Abstract
Objective
Hepatitis C virus (HCV) is the most frequent cause of mixed cryoglobulinemia (MC), which is characterized by endothelial deposition of rheumatoid factor (RF)–containing immune complexes and end-organ vasculitis. MC is a lymphoproliferative disorder in which B cells express RF-like Ig, yet its precise antigenic stimulus is unknown. We have proposed that IgG–HCV immune complexes stimulate B cell expansion and somatic hypermutation (SHM)–induced affinity maturation in part via engagement of an RF-like B cell receptor. This study was undertaken to test the hypothesis that SHM augments RF activity.
Methods
RFs cloned from single B cells from 4 patients with HCV-associated MC (HCV-MC) were expressed as IgM, IgG, or IgG Fab. Selected Ig were reverted to germline. RF activity of somatically mutated Ig and germline-reverted Ig was determined by enzyme-linked immunosorbent assay.
Results
Ig with SHM had RF activity, with the preference for binding being highest for IgG1, followed by IgG2 and IgG4, and lowest for IgG3, where there was no detectable binding. In contrast, reverted germline IgG exhibited markedly diminished RF activity. Competition with 1 μg/ml of protein A abrogated RF activity, suggesting specificity for IgG Fc. Swapping of mutated heavy-chain pairs and light-chain pairs also abrogated RF activity, suggesting that context-specific pairing of appropriate IgH and Igκ, in addition to SHM, is necessary for RF activity.
Conclusion
SHM significantly contributes to RF activity in HCV-MC patients, suggesting that autoreactivity in these patients arises through antigen-dependent SHM, as opposed to nondeletion of autore-active germline Ig.
Chronic hepatitis C virus (HCV) infection is a major global health burden, affecting [H11011]170 million persons worldwide, and it is the leading indication for liver transplantation in the US and Europe (1). Up to 20% of individuals with chronic HCV develop serious liver-related complications, such as cirrhosis and hepatocellular carcinoma. Although hepatocytes are the primary target for HCV infection, extrahepatic manifestations of HCV infection occur in up to 40% of patients (2). One of the most prominent of these manifestations is a small vessel vasculitis associated with mixed cryoglobulinemia (MC) (3). MC is characterized by the aberrant clonal expansion of B cells that produce rheumatoid factor (RF)–like IgM (4,5). This RF forms immune complexes (ICs) containing polyclonal HCV– specific IgG and HCV RNA; these complexes in turn deposit on the vascular endothelium of organs such as skin, kidneys, and peripheral nerves, eliciting a complement C1q–mediated vasculitis (6).
HCV is also associated with an increased risk of B cell non-Hodgkin's lymphoma (NHL) (7), frequently of the low-grade marginal zone (MZ) or mucosa-associated lymphoid tissue (MALT) subtypes. The continued presence of HCV is necessary for abnormal B cell lymphoproliferation in both HCV-associated MC (HCV-MC) and NHL, since eradication of HCV typically results in resolution of both diseases (8). However, it remains unclear why B cells undergo clonal proliferation during chronic HCV infection. It is likely that HCV-induced B cell lymphoproliferation arises from chronic antigenic stimulation of an initially limited pool of preexisting autoreactive B cells. Persistently high levels of HCV-containing ICs may stimulate the proliferation of RF-bearing B cells, but the precise antigen(s) and stimulatory mechanisms have remained elusive. A striking feature of these expanded B cells is their preferential use of the Ig gene segments VH1-69 and Vκ3-20 (5,9,10), which together frequently encode RF of the Wa cross-reactive idiotype (11). We have previously shown that these antibodies have low-to-moderate levels of somatic hypermutation (SHM), and our phylogenetic analyses have suggested that they have acquired SHM as a result of antigen-directed affinity maturation (5).
Expansions of B cells expressing VH1-69/Vκ3-20–encoded RFs have been found in other diseases without clearly defined infectious etiologies, including primary Sjögren's syndrome (SS) (12), chronic lymphocytic leukemia (CLL) (13,14), and non–HCV-related MALT NHL (12). It has been proposed that a VH1-69–restricted repertoire has been shaped by evolution to recognize common antigens and that continued proliferation predisposes to clonal B cell expansion (15). Moreover, it has been suggested that polyreactive IgM “natural antibodies” rapidly form a first line of defense against invading pathogens, as they broadly recognize microbial epitopes independently of SHM. Interestingly, some humoral immune responses to microbial infections are associated with autoreactivity. For example, it has recently been shown that anti–human immunodeficiency virus (anti-HIV) neutralizing antibodies are polyreactive to both HIV and self antigens (16). Whereas the HIV reactivities of these (highly mutated) antibodies depend upon SHM, their self reactivities do not. This implies that in the case of HIV, antibodies are positively selected on the basis of germline-encoded polyreactivity, and that this polyreactivity is retained throughout affinity maturation despite SHM.
Given the above, a central question in HCV-MC pathogenesis is what causes the expansion of autoreactive B cells, and whether RF activity is present in the germline or arises as a result of SHM. A finding that HCV-related RFs depend upon SHM for their activity would strongly suggest that during MC pathogenesis, certain VH1-69/Vκ3-20–expressing B cells may have an initial low affinity for IgG, but this affinity increases as a result of IgG-directed SHM. Such a finding would imply that IgG coupled to HCV, not HCV by itself, is a major driver of B cell lymphoproliferation. This result has implications not only for HCV-MC, but also for other IC-mediated diseases.
PATIENTS AND METHODS
Patients
This study was approved by the Institutional Review Boards at Rockefeller University Hospital (RUH) and New York–Presbyterian Hospital (NYPH). Volunteers with chronic HCV and laboratory-confirmed MC were recruited through the RUH outpatient clinic and the hepatology clinic at NYPH. All donors provided written informed consent according to the Declaration of Helsinki, before enrollment. Four subjects were selected for this study. Subject LDU 125 was a 60-year-old white man with an HCV genotype 3 vial load of 200,000 IU/ml; he had not received previous treatment. Subject 773 was a 29-year-old white woman with an HCV genotype 1b viral load of 380,000 IU/ml; she was considered a nonresponder to PEGylated interferon-[H9251] and ribavirin. Subject 1403 was a 44-year-old white Hispanic woman with an HCV geno-type 1 viral load of 500,000 IU/ml; she had not received previous treatment. Subject 1432 was a 69-year-old white woman with an HCV genotype 1 viral load of 30,000 IU/ml; she had not received previous treatment.
