Abstract
Relatively rare variants with a moderate-to-high biological effect may contribute to the genetic predisposition of common disorders. To investigate this for obesity, we performed exome sequencing for 30 young (mean age: 29.7 years) extremely obese Caucasian subjects (mean body mass index: 51.1 kg/m2; m/f = 11/29). Rare variants with a moderate-to-high predicted biological effect were assembled and subjected to functional clustering analysis. It showed that the 55 clustered protocadherin genes on chromosome 5q31 have a significantly (P = 0.002) higher frequency of rare variants than a set of 325 reference genes. Since the protocadherin genes are expressed in the hypothalamus, we tested another 167 genes related to the function of the hypothalamus, but in those genes, the frequency of rare variants was not different from that of the reference genes. To verify the relation of variation in the protocadherin genes with extreme obesity, we analyzed data from more than 4,000 European Americans present on the Exome Variant Server, representing a sample of the general population. The significant enrichment of rare variants in the protocadherin genes was only observed with the group of extremely obese individuals but not in the “general population”, indicating an association between rare variants in the protocadherin cluster genes and extreme obesity.
Keywords: Extreme obesity, Genetic variation, Functional clustering analysis, Protocadherins, Neuronal plasticity
Introduction
Common traits and diseases in humans often have a considerable heritability indicating that genetic factors account for a major part of their variance. For obesity, the heritability is about 0.4–0.7 (Berndt et al. 2013), whereas also for obesity-related parameters like body mass index (BMI), it has been shown that the variance is to a significant extent genetically determined (Farooqi and O’Rahilly 2007). Two types of studies are intensively performed to find the genetic factors: detailed analysis of candidate genes in relatively severe, early onset cases, and genome-wide association studies (GWAS) with large cohorts of overweight and normal weight subjects. Looking at single nucleotide polymorphisms (SNPs) and copy number variants (CNVs), the GWAS approach has led already to the detection of 56 different loci (Wheeler et al. 2013). According to this study, the risk contribution of each locus may differ between phenotypically different groups, i.e., with more or less severe obesity.
Remarkably, all the variation detected by GWAS together with the highly penetrant single gene mutations so far explains only a small fraction of the genetic background of obesity (Speliotes et al. 2010; Loos 2012). This led scientists to speculate on the nature of the missing variation such as CNVs and epigenetic influences. Based on our previous research on neural tube defects, we, as others, proposed a contribution of relatively rare genetic variants with a moderate-to-high biological effect (Mariman 2009; Manolio et al. 2009). The low population frequency and incomplete penetrance of these variants prevent their identification by association and linkage studies. Persons with the same phenotypic outcome, in this case obesity, may have largely different sets of such rare predisposing variants. Yet part of this variation may occur in genes of the same functional cluster or belong to particular pathways. According to this assumption, we used exome sequencing and functional clustering analysis on a selected cohort of extremely obese subjects and compared the outcome to available population data to try to identify sets of genes enriched for rare variation potentially involved in predisposing to extreme obesity.
Methods and procedures
Subjects
Subjects belonged to a cohort of 561 obese people, who were referred by their family doctor to a private obesity clinic (CO-EUR center for obesity in Europe, http://www.co-eur.eu/) for advice on lifestyle. Thrity extremely obese Caucasian subjects (19 females, 11 males) were selected from this cohort based on a relatively high body mass index (BMI, average 51.1 kg/m2, range 45.3–65.1 kg/m2) and relatively young age (29.7 years, range 19–40.4 years). Eight of them had reported overweight during childhood (average BMI 54.2 kg/m2), but for the others such information was lacking. Informed consent for genetic studies was obtained from all participants, and permission was granted by the ethical committee of Maastricht University Medical Center.
DNA isolation and sequencing
Genomic DNA was isolated from peripheral blood leukocytes using the QIAamp DNA blood kit from Qiagen (Amsterdam, The Netherlands) and was then outsourced for exome sequencing in a CLIA-certified laboratory (EdgeBio, http://www.edgebio.com/). The Nimblegen capture kit was used, followed by sequencing on the Ilumina HiSeq2000. Sequence results were compared with GRCh37 (b37) as the reference genome (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/). For every subject, data were returned as a SNP-file and an indel-file (insertion/deletion). For every variant, information was provided including genetic data (chromosome, cytoband, reference position, gene, zygosity, rs-number, alleles, etc.), quality information on the sequencing (quality score, depth), and information on predicted biological effect by various methods [SNPeffect (Reumers et al. 2005), SIFT score (http://sift.jcvi.org/), Polyphen2 class and score (http://genetics.bwh.harvard.edu/pph2/), etc.].
Selection of rare variants
From the files with SNPs and indels (insertion/deletion), rare genetic variants with a moderate-to-high predicted biological effect were first selected on the absence of an rs-number and a read depth of at least 20. Next, from the remaining list relevant SNPs were selected as follows: either a high biological impact based on SNPeffect or a SIFT score of 0.06 or less in case of a missense variant; relevant indels were selected in the following way: either a high biological impact, or a codon deletion or in-frame insertion that was marked as moderate impact. Absence of rs-number as a marker for low frequency was checked by searching variants in the Exome Variant Server (EVS, http://evs.gs.washington.edu/EVS/). Of 50 randomly chosen variants for which there was no rs-number indicated by EdgeBio, only two were found in the EVS with an rs-number and an allele frequency of 1/8,586 and 1/8,600, respectively, indicating that our assumption was correct.
