Abstract
Adding nonstandard amino acids to the genetic code of E. coli expands the chemical and biological functional space for proteins. This is accomplished with engineered, orthogonal aminoacyl-tRNA synthetase and tRNA pairs that require a nonstandard amino acid in sufficient intracellular quantities to support protein synthesis. While cotranslational insertion of phosphoserine into proteins has been accomplished, conditions that modulate intracellular phosphoamino acid concentrations are still poorly understood. Here we used genetic and metabolic engineering to increase the free intracellular levels of phosphoserine in E. coli. We show that deletion of the phosphoserine phosphatase serB elevates the intracellular levels of phosphoserine within ranges comparable to those of standard amino acids. These new conditions improved insertion of phosphoserine into recombinant proteins. Surprisingly, we also observed dramatic increases in intracellular levels of phosphothreonine and phosphotyrosine when WT cells were grown in LB with supplemented phosphothreonine and serB deficient cells were grown in low phosphate media with supplemented phosphotyrosine, respectively. These findings remove a major barrier for further expansion of the genetic code with additional phosphorylated amino acids.
Site-directed incorporation of nonstandard amino acids (NSAA) to produce novel proteins in vivo has been the aim of many studies with potential applications for biomedical research, human health, and the biotechnology industry. To date, the methodology developed for this purpose relies on protein engineering efforts to generate orthogonal translation systems (OTS) that utilize reassigned stop codons.1,2 Over 70 NSAA have been site-specifically inserted into proteins.1 This was accomplished with a number of strategies including orthogonal tRNA:aminoacyl-tRNA synthetase pairs,1 orthogonal ribosomes,3 and more recently, elongation factor (EF-Tu) variants.4 Recently we reported using a combination of these strategies to incorporate the natural NSAA phosphoserine (pSer).4,5
Serine phosphorylation is among the most abundant posttranslational modifications in eukaryotic cells, and phosphorylated protein networks form the basis for regulating most physiological processes. The kinase component of the human genome is known, and tens of thousands of phosphorylation sites on human proteins are being identified.6,7 However, the corresponding kinase responsible for protein phosphorylation is often not obvious from phosphoproteomics data alone. Genetically encoded phosphorylated amino acids would enable researchers to synthesize natural phosphoproteins without a priori knowledge of their natural posttranslational modification pathway.
Despite many advances in the field, cellular NSAA levels are not typically quantitatively measured in connection with OTS development. This lack of information leads to questions of whether new cotranslationally incorporated NSAA systems may be limited by NSAA bioavailability. Previously we established a pSer OTS without directly examining levels of pSer in the cell.4,5 Questions about the levels of pSer in the cell and optimum OTS function were left unanswered. Here, we determined the intracellular levels of pSer in E. coli and explored the possibility of adding phosphothreonine (pThr) and phosphotyrosine (pTyr) to the intracellular amino acid pool. We accomplished this by developing a quantitative mass spectrometry (MS) assay enabling the quantitation of phosphorylated amino acid levels in E. coli extracts. We then used this method to show that media conditions combined with genomic and metabolomic engineering can elevate pSer to intracellular levels comparable to, or exceeding, other standard amino acids. These conditions improved cotranslational insertion of pSer via the pSer OTS. Furthermore, the same approaches yielded significant elevation of pThr and pTyr in the intracellular amino acid pool. The conditions reported here can be used as a starting point for further expansion of the genetic code with additional phosphorylated amino acids.
Results and Discussion
Expanding the genetic code with any NSAA requires the expansion of the natural pool of free amino acids available for protein synthesis. It has been known for decades that E. coli can import NSAA from the media and even incorporate these new amino acids into proteins.8 We were interested in understanding how phosphorylated NSAA levels would compare to the levels of standard amino acids in the cell. Recent metabolomics studies in E. coli have made quantitative surveys of hundreds of small molecules and metabolites, including amino acids.9,10 However, naturally occurring phosphorylated amino acids, such as pSer, were not considered in these studies despite evidence that pSer is part of serine biosynthetic pathways in most cells.11−13 Mass spectrometry (MS) has increasingly become the method of choice for metabolomics, and new methods have recently been described for quantification of underivatized amino acids from cellular extracts and other complex matrices.14−16 We therefore sought to develop a method to quantitate pSer, pThr, pTyr, and standard amino acids from E. coli cellular extracts. For this purpose we developed a liquid chromatography tandem MS (LC–MS/MS) method on a triple quadrupole system for precise label-free quantitation using multiple reaction monitoring (MRM). This workflow offers high sensitivity, specificity, linear dynamic range, and throughput while multiplexing quantitation of amino acids from a single LC–MS/MS experiment.
We began our study by developing a method to extract amino acids from whole E. coli cells. Our extraction method allowed us to isolate amino acids while removing salts, lipids, and proteins (Figure 1a). Cells lysis in 1 M HCl precipitated out many lipids and most proteins without degrading O-linked phosphorylated amino acids in the solution.17 A chloroform extraction step subsequently removed the majority of the remaining lipids, thereby extending the lifetime of the HPLC column. Amino acids were directly analyzed by LC–MS/MS after a filtration step and normalized for the number of cells in the original cell culture. This method was validated with chemically pure amino acid standards that were spiked into cell extracts to validate efficient recovery. We next developed a MRM method to quantitate relative levels of pSer, pThr, pTyr, serine (Ser), threonine (Thr), tyrosine (Tyr), phenylalanine (Phe), lysine (Lys), histidine (His), and arginine (Arg) in our E. coli extracts. Direct infusion of 30 pmol/μL solutions of each pure amino acid standard into the MS was performed to identify specific transitions for each amino acid and to optimize collision energies for sensitive detection of the amino acids (see Methods). On average, 5 product ions were observed for each singly charged amino acid precursor, and the most sensitive transitions (precursor and product ion pairs) for each amino acid were selected (Supplementary Table 1). The optimized transitions were then used to develop our LC–MS/MS workflow, which included assigning specific retention times for each amino acid (Figure 1a and Supplementary Table 1). Most amino acids in the assay were chromatographically resolved (Figure 1a and Supplementary Table 1) except for moderate overlap of pSer with pThr peaks (0.68 and 0.74 min, respectively) and His, Lys, and Arg peaks (11.43, 11.43, and 11.77 min respectively). A dynamic MRM method with a total dwell time of 800 ms was used for monitoring at least 2 specific transitions for each amino acid. This resulted in at least 1 quantitative transition and 1–3 qualitative transitions for each amino acid (Supplementary Table 1).
Figure 1.