Ig cloning and expression
Ig VH/Ig VL pairs were cloned from singly sorted IgM+κ+CD27[H11001] B cells from patients with HCV-MC and expressed as IgM, as previously described (17). GenBank accession numbers for the cloned Ig nucleotide sequences are EF624068–EF624214 and JF944897. For selected clones, Ig VH/Ig VL pairs were expressed as IgG, as previously described (18). For Fab generation, selected Ig VH were ligated into an expression vector encoding IgG CH2 with a COOH-terminal 6× His-tag and transfected into 293T cells along with appropriate Ig Vκ constructs. For Fab purification, 0.5M NaCl, 10 mM HEPES, and 20 mM imidazole were added to transfection supernatants, which were then purified with HisTrap HP columns (GE Healthcare) using 500 mM imidazole in the elution buffer. Samples were desalted into 20 mM NaHPO4 and 0.5M NaCl (pH 7.4), using HiTrap Desalting Columns (GE Healthcare). Purity and appropriate size of Fab was confirmed by nonreducing sodium dodecyl sulfate– polyacrylamide gel electrophoresis.
Germline reversion of Ig clones
Ig VH/Ig VL clones were compared to the published germline VH1-69 and Vκ3-20 sequences to identify SHM in the V-region segments as previously described (5). To revert SHM to germline sequences, polymerase chain reaction (PCR) was performed using oligonucleotides encoding the relevant germline VH1-69 and Vκ3-20 sequences. Only the portions of the Ig genes encoded by VH1-69 and Vκ3-20 V gene segments (framework region 1 [FR1], first complementarity-determining region [CDR1], FR2, CDR2, and FR3) were changed. CDR3 sequences, which arise as a result of the immunoglobulin gene recombination process, were left unchanged. Therefore, germline-reverted clones encoded germline V region sequences and wild-type CDR3 sequences. Clones were verified by sequencing.
Human Ig enzyme-linked immunosorbent assay (ELISA)
Nunc MaxiSorp 96-well plates were coated overnight at 4°C with capture goat polyclonal anti-human IgM or anti-human IgG antibodies (Bethyl Laboratories) or goat anti-human IgG Fc antibodies (Southern Biotech). All subsequent steps were performed at room temperature. After blocking with 1% bovine serum albumin (BSA) in coating buffer (140 mM NaCl, 50 mM Tris, 0.05% Tween [pH 8.0]) for 2 hours, antibodies (diluted in coating buffer) were added for 1 hour. After 5 washes with coating buffer, bound Ig was detected by addition of horseradish peroxidase (HRP)–conjugated goat anti-human IgM, IgG, or Igκ for 1 hour. Plates were washed 5 times with coating buffer, and tetramethylbenzidine (TMB) substrate (BioFX Laboratories) was added for 5 minutes. After the reaction was stopped with 1N HCl, absorbance at 450 nm (A450) was measured with a FluoStar Omega microplate reader (BMG). Human myeloma IgM and Fab (Jackson ImmunoResearch) and IgG (Sigma) were used to generate standard curves, and concentrations of IgM, IgG, and Fab were calculated.
RF assays
All steps in the RF assays were performed at room temperature. Nunc MaxiSorp 96-well plates were coated with 1 μg/well human myeloma IgG1λ, IgG2λ, IgG3λ, IgG4λ (Sigma), or human IgG Fc (Jackson ImmunoResearch), and incubated for 1 hour. After washing 3 times, plates were blocked with 1% BSA in coating buffer for 1 hour. Serial dilutions of monoclonal antibodies (mAb) (diluted in coating buffer) were then added in duplicate for 1 hour. Plates were washed 5 times with coating buffer, and HRP-labeled goat anti-human IgM (Bethyl Laboratories) was added for 1 hour.
For comparison of anti-IgG1 activities of IgM, IgG, and Fab, HRP-labeled anti-human Igκ was used instead of anti-IgM as the detection antibody. After plates were washed 5 times with coating buffer, 50 μl TMB was added. After 3 minutes, reactions were stopped with 50 μl 1N HCl, and A450 was measured.
Protein A competition assay
Experiments were carried out as described above for the anti-IgG1 IgM-RF assay, with the following exceptions. Serial dilutions of Staphylococcus aureus protein A (Sigma) were added to IgG1λ-coated micro-well plates. After incubating for 1 hour, IgM mAb (5 μg/ml) were added, and ELISA was carried out as described above.
Statistical analysis
Data were analyzed using Graph-Pad Prism software. Results are presented as the mean ± SD of duplicate or triplicate measurements. Fisher's exact test was used to test the null hypothesis that the number of clones with RF activity is not different in the SHM or germline-reverted groups.
RESULTS
Since B cells that have undergone clonal expansion in patients with HCV-MC express RF-like IgM encoded by predominantly hypermutated VH1-69 and Vκ3-20 gene segments (5), we decided to test whether the hypermutated IgM had RF activity. Also, since phylogenetic evidence has suggested that these B cells acquired SHM in an antigen-dependent manner (5), we questioned whether SHM was essential for RF activity.
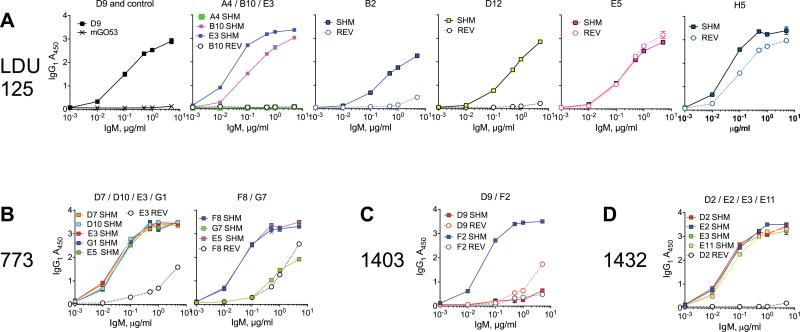
In a previous study, we singly sorted IgM+κ+ memory B cells from HCV-MC patients, and cloned the expressed IgM heavy chain and light chain variable regions by IgV reverse transcription–PCR (5). Four subjects (LDU 125, 773, 1403, and 1432) were selected for the current study. In these patients, all of the cloned Ig used VH1-69 and Vκ3-20. For IgM expression, we selected the most and least hypermutated Ig from each subject's major heavy chain CDR3 (HCDR3) clonal populations. The Ig VH–translated amino acid sequences of clones that we selected for expression are shown in Figure 1. Subjects 1403 and LDU 125 had major clonal HCDR3 populations with unmutated Ig VH. The remaining clones contained SHM, and amino acid replacements tended to be more frequent in HCDR1, HCDR2, and toward the C-terminal end of heavy chain FR3 (HFR3). One residue in particular, S32, was frequently mutated in clonal populations from 2 subjects. Similarly, amino acid sequences for each corresponding Ig Vκ are shown in Figure 2. All of these sequences were mutated, and they contained between 1 and 7 amino acid changes. Substitutions were most common in light chain CDR1 (LCDR1), light chain FR2 (LFR2), LCDR2, and LFR3. Three residues were frequently mutated: S32, A57, and S66.
