Abstract
The assembly of centromeric nucleosomes is mediated by histone variant-specific chaperones. In budding yeast, the centromere-specific histone H3 variant is Cse4, and the histone chaperone Scm3 functions as a Cse4-specific nucleosome assembly factor. Here, we show that Scm3 exhibits specificity for Cse4–H4, but also interacts with major-type H3–H4 and H2A–H2B. Previously published structures of the Scm3 histone complex demonstrate that Scm3 binds only one copy of Cse4–H4. Consistent with this, we show that Scm3 deposits Cse4–H4 through a dimer intermediate onto deoxyribonucleic acid (DNA) to form a (Cse4–H4)2–DNA complex (tetrasome). Scm3-bound Cse4–H4 does not form a tetramer in the absence of DNA. Moreover, we demonstrate that Cse4 and H3 are structurally compatible to be incorporated in the same nucleosome to form heterotypic particles. Our data shed light on the mechanism of Scm3-mediated nucleosome assembly at the centromere.
INTRODUCTION
The centromere is a defined region on each eukaryotic chromosome that provides a platform for kinetochore assembly and plays a critical role in proper chromosome segregation during cell division. The deoxyribonucleic acid (DNA) sequence at the centromere is not conserved between species, and ranges in size from kilobases to megabases. It is for the most part dispensable for centromere function and identity (1). The exception is Saccharomyces cerevisiae, where the centromere is defined by a single 125 bp stretch of DNA that accommodates a single nucleosome (2–6). Despite these differences, the common mark of centromeres in all eukaryotic organisms is the centromere-specific histone H3 variant (generically referred to as CENP-A, after its first discovery in humans (7), and Cse4 in S. cerevisiae (8,9)). Centromere-specific H3 variants are essential in all eukaryotes. They substitute for H3 in centromeric nucleosomes, are required for kinetochore formation (9,10), and are thought to be the epigenetic mark for centromere identity (1,11,12).
The CENP-A/Cse4-containing nucleosome provides the structural basis for centromere identity and function. Several studies demonstrate that octameric CENP-A/Cse4-containing nucleosomes with stoichiometric amounts of the four histones, are assembled in vitro (13–19). Moreover, in vitro reconstituted octameric CENP-A-containing nucleosomal arrays support the binding of centromeric and kinetochore proteins (20), suggesting that the octameric CENP-A nucleosome indeed supports centromere function. However, several different models have been proposed based on in vivo data generated from various organisms and cell types, using a variety of experimental approaches (21–26). These include an octameric nucleosome (as also demonstrated in vitro), a ‘tetrasome’ (two copies of CENP-A–H4, no H2A–H2B), a ‘hemisome’ (one copy each of H2A, H2B, CENP-A and H4), and in yeast, a ‘hexasome’ (two copies of CENP-A–H4, and two copies of the non-histone protein Scm3 replacing the H2A–H2B dimers). In budding yeast, it has also been suggested that both H3 and Cse4 histones co-exist in a single nucleosome on CEN DNA, forming an octameric hybrid (heterotypic) nucleosome together with two copies each of H4, H2A and H2B (27). Recent work attempted to reconcile these conflicting results by suggesting that CENP-A nucleosomes undergoes structural transitions in a cell-cycle-specific manner (28,29); however, this was disputed by subsequent reports (25,26,30). As such, the structure of centromeric nucleosomes in vivo throughout various stages of the cell cycle remains controversial.
The targeting of CENP-A/Cse4 and its deposition at the centromere is mediated by the CENP-A-specific histone chaperone HJURP in mammals, and by its functional homolog Scm3 in fungi (22,23,31–35). In vitro data shows that HJURP/Scm3 binds CENP-A/Cse4 and exhibits CENP-A-/Cse4-specific nucleosome assembly activity (16,36–38). This is in contrast with many general histone chaperones, which often lack specificity for histones and assemble the different histone variants into nucleosomes with equal efficiency. For example, the histone chaperone Nucleosome Assembly Protein 1 (Nap1) binds two copies of either H3–H4 or H2A–H2B with similarly high affinity (low nanomolar Kd; (39–41)), and assembles both CENP-A and H3 nucleosomes in vitro (13,16,29,37).
The structures of HJURP–CENP-A–H4 and Scm3–Cse4–H4 complexes show that both chaperones interact with one copy of CENP-A/Cse4–H4 to form a heterotrimeric complex. This mode of interaction is clearly incompatible with (CENP-A/Cse4–H4)2 heterotetramer formation and with the subsequent interaction of the heterotetramer with DNA (42–45). However, Cse4 nucleosomes assembled by Scm3 in vitro contain stoichiometric amounts of all four histones (Cse4, H4, H2A and H2B) (16,29), raising the question where and how the Cse4–H4 tetramer is assembled. Additionally, we wanted to follow up on the observation that H3 and Cse4 co-localize in a single nucleosome at least under certain conditions (27).
Here, we have applied quantitative assays to show that Scm3 binds Cse4–H4 with high affinity and with a more than 10-fold preference over H3–H4. Scm3 assembles a (Cse4–H4)2 tetramer from two Cse4–H4 dimers on DNA, but not in the absence of DNA. This stepwise assembly of two Cse4–H4 dimers to form a DNA-bound (Cse4–H4)2 tetrasome is likely relevant to the assembly and maintenance of the centromere in higher organisms, and to the assembly of other H3 histone variants. Moreover, we present evidence that Cse4 and H3 are structurally compatible to form a heterotypic nucleosome consisting of a single copy of H3 and Cse4 and two copies of H4, H2A and H2B.
MATERIALS AND METHODS
DNA preparation
The 147 bp ‘601’ DNA (46) and 207 bp CEN3 DNA fragments were prepared by restriction enzyme digestion of the appropriate plasmids. The 79 bp DNA fragment corresponding to the (H3–H4)2 tetramer binding region of the ‘601’ DNA sequence was prepared by annealing two complementary oligonucleotides.
Protein purification and refolding
Histones Cse4, Cse4ΔN, H3, His6·H3, H4, H2A and H2B, and the histone chaperone Scm3 (wild-type and Scm363–189 mutant) were purified as described previously (16,47). Scm3 mutants I111D I117N and V158G L159G I161G were generated by site-directed mutagenesis, and purified and refolded as described for wild-type Scm3 (16). Mutant histones H4 E63C, H2A T118C, and the endogenous cysteine at position 41 of Scm3 were used for labeling the proteins with fluorescent dye. A yeast Nap1 labeling mutant that contains a single endogenous cysteine at position 414 (all other cysteine residues were mutated to alanine) was expressed and purified as previously described (39).
Fluorescent labeling
Histone labeling mutants (H4 E63C or H2A T118C) in 7 M guanidinium–HCl, 20 mM Tris–HCl pH 7.5 and 1 mM Tris(2-carboxyethyl)phosphine (TCEP) were incubated with a 2-fold molar excess of Alexa-488 or Atto-647N maleimide dye on ice for 3 h. Excess dye was removed by gel filtration (PD-10 desalting column, GE Healthcare). Labeled H4 E63C was refolded into tetramer with Cse4, Cse4ΔN or H3, and labeled H2A T118C was refolded with H2B into dimer, and purified by gel filtration (48). Similarly, labeled H4 E63C was refolded with Cse4ΔN and Scm3 to form the Scm3–Cse4ΔN–H4 complex, as described (16,23). Refolded Scm3 was labeled with Atto-647N at the endogenous cysteine at position 41 (C41) and the free dye was removed by gel filtration.
