Abstract
Long 3′ untranslated regions (3′UTRs) are common in eukaryotic mRNAs. In contrast, long 3′UTRs are rare in bacteria, and have not been characterized in detail. We describe a 3′UTR of 310 nucleotides in hilD mRNA, a transcript that encodes a transcriptional activator of Salmonella enterica pathogenicity island 1 (SPI-1). Deletion of the hilD 3′UTR increases the hilD mRNA level, suggesting that the hilD 3′UTR may play a role in hilD mRNA turnover. Cloning of the hilD 3′UTR downstream of the green fluorescent protein (gfp) gene decreases green fluorescent protein (GFP) activity in both Escherichia coli and S. enterica, indicating that the hilD 3′UTR can act as an independent module. S. enterica mutants lacking either ribonuclease E or polynucleotide phosphorylase contain similar amounts of hilD and hilD Δ3′UTR mRNAs, suggesting that the hilD 3′UTR is a target for hilD mRNA degradation by the degradosome. The hilD 3′UTR is also necessary for modulation of hilD and SPI-1 expression by the RNA chaperone Hfq. Overexpression of SPI-1 in the absence of the hilD 3′UTR retards Salmonella growth and causes uncontrolled invasion of epithelial cells. Based on these observations, we propose that the S. enterica hilD 3′UTR is a cis-acting element that contributes to cellular homeostasis by promoting hilD mRNA turnover.
INTRODUCTION
Posttranscriptional regulation of gene expression plays essential roles in cellular physiology in all domains of life. During the last few decades, special attention has been paid to mRNA regulation, and a variety of regulatory mechanisms have been described in both prokaryotes and eukaryotes. A difference in posttranscriptional regulation between eukaryotes and prokaryotes is the region of the mRNA molecule involved: while in eukaryotes the interaction of proteins or RNA molecules occurs with sequences located either in the 5′ untranslated region (5′UTR) or in the 3′ untranslated region (3′UTR) (1), posttranscriptional regulation in prokaryotes is most commonly exerted by targeting either the 5′UTR or the promoter-proximal region of the coding sequence (2–4).
Salmonella enterica is a Gram-negative bacterium that has played a historic role in bacterial genetics and in the study of host–pathogen interactions. More recently, Salmonella has also become a model organism for the study of RNA-mediated regulation (5). A relevant trait of Salmonella virulence is the ability of the pathogen to enter epithelial cells in the animal intestine. This process, known as invasion, requires translocation of bacterial proteins from the bacterial cytoplasm to the epithelial cell cytoplasm through a type 3 secretion system (TTSS) (6,7). The translocated proteins or ‘effectors’ interact with specific targets inside the epithelial cell, triggering a cascade of molecular events that permit Salmonella invasion (6).
Invasion effectors and components of the cognate TTSS are encoded in a 40 kb region of the Salmonella chromosome known as Salmonella pathogenicity island 1 (SPI-1) (7–9). Expression of SPI-1 genes is coordinately regulated by environmental signals and cellular factors through a regulatory network of SPI-1-encoded transcriptional activators, and one of the main transcription factors is HilD (8–10). A number of regulators seem to control hilD expression at the posttranscriptional or the posttranslational level, rather than at the level of transcription initiation (10), and the underlying mechanisms are known in certain cases only. For instance, HilE, a negative SPI-1 regulator (11), physically interacts with HilD (12), likely interfering with its function. Another regulator, FliZ, may activate SPI-1 expression by controlling HilD activity (13). Regulation of hilD expression at the mRNA level has also been proposed: overproduction of the RNA binding protein CsrA represses SPI-1 expression (14,15), and CsrA has been shown to bind a hilD mRNA region that overlaps with the ribosome binding sequence, likely preventing translation and accelerating mRNA decay (15). DNA adenine (Dam) methylation contributes to sustain high levels of SPI-1 expression by decreasing hilD mRNA turnover but the mechanism remains to be identified (16).
Here we show that hilD mRNA possesses a long 3′UTR of 310 nucleotides, and provide evidence that the hilD 3′UTR is a target for the RNA degradosome and for the RNA chaperone Hfq. Lack of the hilD 3′UTR permits SPI-1 expression under conditions that are not favorable for invasion and retards Salmonella growth, suggesting that the hilD 3′UTR may have physiological significance.
MATERIALS AND METHODS
Bacterial strains, bacteriophages and standard strain construction
All the S. enterica strains listed in Supplementary Table S1 belong to serovar Typhimurium, and derive from the mouse-virulent strain ATCC 14028. For simplicity, S. enterica serovar Typhimurium is usually abbreviated as S. enterica. Two Escherichia coli strains were used: DH5α [F− Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK−, mK+) phoA supE44 λ− thi-1 gyrA96 relA1] and CC118 λ pir [phoA20 thi-1 rspE rpoB argE(Am) recA1 (λ pir)]. Targeted gene disruption was achieved using pKD4 or pKD13 (17). Antibiotic resistance cassettes introduced during strain construction were excised by recombination with plasmid pCP20 (17). The oligonucleotides used for disruption (labeled ‘UP’ and ‘DO’) are listed in Supplementary Table S2, together with the oligonucleotides (labeled ‘E’) used for allele verification by the polymerase chain reaction. For construction of lac fusions in the Salmonella chromosome, FLP recombinase recognition target (FRT) sites generated by excision of Kmr cassettes (17) were used to integrate either pCE37 or pCE40 (18). Unless indicated otherwise, all lac fusions used in this study are translational. Addition of a 3xFLAG epitope tag to protein-coding DNA sequences was carried out using plasmid pSUB11 (Kmr, 3xFLAG) as template (19). Transductional crosses using phage P22 HT 105/1 int201 [(20) and Roberts,G., unpublished data] were used for strain construction operations involving chromosomal markers. The transduction protocol was described elsewhere (21). To obtain phage-free isolates, transductants were purified by streaking on green plates. Phage sensitivity was tested by cross-streaking with the clear-plaque mutant P22 H5.
Media and growth conditions
Luria-Bertani (LB) broth was used as standard liquid medium. For standard conditions (SPI-1 induction) (22), cultures were grown in LB at 37°C with shaking, and samples were taken when the cultures had reached stationary phase (O.D. = 2–2.5). To measure SPI-1 expression under non-invasive conditions, saturated cultures were diluted 1:50 in two different conditions: (i) LB and incubation at 37°C with shaking (200 rpm) until O.D.600 = 0.6; (ii) LB without salt and incubation at 37°C with shaking (200 rpm) until O.D.600 = 2–2.5. When required, kanamycin sulfate (50 μg/ml) or chloramphenicol (20 μg/ml) were added to the culture medium. Green plates were prepared according to Chan et al. (23), except that methyl blue (Sigma Chemical Co., St Louis, MO) substituted for aniline blue.
Construction of relevant strains
For construction of the hilD Δ3′UTR allele, the Kmr gene of pKD13 was amplified with primers JVO5462 and JVO5463 (Supplementary Table S2) and inserted in the Salmonella chromosome by λ Red recombination, deleting a 230 nt fragment of the hilD 3′UTR starting at the first nucleotide after the hilD stop codon. The Kmr gene was removed by recombination of flanking FRT sequences mediated by pCP20-encoded FLP recombinase, leaving a scar of 82 nt. As a result, a shorter 3′UTR of 162 nt was produced without eliminating the native Rho-independent transcriptional terminator. The same strategy was used to construct hilD 3′UTR deletion alleles, using different forward primers (Supplementary Table S2) and keeping JVO5463 as the reverse oligo for amplification. The hilD 3′UTR50end allele was constructed using primer pairs JVO5462 and hilD50endDO.