Figure 1.
Ig VH–complementarity-determining region 3 (CDR3) amino acid sequences of singly sorted IgM+κ+CD27+ B cells from patients with hepatitis C virus–associated mixed cryoglobulinemia (HCV-MC). Ig VH and Ig Vκ reverse transcription–polymerase chain reaction was performed on the singly sorted B cells from 4 patients (LDU 125, 773, 1403, and 1432). Depicted are representative clones of each patient's predominant clonal heavy chain CDR3 (HCDR3) populations. Germline VH1-69 amino acid sequences of framework regions (FRs) and CDRs are shown, and sequences are listed by subject and B cell clone. Sequences are compared to those of germline VH1-69 sequences; identity is shown by dashed lines. The frequency of each clone among the total number of clones with identical HCDR3 is listed (at the far right). Complete nucleotide sequences are available from the GenBank database (accession nos. EF624068–EF624214 and JF944897).
Figure 2.
Ig Vκ–CDR3 amino acid sequences of singly sorted IgM+κ+CD27+ B cells from patients with HCV-MC. Ig light chain sequences corresponding to clones listed in Figure 1 are depicted. Sequences are compared to the germline Vκ3-20 sequence; identity is shown by dashed lines. Complete nucleotide sequences are available from the GenBank database (accession nos. EF624068–EF624214 and JF944897). See Figure 1 for definitions and further details.
Sixteen of 21 tested wild-type clones with SHM isolated from HCV-MC patients bound IgG1, with an A450 (1 μg/ml mAb) of >2. Of 10 germline-reverted clones, 2 bound IgG1 with an A450 (1 μg/ml mAb) of >2 (P = 0.0057 by Fisher's exact test) (Figure 3). For subject LDU 125 (Figure 3A), only clone A4 (which had a germline Ig VH) did not bind IgG1. However, germline Ig VH did not by itself preclude anti-IgG1 activity, as another clone with germline Ig VH, clone H5, bound IgG1. The clone with the most Ig VH amino acid replacements and the longest HCDR3 (clone B2) bound IgG1 moderately. Interestingly, clones A4 and B2 shared a minor LCDR3 sequence. In 4 of 5 clonal HCDR3 populations, reversion of Ig VH and Ig Vκ reduced reactivity to IgG1. The exception was clone E5, which had 1 Ig VH and 2 Ig Vκ amino acid changes. Taken together, it is likely that a combination of particular HCDR3 and LCDR3, as well as Ig VH and Ig Vκ SHM, are necessary for anti-IgG1 activity.
Figure 3.
Anti-IgG1 activity of IgM positive for VH1-69/Vκ3-20 is dependent upon the presence of somatic hypermutation (SHM). Anti-IgG1 activity of wild-type, somatically hypermutated IgM (squares) and germline-reverted (REV) IgM (circles) is depicted for subject LDU 125 (A), subject 773 (B), subject 1403 (C), and subject 1432 (D). In A, mGO53 IgM is a negative control. Values are the mean ± SD.
For subject 773 (Figure 3B) none of the clones of the major HCDR3 clonal population had germline Ig VH or Ig Vκ, and, except for clone G7, all had high anti-IgG1 activity that was abrogated upon reversion of Ig VH and Ig Vκ to germline. Clones F8 and G7, from the second largest HCDR3 population, had high and low-to-moderate anti-IgG1 activity, respectively. Thus, 1 μg of recombinant mAb F8 bound to IgG1 had an A450 value above 3; for mAb G7, the A450 value was between 1 and 2. The anti-IgG1 activity of clone F8 was reduced upon reversion to germline, whereas reversion of clone G7 (which was relatively more hypermutated and shared no amino acid replacements with F8) had no effect on anti-IgG1 activity. This suggests that this particular Ig VH/Ig Vκ pair has low-to-moderate anti-IgG1 activity that does not depend upon SHM.
In subject 1403 (Figure 3C), clone D9 was representative of the major clonal population, which utilized a germline Ig VH paired with Ig Vκ containing between 2 and 3 amino acid changes. This clone had no detectable anti-IgG1 activity (A450 [1 μg/ml] <0.5). However, clone F2, representative of the minor clonal population, utilized a unique HCDR3/LCDR3 pair with hypermutated Ig VH and Ig Vκ. Clone F2 had strong anti-IgG1 activity (A450 [1 μg/ml] >3) that was abrogated upon germline reversion.
All 22 B cells sorted from subject 1432 (Figure 3D) utilized the same HCDR3/LCDR3 pair. Individual clones contained between 1 and 6 and between 1 and 2 amino acid Ig VH and Ig Vκ changes, respectively. Of the 4 clones tested, all had strong anti-IgG1 activity; this activity was completely abolished upon germline reversion.
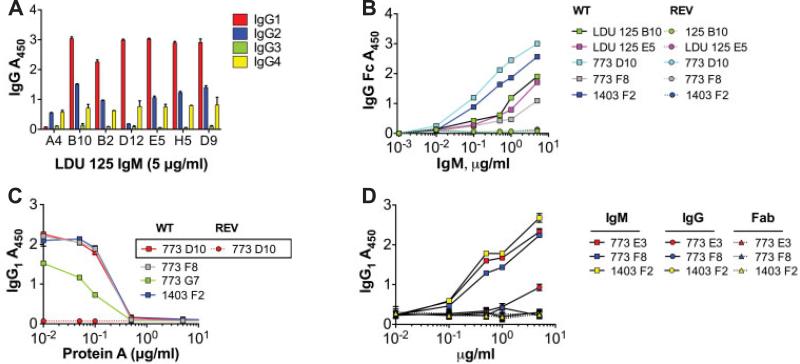
We next tested whether clonal HCV-MC IgM shared reactivity patterns with previously well-defined RFs (Figure 4). It has been established that IgM-RFs from rheumatoid arthritis patients bind IgG1, IgG2, and IgG4 strongly and bind IgG3 weakly (19). Similar results have been reported for monoclonal RFs from patients with Waldenström's macroglobulinemia (20) and cryoglobulinemia (21); it has been shown that these RFs bind a discontinuous epitope spanning the heavy chain C region domains 2 and 3 (20).