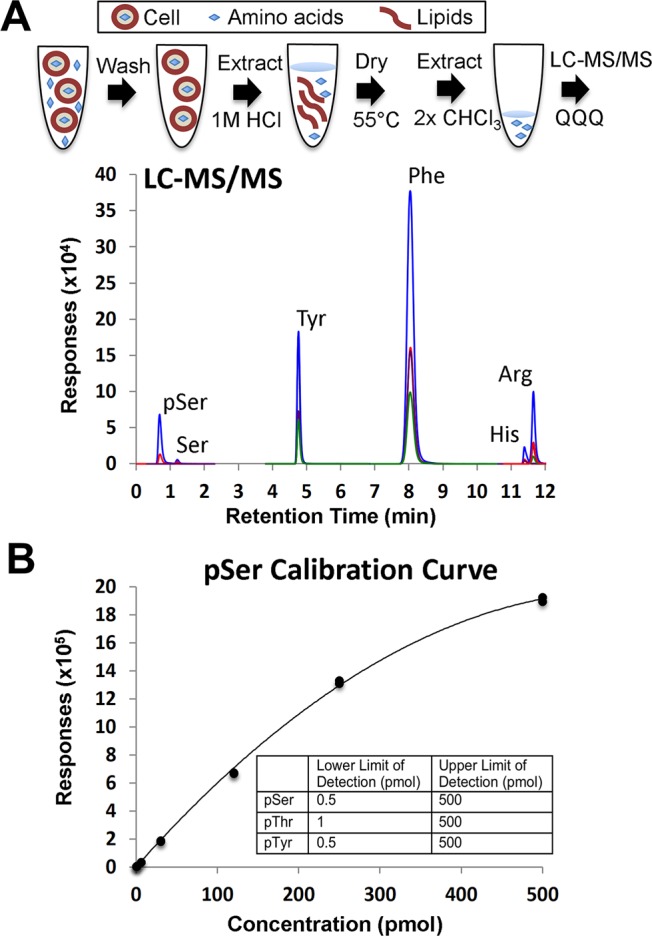
LC–MS/MS analysis of phosphorylated amino acids. (A) Scheme showing the preparation of cell extracts for LC–MS/MS analysis. A typical chromatogram from an LC–MS/MS run of a 90 pmol amino acid standard is shown. Unique MRM transitions are plotted for the amino acids indicated above the individual peaks. (B) Representative calibration curve for pSer. Calibration curves for all analytes were acquired daily in duplicate, and calibration curves were obtained after applying a weighted (1/x) quadratic curve fit. The fitted calibration function for this curve was y = −5.095375x2 + 6398.390847x – 673.466721, and the maximum % residual was 3.9. Experimentally determined lower and upper limits of quantitation for pSer, pThr, and pTyr are listed in the table.
The LC–MS/MS workflow was used to create calibration curves for each amino acid (Figure 1b and Supplementary Figures 1b and 2b). The calibration curve for pSer showed good linearity over a range of concentrations, with similar results for all three phosphorylated amino acids. Calibration curves were fit with a weighted (1/x) quadratic calibration curve ignoring the origin. The quality of the fit for assessing calibration curves was the maximum percent residual as calculated by MassHunter software. All of the amino acids in our study formed calibration curves that were linear up to 250 pmol and began to saturate by 500 pmol, with the exception of pThr, which was linear up to 500 pmol. We used our calibration curves to empirically determine a lower limit of detection (LLOD) and upper limit of detection (ULOD) for each amino acid (Figure 1b and Supplementary Table 2). The lower limit of quantitation (LLOQ) was established in the E. coli matrix assuming a minimum signal/noise ratio of 9:1 for each analyte peak. The upper limit of quantitation (ULOQ) was fixed at 500 pmol for all amino acids, providing good analytical sensitivity and dynamic range for detection of analytes. We used the individual amino acid standard curves and 3–5 replicate measurements from our E. coli extracts to estimate the relative amounts of amino acids in each sample. The measured levels for the standard amino acids were comparable to previous reports.9,10
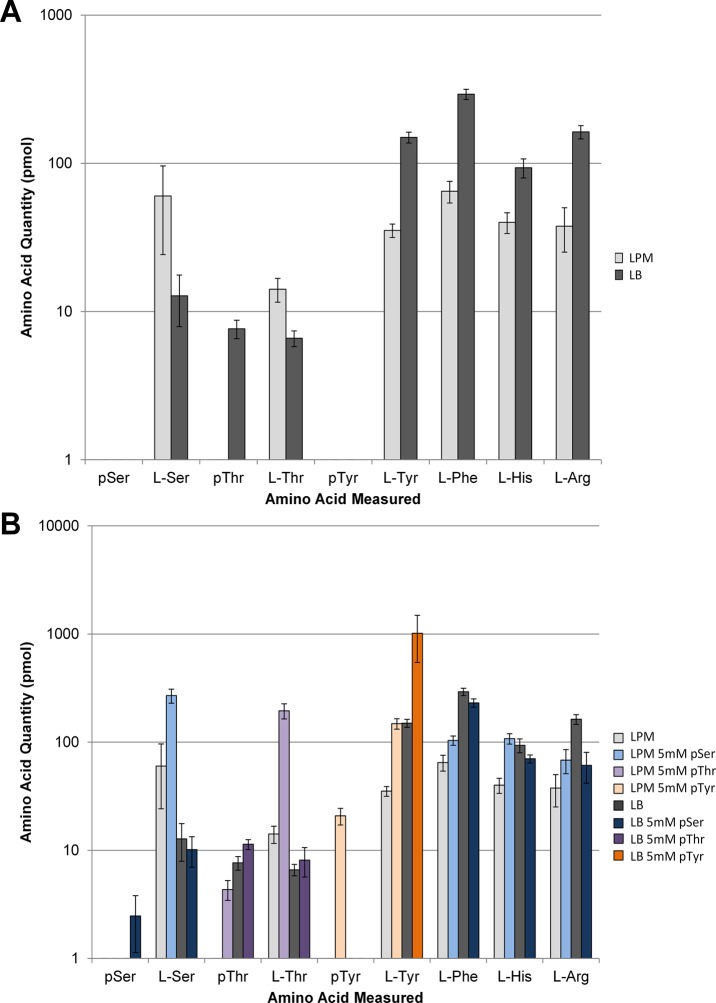
We first explored the steady-state levels of phosphorylated amino acids found in wild type E. coli K12 cells grown in rich Luria–Bertani media (LB) and low phosphate minimal media (LPM). Phosphate concentrations used in LPM were based on previous research conducted to induce the PHO regulon.18 Wild type cells in minimal or rich media were generally devoid of phosphorylated amino acids, while standard amino acids levels were easily detected and showed similar concentrations (Figure 2a). The one unexpected exception was pThr, which was present only in cells cultured in LB and was 10-fold less abundant than standard amino acids. To our knowledge there have been no previous reports of steady-state pools of pThr in any cell. We next tested whether supplementing the media with 5 mM concentration of each phosphorylated amino acid separately would elevate intracellular levels in a wild type background. Supplementing LB media with 5 mM pSer increases steady-state levels in the cell, but the detected concentration is well below the concentration of canonical amino acids (Figure 2b). LPM supplemented with 5 mM pSer produced no detectable levels of pSer in the cell extracts but rather produced an increase in free Ser levels (Figure 2b). This suggested an increase in phosphoserine phosphatase activity consistent with previous studies of E. coli cultured in low phosphate media.19 A slightly more dramatic effect was observed with pThr supplementation. In LB, pThr levels were similar regardless of supplementation, yet in LPM, addition of 5 mM pThr caused a dramatic spike in Thr levels and suggested that, similar to pSer, a phosphate scavenging mechanism is induced and degrades pThr19 (Figure 2b). This mechanism also explains the depletion of the natural pThr pool in LPM without pThr supplementation (Figure 2a). We next examined pTyr by supplementing LB with 5 mM pTyr and observed no detectable levels in the cell. In contrast, a dramatic increase in intracellular pTyr concentration was observed in cells cultured in LPM supplemented with 5 mM pTyr, even greater than pSer/pThr, proportionately (Figure 2b). It is unclear why this might be the case; however, the increase in l-Ser and l-Thr levels after adding pSer and pThr, respectively, relative to l-Tyr might suggest that degradation is the main cause of this difference and not entry into the cell. The most striking effect was a 10-fold rise in Tyr levels after pTyr supplementation in LB (Figure 2b). All three phosphoamino acids are very stable in both media after overnight culture (data not shown) suggesting that pTyr enters the cell and is degraded enzymatically or chemically by endogenous pathways. Further exploration is necessary to understand why pTyr is turned over in cells grown in LB but not in LPM. One possibility is that pTyr is less utilized than pSer/pThr in phosphate scavenging but is degraded for tyrosine scavenging when the cell is in rich media where Tyr is necessary for rapid growth. This suggests that there are divergent mechanisms for pTyr and pSer/pThr turnover in the cell with a previously unrecognized pathway for pTyr turnover in rich media with pThr-sparing properties.