The hilD Δ3′UTRsl (scarless) allele was constructed by overlapping polymerase chain reactions using oligonucleotides EPF097, EPF100, EPF103 and EPF104 (Supplementary Table S2). The resulting amplicon contained the hilD coding sequence followed by the 3′UTR transcriptional terminator (without the 3′UTR) and approximately 500 bp of nearby DNA, flanked by SacI and XbaI restriction sites. The polymerase chain reaction (PCR) product was cloned into the SacI–XbaI sites of the suicide plasmid pDMS197, and propagated in E. coli CC118 lambda pir. The pDMS197 derivative was transformed into E. coli S17-1 lambda pir, and several transformants were used as donors in matings with S. enterica 14028 harboring a KmR cassette in place of the hilD 3′UTR (strain SV6189). TcR transconjugants were selected on E plates supplemented with tetracycline. Several TcR transconjugants were grown in nutrient broth (without NaCl) containing 5% sucrose. Individual tetracycline-sensitive segregants were then screened for kanamycin sensitivity and examined for the incorporation of the mutant hilD allele (hilD Δ3′UTRsl) by PCR amplification with oligonucleotides EPF097 and EPF100 and sequencing of the PCR product.
Expression of hilD from a heterologous promoter was achieved by replacing its native promoter with the PLtetO promoter (24). A fragment containing the cat gene and PLtetO was amplified by PCR using pXG1 as template (25). The primers were labeled PLtetO-hilD UP and PLtetO-hilD DO (Supplementary Table S2). The PCR product was treated with DpnI to remove template traces. The construction was then inserted in the S. enterica chromosome by λ Red recombination (17) and Cmr colonies were selected. Insertion of the construction was verified by PCR, using a pair of primers specific for the cat gene and the target gene (Supplementary Table S2).
Expression of gfp alleles was achieved by electroporation of pXG1-derived plasmids into S. enterica ATCC 14028. To generate the gfp allele carrying a hilD 3′UTR, a PCR product containing the hilD 3′UTR coding sequence downstream of gfp was generated by overlapping PCRs. Two PCR products were obtained using primer pairs EPF043–EPF044 (Supplementary Table S2) and pXG1 as template, and primers EPF045 and EPF046 (Supplementary Table S2) and genomic DNA from S. enterica ATCC 14028 as template. Purified PCR products were subsequently used as templates for amplification with primers EPF043 and EPF046. The resulting product was cloned into Nhe-I digested pXG1 vector, rendering plasmid pIZ1988. To generate the gfp allele with a 3′UTR deletion (Δ3′UTR) the same procedure was used. The primer pairs used were EPF043 and EPF047 (Supplementary Table S2), with pXG1 as template, and primers EPF048 and EPF046 (Supplementary Table S2) with SV6190 genomic DNA as template. The final PCR product was cloned into pXG1 to obtain pIZ1989. Plasmids pIZ1988 and pIZ1999 were constructed using E. coli DH5α as host. Transfer to S. enterica was achieved by electrotransformation of ATCC 14028, selecting Cmr colonies. Introduction of pIZ1988 and pIZ1999 generated strains SV7440 and SV7441, respectively.
RNA extraction procedures
A 2 ml aliquot from a culture of the appropriate strain grown to stationary phase (O.D.600 ∼ 2) was centrifuged at 16 000 g, 4°C, for 5 min. The pellet was resuspended in 100 μl of a solution of lysozyme (3 mg/ml, Sigma Chemical Co.). Cell lysis was facilitated by three consecutive freeze-thaw cycles. After lysis, RNA was extracted using 1 ml of Trizol reagent (Invitrogen Co., Carlsbad, CA), according to manufacturer's instructions. Finally, total RNA was resuspended in 30 μl of RNase-free water for subsequent uses. The quality of the preparation and the RNA concentration were determined using a ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
3′RACE
Rapid amplification of cDNA 3' ends (3′RACE) was performed as described by Argaman et al. (26). The hilD-specific primer used for PCR was JVO5527. Specific PCR products were cloned using the TOPO TA cloning (Invitrogen Co., Carlsbad, CA), and four independent clones of each product were sequenced using external primers.
Northern blotting
Ten micrograms of RNA was loaded per well and electrophoresed in denaturing 1% agarose formaldehyde–3-(N-morpholino)propanesulfonic acid (MOPS) gels. Transfer and fixation to Hybond-N+ membranes (GE Healthcare, Little Chalfont, UK) were performed by vacuum using 0.05 M NaOH. Filters were then hybridized using an internally labeled [(α-32P)UTP] riboprobe specific for the first (5′) 300 nucleotides of the hilD coding sequence. Hybridization was carried out at 65°C. As a control of RNA loading and transfer efficiency, the filters were hybridized with a riboprobe for the RNase P mRNA gene (rnpB). Images of radioactive filters were obtained with a FLA-5100 imaging system (Fujifilm, Tokyo, Japan), and quantification was performed using the MultiGauge software (Fujifilm).
β-Galactosidase assays
Levels of β-galactosidase activity were assayed using the CHCl3–sodium dodecyl sulfate (SDS) permeabilization procedure (27). Unless otherwise indicated, the results shown are averages and standard deviations from three independent experiments.
Protein extracts and western blot analysis
Total protein extracts were prepared from bacterial cultures grown at 37°C in LB until stationary phase (final O.D.600 ∼ 2.5). Approximately 3.5 × 108 bacterial cells were collected by centrifugation (16 000 g, 2 min, 4°C) and suspended in 100 μl of Laemmli sample buffer [1.3% SDS, 10% (v/v) glycerol, 50 mM Tris–HCl, 1.8% β-mercaptoethanol, 0.02% bromophenol blue, pH 6.8]. Proteins were resolved by Tris–Tricine–polyacrylamide gel electrophoresis (PAGE) using 12% gels. Conditions for protein transfer have been described elsewhere (28). Optimal dilutions of primary antibodies were as follows: anti-Flag M2 monoclonal antibody (1:5000, Sigma Chemical Co.), and anti-GroEL polyclonal antibody (1:20 000, Sigma Chemical Co.). Goat anti-mouse horseradish peroxidase-conjugated antibody (1:5000, BioRad, Hercules, CA) or goat anti-rabbit horseradish peroxidase conjugated antibody (1:20 000, Santa Cruz Biotechnology, Heidelberg, Germany) were used as secondary antibodies. Proteins recognized by the antibodies were visualized by chemoluminescence using the luciferin–luminol reagents (Pierce Reagents), and a LAS3000 mini imaging system (Fujifilm). For quantification, the intensity of the bands was determined using MultiGauge software (Fujifilm).