Figure 4.
The modestly hypermutated VH1-69+/Vκ3-20+ IgM from patients with hepatitis C virus are rheumatoid factors. A, Monoclonal antibodies (mAb) from subject LDU 125 were expressed as IgM, and serial dilutions were incubated on enzyme-linked immunosorbent assay (ELISA) plates coated with IgG1, IgG2, IgG3, or IgG4 and were detected with anti-IgM. Binding was strongest for IgG1 followed by IgG2 and IgG4, and was weakest for IgG3. B, Serial dilutions were incubated as in A, except ELISA plates were coated with IgG Fc. The results demonstrated that IgM bound to the Fc portion of IgG. C, Before incubation with IgG1-coated plates, IgM (1 μg/ml) were incubated with serial dilutions of protein A. For all clones tested, preincubation with 0.5–1 μg/ml protein A yielded 50% inhibition of binding. D, Anti-IgG1 activities of Ig expressed as either IgM, IgG, or Fab were compared. These VH1-69+/Vκ3-20+ mAb bound IgG1 when expressed as IgM, but not as IgG or Fab, indicating that the binding to IgG1 is of low affinity and depends upon the avidity conferred by multimeric IgM. Values are the mean ± SD. WT = wild type; REV = germline reverted.
We first compared the abilities of HCV-MC IgM to bind IgG1, IgG2, IgG3, and IgG4. The results obtained from clones from subject LDU 125 are shown in Figure 4A. Consistent with other reports of RFs, binding was highest for IgG1, followed by IgG2 and IgG4, whereas there was no detectable binding to IgG3. We confirmed that the IgM bound to the Fc portion of IgG, using an Fc-specific ELISA (Figure 4B). For clones from each subject, anti-Fc reactivity paralleled that seen for IgG1. We tested whether the binding of IgG1 could be blocked by S aureus protein A, which binds to the CH2/CH3 interface (22). Indeed, we found that 1–5 μg/ml of protein A reduced binding to IgG1 by 50% (Figure 4C). Next, we tested whether the multivalent stoichiometry of IgM was necessary for the binding of these clonal RFs to IgG1 (Figure 4D). When clonal Ig were expressed as IgG or Fab, no significant anti-IgG1 activity was detected by ELISA. This suggests that these RFs have low-to-moderate affinities for IgG1 and that the mutivalency of IgM confers a substantial increase in avidity for IgG1.
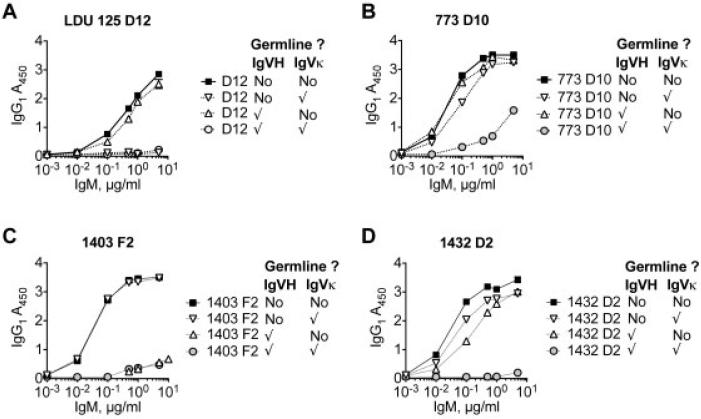
Next, we tested whether Ig VH or Ig Vκ SHM were the main contributors to RF activity. We reverted either, both, or neither of the selected Ig VH and Ig Vκ pairs to germline. We then expressed the pairs as IgM and tested their reactivity to IgG1 (Figure 5). For clone D12 from subject LDU 125, Ig Vκ reversion completely abrogated RF activity, whereas Ig VH reversion did not (Figure 5A). For clone D10 from subject 773, Ig Vκ reversion slightly reduced RF activity, whereas Ig VH reversion did not (Figure 5B). In contrast, for clone F2 from subject 1403, Ig VH reversion completely abrogated RF activity, whereas Ig Vκ reversion did not (Figure 5C). For clone D2 from subject 1432, reversion of either Ig VH or Ig Vκ moderately reduced RF activity, while reversion of both heavy and light chain SHM abrogated RF activity (Figure 5D). Taken together, these results demonstrate that the contributions of Ig VH or Ig Vκ SHM to RF activities are dependent upon particular HCDR3/LCDR3 pairs, as well as upon SHM in the paired Ig Vκ or Ig VH.
Figure 5.
Rheumatoid factor activity is dependent upon both heavy chain and light chain somatic hypermutation. Anti-IgG1 activity of IgM clones from patients with hepatitis C virus–associated mixed cryoglobulinemia (LDU 125 [A], 773 [B], 1403 [C], and 1432 [D]) was determined. IgM expression with either wild-type (WT) Ig VH and Ig Vκ (squares), WT Ig VH and germline Ig Vκ (inverted triangles), germline Ig VH and WT Ig Vκ (upright triangles), or germline Ig VH and Ig Vκ (circles) is shown. Checkmarks indicate germline. Values are the mean ± SD.