Figure 2.
Quantitation of intracellular amino acids in E. coli extracts. (A) Measured amino acid quantities of select amino acids in K12 BW25113 WT E. coli grown in LPM (light gray) and LB (dark gray) media. (B) Relative amino acid concentrations of K12 BW25113 WT E. coli grown in LPM (light gray) and LB (dark gray) media compared to WT E. coli grown in LPM with 5 mM pSer (light blue), LB with 5 mM pSer (dark blue), LPM with 5 mM pThr (light violet), LB with 5 mM pThr (dark violet), LPM with 5 mM pTyr (light orange), and LB with 5 mM pTyr (dark orange). Error bars were calculated using the 95% confidence interval with n = 5.
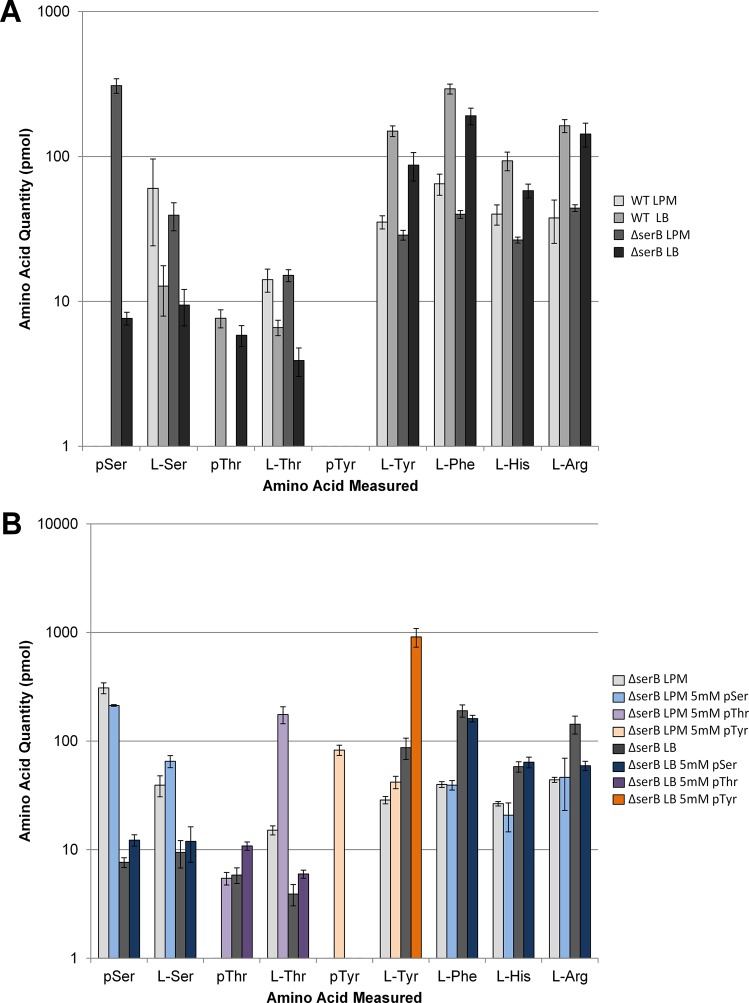
Supplementing LB and LPM with phosphorylated amino acids showed that phosphatase activity may be a critical lynchpin to elevate intracellular levels of phosphorylated amino acids. We have previously shown that deletion of the serB gene is required for our system to genetically encode pSer.4,5 However, we never directly measured the effect of serB deletion (ΔserB) on the steady-state levels of pSer in the cell. We therefore used our LC–MS/MS method to explore the effects of ΔserB on cellular levels of pSer, pThr, and pTyr. We observed a dramatic 10-fold increase in pSer levels in the ΔserB strain compared to wild type cells in LB media (Figure 3a). The same experiment was repeated in LPM and showed an even greater 100-fold increase in pSer levels (Figure 3a). These extreme increases were not influenced by supplementation with pSer and had no dramatic effects on the canonical amino acids monitored in our experiments (Figure 3a,b). Thus, we showed that a mutation in the serine biosynthetic pathway had a more dramatic effect on increasing pSer than supplementing the media with this NSAA.
Figure 3.
Quantitation of intracellular amino acids in a ΔserBE. coli strain extracts. (A) Measured amino acid quantities of select amino acids in K12 BW25113 WT vs ΔserBE. coli grown in LPM media and LB media. (B) Measured amino acid quantities of ΔserBE. coli grown in LPM media (light gray), LB media (dark gray), LPM with 5 mM pSer (light blue), LB with 5 mM pSer (dark blue), LPM with 5 mM pThr (light violet), LB with 5 mM pThr (dark violet), LPM with 5 mM pTyr (light orange), and LB with 5 mM pTyr (dark orange). Error bars were calculated using the 95% confidence interval with n = 5.