Functional clustering analysis
We used the online Panther program (www.pantherdb.org) to analyze the functions of the genes carrying the rare variants. In this program, a gene list is mapped to selected categories/pathways showing representation of molecular functions, biological processes, cellular components, protein class, or biological pathways. We mapped our genes to the biological pathways, which were sorted by the number of mapped genes. The top-ranked pathways were checked manually to search for large gene clusters.
Background variant hit score
In order to assess the basal number of rare variants occurring in a gene, the VeryGene database of tissue- and organ-specific genes (www.verygene.com) was used to extract 325 lung/kidney genes that to our knowledge do not have a relation to obesity. Screening of all 30 persons for rare variants in those 325 genes resulted in 24 hits putting the background hit score at 1/406 variants per gene per person.
Statistics
Statistically significant enrichment was calculated by the Chi-square method with Yates correction (http://www.graphpad.com/quickcalcs/contingency1/).
Population data
For investigating population data from the EVS, missense variants in European Americans with a Polyphen2 class other than ‘benign’ were selected. As we noticed that due to the overlap of transcripts some of the variants were called for more than one gene of the protocadherin cluster, identical repeats of variants were deleted from the final variant list.
Results
Number of rare variants
On average 107,279 variants were detected per obese subject, composed of 98,352 SNPs and 8,927 indels. Of those, 2,693 were without an rs-number and had a read-depth of at least 20. After selection on biological impact or SIFT score as described above, on average 109 (0.1 %) SNP and indel variants per person remained with a total of 3,301 hits in the 30 obese subjects.
The total number of 3,301 hits was present in 1,761 different genes. A total of 1,467 genes were hit only once. Fifty-five genes were carrying variation in more than six subjects. Two variants were detected in all 30 persons, which might be accounted for by (local) polymorphisms, locally fixed alleles differing from the reference genome or sequence errors.
Functional clustering analysis
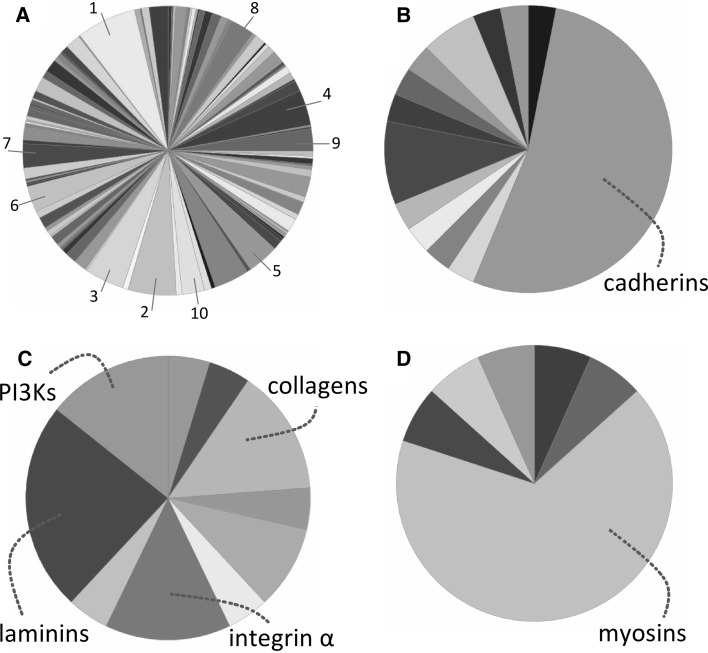
The total number of 1,761 genes with rare variants was analyzed by the Panther program as described in the Methods section. The Wnt signaling pathway scored highest (Fig. 1a; Table 1). Looking in more detail, it appeared that cadherin genes accounted for more than half of the genes carrying the rare variants in the Wnt signaling pathway (Fig. 1b). Focusing on the cadherin genes revealed that 15 variants occurred in 12 of the 55 genes of the human protocadherin cluster (PCDH) on chromosome 5q31 (Table 2) (Wu and Maniatis 2000). Compared with the background hit frequency, the protocadherin cluster displays a significant enrichment (P = 0.0002) of relatively rare genetic variants with a moderate-to-high predicted biological effect in the extremely obese subjects. This cluster was also responsible for the high ranking of the cadherin signaling pathway in the Panther analysis (Table 1). We also found another two clusters in the top ten-ranked pathways, namely the laminin genes as part of the integrin signaling pathway and the myosin genes as part of the nicotinic acetylcholine receptor signaling pathway (Fig. 1c, d). In the 30 subjects, we observed 6 variants in 4 of the 12 human laminin genes (P < 0.0001) and 12 variants in 9 of the 37 human myosin genes (P < 0.0001). The identity of the variants is shown in Table 3. In these three groups of genes, rare variants were detected in both females and males without significant differences between the sexes.
Fig. 1.
Functional clustering analysis of genes with selected rare variants. a Pie chart of the top-ranked pathways with numbers referring to the names of the pathways as indicated in Table 1. b Pie chart of pathway 1 in a, the Wnt signaling pathway. c Pie chart of pathway 3 in a, the integrin signaling pathway. d Pie chart of pathway 6 in a, the nicotinic acetylcholine receptor signaling pathway
Table 1.
