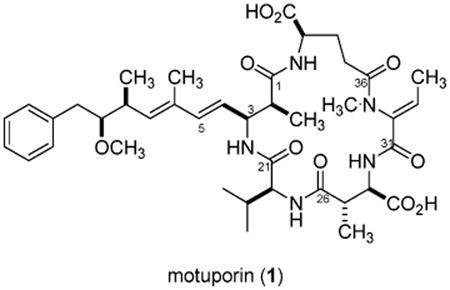
Abstract
An in-depth LCMS examination of 14 different collections of Indo-Pacific Theonella swinhoei sponges resulted in the discovery of four diastereomeric analogues of the cyclic pentapeptide motuporin. These motuporin analogues all contain a novel 2R configuration for the Adda amino acid. Additionally, one analogue has a unique nonoxygenated Adda amino acid. In all, 15 different compounds were observed by LCMS or isolated. The stereochemistries of the constituent amino acids were determined through a combination of the advanced Marfey technique and 1H NMR data.
The sustained worldwide interest over the past 25 years in the chemistry and biology of the sponge Theonella swinhoei (Lithistida, Theonellidae) is astonishing. Kashman was among the first, in the early 1980s, to recognize the potential of this sponge as a source of diverse secondary metabolites with reports on unique sterols1 and complex polyketides headed by swinholide A.2 This remarkably prolific species continues to be a source of powerfully bioactive substances, with motuporin (1)3 being an early example. Currently, the biosynthetic products of T. swinhoei represent more than nine biosynthetic classes4–12 and have been reported from diverse pan-oceanic locals including Papua New Guinea, Indonesia, the Philippines, Palau, the Red Sea, Japan, and Mozambique.
Careful examination of the literature along with scanning of our sponge repository indicated there were at least three phenotypes13 of T. swinhoei with features shown in Figure 1. The first phenotype is defined by a characteristic red-purple ectosome and a cream-colored endosome, and more than 15 such samples have been examined by our group.14 We found that the major constituents varied among the swinholides, motuporin or theonellapeptolide Id, but not all compounds were present in all collections. The second also possesses a red-purple ectosome but with a yellow to orange interior. Fusetani13b has indicated there are eight molecular frameworks isolable from such specimens including polytheonamides, cyclotheonamides, nuzumamide A, pseudotheonamides, onnamides, theopederins, orbiculamide A, and the aurantosides. Recently, both the UCSC lab and the Ireland group13c have observed aurantosides from the third phenotype, which is red-orange throughout. The preceding scenario represents a confounding circumstance that stimulated the research pursued in this study. However, it must be underscored that attempts to gain an understanding of the changing chemical signatures of T. swinhoei are inherently problematic because of the complex microbial communities that are always associated with it.15
Figure 1.
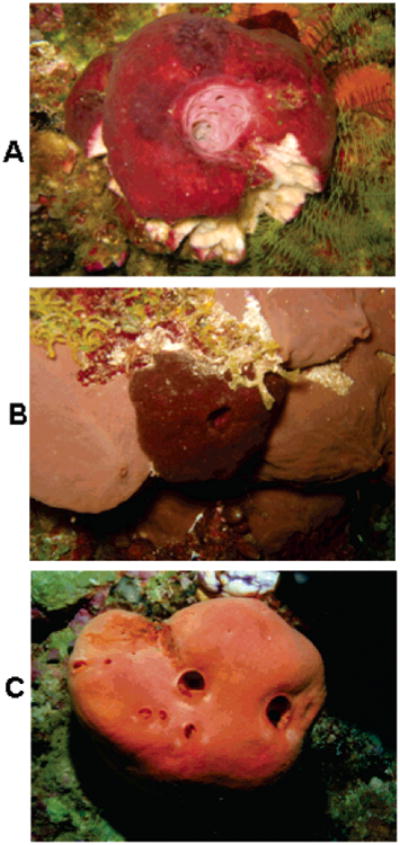
The three Theonella swinhoei phenotypes: (A) type I; (B) type II; and (C) type III.
