Abstract
Regenerative therapies for cartilage defects have been greatly advanced by progress in both the stem cell biology and tissue engineering fields. Despite notable successes, significant barriers remain including shortage of autologous cell sources and generation of a stable chondrocyte phenotype using progenitor cells. Increasing demands for the treatment of degenerative diseases, such as osteoarthritis and rheumatoid arthritis, highlight the importance of epigenetic remodeling in cartilage regeneration. Epigenetic regulatory mechanisms, such as microRNAs, DNA methylation, and histone modifications, have been intensively studied due to their direct regulatory role on gene expression. However, a thorough understanding of the environmental factors that initiate these epigenetic events may provide greater insight into the prevention of degenerative diseases and improve the efficacy of treatments. In other words, if we could identify a specific factor from the environment and its downstream signaling events, then we could stop or retard degradation and enhance cartilage regeneration. A more operational definition of epigenetic remodeling has recently been proposed by categorizing the signals during the epigenetic process into epigenators, initiators, and maintainers. This review seeks to compile and reorganize the existing literature pertaining to epigenetic remodeling events placing emphasis on perceiving the landscape of epigenetic mechanisms during cartilage regeneration with the new operational definition, especially from the environmental factors' point of view. Progress in understanding epigenetic regulatory mechanisms could benefit cartilage regeneration and engineering on a larger scale and provide more promising therapeutic applications.
Introduction
Articular cartilage defects are common disorders that affect people of all ages; treatment of this disorder remains challenging. The incidence of cartilage defects has been reported to be as high as 65% in routine knee arthroscopies [1,2]. Trauma; degenerative joint diseases; metabolic factors, such as obesity and diabetes; and mechanical factors, such as joint instability and misalignment, have been implicated in the etiology of cartilage defects [3]. Cartilage is an avascular tissue composed of chondrocytes and extracellular matrix (ECM); it possesses limited repair capacities. Current solutions for cartilage irregularities include nonoperative treatment, which focuses primarily on pain relief and traditional operative treatment and the utilization of allografts and autografts, which predominately focuses on cartilage resurfacing [4,5].
Despite moderate success, limitations clearly exist. The shortage of autologous chondrocytes is one of the major hurdles. Fortunately, stem cells, especially mesenchymal stem cells (MSCs), have become a promising alternative source in the tissue engineering field and have been applied in autologous transplantation and cartilage regeneration [6]. Tissue-specific MSCs can be obtained from various sources based on criteria of availability, as for adipose tissue, or of proximity to cartilage and the joint environment in vivo, as for bone marrow and synovial tissues [7]. The induction of chondrogenesis in MSCs and the production of a stable cartilaginous tissue is another hurdle. Although pivotal signaling pathways and mechanisms involved in chondrogenesis have been continuously defined, important issues surrounding the primary steps in chondrogenic commitment and differentiation remain to be elucidated.
Epigenetics is the study of changes in gene expression or cellular phenotype caused by mechanisms such as methylation and histone modification, while excluding changes that may occur in the underlying DNA sequence. It results in heritable and reversible changes of gene expression. Both epigenetic mechanisms, such as methylation and histone modification, appear to be important factors for tissue- and cell-specific differentiation, specifically chondrogenic differentiation [8–10]. Also, epigenetic mechanisms arise in mature humans and mice, either by random change or under the influence of the environment [11]. In other words, epigenetic mechanisms allow an organism to respond to environmental stimuli through changes in gene expression. Epigenetic mechanisms during cartilage development and onset of joint diseases have potential value in the treatment of degenerative joint diseases and have been recently reviewed [12–14]. To explore the precise mechanism of action, a more operational definition of epigenetics was proposed to promote the understanding of epigenetic regulatory mechanisms. By categorizing epigenetic events into epigenators, initiators, and maintainers, the full aspects of epigenetic control of genomic function are delineated [15].
In this review, we first summarize environmental factors that initiate epigenetic influences and non-coding RNA (ncRNA) changes during cartilage regeneration. DNA methylations, histone modifications, and nucleosome dynamics are reviewed according to their contribution in the regulation of proliferation, chondrogenic differentiation, and hypertrophy. Epigenetic rejuvenation using decellularized stem cell matrix (DSCM) has been proposed. The increasing knowledge and new discoveries of epigenetic mechanisms regarding the onset and development of osteoarthritis (OA) and rheumatoid arthritis (RA) provide targets for therapeutic applications to combating the deleterious pathologies of cartilage diseases.
Environmental Factor: Initiated Epigenetic Modifications on Cartilage Regeneration
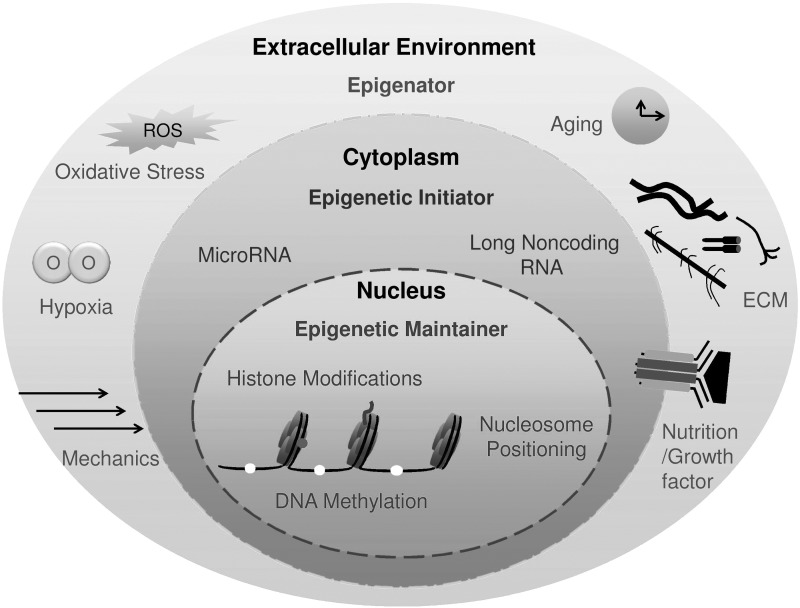
According to Berger et al., an epigenator is a signal that emanates in the cellular environment and initiates an intracellular pathway that is most likely to trigger the expression of an epigenetic phenotype [15]. Environmental factors, such as aging and diet, can modify epigenetic states and contribute to the development of abnormal phenotypes and diseases [16–18]. Similarly, exercise, nutrition, and a variety of other environmental factors can accelerate or delay the process of cartilage development and regeneration following injury [19]. Thus, the connections of environmental cues to chromatin and associated signaling factors that are involved in early epigenetic regulation of cartilage regeneration need to be determined. Based on the new definition of epigenetics, we propose several potentially related environmental factors (listed in the following headings in an alphabetical order) that may serve as epigenators in cartilage regeneration (Fig. 1).
FIG. 1.
The landscape view of epigenetic events during cartilage regeneration.
Aging
Certain epigenetic factors are thought to play a role in mediating aging. Changes in DNA methylation and histone modifications were investigated as markers of aging. Four hypermethylated CpG sites (associated with genes NPTX2, TRIM58, GRIA2, and KCNQ1DN) have been identified and an additional hypomethylated CpG site (BIRC4BP) was found to be an epigenetic-aging-signature across different tissues [20]. Similarly, hypermethylated promoters of protease genes [matrix metalloproteinase 3 (MMP3), MMP9, MMP13, and aggrecanase-1 (ADAMTS4)] expressed by superficial zone chondrocytes were found in aged cartilage compared with the young control human cartilage [21]. Specifically, the methylation of osteogenic protein-1 (OP-1), detected in chondrocytes from older adults, was associated positively with age [22]. Sirtuin (SirT) proteins are a family of nicotinamide-adenine-dinucleotide-dependent protein deacetylases. These proteins can extend the life span in lower organisms and are important in mediating diseases of aging [23]. Among the SirT proteins, SirT1 has been recognized as a critical regulator of stress responses, replicative senescence, inflammation, metabolism, and aging in chondrocytes [24–27]. Knockout of SIRT1 in mice resulted in an altered cartilage phenotype, with an elevated apoptotic process and a potential degradative cartilage process [24]. Both activation (via resveratrol) and overexpression (via gene transfection) of SIRT1 exhibited similar anti-inflammatory effects by inhibiting nuclear factor kappa B (NF-κB) in human chondrocytes [25]. The activation of SIRT1 by resveratrol decreased apoptosis in human chondrocytes, possibly through regulating mitochondria-related apoptotic signals [26]; the overexpression of SIRT1 rescued apoptosis through downregulating protein tyrosine phosphatase 1B (PTP1B) [27]. Cellular senescence is mainly caused by aging or accumulation of oxidative stress and presents a considerable challenge in cartilage engineering and regeneration [28]. Thorough understanding of epigenetic mechanisms implicated in aging could promote the progress of cartilage engineering and regenerative medicine.
Growth factors or nutrition supplements
Transforming growth factor-β (TGF-β) is one of the cytokines involved in chondrogenesis. It is a potent cytokine that enhances cell signaling through the downstream small mother against decapentaplegic (SMAD) family of proteins. In cancer research, TGF-β has been associated with alterations in the epigenetic profile of cells [29]. Bone morphogenetic protein-2 (BMP-2), an osteochondrogenic factor, also induces histone hyperacetylation at the SRY (sex determining region Y)-box 9 (SOX9) gene on chromatin [30].
To study the effects of malnutrition, food-restricted rat models were used. Diet intervention was reported to significantly affect the DNA methylation levels of several CpGs on the WT1 and ATP10A gene promoters in patients with obesity [31]. Vitamin C (ascorbate) induces DNA demethylation of specific gene sets, which may have an impact on pluripotency and reprogramming pathways in human embryonic stem cells (ESCs) [32]. Likewise, vitamin C supplementation also enhances the function of epigenetic modifiers during generation of induced pluripotent stem cells (iPSCs) [33]. These studies suggest that the manipulation of growth factors and nutrition could alter epigenetic states and the chondrogenic process.
Hypoxia
In normal cartilage, resident chondrocytes are adapted to the hypoxic environment. Thoms et al. found that hypoxia suppressed the destruction and induced production of human cartilage through the regulation of hypoxia-inducible factor-1α (HIF-1α) and HIF-2α and their interaction with SOX9 [34]. HIF-2α has been reported to mediate the transcriptional activity of the catabolic gene MMP13 under different methylation statuses in chondrocytes [35]. HIFs can also alternatively interact with histone deacetylase (HDAC) inhibitors and modulate their activities, resulting in the determination of stem cell fate [36]. Likewise, histone demethylase enzymes have been shown to function in the presence of oxygen. In some cases, these enzymes are induced by hypoxia in an HIF-α-dependent manner [37]. The just-discussed findings suggest a hitherto uncharacterized role for epigenetic mechanisms mediating the effect of oxygen in cartilage development.
Inflammation
Stress and proinflammatory mechanisms participate in the pathogenesis of OA. Jmjd3 is a histone demethylase expressed in macrophages in response to bacterial products and inflammatory cytokines. Jmjd3 binds polycomb-group (PcG) protein target genes and regulates their trimethylation levels of lysine 27 on histone 3 (H3K27me3) as well as their transcriptional activity [38]. This study provided the first link between inflammation and epigenetics. Inflammation-activated signaling [tumor necrosis factor (TNF)/p38α] in muscle stem (satellite) cells can promote the polycomb repressive complex 2 (PRC2), one of the two classes of PcG proteins, suppressing Pax7 and impairing satellite cell proliferation [39]. Though these pathways provide significant evidence of inflammation-induced epigenetic regulatory mechanisms, direct evidence of epigenetic changes in chondrogenesis due to inflammation remains to be identified.