Invasion assays
HeLa cells (ATCC CCL2) were cultured in tissue culture medium (Dulbecco's modified essential medium supplemented with 10% fetal calf serum and 2 mM l-glutamine). For routine cultivation, 60 μg/ml penicillin and 100 μg/ml streptomycin were added to the culture medium. The day before infection, ∼1.5 × 105 HeLa cells were seeded, using 24-well plates (Costar, Corning, New York, NY). Each well contained 1 ml of tissue culture medium without antibiotics. Cells were grown at 37°C, 5% CO2 to obtain 80% confluency. One hour before infection, the culture medium was removed and replaced by 0.5 ml fresh tissue culture medium without antibiotics. Bacteria were grown in LB to an O.D. ≈ 0.4 and added to reach a multiplicity of infection (MOI) of 50:1 bacteria/HeLa cell. HeLa cells were infected for 30 min, washed three times with PBS, incubated in fresh tissue culture medium containing 100 μg/ml gentamicin for 1.5 h, and washed three times with PBS. Numbers of viable intracellular bacteria were obtained by lysing infected cells with 1% Triton X-100 (prepared in PBS) and subsequent plating. Invasion rates were determined as the ratio between viable intracellular bacteria and viable bacteria added to infect the HeLa cells. Each assay was run in triplicate, and data shown are the relative invasion rates and standard deviations of five independent experiments. The Student's t test was used to determine the statistical significance of the differences observed.
Flow cytometry
GFP expression was monitored by flow cytometry of live cells. Bacteria were grown overnight at 37°C in LB with shaking, diluted into fresh medium (1:100), and incubated at 37°C to reach and O.D.600 ∼ 2. For each sample, the GFP fluorescence of at least 50 000 cells was measured using a Cytomics FC500-MPl flow cytometer (Beckman Coulter, Brea, CA).
RESULTS
hilD mRNA has a long 3′UTR
The hilD transcription start point was identified previously (29), defining a 5′UTR of 35 nucleotides. In turn, in silico annotation of the S. enterica genome identified a putative long intergenic region between hilD and the neighbor gene hilA (http://www.xbase.ac.uk/colibase/). We determined the hilD transcription stop point by 3′RACE and mapped the hilD transcriptional stop around 300 nucleotides downstream the hilD stop codon, immediately after a U-rich region (Figure 1A; Supplementary Figure S1). U-rich tracts are typically found at the 3′ end of Rho-independent transcriptional terminators (30,31). Furthermore, sequence analysis with the RNAfold software (32–34) predicts the existence of a secondary structure similar to a Rho-independent transcriptional terminator which includes the U-rich region defined by 3′RACE in the hilD transcript (Figure 1B). Altogether, these data provide evidence that hilD mRNA may be 1275 nucleotides long, including a 5′UTR of 35 nucleotides, a coding sequence of 930 nucleotides, and an unusually long 3′UTR of 310 nucleotides. Northern blot analysis in a 4.5% polyacrylamide gel with a riboprobe specific for hilD identified an RNA molecule of ∼1.3 kb, consistent with the estimated size of hilD mRNA (Supplementary Figure S2). In silico analysis of the 3′UTR (35) did not reveal the existence of any potential coding sequence: the longest open reading frame (ORF), 57 nucleotides long, is not preceded by a putative ribosome binding site (data not shown). Nevertheless, the region is conserved among Salmonella serovars (Supplementary Figure S3), which might indicate a functional role for this non-coding RNA region.
Figure 1.
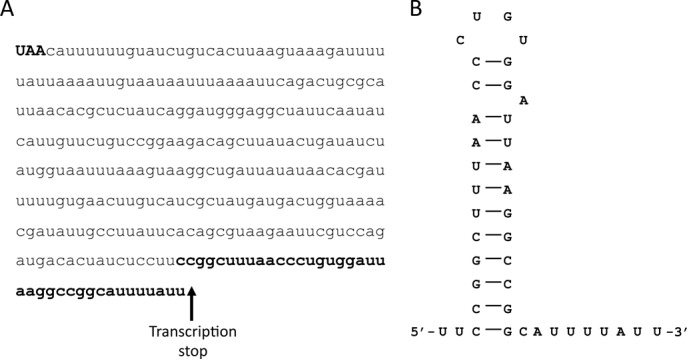
(A) Nucleotide sequence of the hilD 3′UTR. The first three nucleotides (in bold capital letters) correspond to the hilD stop codon. Nucleotides in bold at the 3′ end of the sequence constitute the Rho-independent transcriptional terminator depicted in panel (B). The arrow indicates the hilD transcription termination point determined by 3′RACE. (B) Stem-loop structure of the Rho-independent transcriptional terminator predicted by RNAfold software (bold nucleotides in panel (A)).
Deletion of the hilD 3′UTR increases the level of hilD mRNA
Long 3′UTRs have been poorly studied in prokaryotes, while eukaryotic 3′UTRs often play physiological roles (36). We thus considered the possibility that the hilD 3′UTR might play a role in hilD expression. To test this hypothesis, we constructed a strain carrying a modified hilD 3′UTR (hilD Δ3′UTR). In this strain, a 230 nt fragment of the hilD 3′UTR, starting at the first nucleotide after the hilD stop codon, was deleted leaving the Rho-independent transcriptional terminator intact. The deleted fragment was replaced with the pKD13 scar (82 nucleotides) (17), resulting in a shorter 3′UTR with different sequence but conserving the native transcriptional terminator (Figure 2A). To test whether the presence of the hilD 3′UTR altered the hilD mRNA level, we isolated total RNA from the wild type and the hilD Δ3′UTR strain and monitored hilD expression by northern blotting using a 32P-labeled riboprobe specific for the upstream (5′) 300 nucleotides of the hilD coding sequence. As shown in Figure 2B, the hilD Δ3′UTR mRNA is shorter than the hilD native mRNA. More importantly, the level of hilD mRNA was found to be ∼11-fold higher in the absence of the 3′UTR than in the wild-type strain, suggesting that the presence of the 3′UTR somehow reduces the hilD mRNA level. To exclude the possibility that the hilD mRNA increase detected in the hilD Δ3′UTR strain could be caused by a stabilizing effect of the scar sequence, we constructed a strain in which the 3′UTR was cleanly deleted. Higher levels of hilD mRNA were detected in the scarless strain (Supplementary Figure S4). The specificity of the hilD probe was confirmed using a strain carrying a SPI-1 deletion, and no signal was detected by northern blot (Supplementary Figure S4).
Figure 2.
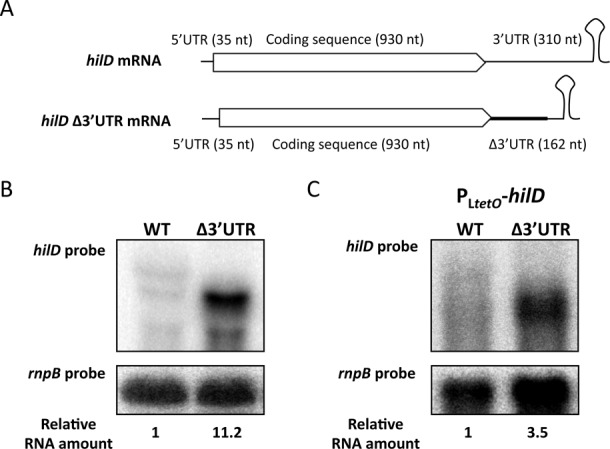
(A) Diagrams of hilD mRNA and hilD Δ3′UTR mRNA. The thick line in the 3′ region of hilD Δ3′UTR mRNA represents the 82-nt pKD13 scar. The Rho-independent transcriptional terminator is left intact in the constructs. (B and C) Northern blots of hilD and hilD Δ3′UTR mRNAs when hilD is transcribed from its own promoter (B), and when transcription is driven by the PLtetO promoter (C). Three discrete bands are detected with a riboprobe when hilD is transcribed from its native promoter. rnpB mRNA has been used as loading control. For quantification, the ratio hilD/rnpB was normalized to 1 in hilD mRNA carrying a native 3′UTR.
