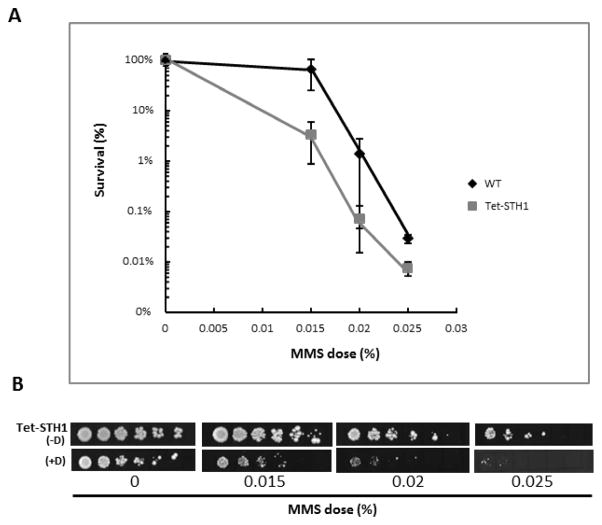
To assess the potential role of the RSC complex in BER, we first analysed the sensitivity of the RSC mutant cells to the DNA alkylating agent, MMS. Both, Tet-STH1 mutant and the isogenic WT strains were incubated in liquid cultures in the presence of doxocycline for approximately 20 hour shr to deplete STH1 in the mutant. As discussed above, the majority of RSC-deficient cells are arrested at G2/M, whereas WT cells continue growing, making a direct comparison of MMS sensitivity in both strains difficult. To allow for a better comparison of cell sensitivity, both mutant and WT cells were spread (or spotted) on YPD plates containing MMS only (i.e. without doxocycline). Under these conditions, G2/M arrested cells which survived prior to MMS damage, have the ability to restore functional expression of STH1 and release from G2/M arrest to divide and form colonies. The number of colonies on YPD plates (free of doxocycline and MMS) was comparable between WT and Tet-STH1 mutant cells [Fig. S2 A and Fig. 1B; Tet-STH1(−D)/(+D)], suggesting that STH1-depleted cells are viable and able to restore functional expression of STH1, when grown on media lacking doxocycline. Significantly, mutant cells depleted of STH1 demonstrated increased sensitivity to MMS, suggesting an important role of the catalytic subunit of RSC in promoting resistance to MMS-induced DNA damage (Fig. 1A,B).
Figure 1. A,B. Conditional loss of STH1 in yeast cells results in hypersensitivity to MMS.
A. Yeast cells were grown in liquid culture in the presence of doxocycline, as described in Material and Methods. Equal numbers of cells were spread on YPD pates supplemented with increasing doses of MMS. Plates were incubated for 3–4 days at 30° C, colonies counted, and % survival was determined. B. Yeast cells were grown in the presence (+D) or absence (−D) of doxocycline and aliquots were spotted on plates supplemented with MMS.
3.2. Loss of STH1 results in decreased BER genome- wide
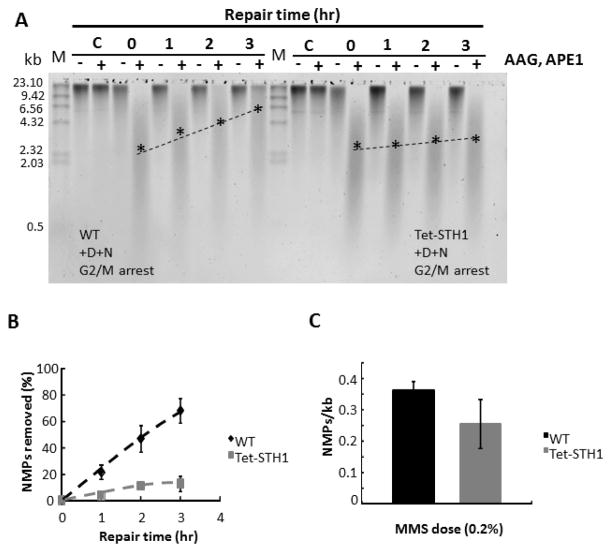
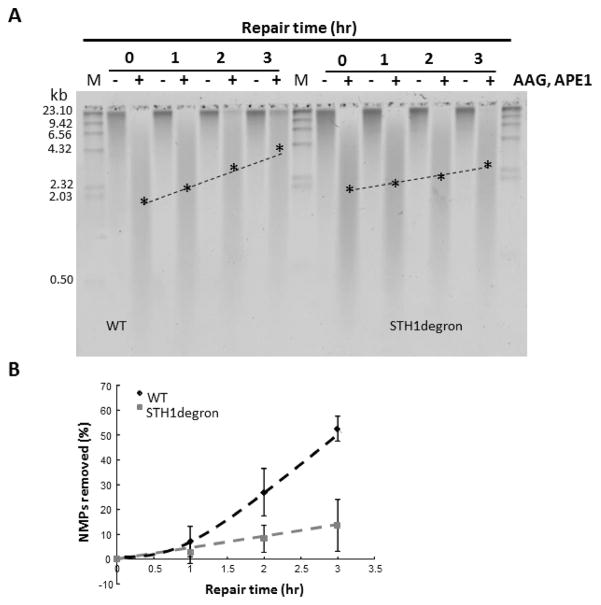
The genomic DNA from both WT and STH1-depleted cells, exposed to MMS and incubated for different repair times, was analysed for the rate in global genomic BER, as previously described [43]. As expected, WT cells efficiently remove NMPs during the repair time course, where ~ 60% of the NMPs are removed in 3 hours (Fig. 2A,B and Fig 3A,B). In contrast, STH1-depleted cells demonstrate a significant inhibition in BER, with the repair efficiency being only ~ 10% at the 3 hour repair time point. This phenotype is observed in two different strain backgrounds and protein depletion systems. Importantly, the frequency of NMP formation in both WT and mutant strains, measured directly by this method [43, 44], is not significantly different (Fig. 2C). Therefore, depletion of STH1 does not change the overall susceptibility of chromatin to MMS-induced DNA damage.. Additionally, doxocycline treatment does not affect the rate of BER following MMS treatment in WT strains (Fig. S4).