We next examined the levels of pTyr and pThr in the ΔserB strain. While the ΔserB background stabilized pSer levels, free pThr levels were not protected in LPM and suggested that another phosphatase scavenges pThr in low phosphate conditions (Figure 3a). Supplementing LPM with pThr stabilized free pThr in the cell but with an accompanying 10-fold increase in Thr levels. Furthermore, this provides evidence for a pSer-sparing pThr phosphatase activity in the ΔserB cells (Figure 3b). We conducted a similar set of experiments in a ΔphnE1 background and saw a dramatic reduction of Ser and Thr levels in cells grown in LPM+pThr or pSer compared with WT cells (Supplementary Figure 3). This observation is again consistent with a typical PHO regulon response which includes the induction of transport pathways (that require phnE) to bring phosphate-containing molecules into the cell. Interestingly, serB disruption also had no effect on the striking pTyr turnover in LB but produced 10-fold more intracellular pTyr in LPM supplemented with pTyr (Figure 2b). These results suggest the pTyr- and pSer-sparing effects of the serB deletion are connected in low phosphate conditions but suggest a more complicated relationship between the three phosphorylated amino acids in LPM vs LB. Taken together, our data showed that genetic lesions in amino acid metabolism combined with supplementation can be used to elevate the levels of three different phosphorylated amino acids in E. coli.
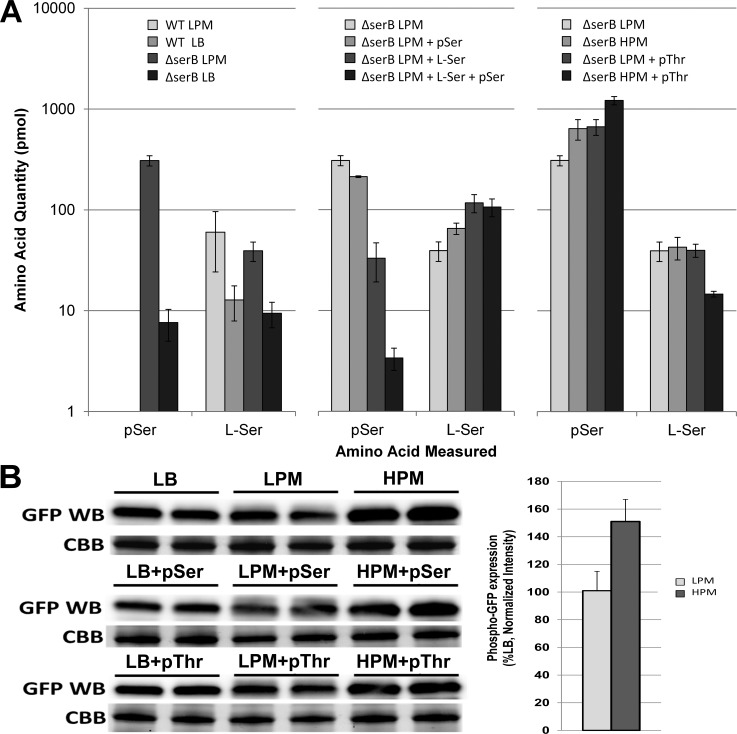
Removing serB had the greatest pSer-sparing effect in the cell; however, a shift to LPM clearly elevated additional native pSer/pThr phosphatase pathways and suggested it might be possible to further elevate pSer levels by deleting nonessential phosphatase enzymes. We observed an increase in Ser levels in LPM media in both WT and serB-deficient backgrounds, which suggested that LPM was stimulating serine biosynthesis (Figure 4a). Since free Ser is a direct inhibitor of serA, which is upstream of serC-dependent pSer formation, we added Ser to LPM and examined the effect on steady-state pSer levels in the cell. LPM+Ser reduced the pSer levels, while pSer supplementation had little effect (Figure 4a). Interestingly, LPM+Ser+pSer drove pSer levels down even further. The mechanism behind the additive effect of pSer and Ser on the serine biosynthesis pathway is unknown, but the serB deficient background suggests cooperativity between serA and serC in this scenario and points to a possible negative feedback contribution of pSer in this pathway.
Figure 4.
Dynamic levels of pSer in the cell and in protein synthesis. (A) pSer and Ser levels in K12 BW25113 WT extracts and the corresponding ΔserB strain grown in LPM, LB and HPM media supplemented with pSer and pThr and l-Ser. (B) Western blot analysis of E17TAG-GFP (GFP) expressed in EcAR7(ΔserB) with the indicated media conditions. Coomassie brilliant blue (CBB) gels are shown as a loading control and for normalizing expression ratios. Expression ratios were calculated for E17TAG-GFP expression in LPM and HPM without supplementation of phosphoamino acids and plotted as a percentage of LB expression. Error bars were calculated using the 95% confidence interval with n = 3.
Since the LPM conditions stimulate phosphate scavenging pathways via the PHO regulon, we added phosphate to create a high phosphate minimal media (HPM). The HPM conditions increased the free levels of pSer compared to LPM and were more stabilizing than adding pSer to LPM (Figure 4a and Supplementary Figure 4). Since our previous experiments suggested that pSer/pThr phosphatase pathways were still active in the ΔserB cells, we added pThr to both LPM and HPM. pThr in LPM had a similar pSer-sparing effect as HPM, while HPM+pThr had a striking additive effect and pushed pSer levels to the highest measured for any amino acid in the entire study. Although pThr contribution is unclear, the addition of pThr appears to drive down l-Ser levels by an unknown mechanism and thus, indirectly, to accelerate the serine biosynthesis pathway (Figure 4a). Finally, we tested the effect of elevated free pSer on protein synthesis. We used an established system in which pSer is incorporated into position 17 of GFP (GFP-E17TAG) and GFP synthesis is dependent on robust insertion of pSer via our pSer OTS.4,5 Western blots of pSer-dependent GFP-E17TAG synthesis showed a 50% increase in GFP synthesis in HPM conditions, regardless of pSer or pThr supplementation (Figure 4b). This provides direct evidence that the conditions identified in this study elevate the levels of endogenous phosphorylated amino acids to ranges that improve pSer incorporation into recombinant proteins.
Nature has evolved complex networks of posttranslational modification to ensure accurate and stoichiometric phosphorylation of proteins in vivo. The majority of these networks are not characterized in sufficient detail to provide the information needed to accurately synthesize phosphoproteins for biochemical analysis. Furthermore, reconstituting these complex pathways might not be possible for the production of recombinant phosphoproteins in E. coli. To solve this problem we added phosphoserine to the genetic code of E. coli to enable the de novo synthesis of important phosphoserine-containing proteins.4,5 This system required adequate intracellular phosphoserine concentrations to support protein synthesis, yet the concentrations of free phosphorylated amino acids in E. coli was not known. Of primary importance, we address that pSer, pThr, and pTyr levels can be measured and altered by manipulating metabolic pathways genetically and through media conditions. Our results suggest insufficient pSer concentrations in E. coli was a limiting factor in recombinant phosphoprotein synthesis and increasing intracellular pSer concentration appears to increase the yield of recombinant phosphoproteins. Furthermore, by understanding the underlying principles for phosphoserine levels in the cell, we may gain important insight into controlling other phosphorylated amino acids in E. coli for future development of orthogonal pThr and pTyr incorporation systems. It is important to note that although increasing pSer might aid in pSer incorporation efficiency, each modification in the media and genome can have opposite effects on the health of cells. This might explain why HPM+pThr did not result in the highest yield of protein although it had the greatest proportion of pSer (Figure 4b). Thus, there is a balance between both ensuring pSer levels sufficient for protein synthesis while optimizing cell fitness in order to yield the greatest amount of phosphoprotein.