High-throughput interactions by fluorescence intensity (HI-FI)
Scm3 and Nap1 interactions with the various histone complexes was measured by HI-FI Fluorescence Resonance Energy Transfer (FRET) as described (49,50). Refolded Cse4 (Cse4ΔN)–H4, H3–H4 or H2A–H2B labeled with Alexa-488 (donor) was kept constant at 1–4 nM, and Atto-647N (acceptor) labeled Scm3 or Nap1 was titrated from 0 to 1000 nM in buffer containing 10 mM Tris–HCl (pH 7.5), 5% glycerol, 300 mM NaCl, 0.01% NP40 and 0.01% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), and incubated at room temperature for 30 min. Binding affinities from the FRET signal were derived as described (50). A FRET competition assay was used to measure the relative affinity of wild-type and Scm3 mutants. To this end, a 5-fold molar excess of Scm3 (50 μM) labeled with Atto-647N (acceptor) was added to Cse4ΔN–H4 (10 μM) labeled with Alexa-488 (donor). Unlabeled wild-type or mutant Scm3 was titrated and the reduction in FRET signal was measured as a function of unlabeled Scm3 concentration using the equation described in (50).
To measure (Cse4ΔN–H4)2 tetramer and tetrasome formation from preformed Scm3–Cse4ΔN–H4 complex we used a variation of the HI-FI assay. Alexa-488 labeled Scm3–Cse4ΔN–H4 (10 nM) was held constant and Atto-647N labeled Scm3–Cse4ΔN–H4 was titrated (0–1000 nM) with or without equimolar amounts of DNA (147 bp ‘601’ or 207 CEN3 DNA). The FRET signal was quantified using equations described previously (50), and the data was fit using a homodimerization equation (51).
Cse4–H4 deposition by Scm3
Deposition of Scm3-bound Cse4ΔN–H4 onto DNA was examined using an in-gel FRET assay. 0.33–1.0 μM preformed Scm3–Cse4ΔN–H4 (or Scm363–189–Cse4ΔN–H4) complex labeled at H4 E63C with either Alexa-488, or with Atto-647N was combined with 1.7–5 μM of ‘601’ DNA (either 79 or 147 bp, as indicated) and incubated at room temperature for an hour in 20 mM Tris–HCl (pH 7.5), 300 mM NaCl, 100 ng/ml BSA and 1 mM ethylenediaminetetraacetic acid (EDTA). For most experiments, Scm3–Cse4ΔN–H4 labeled with Atto-647N or Alexa-488 at H4 E63C. For some lanes in Supplementary Figure S1B, Scm3 C41 was labeled with Atto-647N. Similarly, 1 μM donor labeled (H3–H4)2 tetramer was mixed with equimolar amounts of acceptor labeled (H3–H4)2 tetramer in the presence of a 4-fold molar excess of Nap1 (as a monomer), and incubated with 5 μM of 79 bp DNA as described above. To adjust for the variability in the degree of modification by fluorescent dyes, and to ensure equal level of fluorescent signal, the labeled Scm3–Cse4ΔN–H4 and H3–H4 were adjusted to the same degree of fluorescence substitution with unlabeled corresponding complexes. Samples were analyzed by 5–6% native polyacrylamide gel electrophoresis (PAGE) and scanned for donor, acceptor and FRET signal.
Tetrasome and nucleosome reconstitution by salt dilution
A 10 μl solution of 147 bp ‘601’ DNA (12 μM) and 24 μM tetramer ((Cse4ΔN–H4)2, (H3–H4)2, or Cse4ΔN–His6·H3–H42, or (His6·H3–H4)2), with or without 35 μM H2A–H2B dimer was made in buffer consisting of 25 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 mM β-mercaptoethanol and 2 M NaCl, and reconstituted into tetrasome or nucleosome at 37°C using the rapid salt dilution method (52). The reaction was diluted with buffer (25 mM Tris–HCl, pH 7.5, 1 mM EDTA, 1 mM β-mercaptoethanol) to 1.3, 0.95, 0.57 and 0.3 M NaCl, with a 30-min incubation at 37°C for each step, and the samples were analyzed by native PAGE and ethidium bromide staining.
Refolding and purification of heterotypic tetramer
All histones were refolded using the histone octamer refolding protocol (53). The refolded Cse4, His6·H3 (H3) and H4 samples were bound to Ni-NTA resin equilibrated with 20 mM buffer Tris–HCl pH 7.5, 1 M NaCl and 10 mM imidazole. After washing with buffer containing 20 mM imidazole, the samples were eluted by increasing imidazole concentration to 250 mM, and analyzed by sodium dodecylsulphate (SDS)-PAGE and Coomassie staining. In a separate experiment, refolded Cse4ΔN, His6·H3 and H4 complex was purified by gel filtration and the fractions containing Cse4ΔN, His6·H3 and H4 were used for tetrasome and nucleosome assembly. Tetrasomes or nucleosomes assembled with refolded Cse4ΔN, His6·H3 (or untagged H3) and H4 on 147 bp DNA were purified by Ni-NTA similarly except the salt concentration used here was 150 mM NaCl. In some cases (as indicated), the nucleosomes were purified by 5–25% sucrose gradient before purifying by Ni-NTA to remove the unbound (free) histone proteins.
RESULTS
Scm3 binds Cse4–H4 with high affinity
To understand how Scm3 acts as an assembly factor specific to centromeric nucleosomes, we first quantified the affinity of Scm3 for Cse4–H4 and H3–H4 using the HI-FI FRET assay (49). Full length Scm3 was labeled with Atto-647N (acceptor, A) at its endogenous cysteine 41, and titrated into a constant amount of refolded Cse4–H4 or H3–H4, where H4 was labeled with Alexa-488 (donor, D) at cysteine 63. Cysteine 41 of Scm3 is outside the critical region for Cse4–H4 interaction (43), and labeling this residue with a fluorophore is unlikely to affect Scm3 interactions with Cse4–H4. The binding curves shown in Figure 1A (resulting affinity data summarized in Table 1) demonstrate that Scm3 interacts with refolded Cse4–H4 (which had been purified at high concentration as a (Cse4–H4)2 tetramer) with nanomolar affinity (Kd ∼ 18 nM). This is within the range of affinities exhibited by other, generic histone chaperones for various histone complexes (39,54–57). The long N-terminal tail of Cse4 does not contribute to the interaction with Scm3, since a version of Cse4 in which the first N-terminal 127 amino acids have been deleted (Cse4ΔN) binds with the same affinity (Table 1); thus, in the majority of subsequent experiments Cse4ΔN was used unless specified as full length Cse4.
Figure 1.
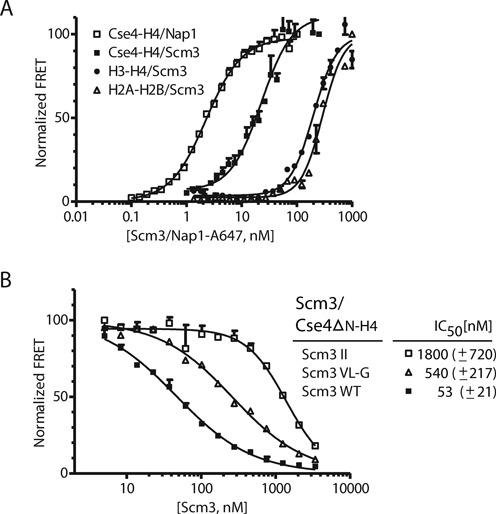
Scm3 binds to Cse4–H4 with nanomolar affinity. (A) Affinities of Scm3 for various histone complexes were measured using the HI-FI assay (50). ‘Histone complexes, as indicated, were labeled with Alexa-488 (donor, held constant at 1–4 nM), and incubated with increasing amounts of Atto-647N labeled Scm3 or Nap1 (acceptor) and scanned for donor, acceptor and FRET fluorescent signal. Representative binding curves are shown; all data are summarized in Table 1. (B) Competition assay to measure the relative affinity of wild-type and Scm3 mutants. Acceptor labeled Scm3 and donor labeled Cse4ΔN–H4 were kept constant at 50 and 10 nM, respectively. Unlabeled wild-type or mutant Scm3 was titrated and the FRET signal was monitored. Wild-type Scm3: solid squares (scm3 WT); double Scm3 mutations I111D I117N (Scm3 II): open squares; triple Scm3 mutations V158G L159G I161G (Scm3 VL-G): open triangles. The inset lists the derived IC50 values with standard deviations; derived from at least three independent experiments.