Pathways with the highest number of genes carrying rare variants with a high-to-moderate predicted biological effect ranked by Panther
| Pathway | Genes | |
|---|---|---|
| 1 | Wnt signaling | 32 |
| 2 | Inflammation mediated by chemokine and cytokine signaling | 26 |
| 3 | Integrin signaling pathway | 21 |
| 4 | Cadherin signaling pathway | 19 |
| 5 | Gonadotropin releasing hormone receptor | 17 |
| 6 | Nicotinic acetylcholine receptor signaling | 14 |
| 7 | PDGF signaling | 13 |
| 8 | Angiogenesis | 13 |
| 9 | Cytoskeletal regulation by Rho GTPase | 12 |
| 10 | Huntington disease | 12 |
Table 2.
Rare variants in the clustered protocadherin genes
| Gene | Subject code | Variant | Protein | SIFT/SNPeffect/Indel impact | Polyphen2/SNPeffect/Indel impact |
|---|---|---|---|---|---|
| PCDHA6 | 698 | GAC/CAC | D376H | 0.00 | D |
| PCDHA8 | 869 | –/−65 | High | High | |
| PCDHA12 | 841 | GAA/GAC | E253D | 0.00 | D |
| PCDHB2 | 975 | AAC/AACATCACC | N420NIT | Moderate | Moderate |
| PCDHB3 | 692 | –/−577 | High | High | |
| PCDHB4 | 1321 | GAG/GGG | E648G | 0.00 | P |
| PCDHB8 | 816 | –/ACAGAGACACC | −494TET (?) | High | High |
| PCDHB10 | 698 | GAC/GAG | D443E | 0.00 | D |
| PCDHB14 | 698 | CCC/TCC | P449S | 0.00 | D |
| PCDHB16 | 698 | GGC/AGC | G665S* | 0.45 | P |
| PCDHGA8 | 709 | GGT/GTT | G70 V | 0.01 | D |
| 852 | TCC/CCC | S179P | 0.06 | D | |
| PCDHGB1 | 816 | CAT/CGT | H755R* | 0.04 | B |
| 1255 | CGA/CCA | R89P | 0.00 | D | |
| PCDHGB7 | 588 | CCC/– | P679− | Moderate | Moderate |
| 629 | –/−105 | High | High |
(?) Unable to make the exact prediction for the protein
* Variant scored only by SIFT or by Polyphen2
Table 3.
Rare variants in the laminin and myosin genes
| Gene | Subject code | Variant | Protein | SIFT/SNP effect/Indel impact |
|---|---|---|---|---|
| Laminins | ||||
| LAMA2 | 633 | GGT/GTT | G420V | 0.04 |
| LAMB3 | 692 | CGC/CAC | R1143H | 0.05 |
| LAMC1 | 841 | GCT/ACT | A610T | 0.02 |
| LAMC3 | 628 | TTT/TTG | F794L | 0.01 |
| 1255 | CCC/CTC | P382L | 0.00 | |
| 1321 | CGC/CTC | R311L | 0.04 | |
| Myosins | ||||
| MYH4 | 836 | GAG/− | E1149− | Moderate |
| 869 | –/A | −990(?) | High | |
| MYH7B | 709 | ACA/CCA | T1824P | 0.06 |
| MYH11 | 633 | AAG/− | K1386− | Moderate |
| 692 | GCA/GTA | A1896V | 0.01 | |
| 836 | GGC/TGC | G1125C | 0.00 | |
| MYH15 | 1003 | TGG/TGA | W503stop | High |
| MYO1F | 1003 | CGT/TGT | R798C | 0.03 |
| MYO1H | 612 | GAG/TAG | E455stop | High |
| MYO7A | 811 | CGC/CAC | R900H | 0.05 |
| MYO10 | 975 | GTG/ATG | V1468 M | 0.00 |
| MYO18A | 1255 | Splice donor | High | |
(?) Unable to make the exact prediction for the protein
Variants in the genome arise by mutation, and the chance that a mutation occurs is a function of the length of the DNA, i.e., the number of nucleotides. As such, the chance to find a variant depends in general on the length of the DNA that is examined. When comparing genes, a correction for difference in length should be made. Therefore, we corrected our data for the length of the protein coding region of the genes based on UniProt data (www.uniprot.org). The average coding length of the lung/kidney genes was 1,641 nucleotides (547 aminoacids), which is close to the reported average length (509.5 aminoacids) of human proteins (Sakharkar et al. 2006). When taking the coding length into account, the results for both the laminin and myosin genes were no longer significant (P = 0.16, 0.29, respectively). However, the enrichment of the protocadherin cluster remained significant (P = 0.002).
The pathway with the second highest number of genes carrying rare variants in Table 1 is the inflammation mediated by chemokine and cytokine signaling pathway. Obesity is associated with chronic inflammation and is sometimes regarded as an inflammatory disorder (Bluher 2008). However, no substantial functional cluster was indicated by the Panther program for this pathway. Most of the genes that carried rare variants and caused the ranking of this pathway had more general functions overlapping with other pathways like protein kinases (CAMK2D, CAMK2G, PAK6, PTK2B, TYK2) and genes involved in inositol triphosphate signaling (ITPR1, PLCB1). Yet, genes with an obvious relation to inflammation were also present like the cytokine fractalkine (CX3CL1), the chemokine receptors C5AR1 and CCR8, and the nuclear factor NFATC4 that in T-cells induces IL2 and IL4 but also may promote adipogenesis (Yang et al. 2002).