Figure 2. A,B,C. Depletion of RSC complex in Tet-STH1 strain dramatically inhibits BER genome-wide.
The Tet-STH1 and WT strains were grown in the presence of doxocycline for 20 hours, arrested at G2/M phase with nocodazole and treated with MMS. Cells were maintained at G2/M through the repair time course. For each time point cell aliquots were collected and genomic DNA was purified. Genomic DNA was processed with (+) or without (−) enzymatic cocktail composed of glycosylase (AAG) and endonuclease (APE1). Enzymes convert methylated purines to single strand breaks. Single strand breaks were separated on the alkaline gel and DNA was visualized with SYBR gold staining. A. A representative gel showing repair time course in WT and STH1- depleted Tet-STH1 strains. M: marker DNA size standard (λ/HindIII); C: untreated cells; 0: cells damaged with MMS; 1–3: repair time course in hours. Positions of the approximate median migration distance of the fragments in each lane are shown with asterisks (*) and are connected by a dotted line. B. Repair time course of N-methylpurines (NMPs). Data represent mean ± 1 s.d. from three independent experiments. C. Formation of NMPs in the genomes of WT and STH1- depleted cells. STH1- depleted and G2/M synchronized cells were damaged with MMS. Genomic DNA was purified and number of NMPs was analysed as described in materials and methods. Data represent mean ± 1 s.d. from three independent experiments.
Figure 3. A,B. Depletion of RSC complex in STH1-degron strain results in major deficiency of BER genome- wide.
The STH1-degron and WT strains were grown and MMS treated as described in materials and methods. To analyse NMPs removal genomic DNA was processed as described above. A. A representative gel showing repair time course in WT and STH1- depleted STH1-degron strains. M: marker DNA size standard (λ/HindIII); 0: cells damaged with MMS; 1–3: repair time course in hours. Positions of the approximate median migration distance of the fragments in each lane are shown with asterisks (*) and are connected by a dotted line. B. Repair time course of NMPs. Data represent mean ± 1 s.d. from three independent experiments.
3.3. Loss of STH1 inhibits repair at individual NMP sites in the non-transcribed GAL1 locus
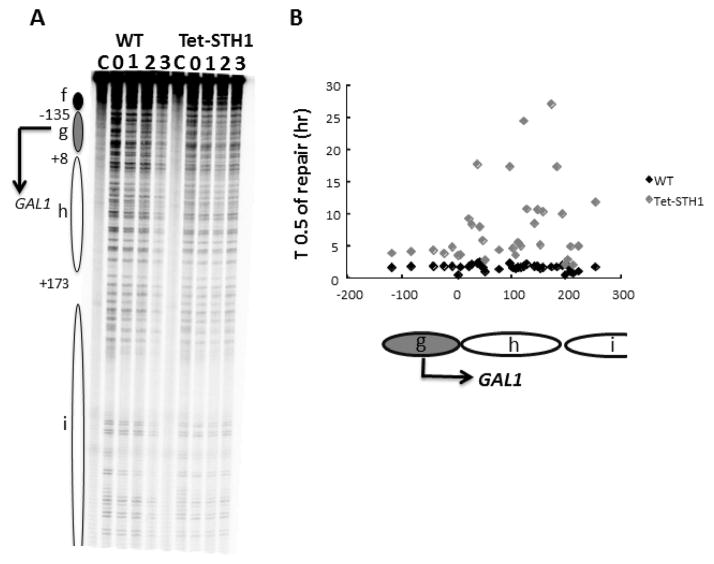
To examine repair at individual NMP sites within a region of known chromatin structure, the efficiency of BER in the STH1 depleted cells was analysed in the GAL1 gene locus. Chromatin structure at this locus has been studied extensively and the positions of nucleosomes are well determined [52–54]. We examined repair of NMPs in the non-transcribed strand of the repressed GAL1 gene of STH1depleted cells. Typically, the rate of BER is correlated with the nucleosome position, with slow and fast repair occurring in the nucleosome core and linker DNA respectively [54]. We showed that the repair pattern in STH1 mutant cells follows a similar trend, however the rate of repair is significantly slower at most NMP sites in the GAL1 gene, in both nucleosome core and linker DNA regions (Fig. 4A,B). This data suggests that activity of the RSC complex is important for efficient repair of NMPs at the GAL1 locus.