Mechanics
The mechanical environment plays a definitive role in regulating chondrogenesis of MSCs [40]. The cytoskeleton, which provides structural integrity to the cell, senses mechanical interactions with the external environment allowing the cytoskeleton to be involved mechanically and biochemically with cellular processes [41]. It is shown that biophysical cues can modulate nuclear shape through lamin A/C, the nuclear matrix protein, and regulate HDAC activity and histone acetylation in bone-marrow-derived mesenchymal stem cells (BMSCs) [42]. Mechanical stimulation can also alter the epigenetic state by reducing DNA methylation, therefore resulting in the upregulation of osteogenic gene expression [43]. In the inner meniscus cells, cyclic tensile strain can increase COL2A1 and SOX9 expression, stimulate nuclear translocation, and cause the phosphorylation of SOX9 to increase the association between SOX9 and related transcriptional complexes on chromatin [44].
Oxidative stress
Free-radical-derived reactive oxygen species (ROS), constantly produced in living organisms, have the potential to damage DNA, proteins, and lipids, while contributing to the aging process. Recently, striking evidence revealed the relationship between ROS-induced reversible cell signaling and epigenetic changes. Forced expression of SIRT3, a deacetylase that activates ROS scavengers to reduce oxidative stress, functionally rejuvenates mouse hematopoietic stem cells, supporting the assertion that reversible processes such as aberrant signaling and epigenetic drift are relevant to oxidative-stress-associated cellular aging [45]. Oxidative stress also plays an important epigenetic modification role in the progression of chronic diabetic complications and cardiovascular diseases. Increased superoxide production causes the activation of several signaling pathways, resulting in epigenetic changes, including augmented production of histone acetyltransferase p300, alterations of HDACs, and modifications in microRNAs (miRNAs) [46,47]. The just-mentioned evidence suggests that epigenetic changes induced by oxidative stress might interfere with cartilage regeneration, which also remains to be elucidated.
Initiation of Epigenetic Events During Cartilage Regeneration
After receiving signals generated by the environment, the epigenetic initiator translates the epigenator signal to mediate in the establishment of local chromatin at a precise location. The initiator can be a DNA-binding protein, an ncRNA, or any other entity that defines the coordinates of the chromatin structure assembly [15]. NcRNAs, including miRNAs and long ncRNAs (lncRNAs), have been accepted as a new major gene class with epigenetic regulating functions [48]. Increasing evidences suggest that the miRNAs and lncRNAs play an integral part in regulating chondrogenic differentiation and cartilage function through their targeted genes [49,50]. However, their role as epigenetic initiators has not been fully described. Table 1 provides a brief summary of recognized and recently discovered DNA-binding proteins, miRNAs, and lncRNAs sorted by their possible upstream environmental factors and their role in initiation of epigenetic events during chondrogenesis.
Table 1.
Summary of Epigenetic Remodeling Events in Cartilage Regeneration
| Environmental factors | Initiators | Target genes | Effect | Cell type | Refs. |
|---|---|---|---|---|---|
| Environmental epigenators and epigenetic initiators | |||||
| Aging/replicative senescence | ↑ miR-766↑miR-558↓miR-let-7f↓miR-125b↓miR-222↓miR-199-3p↓miR-23a↓miR-221 | Unknown, but not global, histone modification profiles | ↓ Proliferation | Rhesus macaque BMSCs | [53] |
| ↓ Differentiation | |||||
| ↓ miR-17↓miR-19b↓miR-20a↓miR-106a | ↑ p21/CDKN1A | ↑ Aging | Seven aging modelsa | [54] | |
| ↑ miR-371↑miR-369-5p↑miR-29c↑miR-499↑miR-let7f | Senescent-related genes located on chromosome 4q21 | ↓ Proliferation | Human BMSCs | [55] | |
| ↓ Adipogenesis | |||||
| ↑ Osteogenesis | |||||
| ↑ miR-199a-3p↑miR-193b↓miR-320c | ↓ COL2A1↓SOX9↓ACAN↑ADAMTS5 | ↑ Chondrocyte aging | Human chondrocytes | [59] | |
| Growth factors/nutrition | |||||
| TGF-β1 | ↓ miR-221↓miR-222↑miR-143↑miR-145 | Unknown | ↓ Chondrocyte dedifferentiation and redifferentiation | Bovine chondrocytes | [49] |
| TGF-β | ↑ miR-455-3p | ↓ ACVR2B↓SMAD2↓CHRDL1 | ↓ TGF-β signaling | Human OA cartilage | [62] |
| ↓ Cartilage destruction | |||||
| ↓ miR-140 | ↑ SMAD3 | ↑ TGF-β signaling | C3HT101/2 cells | [63] | |
| Food restriction | ↓ miR-140 | ↑ SIRT1 | ↑ Growth attenuation | Rat epiphyseal growth plate | [64] |
| High glucose | ↑ miR-486-5p | ↓ SIRT1 | ↑ Senescence↓Proliferation | Human ASCs | [57] |
| ↓ Osteogenesis↓Adipogenesis | |||||
| Hypoxia | |||||
| +FGF-2 | ↑ miR-302 | ↑ OCT4↑NANOG | ↑ Doubling rates | Human primary and immortalized BMSCs | [66] |
| ↓ Senescence | |||||
| HIF-1α | ↑ miR-210 | ↑ HIF-1α | ↑ Cell survival | Human BMSCs | [67] |
| ↑ miR-210 | ↓ FLASH/CASP8AP2 | ↓ Apoptosis | Rat BMSCs | [69] | |
| Inflammation | |||||
| IL-17 | ↑ miR-146a/b↑let-7a↑miR-26↑miR-150↑miR-155 | 17 Validated genesb | ↑ RA pathogenesis | PBMC and RA synovium | [72] |
| IL-1β | ↑ miR-101 | ↓ SOX9 | ↑ ECM degradation | Rat chondrocytes | [73] |
| ↓ miR-140 | Unknown | ↑ ADAMTS5↓ACAN | Human chondrocytes | [74] | |
| ↑ miR-140 | ↓ MMP13 | ↓ ECM degradation | Human cartilage cell line C28/I2 | [75] | |
| ↓ miR-125b | ↑ ADAMTS4 | ↓ Aggrecan degradation | Human OA chondrocytes | [76] | |
| ↓ miR-199aa | ↑ COX-2 | ↑ mPGES1 | Human chondrocytes | [77] | |
| ↑ Prostaglandin E(2) production | |||||
| ↑ miR-34 | ↓ EPHA5 | ↓ COL2A1 | Rat/human chondrocytes | [78,79] | |
| ↑ iNOS↑Apoptosis | |||||
| IL-1β+TNF-α | ↓ H19-miR-675 | ↓ COL2A1 | ↓ Chondrocytes metabolic | Human chondrocytes | [75] |
| Mechanical | |||||
| Cyclic loading | ↑ miR-365 | ↓ HDAC4 | ↑ Proliferation | Primary chicken chondrocytes | [81] |
| ↑ Chondrogenic differentiation | |||||
| Oxidative stress | |||||
| Hydrogen peroxide | ↑ miR-1 | ↓ IGF-1 | ↓ Apoptosis | Human foreskin fibroblast–induced iPSCs | [87] |
| ↓ Mitochondrial dysfunction | |||||
| ↓ Cytochrome-c release | |||||
| ROS | ↑ miR-210 | ↓ ISCU2↓PTPN2 | ↑ Proliferation↑Migration | Human ASCs | [88] |
| Maintenance of epigenetic events during cartilage regeneration | |||||
| DNA methylation | + 5AC | ↑ PCNA | ↑ Chondrocyte proliferation | Fetal rat epiglottis | [94] |
| + 5-aza-dC | Unknown | ↑ Osteogenesis↑Chondrogenesis | Human BMSCs | [102] | |
| Histone modifications | |||||
| Histone methylation | ↑ Twist-1 | ↑ EZH2 | ↑ H3K27me3↓p16/p14↓Senescence | Human BMSCs | [99] |
| ↑ H3K4me2 | ↑ NFAT | ↑ Chondrocyte homeostasis | Mice chondrocytes | [106] | |
| ↑ H3K9MTases | Unknown | ↑ Hypertrophic differentiation | Mouse growth plate | [110] | |
| Histone acetylation | + TSA | Unknown | ↑ Cartilage matrix formation | Human BMSCs | [102] |
| + TSA | ↑ SOX9 | ↑ Cartilage matrix genes | Human chondrocytes | [108] | |
| + Largazole or TSA | H3K9 or H3K14 | ↑ Proliferation↓Differentiation | Human UC-MSCs | [97] | |
| HDAC4 | Unknown | ↑ Chondrogenesis↓Hypertrophy | Porcine SDSCs | [112] | |
| Relocation of HDAC4 by CaMKIV | ↑ RUNX2 | ↑ Hypertrophy | Chicken chondrocytes | [113] | |
| ↑ COL10A1 | |||||
Seven aging models include endothelial cells, replicated CD8(+) T cells, renal proximal tubular epithelial cells, skin fibroblasts, foreskin, MSCs, and CD8(+) T cells from old and young donors.
Seventeen validated genes include AID, c-Myb, Ezh2, FADD, GSK-3β, Hbl-1, IKK, IRAK1, IRAK2, Lin-41, PU.1, RAS, Ripk1, SOCS1, TAB2, TRAF6, and TRIM71.
ACVR2B, activin receptor type-2B; ADAMTS4, aggrecanase-1; ADAMTS5, a disintegrin and metalloproteinase with thrombospondin motifs 5; AID, activation-induced cytidine deaminase; ARPE-19, a human retinal pigment epithelial cell line; ASCs, adipose-derived mesenchymal stem cells; BMSCs, bone-marrow-derived mesenchymal stem cells; CaMKIV, Ca2+/calmodulin-dependent kinase IV; CDKN1A, cyclin-dependent kinase inhibitor 1A; CHRDL1, chordin-like 1; COX-2, cyclooxygenase-2; DNMT, DNA methyltransferases; EPHA5, Ephrin type A receptor 5; EZH2, enhancer of zeste homolog 2 (Drosophila); FADD, Fas-associated death domain protein; FLASH/Casp8ap2, FLICE-associated huge protein/caspase-8-associated protein 2; GSK-3β, glycogen synthase kinase-3β; H3K4me2, histone 3 lysine 4 dimethylation; H3K9MTases, histone 3 lysine 9 methyltransferases; HDAC, histone deacetylase; HIF, hypoxia-inducible factor; IGF-1, insulin-like growth factor-1; IKK, IκB kinase; IL, interleukin; iNOS, inducible nitric oxide synthase; IRAK1, interleukin-1 receptor-associated kinase 1; IRAK2, interleukin-1 receptor-associated kinase 2; ISCU, iron–sulfur cluster scaffold homolog 2; MMP13, matrix metalloproteinase 13; mPGES, microsomal prostaglandin E synthase-1; NFAT, nuclear factor of activated T-cells; OA, osteoarthritis; OCT4, octamer-binding transcription factor 4; PBMCs, peripheral blood mononuclear cells; PCNA, proliferation cell nuclear antigen; PTPN2, protein tyrosine phosphatase, non-receptor type 2; PU.1, spleen focus forming virus (SFFV) proviral integration oncogene; RA, rheumatoid arthritis; Ripk1, receptor interacting serine-threonine kinase 1; ROS, reactive oxygen species; SDSCs, synovium-derived stem cells; SIRT1, sirtuin 1; SOCS1, suppressor of cytokine signaling 1; TAB2, TGF-β-activated kinase 1/MAP3K7 binding protein 2; TNF-α, tumor necrosis factor α; TSA, Trichostatin A; TRAF6, tumor necrosis factor receptor-associated factor 6; TRIM71, tripartite motif-containing 71; Twist-1, basic helix-loop-helix transcription factor 1; UC-MSCs, umbilical-cord-derived mesenchymal stem cells; VSMCs, vascular smooth muscle cells; ECM, extracellular matrix; TGF-β, transforming growth factor-β; SMAD, small mother against decapentaplegic; SOX9, SRY (sex determining region Y)-box 9; iPSC, induced pluripotent stem cell; 5AC, 5-azacytidine; FGF, fibroblast growth factor.