To test whether the hilD 3′UTR is involved in hilD expression, we compared the levels of hilD mRNA in isogenic hilD 3′UTR+ and hilD Δ3′UTR strains in which hilD was transcribed from a heterologous promoter, PLtetO (24). Absence of the native 3′UTR increased the hilD mRNA level 3.5-fold (Figure 2C). Because this observation was made in strains that expressed hilD from a heterologous promoter, we tentatively concluded that the 3′UTR influences hilD expression at the posttranscriptional level. Note that hilD expression upon deletion of the 3′UTR is higher when hilD is transcribed from its own promoter than when transcription is driven by PLtetO (Figure 2B and C). This observation is consistent with the occurrence of autogenous control of hilD transcription (37), which can be expected to create a positive feedback loop that amplifies the regulatory effect (38).
The hilD 3′UTR reduces expression of a heterologous gene
To further confirm that the hilD 3′UTR has functional capacity, we analyzed the effect of the 3′UTR on the expression of a heterologous gene. A synthetic mRNA was generated by cloning the hilD 3′UTR downstream of the green fluorescent protein (gfp) gene on plasmid pXG1 (25), yielding plasmid pIZ1998. The synthetic mRNA was expressed constitutively from the PLtetO promoter (24). As a control, a plasmid containing the hilD Δ3′UTR downstream of gfp (pIZ1989) was also constructed. Both plasmids were introduced in S. enterica and E. coli, and GFP levels were monitored in single bacterial cells by flow cytometry. A representative experiment is shown in Figure 3. Salmonella enterica cells presented a mean fluorescence of 58.4 for cells expressing GFP-3′UTR and 85.3 for cells expressing GFP-Δ3′UTR. In turn, E. coli cells showed a mean fluorescence of 96.9 for cells containing GFP-3′UTR and 132.0 for cells lacking the 3′UTR (GFP-Δ3′UTR). Thus, the presence of the hilD 3′UTR downstream of gfp resulted in lower fluorescence intensity (indicating lower GFP production) in both S. enterica and E. coli. This ability of the hilD 3′UTR to reduce expression of a heterologous gene supports the view that the 3′UTR can act as an independent module.
Figure 3.
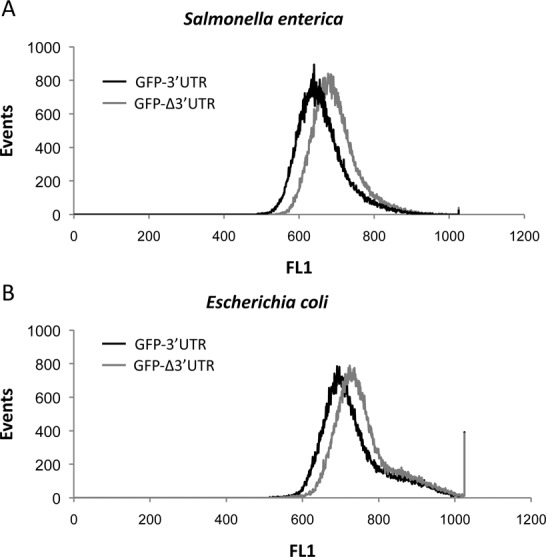
Histogram plots showing the fluorescence of Salmonella enterica (A) or Escherichia coli cells (B) carrying the gfp gene with either the hilD 3′UTR (GFP-3′UTR, solid line) or the hilD Δ3′UTR (GFP-Δ3′UTR, dashed line) downstream of the gfp stop codon. Over 50 000 particles were analyzed for each sample using a Cytomics FC500-MPl flow cytometer. Three independent experiments were performed, and the histogram plots depicted are representative of the results obtained. Analysis of data was performed using the CXP software.
Deletion of the hilD 3′UTR increases SPI-1 expression
Because hilD mRNA is more abundant in the absence of its native 3′UTR and HilD is a positive regulator of SPI-1, we wondered whether increased level of hilD mRNA resulted in increased SPI-1 expression. For this purpose, we analyzed the expression of four SPI-1 genes (invF, invH, sipB and sipC) in isogenic hilD 3′UTR+ and hilD Δ3′UTR strains. Western blotting was used to monitor InvF, InvH, SipB and SipC levels using protein variants tagged with the 3xFLAG epitope. The levels of all four proteins were higher in hilD Δ3′UTR strains compared with the hilD 3′UTR+ background (Figure 4A), suggesting that the increased hilD mRNA level observed in the hilD Δ3′UTR strain yields a higher amount of HilD protein, which in turn results in increased SPI-1 expression. These observations were further confirmed by monitoring β-galactosidase activities of invF::lac, invH::lac, sipB::lac and sipB::lac fusions in hilD 3′UTR+ and hilD Δ3′UTR strains (Supplementary Figure S5). Hence, overproduction of hilD mRNA in the absence of its native 3′UTR correlates with SPI-1 overexpression, suggesting that the hilD 3′UTR may promote hilD mRNA turnover.
Figure 4.
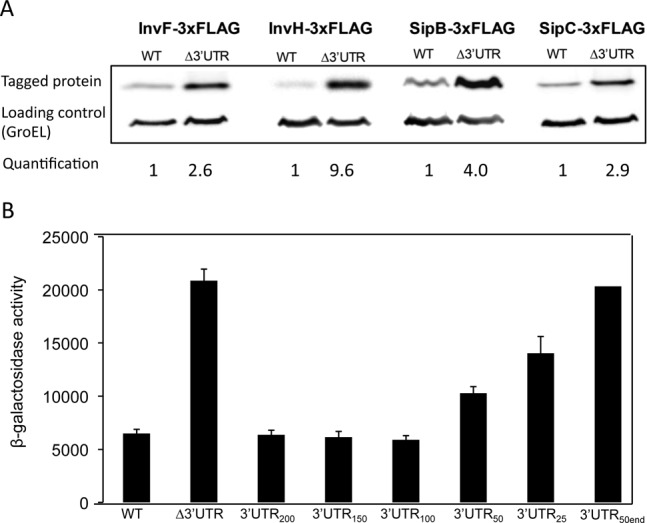
(A) Western blots of InvF-3xFLAG, InvH-3xFLAG, SipB-3xFLAG and SipC-3xFLAG in protein extracts from hilD 3′UTR+ and hilD 3′UTR− hosts. GroEL was used as loading control. For quantification, the ratio 3xFLAG-tagged protein/GroEL was relativized to 1 in the hilD 3′UTR+ background. (B) β-Galactosidase activity of a sipC::lac fusion in Salmonella enterica strains containing a native hilD 3′UTR (wt) or alleles with deletions of the hilD 3′UTR (Δ3′UTR, 3′UTR200, 3′UTR150, 3′UTR100, 3′UTR50, 3′UTR25 and 3′UTR50end).