Figure 4. A, B,C. Loss of STH1 dramatically inhibits repair of NMPs in the GAL1 locus.
Repair of NMPs in WT and Tet-STH1 strains was analysed at the nucleotide resolution as described in materials and methods. A. Panel shows representative sequencing gel of DNA damage and repair profile in yeast cells. Each band represents a single NMP site in a given DNA fragment, band intensity corresponds to number of DNA fragments containing NMP site in the same location. Decreasing intensity of band over the repair time course indicates successful repair of NMPs. C: untreated control, the ovals represent nucleosome locations in glucose cultures, where the intensity of shading refers to variability of the nucleosome positions (darkest, least variable; lightest, most variable). The arrow on the left represents transcription start site (TSS). Nucleotide positions are allocated in relation to the start of GAL1 gene. B. Graph of the time required to repair 50% of the NMPs (T1/2) at individual sites in the GAL1 region in glucose in WT and Tet-STH1 mutant.
3.4. BER deficiency in STH1 depleted cells is not likely caused by changes in the expression of BER genes
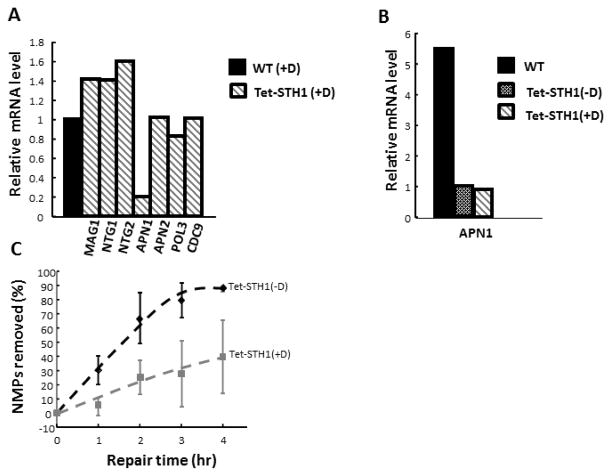
A published genome-wide location analysis suggests that RSC complex regulates transcriptional expression of approximately 700 yeast genes, including genes encoding histone and chromatin assembly proteins [33]. To examine the possibility that the observed deficiency in BER of MMS-induced damage does not result from down regulation of genes encoding proteins in the BER pathway, we analysed the expression of a set of key BER genes in STH1-depleted cells.. We showed that doxocycline treatment has little or no effect on the transcriptional expression of these genes in WT cells (e.g., see Fig. S5). We found that expression of theMAG1 gene, which encodes the major glycosylase for recognition and excision of NMPs, is slightly elevated in the mutant cells as compared to wild type (Fig. 5A). Similarly, expression of other glycosylase genes, such as NTG1 and NTG2, was elevated in the mutant as well. Genes encoding endonuclease APN2, polymerase POL3, and ligase CDC9 showed no significant changes in the expression in WT and STH1- depleted cells. Interestingly, expression of the major endonuclease APN1 was substantially suppressed in the STH1 depleted cells (Fig. 5A), which is consistent with the previous microarray studies, showing down-regulation of APN1 inrsc2 mutant cells [55]. APN1 and APN2 are endonucleases representing alternate pathways for repair of the AP sites [56], and they play complementary roles.
Figure 5. A,B,C. Change in the expression of BER genes in STH1- depleted cells does not contribute to deficiency of BER.
Total RNA was extracted from G2/M synchronized cells of WT (−D), WT (+D), Tet-STH1(−D) and Tet-STH1(+D). RNA was purified and processed for qPCR. A. Differential expression of BER genes in STH1- depleted cells relative to WT (control). Data represents average value of two independent experiments B. Expression of APN1 gene in WT(+D), Tet-STH1 (+D), Tet-STH1 (−D). Data represents average value of two independent experiments C. Repair time-course data in Tet-STH1(−D) and Tet-STH1(+D). Data represent means ± s.d. from three independent experiments.