Comparison to other hypothalamus-related genes
The high expression of the clustered protocadherins in the hypothalamus prompted us to analyze other genes expressed in or functionally related to the hypothalamus. A list of 167 hypothalamus genes was composed from VeryGene data, specific literature (St-Amand et al. 2011), and by text mining of titles and abstracts of PubMed articles selected on ‘obesity’ and ‘hypothalamus’. In the extreme obese subjects, 14 rare variants selected by our protocol were scored in those genes, which had an average length of 1,626 nucleotides (542 aminoacids). We found that hypothalamus-related genes other than the clustered protocadherin genes do not display an increased frequency of rare variants (P = 0.63).
Comparison with a general population
To compare the frequency of rare variants with a moderate-to-high predicted biological effect in the protocadherin cluster of extremely obese people with that of a general population, we decided to perform a similar analysis on the European American population using data from the EVS focusing on missense variants in approximately 4,300 subjects.
In EVS, biological effect of variants is based on Polyphen2 analysis instead of SIFT. Although there is a moderate correlation between those methods (Wei et al. 2011), we decided to first redo the analysis in our extreme obese cohort for the protocadherin genes. For this, we defined the rare variants using a Polyphen2 class other than ‘benign’ instead of a SIFT score of 0.06 or less in case of a missense variant. The other criteria for selection were the same as aforementioned. In this way, 15 hits were scored in the protocadherin genes (Table 2) and 28 hits in the reference genes. This showed as before a significant enrichment of rare variants in the protocadherin gene cluster (P = 0.01). Taking only the missense variants in account, the analysis with Polyphen2 still resulted in a significant enrichment (P = 0.05).
In the general population, the rare variants in the missense dataset were selected based on an allele frequency of no more than 2/8,600 and a non-benign Polyphen2 class. The 55 protocadherin genes on chromosome 5q31 scored 1,068 rare non-benign missense variants, whereas in the lung/kidney reference genes, 4,347 rare non-benign missense variants were scored. It shows that in the general population, we found a 1.14-fold lower abundance of rare missense variants in the protocadherin cluster (P < 0.001) in comparison with the reference genes, whereas in extremely obese subjects, there was a 1.9-fold higher (analyzed with Polyphen2 class) or 2.5-fold higher (analyzed with SIFT score) abundance of such variants in the protocadherin genes (P = 0.05, 0.007, respectively).
Discussion
Exome sequencing is a new powerful tool for studying genetic variation in common traits and disorders. Recently, it was shown by this approach that coding variants with a minor allele frequency of >1 % do not have a strong effect on metabolic traits including obesity, BMI, and waist circumference (Albrechtsen et al. 2013). Here, we looked into the relevance of rare variants with an allele frequency of (far) <1 %. Exome sequencing of the DNA of extreme obese subjects followed by functional clustering analysis revealed a significantly higher than expected frequency of relatively rare variants with a moderate-to-high predicted biological effect in the clustered protocadherin genes on 5q31. These protocadherin genes have attracted attention because of their peculiar genomic organization with the genes in the α- and γ-subclusters sharing constant exons coding for the cytoplasmic part of the proteins (Wu and Maniatis 2000). The genes are expressed in the central nervous system, some of which in a monoallelic and combinatorial manner (Esumi et al. 2005), and probably function in (re)forming neuronal circuitries (Lefebvre et al. 2012). Clustered protocadherins have been shown to be necessary for the development of the central nervous system in mice during the late-embryonic stage (Wang et al. 2002).
Although it was found that 11 % of Europeans carry a 16.7-Kb deletion in the alpha-cluster without an apparent phenotype (Noonan et al. 2003), an involvement in obesity was not examined. Our observations do point to a link of protocadherin genes with (extreme) obesity.
Interestingly, a GWAS study (www.ashg.org/2009meeting/abstracts/fulltext/f10729.htm) also showed the strongest association between extreme obesity (BMI > 35 kg/m2) and the protocadherin cluster. Besides genetic association, functional evidence for involvement of the protocadherin gene cluster in obesity also supports our observation. A conditional knockout of the protocadherin-gamma subcluster in the mouse resulting in the absence of gamma-protocadherins in hypothalamic neurons was accompanied by hyperphagia and obesity as the phenotype (Su et al. 2010). Since we did not find that hypothalamus-related genes other than the clustered protocadherin genes displayed an increased frequency of rare variants, we suspect that a role of the protocadherin cluster in extreme obesity might go beyond weight regulation through the hypothalamus.
Interesting information further linking the protocadherin cluster to obesity is provided by the observation that obesity is associated with the restless leg syndrome and both are associated with reduced dopamine activity in the central nervous system (Gao et al. 2009). The restless leg syndrome was diagnosed in about 5 % of adults with a 42 % higher risk for the obese, especially with a high BMI in early adulthood. Linkage analysis with exome sequencing in a German family with the restless leg syndrome revealed a heterozygous Ser584Pro mutation in the PCDHA3 gene co-segregating with the disorder (Lod score 1.8) (Weissbach et al. 2012). Two additional rare heterozygous missense variants in this gene were detected in 64 non-related patients (3.1 %) and only one in 250 healthy controls (0.4 %).
