Abstract
Osteoporosis is a common bone disease that has a strong genetic component. Genome-wide linkage studies have identified the chromosomal region 3p14-p22 as a quantitative trait locus for bone mineral density (BMD). We have previously identified associations between variation in two related genes located in 3p14-p22, ARHGEF3 and RHOA, and BMD in women. In this study we performed knockdown of these genes using small interfering RNA (siRNA) in human osteoblast-like and osteoclast-like cells in culture, with subsequent microarray analysis to identify genes differentially regulated from a list of 264 candidate genes. Validation of selected findings was then carried out in additional human cell lines/cultures using quantitative real-time PCR (qRT-PCR). The qRT-PCR results showed significant down-regulation of the ACTA2 gene, encoding the cytoskeletal protein alpha 2 actin, in response to RHOA knockdown in both osteoblast-like (P<0.001) and osteoclast-like cells (P = 0.002). RHOA knockdown also caused up-regulation of the PTH1R gene, encoding the parathyroid hormone 1 receptor, in Saos-2 osteoblast-like cells (P<0.001). Other findings included down-regulation of the TNFRSF11B gene, encoding osteoprotegerin, in response to ARHGEF3 knockdown in the Saos-2 and hFOB 1.19 osteoblast-like cells (P = 0.003–0.02), and down-regulation of ARHGDIA, encoding the Rho GDP dissociation inhibitor alpha, in response to RHOA knockdown in osteoclast-like cells (P<0.001). These studies identify ARHGEF3 and RHOA as potential regulators of a number of genes in bone cells, including TNFRSF11B, ARHGDIA, PTH1R and ACTA2, with influences on the latter evident in both osteoblast-like and osteoclast-like cells. This adds further evidence to previous studies suggesting a role for the ARHGEF3 and RHOA genes in bone metabolism.
Introduction
Osteoporosis is a common and debilitating bone disease that is characterised by a low bone mineral density (BMD), which leads to an increased risk of fracture [1]. The disease is particularly prevalent in postmenopausal women due to a reduction in oestrogen production, with subsequent effects on bone as well as intestinal and renal calcium handling [2]. In addition to the effects of oestrogen, calcium and other environmental factors on bone structure, there is a strong genetic effect on peak bone mass (attained in early adult life), bone loss and fracture rates [3], [4]. Twin and family studies suggest that 50–90% of the variation in peak bone mass [5]–[7] and 25–68% of the variance in osteoporotic fracture is heritable [4], [8], [9]. The genome-wide linkage scanning approach has identified at least 11 replicated quantitative trait loci (QTL) for BMD [10]–[12], including the 3p14-p22 region of the human genome (LOD 1.1–3.5) [11]–[14].
We have previously identified significant associations between variation in the RHOA and ARHGEF3 genes, which are both located within the 3p14-p22 region, and BMD in women [15], [16]. The functions of these genes are related, with the product of the ARHGEF3 gene (the Rho guanine nucleotide exchange factor (GEF) 3) specifically activating two members of the RhoGTPase family: RhoA (encoded for by the RHOA gene) and RhoB [17]. RhoA is involved with regulating cytoskeletal dynamics and actin polymerisation [18] and has been shown to have a role in osteoblast differentiation [19], [20] and osteoclastic bone resorption [21].
Given the associations that we have previously identified between the RHOA and ARHGEF3 genes and BMD, coupled with the evidence in the literature suggesting a role for RhoA in osteoblasts and osteoclasts, we decided to further investigate the role of these genes in these particular cell types. Knockdown of the RHOA and ARHGEF3 genes was achieved using small interfering RNA (siRNA) in a human osteoblast-like cell line and in osteoclast-like cells derived from a donor, with subsequent microarray analysis to identify genes that were differentially regulated. Replication of selected significant findings was then conducted in additional human osteoblast-like cell lines and in osteoclast-like cells from additional donors.
Materials and Methods
Ethics Statement
All subjects that donated blood samples for isolation of peripheral blood mononuclear cells (PBMCs) provided written informed consent and the institutional ethics committee of Curtin University approved the experimental protocol.
Experimental Approach
To identify genes involved in osteoblast and osteoclast function that are potentially influenced by the RHOA and ARHGEF3 genes, we examined the influence of knockdown of these two genes on 264 candidate genes in an osteoblast-like cell line and osteoclast-like cells obtained from a donor, in triplicate, by microarray analysis. The microarray results showed significant alterations in the expression of a number of the candidate genes, 7 of which were studied in greater detail to validate the findings, based on quantitative real-time PCR (qRT-PCR) studies of the 7 genes in 3 additional osteoblast-like and osteoclast-like cell cultures/lines.
Cell Culture
The osteoblast-like cell lines used for the gene knockdown experiments included: Saos-2, derived from osteosarcoma tissue (American Type Culture Collection (ATCC) N° HTB-85) [22]; hFOB 1.19, derived from immortalised foetal osteoblasts (ATCC N° CRL-11372) [23]; and MG-63, derived from osteosarcoma tissue (ATCC N° CRL-1427) [24]. These cell lines are all human in origin and were cultured in DMEM (Sigma-Aldrich, St. Louis, USA) pH 7.4 supplemented with 4.77 g/l HEPES, 3.7 g/l NaHCO3, 10% (v/v) foetal bovine serum (FBS) and 1% (v/v) penicillin/streptomycin (100 units penicillin and 100 µg streptomycin per ml of media). The osteoclast-like cells used in these studies were differentiated from PBMCs (process described below) and were cultured in α-MEM (Invitrogen, Carlsbad, USA) pH 7.4 supplemented with 2.2 g/l NaHCO3, 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin. All cells were cultured at 37°C with 5% CO2 and the medium was changed every 2–3 days. Total RNA was harvested from each culture using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and reverse transcription of the RNA was performed using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). Quantitation of total RNA was performed using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, USA).