To investigate a possible correlation between decreased expression of APN1 and deficient BER in STH1 mutants, we examined the expression level of APN1 in Tet-STH1 cells grown in the presence [Tet-STH1(+D), mutant] and absence [Tet-STH1(−D), wild type] of doxocycline. In the Tet-STH1 cells, expression of STH1 is driven by the Tet promoter. We observed that APN1 expression is higher in the WT strain, where STH1 is driven by its endogenous promoter, as compared to the Tet-STH1(−D) strain, where STH1 is driven by the Tet promoter (Fig 5A). Importantly, there is no significant difference in the expression of APN1 in Tet-STH1(+D) cells, as compared to Tet-STH1(−D) cells (Fig. 5B). Furthermore, analysis of global genomic repair showed that the Tet-STH1(+D) is deficient in BER, whereas repair in the wild type equivalent, Tet-STH1(−D), is proficient (Fig. 5C). Hence, even though the level of APN1 is low in the Tet-STH1(±D) compared to WT, this does not seem to have an effect on the BER efficiency in Tet-STH1(−D) cells. This result indicates that the changes in expression ofAPN1likely do not contribute to the deficient BER in STH1 mutant cells.
3.5. Cells depleted of STH1 exhibit decreased chromatin accessibility
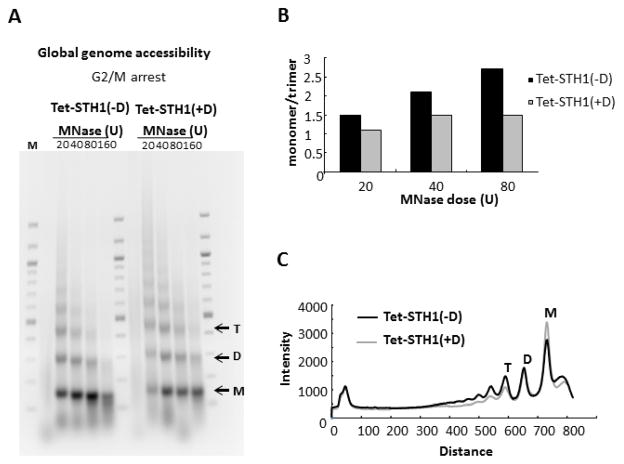
The RSC complex has been shown to modulate chromatin structure locally and globally [25, 28, 40, 57], and has been implicated in nucleosome mobilization to improve access of DNA repair proteins to DSBs in chromatin [37]. To address the possibility that inhibition of BER is linked with a change in chromatin structure, we examined the accessibility of bulk chromatin by assessing MNase digestion patterns for WT and STH1 mutant cells. The Tet-STH1(−D) and Tet-STH1(+D) strains were grown in the presence or absence of doxocycline, cells were synchronized at G2/M with nocodazole, and spheroplasts were isolated from both strains (see Materials and Methods). Spheroplasts were then treated with increasing concentrations of MNase and the resulting DNA fragments separated on agarose gels. As shown in Fig. 6A and S6A, chromatin from Tet-STH1(+D) cells is more resistant to MNase digestion (i.e. shows a slower release of nucleosome multimers) compared to chromatin from Tet-STH1(−D) cells. Furthermore, this enhanced MNase resistance is observed regardless of the synchronization of WT cells at G2/M.
Figure 6. A,B,C. The global chromatin is less accessible in STH1- depleted mutant.
Tet-STH1 strain was split into two liquid cultures and grown over night in the presence and absence of doxocycline Tet-STH1 (+D), Tet-STH1 (−D). Cells were arrested at the G2/M phase of cell cycle. Spheroplasts were isolated, treated with MNase and genomic DNA was purified, separated on agarose gel and stained with ethidium bromide. A. MNase digestion patterns in wild type Tet-STH1(−D) and mutant Tet-STH1(+D) synchronized at G2/M phase of the cell cycle. Gel shows representative data of at least two independent experiments. M: DNA size standard, T: tri-nucleosome, D: di-mucleosome, M: mono-nucleosome. B. Quantitative analysis of MNase accessibility expressed as the ratio of mono- to tri-nucleosome signal at different concentrations of MNase. C. Band intensity profiles of the 40U MNase lanes for Tet-STH1(−D) and Tet-STH1(+D).
To quantify these observations, band intensities corresponding to mono-, di-, and tri-nucleosomes were determined and the ratio of monomer to trimer band intensities compared. The data show that there is less of an increase in the ratio of mono- to tri-nucleosome formation with increased digestion of Tet-STH1(+D) chromatin, compared to Tet-STH1(−D) chromatin (Fig. 6B), indicating that monomer nucleosomes are released slower from the Tet-STH1(+D) chromatin. Furthermore, scans of the bulk DNA banding patterns of the two strains indicate there is no significant difference in the overall nucleosome repeat length upon addition of doxocycline (Fig. 6C). Finally, analysis of the MNase digestion pattern of transcriptionally silent HML gene chromatin in these cells indicates that this locus is also significantly less accessible to MNase in the RSC mutant (Fig. S7A,B). Therefore, these results suggest that RSC activity is important for maintaining a more “open” and accessible chromatin landscape in yeast cells.
4. DISCUSSION
The BER pathway has been extensively studied for almost 40 years [58, 59], yet little is known about how this DNA repair mechanism operates in the context of tightly folded nuclear chromatin, and whether chromatin remodeling regulates efficiency of BER. Our studies indicate that RSC, a conserved and essential ATP dependent remodeling complex, promotes BER in chromatin of intact yeast cells.