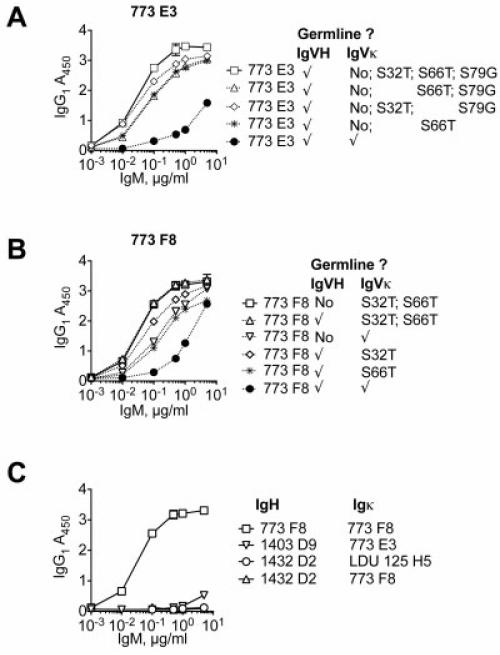
We next tested the effects of particular, isolated Ig Vκ SHM upon RF activity (Figure 6). Replacement of Ig Vκ S32 and S66 frequently occurred in clones from subject LDU 125 and subject 773. Clone E3 from subject 773 contained Ig Vκ S32T, S66T, and S79G; we reverted either S32, S66, or both S32 and S79, and we paired these to germline E3 Ig VH. Clone F8 from subject 773 contained S32 and S66 Ig Vκ; we reverted both mutations to germline, and we paired these to germline F8 Ig VH. For clone E3 from subject 773, reversion of Ig Vκ S66 had less of an impact upon RF activity than did reversion of S32 or of S32 and S79 (Figure 6A). Similarly, for clone F8 from subject 773, reversion of S66 had less of an impact than S32 reversion upon RF activity (Figure 6B). Finally, we tested whether RF activity was preserved upon interchanging Ig VH and Ig Vκ from different patients. For all of the pairs that we tested, swapping of Ig VH and Ig Vκ completely abrogated RF activity (Figure 6C).
Figure 6.
Rheumatoid factor (RF) activity depends upon light chain somatic hypermutation (SHM), as well as particular heavy chain complementarity-determining region 3 (HCDR3) and light chain CDR3 combinations. A and B, Ig VH was paired with wild-type Ig Vκ for which some, all, or none of the SHMs were reverted to germline. C, Ig VH and Ig Vκ from RF from different patients were coexpressed and tested for RF activity. Swapping of Ig VH and Ig Vκ abrogated RF activity. Values are the mean ± SD.
DISCUSSION
This work is, to our knowledge, the first to show that SHM contributes to the RF activity of HCV-MC– derived clonal IgM. We believe our data to be robust, given the number of mAb studied, the expression of mAb as polymeric IgM, and the direct comparison of patient-derived IgM and germline IgM. Taken together, our results suggest that IgG (likely complexed to HCV) drives the affinity maturation required for RF activity in HCV-MC. We speculate that this initial binding event is of low affinity, since we frequently could not detect RF activity in germline Ig.
In our earlier study (5), we found that clonal B cells from HCV-MC patients were often restricted toward VH1-69/Vκ3-20 usage, and that for each patient, B cells were predominantly monoclonal or oligoclonal, based upon CDR3 sequences. Also, Ig expressed by these B cells were modestly hypermutated, although 2 subjects had significant clonal populations of B cells expressing unmutated Ig VH. Notably, S32 in both Ig VH and Ig VL was frequently mutated, as were A57 and S66 in Ig VL. Of note, Ig VH S32 mutations occur in other NHL-associated antibodies (clone N.1 was an exact HCDR3 match with clone B10 from subject LDU 125 [23], and clone HA10 was an exact HCDR3 match with clone G7 from subject 773 [24]). In subject 773, the major clonal population had HCDR3 and LCDR3 exactly identical to those found in mAb M11, isolated by another group from the parotid gland of a patient with MALT NHL with unknown HCV status (GenBank accession nos. AY281329.1 and AY281341.1). Interestingly, mAb M11 contained only 1 amino acid change (S89N), but exhibited high RF activity (12).
Historically, monoclonal IgM-RFs from MC patients have been classified into 3 different groups based upon cross-reactive idiotype: Wa, Po, and Bla (25,26). RFs of the Wa group were shown to have a marked restriction in Vκ3-20 usage (27) and in VH1-69 usage as defined by reactivity with the G6 mAb (28), as well as frequent JH4 usage. Only a small proportion of RFs in rheumatoid arthritis patients use the VH and Vκ segments commonly found in RF from patients with MC (29,30), implying that MC-related and rheumatoid arthritis–related RFs arise through different pathogenic mechanisms. Indeed, a previous study showed that SHM decreased RF activity of a VH1-69/Vκ3-20–encoded mAb (expressed as murine IgG1) derived from the synovium of rheumatoid arthritis patients (31). Consistent with this, it has been thought that the germline VH1-69/Vκ3-20 pair is biased toward intrinsic polyreactivity.
Moreover, since RFs from patients with HCV-MC have low levels of SHM, it has been proposed that these SHMs may not have arisen as a result of antigen-dependent affinity maturation and that SHM contributed little to RF activity (32). In contrast, we have found that a small number of SHMs confer considerable RF activity to VH1-69/Vκ3-20 pairs. In 18 of 21 tested clone pairs, the presence of modest numbers of SHMs conferred increased RF activity. Moreover, depending upon the clone, SHM in Ig VH, Ig Vκ, or both was important for RF activity. Comparison of mAb with selectively reverted Ig Vκ amino acid replacements suggests that the common S32T substitution may be a major contributor to RF activity. However, our data also show that occasionally certain germline VH1-69/Vκ3-20 pairs, with appropriate HCDR3 and LCDR3, can encode antibodies with low-to-moderate RF activity.
The critical importance of CDR3 configuration for RF activity is not surprising and has been corroborated by studies in which a single amino acid change in HCDR3 alters RF activity (33). Previous studies have demonstrated that both IgH and Igκ are required for the RF activity of RF Wa (34). Our study extends this observation by showing that swapping of IgH and Igκ from different RFs abrogates RF activity. Importantly, although SHMs confer a significant boost in RF activity, IgM structure is also required. When we expressed clones as either IgG or IgG Fab, RF activity was barely detectable, suggesting that the observed IgM anti-IgG activity is likely to be of moderate affinity and that the polymeric stoichiometry of IgM confers significant avidity for IgG.
Surprisingly little is known about the epitope specificities of HCV-MC RFs. The mutated RFs in this study had similar reactivities to those that have already been reported for other Wa cross-reactive idiotype RFs. Preference for IgG1, IgG2, and IgG4 was seen, and the RFs did not react with IgG3. Moreover, they bound IgG1 Fc, and their interaction with IgG1 could be blocked by S aureus protein A. As protein A binds to the IgG CH2–CH3 interface (22), it is likely that HCV-MC RFs bind to an epitope in this region as well. The crystal structure of Fab of AN (a non–Wa cross-reactive idio-type VH3/Vλ2-20–utilizing RF isolated from a rheumatoid arthritis patient) complexed with IgG4 Fc reveals binding to the CH2/CH3 interface of Fc. One of the contact residues is an SHM, implicating antigen-driven selection in the production of pathogenic RFs (35).