To define the region(s) of the hilD 3′UTR necessary for mRNA turnover, we constructed a set of mutants containing partial deletions of the 3′UTR. The mutants conserved 200, 150, 100, 50 and 25 nucleotides of the region immediately downstream of the hilD stop codon, followed by the 82 nt pKD13 scar and the Rho-independent transcriptional terminator (Supplementary Figure S6). We also constructed a mutant containing the 82-nt pKD13 scar immediately after the hilD stop codon, followed by the last (3′) 50 nucleotides of the 3′UTR and the transcriptional terminator (Supplementary Figure S6). SPI-1 expression was monitored by measuring the β-galactosidase activity of a sipC::lac fusion in the 3′UTR deletion mutants. Results shown in Figure 4B can be summarized as follows:
The mutants containing 200, 150 and 100 nucleotides from the 5′ region of the 3′UTR showed sipC expression levels similar or identical to the wild type, indicating that deletion of downstream regions does not impair hilD mRNA turnover. Hence, signals for 3′UTR-mediated mRNA decay must be located within nucleotides 1–100.
The mutants containing only 50 and 25 nucleotides of the 5′ region of the 3′UTR showed moderate increases in sipC::lac expression (2-fold and 3-fold, respectively), suggesting that some signals for hilD mRNA turnover had been lost upon deletion of nucleotides 25–50 and 50–100.
In the mutant containing 25 nucleotides of the 3′UTR, sipC::lac expression was higher than in the wild type but lower than in the mutant lacking the whole 3′UTR (hilD Δ3′UTR) (Figure 4B), suggesting that signals for hilD mRNA turnover exist in the upstream (5′) 25 nucleotides of the 3′UTR.
The sipC::lac expression level detected in the mutant containing the last 50 nucleotides of the 3′UTR (3′UTR50end) was similar to that of the whole-deletion mutant (hilD Δ3′UTR), supporting the view that downstream regions of the hilD 3′UTR do not play a relevant role in hilD mRNA turnover.
A side, less relevant observation was that the length of the 3′UTR does not seem to be relevant for hilD mRNA turnover: mutants containing 50 nucleotides of the 3′UTR, either from the upstream region (3′UTR50) or the downstream region (3′UTR50end), have deletions of the same length but undergo disparate levels of sipC expression (Figure 4B and Supplementary Figure S6).
The levels of hilD mRNA in the deletion mutants were analyzed by northern blotting (Supplementary Figure S7), and clear-cut correlations between mRNA abundance and SPI-1 expression levels were found.
In summary, conservation of the upstream (5′) 100 nucleotides of the 3′UTR is sufficient to keep sipC::lac expression at low levels, similar to those of the wild type, suggesting that signals for mRNA decay are contained within nucleotides 1–100 after the hilD stop codon.
The hilD 3′UTR is a target for mRNA degradation by the RNA degradosome
Because eukaryotic mRNA degradation often depends on long 3′UTRs (39,40), we hypothesized that the long 3′UTR of hilD mRNA might be a target for mRNA degradation by the RNA degradosome. If such was the case, the increased hilD mRNA level detected in the absence of the 3′UTR might be the consequence of reduced mRNA turnover. Hence, we reasoned, inactivation of factors involved in RNA degradation might suppress the differences observed in the level of hilD mRNA with and without its native 3′UTR. Salmonella enterica mutants carrying deletions in genes encoding ribonuclease E (RNase E), ribonuclease G (RNase G), polynucleotide phosphorylase (Pnp), or poly(A) polymerase I (PapI) were used. Because RNase E is essential in S. enterica, we used a mutant carrying a deletion that eliminates only the C-terminal region of the protein (41). The levels of hilD and hilD Δ3′UTR mRNAs were monitored in strains lacking RNase E, RNase G, Pnp or PapI. Increased levels of hilD Δ3′UTR mRNA were detected in the RNase G− and PapI− backgrounds (Figure 5), suggesting that the hilD 3′UTR is not involved in hilD mRNA degradation by RNase G or polyadenylation-dependent pathways. However, similar levels of hilD and hilD Δ3′UTR mRNAs were detected in strains lacking either RNase E or Pnp (Figure 5). These proteins are components of the RNA degradosome (42), suggesting that the hilD 3′UTR may be a target for mRNA degradation by the degradosome. Furthermore, the levels of hilD mRNA in mutants lacking RNase E and Pnp were higher than in the wild type (Figure 5).
Figure 5.
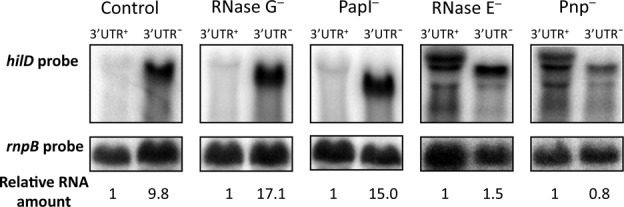
Northern blots of hilD and hilD Δ3′UTR mRNAs in Salmonella enterica strains lacking RNase G, PapI, RNase E, or Pnp. For simplicity, hilD and hilD Δ3′UTR mRNAs have been labeled 3′UTR+ and 3′UTR−, respectively. A riboprobe specific for hilD was used, and rnpB mRNA was used as an internal control. For quantification, the hilD/rnpB ratio was relativized to 1 in a 3′UTR+ background.
In silico analysis of the hilD RNA sequence revealed six typical cleavage sites for RNase E [′-(A/G)N↓AU, where N identifies any nucleotide, and ↓ indicates the cleavage site (43)] in the upstream (5′) 100 nucleotides of the 3′UTR, with two putative sites contained in the upstream 25 nucleotides (Supplementary Figure S8). Altogether, our observations suggest that targeting of the upstream region of the hilD 3′UTR by the degradosome may contribute to maintain steady-state levels of hilD mRNA in the cell.
The hilD mRNA level is modulated by Hfq in a 3′UTR-dependent manner
A possibility suggested by the above results was that the 3′UTR of hilD mRNA might be a target for modulation of SPI-1 expression. On these grounds, we studied the role of the RNA chaperone Hfq. It has been reported that SPI-1 expression is repressed in mutants lacking Hfq (44). In addition, Hfq co-immunoprecipitation experiments have shown that Hfq binds hilD mRNA (45). To determine whether hilD expression was under Hfq control, we examined the levels of hilD mRNA in isogenic Hfq+ and Hfq− strains by northern blotting (Figure 6A, lanes 1 and 3). Lack of Hfq reduced the level of hilD mRNA almost 5-fold. However, when the 3′UTR was deleted, similar levels of hilD Δ3′UTR mRNA were detected in Hfq+ and Hfq− backgrounds (Figure 6A, lanes 2 and 4), indicating that the 3′UTR of hilD mRNA is necessary for modulation of hilD expression by Hfq. Next, we examined whether modulation of SPI-1 expression by Hfq depended on the hilD 3′UTR. For this purpose, we measured InvF, SipB and SipC protein levels in Hfq+ and Hfq− backgrounds, either in the presence or in the absence of the hilD 3′UTR. As expected, protein levels were lower in Hfq− hosts when the hilD 3′UTR was intact (Figure 6B). However, when the hilD 3′UTR was deleted, similar protein levels were detected in Hfq+ and Hfq− backgrounds (Figure 6B), indicating that the 3′UTR of hilD mRNA is involved in modulation of SPI-1 expression by Hfq.