Several in vitro studies have demonstrated the inhibitory role of chromatin on the rate of BER. Several reports have shown a reduced activity of BER enzymes on model nucleosome substrates [60–62]. These studies demonstrated that the rotational position of lesions within nucleosome DNA significantly affects the recognition step by DNA glycosylase/AP endonuclease. Specifically, lesions facing the solution (or ‘out’) are easily accessible and cleaved, while cleavage at lesions facing the histone octamer (or ‘in’) is inhibited [15]. Moreover, we have recently shown that, although outwardly oriented uracils near the nucleosome center are efficiently cleaved by uracil DNA glycosylase/AP endonuclease activity, polymerase β extension of the resulting gaps is strongly inhibited at these sites [63]. These studies indicate that glycosylase, AP endonuclease, and polymerase β, enzymes that catalyse the first three steps of BER, are able to process lesions with various efficiencies without irreversibly disrupting the nucleosome structure. In contrast, the DNA ligase IIIα-XRCC1 complex, which completes BER, is able to bind and disrupt nucleosome substrates to enhance its own activity and the activity of Pol β. Therefore, nucleosome disruption by this ligase promotes BER [16, 64].
Higher order chromatin structure may also interfere with at least the late steps of BER. For example, our lab showed previously that the level of compaction of oligonucleosome templates did not interfere with the activity of uracil DNA glycosylase and APE1 endonuclease; however, the degree of compaction greatly inhibited (up to 80%) the function of Pol β [62]. Importantly, in the same report, we also demonstrated that ACR complexes ISW1 and ISW2 significantly enhanced Pol β activity in the oligonucleosome templates in vitro [62]. Moreover, a recent in vitro study by Menoni and coworkers, using di-nucleosomes assembled with linker histone H1, revealed that concerted action of chaperone NAP1 with ACR complex RSC was necessary for efficient repair of 8-oxoG by 8-oxoguanine glycosylase OGG1 [38]. Finally, studies by Amouroux et al., indicate that chromatin compaction induces strong inhibition of the recognition and excision of the damaged bases by BER in intact human cells [65]. Therefore, chromatin remodeling might be necessary to promote fast and efficient BER in at least certain (e.g., highly condensed) regions of chromatin in vivo.
In the present manuscript, we investigate the role of ATP-dependent remodeling complex RSC in the BER of intact yeast cells. The abundant, essential and conserved RSC complex [24, 66], has been implicated in several DNA-templated processes, such as replication, transcription, and DNA repair [32, 67]. We conditionally knockdown STH1, the essential catalytic subunit of RSC and analysed its role in the repair of MMS-induced N-methylated DNA purines via BER. Previous studies showed that conditional depletion of STH1 protein results in the sensitivity of yeast cells to various DNA damaging agents, including MMS [68]. Consistent with these findings, we have also found an increased sensitivity to MMS in cells depleted of STH1 (Fig. 1A,B). Importantly, the Tet-STH1 depleted cells are viable and when plated on doxocycline-free and MMS-free media, cells recover quickly and the growth rate is similar to WT (Fig. S2A,C). Therefore the abundance of the RSC complex appears to be important for cells to repair methylated DNA damage.
We have further demonstrated that loss of the RSC complex significantly decreases the rate of BER genome-wide. Our data show that WT cells efficiently remove NMPs during the repair time course used, where approximately 60% of the NMPs are removed by 3 hours. In contrast, RSC-depleted cells have a significantly reduced rate of BER, with only about 10% of the lesions being removed at the 3 hour repair time point (Fig. 2A, B and 3A,B). This phenotype was consistently observed in two different strain backgrounds and in both STH1-protein depletion systems (Tet-off and STH1-degron), suggesting a specific role of RSC in BER. It is noted that there appears to be a slight difference in the time course of BER when comparing Tet-STH1 with STH1-degron mutant cells; however, a strong inhibition of BER is significant in both mutant types. The differences are most likely attributable to differences in genetic background of these strains or experimental conditions associated with different protein depletion timelines and/or temperature of the repair time course (see Materials and Methods). Additionally, we verified that depletion of STH1 did not significantly affect the frequency of NMP formation, as similar amounts of methylated bases were formed in WT and STH1 mutant cells (Fig. 2C).
Importantly, we found that the deficient BER in STH1 mutant cells does not appear to be caused by changes in the transcriptional expression of BER genes (Fig. 5A,B,C). Additionally, we note that even though there might be a general decline in new transcription [40], the Tet- STH1depleted cells retain transcriptional competence, transcript abundance and stability during the time period used in our experiments.