Clonal expansions of B cells expressing the mAb G6–reactive cross-reactive idiotype are seen in various other disease states, such as CLL (13,14), primary SS (36), Helicobacter pylori–associated MALT NHL (12), and infection with Borrelia burgdorferi (37). In the case of CLL, unmutated VH1-69–encoded Ig are polyreactive, low-affinity antibodies, whereas mutated VH1-69/Vκ3-20 Ig are monoreactive, high-affinity antibodies (33,38). It has been proposed that a VH1-69–restricted repertoire has been shaped by evolution to recognize common antigens and that continued proliferation in response to particular microbes predisposes to CLL (15). Indeed, it has recently been shown that unmutated VH1-69–encoded mAb from CLL patients and healthy donors recognize cytomegalovirus phosphoprotein pUL 32 (39). We are working to define the reactivities of HCV-MC mAb toward HCV and other viral antigens.
Expansion of B cells expressing VH1-69 has been noted in patients with other autoimmune diseases such as thrombotic thrombocytopenic purpura (40), in patients who have undergone renal transplant (41), and in patients with viral infections such as HIV (16) and influenza (42). This has prompted speculation that the VH1-69 gene segment has germline-encoded, weak poly-reactivity that may function as a first-line, rapidly evoked defense against invading pathogens. A number of HIV-reactive mAb are polyreactive, bind to autoantigens, and are encoded by VH1-69 (43,44). In one study, when autoreactive anti-HIV mAb were reverted to germline, they retained self-reactivity, yet lost anti-HIV activity (16). Because of this, it was postulated that polyreactive antibodies are positively selected before the germinal center reaction and before SHM, and polyreactivity is retained throughout affinity maturation despite hyper-mutation.
An implication of this is that foreign agents, rather than autoantigens, drive the acquisition of SHM. However, our study suggests that in the case of HCVMC, self IgG can drive selection of SHM. Of course, it remains formally possible that anti-IgG activity arises through HCV-driven affinity maturation and epitope mimicry. Consistent with our findings, it has been observed in other disease processes that the accumulation of SHM on a polyreactive backbone does not lead to loss of autoreactivity. In patients with primary SS and parotid MALT NHL, the lymphoma cells express mutated, VH1-69/Vκ3-20–encoded, monospecific, high-affinity RF (12), and in patients with H pylori–associated gastric MALT lymphoma, the lymphomas express mutated, VH1-69/Vκ3-20–encoded, polyreactive RF (45). Moreover, some anti-DNA antibodies lose reactivity when reverted to germline (46), and in a mouse model of lupus, antinuclear antibodies were found to arise from SHM of nonautoreactive germline antibodies (47).
The biologic role of moderately active RF in microbial clearance remains unclear. Evidence suggests that VH1-69–expressing B cells are expanded among the naive B cell compartment in individuals in whom the HCV infection spontaneously clears, as compared to chronically infected or healthy donors (48). It is uncertain whether this expansion relates directly to HCV clearance, and if so, whether clearance is due to moderately reactive RF or high-affinity, HCV-specific antibodies. It is unlikely that these RFs neutralize HCV by directly blocking viral hepatocyte interactions. It is more likely that their role is to further immunogenize HCV through IC formation. Indeed, it has been shown that RF-expressing B cells can efficiently present tetanus toxin antibody ICs (49). It is likely that clearance of ICs involves complement, particularly C1q, which is avidly fixed by IgM. Consistent with this, attachment of RF to herpes simplex virus IgG ICs does not lead to neutralization, unless complement is added (50).
Although this study provides evidence that HCVMC–related RFs develop as a consequence of affinity maturation to self antigens, it remains formally possible that RF activity is a result of cross-reactivity with HCV. However, we believe that it is unlikely that HCV is the sole driver of RF activity, as we have not detected RF activity in several VH1-69/Vκ3-20–encoded mAb that neutralize HCV (Charles ED, Dustin LB: unpublished observations). Future translational studies of HCV-MC patients are needed to elucidate the mechanisms of tolerance loss, dissect shared recognition epitopes, and define their potential roles in microbial control.
ACKNOWLEDGMENTS
We gratefully acknowledge the generosity of the patient volunteers. We thank Donna Brassil, Veronica Whalen, and Rhonda Kost of the Rockefeller University Center for Clinical and Translational Science. We also thank Santa Maria Di Vittorio for administrative assistance.
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Drs. Charles and Dustin had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Charles, Orloff, Nishiuchi, Marukian, Rice, Dustin.
Acquisition of data. Charles, Orloff, Nishiuchi.
Analysis and interpretation of data. Charles, Orloff, Nishiuchi, Marukian, Rice, Dustin.