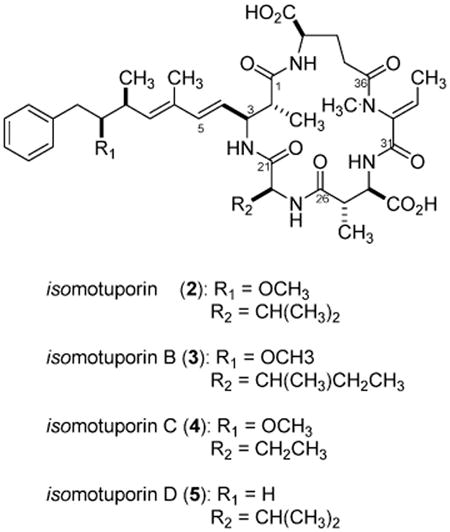
We believed that a careful screening study by LCMS of Indo-Pacific-derived T. swinhoei could allow refinement of observations published in 1998.14 In contrast to the previous study, we sought definition of both major and minor components. Another goal was to be able to reliably locate populations that contained motuporin (1) or the swinholides or both because there is continuing speculation that these compounds could be arising from associated cyanobacteria.16 None of the 17 specimens14 we previously examined contained the swinholides, yet 1 was present in 14 of them. Thirteen new collections of T. swinhoei were investigated and to our surprise none of them contained 1, but two were a source of a motuporin diastereomer. This new compound, isomotuporin (2), along with three congeners, isomotuporins B (3), C (4), and D (5), plus 11 known compounds were either observed or isolated, and these results are described next.
Results and Discussion
The extracts of 14 T. swinhoei collections, obtained from different Indo-Pacific sites, were processed as described in the Experimental Section and then screened using LCMS. One of these (coll. no. 94590), though previously discussed,14 was not evaluated by the methods used herein. There were 14 compounds profiled as shown in Table 1. Comparisons of our observed MS m/z values, UV curves, and diagnostic 1H NMR shifts to those in the literature were used to dereplicate compounds but not their stereochemical details, including swinholide A2 or F;17 swinholide B,17b C,17b D,17 or G;17 swinholide E;17 swinhoeiamide A;7 theonellamine B;18 theonellamide A;19 papuamide D;9 theopaluamide;10 misakinolide A;20 theoneberine;21 and theopederin J11 (see Figure S8). Only the planar structures of these compounds were determined, as their optical properties were not explored. Two of the samples (coll. nos. 02141 and 02142, 0.5 kg wet weight each) contained ESI [M + H]+ m/z at 768.4 amu, diagnostic of motuporin (1). Purification of this compound followed by 1H NMR analysis indicated variations in seven chemical shifts, indicating that a new diastereomer, isomotuporin (2), was in hand (Figure S1). The substantial differences (>0.1 ppm) in the shifts for seven protons can easily be seen by the plot in Figure 2. Alternatively, a similar comparison of the 1H NMR data of natural 13 versus synthetic22 demonstrated that no shifts differed by more than 0.05 ppm. Further examination of the LCMS profiles for these two sponges indicated the presence of small amounts of three other motuporin analogues, B–D (as indicated by m/z values of 782.4, 754.4, and 738.4, respectively). Repeated semipreparative reversed-phase HPLC was used to purify these compounds as amorphous, white solids from the methanol and dichloromethane extracts.
Table 1. Theonella swinhoei Collections and Their Respective Secondary Metabolitesa.
| compound | collection numbers/locationb | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||
| 94590 Indo |
94647 Indo |
94648 Indo |
00342 PNG |
00356 PNG |
00365 PNG |
00399 PNG |
02141 PNG |
02142 PNG |
03506 PNG |
05221 PNG |
05254 PNG |
05259A PNG |
05259B PNG |
|
| swinholides A, F | • | • | • | • | • | |||||||||
| swinholides B, C, D, G | • | |||||||||||||
| swinholide E | • | • | ||||||||||||
| swinhoeiamide A | • | • | • | • | • | |||||||||
| theonellamine B | • | • | • | • | ||||||||||
| theonellamide A | • | • | • | |||||||||||
| papuamide D | • | |||||||||||||
| theopalauamide | • | • | • | • | • | • | • | |||||||
| misakinolide A | • | |||||||||||||
| theoneberine | • | |||||||||||||
| theopederin J | • | |||||||||||||
| motuporin isomotuporin | • | • | ||||||||||||
Structures of named compounds and above and/or underwater photographs of all the sponges studied can bo found in Figures S8 and S9, respectively.