Data are accumulating that the protocadherin cluster genes are subject to epigenetic regulation. The protocadherin cluster is known to change its methylation status in tumors (Dallosso et al. 2009), and recently, it was shown that the methylation status of this cluster in adults is associated with early-life socioeconomic status (Borghol et al. 2012). Notably, several studies have shown that childhood socioeconomic status is inversely related with long-term weight gain and adult BMI, particularly in women (Baltrus et al. 2007; Giskes et al. 2008; Gustafsson et al. 2012). Moreover, it was observed that in the mouse the Smchd1 gene, which is involved in silencing clusters of genes on the inactive X-chromosome, was also important for silencing genes from the alpha- and beta-protocadherin subclusters as well as a cluster of four imprinted genes in the Prader–Willi syndrome locus, a well-known hyperphagia syndrome (Gendrel et al. 2013).
It should be noted that the present study may have some limitations. One limitation might be the relatively small cohort size of 30 extreme obese subjects. On the other hand, those subjects enter 3,300 clustered protocadherin gene-copies into the analysis. Secondly, genetic differences between the Dutch and European American population will occur and also the reference genome may be of influence on the analysis. In order to minimize the effect of population differences, we have compared the hit frequency of the protocadherin cluster genes with that of a set of reference genes within each population. Finally, the way in which we used the Panther program might favor the detection of relatively large functional gene clusters. Therefore, we may have missed other genes of which the genetic variation could be involved in extreme obesity. Nonetheless, our observation is in line with other genetic and functional findings pointing to a role of the protocadherin cluster in the development of (extreme) obesity.
Although the laminin and myosin genes were rendered statistically non-significant after correcting for length of the coding region, they remain interesting candidates for further research into a central role in extreme obesity, since genes predisposing to disease are known to be relatively large on average (Sakharkar et al. 2006). SNPs in the LAMA5 gene have been shown to be associated with adiposity parameters in European and African Americans (De Luca et al. 2008). Intriguingly, four of the six variants that we detected in the laminin genes, are present in the genes coding for LAMC1 and LAMC3, which have been shown to interact with NTN4 (Schneiders et al. 2007), of which the gene also has a rare variant (R225H, SIFT score = 0.00) in another of the investigated subjects. As such, five of the 30 extreme obese subjects have a dramatic variant in a gene for this laminin-netrin complex. Both laminins and netrins play critical roles in guiding the growth of peripheral and central axons [refs in (Yin et al. 2002)]. Also, MYO10 has been shown to mediate laminin-induced growth of neurites after peripheral nerve injury (Plantman et al. 2013), whereas laminin with myosin II is able to mediate axon branching (Liu et al. 2013). Together with what is known about the function of the PCDH-genes, this suggests that rare variants with a moderate-to-high biological effect have an impact on central nervous system development and function. Recently, Wheeler et al. examined 490 genes of which the coding part was affected by an obesity-specific deletion (Wheeler et al. 2013). Functional enrichment analysis of those genes revealed “Nervous System Development and Function” as the most significant geneset with as the most important subsets: neuritogenesis, development of brain, synaptic transmission, and synaptic transmission of neurons. It seems that our findings and those of Wheeler point to the same predisposing mechanism for obesity with a genetic impediment of brain development and function.
Knocking out the protocadherin gamma subcluster leads to a change in eating behavior in the mouse (Su et al. 2010). It suggests that rare variants in the PCDH-genes may also have an impact on food intake in humans. However, so far, no research has been done in this direction. On the other hand, inter-individual differences in food intake are undoubtedly influenced by genetic variation as for instance by polymorphisms of taste receptor genes and mutations in genes regulating feelings of hunger and satiety (Grimm and Steinle 2011; Mariman 2009). The latter refers to homozygous or compound heterozygous cases of early onset obesity, but evidence is accumulating that heterozygous mutations in those genes can influence food intake as well. In a three-generation family, a heterozygous mutation in the pro-opiomelanocortin gene (POMC) was found to segregate and all carriers had hyperphagia and obesity but no red hair or adrenal impairment, which are characteristic for homozygous POMC mutations (Challis et al. 2002). Recently, an intermediate-frequency variant of the leptin receptor gene (LEPR) was found by GWAS among subjects with severe early onset obesity. This variant in monocytes is associated with lower LEPR expression (Wheeler et al. 2013). It shows that merely the fact that all variants reported here are heterozygous, does not rule out an effect on the phenotype, presumably on food intake. Hopefully, our results will initiate profound research into the role of PCDH-genes in food intake. Genetic variation in those genes may then be used to identify persons at risk for overeating, allowing them to receive proper guidance to prevent severe overweight and associated health complications.
Here, we have provided data indicating that in extremely obese subjects relatively rare variants with a moderate-to-high predicted biological effect occur with an increased frequency in the protocadherin gene cluster on chromosome 5q31, which is supported by other genetic and functional observations. In addition, also the lamininC1/3-netrin4 complex is a suspected target for such variation with relevance to obesity. Together with reported studies, our findings suggest a role for early life formation or adaptation of neuronal networks in the development of (extreme) obesity. In this respect, it is tempting to speculate that genetically modified neuronal plasticity relates to childhood tracking of eating behavior and body weight (Craigie et al. 2011).
Acknowledgments
Conflict of interest
All authors state that there is no conflict of interest.