REFERENCES
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–67. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Galossi A, Guarisco R, Bellis L, Puoti C. Extrahepatic manifestations of chronic HCV infection. J Gastrointestin Liver Dis. 2007;16:65–73. [PubMed] [Google Scholar]
- 3.Agnello V, Chung RT, Kaplan LM. A role for hepatitis C virus infection in type II cryoglobulinemia. N Engl J Med. 1992;327:1490–5. doi: 10.1056/NEJM199211193272104. [DOI] [PubMed] [Google Scholar]
- 4.Charles ED, Dustin LB. Hepatitis C virus-induced cryoglobulinemia. Kidney Int. 2009;76:818–24. doi: 10.1038/ki.2009.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charles ED, Green RM, Marukian S, Talal AH, Lake-Bakaar GV, Jacobson IM, et al. Clonal expansion of immunoglobulin M+CD27+ B cells in HCV-associated mixed cryoglobulinemia. Blood. 2008;111:1344–56. doi: 10.1182/blood-2007-07-101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sansonno D, Dammacco F, Hepatitis F. C virus, cryoglobulinaemia, and vasculitis: immune complex relations. Lancet Infect Dis. 2005;5:227–36. doi: 10.1016/S1473-3099(05)70053-0. [DOI] [PubMed] [Google Scholar]
- 7.Giordano TP, Henderson L, Landgren O, Chiao EY, Kramer JR, El-Serag H, et al. Risk of non-Hodgkin lymphoma and lymphoproliferative precursor diseases in US veterans with hepatitis C virus. JAMA. 2007;297:2010–7. doi: 10.1001/jama.297.18.2010. [DOI] [PubMed] [Google Scholar]
- 8.Hermine O, Lefrere F, Bronowicki JP, Mariette X, Jondeau K, Eclache-Saudreau V, et al. Regression of splenic lymphoma with villous lymphocytes after treatment of hepatitis C virus infection. N Engl J Med. 2002;347:89–94. doi: 10.1056/NEJMoa013376. [DOI] [PubMed] [Google Scholar]
- 9.Ivanovski M, Silvestri F, Pozzato G, Anand S, Mazzaro C, Burrone OR, et al. Somatic hypermutation, clonal diversity, and preferential expression of the VH 51p1/VL kv325 immunoglobulin gene combination in hepatitis C virus-associated immunocytomas. Blood. 1998;91:2433–42. [PubMed] [Google Scholar]
- 10.Carbonari M, Caprini E, Tedesco T, Mazzetta F, Tocco V, Casato M, et al. Hepatitis C virus drives the unconstrained monoclonal expansion of VH1-69-expressing memory B cells in type II cryoglobulinemia: a model of infection-driven lymphomagenesis. J Immunol. 2005;174:6532–9. doi: 10.4049/jimmunol.174.10.6532. [DOI] [PubMed] [Google Scholar]
- 11.Knight GB, Agnello V, Bonagura V, Barnes JL, Panka DJ, Zhang QX. Human rheumatoid factor cross-idiotypes. IV. Studies on WA XId-positive IgM without rheumatoid factor activity provide evidence that the WA XId is not unique to rheumatoid factors and is distinct from the 17.109 and G6 XIds. J Exp Med. 1993;178:1903–11. doi: 10.1084/jem.178.6.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bende RJ, Aarts WM, Riedl RG, de Jong D, Pals ST, van Noesel CJ. Among B cell non-Hodgkin's lymphomas, MALT lymphomas express a unique antibody repertoire with frequent rheumatoid factor reactivity. J Exp Med. 2005;201:1229–41. doi: 10.1084/jem.20050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kipps TJ, Fong S, Tomhave E, Chen PP, Goldfien RD, Carson DA. High-frequency expression of a conserved κ light-chain variable-region gene in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 1987;84:2916–20. doi: 10.1073/pnas.84.9.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kipps TJ, Tomhave E, Pratt LF, Duffy S, Chen PP, Carson DA. Developmentally restricted immunoglobulin heavy chain variable region gene expressed at high frequency in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 1989;86:5913–7. doi: 10.1073/pnas.86.15.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forconi F, Potter KN, Wheatley I, Darzentas N, Sozzi E, Stamatopoulos K, et al. The normal IGHV1-69-derived B-cell repertoire contains stereotypic patterns characteristic of unmutated CLL. Blood. 2010;115:71–7. doi: 10.1182/blood-2009-06-225813. [DOI] [PubMed] [Google Scholar]
- 16.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–5. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charles ED, Orloff MI, Dustin LB. A flow cytometry-based strategy to identify and express IgM from VH1-69+ clonal peripheral B cells. J Immunol Methods. 2011;363:210–20. doi: 10.1016/j.jim.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–7. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 19.Artandi E, Canfield SM, Tao MH, Calame KL, Morrison SL, Bonagura VR. Molecular analysis of IgM rheumatoid factor binding to chimeric IgG. J Immunol. 1991;146:603–10. [PubMed] [Google Scholar]
- 20.Artandi SE, Calame KL, Morrison SL, Bonagura VR. Monoclonal IgM rheumatoid factors bind IgG at a discontinuous epitope comprised of amino acid loops from heavy-chain constant-region domains 2 and 3. Proc Natl Acad Sci U S A. 1992;89:94–8. doi: 10.1073/pnas.89.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mageed RA, Carson DA, Jefferis R. Immunogenic and antigenic epitopes of immunoglobulins. XXIII. Idiotypy and molecular specificity of human rheumatoid factors: analysis of cross-reactive idiotype of rheumatoid factor paraproteins from the Wa idiotype group in relation to their IgG subclass specificity. Scand J Immunol. 1988;28:233–40. doi: 10.1111/j.1365-3083.1988.tb02436.x. [DOI] [PubMed] [Google Scholar]
- 22.Graille M, Stura EA, Corper AL, Sutton BJ, Taussig MJ, Char-bonnier JB, et al. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci U S A. 2000;97:5399–404. doi: 10.1073/pnas.97.10.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marasca R, Vaccari P, Luppi M, Zucchini P, Castelli I, Barozzi P, et al. Immunoglobulin gene mutations and frequent use of VH1-69 and VH4-34 segments in hepatitis C virus-positive and hepatitis C virus-negative nodal marginal zone B-cell lymphoma. Am J Pathol. 2001;159:253–61. doi: 10.1016/S0002-9440(10)61691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahler DW, Miklos JA, Swerdlow SH. Ongoing Ig gene hypermutation in salivary gland mucosa-associated lymphoid tissue-type lymphomas. Blood. 1997;89:3335–44. [PubMed] [Google Scholar]
- 25.Kunkel HG, Agnello V, Joslin FG, Winchester RJ, Capra JD. Cross-idiotypic specificity among monoclonal IgM proteins with anti-γ-globulin activity. J Exp Med. 1973;137:331–42. doi: 10.1084/jem.137.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agnello V, Arbetter A, Ibanez de Kasep G, Powell R, Tan EM, Joslin F. Evidence for a subset of rheumatoid factors that cross-react with DNA-histone and have a distinct cross-idiotype. J Exp Med. 1980;151:1514–27. doi: 10.1084/jem.151.6.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radoux V, Chen PP, Sorge JA, Carson DA. A conserved human germline Vκ gene directly encodes rheumatoid factor light chains. J Exp Med. 1986;164:2119–24. doi: 10.1084/jem.164.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mageed RA, Dearlove M, Goodall DM, Jefferis R. Immunogenic and antigenic epitopes of immunoglobulins. XVII. Monoclonal antibodies reactive with common and restricted idiotopes to the heavy chain of human rheumatoid factors. Rheumatol Int. 1986;6:179–83. doi: 10.1007/BF00541285. [DOI] [PubMed] [Google Scholar]