Indo = Indonesia, PNG = Papua New Guinea.
Figure 2.
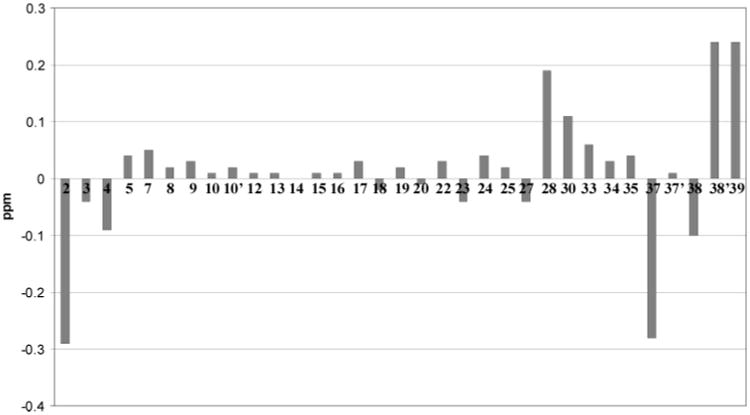
1H NMR chemical shift differences of motuporin (1)3 versus isomotuporin (2).
The confirmation that isomotuporin (2), of molecular formula C40H57N5O10 by HRESIMS (m/z 768.4122 [M + H]+), possessed the planar structure shown came from diagnostic 2D NMR correlations including gCOSY, gHMQC, gHMBC, and 15N-HMBC (see Table 2 and Figure S1). While the 13C NMR shifts of 1 and 2 were not very different, the 1H NMR data of Figure 2 along with the observed rotation for 2 ([(α]25D = −38.4) versus that in the literature for 1 ([(α]25D = −83.8)3 represented distinctive properties. The stereochemistry at the eight chiral centers (2S, 3S, 8S, 9S, 22S, 27S, 28R, 39R) and geometry across the three double bonds (4E, 6E, 32Z) have been rigorously proven for 1.3,22 A systematic approach was employed to evaluate these features in 2. The identical vinylic proton shift values and coupling constants for 2 versus 1 confirmed the 4E and 6E geometries of the former. In addition, a strong NOE from Me-34 to Me-35 confirmed a 32Z geometry.
Table 2. NMR Dataa for Compound 2 in MeOD-d4.
| position | δC | δNb | δH (J in Hz) | gCOSY | gHMBC | NOE | 13N-HMBC |
|---|---|---|---|---|---|---|---|
| ADDA | |||||||
| 1 | 176.1 | ||||||
| 2 | 45.4 | 2.73 dq (10.7, 7.0) | 3, 17 | 1, 3, 17 | 3, 17 | ||
| 3 | 56.2 | 4.57 dd (9.0, 10.7) | 2, 4 | 2, 4, 5 | 2, 4, 5, 17 | ||
| 4 | 126.2 | 5.52 dd (9.0, 15.5) | 3, 5 | 3, 6 | 3, 5, 17, 18 | ||
| 5 | 139.4 | 6.26 d (15.5) | 4 | 3, 6, 7, 18 | 3, 4, 7, 17, 19 | ||
| 6 | 133.9 | ||||||
| 7 | 137.4 | 5.44 d (10.0) | 8, 18 | 5, 8, 9, 18 | 5, 8, 9, 10a, 10b, 19 | ||
| 8 | 37.9 | 2.59 d pent (10.0, 6.7) | 7, 9, 19 | 6, 7, 9, 10, 19 | 18, 19 | ||
| 9 | 88.5 | 3.26 m | 8, 10a, 10b | 7, 10, 11, 19, 20 | Ar, 7, 19 | ||
| 10a | 39.1 | 2.82 dd (5.0, 14.1) | 9, 10b | 8, 9, 11, 12, 16 | 12, 16, 19 | ||
| 10b | 2.67 dd (7.3, 14.1) | 9, 10a | 8, 9, 11, 12, 16 | 12, 16 | |||
| 11 | 140.6 | ||||||
| 12 | 130.6 | 7.18 m | 10, 14, 16 | 10a, 10b, 18, 19, 20 | |||
| 13 | 129.3 | 7.24 m | 11, 15 | ||||
| 14 | 127.1 | 7.17 m | 12, 16 | ||||
| 15 | 129.3 | 7.24 m | 11, 13 | ||||
| 16 | 130.6 | 7.18 m | 10, 12, 14 | 10a, 10b, 18, 19, 20 | |||
| 17 | 16.4 | 1.05 d (7.0) | 2 | 1, 2, 3 | 2, 17 | ||
| 18 | 13.0 | 1.60 d (1.1) | 7 | 5, 6, 7 | 8, 12, 16, 19, 20 | ||
| 19 | 16.7 | 1.01 d (6.7) | 8 | 7, 8, 9 | 8, 10a, 12, 16, 18 | ||
| 20 | 58.9 | 3.22 s | 9 | 12, 16, 18 | |||
| Val | |||||||
| 21 | 171.3 | ||||||