Senescence responsive initiators
Replicative senescence, which has a detrimental impact on MSCs, is associated with continuous epigenetic modifications [51,52]. Senescence influences the overall expression of coding genes and miRNAs in MSCs [53,54]. The upregulation of miR-371, miR-369-5p, miR-29c, miR-499, and miR-let7f has been implicated in replicative senescence of human BMSCs [55]. In a similar but inconsistent fashion, an upregulation of miR-766 and miR-558 and a downregulation of miR-let-7f, miR-125b, miR-222, miR-199-3p, miR-23a, and miR-221 were found in older monkey BMSCs [53]. DNA methyltransferases (DNMTs) were specifically predicted to be the direct targets of miR-29c, miR-499, and miR-371 in lung cancer cells [56]. SirT1 is also a target of miR-486-5p that can manipulate the senescence of human adipose-derived mesenchymal stem cells (ASCs) [57]. Cyclin-dependent kinase (CDK) inhibitor p21 was established to be the target gene due to its increases correlated with decreases in miR-17, miR-19b, miR-20a, and miR-106a in different aging models [54]. It was also found in human diploid fibroblasts that miR-519 triggered replicative senescence by repressing an RNA-binding protein known as HuR [58]. Finally, miR-199a-3p, miR-193b, and miR-320c were specifically identified to correlate to the senescence of chondrocytes [59]. The changes in miRNAs may lead to interactions with methylation and histone modifications of target genes, consequently influencing human MSC self-renewal and differentiation [60].
Growth-factor- and nutrition-related miRNAs
TGF-β can regulate miRNA directly and through SMADs [61]. ACVR2B, SMAD2, and CHRDL1 were direct targets of miR-455-3p, which can also regulate TGF-β signaling during chondrogenesis [62]. Smad3 was identified as a direct target of miR-140; TGF-β signaling can be inhibited by miR-140 through suppression of Smad3 [63]. The set of miRNAs, including miR-221, miR-222, miR-140, miR-143, and miR-145, was enriched in the artificial zone of articular cartilage and exhibited expression changes with zonal differentiation. Interestingly, these areas were also involved in the regulation of homeostasis and could be modulated by TGF-β1 [49]. Several miRNAs, including chondrocyte-specific miR-140 and its target gene SIRT1, were reduced in a food-restriction model [64]. A high-glucose diet increased the expression of miR-486-5p and inhibited SIRT1 expression, which further promoted human ASC senescence [57].
Hypoxamirs
A specific set of miRNAs that has been induced in hypoxic conditions is defined as “hypoxamirs” [65]. An ESC-specific cluster, miR-302, was induced upon hypoxic culture and may explain the improved reprogramming of primary and immortalized MSCs [66]. A highly conserved and ubiquitously expressed miRNA, miR-210, was also induced in hypoxic human MSCs; miR-210 positively regulated HIF-1α to maintain the survival of MSCs under hypoxic conditions [67]. Reciprocally, HIF-1α induced miR-210 in differentiating myoblasts; blockage of miR-210 greatly increased myotube sensitivity to oxidative stress and mitochondrial dysfunction [68]. In an ischemic model, miR-210 also suppressed apoptosis in BMSCs through caspase-8-associated protein 2 [69]. Under hypoxic signaling, the culture of human chondrocytes showed that lncRNA H19 and its encoded miR-675 as well as COL2A1 were upregulated in close relation [70].
Inflammation-regulated initiators
Proinflammatory cytokines were reported to be responsible for the upregulation and downregulation of a number of miRNAs. Interleukin (IL)-1α elevated expression of miR-146a/b as a compensatory response of senescence to reduce inflammatory signals in human neonatal foreskin fibroblasts [71]. However, increases in miR-146a were later found to be associated with the production of proinflammatory cytokine IL-17 in the synovium of RA patients [72]. Upregulated miR-101 participated in IL-1β-induced ECM degradation in chondrocytes, likely doing so by directly targeting SOX9 [73]. In vitro treatment of chondrocytes with IL-1β suppressed the expression of miR-140; the silencing of miR-140 downregulated IL-1β, induced a disintegrin and metalloproteinase with thrombospondin motifs 5 (ADAMTS5) expression, and rescued the IL-1β-dependent repression of ACAN expression [74]. Similarly, the increase of miR-140 expression was also a negative feedback regulator to decrease MMP13 expression in IL-1β-stimulated human articular chondrocyte C28/I2 cells [75].
Both IL-1β and TNF-α downregulated H19, COL2A1, and miR-675 expression [70]. The stress-induced regulation of H19 by hypoxic signaling and inflammation suggested that H19 acted as a metabolic correlate in cartilage and cultured chondrocytes; there may be an indirect influence of miR-675 on COL2A1 levels in OA-affected cartilage [70]. Overexpression of miR-125b suppressed IL-1β-induced ADAMTS4 expression [76]; downregulation of miR-199a* was involved in the IL-1β induction of cyclooxygenase-2 (COX-2) in human osteoarthritic chondrocytes [77]. In another in vitro OA model, miR-34 was upregulated in chondrocytes by the induction of IL-1β. Silencing of miR-34 significantly prevented IL-1β-induced downregulation of COL2A1 and upregulation of inducible nitric oxide synthase (iNOS), and reduced chondrocyte apoptosis [78] by the targeting of EPHA5 [79].
Mechanoresponsive miRNAs
By profiling miRNAs in bovine cartilage, miR-222 was recognized as a potential regulator for mechnotransduction pathways [80]. Recently, miR-365 was identified as a mechanically responsive miRNA that regulated chondrocyte differentiation under cyclic loading via directly targeting HDAC4 [81]. Both mechanical loading and TGF-β treatment modulated the expression of several miRNAs that regulate tendon fibroblast proliferation and ECM synthesis [82]. Mechanical stimulation altered the epigenetic state of BMSCs by reducing DNA methylation, which increased the expression of osteogenic genes [43]. A more complete understanding of the translation of extracellular biophysical signals into biochemical signaling events will significantly improve the understanding of epigenetic regulation in MSCs.
Oxidative-stress-induced initiators
Some miRNAs induced by oxidative stress were involved in the regulation of SIRT1 in the endothelium [83]. In a human retinal pigment epithelial cell line, hydrogen peroxide downregulated 18 miRNAs and upregulated 29 miRNAs; the addition of curcumin (diferuloylmethane) altered the expression of hydrogen-peroxide-modulated miRNAs, increased the expression of antioxidant genes, and reduced activity of the renin-angiotensin systems; these effects suggest many possible regulatory roles of miRNAs [84]. Specifically, hydrogen-peroxide-treated vascular smooth muscle cells aberrantly expressed miR-21 [85]. In arsenite-induced human embryo lung fibroblast transformations, miR-21 was regulated by ROS-activated extracellular signal-regulated kinase (ERK)/NF-κB pathways [86]. In iPSCs, hydrogen peroxide also induced oxidative stress and miR-1 expression. Insulin-like growth factor-1 (IGF-1) provided protection from oxidative-stress-induced apoptosis in iPSCs partially via miR-1 [87]. ROS from various sources induced miR-210 expression in human ASCs via platelet-derived growth factor receptor beta (PDGFR-β), Akt, and ERK pathways, and miR-210 increased the proliferation and migration of ASCs by downregulating ISCU2 and PTPN2 [88]. Although most research on oxidative-stress-responsive miRNAs is conducted within the fields of vascular disorders and neurontoxicology, an understanding of this topic will provide useful insight in cartilage regeneration.
Maintenance of Epigenetic Remodeling in Cartilage Regeneration
Following epigenetic initiation mechanisms, maintaining and passing on the heritable epigenetic effects to the next generation is completed via DNA methylation, histone modifications, and nucleosome dynamics. To dissect the maintenance of epigenetic regulation mechanisms in regenerating cartilage, epigenetic evidence on both chondrocyte/progenitor cell proliferation and chondrogenic differentiation are summarized in the following headings.
Epigenetic regulation of proliferation in chondrocytes and progenitor cells
Nanog, Sox2, and Oct4 (also known as Pou5f1) are three transcription factors essential for the maintenance of stem cell pluripotency. The expression of NANOG and SOX2 is inversely correlated with DNA methylation pattern in the promoter region of equine BMSCs [89]. During long-term culture, a significant decrease in the DNA methylation levels could be responsible for the enlarged morphology, the decreased number of cell divisions, the random loss of genomic regions, and the shortening of the telomeres in human BMSCs [90]. Additionally, there is increasing evidence that loss of ex vivo differentiation potentials, especially toward the osteochondrogenic lineage, was in late-passage porcine BMSCs [91], and reduced expression of SOX2 and OCT4 was in human-placenta-derived MSCs [92]. A recent report indicated that overexpression of both NANOG and OCT4 could improve stemness and chondrogenesis in human BMSCs [93]. Intriguingly, an in vivo study showed that, after DNA methylation inhibitor 5-azacytidine (5AC) was injected intraperitoneally, it enhanced the proliferation of elastic cartilage in transplanted rat fetal epiglottis tissue [94].
The important role of histone acetylation in cell replication has long been recognized [95]. As human-placenta-derived MSCs age, alterations of gene expression were observed with epigenetic dysregulation of acetylation in H3K9 and H3K14 [92]. Also, HDAC1 was essential for unlimited cell proliferation in mouse ESCs by repressing the expression of selective cell cycle inhibitors [96]. HDAC inhibitors can significantly improve human umbilical cord MSC proliferation. The inhibitors can also delay their aging at low concentrations by modulating histone H3 acetylation and methylation in pluripotent and proliferative genes [97].
In iPSCs, histone methyltransferase Ezh2 (a catalytic subunit of PRC2) influenced the expression of NANOG, a transcription factor critically involved with self-renewal of undifferentiated ESCs [98]. A basic helix-loop-helix transcription factor, Twist-1, can also induce Ezh2. This induction resulted in an increase in H3K27me3, repression of the transcription of p16/p14, and the senescence of human BMSCs [99]. DNMTs regulated the cellular senescence of human umbilical cord MSCs through the histone marks at genomic regions of Ezh2-targeting miRNAs and p16 and p21 promoter regions [100].
Epigenetic regulation during chondrogenic differentiation
DNA methylation and cartilage regeneration
Compared with ESCs, MSCs had more methylations on OCT4 and NANOG promoters. The increased methylation suggested that pluripotency was restricted in MSCs [10]. Cell-type-specific DNA methylation patterns are thought to be established prior to the terminal differentiation of adult progenitor cells. The removal of DNA methylation by 5AC treatment altered the myogenic lineage commitment of C2C12 myoblasts and induced spontaneous osteogenic and adipogenic differentiation [101]. Similarly, another DNA demethylating agent, 5-aza-2′-deoxycytidine (Decitabine), also stimulated osteogenic differentiation in human BMSCs [102].
The SOX trio (SOX5, SOX6, and SOX9) plays an essential role in chondrogenic differentiation. During chondrogenic induction of human synovium-derived stem cells (SDSCs), DNA methylation levels of CpG-rich promoters related to chondrocyte phenotypes were largely hypomethylated [103]. Correspondingly, increased methylation in promoter regions of SOX5 and SOX9 genes explains the low expression of these respective genes in a surgically induced rat OA model [104].
Histone modifications and cartilage regeneration
Recently, genome-wide chromatin immunoprecipitation and deep sequencing were performed to quantify epigenetic changes during in vitro chondrogenesis in human primary MSCs. Results suggested that histone modifications, rather than DNA methylation, provide the primary mechanism of control of early chondrogenic differentiation of MSCs [8]. Histone lysine methylation is an epigenetic event that establishes cell-specific lineage differentiation; specifically, the methylation of H3K9 has been shown to be the primary determinant for reprogramming somatic cells into iPSCs [105]. Nuclear factor of activated T-cells (NFAT) is an important transcription factor that regulates chondrocyte homeostasis. The age-dependent NFAT-1 expression in articular chondrocytes was also regulated by dynamic histone methylation [106]. Polycomb-associated H3K27me3 blocks chromatin access to early growth response-1 (EGR-1). The ablation of EGR-1 results in abnormal EZH2 (H3K27me3 histone methyltransferase) and BMI1 (E3-ubiquitin ligase for H2A) expression. This relationship suggests that there is an important role for EGR-1 in early chondrogenic epigenetic programming to accommodate early gene–environment interactions in chondrogenesis [107].