References
- Albrechtsen A, Grarup N, Li Y, Sparso T, Tian G, Cao H, Jiang T, Kim SY, Korneliussen T, Li Q, Nie C, Wu R, Skotte L, Morris AP, Ladenvall C, Cauchi S, Stancakova A, Andersen G, Astrup A, Banasik K, Bennett AJ, Bolund L, Charpentier G, Chen Y, Dekker JM, Doney AS, Dorkhan M, Forsen T, Frayling TM, Groves CJ, Gui Y, Hallmans G, Hattersley AT, He K, Hitman GA, Holmkvist J, Huang S, Jiang H, Jin X, Justesen JM, Kristiansen K, Kuusisto J, Lajer M, Lantieri O, Li W, Liang H, Liao Q, Liu X, Ma T, Ma X, Manijak MP, Marre M, Mokrosinski J, Morris AD, Mu B, Nielsen AA, Nijpels G, Nilsson P, Palmer CN, Rayner NW, Renstrom F, Ribel-Madsen R, Robertson N, Rolandsson O, Rossing P, Schwartz TW, Group DESIRS, Slagboom PE, Sterner M, Consortium D, Tang M, Tarnow L, Tuomi T, van’t Riet E, van Leeuwen N, Varga TV, Vestmar MA, Walker M, Wang B, Wang Y, Wu H, Xi F, Yengo L, Yu C, Zhang X, Zhang J, Zhang Q, Zhang W, Zheng H, Zhou Y, Altshuler D, t Hart LM, Franks PW, Balkau B, Froguel P, McCarthy MI, Laakso M, Groop L, Christensen C, Brandslund I, Lauritzen T, Witte DR, Linneberg A, Jorgensen T, Hansen T, Wang J, Nielsen R, Pedersen O (2013) Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia 56(2):298–310. doi:10.1007/s00125-012-2756-1 [DOI] [PMC free article] [PubMed]
- Baltrus PT, Everson-Rose SA, Lynch JW, Raghunathan TE, Kaplan GA. Socioeconomic position in childhood and adulthood and weight gain over 34 years: the Alameda County Study. Ann Epidemiol. 2007;17(8):608–614. doi: 10.1016/j.annepidem.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt SI, Gustafsson S, Magi R, Ganna A, Wheeler E, Feitosa MF, Justice AE, Monda KL, Croteau-Chonka DC, Day FR, Esko T, Fall T, Ferreira T, Gentilini D, Jackson AU, Luan J, Randall JC, Vedantam S, Willer CJ, Winkler TW, Wood AR, Workalemahu T, Hu YJ, Lee SH, Liang L, Lin DY, Min JL, Neale BM, Thorleifsson G, Yang J, Albrecht E, Amin N, Bragg-Gresham JL, Cadby G, den Heijer M, Eklund N, Fischer K, Goel A, Hottenga JJ, Huffman JE, Jarick I, Johansson A, Johnson T, Kanoni S, Kleber ME, Konig IR, Kristiansson K, Kutalik Z, Lamina C, Lecoeur C, Li G, Mangino M, McArdle WL, Medina-Gomez C, Muller-Nurasyid M, Ngwa JS, Nolte IM, Paternoster L, Pechlivanis S, Perola M, Peters MJ, Preuss M, Rose LM, Shi J, Shungin D, Smith AV, Strawbridge RJ, Surakka I, Teumer A, Trip MD, Tyrer J, Van Vliet-Ostaptchouk JV, Vandenput L, Waite LL, Zhao JH, Absher D, Asselbergs FW, Atalay M, Attwood AP, Balmforth AJ, Basart H, Beilby J, Bonnycastle LL, Brambilla P, Bruinenberg M, Campbell H, Chasman DI, Chines PS, Collins FS, Connell JM, Cookson WO, de Faire U, de Vegt F, Dei M, Dimitriou M, Edkins S, Estrada K, Evans DM, Farrall M, Ferrario MM, Ferrieres J, Franke L, Frau F, Gejman PV, Grallert H, Gronberg H, Gudnason V, Hall AS, Hall P, Hartikainen AL, Hayward C, Heard-Costa NL, Heath AC, Hebebrand J, Homuth G, Hu FB, Hunt SE, Hypponen E, Iribarren C, Jacobs KB, Jansson JO, Jula A, Kahonen M, Kathiresan S, Kee F, Khaw KT, Kivimaki M, Koenig W, Kraja AT, Kumari M, Kuulasmaa K, Kuusisto J, Laitinen JH, Lakka TA, Langenberg C, Launer LJ, Lind L, Lindstrom J, Liu J, Liuzzi A, Lokki ML, Lorentzon M, Madden PA, Magnusson PK, Manunta P, Marek D, Marz W, Mateo Leach I, McKnight B, Medland SE, Mihailov E, Milani L, Montgomery GW, Mooser V, Muhleisen TW, Munroe PB, Musk AW, Narisu N, Navis G, Nicholson G, Nohr EA, Ong KK, Oostra BA, Palmer CN, Palotie A, Peden JF, Pedersen N, Peters A, Polasek O, Pouta A, Pramstaller PP, Prokopenko I, Putter C, Radhakrishnan A, Raitakari O, Rendon A, Rivadeneira F, Rudan I, Saaristo TE, Sambrook JG, Sanders AR, Sanna S, Saramies J, Schipf S, Schreiber S, Schunkert H, Shin SY, Signorini S, Sinisalo J, Skrobek B, Soranzo N, Stancakova A, Stark K, Stephens JC, Stirrups K, Stolk RP, Stumvoll M, Swift AJ, Theodoraki EV, Thorand B, Tregouet DA, Tremoli E, Van der Klauw MM, van Meurs JB, Vermeulen SH, Viikari J, Virtamo J, Vitart V, Waeber G, Wang Z, Widen E, Wild SH, Willemsen G, Winkelmann BR, Witteman JC, Wolffenbuttel BH, Wong A, Wright AF, Zillikens MC, Amouyel P, Boehm BO, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Cupples LA, Cusi D, Dedoussis GV, Erdmann J, Eriksson JG, Franks PW, Froguel P, Gieger C, Gyllensten U, Hamsten A, Harris TB, Hengstenberg C, Hicks AA, Hingorani A, Hinney A, Hofman A, Hovingh KG, Hveem K, Illig T, Jarvelin MR, Jockel KH, Keinanen-Kiukaanniemi SM, Kiemeney LA, Kuh D, Laakso M, Lehtimaki T, Levinson DF, Martin NG, Metspalu A, Morris AD, Nieminen MS, Njolstad I, Ohlsson C, Oldehinkel AJ, Ouwehand WH, Palmer LJ, Penninx B, Power C, Province MA, Psaty BM, Qi