| 22 | 58.1 | 4.43 d (4.1) | 23, 22NH | 21, 23, 24, 26 | 23, 24, 25 | 22-N | |
| 23 | 30.1 | 2.45 m | 22, 24, 25 | 22, 24, 25 | 22-N | ||
| 24 | 17.0 | 0.80 d (7.1) | 23 | 22, 23, 25 | 22, 23 | ||
| 25 | 19.9 | 0.86 d (7.1) | 23 | 22, 23, 24 | 22, 23 | ||
| 22-N | 119.5 | 8.32 d (10.2) | 22 | ||||
| βMeASP | |||||||
| 26 | 176.8 | ||||||
| 27 | 39.8 | 3.19 dq (2.5, 7.0) | 28, 30 | 26/29 | 28, 30 | 28-N | |
| 28 | 57.7 | 4.39 d (2.5) | 27 | 27, 29, 30 | 27, 30 | ||
| 29 | 176.7 | ||||||
| 30 | 16.9 | 1.31 d (7.0) | 27 | 26, 27, 28 | 27, 28 | ||
| 28-N | 110.6 | 8.89 d (6.8) | |||||
| NMeΔBUT | |||||||
| 31 | 165.7 | ||||||
| 32 | 137.2 | ||||||
| 33 | 138.1 | 7.00 q (7.1) | 34 | 31, 32, 34 | 32-N | ||
| 34 | 13.4 | 1.77 d (7.1) | 33 | 31, 32 | 35 | ||
| 35 | 35.2 | 3.11 s | 32, 36 | 34 | 32-N | ||
| 32-N | 112.5 | ||||||
| GLU | |||||||
| 36 | 174.3 | ||||||
| 37a | 29.5 | 2.45 m | 37b, 38a, 38b | ||||
| 37b | 2.05 m | 37a, 38a | |||||
| 38a | 27.8 | 2.17 m | 38b, 39, 37a | 39-N | |||
| 38b | 2.05 m | 38a, 39 | |||||
| 39 | 52.0 | 4.69 dd (3.9, 5.7) | 38a, 38b | 38a, 38b/37b | |||
| 40 | 174.6 | ||||||
| 39-N | 122.2 |
Measured at 600 MHz (1H) (also see Figure S4) and 125 MHz (13C).
NH3 used as δ = 0 reference.
Subjecting 2 to acid hydrolysis (6 N HCl, 110 °C, 16 h), derivatization with Marfey's reagent,23 and subsequent HPLC analysis was used to identify S-valine and R-glutamic acid by comparison with standard samples,24 while the advanced Marfey technique25 (Figure S2) was used to identify 3S-Adda and R-β-methyl aspartic acid. These data justified 3S, 22S, 28R, and 39R assignments. An R-erythro-β-methyl aspartic acid was assigned on the basis of the identical H-27 to H-28 J's and δ's observed between 1 and 2, thereby indicating a 27S configuration. Likewise, identical 8S, 9S configurations for 2 versus 1 were deduced on the basis of their identical 1H NMR data at these sites. Finally, by default the C-2 configuration of Adda was considered to be R since all other stereocenters and double-bond geometries in 2 were determined by the above analysis to be identical to those of 1. Also supporting this assignment was that the largest 1H NMR shift difference between 2 and 1 was at H-2 (Δ = 0.29 ppm, Figure 2). It is important to note that the shift differences shown in Figure 2 for 2 versus 1 at H-37 to H-39 accompanied by differences in 3J38–39, and changes at H-28 and H-30 also accompanied by slight 3J27–28 differences, appear due to local conformational variations. Therefore, only the 2R configuration of 1 was inverted to 2S in 2.