Figure 6.
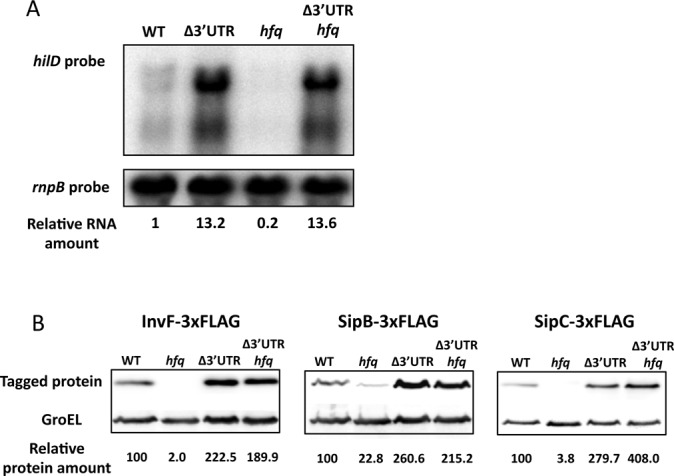
(A) hilD mRNA levels in Hfq+hilD 3′UTR+, Hfq+hilD 3′UTR−, Hfq−hilD 3′UTR+ and Hfq−hilD 3′UTR− isogenic strains. hilD mRNA was detected by northern blot using a specific riboprobe, and rnpB mRNA was used as an internal control. For quantification, the ratio hilD mRNA/rnpB mRNA was relativized to 1 in Hfq+hilD 3′UTR+ background. (B) Western blots of InvF-3xFLAG, SipB-3xFLAG and SipC-3xFLAG in protein extracts from wild type, Δ3′UTR, Hfq− and Δ3′UTR Hfq− hosts. GroEL was used as loading control. For quantification, the ratio 3xFLAG-tagged protein/GroEL was relativized to 1 in the WT background.
The hilD 3′UTR is necessary to keep low levels of SPI-1 expression under non-invasive conditions
A standard practice in studies of Salmonella invasion and SPI-1 expression is growth in LB broth without aeration until stationary phase (22). These conditions intend a reductionist imitation of the animal intestine, and permit efficient activation of SPI-1 expression. Under ‘non-invasive’ conditions, SPI-1 expression is low or absent (8,9). On these grounds, we wondered whether degradosome targeting of the hilD 3′UTR might play a role in SPI-1 repression under non-invasive conditions. We monitored expression of an invF::lac fusion in hilD 3′UTR+ and hilD Δ3′UTR strains grown under two conditions in which SPI-1 is known to be partially or completely repressed: exponential growth with high aeration and growth at low osmolarity (LB without salt). Absence of the hilD 3′UTR caused a strong increase in invF expression under non-invasive conditions (Figure 7A), suggesting that the 3′UTR of hilD mRNA may contribute to keep SPI-1 repressed under conditions that are not appropriate for invasion.
Figure 7.
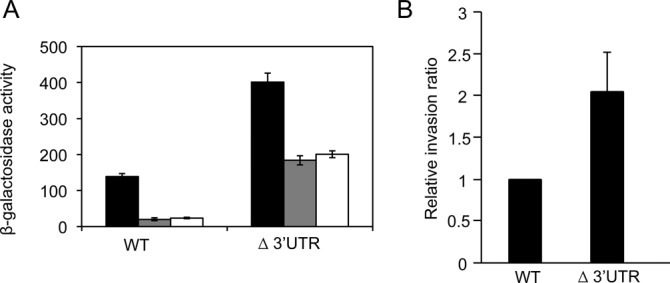
(A) β-Galactosidase activity of an invF::lac fusion in hilD 3′UTR+ and hilD Δ3′UTR hosts grown under different conditions: (i) standard conditions (black bars); (ii) exponentially growing cultures with high aeration (gray bars); and (iii) stationary phase cultures grown under low osmolarity (LB without salt). (B) Relative invasion ratios of epithelial HeLa cells by Salmonella enterica serovar Typhimurium carrying either the native hilD 3′UTR (wild-type strain ATCC 14018) or the hilD Δ3′UTR allele (hilD Δ3′UTR Kmr, strain SV6189). The invasion ratio was determined 2 h post-infection and is expressed as the ratio between viable intracellular bacteria and viable bacteria added to the HeLa cell culture. For simplicity, ratios were normalized to the infection ratio of ATCC 14028. Each experiment was run in triplicate, and the data represented are averages of five independent experiments, with standard deviations shown as error bars. The difference in invasion ratios was significant (P < 0.05) according to Student's t test.
To investigate whether expression of SPI-1 genes under non-invasive conditions permitted Salmonella invasion, we examined the effect of hilD 3′UTR deletion on the invasion rate of epithelial cells in vitro. Cultured HeLa cells were infected with wild-type S. enterica ATCC 14028 and with the hilD Δ3′UTR mutant grown in LB to exponential phase. The infection ratios of ATCC 14028 were extremely low (between 10−4 and 10−6), as expected under non-invasive conditions. However, the hilD Δ3′UTR mutant showed a higher invasion rate (Figure 7B).
DISCUSSION
Certain eukaryotic mRNAs have long 3′UTRs (36) that modulate stability, translation and/or localization (39,46–48). In prokaryotes, the traditional notion is that 3′UTRs mainly harbor a transcriptional terminator that contributes to RNA stabilization, preventing degradation by exonucleases (49–51). However, recent advances in transcriptomic analysis have identified long 3′UTRs in some bacterial transcripts (52–54), raising the possibility that prokaryotic 3′UTRs might have physiological roles like their eukaryotic counterparts (55). For example, in Bacillus subtilis, nine different mRNAs harbor a conserved 3′UTR of around 220 nucleotides whose functional significance remains to be confirmed (53). Furthermore, 3′UTR-derived sRNAs potentially involved in posttranscriptional regulation in trans have been found in S. enterica (56) and E. coli (57). A recent study has shown that translation of a Staphylococcus aureus mRNA is controlled by interaction between the 5′UTR and the 3′UTR (58). In support of the emerging view that prokaryotic 3′UTRs may have functional relevance, we describe that S. enterica hilD mRNA contains a 3′UTR of 310 nucleotides (Figure 1), and provide evidence that the 3′UTR is a cis-acting element for modulation of mRNA stability.
Increased levels of hilD mRNA are found in the absence of the 3′UTR (Figure 2), and hilD mRNA increase is suppressed in the absence of either RNase E or Pnp (Figure 5), suggesting that the hilD 3′UTR is a target for hilD mRNA degradation by the RNA degradosome. This hypothesis is consistent with a previous report indicating that Salmonella mutants lacking functional RNase E undergo increased SPI-1 expression (11). Furthermore, degradation of certain bacterial RNAs has been shown to involve endonucleolytic cleavages downstream the coding sequence (59–62).