Isolation of Peripheral Blood Mononuclear Cells and Osteoclastogenesis
Osteoclast-like cells were differentiated from PBMCs isolated from 4 male donors of European descent aged 48±15 years (mean ± SD). Each batch of cells was isolated from 30 ml whole blood collected in 10 ml K2EDTA Vacutainer tubes (Becton, Dickinson and Company, Franklin Lakes, USA). Anti-coagulated whole blood samples were centrifuged at 2,200 rpm for 10 min at room temperature before buffy coats were collected and diluted to a total volume of 4 ml with 1× phosphate buffered saline (PBS). The cell suspension was then gently layered over 3 ml of Ficoll-Paque (Pfizer, New York, USA) before being centrifuged again at 1,600 rpm for 40 min at room temperature. The PBMC layer was collected and washed by re-suspension in 6 ml 1× PBS and centrifuged at 800 rpm for 10 min at room temperature. The wash step was repeated on the cell pellet before the cells were re-suspended in 5 ml medium supplemented with 10 ng/ml macrophage colony stimulating factor (M-CSF) (Invitrogen, Carlsbad, USA) and seeded directly into either a 24-well tissue culture plate or 25 cm2 tissue culture flask. After two days, the medium was replaced with medium supplemented with 10 ng/ml M-CSF and 100 ng/ml receptor activator of nuclear factor kappa-B ligand (RANKL) (Invitrogen, Carlsbad, USA). The cells were then grown using this medium formulation for 17 days while osteoclastogenesis occurred.
Osteoclast-like cells were stained for tartrate resistant acid phosphatase (TRAP) using a chromogenic TRAP enzyme substrate to confirm production of the TRAP enzyme as an indicator of the osteoclast phenotype. This involved washing the cells with 1× PBS, fixation with 4% (v/v) paraformaldehyde for 15 min, washing 3 times with 1× PBS before incubation with filtered TRAP stain solution at 37°C for 25 min. The stained cells were then washed 3 times with 1× PBS prior to visualisation using light microscopy.
siRNA Knockdown
Transfection of cells with siRNA sequences was used to knockdown expression of the ARHGEF3 and RHOA genes. Transfections were performed using HiPerFect Transfection Reagent (Qiagen, Hilden, Germany). Two different siRNA sequences were used in tandem to knockdown expression of each gene. There is evidence to suggest that the RhoA protein has a half-life of up to 31 h [25], therefore a minimum gene knockdown period of 48 h was used to ensure an effect at the protein level. Negative controls treated with AllStars Negative Control siRNA (Qiagen, Hilden, Germany) were included in each experiment. All knockdown experiments were performed in triplicate. Knockdown of the ARHGEF3 and RHOA genes did not appear to influence the proliferation or viability of any of the cell types studied.
Knockdown in Osteoblast-like Cells
siRNA knockdown experiments were performed in 24-well tissue culture plates. For the Saos-2, hFOB 1.19 and MG-63 osteoblast-like cell lines, each well was seeded with 5×104 cells. Cells were grown for 24 h before fresh medium was added to each culture and transfections were performed using a final siRNA concentration of 30 nM with 6 µL transfection reagent per well. Cells in each well were incubated with the transfection mix for 48 h at 37°C prior to washing with 1× PBS and extraction of total RNA.
Knockdown in Peripheral Blood Mononuclear Cells/Osteoclast-like Cells
500 µL of freshly isolated PBMCs were aliquoted into 24-well tissue culture plates. Osteoclastogenesis was stimulated and confirmed microscopically and biochemically as described previously by TRAP staining. siRNA knockdown experiments were performed using a final siRNA concentration of 100 nM with 6 µL transfection reagent per well. Cells in each well were incubated with the transfection mix for 48 h at 37°C prior to washing with 1× PBS and extraction of total RNA.
RNA Extraction and Microarray Analysis
A total of 18 RNA samples, 9 from Saos-2 and 9 from osteoclast-like cell cultures (donor 1) were used for the microarray analysis. Each set of 9 was comprised of 3 cultures treated with siRNA specific for ARHGEF3, 3 treated with siRNA specific for RHOA and 3 treated with negative control siRNA. Total RNA was extracted from each culture using the RNeasy Mini Kit (Qiagen, Hilden, Germany). The quality and quantity of all RNA samples was checked prior to microarray analysis using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA). 10 µL of each RNA sample was amplified using the TotalPrep RNA Amplification Kit (Applied Biosystems, Foster City, USA) before microarray analysis was performed using the HumanHT-12 v3 Expression BeadChip Kit (Illumina, San Diego, USA). The HumanHT-12 BeadChip profiles the expression of more than 25,000 annotated genes derived from NCBI RefSeq (Build 36.2) [26]. The complete results from the microarray analyses performed in this study have been submitted to The University of Western Australia’s Research Data Online resource.
Gene Selection
While data were generated for most of the >25,000 genes included on the microarray, only 264 candidate genes were selected for statistical analysis in order to limit the potential for false positives. These candidate genes were selected on the basis of the following criteria: genes thought to have potentially important roles in osteoblast (n = 45) or osteoclast function (n = 62), or genes thought to play a role in the RhoA/ARHGEF3 signalling pathway (n = 157).