Table 1. Binding affinity of Scm3 to histone complexes, and equilibrium constants for (Cse4–H4)2 tetramer formation.
| Scm3 | Kdapp (nM) | Hill coefficient | Overall fit (R2) |
|---|---|---|---|
| Cse4–H4 | 17.5 ± 5.2 | 1.1 ± 0.45 | 0.98 |
| Cse4ΔN–H4 | 17.8 ± 2.4 | 1.4 ± 0.23 | 0.98 |
| H3–H4 | 195.8 ± 7.7 | 1.8 ± 0.18 | 0.94 |
| H2A–H2B | 305.0 ± 40.9 | 1.7 ± 0.47 | 0.96 |
| Nap1 | |||
| Cse4–H4 | 2.4 ± 0.33 | 1.3 ± 0.08 | 0.99 |
| H3–H4 | 0.7 ± 0.04 | 1.7 ± 0.15 | 0.99 |
| (Cse4ΔN–H4)2 tetramer formation on DNA from Scm3–Cse4ΔN–H4 | |||
| ‘601’ DNA (147 bp) | 44.1 ± 11.0 | 1.1 ± 0.3 | 0.98 |
| CEN3 DNA (207 bp) | 34.6 ± 10.2 | 1.5 ± 0.6 | 0.96 |
The calculated dissociation constants (Kdapp), Hill coefficients and overall non-linear fit of the data (R2) were calculated from experiments as shown in Figure 1A. Experiments to measure (Cse4ΔN–H4)2 tetramer formation on DNA from a Scm3–Cse4ΔN–H4 trimeric complex are shown in Figure 2C. The apparent Kd for (Cse4ΔN–H4)2 tetramer formation on DNA from a Scm3–Cse4ΔN–H4 trimeric complex was measured using HI-FI. Standard deviations were calculated from at least three replicate experiments.
Scm3 exhibits measurable affinity for H3–H4 and H2A–H2B; however, these affinities are at least one order of magnitude weaker than that for Cse4–H4 (Figure 1A and Table 1). The quantitative binding data presented here is in agreement with previous qualitative data demonstrating Scm3 specificity for Cse4–H4 over H3–H4 (23). The unexpected, significant interaction of Scm3 with histones H2A–H2B demonstrates a generic histone binding property of this Cse4-specific histone chaperone even at 300 mM NaCl.
Using a competition FRET assay (50), we examined the effect of mutations in the Scm3 core motif previously shown to affect Scm3 binding to Cse4–H4 (43). Scm3 residues I110, I111, Y114 and I117 are all part of the αN helix that forms a hydrophobic cluster with Cse4-specific residues in the Scm3–Cse4–H4 complex (43). The mutation of Scm3 residues I111D/I117N in this hydrophobic cluster increased the IC50, and hence reduced its affinity for Cse4ΔN–H4 by a factor of 35 (Figure 1B). Previous isothermal calorimetry (ITC) binding measurements demonstrate that the replacement of I110D and I117N reduced the affinity of Scm3 for Cse4–H4 85-fold (43). According to published work, the αC helix of Scm3 (residues 155–161) interacts with H4, and substitution of amino acids V158G, L159G and I161G increased the IC50 (i.e. reduced the affinity) by a factor of 10 (Figure 1B). The relative affinity difference between wild-type Scm3 and mutant V158G, L159G and I161G is consistent with previously reported ITC data (43) and confirm that residues in the hydrophobic cluster at αN and the αC helix of Scm3 (residues 155–161) are required for Scm3 interaction with Cse4–H4. The values reported here are much more in keeping with the histone chaperone function of Scm3 than the previously published affinity estimates which were in the micromolar range (see ‘Discussion’ section).
In contrast to the high degree of specificity of Scm3 for Cse4–H4 over H3–H4, the general histone chaperone Nap1 from yeast binds both Cse4–H4 and H3–H4 with comparably high affinity (Kd ∼ 2.4 and 0.7 nM, respectively; Figure 1A and Table 1). This is consistent with the reported ability of Nap1 to facilitate the assembly of both canonical and CENP-A/Cse4-containing nucleosomes in vitro (13,16,29,37). Our data suggest that Nap1 and perhaps other general histone chaperones may function in chromatin assembly both at the centromere and at chromosome arms, while the centromere-targeted histone chaperone Scm3 functions specifically in assembling centromeric chromatin.
Scm3 promotes the formation of a Cse4–H4 tetramer on DNA
The structure of the histone-binding region of Scm3 in complex with Cse4–H4 shows that a single Cse4–H4 dimer binds a monomer of Scm3. The interaction of Scm3 with Cse4–H4 prevents the formation of a (Cse4–H4)2 tetramer, and blocks Cse4–H4 DNA binding regions (42,43). How then does Scm3 assemble an octameric nucleosome? More specifically, does Scm3 promote or preclude the formation of a (Cse4–H4)2 tetramer on DNA in vitro? To answer this question, we used an in-gel FRET assay. A preformed Scm3–Cse4ΔN–H4 complex, labeled with Alexa-488 on H4 E63C (D), was incubated with equal amounts of the same complex labeled with Atto-647N on H4 E63C (A), in the presence of a 147 bp ‘601’ DNA fragment. Complex formation was analyzed on a native gel, at wave lengths appropriate to visualize donor and acceptor fluorescence as well as FRET between the two fluorophores (Figure 2A). FRET is observed upon mixing donor and acceptor-labeled Scm3–Cse4ΔN–H4 complex with DNA (bottom panel, lanes 7–9). The complex for which FRET is observed migrates at the same position as a salt-reconstituted Cse4–H4 tetrasome (not shown). Mixing pre-assembled donor- and acceptor-labeled Cse4ΔN–H4–DNA complexes under the same conditions does not result in FRET, indicating that simple exchange of Cse4ΔN–H4 units bound to DNA does not account for our observation (Figure 2A, lane 10). To exclude the possibility that two neighboring tetramers undergo FRET, we repeated the experiment with a 79 bp DNA fragment encompassing the central tetramer-binding region of the ‘601’ DNA sequence, with similar results (Figure 2B, lanes 4–6). This is the minimal length of DNA required for tetramer binding (58). Since FRET is strongly distance-dependent, we conclude that Scm3 indeed assembles a (Cse4ΔN–H4)2 tetramer onto both DNA fragments.
Figure 2.