The ability of the hilD 3′UTR to serve as a degradosome target is supported by its nucleotide composition, which fits well in the preference of RNase E to degrade RNAs rich in A and U residues (42). The hilD 3′UTR contains 64% of A–U nucleotides, a percentage that increases up to 85% in the first (5′) 60 nucleotides. Signals for degradosome-mediated turnover appear to be located within the upstream 100 nucleotides of the hilD 3′UTR, and the upstream (5′) 25 nucleotides seem to be especially relevant (Figure 4B). These tentative conclusions are supported by the occurrence of six putative RNase E cleavage sites in the upstream 100 nucleotides of the hilD 3′UTR (Supplementary Figure S8).
Although the above observations suggest that the hilD 3′UTR might be a direct target for the RNA degradosome, we have not been able to detect differences in the half-lives of hilD mRNA with and without 3′UTR (data not shown), opening the possibility for alternative, more indirect models. One such model is the involvement of an unstable small RNA (sRNA) in the control of hilD mRNA turnover. Stopping transcription with rifampicin would cause fast disappearance of the sRNA, and the sRNA effect on mRNA stability would be masked. It seems also likely that the hilD mRNA increase detected in the absence of the 3′UTR (∼11-fold, Figure 2B) may have a transcriptional component due to autogenous control of hilD transcription and the positive feedback loop that governs SPI-1 expression (37,38). In agreement with this prediction, we observe a smaller increase (3.5-fold) when hilD is transcribed from a heterologous promoter (Figure 2C). Hence, it is conceivable that the 3′UTR may have a modest impact on the stability of hilD mRNA but its effect is amplified by a positive feedback loop at the transcriptional level (38).
This study provides evidence also that the hilD 3′UTR may play an additional role in SPI-1 expression by serving as target for the Hfq RNA chaperone. Deletion of the hilD 3′UTR suppresses regulation of hilD and SPI-1 by Hfq (Figure 6), suggesting that the hilD 3′UTR is necessary for SPI-1 regulation by Hfq. The involvement of Hfq in hilD regulation further strengthens the possibility that the hilD 3′UTR might be the target for a sRNA. A previous study in E. coli has proposed that the sRNA GadY can stabilize gadX mRNA by direct interaction with its 3′UTR (63). If a similar mechanism is involved in control of hilD mRNA stability, it would explain the reduced levels of hilD mRNA found in Hfq− mutants and the suppression of this decrease when the hilD 3′UTR is deleted.
The relevance of the hilD 3′UTR in Salmonella physiology is illustrated by the phenotypic consequences of hilD 3′UTR deletion. Mutants lacking the hilD 3′UTR form small colonies on LB plates, and show retarded growth in liquid medium (Supplementary Figure S9). Growth retardation is a well known consequence of SPI-1 overexpression (64), probably because of the burden of building the TTSS machinery. Like other pathogenicity islands, SPI-1 is a gene cluster acquired by horizontal transfer whose accomodation into the host regulatory network requires the concurrence of multiple controls. Transcriptional regulators encoded in SPI-1 itself and in core regions of the chromosome relieve H-NS-mediated silencing and adjust transcription to appropriate environmental circumstances (8–10). HilD is one of the main transcriptional regulators of SPI-1. Although the hilD gene itself is subjected to transcriptional control, most regulatory systems that control SPI-1 expression through hilD appear to act at the posttranscriptional or the postranslational levels (10), and the underlying mechanisms are incompletely understood. Targeting of the hilD 3′UTR by the degradosome may contribute to maintain steady-state levels of hilD mRNA, thus contributing to cellular homeostasis. Efficient turnover of hilD mRNA may be especially relevant under non-invasive conditions, when the growth retardation caused by SPI-1 expression is not compensated by the possibility of colonizing an animal niche.
The abundance of RNase E targets in the upstream 100 nucleotides of the 3′UTR and the evidence that this region is sufficient for hilD RNA turnover may fuel the speculation that downstream regions of the 3′UTR might contain additional regulatory signals. For instance, Chao et al. have described that the 3′UTR region of hilD mRNA appears enriched upon Hfq co-inmunoprecipitation, defining a candidate region for a new 3′UTR-derived sRNA (56). Hence, the hilD 3′UTR might not only provide a target for hilD mRNA turnover but be also the source of a sRNA. In support of this speculation, certain 3′UTRs are known to be a source of small regulatory RNAs in S. enterica (56). This outlook, at this stage speculative, may further increase the interest of the hilD 3′UTR as a prokaryotic example of a 3′UTR with functional capacity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
We thank Iñigo Lasa and Alejandro Toledo-Arana for helpful discussions, and Jörg Vogel and Kathrin Fröhlich for support with 3′RACE experiments. We are grateful to Fernando Baisón-Olmo for the construction of strain SV6109, to Ignacio Cota for assistance in the construction of the Δ3′UTR scarless mutant, and to the remaining members of the Casadesús laboratory for help in experiments and constructive discussion. We also thank Modesto Carballo, Laura Navarro and Cristina Reyes of the Servicio de Biología (CITIUS, Universidad de Sevilla) for help in experiments performed at the facility.
Footnotes
Both authors contributed equally to this work.
Present addresses: Javier López-Garrido, University of California San Diego, 9500 Gilman Drive, 4117 Natural Sciences Building, La Jolla, CA 92093-0377, USA
Elena Puerta-Fernández, Abengoa Research, Campus Palmas Altas, C/ Energía Solar 1, Edificio G, 41014 Sevilla, Spain.
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
FUNDING
Ministerio de Economía y Competitividad of Spain (MINECO) and the European Regional Fund [BIO2010-15023, CSD2008-00013]; Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía, Spain [P10-CVI-5879]. Source of open access funding: Ministerio de Economía y Competitividad of Spain (MINECO) and the European Regional Fund[CSD2008-00013].
Conflict of interest statement. None declared.