The NMR properties of 2, shown in Table 2, served as an important reference point to enable a concise approach to establishing the structures of the other three related congeners. Thus, isomotuporin B (3) ([α]25D = −29.7), with a molecular formula of C41H59N5O10 established by HRESIMS (m/z 782.4358 [M + H]+), differed from 2 by +14 amu. The addition of a –CH2– unit was easily rationalized by observing signals of a sec-butyl group evident from the data in Table 3, consisting of a methyl triplet at δ 0.86 (J = 7.0 Hz), a methylene at δ 1.33 (m), and a methyl doublet at δ 0.91 (J = 6.7 Hz), which are not present in 1 or 2. Analysis of gHMBC data showed that in 3 an isoleucine side chain was in place of the valine side chain originally found in motuporin. All of the remaining 2D NMR data were unchanged for 3 versus 2, supporting the planar structure shown. Similar logic was used to establish the gross structure for 4 of molecular formula C39H55N5O10 established by HRESIMS (m/z 754.4090 [M + H]+). In this case there was a –14 amu difference relative to 2, indicating the loss of a –CH2– unit. The aliphatic methyl region in 3 also exhibited a methyl triplet at δ 0.86 (J = 7.5 Hz); however, analysis of gHMBC data showed that in 4 a C-22 ethyl group replaced the C-22 isopropyl group of 2.
Table 3. NMR Dataa for Compounds 3–5 in MeOD-d4.
| position | 3 | 4 | 5 | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | |
| ADDA | ||||||
| 1 | 176.0 | 176.1 | 176.1 | |||
| 2 | 45.1 | 2.77 dq (10.7, 6.7) | 45.3 | 2.71 dq (10.7, 6.9) | 45.4 | 2.73 dq (10.7, 6.7) |
| 3 | 55.9 | 4.58 dd (9.3, 10.5) | 56.2 | 4.56 dd (9.1, 10.7) | 56.1 | 4.58 dd (8.9, 10.6) |
| 4 | 126.3 | 5.53 dd (8.8, 15.5) | 126.4 | 5.47 dd (9.1, 15.4) | 125.7 | 5.51 dd (8.9, 15.6) |
| 5 | 139.2 | 6.26 d (15.5) | 139.4 | 6.25 d (15.4) | 139.6 | 6.28 d (15.7) |
| 6 | 133.9 | 133.9 | 133.8 | |||
| 7 | 137.1 | 5.43 d (9.8) | 137.4 | 5.43 d (10.1) | 140.9 | 5.34 d (9.5) |
| 8 | 37.7 | 2.59 ddq (6.6, 10.0, 6.7) | 37.9 | 2.59 ddq (6.9, 10.1, 6.8) | 33.4 | 2.49 m |
| 9 | 88.5 | 3.26 m | 88.5 | 3.26 m | 40.6 | 1.68 m 1.54 m |
| 10a | 39.0 | 2.82 dd (4.8, 13.8) | 39.2 | 2.82 dd (4.7, 13.9) | 35.0 | 2.55 m |
| 10b | 2.67 dd (7.2, 14.1) | 2.67 dd (7.3, 13.9) | ||||
| 11 | 140.6 | 140.6 | 140.4 | |||
| 12 | 130.6 | 7.19 m | 130.6 | 7.19 m | 129.5 | 7.13 m |
| 13 | 129.3 | 7.25 m | 129.4 | 7.25 m | 129.4 | 7.23 m |
| 14 | 127.1 | 7.18 m | 127.3 | 7.18 m | 126.8 | 7.12 m |
| 15 | 129.3 | 7.25 m | 129.4 | 7.25 m | 129.4 | 7.23 m |
| 16 | 130.6 | 7.19 m | 130.6 | 7.19 m | 129.5 | 7.13 m |
| 17 | 16.4 | 1.04 d (6.9) | 16.6 | 1.05 d (6.9) | 16.4 | 1.05 d (6.9) |