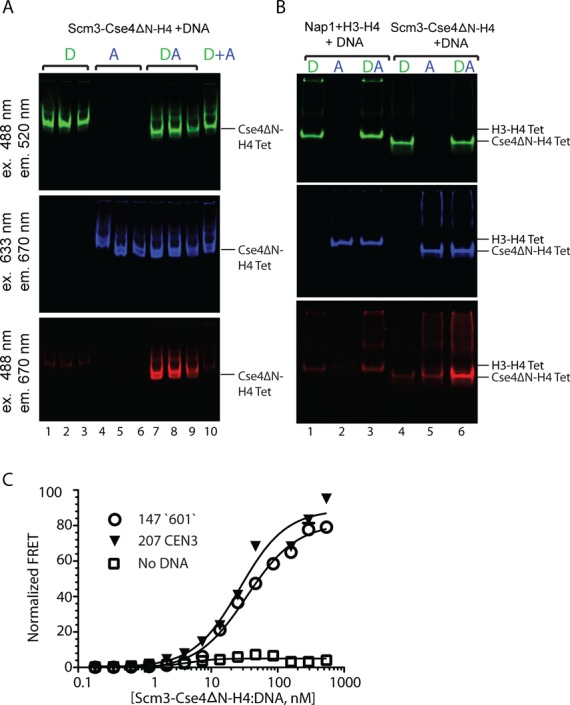
Scm3 promotes (Cse4–H4)2 tetramer formation on DNA. (A) A preformed complex of Scm3–Cse4ΔN–H4 (0.33 μM), labeled at H4 E63C with either Alexa-488 (D), or Atto-647N (A); or a mixture of the differently labeled complexes (DA) as indicated, was incubated with a five molar excess of 147 bp DNA. Protein–DNA complexes were analyzed by 5% native PAGE followed by fluorescence scanning. The reactions were done in triplicates: lanes 1–3, Scm3–Cse4ΔN–H4 (donor labeled) + DNA; lanes 4–6, Scm3–Cse4ΔN–H4 (acceptor labeled) + DNA; lanes 7–9: Scm3–Cse4ΔN–H4 (donor labeled) + Scm3–Cse4ΔN–H4 (acceptor labeled) + DNA. In lane 10, tetrasomes, previously assembled separately under the same conditions with either donor- or acceptor-labeled Cse4–H4 were mixed as a control for spontaneous exchange. Cse4ΔN–H4 Tet and H3-H4 Tet refer to the corrosponding tetrasomes. (B) Different mechanisms for Nap1 and Scm3 tetrasome formation. 1 μM refolded H3–H4 tetramer or Scm3–Cse4ΔN–H4 labeled with Alexa-488 (D) was mixed with an equimolar concentration of Atto-647N labeled tetramer in the presence of Nap1 (4 μM, calculated as a monomer) or Scm3–Cse4ΔN–H4 (A) and incubated with 5 μM 79 bp DNA. The samples were analyzed on a 6% native gel and scanned for fluorescent signal as in (A). Lanes 1–3: H3–H4 (labeled as indicated) with Nap1 and 79 bp DNA. Lanes 4–6: Scm3–Cse4ΔN–H4 (labeled as indicated) and 79 bp DNA. Top, middle and bottom gel scans are for donor, acceptor and FRET, respectively. (C) Cse4–H4 tetramer formation from a Scm3–Cse4–H4 complex is DNA dependent. Scm3–Cse4ΔN–H4–H4 (donor labeled, and at a constant concentration of 10 nM) was incubated with 0–1000 nM acceptor labeled Scm3–Cse4ΔN–H4 with or without DNA. The FRET signal was measured as a function of fluorescence acceptor and the data was fit using homodimerization equation. The apparent Kd values for (Cse4ΔN–H4)2 tetramer formation on DNA are presented in Table 1. Curve with open circles represents data with 147 bp ‘601’ DNA, black triangles with 207 bp CEN DNA and open squares without DNA.
In a control experiment, H3–H4 labeled with Alexa-488 (D), or H3–H4 labeled with Atto-647N (A) was combined with Nap1, and 79 bp DNA was then added, resulting in efficient tetrasome formation (Figure 2B, top and middle panels, lanes 1–3). When the two differently labeled tetramer–Nap1 complexes were mixed and incubated with DNA, tetrasomes were formed, but only background levels of FRET signal were observed (Figure 2B). This is in agreement with the finding that Nap1 deposits a preformed (H3–H4)2 tetramer onto DNA (59). We conclude that the mechanism of Cse4-nucleosome assembly involves the sequential deposition of two Cse4–H4 dimers by Scm3 to form a Cse4–H4 tetrasome, while the generic histone chaperone Nap1 assembles preformed (H3–H4)2 tetramers on DNA.
To quantify tetrasome formation and to examine the role of DNA in this process, we monitored FRET between differently labeled Cse4ΔN–H4 molecules in solution, in the presence and absence of DNA. Donor labeled Scm3–Cse4ΔN–H4 complex was held at a constant low concentration (10 nM), and acceptor labeled Scm3–Cse4ΔN–H4 was titrated either with or without an equimolar amount of 147 bp ‘601’ DNA. An increase in FRET signal was detected only in the presence of DNA, while the signal remained nearly constant throughout the titration range in the absence of DNA (Figure 2C). This demonstrates that Cse4–H4 tetramers are formed only on DNA, but are unable to form in their chaperone-bound form in the absence of DNA. The apparent Kd for Cse4ΔN–H4 tetramer formation on 147 bp ‘601’ DNA, obtained by fitting the data to a homodimerization model, is 44 nM (Figure 2C and Table 1).
The budding yeast centromere is characterized by a unique CEN DNA sequence, which harbors a highly AT-rich stretch in its center. This further destabilizes Cse4- or H3-containing nucleosomes (14,16,60). To test whether the unusual properties of CEN DNA affect Scm3-mediated tetrasome formation either positively or negatively, we repeated the above experiment with a 207 bp CEN3 DNA fragment. The apparent Kd for tetrasome formation on this DNA of 34 nM is within error of the value obtained for 147mer ‘601’ DNA (Figure 2C and Table 1). These data indicate that Scm3 promotes the deposition of Cse4–H4 dimers onto DNA to form a Cse4–H4 tetrasome, and that this process is not influenced by the sequence properties of CEN3.
In addition to its Cse4–H4 binding domain, Scm3 has a distinct DNA binding domain that has been mapped to amino acids 1–103 (60). We examined whether (Cse4–H4)2 tetramer formation on DNA is mediated through this DNA binding domain. We have previously shown that Scm3 in which the first 62 amino acids of DNA binding domain have been deleted (Scm363–189), exhibits Cse4-nucleosome assembly activity comparable to wild-type Scm3 (16). Gel shift experiments clearly show that Scm363–189 lacks DNA binding activity (Supplementary Figure S1A). Our data show that tetrasomes carrying two different labels are formed with equal efficiency by full length Scm3 and by the DNA-binding deficient version (Supplementary Figure S1B, compare lanes 3 and 6). We conclude that the assembly of Cse4–H4 to form a (Cse4–H4)2 tetramer on DNA from a Scm3–Cse4–H4 complex is independent of Scm3 DNA binding activity.
To examine whether Scm3 associates with the Cse4–H4/DNA complex (i.e. the tetrasome), two Scm3–Cse4ΔN–H4 complexes (one with Scm3 labeled with Atto-647N at C41 and the other with H4 E63C labeled with Alexa-488) were combined with 79 bp DNA. Tetrasome assembly was monitored by the appearance of H4 fluorescence. No Scm3 fluorescence was observed to co-migrate with the tetrasome complex thus assembled (Supplementary Figure S1B, compare lanes 7 and 8), consistent with our previous findings (16). We found no evidence for a ‘hexasome’ with two copies of Scm3–Cse4–H4 bound to DNA, as suggested earlier (23).