Quantitative Real-time PCR
qRT-PCR was used to determine the degree of gene knockdown achieved and to validate microarray results for selected targets. Reverse transcription of RNA samples was first performed using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). The resulting cDNA was then amplified using the QuantiFast SYBR Green Kit (Qiagen, Hilden, Germany) in conjunction with an iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, USA). cDNA samples were diluted in 1× TE buffer before analysis. QuantiTect Primer Assays (Qiagen, Hilden, Germany) were used to amplify most gene transcript sequences. Bioinformatics analysis revealed that the QuantiTect Primer Assay for the candidate gene ACTA2 amplifies only one of the two transcript variants for this gene. Therefore, a custom primer pair was designed for this gene using the web-based Primer3 software package [27]. The human 18S ribosomal RNA gene (RRN18S) was selected as an internal reference for this work to allow for normalisation of the data for variations in the quantity of cDNA added to each reaction. The reaction efficiency of each primer pair was calculated by amplifying a 10-fold dilution series of target sequence across 5 orders of magnitude. This was performed to confirm that the amplification efficiency of each gene of interest is no more than 10% from that of the internal reference as recommended by Schmittgen and Livak [28]. The log template dilution (x-axis) was plotted against the cycle threshold (CT) value obtained for each dilution (y-axis) with the slope of the line used for calculation of amplification efficiency using the equation m = −(1/log E), where m is the slope of the line and E is the reaction efficiency. A reaction efficiency of 2.0 equates to a perfect doubling of amplicon product during each PCR cycle. All reactions were performed in triplicate with the mean CT value used in the statistical analysis. Melting-curve analysis was performed on all real-time PCR products to confirm amplification of a single DNA sequence. A random selection of PCR products were also subjected to agarose gel electrophoresis for additional confirmation of the specificity of amplification.
Microarray Statistical Analysis
Differential expression analysis of the microarray data using the Illumina custom error model was performed using the BeadStudio v3.4.0 software package (Illumina, San Diego, USA). Samples treated with the negative control siRNA were specified as the reference group. The raw microarray gene expression data were normalised using the quantile normalisation algorithm [29], which adjusts the sample signals to minimise the influence of variation arising from non-biological factors (eg. pipetting variation) [30]. Background subtraction was performed on the data to minimise the variation in background noise between arrays and to remove signal resulting from non-specific hybridisation [31]. Once background subtraction has been performed on the data, the expected signal for unexpressed targets is zero. The data were corrected for multiple testing using the Benjamini-Hochberg False Discovery Rate algorithm [32].
Real-time PCR Statistical Analysis
Gene expression ratios were calculated using the comparative CT method as described by Schmittgen and Livak [28]. Briefly, the ΔCT (CT of the test gene−CT of the internal reference) was calculated for each gene of interest in each sample in the test and control groups. This figure was then entered into the equation 2−ΔC T with the mean ± standard error calculated for each of the test and control groups. 2−ΔC T values for test and control groups were analysed using an unpaired t-test to determine whether differences in expression were statistically significant. Combined 2−ΔC T values for the osteoclast-like cells were examined by 2-way analysis of variance (ANOVA) (note that this combined analysis was not performed for the osteoblast-like cells due to potential variation in the maturation state and gene expression profile of each cell line). Significant associations are defined as P<0.05.
Results
Osteoblast Microarray Results
Knockdown of the ARHGEF3 and RHOA genes was validated in the Saos-2 cells by qRT-PCR prior to microarray analysis. For the ARHGEF3 and RHOA genes, 81% and 79% knockdown was achieved respectively in these cells (Fig. 1A and B). Of the 202 candidate genes examined in the osteoblast-like cells, gene knockdown resulted in significant changes in expression of 10 genes after adjustment for multiple testing (Table 1). Knockdown of ARHGEF3 resulted in significant changes to the expression of 8 genes: TNFRSF11B, SP7, ALPL, ANGPTL2, GNA11, MYO9B, GNAI2 and PFN1. For RHOA knockdown, 2 genes were affected: PTH1R and ACTA2. Table S1 contains the microarray results for all of the candidate genes examined in the Saos-2 cells (P values corrected for multiple testing).
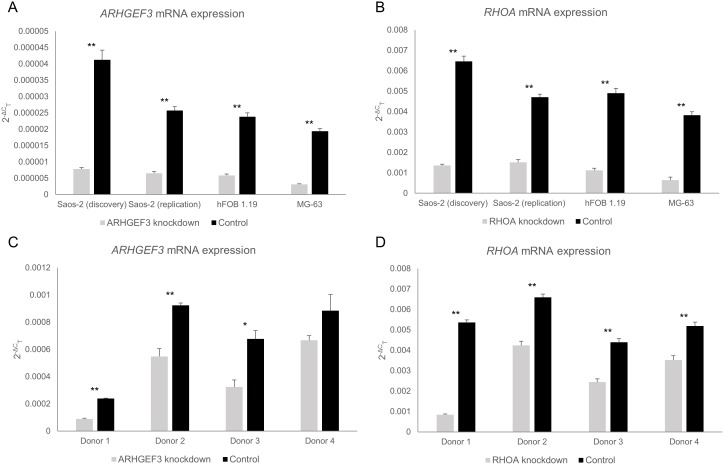
Figure 1. qRT-PCR validation of ARHGEF3 and RHOA gene knockdown in each cell type.
(A) ARHGEF3 mRNA expression in osteoblast-like cells. (B) RHOA mRNA expression in osteoblast-like cells. (C) ARHGEF3 mRNA expression in osteoclast-like cells. (D) RHOA mRNA expression in osteoclast-like cells. Data displayed as mean 2−ΔC T ± SEM from three biological replicates. *P<0.05, **P<0.01.