| 18 | 13.0 | 1.61 d (1.2) | 13.2 | 1.61 d (1.2) | 13.1 | 1.67 d (1.0) |
| 19 | 16.4 | 1.01 d (6.7) | 16.7 | 1.01 d (6.8) | 21.3 | 0.98 d (6.7) |
| 20 | 58.8 | 3.24 s | 58.9 | 3.24 s | ||
| Aba | Ile | Val | ||||
| 21 | n/d | n/d | n/d | |||
| 22 | 58.3 | 4.37 d (2.4) | 54.8 | 4.34 dd (3.6, 10.5) | 58.2 | 4.44 dd (4.1, 9.6) |
| 23 | 29.4 | 2.51 m | 25.4 | 2.04 m | 30.1 | 2.44 m |
| 1.43 ddq (5.6, 10.5, 7.5) | ||||||
| 24 | 12.6 | 0.91 d (6.7) | 10.9 | 0.86 t (7.5) | 16.8 | 0.80 d (6.9) |
| 25 | 23.8 | 1.33 m | 19.8 | 0.86 d (6.9) | ||
| 25-Me | 14.5 | 0.86 t (7.0) | ||||
| βMeASP | ||||||
| 26 | 176.9b | 176.8b | 177.5b | |||
| 27 | 40.0 | 3.18 m | 40.5 | 3.04 dq (1.9, 6.8) | 39.7 | 3.18 m |
| 28 | 58.3 | 4.44 d (3.8) | 57.9 | 4.41 d (1.9) | 57.7 | 4.39 dd (2.6, 6.9) |
| 29 | 176.7b | 176.7b | 176.7b | |||
| 30 | 16.7 | 1.31 d (6.9) | 16.9 | 1.31 d (6.8) | 16.8 | 1.30 d (6.9) |
| NMeΔBUT | ||||||
| 31 | 165.6 | 165.8 | 165.7 | |||
| 32 | 137.3 | 137.1 | 137.2 | |||
| 33 | 137.8 | 6.99 q (7.2) | 138.1 | 7.00 q (7.1) | 138.1 | 7.00 q (7.1) |
| 34 | 13.4 | 1.77 d (7.1) | 13.5 | 1.77 d (7.1) | 13.4 | 1.77 d (7.1) |
| 35 | 35.2 | 3.10 s | 35.4 | 3.11 s | 35.2 | 3.11 s |
| GLU | ||||||
| 36 | 173.6 | 174.4 | 174.2 | |||
| 37a | 29.4 | 2.53 m | 29.6 | 2.47 m | 29.4 | 2.46 m |
| 37b | 2.05 m | 2.06 m | 2.05 m | |||
| 38a | 27.7 | 2.18 m | 28.0 | 2.17 m | 27.8 | 2.18 m |
| 38b | 2.05 m | 2.05 m | 2.05 m | |||
| 39 | 52.5 | 4.67 dd (4.1, 5.7) | 52.1 | 4.70 dd (3.4, 5.4) | 52.1 | 4.69 dd (3.8, 5.7) |
| 40 | n/d | n/d | n/d | |||
Measured at 600 MHz (1H) (also see Figures S5–S7) and 125 MHz (13C).
Interchangeable; n/d = not detected; Aba = 2-aminobutyric acid.
Slightly greater differences were observed when comparing the properties of isomotuporin C (5) ([α]25D = −13.2) with those of 2. The molecular formula of C39H55N5O9 for 5 was established by HRESIMS (m/z 738.4023 [M + H]+). This differs from that of 2 by −30 amu, or a loss of an OCH2 residue. It was immediately evident that the C-9 OCH3 was replaced by an H because (a) no singlet was observed at δ 3.01, (b) the H-10 protons (δ 2.55 m) were isochronous, and (c) the DEPT spectrum showed C-9 (δ 40.6) was a CH2 group. Finally, analysis of gHMBC data confirmed that a C2H4 group connected the aryl ring to C-8. The absolute stereochemistry deduced here for 2 was provisionally assigned to 3–5. This is based on the parallel 1H chemical shifts and coupling constants shown for this set and the supposition that a parallel biosynthetic gene cluster was involved in the production of these metabolites.