Cse4 and H3 can both be incorporated into one ‘heterotypic’ nucleosome in vitro
In principle, the Scm3-mediated deposition of two individual Cse4–H4 heterodimers onto DNA could allow the formation of a heterotypic tetramer consisting of one Cse4–H4 dimer and one H3–H4 dimer (deposited by one of the many histone chaperones that bind to a H3–H4 dimer) each in vivo. Such a heterotypic tetrasome could be a key intermediate in centromeric nucleosome assembly. Alternatively, it might confer specific properties to centromeric chromatin, as proposed recently (27). A structural superposition of the four-helix bundle region from the closely related Kluyveromyces lactis Cse4–H4 tetramer structure (42) onto the corresponding region of the H3-containing yeast nucleosome structure (61) shows that Cse4 is compatible with H3 to form a heterotypic four-helical bundle (Supplementary Figure S2A). The amino acid residues in H3 that are critical for four-helical bundle formation (H3 H113, D/E123, L126 and I/L 130) (58,61) are conserved between H3 and Cse4/CENP-A from yeast to humans (Supplementary Figure S2B).
We therefore asked whether a heterotypic tetramer containing a single molecule each of Cse4 and H3, and two molecules of H4 could be assembled by refolding the three proteins together in the absence of DNA. N-terminally His6-tagged H3 (His6·H3) was refolded with Cse4 and H4 using our standard refolding protocol (53). After refolding, the samples were purified by Ni-NTA instead of size-exclusion. Cse4 co-elutes with His6·H3 and H4 from a Ni-NTA affinity column (Figure 3A), suggesting heterotypic tetramer formation. To test whether a heterotypic tetrasome can be assembled on DNA, we refolded Cse4ΔN, His6·H3 and H4, and purified the tetramer by gel filtration without affinity purification. The peak fraction that contains His6·H3, H4 and Cse4ΔN (likely in the form of a stochastic mixture of (His6·H3–H4)2, (Cse4ΔN–H4)2 and (His6·H3–Cse4ΔN–H42) tetramers; Supplementary Figure S3A, fraction 3), was reconstituted into tetrasomes on 147 bp ‘601’ DNA by salt dilution (Supplementary Figure S3B, lanes 2, 5, 8 and 11). Ni-NTA purification of these tetrasomes revealed that untagged Cse4ΔN co-purifies with His6·H3 and H4 (Supplementary Figure S3C, lane 5). This demonstrates that Cse4 and H3 are structurally compatible to form a heterotypic tetrasome (Cse4–H3–H42–DNA), consistent with in silico predictions of a hybrid structure (Supplementary Figure S2).
Figure 3.
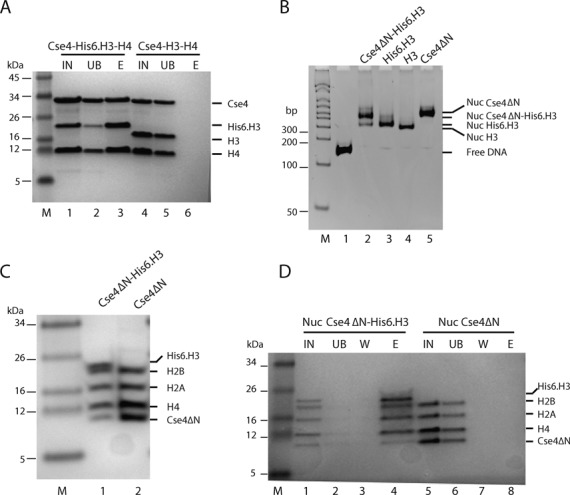
Heterotypic nucleosomes that contain both Cse4 and H3 can be reconstituted in vitro. (A) Cse4 forms a complex with H3 and H4. Full length Cse4, His6·H3 and H4 were refolded to a tetramer complex. The sample was purified by Ni-NTA and analyzed by 4–12% Bis–Tris SDS-PAGE gel and Coomassie staining. Lane 1, refolded Cse4–H3·His6–H4 input (IN); lane 2, unbound Ni-NTA fraction (UB); lane 3, 250 mM imidazole elution (E). Lanes 4, 5 and 6 are samples from refolded untagged Cse4–H3–H4 IN, UB and E, respectively. (B) Refolded H3–H4, Cse4ΔN–H4 or heterotypic tetramers (purified by gel filtration as shown in Supplementary Figure S3A) and H2A–H2B dimers were reconstituted into nucleosomes on 147 bp ‘601’ DNA using salt dilution, and analyzed by 6% native PAGE and ethidium bromide staining. Lane 1, 147 bp ‘601’ DNA; lane 2, Cse4ΔN–His6·H3–H42 tetramers and H2A–H2B; lane 3, His6·H3–H4 tetramer and H2A–H2B; lane 4, canonical (H3–H4)2 tetramer and H2A–H2B dimer; lane 5: (Cse4ΔN–H4)2 tetramer and H2A–H2B dimer. The tetramer used in lane 2 is likely a mixture of His6·H3–H4, Cse4ΔN–H4 and heterotypic Cse4ΔN–His6·H3–H42 tetramer (Supplementary Figure S3A). (C) Heterotypic nucleosomes contain Cse4ΔN, His6·H3, H4, H2A and H2B. The nucleosome bands from (B) were excised from the gel, eluted and analyzed by 4–12% Bis–Tris SDS-PAGE and Coomassie staining. Lanes 1 and 2: samples from the nucleosome bands of lanes 2 and 5 of the gel in (B), respectively. (D) Cse4ΔN and His6·H3 are incorporated into the same nucleosome. Nucleosome samples from (B) were purified from free histones by sucrose-gradient, and the nucleosome fractions were purified by Ni-NTA and analyzed on Bis–Tris 4–12% SDS-PAGE. IN, Ni-NTA input nucleosomes; UB, unbound; W, wash; E, elution fraction. Lanes 1–4: Nuc His6·H3–Cse4ΔN; nucleosomes assembled from Cse4ΔN–His6·H3–H42 tetramers and H2A–H2B dimers. Lanes 5–8: Nuc Cse4ΔN; nucleosome samples reconstituted from (Cse4ΔN–H4)2 tetramer and H2A–H2B dimer.
We next examined whether these heterotypic tetramers can form nucleosomes, by including H2A–H2B dimers in the reconstitution reactions (Figure 3B, also Supplementary Figure S3B, lanes 3 and 4). The nucleosome bands from Figure 3B (lanes 2 and 5) were excised from the gel and their histone content was analyzed by SDS-PAGE (Figure 3C). Nucleosomes reconstituted with heterotypic tetramers and H2A–H2B clearly contain Cse4ΔN in addition to His6·H3, H2A and H2B. As an independent proof for heterotypic nucleosome formation, we removed traces of free histones from the nucleosome preparations by sucrose gradient prior to Ni-NTA purification. As is the case for reconstituted tetrasomes, untagged Cse4ΔN co-purifies with tagged H3 after sucrose gradient and Ni-NTA purification (Figure 3D, compare lanes 4 and 8). Note that the Cse4 band is less intense than the other bands, as expected from the enrichment of homotypic His6–H3 containing nucleosomes during Ni-NTA purification. Together, this demonstrates that Cse4 and H3 can combine in a heterotypic nucleosome after in vitro assembly, and in the absence of histone chaperones. Similar to our previous observations (16), incorporation of Cse4 into nucleosomes alters its migration on a native gel, consistent with a more open structure. The nucleosome containing both Cse4ΔN and His6·H3 migrates slower on the gel than nucleosome containing two copies His6·H3 or H3 (Figure 3B).