A Sox9-related transcriptional apparatus activates its target expression by histone acetylation and p300 mediation [108], suggesting that the epigenetic status, including histone modification and chromatin structure, directly influences Sox9-regulated chondrogenic differentiation. Trichostatin A (TSA), selectively inhibiting the classes I and II mammalian HDAC families of enzymes, enhanced the chondrogenic structure and Sox9-regulated cartilage matrix gene expression of COL2A1 and ACAN in human chondrocytes [102,108].
Epigenetic regulation in chondrocyte hypertrophy
Cartilage hypertrophy is of paramount concern for the application of MSCs in tissue engineering, primarily due to the fact that hypertrophy results in apoptosis and ossification. To generate a stable chondrocyte phenotype, epigenetically controlling chondrocyte hypertrophy is desired. Type X collagen, MMPs, and vascular endothelial growth factor are the most commonly recognized hypertrophic markers and target genes of RUNX2. The demethylation of the COL10A1 promoter correlated with the induction of type X collagen during MSC-derived chondrogenesis [109]. H3K9 methyltransferases were predominantly expressed in prehypertrophic and hypertrophic chondrocytes. The presence of these methyltransferases may represent the progression of chondrocyte differentiation by affecting the methylation state of H3K9 in the mouse growth plate, suggesting a regulatory role of gene expression [110]. This supposition was consistent with Vega et al.'s finding that demonstrated that mice lacking HDAC4 developed abnormal chondrocyte hypertrophy and died perinatally [111]; overexpression of HDAC4 promoted TGF-β1-induced SDSC chondrogenesis, but inhibited hypertrophy [112] which could benefit cartilage regeneration. The relocation of HDAC4 from the nucleus to the cytoplasm and activation of RUNX2 and COL10A1 was regulated by Ca2+/calmodulin-dependent kinase IV to control chondrocyte hypertrophic differentiation [113].
Potential roles of nucleosome positioning in cartilage regeneration
Nucleosomes are the basic packaging units of DNA in eukaryotes. The assembly, mobilization, and disassembly of nucleosomes can influence the regulation of gene expression and other biological processes such as replication in eukaryotic DNA [114,115]. Nucleosome positioning refers to the location of the nucleosome in relation to the expressed genomic DNA sequence. Nucleosome positioning is a dynamic process determined by the combined effects of several factors, such as DNA sequence, DNA-binding proteins, nucleosome remodelers, and the RNA polymerase II transcription machinery [116]. Thus, nucleosome positioning could have a potential role in regulating cartilage regeneration.
Human telomerase reverse transcriptase (hTERT), a catalytic subunit of the enzyme telomerase, is responsible for maintaining the length of telomeres. Without this enzyme, critical shortening of telomeres occurred resulting in genetic instability and cellular senescence [117]. Therefore, hTERT is highly expressed in stem cells, while cells that become gradually differentiated lose hTERT activity. One recent study showed that the expression of hTERT mRNA decreased continuously with differentiation due to the increased prevalence of nucleosomes at the hTERT core promoter regions. As remodeling continued to increase nucleosomes at the promoter region, stable silencing of the hTERT promoter also increased; thus, nucleosomal deposition at the core promoter was determined to be the cause of transcriptional repression of hTERT in differentiated human leukemic HL60 cells [118].
Genome-wide nucleosome positioning was investigated in mouse ESCs and their neural progenitor as well as embryonic fibroblasts to determine the association between nucleosome positioning and lineage commitment. By comparing these three cell types, local and global rearrangements of nucleosome occupancy revealed important roles of nucleosome positioning in cell differentiation [119]. Not surprisingly, active genes have broader and more pronounced nucleosome-depleted regions around transcription start and transcription termination sites. The arrangement and occupancy of nucleosomes in these sites correlated with certain histone methylation or acetylation modifications. Similarly, the average distance between two neighboring nucleosomes, known as the average nucleosome repeat length, increased during differentiation by five to seven base pairs. The alteration of nucleosome repeat length may affect the folding properties of the nucleosomal chain resulting in another mechanism by which nucleosomes may influence stem cell differentiation [119].
DSCM: A Potential In Vitro Model of Epigenetic Application for Cartilage Regeneration
The ECM, which is secreted by resident cells in all tissues, forms a unique tissue-specific 3D microenvironment. ECM proteins are key components in the shaping of the stem cell niche and maintaining stem cell homeostasis. Cell–matrix-interaction-induced signaling constitutes a critical determinant of cell behavior, making the ECM a key factor in determining a stem cell niche. Decellularized ECM (DECM), which is derived from tissues, has proven to be beneficial in regenerative medicine [120]. DECM serves as a biological scaffold for selective cell types allowing increased proliferation. The characterization of decellularized cartilage ECM suggested that it is not only a complex 3D structure full of biochemical signals, but also a mechanotransduction device [121]. This attribute makes decellularized cartilage ECM a perfect candidate for cartilage tissue engineering. Encouraging preliminary animal and clinical data have been reported [122–124].
More interestingly, DECM deposited by human MSCs (DSCM) could be used as an in vitro expansion system [125]. DSCM promoted cell attachment, spreading, migration, proliferation, and the maintenance of responses to differentiation signals [126]. As a tissue-specific stem cell for chondrogenesis [127], SDSCs were chosen to deposit stem cell matrix, on which SDSCs were greatly expanded with enhanced chondrogenic potential in our studies. The rejuvenating effect of DSCM has been observed in not only adult stem cells, such as SDSCs [128–130] and BMSCs [131], but also primary cells, such as articular chondrocytes [132,133] and nucleus pulposus cells [134,135]. Most recently, we found that DSCM deposited by fetal SDSCs provided a robust rejuvenating effect in promoting adult SDSC proliferation and chondrogenic capacities [136]. The increased proliferative and chondrogenic potentials may possibly provide large quantities of high-quality cells in an autologous implantation strategy, which has been encouraged by a recent minipig study in which DSCM-expanded SDSCs were injected intraarticularly to treat partial-thickness cartilage defects [137].
The rejuvenation effect of DSCM is supported by its excellent ability in managing environmental factors. DSCM deposited by fetal SDSCs aided in the protection of expanded cells from senescence [138]. DSCM-expanded SDSCs showed robust resistance to hydrogen-peroxide-induced oxidative stress [130]. Our unpublished data suggested that DSCM-expanded SDSCs exhibited an upregulated ability to resist IL-1β-mediated inflammation. DSCM is also rich in growth factors and acts as a reservoir for the needs and demands of the cell [136]. The addition of hypoxia and basic fibroblast growth factor (FGF)-2 in the DSCM expansion system improved expanded SDSC proliferative and chondrogenic potentials [128]. The presence of hypoxia alone also enhanced the rejuvenating effect of the DECM on nucleus pulposus cells [134].
While the mechanisms involved in DSCM rejuvenation remain unclear, Choi et al. showed that the restoration of senescent human diploid fibroblasts by matrix was regulated by epigenetic mechanisms; both Ku and SIRT1 were induced during restoration and were required for senescent cells to return to a youthful phenotype [139]. Our microarray data also showed that both miR-140 and miR-145 were dramatically downregulated during cell expansion and upregulated during chondrogenic differentiation in DSCM-pretreated SDSCs accompanied with enhanced proliferative and chondrogenic potentials, suggesting a pivotal role of miR-140 and miR-145 in DSCM-mediated SDSC rejuvenation mechanisms (unpublished data). In response to environmental stimuli, miR-140 targeted multiple genes to play different roles during chondrogenic differentiation, endochondral bone formation, and OA pathogenesis, as summarized by Hong and Reddi [140]. Interestingly, miR-140 has been reported to target chemokine (CXC motif) ligand 12 (CXCL12) and ADAMTS5 in equine-cord-blood-derived MSCs [141]; its overexpression protected cartilage from antigen-induced arthritis and maintained the cartilage homeostasis in a knockout mouse model [142]. It also stimulated in vitro chondrogenesis by upregulating SOX9 and ACAN in human MSCs [143]. SOX9 has been suggested as a downstream target gene of miR-145 or miR-449a, which directly or indirectly represses SOX9 and cartilage matrix gene expression in human primary chondrocytes, BMSCs, and a murine embryonic mesenchymal cell line C3H10T1/2 [144–146].
Epigenetic Therapeutic Strategies for the Treatment of Arthritis-Related Cartilage Degradation
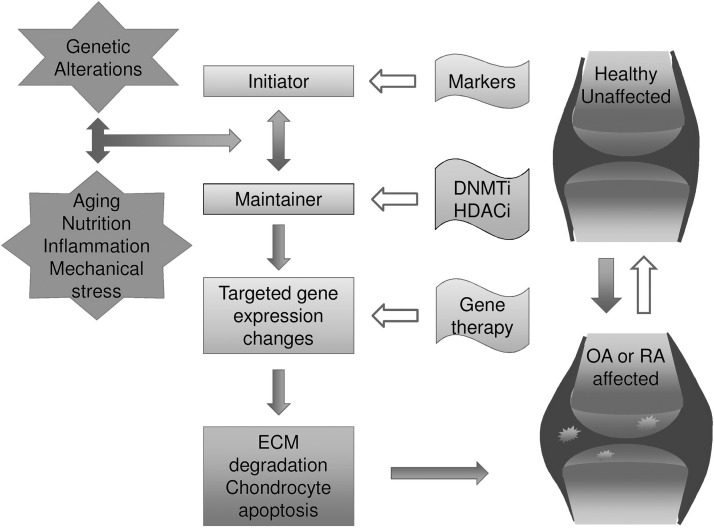
Since heritable epigenetic events are potentially reversible, opportunities for therapeutic intervention arise. Recently, more attention has been paid to epigenetic research of musculoskeletal development and potential treatments for musculoskeletal diseases [147]. OA and RA, both degenerative diseases characterized by cartilage degradation, are not fatal but detrimentally affect the quality of life and cause a huge economic burden. Chondrocytes in healthy states respond to their environment and are responsible for maintaining the balance of synthesis and degradation of the ECM. However, increased degradation of ECM components occurs in arthritic states, resulting in cartilage loss. MMP and aggrecanase are proteolytic enzymes that regulate the turnover and degradation of ECM. A recent study showed that prevention of ECM degradation alleviates cartilage loss in a murine arthritis model [148]. To gain a greater understanding about epigenetic events resulting from environmental factors surrounding cartilage regeneration, recent advances in the epigenetic mechanisms of OA and RA are reviewed as good examples (Fig. 2).
FIG. 2.
Epigenetics in osteoarthritis (OA) and rheumatoid arthritis (RA) and the implications for future therapy.
Osteoarthritis
OA is a complex multifactorial disease with a strong genetic component [149]. However, difficulties remain in the identification of genes that provide full genetic susceptibility to the disease. This is due to the presence of low-penetrance polymorphisms and other mechanisms, such as epigenetic modifications [12]. Recent genome-wide association scans along with several powered candidate gene functional studies have revealed that effects on gene expression, together with epigenetic mechanisms, are likely to be the main mechanisms involved in OA susceptibility [150]. Environmental factors, including aging and nutrition, have been associated with aberrant epigenetic modifications [151,152]. These factors may also have an impact on the onset and progression of OA.
To prevent the onset of OA, an understanding of environmental factors with epigenetic influences is crucial. The recent advancements of many other miRNA functions and molecular targets for treatment in OA have been reviewed [12,153–155]. Among them, miR-140 is tissue specific and is well studied in cartilage tissue and bone development [156]. Though miR-140 was regulated by SOX9, when comparing advanced-stage and early stage OA cartilage, intriguingly, the SOX9 promoter regions showed no difference in epigenetic status [104].