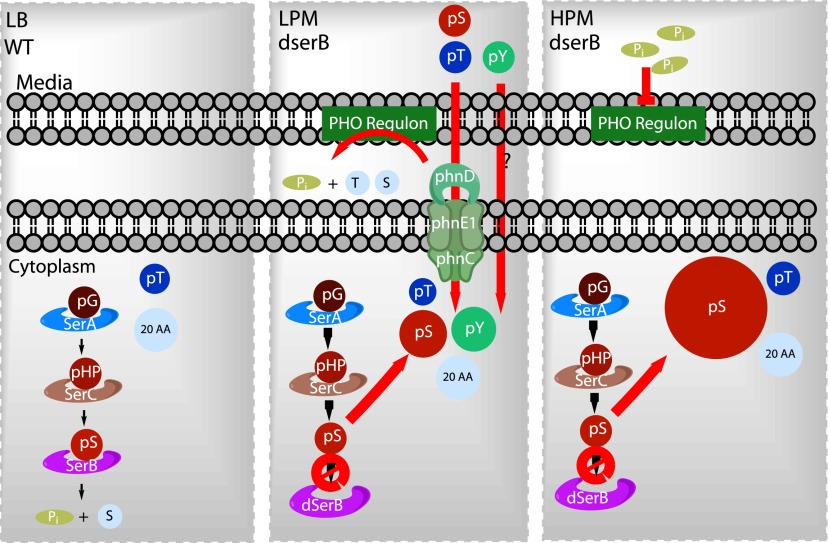
We developed an assay to quantitate phosphorylated amino acids with LC–MS/MS and subsequently discovered conditions by which pSer, pThr, and pTyr could be added to the natural pool of amino acids in E. coli (Figure 5). We found that pThr is naturally present in E. coli and that pSer and pTyr can be imported into the cell via known pathways (Figure 5). We also confirmed that deletion of the phosphoserine phosphatase serB increased intracellular pSer concentration as predicted.4 Surprisingly, serB deletion further stabilized the levels of pTyr in the cell while having only a modest effect on pThr. LPM induces the PHO regulon,20 which stimulates the uptake of pSer, pTyr, and pThr (Figure 2, Figure 3, and Supplementary Figure 4). Cells harboring a serB deletion and cultured separately in pSer-, pTyr-, and pThr-supplemented LPM bring the phosphorylated amino acids within a suitable range for protein synthesis. This was demonstrated by testing HPM that enhanced pSer levels in the cell and enhanced phosphoprotein synthesis (Figure 4). While a serB deletion strain cultured in LB supplemented with 5 mM pTyr elevated all three phosphorylated amino acids in the same cell, we found optimal conditions that enhanced each phosphoamino acid separately (summarized in Figure 5 and Supplementary Table 5).
Figure 5.
A model for steady-state pSer, pThr, and pTyr levels in WT vs ΔserB E. coli strains. A WT cell (left) contains low levels of phosphothreonine (pT), and the serine biosynthetic pathway (SerA, SerC, SerB) catabolizes phosphoserine (pS). Low phosphate conditions (middle) induces the PHO regulon and phosphate uptake machinery. In the absence of SerB, pS accumulates and extracellular pS, phosphothreonine (pT); phosphotyrosine (pY) can elevate intracellular pT/pY levels. In this study, we show that pY is imported by additional unknown transport pathways and various media conditions can support intracellular pS/pT/pY levels on par with standard amino acids (20 aa, represented by circle size). High phosphate minimal media (right) suppresses PHO regulon phosphatase activities and, in the absence of SerB, can result in pS levels 10-fold higher than the standard 20 aa. Abbreviations: d-glyceraldehyde 3-phosphate (pG); 3-phosphohydroxypyruvate (pHP); serine (S); threonine (T); inorganic phosphate (Pi); Organophosphate transporter (phnD,phnE1,phnC).
We investigated the natural pool of amino acids to find preexisting pSer, pThr, pTyr or ways to naturally stabilize these amino acids by modulating known metabolic pathways. We focused on these three amino acids because of their pivotal role in signal transduction, regulation of biological systems, and the need to decipher their function. Incorporation of pSer into the genetic code was enabled by the discovery of a natural system for charging pSer onto tRNA.21 Subsequent engineering efforts then provided a system for incorporation of pSer at amber (UAG) codons.4 Furthermore, we and others have shown that the amber stop codon can be reassigned from a stop to a sense codon by eliminating release factor 1 from E. coli.5,22−26 We therefore had all of the requirements for a UAG codon dedicated for pSer, except for a clear understanding of the levels of pSer in the cell. We reasoned that serB would degrade free pSer and thus be refractory to pSer incorporation into polypeptides. Indeed, this idea was upheld, and in this study we showed that serB was the key factor in elevating pSer levels for protein synthesis. Since most of the components have been worked out for pSer, future studies will focus on engineering orthogonal translation systems for pThr and pTyr. Importantly, while more work needs to be done to add combinations of NSAA into proteins, we have demonstrated, in principle, that all three phosphorylated NSAA can be present in the same cell and could theoretically be added to the same polypeptide (see Supplementary Table 5).
E. coli encodes a complex protein network to ensure phosphate (Pi) is provided to the intracellular environment and made available for vital processes.20 Phosphorylated amino acids and a myriad of other organophosphates can enter the cell during low Pi conditions via the well characterized PHO regulon. The preferred phosphate source is Pi, which enters the cell via either low or high affinity phosphate transporters. Alternatively, organophosphates or phosphonates can be utilized by the cell, and many of the components of their uptake have been described.20 We hypothesized that low phosphate concentrations in the media could increase the uptake of phosphorylated amino acids and could divert or inhibit their subsequent breakdown into Pi to further stabilize the levels of the phosphorylated amino acid (Figure 5). This general hypothesis was confirmed for pSer (Figure 3). We confirmed that known organophosphate uptake pathways played a role by using phnE1 deletion strains (Supplementary Figure 3). This protein comprises the main conduit for organophosphate transport into the cytoplasm18−20 (Figure 5). However, the pathways for pThr and pTyr, once inside the cell, are not well understood. While we were able to elevate intracellular concentrations of pTyr and pThr, there was clear evidence of increased phosphatase activity directed at these two amino acids. Interestingly, this background phosphatase activity was evident even under pSer-sparing conditions (Figure 3b). Oddly, there was clearly an enhanced pTyr turnover in rich media where the actions of the PHO regulon could not easily be explained (Figure 3b). This evidence suggests that other phosphatases might be targeted in future studies which may further stabilize pTyr and pThr levels for expanding the genetic code.