To examine the link between RSC activity and BER in more detail, we analysed NMP repair in the well-characterized GAL1 gene locus. It has been previously shown that RSC plays a specific role in controlling nucleosome organization at the promoter region of the GAL1 gene. RSC is able to bind the upstream activating sequence (UAS) in the promoter and form an RSC-nucleosome complex, which can position and phase flanking nucleosomes [29, 69]. We found that repair of NMPs at this locus is severely impaired in STH1 mutant cells (Fig. 4A,B). Several studies indicate that loss of the essential subunits of the RSC complex, including STH1, has a major impact on the chromatin structure of promoters. Reduced abundance of STH1 decreases the loss of histone H3 from promoters of heat shock genes [70], and loss of the STH1 protein increases histone density in tRNA genes [40]. Furthermore, mutation in RSC3 results in a dramatic increase of nucleosome occupancy in hundreds of promoters in yeast cells [57]. Finally, RSC function has also been implicated in restricting the spread of heterochromatic silencing in yeast [71]. Consistent with these observations, we observe that global chromatin accessibility to MNase declines in the STH1 mutants (Fig. 6A,B,C). Similarly, studies have shown decreased chromatin accessibility in SWI/SNF and INO80 mutant cells have an associated deficiency of NER [72, 73]. Altered chromatin remodeling and increased chromatin condensation can impede accessibility to damaged DNA, recognition of damaged bases, and recruitment/assembly of BER enzymes. RSC might also directly impact the BER pathway by modulating activity of BER enzymes through protein-protein interactions and modifications, stabilizing DNA repair intermediates and/or acting as a scaffolding complex for the proper assembly and retention of BER machinery at damaged DNA sites.
In summary, the consistency of the slow BER phenotype, transcriptional stability and competence, metabolic competence, cell viability and rapid recovery of STH1 depleted cells under normal growth conditions in both Tet-off and degron systems, suggest a specific role of RSC in BER, rather than a nonspecific (or indirect) effect. Our data highlights the important role of RSC in BER in living cells, and agrees with the recent in vitro studies [38]. Our results reveal an intriguing functional connection between ATP-dependent chromatin remodelling and the efficiency of BER in chromatin of eukaryotic cells.
Supplementary Material
Highlights.
Chromatin remodeling complex RSC is required for efficient BER in intact yeast cells.
RSC promotes BER in chromatin genome-wide and at the inactive GAL1 locus.
RSC regulates DNA accessibility in chromatin.
Novel role of ATP-dependent chromatin remodeling for BER in intact cells.
Acknowledgments
We thank Dr. Bradley Cairns for providing yeast degron strains and anti-STH1 antibodies. We also thank Rithy Meas for help with data analyses, and Kathy Dorgan and Dr. John Hinz for critical evaluation of this manuscript.
FUNDING
This work was made possible by National Institutes of Health grant ES002614 from the National Institute of Environmental Health Sciences (NIEHS). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH.
APPENDIX A SUPPLEMENTARY DATA
Supplementary Data are available at DNA repair online: Supplementary Figures S1–S7.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanawalt PC. Genomic instability: environmental invasion and the enemies within. Mutation Research. 1998;400:117–125. doi: 10.1016/s0027-5107(98)00084-0. [DOI] [PubMed] [Google Scholar]
- 2.Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J, Bird A, Schar P. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 3.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeppesen DK, Bohr VA, Stevnsner T. DNA repair deficiency in neurodegeneration. Progress in neurobiology. 2011;94:166–200. doi: 10.1016/j.pneurobio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lahtz C, Pfeifer GP. Epigenetic changes of DNA repair genes in cancer. Journal of molecular cell biology. 2011;3:51–58. doi: 10.1093/jmcb/mjq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weidenheim KM, Dickson DW, Rapin I. Neuropathology of Cockayne syndrome: Evidence for impaired development, premature aging, and neurodegeneration. Mechanisms of ageing and development. 2009;130:619–636. doi: 10.1016/j.mad.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic acids research. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alseth I, Eide L, Pirovano M, Rognes T, Seeberg E, Bjoras M. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Molecular and cellular biology. 1999;19:3779–3787. doi: 10.1128/mcb.19.5.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blank A, Kim B, Loeb LA. DNA polymerase delta is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:9047–9051. doi: 10.1073/pnas.91.19.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boiteux S, Guillet M. A basic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. The EMBO journal. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley MRK, YW, Wilson DM. Disparity between DNA Base Excision Repair in Yeast and Mammals: Translational Implications. Cancer Res. 2003;63:549–554. [PubMed] [Google Scholar]