Table 1. Genes significantly influenced in microarray analysis of ARHGEF3 and RHOA gene knockdown in Saos-2 osteoblast-like cells.
| Knockdown | Gene | Geneproduct | Meanknockdown*FU | Meancontrol*FU | Expressionratio | P |
| ARHGEF3 | TNFRSF11B | Osteoprotegerin | 235 | 308 | 0.76 | <0.001 |
| SP7 | Osterix | 434 | 381 | 1.14 | 0.007 | |
| ALPL | Alkalinephosphatase | 19262 | 17173 | 1.12 | 0.03 | |
| ANGPTL2 | Angiopoietin-like 2 | 81 | 50 | 1.64 | <0.001 | |
| GNA11 | Guanine nucleotide bindingprotein alpha 11 | 352 | 469 | 0.75 | 0.002 | |
| MYO9B | Myosin IXB | 165 | 212 | 0.78 | 0.005 | |
| GNAI2 | Guanine nucleotide bindingprotein alpha inhibiting activity polypeptide 2 | 1106 | 1330 | 0.83 | 0.006 | |
| PFN1 | Profilin 1 | 7458 | 8220 | 0.91 | 0.02 | |
| RHOA | PTH1R | Parathyroid hormone 1 receptor | 1166 | 492 | 2.37 | 0.002 |
| ACTA2 | Alpha 2 actin, smooth muscle | 3059 | 7812 | 0.39 | <0.001 |
Expression ratios are given as expression of the gene in the knockdown cultures relative to the negative control cultures. P adjusted for multiple testing.
*FU, fluorescence units.
qRT-PCR Validation and Replication of Microarray Results for Targeted Genes in Osteoblast-like Cell Lines
Both of the differentially regulated genes in the RHOA knockdown group (PTH1R and ACTA2) and 2 from the ARHGEF3 knockdown group (TNFRSF11B and ALPL) were then selected for confirmatory and replication studies using qRT-PCR. While the microarray results suggested that 8 of the 202 genes examined could potentially be regulated by ARHGEF3, the TNFRSF11B and ALPL genes were selected based on a number of factors including their importance to bone metabolism, their level of expression in the cell type and the size and statistical significance of the regulatory effect. These 4 genes were thus examined in one additional replication study experiment in Saos-2 cells as well as in two additional osteoblast-like cell lines, hFOB 1.19 and MG-63. For the ARHGEF3 and RHOA genes, 75% and 68% knockdown was achieved respectively in the replication batch of Saos-2 cells, 75% and 77% respectively in the hFOB 1.19 cells and 84% and 83% respectively in the MG-63 cells (Fig. 1A and B). The average knockdown achieved across all of the osteoblast-like cell lines as determined by qRT-PCR was 76.8% for RHOA and 78.7% for ARHGEF3 (Fig. 1A and B).
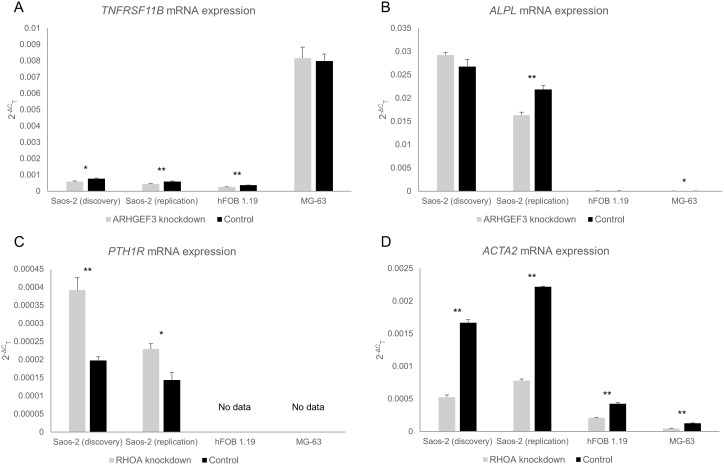
The influence of gene knockdown on TNFRSF11B, ALPL, PTH1R and ACTA2 expression is shown in Fig. 2A–D. A highly significant down regulation of ACTA2 was observed in response to RHOA knockdown in each of the osteoblast-like cell lines examined (P<0.001). The qRT-PCR results also confirmed the up-regulation of the PTH1R gene in response to RHOA knockdown observed in the microarray screen (P<0.001); however, neither the hFOB 1.19 nor the MG-63 cell lines expressed this particular gene. The qRT-PCR studies showed ARHGEF3 knockdown had a significant influence on TNFRSF11B expression in the Saos-2 and hFOB 1.19 cell lines (P = 0.003–0.02), however little influence was seen in the MG-63 cells. ARHGEF3 knockdown had no consistent influence on ALPL expression.
Figure 2. qRT-PCR validation of selected genes in response to ARHGEF3 and RHOA gene knockdown in osteoblast-like cells.
(A) TNFRSF11B mRNA expression in osteoblast-like cells. (B) ALPL mRNA expression in osteoblast-like cells. (C) PTH1R mRNA expression in osteoblast-like cells. (D) ACTA2 mRNA expression in osteoblast-like cells. Data displayed as mean 2−ΔC T ± SEM from three biological replicates. *P<0.05, **P<0.01.
Osteoclast Microarray Results
The osteoclastic phenotype of the cells was confirmed by expression of the genes encoding the osteoclastic biochemical markers TRAP (ACP5), cathepsin K (CTSK) and calcitonin receptor (CALCR) from the microarray output. The ACP5 and CTSK genes were found to be expressed at particularly high levels in this cell type (mean microarray signal >14,000 fluorescence units).
Knockdown of the ARHGEF3 and RHOA genes was validated in the osteoclast-like cells by qRT-PCR prior to microarray analysis. For the ARHGEF3 and RHOA genes, a mean knockdown of 63% and 84% was achieved respectively in the osteoclast-like cells from donor 1 (Fig. 1C and D). Of the 219 candidate genes examined in this cell type, gene knockdown resulted in significant changes in expression of 17 genes after adjustment for multiple testing (Table 2). ARHGEF3 knockdown was found to significantly influence the expression of 12 genes: CCL5, HLA-C, SNCA, TNF, OSCAR, CD44, BIRC3, ITGB7, ITGAE, ITGAL, ITGA3 and ITGAM. For RHOA knockdown, 9 genes were found to be significantly influenced: TNF, THBS2, CCL5, ITGB7, ARHGDIA, IGF1, ACTA2, MYL9 and ITGAE. Of these, the effect of RHOA knockdown on the ACTA2 gene was also observed in the osteoblast-like cells. Table S2 contains the microarray results for all of the candidate genes examined in the osteoclast-like cells (P values corrected for multiple testing).