Genetic code expansion and engineering has been explored for its potential to introduce NSAA into proteins for industrial and medical applications. These efforts have mostly focused on the orthogonal pairs of aminoacyl-tRNA synthetases (aaRS) and tRNAs, and less effort has been devoted to establishing intracellular amino acid pools necessary for stoichiometric incorporation of NSAA into proteins. Indeed, if the NSAA is not freely available at concentrations needed for translation, engineering and laboratory evolution experiments could fail to produce orthogonal translations systems that would be comparable to their natural counterparts.27,28 The uptake of NSAA into the cell has been appreciated and studied far longer than engineering of orthogonal aaRS and tRNA pairs.8 The described LC–MS/MS method has broad applicability for quantitative measurement of NSAA levels in the intracellular pool and will be a useful tool to optimize intracellular NSAA concentrations that support efficient NSAA protein production. Here we leveraged this approach to improve our system for genetically encoded pSer and opened up a new pathway to adding pThr and pTyr to the genetic code.
Methods
Amino Acid Standards
Standards of free amino acids were prepared by weight in HPLC grade water as follows: 10 mM O-phospho-l-tyrosine (Sigma P9405), 10 mM O-phospho-l-threonine (Sigma P1053), 10 mM O-phospho-l-serine (Sigma P0878), 10 mM l-lysine (Sigma L5626), 10 mM l-arginine (Sigma A-5131), 10 mM l-histidine (Sigma H8000), and 10 mM l-phenylalanine (Sigma P-2126) all prepared at 10 mM, 20 mM threonine (Sigma-Aldrich 89179), 20 mM serine (Sigma-Aldrich S4500), and 2 mM tyrosine (Sigma-Aldrich 93829). A combination of these amino acid stocks was prepared at 1 mM. All standards were filtered (0.22 μm, Millipore) and stored at −80 °C.
Strains and Culture Conditions
The E. coli strains MG1655, BW25113, ΔserB(Keio Collection JW4351), ΔphnE1(Keio Collection JW4064), and EcAR75 were used in this study. Twenty-milliliter cultures were grown for 18 h at 37 °C in the appropriate media, and OD600 values were recorded for normalization. LB media contained 5 g/L NaCl (American Bioanalytical NaCl AB01915), 5 g/L yeast extract (BD Bacto Yeast Extract REF212750), and 10 g/L of tryptone (BD Bacto tryptone REF211705). Low Phosphate Minimal Media (LPM) media was made with 200 mL of 5x LPM salts (2.5 g NaCl [43 mM], 5 g NH4Cl [94 mM], 0.07 g KH2PO4 [0.5 mM], 0.015g CaCl2 [0.14 mM], and 60 g 2-amino-2-(hydroxymethyl)-1,3-propanediol (Tris) [500 mM] adjusted to pH 7.0 with conc HCl (J.T. Baker), 1 mL of 1 M MgSO4, 4 mL of 50% d-glucose, 88 mL of 2 mM l-tyrosine (Sigma-Aldrich 93829), 38 mL of 30 mM l-aspartic acid (Sigma-Aldrich A9256), 17 mL of 30 mM l-glutamic acid (Sigma-Aldrich G1251) and 10 mL of 20% pure amino acid mix [50 mL water, 0.44 g l-alanine (Sigma-Aldrich A7627), 0.21 g l-arginine (Sigma-Aldrich A5131), 0.06 g l-cysteine (Sigma-Aldrich C7880), 0.11 g l-glycine (American Bioanalytical AB00730), 0.11 g l-histidine (Sigma-Aldrich H8000), 0.27 g l-isoleucine (Sigma-Aldrich I2752), 0.46 g l-leucine (Sigma-Aldrich L8000), 0.57 g l-lysine (Sigma-Aldrich L5626), 0.12 g l-methionine (Sigma-Aldrich M9625), 0.19 g l-phenylalanine (Sigma-Aldrich P2126), 0.57 g l-proline (Sigma-Aldrich P-0380), 0.21 g l-serine (Sigma-Aldrich S4500), 0.05 g l-threonine (Sigma-Aldrich 89179), 0.34 g l-valine (Sigma-Aldrich V0500)] was added. Finally, the volume was adjusted with DI water to 1000 mL. Amino acid concentrations were based on standard casein preparation. Additional amino acids were added at the concentrations indicated. High Phosphate Minimal Media (HPM) was formulated as the LPM media with 5x M9 salt (56.4 g/L BD Difco M9 salts REF 248510), 28 μL/L of 1 M CaCl2, 1 mL/L of 1 M MgSO4, 4 mL/L of 50% d-glucose, 88 mL of 2 mM l-tyrosine, 38 mL of 30 mM l-aspartic acid, 17 mL of 30 mM l-glutamic acid, and 10 mL/L of 20% pure amino acid mix.
Protein Expression
The E. coli strain EcAR7 was transformed with a plasmid bearing a GFP variant with an amber STOP codon at position 17 (E17TAG-GFP) and an all-in-one OTS plasmid to enable cotranslational insertion of phosphoserine.4,5 The all-in-one OTS plasmid was created with pSepT and pKD-SepRS-EFSep plasmids.4 The 250 bp tRNASep cassette was PCR amplified from the pSepT plasmid using primers tRNASep −F (5′-ACC GCG GCC GCA AAA AAA ATC cttagctttcg-3′) and tRNASep −R (5′-AAA GCG GCC GCG CTT CTT TG agcgaac-3′). The PCR primers added NotI restriction sites to each end of the PCR product. The pKD-SepRS-EFSep plasmid was linearly digested with NotI, and two copies of the tRNASep cassette were ligated sequentially. Phosphoserine insertion at position 17 in GFP was confirmed with mass spectrometry as previously described.5 The transformed EcAR7 precultures were grown at 30 °C overnight in LB media (pH 7.5), with 0.08% glucose and 25 μg/mL of both zeocin and kanamycin, to retain the E17TAG-GFP and all-in-one OTS plasmid; respectively. The preculture was pelleted (1000g for 7 min) and resuspended in LPM (with antibiotics) in a 20:1 ratio. The resuspended cells were inoculated into each of the following media conditions containing the appropriate antibiotics in duplicate: LB, LPM, HPM, LB+5 mM pSer, LPM+5 mM pSer, HPM+5 mM pSer, LB+5 mM pThr, LPM+pThr, and HPM+pThr. The cultures were grown at 30 °C, 230 rpm for 24 h. The cultures were further diluted 1:2 in the corresponding growth media with antibiotics and the addition of 1 mM IPTG, and 100 ng/mL anhydrotetracycline to induce the SepRS, EFSep, and E17TAG-GFP. Protein was overexpressed at 30 °C, 230 rpm for 6 h. After expression, OD600 values were obtained for each culture and the amount of cells harvested was normalized on the basis of optical density. The normalized cell pellets from each condition were resuspended in lysis buffer (50 mM Tris/HCl pH 7.5, 150 mM NaCl, 5% glycerol, 0.5x BugBuster, 1 mM DTT, protease inhibitors (Roche), 25 U/mL benzonase, 50 mM NaF, 1 mM Na3VO4) and incubated at RT for 15 min. The total lysates were diluted with 2x Laemmli sample buffer, boiled at 95 °C for 5 min, and centrifuged to remove insoluble debris. The samples were analyzed with both SDS–PAGE electrophoresis followed by staining with Coomassie brilliant blue (CBB) and parallel Western blot analysis. Western blot analysis was performed with mouse monoclonal anti-GFP (Invitrogen no. 332600) and imaged with an HRP secondary antibody via chemiluminescence on a ChemiDoc system (BioRad). Quantitation was performed with BioRad software, and GFP signals were normalized to reference Coomassie-stained proteins. Data from biological triplicates were exported to Microsoft Excel for generation of the graphs. Error bars were constructed using the 95% confidence interval.