- 29.Pascual V, Victor K, Randen I, Thompson K, Steinitz M, Forre O, et al. Nucleotide sequence analysis of rheumatoid factors and polyreactive antibodies derived from patients with rheumatoid arthritis reveals diverse use of VH and VL gene segments and extensive variability in CDR-3. Scand J Immunol. 1992;36:349–62. doi: 10.1111/j.1365-3083.1992.tb03108.x. [published erratum appears in Scand J Immunol 1994;40:125] [DOI] [PubMed] [Google Scholar]
- 30.Natvig JB, Randen I, Thompson K, Forre O, Mageed RA, Jefferis R, et al. Probing of the rheumatoid factor (RF) V gene repertoire in rheumatoid arthritis (RA) by hybridoma clones. Clin Exp Rheumatol. 1990;8(Suppl 5):75–80. [PubMed] [Google Scholar]
- 31.Carayannopoulos MO, Potter KN, Li Y, Natvig JB, Capra JD. Evidence that human immunoglobulin M rheumatoid factors can be derived from the natural autoantibody pool and undergo an antigen driven immune response in which somatically mutated rheumatoid factors have lower affinities for immunoglobulin G Fc than their germline counterparts. Scand J Immunol. 2000;51:327–36. doi: 10.1046/j.1365-3083.2000.00731.x. [DOI] [PubMed] [Google Scholar]
- 32.Pascual V, Randen I, Thompson K, Sioud M, Forre O, Natvig J, et al. The complete nucleotide sequences of the heavy chain variable regions of six monospecific rheumatoid factors derived from Epstein-Barr virus-transformed B cells isolated from the synovial tissue of patients with rheumatoid arthritis: further evidence that some autoantibodies are unmutated copies of germ line genes. J Clin Invest. 1990;86:1320–8. doi: 10.1172/JCI114841. [published erratum appears in J Clin Invest 1994;94:466] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin T, Crouzier R, Weber JC, Kipps TJ, Pasquali JL. Structure-function studies on a polyreactive (natural) autoantibody: polyre-activity is dependent on somatically generated sequences in the third complementarity-determining region of the antibody heavy chain. J Immunol. 1994;152:5988–96. [PubMed] [Google Scholar]
- 34.Agnello V, Barnes JL. Human rheumatoid factor crossidiotypes. I. WA and BLA are heat-labile conformational antigens requiring both heavy and light chains. J Exp Med. 1986;164:1809–14. doi: 10.1084/jem.164.5.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corper AL, Sohi MK, Bonagura VR, Steinitz M, Jefferis R, Feinstein A, et al. Structure of human IgM rheumatoid factor Fab bound to its autoantigen IgG Fc reveals a novel topology of antibody-antigen interaction. Nat Struct Biol. 1997;4:374–81. doi: 10.1038/nsb0597-374. [DOI] [PubMed] [Google Scholar]
- 36.Martin T, Weber JC, Levallois H, Labouret N, Soley A, Koenig S, et al. Salivary gland lymphomas in patients with Sjögren’s syndrome may frequently develop from rheumatoid factor B cells. Arthritis Rheum. 2000;43:908–16. doi: 10.1002/1529-0131(200004)43:4<908::AID-ANR24>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 37.Axford JS, Rees DH, Mageed RA, Wordsworth P, Alavi A, Steere AC. Increased IgA rheumatoid factor and VH1 associated cross reactive idiotype expression in patients with Lyme arthritis and neuroborreliosis. Ann Rheum Dis. 1999;58:757–61. doi: 10.1136/ard.58.12.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herve M, Xu K, Ng YS, Wardemann H, Albesiano E, Messmer BT, et al. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J Clin Invest. 2005;115:1636–43. doi: 10.1172/JCI24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steininger C, Widhopf GF II, Ghia EM, Morello CS, Vanura K, Sanders R, et al. Recombinant antibodies encoded by IGHV1-69 react with pUL32, a phosphoprotein of cytomegalovirus and B-cell superantigen. Blood. 2012;119:2293–301. doi: 10.1182/blood-2011-08-374058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pos W, Luken BM, Kremer Hovinga JA, Turenhout EA, Scheiflinger F, Dong JF, et al. VH1-69 germline encoded antibodies directed towards ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. J Thromb Haemost. 2009;7:421–8. doi: 10.1111/j.1538-7836.2008.03250.x. [DOI] [PubMed] [Google Scholar]
- 41.Cheng J, Torkamani A, Grover RK, Jones TM, Ruiz DI, Schork NJ, et al. Ectopic B-cell clusters that infiltrate transplanted human kidneys are clonal. Proc Natl Acad Sci U S A. 2011;108:5560–5. doi: 10.1073/pnas.1101148108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–93. doi: 10.1084/jem.20101352. [published erratum appears in J Exp Med 2011;208: 411] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dennison SM, Anasti K, Scearce RM, Sutherland L, Parks R, Xia SM, et al. Nonneutralizing HIV-1 gp41 envelope cluster II human monoclonal antibodies show polyreactivity for binding to phospholipids and protein autoantigens. J Virol. 2011;85:1340–7. doi: 10.1128/JVI.01680-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–8. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 45.Craig VJ, Arnold I, Gerke C, Huynh MQ, Wundisch T, Neubauer A, et al. Gastric MALT lymphoma B cells express polyreactive, somatically mutated immunoglobulins. Blood. 2010;115:581–91. doi: 10.1182/blood-2009-06-228015. [DOI] [PubMed] [Google Scholar]
- 46.Winkler TH, Fehr H, Kalden JR. Analysis of immunoglobulin variable region genes from human IgG anti-DNA hybridomas. Eur J Immunol. 1992;22:1719–28. doi: 10.1002/eji.1830220709. [DOI] [PubMed] [Google Scholar]
- 47.Guo W, Smith D, Aviszus K, Detanico T, Heiser RA, Wysocki LJ. Somatic hypermutation as a generator of antinuclear antibodies in a murine model of systemic autoimmunity. J Exp Med. 2010;207:2225–37. doi: 10.1084/jem.20092712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Racanelli V, Brunetti C, De Re V, Caggiari L, De Zorzi M, Leone P, et al. Antibody Vh repertoire differences between resolving and chronically evolving hepatitis C virus infections. PLoS One. 2011;6:e25606. doi: 10.1371/journal.pone.0025606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roosnek E, Lanzavecchia A. Efficient and selective presentation of antigen-antibody complexes by rheumatoid factor B cells. J Exp Med. 1991;173:487–9. doi: 10.1084/jem.173.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Notkins AL. Infectious virus-antibody complexes: interaction with anti-immunoglobulins, complement, and rheumatoid factor. J Exp Med. 1971;134:41–51. [PMC free article] [PubMed] [Google Scholar]