The OA model based on inflammatory stimulation provides more insight into the role of environmental factors with epigenetic influences. After induction by IL-1β or TNF-α, miR-27b and miR-149 became downregulated in human chondrocytes. As a result, their downstream target genes, such as MMP13 and proinflammatory cytokines, were affected, which contributed to the progression of OA [157,158]. TNF-α, a cytokine that mediates joint inflammation in arthritis, can cleave and deactivate SirT1 [159]. SirT proteins were involved in OA through their regulation of cellular energy and metabolism [160]. The decrease in SirT1 in heterozygous SIRT1+/− mice correlated with the development of premature OA-like phenotypes and increased chondrocyte apoptosis [161]. The administration of resveratrol has been reported to protect cartilage from degradation in rabbit OA models [162]. Resveratrol can not only inhibit nitric-oxide-induced apoptosis in rabbit chondrocytes [163] but can also transiently promote proliferation in human BMSCs [164]. Resveratrol exerts its chondroprotective function in human chondrocytes in vitro by deactivating p53-induced apoptosis [165] or inhibiting IL-1β-induced apoptosis [166]. Evidence shows that oxidized hyaluronic acid/resveratrol hydrogel is a promising cell carrier for chondrocytes to repair cartilage defects [167].
Targeting and inhibiting the expression of enzymes, such as MMPs and aggrecanases, is a potential strategy to treat OA. In osteoarthritic dogs, increased MMP2 and MMP9 activities correlated with the onset and progression of OA [168]. The epigenetic regulation of MMP9 gene expression helped prevent and manage its role in degenerative diseases and cancer [169]. Similarly, the demethylation of the MMP13 promoter in OA chondrocytes was found to be responsible for the increase of MMP13 expression [170]. Control of this enzyme may have analogous effects in deterring the progression of OA.
Glucosamine is a commonly prescribed drug for alleviating OA. The mechanism of action behind its therapeutic efficacy is not yet clear. However, it was demonstrated that glucosamine and an NF-κB inhibitor prevent cytokine-induced demethylation of a specific CpG site in the IL-1β promoter resulting in the decreased expression of IL-1β [171]. Many potential treatments for OA will be identified with the integration of genetic and epigenetic data.
Rheumatoid arthritis
RA is a chronic autoimmune inflammatory disease characterized by progressive joint destruction due to the aggressive phenotype of synovial fibroblasts. Its etiopathology was attributed to the crosstalk between genetic predisposition and environmental factors [172]. Changes in both autoimmune-related genes and the environment generated aberrant epigenetic profiles in a cell-specific manner, ultimately resulting in dysregulated gene expression [172].
The expression of a number of miRNAs, such as miR-16, miR-132, miR-146a, and miR-155, has been shown to be dysregulated during the inflammatory response in RA [173,174]. Elevated levels of miR-115 and miR-203 have been observed to correlate with increases of MMP1 and IL-6 expression [175,176]. The upregulated expression of miRNAs may possibly serve as biomarkers, since they are detectable in serum or plasma [173]. The pathogenesis of RA will be more thoroughly understood with more complete identification of miRNAs and a greater awareness of crosstalk between the epigenetic regulators [173,177,178].
IL-1, a proinflammatory cytokine, altered DNA methylation in RA synovial fibroblasts by decreasing the expression and function of DNMTs [179]. The expression of synovial fibroblast genes contributed to the pathogenesis of RA due to the changes in methylated genes [180]. Key hypomethylated and hypermethylated gene loci have been identified by use of genome-wide evaluation of DNA methylation in RA synovial fibroblasts [181,182]. By performing both DNA methylation and miRNA expression screenings of RA synovial fibroblasts compared with OA patients with normal phenotypes, four dysregulated target genes (IL-6R, CAPN8, DPP4, and HOX) were identified as potential clinical markers [183]. The integrative approach of involving genetics and epigenetics provides more insights about the intricate connections between various mechanisms and helps to identify novel targets for future therapeutic options in RA patients.
SIRT1, which is overexpressed in RA tissues, promoted proinflammatory cytokine production in monocytes and RA synovial fibroblasts [184]. In the past 10 years, the systemic administration of HDAC inhibitors has been shown to decrease the severity of the disease. However, the underlying mechanisms in vitro and in vivo are still obscure [172,185]. Continued interest in epigenetic research will advance the discovery of new treatments for RA [174,185].
Concluding Remarks and Future Perspectives
In this review, we emphasized the importance of involving the environmental context in order to better understand the nuances and complexities of epigenetic remodeling events on chondrogenesis. The numerous ways in which epigenetic events may alter gene expression were discussed. Although much has been learned about the variety of roles and mechanisms involved in the epigenetics of chondrogenesis, more information and details still need to be elucidated. Epigenetic events can provide an avenue for more precise and stable control of gene expression and genomic regulation through multiple generations. A deeper understanding of the underlying mechanisms of epigenetic regulation will allow us to actively manipulate cell fate conversion, including senescence, differentiation, reprogramming, and transdifferentiation, which will undoubtedly lead to unlimited therapeutic applications.
Acknowledgments
The authors thank Suzanne Danley and Tyler Pizzute for help in editing the manuscript. This project was partially supported by research grants from the West Virginia University Senate Research Grant Award (R-12-010), the AO Foundation (S-12-19P), and the National Institutes of Health (NIH) (1 R03 AR062763-01A1 and 5 R03 DE021433-02).
Author Disclosure Statement
The authors declare no potential conflict of interest.
References
- 1.Arøen A, Løken S, Heir S, Alvik E, Ekeland A, Granlund OG. and Engebretsen L. (2004). Articular cartilage lesions in 993 consecutive knee arthroscopies. Am J Sports Med 32:211–215 [DOI] [PubMed] [Google Scholar]
- 2.Curl WW, Krome J, Gordon ES, Rushing J, Smith BP. and Poehling GG. (1997). Cartilage injuries: a review of 31,516 knee arthroscopies. Arthroscopy 13:456–460 [DOI] [PubMed] [Google Scholar]
- 3.Mandelbaum BR, Browne JE, Fu F, Micheli L, Mosely JB, Erggelet C, Minas T. and Peterson L. (1998). Articular cartilage lesions of the knee. Am J Sports Med 26:853–861 [DOI] [PubMed] [Google Scholar]
- 4.Alford JW. (2005). Cartilage restoration, part 1: basic science, historical perspective, patient evaluation, and treatment options. Am J Sports Med 33:295–306 [DOI] [PubMed] [Google Scholar]
- 5.Alford JW. (2005). Cartilage restoration, part 2: techniques, outcomes, and future directions. Am J Sports Med 33:443–460 [DOI] [PubMed] [Google Scholar]
- 6.Nejadnik H, Hui JH, Feng Choong EP, Tai BC. and Lee EH. (2010). Autologous bone marrow-derived mesenchymal stem cells versus autologous chondrocyte implantation: an observational cohort study. Am J Sports Med 38:1110–1116 [DOI] [PubMed] [Google Scholar]
- 7.Boeuf S. and Richter W. (2010). Chondrogenesis of mesenchymal stem cells: role of tissue source and inducing factors. Stem Cell Res Ther 1:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herlofsen SR, Bryne JC, Høiby T, Wang L, Issner R, Zhang X, Coyne MJ, Boyle P, Gu H, et al. (2013). Genome-wide map of quantified epigenetic changes during in vitro chondrogenic differentiation of primary human mesenchymal stem cells. BMC Genomics 14:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson KA, Teteak CJ, Zou J, Hacquebord J, Ghatan A, Zielinska-Kwiatkowska A, Fernandes RJ, Chansky HA. and Yang L. (2013). Mesenchymal-specific knockout of ESET histone methyltransferase causes ectopic hypertrophy and terminal differentiation of articular chondrocytes. J Biol Chem 288:32119–32125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yannarelli G, Pacienza N, Cuniberti L, Medin J, Davies J. and Keating A. (2013). Brief report: The potential role of epigenetics on multipotent cell differentiation capacity of mesenchymal stromal cells. Stem Cells 31:215–220 [DOI] [PubMed] [Google Scholar]
- 11.Issa JP. (2000). CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol 249:101–118 [DOI] [PubMed] [Google Scholar]
- 12.Barter MJ, Bui C. and Young DA. (2012). Epigenetic mechanisms in cartilage and osteoarthritis: DNA methylation, histone modifications and microRNAs. Osteoarthritis Cartilage 20:339–349 [DOI] [PubMed] [Google Scholar]
- 13.Reynard LN. and Loughlin J. (2012). Genetics and epigenetics of osteoarthritis. Maturitas 71:200–204 [DOI] [PubMed] [Google Scholar]
- 14.Viatte S, Plant D. and Raychaudhuri S. (2013). Genetics and epigenetics of rheumatoid arthritis. Nat Rev Rheumatol 9:141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger SL, Kouzarides T, Shiekhattar R. and Shilatifard A. (2009). An operational definition of epigenetics. Genes Dev 23:781–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaenisch R. and Bird A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33Suppl:245–254 [DOI] [PubMed] [Google Scholar]
- 17.Feinberg AP. (2007). Phenotypic plasticity and the epigenetics of human disease. Nature 447:433–440 [DOI] [PubMed] [Google Scholar]
- 18.Barros SP. and Offenbacher S. (2009). Epigenetics: connecting environment and genotype to phenotype and disease. J Dent Res 88:400–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ntanasis-Stathopoulos J, Tzanninis JG, Philippou A. and Koutsilieris M. (2013). Epigenetic regulation on gene expression induced by physical exercise. J Musculoskelet Neuronal Interact 13:133–146 [PubMed] [Google Scholar]
- 20.Koch CM. and Wagner W. (2011). Epigenetic-aging-signature to determine age in different tissues. Aging (Albany NY) 3:1018–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Da Silva MA, Yamada N, Clarke NMP. and Roach HI. (2009). Cellular and epigenetic features of a young healthy and a young osteoarthritic cartilage compared with aged control and OA cartilage. J Orthop Res 27:593–601 [DOI] [PubMed] [Google Scholar]
- 22.Loeser RF, Im HJ, Richardson B, Lu Q. and Chubinskaya S. (2009). Methylation of the OP-1 promoter: potential role in the age-related decline in OP-1 expression in cartilage. Osteoarthritis Cartilage 17:513–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donmez G. and Guarente L. (2010). Aging and disease: connections to sirtuins. Aging Cell 9:285–290 [DOI] [PubMed] [Google Scholar]
- 24.Gabay O, Zaal KJ, Sanchez C, Dvir-Ginzberg M, Gagarina V, Song Y, He XH. and McBurney MW. (2013). Sirt1-deficient mice exhibit an altered cartilage phenotype. Joint Bone Spine 80:613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moon MH, Jeong JK, Lee YJ, Seol JW, Jackson CJ. and Park SY. (2013). SIRT1, a class III histone deacetylase, regulates TNF-α-induced inflammation in human chondrocytes. Osteoarthritis Cartilage 21:470–480 [DOI] [PubMed] [Google Scholar]
- 26.Takayama K, Ishida K, Matsushita T, Fujita N, Hayashi S, Sasaki K, Tei K, Kubo S, Matsumoto T, et al. (2009). SIRT1 regulation of apoptosis of human chondrocytes. Arthritis Rheum 60:2731–2740 [DOI] [PubMed] [Google Scholar]
- 27.Gagarina V, Gabay O, Dvir-Ginzberg M, Lee EJ, Brady JK, Quon MJ. and Hall DJ. (2010). SirT1 enhances survival of human osteoarthritic chondrocytes by repressing protein tyrosine phosphatase 1B and activating the insulin-like growth factor receptor pathway. Arthritis Rheum 62:1383–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J. and Pei M. (2012). Cell senescence: a challenge in cartilage engineering and regeneration. Tissue Eng Part B Rev 18:270–287 [DOI] [PubMed] [Google Scholar]