Amino Acid Extraction
E. coli were grown overnight in 20-mL cultures, and cells were harvested by centrifugation for 10 min at 4,000g at 4 °C. The cell pellets were transferred into a 1.5-mL PCR tube and washed three times with 1 mL of ice cold 1x LPM salt solution. The cell pellets were then lysed in 1 mL of 1 M HCl17,29 and centrifuged for 20 min at 19,000g at RT. The resulting clear amino acid extract was transferred into a new 1.5-mL Eppendorf tube and dried in a rotary vacuum centrifuge operated at 2500g for 3 h at 55 °C and an additional 90 min at 30 °C. The dried pellets were frozen at −80 °C until use. The dry pellet was reconstituted in 300 μL of water, and lipids were removed by serial liquid–liquid extraction with 500 and 300 μL of chloroform, respectively. Extractions were performed by vortex (30 s), and phase separation was facilitated by centrifugation for 30s at 13000g. The upper aqueous layer containing the amino acids was collected. The two chloroform phases were extracted once more with 300 μL of water. The combined amino acid extracts were clarified through a 0.22 μm pore size Durapure PDVF centrifugal filter (2 min at 12,000g at 4 °C), and the filter was washed 1x with 100 μL of water. The combined aqueous phases were dried in a rotary vacuum centrifuge with the following program: 80 min at 2,500g and 65 °C and then 80 min at 55 °C. The dried pellet was stored at −80 °C until use.
LC–MS
Frozen pellets were thawed on ice, dissolved by vortex in 50 μL of HPLC grade water, and then centrifuged for 3 min at 17,500g at RT. The clarified solution was diluted for LC–MS/MS according to the OD600 of the cell culture. As an example 8.4 μL extract of a OD 1.00 culture was mixed with 21.6 μL eluent A (see below). Deactivated glass inserts (Agilent no. 5182-0720) were found critical for the performance of the method as polypropylene vials resulted in significant analyte loss (data not shown). Calibration curves were obtained by dilution of the 1 mM amino acid master mix in eluent A. LC–MS/MS was performed on an Agilent 6490 triple quadrupole instrument equipped with a JetStream ion source operated at 350 °C and 2500 V spray voltage. Transitions for quantitation of amino acids were obtained by direct infusion of amino acid standards prepared at 2–30 pmol/μL in HPLC eluent B (see below). The flow rate was 200 μL/min, and collision energies for up to 6 transitions were obtained using the optimizer software package provided with the instrument. Reversed phase HPLC was performed on a Agilent 1260 system equipped with a temperature-controlled autosampler operated at 4 °C. The column was an Acuity UPLC BEH C18 2.1 × 50 mm (Waters, Millford MA) that was packed with 1.7 μm C18 particles. Chromatography was performed at RT with a flow rate of 0.2 mL/min unless noted otherwise. The injection volume was 3 μL for all experiments. Eluent A consisted of 0.5% acetonitrile (ACN) containing 0.1% formic acid and 0.1% perfluorooctanoic acid (PFOA), and eluent B was 90% ACN with 0.1% formic acid and 0.1% PFOA. Gradient conditions were optimized by injecting an amino acids standard mix prepared at 30 pmol/μL into eluent A. This resulted in the following optimized linear gradient: 0–2 min, 0% B; 3 min, 30% B; 15 min, 62% B; 15.5 min, 100% B (0.3 mL/min); 21.5 min, 100% B (0.35 mL/min); 22.5 min, 0% B; and 30 min, 0% B. The final dynamic MRM method had a cycle time of 800 ms. Details of this method can be found in the supplemental material (Supplementary Table 4). Standards for obtaining the calibration curve were analyzed in triplicate. Samples were analyzed in a randomized fashion, and blanks and quality control standards were run after sets of no more than 10 samples. Between 3 and 5 biological replicates were analyzed for each experimental condition tested.
Data Processing and Statistical Analysis
Quantitation of MRM data was performed with Agilent MassHunter Software v. 5.00 considering 1 quantifier and up to 3 qualifier ions for each analyte. Peaks below the limit of quantitation (signal-to-noise ratios <9:1) were excluded from the analysis. The ratio between quantifier and qualifier transitions had to be within 20% of the ratio obtained from the analysis of a corresponding pure amino acid standard. Automated peak integration was manually checked and if necessary adjusted to ensure consistent peak integration. Finally, data were exported to Microsoft Excel for generation of the graphs. Error bars were constructed using the 95% confidence interval.
Acknowledgments
We thank D. Söll for helpful discussion and support for J. Steinfeld and Y. Liu during preliminary phases of this study (Grants to D.S.: NIH GM22854, Defense Advanced Research Projects Agency contract N66001-12-C-4020, and NSF MCB-0950474). We thank N. Pirman, L. Cheng, and J. Ling for critical review of the manuscript and J. Lynch, D. Postl, S. Kulkarni, and G. Alexis for helpful discussion and support of the LC–MS platform. Funding to J.R. by NIH NIDDK-K01DK089006 and DARPA contract N66001-12-C-4211.