Table 2. Genes significantly influenced in microarray analysis of ARHGEF3 and RHOA gene knockdown in osteoclast-like cells from donor 1.
| Knockdown | Gene | Gene product | Meanknockdown*FU | Meancontrol*FU | Expressionratio | P |
| ARHGEF3 | CCL5 | Chemokine ligand 5 | 4402 | 588 | 7.48 | <0.001 |
| HLA-C | Major histocompatibility complex,class I, C | 235 | 87 | 2.71 | <0.001 | |
| SNCA | Synuclein, alpha | 129 | 260 | 0.49 | <0.001 | |
| TNF | Tumour necrosisfactor alpha | 420 | 156 | 2.69 | <0.001 | |
| OSCAR | Osteoclast associatedIg-like receptor | 1950 | 1244 | 1.57 | 0.01 | |
| CD44 | CD44 molecule | 2279 | 3360 | 0.68 | 0.03 | |
| BIRC3 | Baculoviral IAP repeat-containing 3 | 496 | 321 | 1.55 | 0.04 | |
| ITGB7 | Integrin, beta 7 | 588 | 106 | 5.54 | <0.001 | |
| ITGAE | Integrin, alpha E | 256 | 483 | 0.53 | <0.001 | |
| ITGAL | Integrin, alpha L | 145 | 79 | 1.85 | 0.003 | |
| ITGA3 | Integrin, alpha 3 | 107 | 221 | 0.48 | 0.003 | |
| ITGAM | Integrin, alpha M | 1033 | 1699 | 0.61 | 0.004 | |
| RHOA | TNF | Tumour necrosisfactor alpha | 345 | 156 | 2.21 | <0.001 |
| THBS2 | Thrombospondin 2 | 89 | 43 | 2.05 | <0.001 | |
| CCL5 | Chemokine ligand 5 | 891 | 588 | 1.52 | 0.006 | |
| ITGB7 | Integrin, beta 7 | 246 | 106 | 2.32 | <0.001 | |
| ARHGDIA | Rho GDP dissociationinhibitor alpha | 579 | 998 | 0.58 | <0.001 | |
| IGF1 | Insulin-like growth factor 1 | 121 | 42 | 2.86 | <0.001 | |
| ACTA2 | Alpha 2 actin, smooth muscle | 545 | 965 | 0.56 | 0.001 | |
| MYL9 | Myosin, light chain 9, regulatory | 22 | 49 | 0.46 | 0.04 | |
| ITGAE | Integrin, alpha E | 342 | 483 | 0.71 | 0.04 |
Expression ratios are given as expression of the gene in the knockdown cultures relative to the negative control cultures. P adjusted for multiple testing.
*FU, fluorescence units.
qRT-PCR Validation and Replication of Microarray Results for Targeted Genes in Osteoclast-like Cells
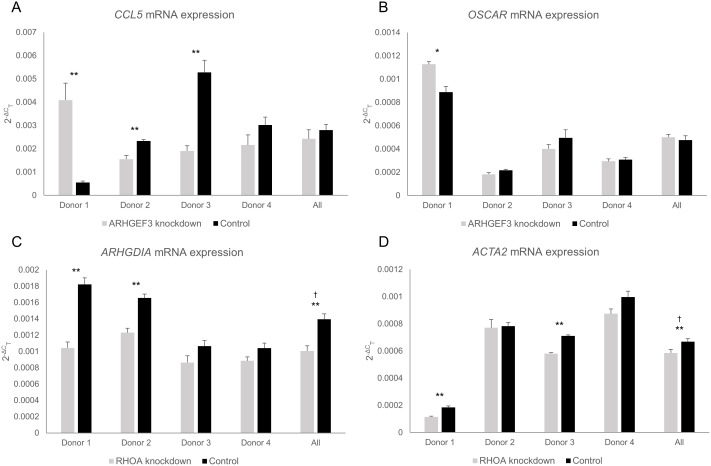
In the osteoclast studies, two of the differentially regulated genes from each of the knockdown experiments were selected for validation and replication analysis by qRT-PCR in osteoclast-like cells from 3 additional donors. These included the CCL5 and OSCAR genes for ARHGEF3 knockdown, and ARHGDIA and ACTA2 genes for RHOA knockdown (Fig. 3A–D).
Figure 3. qRT-PCR validation of selected genes in response to ARHGEF3 and RHOA gene knockdown in osteoclast-like cells.
(A) CCL5 mRNA expression in osteoclast-like cells. (B) OSCAR mRNA expression in osteoclast-like cells. (C) ARHGDIA mRNA expression in osteoclast-like cells. (D) ACTA2 mRNA expression in osteoclast-like cells. Data displayed as mean 2−ΔC T ± SEM from three biological replicates. *P<0.05, **P<0.01, † determined as significant by ANOVA.
For the ARHGEF3 and RHOA genes, 41% and 36% knockdown was achieved respectively in the donor 2 cells, 52% and 45% respectively in the donor 3 cells and 25% and 32% respectively in the donor 4 cells (Fig. 1C and D). The efficiency of knockdown of ARHGEF3 and RHOA averaged 45.3% and 49.3% respectively in this cell type, substantially lower than that observed in the osteoblast-like cells (76.8% vs 49.3% for RHOA, P = 0.07; 78.7% vs 45.3% for ARHGEF3, P = 0.007). The knockdown was considerably lower than desired, however there was some evidence from the overall analysis to suggest that knockdown of RHOA reduces the expression of ACTA2 in this cell type (P = 0.002 by ANOVA). RHOA knockdown also caused a significant overall reduction in ARHGDIA expression in the osteoclast-like cells (P<0.001 by ANOVA). While some significant changes were seen for cells from particular donors, the influence of ARHGEF3 knockdown on CCL5 and OSCAR was inconsistent.