- 29.You H, Ding W. and Rountree CB. (2010). Epigenetic regulation of cancer stem cell marker CD133 by transforming growth factor-beta. Hepatology 51:1635–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan Q, Wu Y, Lin T, Yao H, Yang Z, Gao G, Song E. and Shen H. (2009). Bone morphogenetic protein-2 induces chromatin remodeling and modification at the proximal promoter of Sox9 gene. Biochem Biophys Res Commun 379:356–361 [DOI] [PubMed] [Google Scholar]
- 31.Milagro FI, Campión J, Cordero P, Goyenechea E, Gómez-Uriz AM, Abete I, Zulet MA. and Martínez JA. (2011). A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J 25:1378–1389 [DOI] [PubMed] [Google Scholar]
- 32.Chung TL, Brena RM, Kolle G, Grimmond SM, Berman BP, Laird PW, Pera MF. and Wolvetang EJ. (2010). Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells 28:1848–1855 [DOI] [PubMed] [Google Scholar]
- 33.Esteban MA. and Pei D. (2012). Vitamin C improves the quality of somatic cell reprogramming. Nat Genet 44:366–367 [DOI] [PubMed] [Google Scholar]
- 34.Thoms BL, Dudek KA, Lafont JE. and Murphy CL. (2013). Hypoxia promotes the production and inhibits the destruction of human articular cartilage. Arthritis Rheum 65:1302–1312 [DOI] [PubMed] [Google Scholar]
- 35.Hashimoto K, Otero M, Imagawa K, de Andrés MC, Coico JM, Roach HI, Oreffo ROC, Marcu KB. and Goldring MB. (2013). Regulated transcription of human matrix metalloproteinase 13 (MMP13) and interleukin-1β (IL1B) genes in chondrocytes depends on methylation of specific proximal promoter CpG sites. J Biol Chem 288:10061–10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okazaki K. and Maltepe E. (2006). Oxygen, epigenetics and stem cell fate. Regen Med 1:71–83 [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Ledaki I, Turley H, Gatter KC, Montero JC, Li JL. and Harris AL. (2009). Role of hypoxia-inducible factors in epigenetic regulation via histone demethylases. Ann N Y Acad Sci 1177:185–197 [DOI] [PubMed] [Google Scholar]
- 38.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G. and Natoli G. (2007). The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130:1083–1094 [DOI] [PubMed] [Google Scholar]
- 39.Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, et al. (2010). TNF/p38α/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 7:455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly DJ. and Jacobs CR. (2010). The role of mechanical signals in regulating chondrogenesis and osteogenesis of mesenchymal stem cells. Birth Defects Res C Embryo Today 90:75–85 [DOI] [PubMed] [Google Scholar]
- 41.Fletcher DA. and Mullins RD. (2010). Cell mechanics and the cytoskeleton. Nature 463:485–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Chu JS, Kurpinski K, Li X, Bautista DM, Yang L, Sung KL. and Li S. (2011). Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys J 100:1902–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnsdorf E, Tummala P. and Castillo A. (2010). The epigenetic mechanism of mechanically induced osteogenic differentiation. J Biomech 43:2881–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanazawa T, Furumatsu T, Hachioji M, Oohashi T, Ninomiya Y. and Ozaki T. (2012). Mechanical stretch enhances COL2A1 expression on chromatin by inducing SOX9 nuclear translocalization in inner meniscus cells. J Orthop Res 30:468–474 [DOI] [PubMed] [Google Scholar]
- 45.Mendelsohn AR. and Larrick JW. (2013). Rejuvenation of adult stem cells: is age-associated dysfunction epigenetic? Rejuvenation Res 16:152–157 [DOI] [PubMed] [Google Scholar]
- 46.Feng B, Ruiz MA. and Chakrabarti S. (2013). Oxidative-stress-induced epigenetic changes in chronic diabetic complications. Can J Physiol Pharmacol 91:213–220 [DOI] [PubMed] [Google Scholar]
- 47.Kim GH, Ryan JJ. and Archer SL. (2013). The role of redox signaling in epigenetics and cardiovascular disease. Antioxid Redox Signal 18:1920–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mercer TR. and Mattick JS. (2013). Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20:300–307 [DOI] [PubMed] [Google Scholar]
- 49.Hong E. and Reddi AH. (2013). Dedifferentiation and redifferentiation of articular chondrocytes from surface and middle zones: changes in microRNAs-221/-222, -140, and -143/145 expression. Tissue Eng Part A 19:1015–1022 [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, Kang Y, Zhang H, Duan X, Liu J, Li X. and Liao W. (2012). Expression of microRNAs during chondrogenesis of human adipose-derived stem cells. Osteoarthritis Cartilage 20:1638–1646 [DOI] [PubMed] [Google Scholar]
- 51.Wagner W, Bork S, Lepperdinger G, Joussen S, Ma N, Strunk D. and Koch C. (2010). How to track cellular aging of mesenchymal stromal cells? Aging (Albany NY) 2:224–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schellenberg A, Lin Q, Schüler H, Koch CM, Joussen S, Denecke B, Walenda G, Pallua N, Suschek C V, Zenke M. and Wagner W. (2011). Replicative senescence of mesenchymal stem cells causes DNA-methylation changes which correlate with repressive histone marks. Aging (Albany NY) 3:873–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu JM, Wu X, Gimble JM, Guan X, Freitas MA. and Bunnell BA. (2011). Age-related changes in mesenchymal stem cells derived from rhesus macaque bone marrow. Aging Cell 10:66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Mück C, Laschober GT, Lepperdinger G, Sampson N, et al. (2010). miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell 9:291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R, Benes V, Blake J, Pfister S, Eckstein V. and Ho AD. (2008). Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One 3:e2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, et al. (2007). MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A 104:15805–15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim YJ, Hwang SH, Lee SY, Shin KK, Cho HH, Bae YC. and Jung JS. (2012). miR-486-5p induces replicative senescence of human adipose tissue-derived mesenchymal stem cells and its expression is controlled by high glucose. Stem Cells Dev 21:1749–1760 [DOI] [PubMed] [Google Scholar]
- 58.Marasa BS, Srikantan S, Martindale JL, Kim MM, Lee EK, Gorospe M. and Abdelmohsen K. (2010). MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging (Albany NY) 2:333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ukai T, Sato M, Akutsu H, Umezawa A. and Mochida J. (2012). MicroRNA-199a-3p, microRNA-193b, and microRNA-320c are correlated to aging and regulate human cartilage metabolism. J Orthop Res 30:1915–1922 [DOI] [PubMed] [Google Scholar]
- 60.Yu Z, Li Y, Fan H, Liu Z. and Pestell RG. (2012). miRNAs regulate stem cell self-renewal and differentiation. Front Genet 3:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Butz H, Rácz K, Hunyady L. and Patócs A. (2012). Crosstalk between TGF-β signaling and the microRNA machinery. Trends Pharmacol Sci 33:382–393 [DOI] [PubMed] [Google Scholar]
- 62.Swingler TE, Wheeler G, Carmont V, Elliott HR, Barter MJ, Abu-Elmagd M, Donell ST, Boot-Handford RP, Hajihosseini MK, et al. (2012). The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum 64:1909–1919 [DOI] [PubMed] [Google Scholar]
- 63.Pais H, Nicolas FE, Soond SM, Swingler TE, Clark IM, Chantry A, Moulton V. and Dalmay T. (2010). Analyzing mRNA expression identifies Smad3 as a microRNA-140 target regulated only at protein level. RNA 16:489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pando R, Even-Zohar N, Shtaif B, Edry L, Shomron N, Phillip M. and Gat-Yablonski G. (2012). MicroRNAs in the growth plate are responsive to nutritional cues: association between miR-140 and SIRT1. J Nutr Biochem 23:1474–1481 [DOI] [PubMed] [Google Scholar]
- 65.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu C-G, Croce CM, et al. (2007). A microRNA signature of hypoxia. Mol Cell Biol 27:1859–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foja S, Jung M, Harwardt B, Riemann D, Pelz-Ackermann O, and Schroeder IS. (2013). Hypoxia supports reprogramming of mesenchymal stromal cells via induction of embryonic stem cell-specific microRNA-302 cluster and pluripotency-associated genes. Cell Reprogram 15:68–79 [DOI] [PubMed] [Google Scholar]
- 67.Chang W, Lee CY, Park JH, Park MS, Maeng LS, Yoon CS, Lee MY, Hwang KC. and Chung YA. (2013). Survival of hypoxic human mesenchymal stem cells is enhanced by a positive feedback loop involving miR-210 and hypoxia-inducible factor 1. J Vet Sci 14:69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cicchillitti L, Di Stefano V, Isaia E, Crimaldi L, Fasanaro P, Ambrosino V, Antonini A, Capogrossi MC, Gaetano C, Piaggio G. and Martelli F. (2012). Hypoxia-inducible factor 1-α induces miR-210 in normoxic differentiating myoblasts. J Biol Chem 287:44761–44771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim HW, Haider HK, Jiang S. and Ashraf M. (2009). Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J Biol Chem 284:33161–33168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steck E, Boeuf S, Gabler J, Werth N, Schnatzer P, Diederichs S. and Richter W. (2012). Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J Mol Med (Berl) 90:1185–1195 [DOI] [PubMed] [Google Scholar]
- 71.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Orjalo A V, Rodier F, Lithgow GJ. and Campisi J. (2009). MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY) 1:402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Niimoto T, Nakasa T, Ishikawa M, Okuhara A, Izumi B, Deie M, Suzuki O, Adachi N. and Ochi M. (2010). MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC Musculoskelet Disord 11:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dai L, Zhang X, Hu X, Zhou C. and Ao Y. (2012). Silencing of microRNA-101 prevents IL-1β-induced extracellular matrix degradation in chondrocytes. Arthritis Res Ther 14:R268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK. and Asahara H. (2009). MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum 60:2723–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liang ZJ, Zhuang H, Wang GX, Li Z, Zhang HT, Yu TQ. and Zhang BD. (2012). MiRNA-140 is a negative feedback regulator of MMP-13 in IL-1β-stimulated human articular chondrocyte C28/I2 cells. Inflamm Res 61:503–509 [DOI] [PubMed] [Google Scholar]
- 76.Matsukawa T, Sakai T, Yonezawa T, Hiraiwa H, Hamada T, Nakashima M, Ono Y, Ishizuka S, Nakahara H, et al. (2013). MicroRNA-125b regulates the expression of aggrecanase-1 (ADAMTS-4) in human osteoarthritic chondrocytes. Arthritis Res Ther 15:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Akhtar N. and Haqqi TM. (2012). MicroRNA-199a* regulates the expression of cyclooxygenase-2 in human chondrocytes. Ann Rheum Dis 71:1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abouheif MM, Nakasa T, Shibuya H, Niimoto T, Kongcharoensombat W. and Ochi M. (2010). Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology (Oxford) 49:2054–2060 [DOI] [PubMed] [Google Scholar]
- 79.Kim D, Song J, Kim S, Chun CH. and Jin EJ. (2011). MicroRNA-34a regulates migration of chondroblast and IL-1β-induced degeneration of chondrocytes by targeting EphA5. Biochem Biophys Res Commun 415:551–557 [DOI] [PubMed] [Google Scholar]
- 80.Dunn W, DuRaine G. and Reddi AH. (2009). Profiling microRNA expression in bovine articular cartilage and implications for mechanotransduction. Arthritis Rheum 60:2333–2339 [DOI] [PubMed] [Google Scholar]