Supporting Information Available
Additional figures and tables as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
∥ These authors contributed equally to this work.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Liu C. C.; Schultz P. G. (2010) Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444. [DOI] [PubMed] [Google Scholar]
- Chin J. W. (2011) Reprogramming the genetic code. EMBO J. 30, 2312–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.; Neumann H.; Peak-Chew S. Y.; Chin J. W. (2007) Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 25, 770–777. [DOI] [PubMed] [Google Scholar]
- Park H. S.; Hohn M. J.; Umehara T.; Guo L. T.; Osborne E. M.; Benner J.; Noren C. J.; Rinehart J.; Söll D. (2011) Expanding the genetic code of Escherichia coli with phosphoserine. Science 333, 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann I. U.; Rovner A. J.; Aerni H. R.; Rogulina S.; Cheng L.; Olds W.; Fischer J. T.; Soll D.; Isaacs F. J.; Rinehart J. (2012) Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett. 586, 3716–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G.; Whyte D. B.; Martinez R.; Hunter T.; Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- Grimsrud P. A.; Swaney D. L.; Wenger C. D.; Beauchene N. A.; Coon J. J. (2010) Phosphoproteomics for the masses. ACS Chem. Biol. 5, 105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budisa N. (2004) Prolegomena to future experimental efforts on genetic code engineering by expanding its amino acid repertoire. Angew. Chem., Int. Ed. 43, 6426–6463. [DOI] [PubMed] [Google Scholar]
- Bennett B. D.; Kimball E. H.; Gao M.; Osterhout R.; Van Dien S. J.; Rabinowitz J. D. (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J.; Fowler W. U.; Kimball E.; Lu W.; Rabinowitz J. D. (2006) Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nat. Chem. Biol. 2, 529–530. [DOI] [PubMed] [Google Scholar]
- Kung C.; Hixon J.; Choe S.; Marks K.; Gross S.; Murphy E.; DeLaBarre B.; Cianchetta G.; Sethumadhavan S.; Wang X.; Yan S.; Gao Y.; Fang C.; Wei W.; Jiang F.; Wang S.; Qian K.; Saunders J.; Driggers E.; Woo H. K.; Kunii K.; Murray S.; Yang H.; Yen K.; Liu W.; Cantley L. C.; Vander Heiden M. G.; Su S. M.; Jin S.; Salituro F. G.; Dang L. (2012) Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. ACS Chem. Biol. 19, 1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K. H.; Farley J. R.; Baylink D. J. (1989) Phosphotyrosyl protein phosphatases. Biochem. J. 257, 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizer L. I. (1963) Pathway and control of serine biosynthesis in Escherichia coli. J. Biol. Chem. 238, 3934–&. [PubMed] [Google Scholar]
- Armstrong M.; Jonscher K.; Reisdorph N. A. (2007) Analysis of 25 underivatized amino acids in human plasma using ion-pairing reversed-phase liquid chromatography/time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 21, 2717–2726. [DOI] [PubMed] [Google Scholar]
- Piraud M.; Vianey-Saban C.; Petritis K.; Elfakir C.; Steghens J. P.; Bouchu D. (2005) Ion-pairing reversed-phase liquid chromatography/electrospray ionization mass spectrometric analysis of 76 underivatized amino acids of biological interest: a new tool for the diagnosis of inherited disorders of amino acid metabolism. Rapid Commun. Mass Spectrom. 19, 1587–1602. [DOI] [PubMed] [Google Scholar]
- Qu J.; Wang Y.; Luo G.; Wu Z.; Yang C. (2002) Validated quantitation of underivatized amino acids in human blood samples by volatile ion-pair reversed-phase liquid chromatography coupled to isotope dilution tandem mass spectrometry. Anal. Chem. 74, 2034–2040. [DOI] [PubMed] [Google Scholar]
- Niedbalski J. S.; Ringer D. P. (1986) Separation and quantitative analysis of O-linked phosphoamino acids by isocratic high-performance liquid chromatography of the 9-fluorenylmethyl chloroformate derivatives. Anal. Biochem. 158, 138–145. [DOI] [PubMed] [Google Scholar]
- Wanner B. L.; Metcalf W. W. (1992) Molecular genetic studies of a 10.9-kb operon in Escherichia coli for phosphonate uptake and biodegradation. FEMS Microbiol. Lett. 79, 133–139. [DOI] [PubMed] [Google Scholar]
- Rao N. N.; Wang E.; Yashphe J.; Torriani A. (1986) Nucleotide pool in pho regulon mutants and alkaline phosphatase synthesis in Escherichia coli. J. Bacteriol. 166, 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L. (1993) Gene-regulation by phosphate in enteric bacteria. J. Cell. Biochem. 51, 47–54. [DOI] [PubMed] [Google Scholar]
- Sauerwald A.; Zhu W. H.; Major T. A.; Roy H.; Palioura S.; Jahn D.; Whitman W. B.; Yates J. R.; Ibba M.; Soll D. (2005) RNA-dependent cysteine biosynthesis in archaea. Science 307, 1969–1972. [DOI] [PubMed] [Google Scholar]
- Mukai T.; Hayashi A.; Iraha F.; Sato A.; Ohtake K.; Yokoyama S.; Sakamoto K. (2010) Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 38, 8188–8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. B. F.; Xu J. F.; Shen Z. X.; Takimoto J. K.; Schultz M. D.; Schmitz R. J.; Xiang Z.; Ecker J. R.; Briggs S. P.; Wang L. (2011) RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat. Chem. Biol. 7, 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. B. F.; Wang C.; Xu J. F.; Schultz M. D.; Schmitz R. J.; Ecker J. R.; Wang L. (2012) Release factor one is nonessential in Escherichia coli. ACS Chem. Biol. 7, 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake K.; Sato A.; Mukai T.; Hino N.; Yokoyama S.; Sakamoto K. (2012) Efficient decoding of the UAG triplet as a full-fledged sense codon enhances the growth of a prfA-deficient strain of Escherichia coli. J. Bacteriol. 194, 2606–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie M. J.; Rovner A. J.; Goodman D. B.; Aerni H. R.; Haimovich A. D.; Kuznetsov G.; Mercer J. A.; Wang H. H.; Carr P. A.; Mosberg J. A.; Rohland N.; Schultz P. G.; Jacobson J. M.; Rinehart J.; Church G. M.; Isaacs F. J. (2013) Genomically recoded organisms expand biological functions. Science 342, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L. T.; Helgadottir S.; Soll D.; Ling J. Q. (2012) Rational design and directed evolution of a bacterial-type glutaminyl-tRNA synthetase precursor. Nucleic Acids Res. 40, 7967–7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara T.; Kim J.; Lee S.; Guo L. T.; Soll D.; Park H. S. (2012) N-Acetyl lysyl-tRNA synthetases evolved by a CcdB-based selection possess N-acetyl lysine specificity in vitro and in vivo. FEBS Lett. 586, 729–733. [DOI] [PubMed] [Google Scholar]
- de Witte P. A.; Cuveele J. F.; Merlevede W. J.; Vandenheede J. R. (1995) Analysis of phosphorylhydroxyamino acids present in hydrolyzed cell extracts using dabsyl derivatization. Anal. Biochem. 226, 1–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.