Discussion
We previously reported associations between polymorphism in the RHOA and ARHGEF3 genes and bone density in women and in this study investigated the potential role of these genes in the regulation of bone cells. The knockdown of these two genes showed clear effects on the expression of a number of potentially relevant genes and pathways in two of the major bone cell types – osteoblasts and osteoclasts. Greater gene knockdown levels were achieved in the osteoblast-like cells than in the osteoclast-like cells.
Concerning the studies performed in the osteoblast-like cells, expression of the ACTA2 gene was found to be significantly down-regulated by RHOA knockdown in all three osteoblast-like cell lines examined (Saos-2, hFOB 1.19 and MG-63), with an average expression ratio of 0.35 seen in knockdown cell cultures relative to control cell cultures by qRT-PCR. The ACTA2 gene encodes the alpha 2 actin cytoskeletal protein, which is a major component of the smooth muscle cell contractile apparatus and accounts for around 40% of the total protein and around 70% of the total actin in smooth muscle cells [33], [34]. There have been few studies on the role of the ACTA2 gene product in bone metabolism, however there is evidence in the literature to suggest that the ACTA2 gene is regulated by RhoA signalling. Mack et al. [35] found that expression of constitutively active RhoA in rat smooth muscle cell cultures increased the activity of the Acta2 promoter, whereas inhibition of RhoA decreased the activity of the promoter. They also found that stimulation of actin polymerisation in these smooth muscle cells increased the activity of the Acta2 promoter by 13-fold [35]. In addition, Zhao et al. [36] reported that static tensile forces applied to rat fibroblasts stimulates the promoter activity of the Acta2 gene through the Rho signalling pathway. Collectively, these data suggest that expression of the ACTA2 gene may be regulated through the RhoA signalling pathway, and the results presented here support this.
Knockdown of the ARHGEF3 gene in both the discovery and replication experiments with Saos-2 cells resulted in significant down-regulation of the levels of TNFRSF11B (osteoprotegerin) mRNA. This effect was replicated in the hFOB 1.19 cells, but not in the MG-63 cell line. It is not clear why this effect was not seen in the MG-63 cells, it may be an effect specific to that cell line. TNFRSF11B mRNA levels were significantly higher in the MG-63 cells than in the hFOB 1.19 and Saos-2 cells, in line with studies by Pautke et al. [37]. There are well described differences in the expression patterns between osteoblast-like cell lines reported elsewhere [37]–[39].
In addition to these findings, knockdown of the RHOA gene in both the discovery and replication batches of Saos-2 cells resulted in significant up-regulation of PTH1R (parathyroid hormone 1 receptor) mRNA levels, although expression of this gene was not detected in the hFOB 1.19 or MG-63 cell lines. Both PTH1R and TNFRSF11B have a major role in the stimulation of osteoclastogenesis upon exposure to parathyroid hormone (PTH), suggesting that the ARHGEF3 and RHOA genes may be involved in this process. Radeff et al. [40] found that treatment of UMR-106 rat osteoblast-like cells with Clostridium difficile toxin B, which specifically inhibits the Rho proteins (including RhoA) through glucosylation of the nucleotide binding site [41], reduced PTH-induced expression of the Il6 gene, the product of which has been shown to promote osteoclastogenesis [42]. The authors concluded that the Rho proteins are an important component of PTH signalling in osteoblasts and may have a role in the activation of the intracellular messenger protein kinase C alpha [40]. Another study published by Wang and Stern [43] found that UMR-106 rat osteoblast-like cells transfected with dominant negative RhoA and treated with PTH and/or calcitriol increased production of TNFSF11 mRNA (encoding RANKL) and reduced production of TNFRSF11B mRNA, stimulating osteoclastogenesis of co-cultured RAW 264.7 mouse monocyte/macrophage-like cells [43]. However, when these cells were transfected with constitutively active RhoA and treated with PTH and/or calcitriol, the levels of TNFSF11 and TNFRSF11B mRNA did not change significantly and osteoclastogenesis of the RAW 264.7 cells failed to occur [43]. These results led the authors to suggest that RhoA signalling can inhibit hormone-stimulated osteoclastogenesis through effects on RANKL and osteoprotegerin expression in osteoblasts [43]. No consistent effect of ARHGEF3 knockdown on ALPL expression could be found, however a higher expression of this gene in Saos-2 cells was found compared to the other cells investigated, including the MG-63 cells, in line with the findings of Pautke et al. [37].
One limitation of the gene expression data in the osteoclast-like cells was that consistently high gene knockdown (>60%) was not achieved in some of our experiments, and a greater level of knockdown may show more substantial changes than seen in our studies. Nevertheless, some interesting results were obtained. Expression of the ARHGDIA and ACTA2 genes was found to be significantly reduced in response to RHOA gene knockdown. The product of the ARHGDIA gene is a Rho GDP dissociation inhibitor (GDI) which acts as a negative regulator of several of the RhoGTPases [44]. RhoGDIs maintain the Rho proteins in their inactive GDP-bound state by inhibiting the exchange of GDP for GTP [45] and by restricting membrane anchoring [46]. The down-regulation of ARHGDIA expression seen in the RHOA knockdown osteoclast-like cells in this study could be a compensatory mechanism for the reduced expression of the RHOA gene. The influence of RHOA knockdown on expression of the ACTA2 gene adds further support to the earlier suggestion that expression of this gene is regulated by the RhoA signalling pathway.