Conclusions
Some of the findings outlined above will be relevant to those seeking further understanding of factors that control PKS-mediated production of the swinholides and mixed PKS–NRPS elaboration of motuporin. The selection of the proper sponge phenotype using the natural history information of Figure 1 is an essential first step to probe for the swinholide or motuporin pathways. To date, no populations of this sponge have been identified that have both pathways simultaneously expressed. Until now all compounds known containing the Adda amino acid, including (−)-nodularin and microcystin-LR (both of cyanobacterial origin) and motuporin (1), carried a consistent 2S stereochemistry. The discovery of distinct sponge populations that produce either 2S-motuporin (1) or 2R-motuporin (2) is significant. Additionally, the loss of the C-9 OCH3 in 5 is the first report of such a variation on the Adda side chain from natural sources. The results herein should now provide the impetus to stimulate further investigations on the ecology and molecular genetics of T. swinhoei.
Experimental Section
General Experimental Procedures
These are according to those previously published,26 except 1H NMR spectra were recorded on a Varian Inova 600 spectrometer operating at 599.8 MHz.
Animal Material
Specimens of Theonella swinhoei were collected in the waters near the following locations: coll. no. 94590, depth 90 ft, Sangihe Island, Indonesia; coll. nos. 94647 and 94648, Indonesia; coll. nos. 00342 and 00356, depth 60 ft, New Britain, Papua New Guinea; coll. nos. 00365 and 00399, depth 50 ft, New Britain, Papua New Guinea; coll. nos. 02141 and 02142, depth 50 ft, Normanby, Papua New Guinea; coll. no. 03506, depth 50 ft, Milne Bay, Papua New Guinea; coll. no. 05221, depth 50 ft, Rabaul and New Ireland, Papua New Guinea; coll. no. 05254, depth 50 ft, New Ireland, Papua New Guinea; coll. nos. 05259A and 05259B (blanched specimen), depth 55 ft, New Ireland, Papua New Guinea. Photographs of the sponges are available from the Crews laboratory.
Extraction and Isolation
All sponges were immediately preserved after collection according to our standard procedure14 and transported back to the laboratory at ambient temperature. Analytical reversed-phase LCMS using a gradient of 10:90 CH3CN/H2O to 100% CH3CN over 35 min was performed on organic solvent partition extracts14 (coll. nos. 94590, 94647, 94648, 00342, 00356, and 00399) and ASE solvent extracts26 (coll. nos. 02141, 02142, 03506, 05221, 05254, 05259A, and 05259B). The LCMS profiles of these extracts exhibited molecular ions (see Table 1) attributable to swinholide A (m/z 1389.8 [M + H]+), swinholides B, C, or G (m/z 1375.9 [M + H]+), swinholide E (m/z 1405.8 [M + H]+), swinhoeiamide A (m/z 797.5 [M + H]+), theonellamine B (m/z 1405.0 [M + H]+), theonellamide A (m/z 1765.4 [M + H]+), papuamide D (m/z 1385.7 [M + H]+), theopalauamide (m/z 1750.5 [M + H]+), misakinolide A (m/z 1338.0 [M + H]+), theoneberine (m/z 779.8 [M + H]+), theopederin J (m/z 626.4 [M + H]+), and motuporin/isomotuporin (m/z 768.4 [M + H]+). After ASE extraction of coll. no. 02142, the CH3OH extract was subjected to preparative RP HPLC (20:80 CH3CN/H2O to 100% CH3CN) to yield 11 fractions. Fractions 8–11 were further purified using semipreparative RP HPLC (65:35 CH3CN/H2O to 80:20 CH3CN/H2O) to yield 2 (10.4 mg), 3 (2.0 mg), 4 (2.2 mg), and 5 (3.0 mg).
Isomotuporin (2)
colorless solid, [α]25D −38.4 (c0.1, MeOH); UV (MeOH) λmax (log ε) 238 (4.48) nm; 1H and 13C NMR data, see Table 2; HRESIMS m/z 768.4122 [M + H]+ (calcd for C40H57N5O10 + H, 768.4105).