- 81.Guan Y, Yang X, Wei L. and Chen Q. (2011). MiR-365: a mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4. FASEB J 25:4457–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mendias CL, Gumucio JP. and Lynch EB. (2012). Mechanical loading and TGF-β change the expression of multiple miRNAs in tendon fibroblasts. J Appl Physiol 113:56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Z, Shentu TP, Wen L, Johnson DA. and Shyy JY. (2013). Regulation of SIRT1 by oxidative stress-responsive miRNAs and a systematic approach to identify its role in the endothelium. Antioxid Redox Signal 19:1522–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Howell JC, Chun E, Farrell AN, Hur EY, Caroti CM, Iuvone PM. and Haque R. (2013). Global microRNA expression profiling: curcumin (diferuloylmethane) alters oxidative stress-responsive microRNAs in human ARPE-19 cells. Mol Vis 19:544–560 [PMC free article] [PubMed] [Google Scholar]
- 85.Lin Y, Liu X, Cheng Y, Yang J, Huo Y. and Zhang C. (2009). Involvement of MicroRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J Biol Chem 284:7903–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ling M, Li Y, Xu Y, Pang Y, Shen L, Jiang R, Zhao Y, Yang X, Zhang J, et al. (2012). Regulation of miRNA-21 by reactive oxygen species-activated ERK/NF-κB in arsenite-induced cell transformation. Free Radic Biol Med 52:1508–1518 [DOI] [PubMed] [Google Scholar]
- 87.Li Y, Shelat H. and Geng YJ. (2012). IGF-1 prevents oxidative stress induced-apoptosis in induced pluripotent stem cells which is mediated by microRNA-1. Biochem Biophys Res Commun 426:615–619 [DOI] [PubMed] [Google Scholar]
- 88.Kim JH, Park SG, Song SY, Kim JK. and Sung JH. (2013). Reactive oxygen species-responsive miR-210 regulates proliferation and migration of adipose-derived stem cells via PTPN2. Cell Death Dis 4:e588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hackett CH, Greve L, Novakofski KD. and Fortier LA. (2012). Comparison of gene-specific DNA methylation patterns in equine induced pluripotent stem cell lines with cells derived from equine adult and fetal tissues. Stem Cells Dev 21:1803–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Redaelli S, Bentivegna A, Foudah D, Miloso M, Redondo J, Riva G, Baronchelli S, Dalprà L. and Tredici G. (2012). From cytogenomic to epigenomic profiles: monitoring the biologic behavior of in vitro cultured human bone marrow mesenchymal stem cells. Stem Cell Res Ther 3:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vacanti V, Kong E, Suzuki G, Sato K, Canty JM. and Lee T. (2005). Phenotypic changes of adult porcine mesenchymal stem cells induced by prolonged passaging in culture. J Cell Physiol 205:194–201 [DOI] [PubMed] [Google Scholar]
- 92.Li Z, Liu C, Xie Z, Song P, Zhao RC, Guo L, Liu Z. and Wu Y. (2011). Epigenetic dysregulation in mesenchymal stem cell aging and spontaneous differentiation. PLoS One 6:e20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu TM, Wu YN, Guo XM, Hui JH, Lee EH. and Lim B. (2009). Effects of ectopic Nanog and Oct4 overexpression on mesenchymal stem cells. Stem Cells Dev 18:1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marinovic-Kulisic S, Juric-Lekic G, Vikic-Topic M, Lokosek V, Radujkovic V, Bulic-Jakus F, Katusic A, Vlahovic M, Serman L. and Sincic N. (2011). 5-azacytidine enhances proliferation in transplanted rat fetal epiglottis. Front Biosci (Elite Ed) 3:581–590 [DOI] [PubMed] [Google Scholar]
- 95.Vogelauer M, Rubbi L, Lucas I, Brewer BJ. and Grunstein M. (2002). Histone acetylation regulates the time of replication origin firing. Mol Cell 10:1223–1233 [DOI] [PubMed] [Google Scholar]
- 96.Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T. and Seiser C. (2002). Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J 21:2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y, Chen T, Yan H, Qi H, Deng C, Ye T, Zhou S. and Li FR. (2013). Role of histone deacetylase inhibitors in the aging of human umbilical cord mesenchymal stem cells. J Cell Biochem 114:2231–2239 [DOI] [PubMed] [Google Scholar]
- 98.Villasante A, Piazzolla D, Li H, Gomez-Lopez G, Djabali M. and Serrano M. (2011). Epigenetic regulation of Nanog expression by Ezh2 in pluripotent stem cells. Cell Cycle 10:1488–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cakouros D, Isenmann S, Cooper L, Zannettino A, Anderson P, Glackin C. and Gronthos S. (2012). Twist-1 induces Ezh2 recruitment regulating histone methylation along the Ink4A/Arf locus in mesenchymal stem cells. Mol Cell Biol 32:1433–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.So AY, Jung JW, Lee S, Kim HS. and Kang KS. (2011). DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs. PLoS One 6:e19503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hupkes M, Van Someren EP, Middelkamp SH, Piek E, Van Zoelen EJ. and Dechering KJ. (2011). DNA methylation restricts spontaneous multi-lineage differentiation of mesenchymal progenitor cells, but is stable during growth factor-induced terminal differentiation. Biochim Biophys Acta 1813:839–849 [DOI] [PubMed] [Google Scholar]
- 102.El-Serafi AT, Oreffo RO. and Roach HI. (2011). Epigenetic modifiers influence lineage commitment of human bone marrow stromal cells: differential effects of 5-aza-deoxycytidine and trichostatin A. Differentiation 81:35–41 [DOI] [PubMed] [Google Scholar]
- 103.Ezura Y, Sekiya I, Koga H, Muneta T. and Noda M. (2009). Methylation status of CpG islands in the promoter regions of signature genes during chondrogenesis of human synovium-derived mesenchymal stem cells. Arthritis Rheum 60:1416–1426 [DOI] [PubMed] [Google Scholar]
- 104.Kim SY. and Im GI. (2011). The expressions of the SOX trio, PTHrP (parathyroid hormone-related peptide)/IHH (Indian hedgehog protein) in surgically induced osteoarthritis of the rat. Cell Biol Int 35:529–535 [DOI] [PubMed] [Google Scholar]
- 105.Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Chen J, et al. (2013). H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet 45:34–42 [DOI] [PubMed] [Google Scholar]
- 106.Rodova M, Lu Q, Li Y, Woodbury BG, Crist JD, Gardner BM, Yost JG, Zhong XB, Anderson HC. and Wang J. (2011). Nfat1 regulates adult articular chondrocyte function through its age-dependent expression mediated by epigenetic histone methylation. J Bone Miner Res 26:1974–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Spaapen F, van den Akker GG, Caron MM, Prickaerts P, Rofel C, Dahlmans VE, Surtel DA, Paulis Y, Schweizer F, et al. (2013). The immediate early gene product EGR1 and polycomb group proteins interact in epigenetic programming during chondrogenesis. PLoS One 8:e58083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Furumatsu T, Tsuda M, Yoshida K, Taniguchi N, Ito T, Hashimoto M, Ito T. and Asahara H. (2005). Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J Biol Chem 280:35203–35208 [DOI] [PubMed] [Google Scholar]
- 109.Zimmermann P, Boeuf S, Dickhut A, Boehmer S, Olek S. and Richter W. (2008). Correlation of COL10A1 induction during chondrogenesis of mesenchymal stem cells with demethylation of two CpG sites in the COL10A1 promoter. Arthritis Rheum 58:2743–2753 [DOI] [PubMed] [Google Scholar]
- 110.Ideno H, Shimada A, Imaizumi K, Kimura H, Abe M, Nakashima K. and Nifuji A. (2013). Predominant expression of H3K9 methyltransferases in prehypertrophic and hypertrophic chondrocytes during mouse growth plate cartilage development. Gene Expr Patterns 13:84–90 [DOI] [PubMed] [Google Scholar]
- 111.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, et al. (2004). Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119:555–566 [DOI] [PubMed] [Google Scholar]
- 112.Pei M, Chen D, Li J. and Wei L. (2009). Histone deacetylase 4 promotes TGF-beta1-induced synovium-derived stem cell chondrogenesis but inhibits chondrogenically differentiated stem cell hypertrophy. Differentiation 78:260–268 [DOI] [PubMed] [Google Scholar]
- 113.Guan Y, Chen Q, Yang X, Haines P, Pei M, Terek R, Wei X, Zhao T. and Wei L. (2012). Subcellular relocation of histone deacetylase 4 regulates growth plate chondrocyte differentiation through Ca2+/calmodulin-dependent kinase IV. Am J Physiol Cell Physiol 303:C33–C40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Henikoff S. (2008). Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet 9:15–26 [DOI] [PubMed] [Google Scholar]
- 115.Dorn ES. and Cook JG. (2011). Nucleosomes in the neighborhood: New roles for chromatin modifications in replication origin control. Epigenetics 6:552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Struhl K. and Segal E. (2013). Determinants of nucleosome positioning. Nat Struct Mol Biol 20:267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Herbig U, Jobling WA, Chen BP, Chen DJ. and Sedivy JM. (2004). Telomere shortening triggers senescence of Human Cells through a Pathway Involving ATM, p53, and p21(CIP1), but Not p16(INK4a). Mol Cell 14:501–513 [DOI] [PubMed] [Google Scholar]
- 118.Wang S, Hu C. and Zhu J. (2010). Distinct and temporal roles of nucleosomal remodeling and histone deacetylation in the repression of the hTERT gene. Mol Biol Cell 21:821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Höfer T. and Rippe K. (2012). Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol 19:1185–1192 [DOI] [PubMed] [Google Scholar]
- 120.Badylak SF, Weiss DJ, Caplan A. and Macchiarini P. (2012). Engineered whole organs and complex tissues. Lancet 379:943–952 [DOI] [PubMed] [Google Scholar]
- 121.Benders KE, van Weeren PR, Badylak SF, Saris DB, Dhert WJ. and Malda J. (2013). Extracellular matrix scaffolds for cartilage and bone regeneration. Trends Biotechnol 31:169–176 [DOI] [PubMed] [Google Scholar]
- 122.Yang Q, Peng J, Guo Q, Huang J, Zhang L, Yao J, Yang F, Wang S, Xu W, Wang A. and Lu S. (2008). A cartilage ECM-derived 3-D porous acellular matrix scaffold for in vivo cartilage tissue engineering with PKH26-labeled chondrogenic bone marrow-derived mesenchymal stem cells. Biomaterials 29:2378–2387 [DOI] [PubMed] [Google Scholar]
- 123.Cole BJ, Farr J, Winalski CS, Hosea T, Richmond J, Mandelbaum B. and De Deyne PG. (2011). Outcomes after a single-stage procedure for cell-based cartilage repair: a prospective clinical safety trial with 2-year follow-up. Am J Sports Med 39:1170–1179 [DOI] [PubMed] [Google Scholar]
- 124.Kang H, Peng J, Lu S, Liu S, Zhang L, Huang J, Sui X, Zhao B, Wang A, et al. (2012). In vivo cartilage repair using adipose-derived stem cell-loaded decellularized cartilage ECM scaffolds. J Tissue Eng Regen Med DOI: 10.1002/term.1538 [DOI] [PubMed] [Google Scholar]
- 125.Pei M, Li JT, Shoukry M. and Zhang Y. (2011). A review of decellularized stem cell matrix: a novel cell expansion system for cartilage tissue engineering. Eur Cell Mater 22:333–343; discussion 343. [DOI] [PubMed] [Google Scholar]
- 126.Lin H, Yang G, Tan J. and Tuan RS. (2012). Influence of decellularized matrix derived from human mesenchymal stem cells on their proliferation, migration and multi-lineage differentiation potential. Biomaterials 33:4480–4489 [DOI] [PubMed] [Google Scholar]
- 127.Jones BA. and Pei M. (2012). Synovium-derived stem cells: a tissue-specific stem cell for cartilage engineering and regeneration. Tissue Eng Part B Rev 18:301–311 [DOI] [PubMed] [Google Scholar]
- 128.Li J. and Pei M. (2011). Optimization of an in vitro three-dimensional microenvironment to reprogram synovium-derived stem cells for cartilage tissue engineering. Tissue Eng Part A 17:703–712 [DOI] [PubMed] [Google Scholar]
- 129.He F, Chen X. and Pei M. (2009). Reconstruction of an in vitro tissue-specific microenvironment to rejuvenate synovium-derived stem cells for cartilage tissue engineering. Tissue Eng Part A 15:3809–3821 [DOI] [PubMed] [Google Scholar]