In conclusion, knockdown of the ARHGEF3 and RHOA genes in bone cells of human origin reveals important regulatory changes including significant down-regulation of the ACTA2 gene, encoding the cytoskeletal protein alpha 2 actin, in both osteoblast-like and osteoclast-like cells in response to RHOA knockdown. RHOA knockdown also resulted in up-regulation of the PTH1R gene in the Saos-2 osteoblast-like cell line and down-regulation of ARHGDIA in osteoclast-like cells, whereas ARHGEF3 knockdown caused down-regulation of the TNFRSF11B gene in the Saos-2 and hFOB 1.19 osteoblast-like cells. These findings add further evidence to previous studies suggesting a role for the RHOA and ARHGEF3 genes in bone metabolism. Future work in this area could include confirmatory studies investigating the influence of over-expression of the ARHGEF3 and RHOA genes in these cell types and examination of effects at the protein level.
Supporting Information
Microarray results including P values adjusted for multiple-testing for all of the candidate genes examined in the Saos-2 cells.
(XLSX)
Microarray results including P values adjusted for multiple-testing for all of the candidate genes examined in the osteoclast-like cells.
(XLSX)
Acknowledgments
Thanks to Suzanne Brown for her expert advice relating to statistical analysis of the microarray data.
Funding Statement
This study was supported by Arthritis Australia (Grant 0402067) and Curtin University Postgraduate Scholarships. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kanis JA, Melton LJ 3rd, Christiansen C, Johnston CC, Khaltaev N (1994) The diagnosis of osteoporosis. J Bone Miner Res 9: 1137–1141. [DOI] [PubMed] [Google Scholar]
- 2. Prince RL, Dick I (1997) Oestrogen effects on calcium membrane transport: a new view of the inter-relationship between oestrogen deficiency and age-related osteoporosis. Osteoporos Int 7 Suppl 3 S150–154. [DOI] [PubMed] [Google Scholar]
- 3. Flicker L, Hopper JL, Rodgers L, Kaymakci B, Green RM, et al. (1995) Bone density determinants in elderly women: a twin study. J Bone Miner Res 10: 1607–1613. [DOI] [PubMed] [Google Scholar]
- 4. Michaelsson K, Melhus H, Ferm H, Ahlbom A, Pedersen NL (2005) Genetic liability to fractures in the elderly. Arch Intern Med 165: 1825–1830. [DOI] [PubMed] [Google Scholar]
- 5. Evans RA, Marel GM, Lancaster EK, Kos S, Evans M, et al. (1988) Bone mass is low in relatives of osteoporotic patients. Ann Intern Med 109: 870–873. [DOI] [PubMed] [Google Scholar]
- 6. Pocock NA, Eisman JA, Hopper JL, Yeates MG, Sambrook PN, et al. (1987) Genetic determinants of bone mass in adults. A twin study. J Clin Invest 80: 706–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seeman E, Hopper JL, Young NR, Formica C, Goss P, et al. (1996) Do genetic factors explain associations between muscle strength, lean mass, and bone density? A twin study. Am J Physiol 270: E320–327. [DOI] [PubMed] [Google Scholar]
- 8. Andrew T, Antioniades L, Scurrah KJ, Macgregor AJ, Spector TD (2005) Risk of wrist fracture in women is heritable and is influenced by genes that are largely independent of those influencing BMD. J Bone Miner Res 20: 67–74. [DOI] [PubMed] [Google Scholar]
- 9. Deng HW, Chen WM, Recker S, Stegman MR, Li JL, et al. (2000) Genetic determination of Colles’ fracture and differential bone mass in women with and without Colles’ fracture. J Bone Miner Res 15: 1243–1252. [DOI] [PubMed] [Google Scholar]
- 10. Ioannidis JP, Ng MY, Sham PC, Zintzaras E, Lewis CM, et al. (2007) Meta-analysis of genome-wide scans provides evidence for sex- and site-specific regulation of bone mass. J Bone Miner Res 22: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee YH, Rho YH, Choi SJ, Ji JD, Song GG (2006) Meta-analysis of genome-wide linkage studies for bone mineral density. J Hum Genet 51: 480–486. [DOI] [PubMed] [Google Scholar]
- 12. Xiao P, Shen H, Guo YF, Xiong DH, Liu YZ, et al. (2006) Genomic regions identified for BMD in a large sample including epistatic interactions and gender-specific effects. J Bone Miner Res 21: 1536–1544. [DOI] [PubMed] [Google Scholar]
- 13. Wilson SG, Reed PW, Bansal A, Chiano M, Lindersson M, et al. (2003) Comparison of genome screens for two independent cohorts provides replication of suggestive linkage of bone mineral density to 3p21 and 1p36. Am J Hum Genet 72: 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wynne F, Drummond FJ, Daly M, Brown M, Shanahan F, et al. (2003) Suggestive linkage of 2p22-25 and 11q12-13 with low bone mineral density at the lumbar spine in the Irish population. Calcif Tissue Int 72: 651–658. [DOI] [PubMed] [Google Scholar]
- 15. Mullin BH, Prince RL, Dick IM, Hart DJ, Spector TD, et al. (2008) Identification of a role for the ARHGEF3 gene in postmenopausal osteoporosis. Am J Hum Genet 82: 1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mullin BH, Prince RL, Mamotte C, Spector TD, Hart DJ, et al. (2009) Further genetic evidence suggesting a role for the RhoGTPase-RhoGEF pathway in osteoporosis. Bone 45: 387–391. [DOI] [PubMed] [Google Scholar]
- 17. Arthur WT, Ellerbroek SM, Der CJ, Burridge K, Wennerberg K (2002) XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC. J Biol Chem 277: 42964–42972. [DOI] [PubMed] [Google Scholar]
- 18. Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420: 629–635. [DOI] [PubMed] [Google Scholar]
- 19. McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS (2004) Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell 6: 483–495. [DOI] [PubMed] [Google Scholar]
- 20. Meyers VE, Zayzafoon M, Douglas JT, McDonald JM (2005) RhoA and cytoskeletal disruption mediate reduced osteoblastogenesis and enhanced adipogenesis of human mesenchymal stem cells in modeled microgravity. J Bone Miner Res 20: 1858–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chellaiah MA, Soga N, Swanson S, McAllister S, Alvarez U, et al. (2000) Rho-A is critical for osteoclast podosome organization, motility, and bone resorption. J Biol Chem 275: 11993–12002. [DOI] [PubMed] [Google Scholar]
- 22. Fogh J, Fogh JM, Orfeo T (1977) One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J Natl Cancer Inst 59: 221–226. [DOI] [PubMed] [Google Scholar]
- 23. Harris SA, Enger RJ, Riggs BL, Spelsberg TC (1995) Development and characterization of a conditionally immortalized human fetal osteoblastic cell line. J Bone Miner Res 10: 178–186. [DOI] [PubMed] [Google Scholar]
- 24. Billiau A, Edy VG, Heremans H, Van Damme J, Desmyter J, et al. (1977) Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother 12: 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Backlund PS Jr (1997) Post-translational processing of RhoA. Carboxyl methylation of the carboxyl-terminal prenylcysteine increases the half-life of Rhoa. J Biol Chem 272: 33175–33180. [DOI] [PubMed] [Google Scholar]
- 26.Illumina (2008) HumanHT-12 v3 Expression BeadChip. Illumina Technical Bulletin Pub. No.470-2008-005.