Having established that Cse4 and H3 are structurally compatible to be refolded into a heterotypic H3–Cse4–H42 tetramer that can be assembled into nucleosomes by salt deposition, we wanted to test whether chaperone-mediated assembly has a similar outcome. Equal concentrations of donor labeled Scm3–Cse4ΔN–H4 and acceptor labeled H3–H4 (pre-incubated with Nap1) were mixed in the presence of a 5-fold molar excess of 79 bp DNA at physiologically relevant ionic strength and incubated for an hour. The samples were analyzed on a native gel and in-gel FRET was visualized (Figure 4A). In the absence of Nap1, little (H3–H4)2 tetrasome is formed on DNA (lane 4), while Nap1 facilitates the formation of donor-only and acceptor-only tetrasomes with little FRET (lane 3). Scm3–Cse4ΔN–H4 incubation with DNA (lane 7) results in FRET within a tetrasome, as also shown in Figure 2 and Supplementary Figure S1B. When combining Scm3–Cse4ΔN–H4 and Nap1–H3–H4, heterotypic tetrasomes are formed but less efficiently than homotypic tetrasomes, as the FRET signal from combining Scm3–Cse4ΔN–H4 (donor) and Nap1–H3–H4 (acceptor) in the presence of DNA is lower than the FRET signal obtained from incubating donor- and acceptor-labeled Scm3–Cse4ΔN–H4 with DNA (Figure 4A, compare lanes 7 and 9). (Cse4–H4)2 tetrasomes assembled on 79 bp DNA migrate faster than (H3–H4)2 tetrasomes (Figure 4A). This is more obvious in a gel stained with ethidium bromide (Figure 4B, lane 8; the heterotypic tetrasome is indicated by an asterisk). Together, the data presented in Figure 4 show that in the presence of Nap1, H3–H4 is deposited mainly as a tetramer and this likely reduces its propensity to form a heterotypic tetrasome with Cse4–H4. Because of the low affinity of Scm3 for H3–H4, Scm3 is unlikely to play a role in its deposition onto DNA. Overall, our data show that Scm3 deposits Cse4–H4 through a dimer intermediate while Nap1 mainly deposits H3–H4 as a tetramer.
Figure 4.
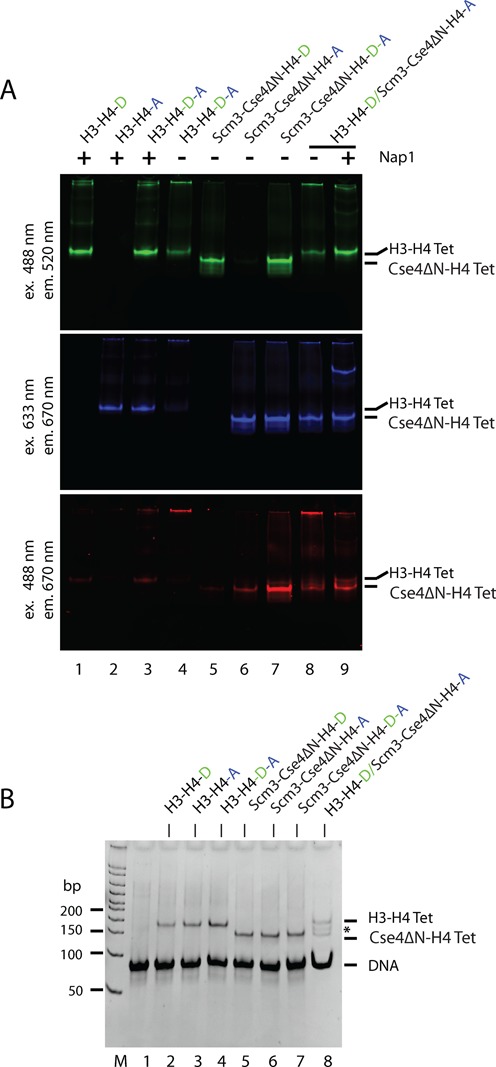
The deposition H3–H4 on DNA as a tetramer reduces the propensity of heterotypic Cse4–H3–H42 tetrasome formation. (A) 1 μM refolded (H3–H4)2 tetramer labeled with Alexa-488 was mixed with an equimolar concentration of Atto-647N labeled (H3–H4)2 tetramer (in presence or absence of 4 μM Nap1, calculated as a monomer) or Scm3–Cse4ΔN–H4 and incubated with 5 μM 79 bp DNA. Samples were incubated for 1 h at 25°C, then analyzed on a 6% native gel and scanned for fluorescence as in Figure 2A. (B) Tetrasomes assembled on 79 bp DNA as in (A), were analyzed on 6% native gel and stained with ethidium bromide. The band indicated by (*) is a heterotypic tetramer.
DISCUSSION
Scm3 is a histone chaperone that assembles Cse4-containing nucleosomes at the centromere in budding yeasts. Here, we have quantified the preferential interaction of Scm3 with the histone variant Cse4–H4 over H3–H4 and H2A–H2B, and have investigated the mechanism by which Scm3 assembles Cse4–H4 on DNA during the first step of centromeric nucleosome assembly. Scm3 binds Cse4–H4 with low nanomolar affinity, and with a 10–15-fold preference over canonical histones H3–H4 and H2A–H2B. In agreement with previous reports (43), mutations in Scm3 residues that are critical for the interaction with Cse4–H4 reduced the affinity significantly. The affinity of wild-type Scm3 for Cse4–H4 reported here is about 30-fold higher than previously published values measured by ITC (43). This difference is likely due to a markedly different experimental setup, as well as differences in conditions and proteins constructs used. Notably, the previous study was done at a pH 5.4, and a single-chain comprising partial sequences of Cse4–H4 was used, whereas our study used more physiological conditions and substrates. As such, the affinities reported here are within range reported for other histone chaperones under similar conditions (39,55–57,62).
The affinity of Scm3 for Cse4–H4 reported here is thermodynamically consistent with a role of Scm3 in ‘depositing’ histones onto DNA. We find that a (Cse4–H4)2 tetramer is assembled on DNA from two Scm3-bound Cse4–H4 dimers. The tetramerization of Cse4–H4 under physiological conditions happens only in the presence of DNA, and is not dependent on the DNA binding activity of Scm3. Two mutually non-exclusive mechanisms are possible. First, free Cse4–H4 (in equilibrium with Scm3-bound Cse4–H4) could form a transient (Cse4–H4)2 tetramer which is then stabilized in the form of a tetrasome upon DNA binding (Figure 5A). Second, it is also possible that the free Cse4–H4 dimer binds to DNA and the binding of a second Cse4–H4 dimer to form a tetrasome in turn stabilizes the complex (Figure 5B). In both cases, the association H2A/H2B dimers to (Cse3–H4)2 tetrasomes complete the assembly of the Cse4 nucleosome. Our data argues against a mechanism in which two Scm3 molecules dimerize to bring together two copies of Cse4–H4 before delivering them to DNA. Recent data demonstrate that dimerization of the human homolog HJURP through its C-terminal region is required for CENP-A deposition at the centromeres (63), suggesting that this mechanism may not be conserved between budding yeast and human centromeric histone chaperones.
Figure 5.
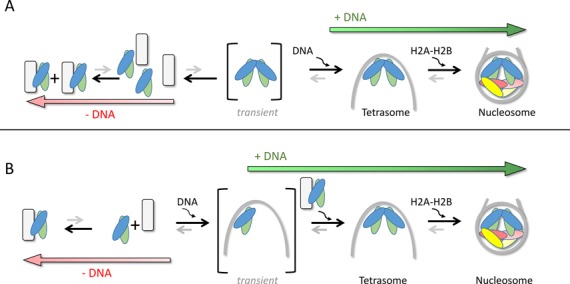
Model for Cse4-nucleosome assembly. In absence of DNA, the majority of Cse4–H4 forms a complex with Scm3. Any free Cse4–H4 dimer may bind a second Cse4–H4 dimer to form a tetramer, which is stabilized in the presence of DNA (A). Alternatively, a Cse4–H4 dimer binds to DNA and the binding of the second Cse4–H4 dimer leads a formation of a tetrasome (B). The tetrasome is further stabilized upon incorporation of H2A/H2B dimers to form a nucleosome. The formation of tetrasome and then nucleosomes shifts the equilibrium from Scm3–Cse4–H4 complex toward Cse4-nucleosome assembly.