- 13.Tan S, Davey CA. Nucleosome structural studies. Current opinion in structural biology. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlick T, Hayes J, Grigoryev S. Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. The Journal of biological chemistry. 2012;287:5183–5191. doi: 10.1074/jbc.R111.305763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinz JM, Rodriguez Y, Smerdon MJ. Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4646–4651. doi: 10.1073/pnas.0914443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odell ID, Barbour JE, Murphy DL, Della-Maria JA, Sweasy JB, Tomkinson AE, Wallace SS, Pederson DS. Nucleosome disruption by DNA ligase III-XRCC1 promotes efficient base excision repair. Molecular and cellular biology. 2011;31:4623–4632. doi: 10.1128/MCB.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hildrestrand GA, Rolseth V, Bjoras M, Luna L. Human NEIL1 localizes with the centrosomes and condensed chromosomes during mitosis. DNA Repair (Amst) 2007;6:1425–1433. doi: 10.1016/j.dnarep.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Okano S, Lan L, Tomkinson AE, Yasui A. Translocation of XRCC1 and DNA ligase IIIalpha from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic acids research. 2005;33:422–429. doi: 10.1093/nar/gki190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lans H, Marteijn JA, Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics & chromatin. 2012;5:4. doi: 10.1186/1756-8935-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nature reviews Molecular cell biology. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green CM, Almouzni G. When repair meets chromatin. First in series on chromatin dynamics. EMBO reports. 2002;3:28–33. doi: 10.1093/embo-reports/kvf005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao Y. Chromatin response to DNA double-strand break damage. Epigenomics. 2011;3:307–321. doi: 10.2217/epi.11.14. [DOI] [PubMed] [Google Scholar]
- 23.Czaja W, Mao P, Smerdon MJ. The Emerging Roles of ATP-Dependent Chromatin Remodeling Enzymes in Nucleotide Excision Repair. International journal of molecular sciences. 2012;13:11954–11973. doi: 10.3390/ijms130911954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 25.Hsu Jm, Huang J, Meluh PB, Laurent BC. The Yeast RSC Chromatin-Remodeling Complex Is Required for Kinetochore Function in Chromosome Segregation. Molecular and cellular biology. 2003;23:3202–3215. doi: 10.1128/MCB.23.9.3202-3215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasten MM, Clapier CR, Cairns BR. SnapShot: Chromatin remodeling: SWI/SNF. Cell. 2011;144:310, e311. doi: 10.1016/j.cell.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Molecular cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaban Y, Ezeokonkwo C, Chung WH, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nature structural & molecular biology. 2008;15:1272–1277. doi: 10.1038/nsmb.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchler NE, Bai L. Chromatin: bind at your own RSC. Current biology: CB. 2011;21:R223–225. doi: 10.1016/j.cub.2011.01.060. [DOI] [PubMed] [Google Scholar]
- 30.Wippo CJ, Israel L, Watanabe S, Hochheimer A, Peterson CL, Korber P. The RSC chromatin remodelling enzyme has a unique role in directing the accurate positioning of nucleosomes. The EMBO journal. 2011;30:1277–1288. doi: 10.1038/emboj.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andress EJ, Holic R, Edelmann MJ, Kessler BM, Yu VP. Dia2 controls transcription by mediating assembly of the RSC complex. PloS one. 2011;6:e21172. doi: 10.1371/journal.pone.0021172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charles GM, Chen C, Shih SC, Collins SR, Beltrao P, Zhang X, Sharma T, Tan S, Burlingame AL, Krogan NJ, Madhani HD, Narlikar GJ. Site-specific acetylation mark on an essential chromatin-remodeling complex promotes resistance to replication stress. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10620–10625. doi: 10.1073/pnas.1019735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chambers AL, Brownlee PM, Durley SC, Beacham T, Kent NA, Downs JA. The two different isoforms of the RSC chromatin remodeling complex play distinct roles in DNA damage responses. PloS one. 2012;7:e32016. doi: 10.1371/journal.pone.0032016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang B, Qiu J, Ratnakumar K, Laurent BC. RSC functions as an early double-strand-break sensor in the cell’s response to DNA damage. Current biology: CB. 2007;17:1432–1437. doi: 10.1016/j.cub.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Molecular and cellular biology. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menoni H, Shukla MS, Gerson V, Dimitrov S, Angelov D. Base excision repair of 8-oxoG in dinucleosomes. Nucleic acids research. 2012;40:692–700. doi: 10.1093/nar/gkr761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 40.Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. The EMBO journal. 2008;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amberg DCBDJ, Strathern JN. Methods in Yeast Genetics. 2005. Cold Spring Harbor Laboratory Press; New York: 2005. [Google Scholar]
- 42.Conconi A, Jager-Vottero P, Zhang X, Beard BC, Smerdon MJ. Mitotic viability and metabolic competence in UV-irradiated yeast cells. Mutat Res. 2000;459:55–64. doi: 10.1016/s0921-8777(99)00057-9. [DOI] [PubMed] [Google Scholar]
- 43.Czaja W, Bespalov VA, Hinz JM, Smerdon MJ. Proficient repair in chromatin remodeling defective ino80 mutants of Saccharomyces cerevisiae highlights replication defects as the main contributor to DNA damage sensitivity. DNA Repair (Amst) 2010;9:976–984. doi: 10.1016/j.dnarep.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bespalov VA, Conconi A, Zhang X, Fahy D, Smerdon MJ. Improved method for measuring the ensemble average of strand breaks in genomic DNA. Environmental and molecular mutagenesis. 2001;38:166–174. doi: 10.1002/em.1068. [DOI] [PubMed] [Google Scholar]