- 27. Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386. [DOI] [PubMed] [Google Scholar]
- 28. Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 29. Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- 30.Illumina (2007) BeadStudio Normalization Algorithms for Gene Expression Data. Illumina Technical Bulletin Pub. No.470-2007-005.
- 31. Dunning MJ, Barbosa-Morais NL, Lynch AG, Tavare S, Ritchie ME (2008) Statistical issues in the analysis of Illumina data. BMC Bioinformatics 9: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B 57: 289–300. [Google Scholar]
- 33. Fatigati V, Murphy RA (1984) Actin and tropomyosin variants in smooth muscles. Dependence on tissue type. J Biol Chem 259: 14383–14388. [PubMed] [Google Scholar]
- 34. Owens GK (1995) Regulation of differentiation of vascular smooth muscle cells. Physiol Rev 75: 487–517. [DOI] [PubMed] [Google Scholar]
- 35. Mack CP, Somlyo AV, Hautmann M, Somlyo AP, Owens GK (2001) Smooth muscle differentiation marker gene expression is regulated by RhoA-mediated actin polymerization. J Biol Chem 276: 341–347. [DOI] [PubMed] [Google Scholar]
- 36. Zhao XH, Laschinger C, Arora P, Szaszi K, Kapus A, et al. (2007) Force activates smooth muscle alpha-actin promoter activity through the Rho signaling pathway. J Cell Sci 120: 1801–1809. [DOI] [PubMed] [Google Scholar]
- 37. Pautke C, Schieker M, Tischer T, Kolk A, Neth P, et al. (2004) Characterization of osteosarcoma cell lines MG-63, Saos-2 and U-2 OS in comparison to human osteoblasts. Anticancer Res 24: 3743–3748. [PubMed] [Google Scholar]
- 38. Bilbe G, Roberts E, Birch M, Evans DB (1996) PCR phenotyping of cytokines, growth factors and their receptors and bone matrix proteins in human osteoblast-like cell lines. Bone 19: 437–445. [DOI] [PubMed] [Google Scholar]
- 39. Subramaniam M, Jalal SM, Rickard DJ, Harris SA, Bolander ME, et al. (2002) Further characterization of human fetal osteoblastic hFOB 1.19 and hFOB/ER alpha cells: bone formation in vivo and karyotype analysis using multicolor fluorescent in situ hybridization. J Cell Biochem 87: 9–15. [DOI] [PubMed] [Google Scholar]
- 40. Radeff JM, Nagy Z, Stern PH (2004) Rho and Rho kinase are involved in parathyroid hormone-stimulated protein kinase C alpha translocation and IL-6 promoter activity in osteoblastic cells. J Bone Miner Res 19: 1882–1891. [DOI] [PubMed] [Google Scholar]
- 41. Wilkins TD, Lyerly DM (1996) Clostridium difficile toxins attack Rho. Trends Microbiol 4: 49–51. [DOI] [PubMed] [Google Scholar]
- 42. Lowik CW, van der Pluijm G, Bloys H, Hoekman K, Bijvoet OL, et al. (1989) Parathyroid hormone (PTH) and PTH-like protein (PLP) stimulate interleukin-6 production by osteogenic cells: a possible role of interleukin-6 in osteoclastogenesis. Biochem Biophys Res Commun 162: 1546–1552. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Stern PH (2010) Osteoclastogenic activity and RANKL expression are inhibited in osteoblastic cells expressing constitutively active Galpha(12) or constitutively active RhoA. J Cell Biochem. [DOI] [PMC free article] [PubMed]
- 44. DerMardirossian C, Bokoch GM (2005) GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol 15: 356–363. [DOI] [PubMed] [Google Scholar]
- 45. Fukumoto Y, Kaibuchi K, Hori Y, Fujioka H, Araki S, et al. (1990) Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene 5: 1321–1328. [PubMed] [Google Scholar]
- 46. Olofsson B (1999) Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell Signal 11: 545–554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microarray results including P values adjusted for multiple-testing for all of the candidate genes examined in the Saos-2 cells.
(XLSX)
Microarray results including P values adjusted for multiple-testing for all of the candidate genes examined in the osteoclast-like cells.
(XLSX)