The equilibrium constant of Scm3-mediated tetrasome formation is not affected by DNA sequence; in particular, we did not observe a difference between the centromeric CEN DNA and the strong ‘601’ positioning sequence. This indicates that the unusual sequence composition of CEN DNA does not contribute to the targeting of Cse4 to the yeast centromere. Rather, the DNA binding activity of the Scm3 N-terminal domain (60), and the ability of Scm3 to interact with other centromeric proteins such as the CBF3 subunit Ndc10 (22,64) might help target Scm3 (and with it, Cse4–H4) to the centromere. Previous results suggest that the N-terminal DNA binding domain of Scm3 is not required for the interaction with Cse4–H4, but is critical for Cse4 deposition at the centromere (60).
Our finding that the general histone chaperone Nap1 binds Cse4–H4, H3–H4 and H2A–H2B with equally high affinity suggests that Nap1 and other general histone chaperones might participate in the assembly/disassembly of centromeric nucleosomes in vivo. Previous reports showed that Nap1 has the ability to assemble both regular and centromeric nucleosomes in vitro (13,16,29,37). The lethal phenotype resulting from a scm3 deletion in yeast can be rescued by the overexpression of Cse4 (14), suggesting that other general histone chaperones may function in centromeric nucleosomes assembly. In other organisms, several general histone chaperones have been proposed to double as CENP-A histone chaperones. For example, RbAp48 forms a complex with Drosophila CENP-A–H4 in vivo and functions as CENP-A nucleosome assembly factor in vitro (65). FACT and nucleoplasmin-1 (Npm1) co-purify with CENP-A nucleosomes in human cells (66). In vitro, Npm1 assembles both CENP-A and H3 nucleosomes (37). Sim3 (a human NASP homolog) deposits CENP-A in Schizosaccharomyces pombe (67), and human NASP exhibits CENP-A nucleosome assembly activity in vitro (68).
The deposition of newly synthesized CENP-A at centromeres is restricted to a discrete time that varies among different organisms from metaphase to early G1 (69–74). In contrast, replication-coupled assembly of bulk chromatin occurs during S phase. It has been suggested that the gaps left after the segregation of parental CENP-A molecules are temporarily filled by the histone variant H3.3, to be replaced later with CENP-A (75). It is not clear at this point whether the newly synthesized CENP-A–H4 loaded by Scm3/HJURP combines with existing DNA-bound CENP-A–H4 during replication-independent centromeric nucleosome assembly, or whether new CENP-A–H4 replaces the H3.3 nucleosomes that fill the gaps after DNA duplication. Our data show that Cse4 and H3 are structurally compatible to form heterotypic nucleosomes, and these possibly exist in vivo (27). However, the temporal separation of bulk chromatin assembly during S phases, and CENP-A nucleosome assembly that occurs outside S phase likely limits the formation of heterotypic nucleosomes at the centromeres. Reports have shown that CENP-A is deposited in non-centromeric regions (14,76,77). Further investigation will determine whether heterotypic nucleosomes assemble in centromere or other regions of the chromosome and have any functional roles in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online, including [1,2].
ACKNOWLEDGMENTS
We thank H. Scherman and D. Noblitt from the CSU Protein Expression and Purification Facility, as well as P. Dyer and A. White for reagents. We also thank A. Hieb for help with FRET experiments and K. Brown for critically reading the manuscript.
FUNDING
National Institutes of Health [GM067777 to K.L.]; American Heart Association [AHA-10POST4190042 postdoctoral fellowship to M.L.D.]; Howard Hughes Medical Institute. Source of open access funding: NIH, HHMI.
Conflict of interest statement None declared.
REFERENCES
- 1.Cleveland D.W., Mao Y., Sullivan K.F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 3.Cottarel G., Shero J.H., Hieter P., Hegemann J.H. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 1989;9:3342–3349. doi: 10.1128/mcb.9.8.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furuyama S., Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. PNAS. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henikoff S., Henikoff J.G. “Point” centromeres of Saccharomyces harbor single centromere-specific nucleosomes. Genetics. 2012;190:1575–1577. doi: 10.1534/genetics.111.137711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole H.A., Nagarajavel V., Clark D.J. Perfect and imperfect nucleosome positioning in yeast. Biochim. Biophys. Acta. 2012;1819:639–643. doi: 10.1016/j.bbagrm.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earnshaw W.C., Allshire R.C., Black B.E., Bloom K., Brinkley B.R., Brown W., Cheeseman I.M., Choo K.H., Copenhaver G.P., Deluca J.G., et al. Esperanto for histones: CENP-A, not CenH3, is the centromeric histone H3 variant. Chromosome Res. 2013;21:101–106. doi: 10.1007/s10577-013-9347-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer D.K., O’Day K., Wener M.H., Andrews B.S., Margolis R.L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoler S., Keith K.C., Curnick K.E., Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 10.Howman E.V., Fowler K.J., Newson A.J., Redward S., MacDonald A.C., Kalitsis P., Choo K.H. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. PNAS. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warburton P.E. Epigenetic analysis of kinetochore assembly on variant human centromeres. Trends Genet. 2001;17:243–247. doi: 10.1016/s0168-9525(01)02283-1. [DOI] [PubMed] [Google Scholar]
- 12.Henikoff S., Furuyama T. Epigenetic inheritance of centromeres. Cold Spring Harb. Symp. Quant. Biol. 2010;75:51–60. doi: 10.1101/sqb.2010.75.001. [DOI] [PubMed] [Google Scholar]
- 13.Yoda K., Ando S., Morishita S., Houmura K., Hashimoto K., Takeyasu K., Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. PNAS. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camahort R., Shivaraju M., Mattingly M., Li B., Nakanishi S., Zhu D., Shilatifard A., Workman J.L., Gerton J.L. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekulic N., Bassett E.A., Rogers D.J., Black B.E. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dechassa M.L., Wyns K., Li M., Hall M.A., Wang M.D., Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat. Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kingston I.J., Yung J.S., Singleton M.R. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J. Biol. Chem. 2011;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K., Hayashi-Takanaka Y., Oda T., Sato M., Park S.Y., et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 19.Miell M.D., Fuller C.J., Guse A., Barysz H.M., Downes A., Owen-Hughes T., Rappsilber J., Straight A.F., Allshire R.C. CENP-A confers a reduction in height on octameric nucleosomes. Nat. Struct. Mol. Biol. 2013;20:763–765. doi: 10.1038/nsmb.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guse A., Carroll C.W., Moree B., Fuller C.J., Straight A.F. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalal Y., Wang H., Lindsay S., Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camahort R., Li B., Florens L., Swanson S.K., Washburn M.P., Gerton J.L. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Mizuguchi G., Xiao H., Wisniewski J., Smith M.M., Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 24.Furuyama T., Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasson D., Panchenko T., Salimian K.J., Salman M.U., Sekulic N., Alonso A., Warburton P.E., Black B.E. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 2013;20:687–695. doi: 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padeganeh A., Ryan J., Boisvert J., Ladouceur A.M., Dorn J.F., Maddox P.S. Octameric CENP-A nucleosomes are present at human centromeres throughout the cell cycle. Curr. Biol. 2013;23:764–769. doi: 10.1016/j.cub.2013.03.037. [DOI] [PubMed] [Google Scholar]