- 45.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic acids research. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S, Waters R, Smerdon MJ. Low- and high-resolution mapping of DNA damage at specific sites. Methods. 2000;22:170–179. doi: 10.1006/meth.2000.1058. [DOI] [PubMed] [Google Scholar]
- 47.Nag R, Gong F, Fahy D, Smerdon MJ. A single amino acid change in histone H4 enhances UV survival and DNA repair in yeast. Nucleic acids research. 2008;36:3857–3866. doi: 10.1093/nar/gkn311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan HY, Trotter KW, Archer TK, Kingston RE. Swapping function of two chromatin remodeling complexes. Molecular cell. 2005;17:805–815. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 49.Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, Trochesset M, Morse D, Krogan NJ, Hiley SL, Li Z, Morris Q, Grigull J, Mitsakakis N, Roberts CJ, Greenblatt JF, Boone C, Kaiser CA, Andrews BJ, Hughes TR. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 50.Campsteijn C, Wijnands-Collin AM, Logie C. Reverse genetic analysis of the yeast RSC chromatin remodeler reveals a role for RSC3 and SNF5 homolog 1 in ploidy maintenance. PLoS genetics. 2007;3:e92. doi: 10.1371/journal.pgen.0030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du J, Nasir I, Benton BK, Kladde MP, Laurent BC. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bryant GO, Prabhu V, Floer M, Wang X, Spagna D, Schreiber D, Ptashne M. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS biology. 2008;6:2928–2939. doi: 10.1371/journal.pbio.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nature genetics. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 54.Li S, Smerdon MJ. Nucleosome structure and repair of N-methylpurines in the GAL1-10 genes of Saccharomyces cerevisiae. The Journal of biological chemistry. 2002;277:44651–44659. doi: 10.1074/jbc.M206623200. [DOI] [PubMed] [Google Scholar]
- 55.Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, Ko CW, van Leenen D, Sameith K, van Hooff SR, Lijnzaad P, Kemmeren P, Hentrich T, Kobor MS, Buratowski S, Holstege FC. The specificity and topology of chromatin interaction pathways in yeast. Molecular cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes & Development. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, Gebbia M, Talukder S, Yang A, Mnaimneh S, Terterov D, Coburn D, Li Yeo A, Yeo ZX, Clarke ND, Lieb JD, Ansari AZ, Nislow C, Hughes TR. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Molecular cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindahl T. New class of enzymes acting on damaged DNA. Nature. 1976;259:64–66. doi: 10.1038/259064a0. [DOI] [PubMed] [Google Scholar]
- 60.Beard BC, Wilson SH, Smerdon MJ. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cole HA, Tabor-Godwin JM, Hayes JJ. Uracil DNA glycosylase activity on nucleosomal DNA depends on rotational orientation of targets. The Journal of biological chemistry. 2010;285:2876–2885. doi: 10.1074/jbc.M109.073544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakanishi S, Prasad R, Wilson SH, Smerdon M. Different structural states in oligonucleosomes are required for early versus late steps of base excision repair. Nucleic acids research. 2007;35:4313–4321. doi: 10.1093/nar/gkm436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez Y, Smerdon MJ. The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes. The Journal of biological chemistry. 2013;288:13863–13875. doi: 10.1074/jbc.M112.441444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Odell ID, Wallace SS, Pederson DS. Rules of engagement for base excision repair in chromatin. Journal of cellular physiology. 2013;228:258–266. doi: 10.1002/jcp.24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amouroux R, Campalans A, Epe B, Radicella JP. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic acids research. 2010;38:2878–2890. doi: 10.1093/nar/gkp1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Molecular cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 67.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annual review of biochemistry. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 68.Koyama H, Itoh M, Miyahara K, Tsuchiya E. Abundance of the RSC nucleosome-remodeling complex is important for the cells to tolerate DNA damage in Saccharomyces cerevisiae. FEBS letters. 2002;531:215–221. doi: 10.1016/s0014-5793(02)03504-4. [DOI] [PubMed] [Google Scholar]
- 69.Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, Hicks J, Bryant GO, Ptashne M. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Erkina TY, Zou Y, Freeling S, Vorobyev VI, Erkine AM. Functional interplay between chromatin remodeling complexes RSC, SWI/SNF and ISWI in regulation of yeast heat shock genes. Nucleic acids research. 2010;38:1441–1449. doi: 10.1093/nar/gkp1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jambunathan N, Martinez AW, Robert EC, Agochukwu NB, Ibos ME, Dugas SL, Donze D. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics. 2005;171:913–922. doi: 10.1534/genetics.105.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang Y, Wang X, Bao S, Guo R, Johnson DG, Shen X, Li L. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17274–17279. doi: 10.1073/pnas.1008388107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao Q, Wang QE, Ray A, Wani G, Han C, Milum K, Wani AA. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. The Journal of biological chemistry. 2009;284:30424–30432. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.