L, Rauramaa R, Ridker PM, Ripatti S, Salomaa V, Samani NJ, Snieder H, Sorensen TI, Spector TD, Stefansson K, Tonjes A, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Vollenweider P, Wallaschofski H, Wareham NJ, Watkins H, Wichmann HE, Wilson JF, Abecasis GR, Assimes TL, Barroso I, Boehnke M, Borecki IB, Deloukas P, Fox CS, Frayling T, Groop LC, Haritunian T, Heid IM, Hunter D, Kaplan RC, Karpe F, Moffatt MF, Mohlke KL, O’Connell JR, Pawitan Y, Schadt EE, Schlessinger D, Steinthorsdottir V, Strachan DP, Thorsteinsdottir U, van Duijn CM, Visscher PM, Di Blasio AM, Hirschhorn JN, Lindgren CM, Morris AP, Meyre D, Scherag A, McCarthy MI, Speliotes EK, North KE, Loos RJ, Ingelsson E (2013) Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet 45(5):501–512. doi:10.1038/ng.2606 [DOI] [PMC free article] [PubMed]
- Bluher M. The inflammatory process of adipose tissue. Pediatr Endocrinol Rev PER. 2008;6(1):24–31. [PubMed] [Google Scholar]
- Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M, Hertzman C, Power C, Szyf M. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2012;41(1):62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis BG, Pritchard LE, Creemers JW, Delplanque J, Keogh JM, Luan J, Wareham NJ, Yeo GS, Bhattacharyya S, Froguel P, White A, Farooqi IS, O’Rahilly S. A missense mutation disrupting a dibasic prohormone processing site in pro-opiomelanocortin (POMC) increases susceptibility to early-onset obesity through a novel molecular mechanism. Hum Mol Genet. 2002;11(17):1997–2004. doi: 10.1093/hmg/11.17.1997. [DOI] [PubMed] [Google Scholar]
- Craigie AM, Lake AA, Kelly SA, Adamson AJ, Mathers JC. Tracking of obesity-related behaviours from childhood to adulthood: a systematic review. Maturitas. 2011;70(3):266–284. doi: 10.1016/j.maturitas.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Dallosso AR, Hancock AL, Szemes M, Moorwood K, Chilukamarri L, Tsai HH, Sarkar A, Barasch J, Vuononvirta R, Jones C, Pritchard-Jones K, Royer-Pokora B, Lee SB, Owen C, Malik S, Feng Y, Frank M, Ward A, Brown KW, Malik K. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet. 2009;5(11):e1000745. doi: 10.1371/journal.pgen.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Chambers MM, Casazza K, Lok KH, Hunter GR, Gower BA, Fernandez JR. Genetic variation in a member of the laminin gene family affects variation in body composition in Drosophila and humans. BMC Genet. 2008;9:52. doi: 10.1186/1471-2156-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat Genet. 2005;37(2):171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- Farooqi IS, O’Rahilly S. Genetic factors in human obesity. Obes Rev. 2007;8(Suppl 1):37–40. doi: 10.1111/j.1467-789X.2007.00315.x. [DOI] [PubMed] [Google Scholar]
- Gao X, Schwarzschild MA, Wang H, Ascherio A. Obesity and restless legs syndrome in men and women. Neurology. 2009;72(14):1255–1261. doi: 10.1212/01.wnl.0000345673.35676.1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Tang YA, Suzuki M, Godwin J, Nesterova TB, Greally JM, Heard E, Brockdorff N. Epigenetic functions of smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol Cell Biol. 2013;33(16):3150–3165. doi: 10.1128/MCB.00145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giskes K, van Lenthe FJ, Turrell G, Kamphuis CB, Brug J, Mackenbach JP. Socioeconomic position at different stages of the life course and its influence on body weight and weight gain in adulthood: a longitudinal study with 13-year follow-up. Obesity (Silver Spring, Md) 2008;16(6):1377–1381. doi: 10.1038/oby.2008.54. [DOI] [PubMed] [Google Scholar]
- Grimm ER, Steinle NI. Genetics of eating behavior: established and emerging concepts. Nutr Rev. 2011;69(1):52–60. doi: 10.1111/j.1753-4887.2010.00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson PE, Persson M, Hammarstrom A. Socio-economic disadvantage and body mass over the life course in women and men: results from the Northern Swedish Cohort. Eur J Pub Health. 2012;22(3):322–327. doi: 10.1093/eurpub/ckr061. [DOI] [PubMed] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488(7412):517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xing S, Yuan B, Zheng W, Jiang X (2013) Change of laminin density stimulates axon branching via growth cone myosin II-mediated adhesion. Integr Biol Quant Biosci Nano Macro. doi:10.1039/c3ib40131f [DOI] [PubMed]
- Loos RJF. Genetic determinants of common obesity and their value in prediction. Best Pract Res Clin Endocrinol Metab. 2012;26(2):211–226. doi: 10.1016/j.beem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariman EC. Future nutrigenetics: in search of the missing genetic variation. J Nutr Nutr. 2009;2(4–5):257–262. doi: 10.1159/000297212. [DOI] [PubMed] [Google Scholar]