REFERENCES
- 1.Pesole G., Mignone F., Gissi C., Grillo G., Licciulli F., Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- 2.Winkler W.C., Breaker R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 3.Waters L.S., Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Storz G., Vogel J., Wassarman K.M. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol. Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 6.Darwin K.H., Miller V.L. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 1999;12:405–428. doi: 10.1128/cmr.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lostroh C.P., Lee C.A. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 2001;3:1281–1291. doi: 10.1016/s1286-4579(01)01488-5. [DOI] [PubMed] [Google Scholar]
- 8.Altier C. Genetic and environmental control of Salmonella invasion. J. Microbiol. 2005:85–92. 43 Spec No. [PubMed] [Google Scholar]
- 9.Jones B.D. Salmonella invasion gene regulation: a story of environmental awareness. J. Microbiol. 2005:110–117. 43 Spec No. [PubMed] [Google Scholar]
- 10.Ellermeier J.R., Slauch J.M. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 2007;10:24–29. doi: 10.1016/j.mib.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Fahlen T.F., Mathur N., Jones B.D. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 2000;28:25–35. doi: 10.1111/j.1574-695X.2000.tb01453.x. [DOI] [PubMed] [Google Scholar]
- 12.Baxter M.A., Fahlen T.F., Wilson R.L., Jones B.D. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 2003;71:1295–1305. doi: 10.1128/IAI.71.3.1295-1305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chubiz J.E., Golubeva Y.A., Lin D., Miller L.D., Slauch J.M. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2010;192:6261–6270. doi: 10.1128/JB.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altier C., Suyemoto M., Lawhon S.D. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 2000;68:6790–6797. doi: 10.1128/iai.68.12.6790-6797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez L.C., Yakhnin H., Camacho M.I., Georgellis D., Babitzke P., Puente J.L., Bustamante V.H. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol. Microbiol. 2011;80:1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Garrido J., Casadesus J. Regulation of Salmonella enterica pathogenicity island 1 by DNA adenine methylation. Genetics. 2010;184:637–649. doi: 10.1534/genetics.109.108985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellermeier C.D., Janakiraman A., Slauch J.M. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene. 2002;290:153–161. doi: 10.1016/s0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 19.Uzzau S., Figueroa-Bossi N., Rubino S., Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 21.Garzon A., Cano D.A., Casadesus J. Role of Erf recombinase in P22-mediated plasmid transduction. Genetics. 1995;140:427–434. doi: 10.1093/genetics/140.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song M., Kim H.J., Kim E.Y., Shin M., Lee H.C., Hong Y., Rhee J.H., Yoon H., Ryu S., Lim S., et al. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J. Biol. Chem. 2004;279:34183–34190. doi: 10.1074/jbc.M313491200. [DOI] [PubMed] [Google Scholar]
- 23.Chan R.K., Botstein D., Watanabe T., Ogata Y. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology. 1972;50:883–898. doi: 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- 24.Lutz R., Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urban J.H., Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E.G., Margalit H., Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 27.Miller J.H. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 28.Jakomin M., Chessa D., Baumler A.J., Casadesus J. Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA, and HdfR. J. Bacteriol. 2008;190:7406–7413. doi: 10.1128/JB.01136-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olekhnovich I.N., Kadner R.J. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2002;184:4148–4160. doi: 10.1128/JB.184.15.4148-4160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lesnik E.A., Sampath R., Levene H.B., Henderson T.J., McNeil J.A., Ecker D.J. Prediction of Rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001;29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.d’Aubenton Carafa Y., Brody E., Thermes C. Prediction of Rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J. Mol. Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 32.Schuster P., Fontana W., Stadler P.F., Hofacker I.L. From sequences to shapes and back: a case study in RNA secondary structures. Proc. Biol. Sci. 1994;255:279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- 33.McCaskill J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 34.Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishikawa J., Hotta K. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 1999;174:251–253. doi: 10.1111/j.1574-6968.1999.tb13576.x. [DOI] [PubMed] [Google Scholar]
- 36.Grzybowska E.A., Wilczynska A., Siedlecki J.A. Regulatory functions of 3′UTRs. Biochem. Biophys. Res. Commun. 2001;288:291–295. doi: 10.1006/bbrc.2001.5738. [DOI] [PubMed] [Google Scholar]
- 37.Ellermeier C.D., Ellermeier J.R., Slauch J.M. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2005;57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- 38.Saini S., Ellermeier J.R., Slauch J.M., Rao C.V. The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella. PLoS Pathog. 2010;6:e1001025. doi: 10.1371/journal.ppat.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barreau C., Paillard L., Osborne H.B. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2006;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beelman C.A., Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 41.Viegas S.C., Pfeiffer V., Sittka A., Silva I.J., Vogel J., Arraiano C.M. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–7664. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carpousis A.J. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 43.Mackie G.A. RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol. 2013;11:45–57. doi: 10.1038/nrmicro2930. [DOI] [PubMed] [Google Scholar]
- 44.Sittka A., Pfeiffer V., Tedin K., Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sittka A., Lucchini S., Papenfort K., Sharma C.M., Rolle K., Binnewies T.T., Hinton J.C., Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnston D., St The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 48.Wilkie G.S., Dickson K.S., Gray N.K. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 49.Mott J.E., Galloway J.L., Platt T. Maturation of Escherichia coli tryptophan operon mRNA: evidence for 3′ exonucleolytic processing after Rho-dependent termination. EMBO J. 1985;4:1887–1891. doi: 10.1002/j.1460-2075.1985.tb03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abe H., Aiba H. Differential contributions of two elements of Rho-independent terminator to transcription termination and mRNA stabilization. Biochimie. 1996;78:1035–1042. doi: 10.1016/s0300-9084(97)86727-2. [DOI] [PubMed] [Google Scholar]
- 51.Guarneros G., Montanez C., Hernandez T., Court D. Posttranscriptional control of bacteriophage lambda gene expression from a site distal to the gene. Proc. Natl. Acad. Sci. U.S.A. 1982;79:238–242. doi: 10.1073/pnas.79.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., Loh E., Gripenland J., Tiensuu T., Vaitkevicius K., et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 53.Rasmussen S., Nielsen H.B., Jarmer H. The transcriptionally active regions in the genome of Bacillus subtilis. Mol. Microbiol. 2009;73:1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.ten Broeke-Smits N.J., Pronk T.E., Jongerius I., Bruning O., Wittink F.R., Breit T.M., van Strijp J.A., Fluit A.C., Boel C.H. Operon structure of Staphylococcus aureus. Nucleic Acids Res. 2010;38:3263–3274. doi: 10.1093/nar/gkq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gripenland J., Netterling S., Loh E., Tiensuu T., Toledo-Arana A., Johansson J. RNAs: regulators of bacterial virulence. Nat. Rev. Microbiol. 2010;8:857–866. doi: 10.1038/nrmicro2457. [DOI] [PubMed] [Google Scholar]
- 56.Chao Y., Papenfort K., Reinhardt R., Sharma C.M., Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawano M., Reynolds A.A., Miranda-Rios J., Storz G. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruiz de Los Mozos I., Vergara-Irigaray M., Segura V., Villanueva M., Bitarte N., Saramago M., Domingues S., Arraiano C.M., Fechter P., Romby P., et al. Base pairing interaction between 5′- and 3′-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet. 2013;9:e1004001. doi: 10.1371/journal.pgen.1004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Regnier P., Hajnsdorf E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3′ stabilizing stem and loop structure. J. Mol. Biol. 1991;217:283–292. doi: 10.1016/0022-2836(91)90542-e. [DOI] [PubMed] [Google Scholar]
- 60.Braun F., Hajnsdorf E., Regnier P. Polynucleotide phosphorylase is required for the rapid degradation of the RNaseE-processed rpsO mRNA of Escherichia coli devoid of its 3″hairpin. Mol. Microbiol. 1996;19:997–1005. doi: 10.1046/j.1365-2958.1996.440971.x. [DOI] [PubMed] [Google Scholar]
- 61.Hankins J.S., Zappavigna C., Prud’homme-Genereux A., Mackie G.A. Role of RNA structure and susceptibility to RNase E in regulation of a cold shock mRNA, cspA mRNA. J. Bacteriol. 2007;189:4353–4358. doi: 10.1128/JB.00193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maeda T., Wachi M. 3′ Untranslated region-dependent degradation of the aceA mRNA, encoding the glyoxylate cycle enzyme isocitrate lyase, by RNase E/G in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2012;78:8753–8761. doi: 10.1128/AEM.02304-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Opdyke J.A., Kang J.G., Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sturm A., Heinemann M., Arnoldini M., Benecke A., Ackermann M., Benz M., Dormann J., Hardt W.D. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 2011;7:e1002143. doi: 10.1371/journal.ppat.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