- 27.Lochmann B., Ivanov D. Histone h3 localizes to the centromeric DNA in budding yeast. PLoS Genet. 2012;8:e1002739. doi: 10.1371/journal.pgen.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bui M., Dimitriadis E.K., Hoischen C., An E., Quenet D., Giebe S., Nita-Lazar A., Diekmann S., Dalal Y. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell. 2012;150:317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shivaraju M., Unruh J.R., Slaughter B.D., Mattingly M., Berman J., Gerton J.L. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell. 2012;150:304–316. doi: 10.1016/j.cell.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aravamudhan P., Felzer-Kim I., Joglekar A.P. The budding yeast point centromere associates with two Cse4 molecules during mitosis. Curr. Biol. 2013;23:770–774. doi: 10.1016/j.cub.2013.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoler S., Rogers K., Weitze S., Morey L., Fitzgerald-Hayes M., Baker R.E. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. PNAS. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams J.S., Hayashi T., Yanagida M., Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 34.Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R., 3rd, Bassett E.A., Wood S., Black B.E., Cleveland D.W. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pidoux A.L., Choi E.S., Abbott J.K., Liu X., Kagansky A., Castillo A.G., Hamilton G.L., Richardson W., Rappsilber J., He X., et al. Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuaib M., Ouararhni K., Dimitrov S., Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. PNAS. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnhart M.C., Kuich P.H., Stellfox M.E., Ward J.A., Bassett E.A., Black B.E., Foltz D.R. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shivaraju M., Camahort R., Mattingly M., Gerton J.L. Scm3 is a centromeric nucleosome assembly factor. J. Biol. Chem. 2011;286:12016–12023. doi: 10.1074/jbc.M110.183640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrews A.J., Downing G., Brown K., Park Y.J., Luger K. A thermodynamic model for Nap1-histone interactions. J. Biol. Chem. 2008;283:32412–32418. doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowman A., Ward R., Wiechens N., Singh V., El-Mkami H., Norman D.G., Owen-Hughes T. The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation. Mol. Cell. 2011;41:398–408. doi: 10.1016/j.molcel.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D’Arcy S., Martin K.W., Panchenko T., Chen X., Bergeron S., Stargell L.A., Black B.E., Luger K. Chaperone Nap1 shields histone surfaces used in a nucleosome and can put H2A-H2B in an unconventional tetrameric form. Mol. Cell. 2013;51:662–677. doi: 10.1016/j.molcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho U.S., Harrison S.C. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. PNAS. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Z., Feng H., Zhou B.R., Ghirlando R., Hu K., Zwolak A., Miller Jenkins L.M., Xiao H., Tjandra N., Wu C., et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu H., Liu Y., Wang M., Fang J., Huang H., Yang N., Li Y., Wang J., Yao X., Shi Y., et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong J., Feng H., Zhou Z., Ghirlando R., Bai Y. Identification of functionally conserved regions in the structure of the chaperone/Cen H3/H4 complex. J. Mol. Biol. 2012;425:536–545. doi: 10.1016/j.jmb.2012.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowary P.T., Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 47.Luger K., Rechsteiner T.J., Richmond T.J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 48.Luger K., Rechsteiner T.J., Richmond T.J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 49.Winkler D.D., Luger K., Hieb A.R. Quantifying chromatin-associated interactions: the HI-FI system. Methods Enzymol. 2012;512:243–274. doi: 10.1016/B978-0-12-391940-3.00011-1. [DOI] [PubMed] [Google Scholar]
- 50.Hieb A.R., D’Arcy S., Kramer M.A., White A.E., Luger K. Fluorescence strategies for high-throughput quantification of protein interactions. Nucleic Acids Res. 2012;40:e33. doi: 10.1093/nar/gkr1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodrich J.A., Kugel J.F. Binding and Kinetics for Molecualr Biologists. NY: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 52.Kassabov S.R., Henry N.M., Zofall M., Tsukiyama T., Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dyer P.N., Edayathumangalam R.S., White C.L., Bao Y., Chakravarthy S., Muthurajan U.M., Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 54.Park Y.J., Sudhoff K.B., Andrews A.J., Stargell L.A., Luger K. Histone chaperone specificity in Rtt109 activation. Nat. Struct. Mol. Biol. 2008;15:957–964. doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winkler D.D., Muthurajan U.M., Hieb A.R., Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 2011;286:41883–41892. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winkler D.D., Zhou H., Dar M.A., Zhang Z., Luger K. Yeast CAF-1 assembles histone (H3-H4)2 tetramers prior to DNA deposition. Nucleic Acids Res. 2012;40:10139–10149. doi: 10.1093/nar/gks812. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Liu W.H., Roemer S.C., Port A.M., Churchill M.E. CAF-1-induced oligomerization of histones H3/H4 and mutually exclusive interactions with Asf1 guide H3/H4 transitions among histone chaperones and DNA. Nucleic Acids Res. 2012;40:11229–11239. doi: 10.1093/nar/gks906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 59.Bowman A., Owen-Hughes T. Sulfyhydryl-reactive site-directed cross-linking as a method for probing the tetrameric structure of histones H3 and H4. Methods Mol. Biol. 2012;833:373–387. doi: 10.1007/978-1-61779-477-3_22. [DOI] [PubMed] [Google Scholar]
- 60.Xiao H., Mizuguchi G., Wisniewski J., Huang Y., Wei D., Wu C. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol. Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White C.L., Suto R.K., Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Donham D.C., 2nd, Scorgie J.K., Churchill M.E. The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4-DNA complexes. Nucleic Acids Res. 2011;39:5449–5458. doi: 10.1093/nar/gkr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zasadzinska E., Barnhart-Dailey M.C., Kuich P.H., Foltz D.R. Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 2013;32:2113–2124. doi: 10.1038/emboj.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aravind L., Iyer L.M., Wu C. Domain architectures of the Scm3p protein provide insights into centromere function and evolution. Cell Cycle. 2007;6:2511–2515. doi: 10.4161/cc.6.20.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Furuyama T., Dalal Y., Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. PNAS. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., 3rd, Cleveland D.W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 67.Dunleavy E.M., Pidoux A.L., Monet M., Bonilla C., Richardson W., Hamilton G.L., Ekwall K., McLaughlin P.J., Allshire R.C. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol. Cell. 2007;28:1029–1044. doi: 10.1016/j.molcel.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osakabe A., Tachiwana H., Matsunaga T., Shiga T., Nozawa R.S., Obuse C., Kurumizaka H. Nucleosome formation activity of human somatic nuclear autoantigenic sperm protein (sNASP) J. Biol. Chem. 2010;285:11913–11921. doi: 10.1074/jbc.M109.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jansen L.E., Black B.E., Foltz D.R., Cleveland D.W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schuh M., Lehner C.F., Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 71.Mellone B.G., Grive K.J., Shteyn V., Bowers S.R., Oderberg I., Karpen G.H. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moree B., Meyer C.B., Fuller C.J., Straight A.F. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silva M.C., Bodor D.L., Stellfox M.E., Martins N.M., Hochegger H., Foltz D.R., Jansen L.E. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 74.Bodor D.L., Valente L.P., Mata J.F., Black B.E., Jansen L.E. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol. Biol. Cell. 2013;24:923–932. doi: 10.1091/mbc.E13-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dunleavy E.M., Almouzni G., Karpen G.H. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeitlin S.G., Baker N.M., Chapados B.R., Soutoglou E., Wang J.Y., Berns M.W., Cleveland D.W. Double-strand DNA breaks recruit the centromeric histone CENP-A. PNAS. 2009;106:15762–15767. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lefrancois P., Euskirchen G.M., Auerbach R.K., Rozowsky J., Gibson T., Yellman C.M., Gerstein M., Snyder M. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