- Noonan JP, Li J, Nguyen L, Caoile C, Dickson M, Grimwood J, Schmutz J, Feldman MW, Myers RM. Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin alpha cluster. Am J Hum Genet. 2003;72(3):621–635. doi: 10.1086/368060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantman S, Zelano J, Novikova LN, Novikov LN, Cullheim S. Neuronal myosin-X is upregulated after peripheral nerve injury and mediates laminin-induced growth of neurites. Mol Cell Neurosci. 2013;56C:96–101. doi: 10.1016/j.mcn.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Reumers J, Schymkowitz J, Ferkinghoff-Borg J, Stricher F, Serrano L, Rousseau F. SNPeffect: a database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acids Res. 2005;33(Database issue):D527–D532. doi: 10.1093/nar/gki086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar MK, Kangueane P, Sakharkar KR, Zhong Z. Huge proteins in the human proteome and their participation in hereditary diseases. In Silico Biol. 2006;6(4):275–279. [PubMed] [Google Scholar]
- Schneiders FI, Maertens B, Bose K, Li Y, Brunken WJ, Paulsson M, Smyth N, Koch M. Binding of netrin-4 to laminin short arms regulates basement membrane assembly. J Biol Chem. 2007;282(33):23750–23758. doi: 10.1074/jbc.M703137200. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proenca C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grassler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jorgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, Konig IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaloy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimaki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Magic, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O’Donnell CJ, O’Rahilly S, Ong KK, Oostra B, Pare G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstrale M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tonjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kahonen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Gronberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Procardis C, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 42(11):937–948. doi:10.1038/ng.686 [DOI] [PMC free article] [PubMed]
- St-Amand J, Yoshioka M, Tanaka K, Nishida Y. Transcriptome-wide identification of preferentially expressed genes in the hypothalamus and pituitary gland. Front Endocrinol. 2011;2:111. doi: 10.3389/fendo.2011.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Marcheva B, Meng S, Liang FA, Kohsaka A, Kobayashi Y, Xu AW, Bass J, Wang X. Gamma-protocadherins regulate the functional integrity of hypothalamic feeding circuitry in mice. Dev Biol. 2010;339(1):38–50. doi: 10.1016/j.ydbio.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Weiner JA, Levi S, Craig AM, Bradley A, Sanes JR. Gamma protocadherins are required for survival of spinal interneurons. Neuron. 2002;36(5):843–854. doi: 10.1016/S0896-6273(02)01090-5. [DOI] [PubMed] [Google Scholar]
- Wei P, Liu X, Fu YX. Incorporating predicted functions of nonsynonymous variants into gene-based analysis of exome sequencing data: a comparative study. BMC Proc. 2011;5(Suppl 9):S20. doi: 10.1186/1753-6561-5-S9-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach A, Siegesmund K, Bruggemann N, Schmidt A, Kasten M, Pichler I, Muhle H, Lohmann E, Lohnau T, Schwinger E, Hagenah J, Stephani U, Pramstaller PP, Klein C, Lohmann K. Exome sequencing in a family with restless legs syndrome. Mov Disord. 2012;27(13):1686–1689. doi: 10.1002/mds.25191. [DOI] [PubMed] [Google Scholar]
- Wheeler E, Huang N, Bochukova EG, Keogh JM, Lindsay S, Garg S, Henning E, Blackburn H, Loos RJ, Wareham NJ, O’Rahilly S, Hurles ME, Barroso I, Farooqi IS. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat Genet. 2013;45(5):513–517. doi: 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Maniatis T. Large exons encoding multiple ectodomains are a characteristic feature of protocadherin genes. Proc Natl Acad Sci USA. 2000;97(7):3124–3129. doi: 10.1073/pnas.97.7.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Xiong Q, Enslen H, Davis RJ, Chow CW. Phosphorylation of NFATc4 by p38 mitogen-activated protein kinases. Mol Cell Biol. 2002;22(11):3892–3904. doi: 10.1128/MCB.22.11.3892-3904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Miner JH, Sanes JR. Laminets: laminin- and netrin-related genes expressed in distinct neuronal subsets. Mol Cell Neurosci. 2002;19(3):344–358. doi: 10.1006/mcne.2001.1089. [DOI] [PubMed] [Google Scholar]