- 130.Pei M, Zhang Y, Li J. and Chen D. (2013). Antioxidation of decellularized stem cell matrix promotes human synovium-derived stem cell-based chondrogenesis. Stem Cells Dev 22:889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pei M, He F. and Kish VL. (2011). Expansion on extracellular matrix deposited by human bone marrow stromal cells facilitates stem cell proliferation and tissue-specific lineage potential. Tissue Eng Part A 17:3067–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pei M. and He F. (2012). Extracellular matrix deposited by synovium-derived stem cells delays replicative senescent chondrocyte dedifferentiation and enhances redifferentiation. J Cell Physiol 227:2163–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pei M, He F. and Wei L. (2011). Three-dimensional cell expansion substrate for cartilage tissue engineering and regeneration: a comparison in decellularized matrix deposited by synovium-derived stem cells and chondrocytes. J Tissue Sci Eng 2:104 [Google Scholar]
- 134.Pei M, Shoukry M, Li J, Daffner SD, France JC. and Emery SE. (2012). Modulation of in vitro microenvironment facilitates synovium-derived stem cell-based nucleus pulposus tissue regeneration. Spine (Phila Pa 1976) 37:1538–1547 [DOI] [PubMed] [Google Scholar]
- 135.He F. and Pei M. (2012). Rejuvenation of nucleus pulposus cells using extracellular matrix deposited by synovium-derived stem cells. Spine (Phila Pa 1976) 37:459–469 [DOI] [PubMed] [Google Scholar]
- 136.Li J, Hansen KC, Zhang Y, Dong C, Dinu CZ, Dzieciatkowska M. and Pei M. (2014). Rejuvenation of chondrogenic potential in a young stem cell microenvironment. Biomaterials 35:642–653 [DOI] [PubMed] [Google Scholar]
- 137.Pei M, He F, Li J, Tidwell JE, Jones AC. and McDonough EB. (2013). Repair of large animal partial-thickness cartilage defects through intraarticular injection of matrix-rejuvenated synovium-derived stem cells. Tissue Eng Part A 19:1144–1154 [DOI] [PubMed] [Google Scholar]
- 138.Li J, He F. and Pei M. (2011). Creation of an in vitro microenvironment to enhance human fetal synovium-derived stem cell chondrogenesis. Cell Tissue Res 345:357–365 [DOI] [PubMed] [Google Scholar]
- 139.Choi HR, Cho KA, Kang HT, Bin Lee J, Kaeberlein M, Suh Y, Chung IK. and Park SC. (2011). Restoration of senescent human diploid fibroblasts by modulation of the extracellular matrix. Aging Cell 10:148–157 [DOI] [PubMed] [Google Scholar]
- 140.Hong E. and Reddi AH. (2012). MicroRNAs in chondrogenesis, articular cartilage, and osteoarthritis: implications for tissue engineering. Tissue Eng Part B Rev 18:445–453 [DOI] [PubMed] [Google Scholar]
- 141.Buechli ME, Lamarre J. and Koch TG. (2013). MicroRNA-140 expression during chondrogenic differentiation of equine cord blood-derived mesenchymal stromal cells. Stem Cells Dev 22:1288–1296 [DOI] [PubMed] [Google Scholar]
- 142.Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, et al. (2010). MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev 24:1173–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Karlsen TA, Jakobsen RB, Mikkelsen TS. and Brinchmann JE. (2013). microRNA-140 targets RALA and regulates chondrogenic differentiation of human mesenchymal stem cells by translational enhancement of SOX9 and ACAN. Stem Cells Dev 23:290–304 [DOI] [PubMed] [Google Scholar]
- 144.Yang B, Guo H, Zhang Y, Chen L, Ying D. and Dong S. (2011). MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One 6:e21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Martinez-Sanchez A, Dudek KA. and Murphy CL. (2012). Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145). J Biol Chem 287:916–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Paik S, Jung HS, Lee S, Yoon DS, Park MS. and Lee JW. (2012). miR-449a regulates the chondrogenesis of human mesenchymal stem cells through direct targeting of lymphoid enhancer-binding factor-1. Stem Cells Dev 21:3298–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.De Andrés MC, Kingham E, Imagawa K, Gonzalez A, Roach HI, Wilson DI. and Oreffo RO. (2013). Epigenetic regulation during fetal femur development: DNA methylation matters. PLoS One 8:e54957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Van den Berg WB. (2011). Osteoarthritis year 2010 in review: pathomechanisms. Osteoarthritis Cartilage 19:338–341 [DOI] [PubMed] [Google Scholar]
- 149.Loughlin J. (2005). The genetic epidemiology of human primary osteoarthritis: current status. Expert Rev Mol Med 7:1–12 [DOI] [PubMed] [Google Scholar]
- 150.Reynard LN. and Loughlin J. (2013). The genetics and functional analysis of primary osteoarthritis susceptibility. Expert Rev Mol Med 15:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fraga MF. and Esteller M. (2007). Epigenetics and aging: the targets and the marks. Trends Genet 23:413–418 [DOI] [PubMed] [Google Scholar]
- 152.Skinner MK, Manikkam M. and Guerrero-Bosagna C. (2010). Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab 21:214–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Im G. and Choi Y. (2013). Epigenetics in osteoarthritis and its implication for future therapeutics. Expert Opin Biol Ther 13:713–721 [DOI] [PubMed] [Google Scholar]
- 154.Miyaki S. and Asahara H. (2012). Macro view of microRNA function in osteoarthritis. Nat Rev Rheumatol 8:543–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Goldring MB. and Marcu KB. (2012). Epigenomic and microRNA-mediated regulation in cartilage development, homeostasis, and osteoarthritis. Trends Mol Med 18:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I. and Dalmay T. (2006). The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett 580:4214–4217 [DOI] [PubMed] [Google Scholar]
- 157.Akhtar N, Rasheed Z, Ramamurthy S, Anbazhagan AN, Voss FR. and Haqqi TM. (2010). MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum 62:1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Santini P, Politi L, Vedova PD, Scandurra R. and Scotto d'Abusco A. (2013). The inflammatory circuitry of miR-149 as a pathological mechanism in osteoarthritis. Rheumatol Int DOI: 10.1007/s00296-013-2754-8 [DOI] [PubMed] [Google Scholar]
- 159.Dvir-Ginzberg M, Gagarina V, Lee EJ, Booth R, Gabay O. and Hall DJ. (2011). Tumor necrosis factor α-mediated cleavage and inactivation of SirT1 in human osteoarthritic chondrocytes. Arthritis Rheum 63:2363–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gabay O. and Sanchez C. (2012). Epigenetics, sirtuins and osteoarthritis. Joint Bone Spine 79:570–573 [DOI] [PubMed] [Google Scholar]
- 161.Gabay O, Oppenhiemer H, Meir H, Zaal K, Sanchez C. and Dvir-Ginzberg M. (2012). Increased apoptotic chondrocytes in articular cartilage from adult heterozygous SirT1 mice. Ann Rheum Dis 71:613–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Elmali N, Esenkaya I, Harma A, Ertem K, Turkoz Y. and Mizrak B. (2005). Effect of resveratrol in experimental osteoarthritis in rabbits. Inflamm Res 54:158–162 [DOI] [PubMed] [Google Scholar]
- 163.Eo SH, Cho H. and Kim SJ. (2013). Resveratrol inhibits nitric oxide-induced apoptosis via the NF-kappa B pathway in rabbit articular chondrocytes. Biomol Ther (Seoul) 21:364–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yuan HF, Zhai C, Yan XL, Zhao DD, Wang JX, Zeng Q, Chen L, Nan X, He LJ, et al. (2012). SIRT1 is required for long-term growth of human mesenchymal stem cells. J Mol Med (Berl) 90:389–400 [DOI] [PubMed] [Google Scholar]
- 165.Im HJ, Li X, Chen D, Yan D, Kim J, Ellman MB, Stein GS, Cole B, Kc R, Cs-Szabo G. and van Wijnen AJ. (2012). Biological effects of the plant-derived polyphenol resveratrol in human articular cartilage and chondrosarcoma cells. J Cell Physiol 227:3488–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shakibaei M, John T, Seifarth C. and Mobasheri A. (2007). Resveratrol inhibits IL-1 beta-induced stimulation of caspase-3 and cleavage of PARP in human articular chondrocytes in vitro. Ann N Y Acad Sci 1095:554–563 [DOI] [PubMed] [Google Scholar]
- 167.Sheu SY, Chen WS, Sun JS, Lin FH. and Wu T. (2013). Biological characterization of oxidized hyaluronic acid/resveratrol hydrogel for cartilage tissue engineering. J Biomed Mater Res A 101:3457–3466 [DOI] [PubMed] [Google Scholar]
- 168.Volk SW, Kapatkin AS, Haskins ME, Walton RM. and D'Angelo M. (2003). Gelatinase activity in synovial fluid and synovium obtained from healthy and osteoarthritic joints of dogs. Am J Vet Res 64:1225–1233 [DOI] [PubMed] [Google Scholar]
- 169.Labrie M. and St-Pierre Y. (2013). Epigenetic regulation of mmp-9 gene expression. Cell Mol Life Sci 70:3109–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bui C, Barter MJ, Scott JL, Xu Y, Galler M, Reynard LN, Rowan AD. and Young DA. (2012). cAMP response element-binding (CREB) recruitment following a specific CpG demethylation leads to the elevated expression of the matrix metalloproteinase 13 in human articular chondrocytes and osteoarthritis. FASEB J 26:3000–3011 [DOI] [PubMed] [Google Scholar]
- 171.Imagawa K, de Andrés MC, Hashimoto K, Pitt D, Itoi E, Goldring MB, Roach HI. and Oreffo RO. (2011). The epigenetic effect of glucosamine and a nuclear factor-kappa B (NF-kB) inhibitor on primary human chondrocytes—implications for osteoarthritis. Biochem Biophys Res Commun 405:362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Klein K, Ospelt C. and Gay S. (2012). Epigenetic contributions in the development of rheumatoid arthritis. Arthritis Res Ther 14:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Duroux-richard I, Jorgensen C. and Apparailly F. (2012). What do microRNAs mean for rheumatoid arthritis? Arthritis Rheum 64:11–20 [DOI] [PubMed] [Google Scholar]
- 174.Ammari M, Jorgensen C. and Apparailly F. (2013). Impact of microRNAs on the understanding and treatment of rheumatoid arthritis. Curr Opin Rheumatol 25:225–233 [DOI] [PubMed] [Google Scholar]
- 175.Stanczyk J, Ospelt C, Karouzakis E, Filer A, Raza K, Kolling C, Gay R, Buckley CD, Tak PP, Gay S. and Kyburz D. (2011). Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum 63:373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S. and Kyburz D. (2008). Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum 58:1001–1009 [DOI] [PubMed] [Google Scholar]
- 177.Ceribelli A, Nahid MA, Satoh M. and Chan EK. (2011). MicroRNAs in rheumatoid arthritis. FEBS Lett 585:3667–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Miao C, Yang Y, He X, Xu T, Huang C, Huang Y, Zhang L, Lv XW, Jin Y. and Li J. (2013). New advances of microRNAs in the pathogenesis of rheumatoid arthritis, with a focus on the crosstalk between DNA methylation and the microRNA machinery. Cell Signal 25:1118–1125 [DOI] [PubMed] [Google Scholar]
- 179.Nakano K, Boyle DL. and Firestein GS. (2013). Regulation of DNA methylation in rheumatoid arthritis synoviocytes. J Immunol 190:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nakano K, Whitaker JW, Boyle DL, Wang W. and Firestein GS. (2013). DNA methylome signature in rheumatoid arthritis. Ann Rheum Dis 72:110–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Karouzakis E, Gay RE, Michel BA, Gay S. and Neidhart M. (2009). DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum 60:3613–3622 [DOI] [PubMed] [Google Scholar]
- 182.Ballestar E. (2011). Epigenetic alterations in autoimmune rheumatic diseases. Nat Rev Rheumatol 7:263–271 [DOI] [PubMed] [Google Scholar]
- 183.De la Rica L, Urquiza JM, Gómez-Cabrero D, Islam AB, López-Bigas N, Tegnér J, Toes RE. and Ballestar E. (2013). Identification of novel markers in rheumatoid arthritis through integrated analysis of DNA methylation and microRNA expression. J Autoimmun 41:6–16 [DOI] [PubMed] [Google Scholar]
- 184.Niederer F, Ospelt C, Brentano F, Hottiger MO, Gay RE, Gay S, Detmar M. and Kyburz D. (2011). SIRT1 overexpression in the rheumatoid arthritis synovium contributes to proinflammatory cytokine production and apoptosis resistance. Ann Rheum Dis 70:1866–1873 [DOI] [PubMed] [Google Scholar]
- 185.Klein K. and Gay S. (2013). Epigenetic modifications in rheumatoid arthritis, a review. Curr Opin Pharmacol 13:420–425 [DOI] [PubMed] [Google Scholar]