Isomotuporin B (3)
colorless solid, [α]25D −29.7 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 238 (4.48) nm; 1H and 13C NMR data, see Table 3; HRESIMS m/z 782.4358 [M + H]+ (calcd for C41H59N5O10 + H, 782.4335).
Isomotuporin C (4)
colorless solid, [α]25D −21.8 (c0.1, MeOH); UV (MeOH) λmax (log ε) 238 (4.48) nm; 1H and 13C NMR data, see Table 3; HRESIMS m/z 754.4090 [M + H]+ (calcd for C39H55N5O10 + H, 754.4022).
Isomotuporin D (5)
colorless solid, [α]25D −13.2 (c0.1, MeOH); UV (MeOH) λmax (log ε) 238 (4.48) nm; 1H and 13C NMR data, see Table 3; HRESIMS m/z 738.4023 [M + H]+ (calcd for C39H55N5O9 + H, 738.4073).
Supplementary Material
Acknowledgments
This research was supported by NIH grant RO1-CA047135 and NMR equipment grants from NSF CHE-0342912 and NIH S10-RR19918. Additional support was provided by the GAANN fellowship and the MBRS program. Special thanks to the crew and Skipper (C. DeWitt) of the M/V Golden Dawn for their assistance.
Footnotes
Supporting Information Available: 1H NMR comparison of motuporin (1) and isomotuporin (2), representation of gCOSY, gHMBC, and 15N-HMBC correlations for isomotuporin (2), advanced Marfey analysis chromatograms, 1H NMR spectra for 2–5, and structures for compounds in Table 1. Above and underwater Theonella sponges and photographs of all of the sponges studied. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Kho E, Imagawa DK, Rohmer M, Kashman Y, Djerassi C. J Org Chem. 1981;46:1836–1839. [Google Scholar]
- 2.(a) Carmely S, Kashman Y. Tetrahedron Lett. 1985;26:511–514. [Google Scholar]; (b) Kobayashi M, Tanaka J, Katori T, Matsuura Kitagawa I. Tetrahedron Lett. 1989;30:2963–2966. [Google Scholar]
- 3.Dilip de Silva E, Williams DE, Andersen RJ, Klix H, Holmes CFB, Allen TM. Tetrahedron Lett. 1992;33:1561–1564. [Google Scholar]
- 4.Bisabolene sesquiterpenes: Nakamura H, Kobayashi J, Hirata Y. Tetrahedron Lett. 1984;25:5401–5404.
- 5.Polyketides such as the swinholides: ref 2. The bitungolides: Sirirath S, Tanaka J, Ohtani II, Ichiba T, Rachmat R, Ueda K, Usui T, Osada J, Higa T. J Nat Prod. 2002;65:1820–1823. doi: 10.1021/np0200865.
- 6.Theonelladin pyridine alkaloids: Kobayashi J, Murayama T, Ohoizumi Y. Tetrahedron Lett. 1989;30:4833–4836.
- 7.Linear peptides headed by nazumamide A: Fusetani N, Nakao Y, Matsunaga S. Tetrahedron Lett. 1991;32:7073–7076.. The calyculin analogue swinhoeiamide A: Proksch P, Ebel R, Edrada RA, Schupp P, Lin WH, Sudarsono, Wray V, Steube K. Pure Appl Chem. 2003;75:343–352.
- 8.Forty-eight-residue-containing polytheonamides: see ref 13. The tridecapeptide lactones theonellapeptolides: Doi M, Ishida T, Kobayashi M, Katsuya Y, Mezaki Y, Sasaki M, Terashima A, Taniguchi T, Tanaka C. Biopolymers. 2000;54:27–34. doi: 10.1002/(SICI)1097-0282(200007)54:1<27::AID-BIP30>3.0.CO;2-3.
- 9.The cyclic polypeptides including motuporin: ref 3. Cyclotheonamides: Nakao Y, Matsunaga S, Fusetani N. Biorg Med Chem. 1995;8:1115–1122. doi: 10.1016/0968-0896(95)00106-q.. Orbiculamide A: Fusetani N, Matsunaga S. Chem Rev. 1993;93:1793–1806.. Cyclolithistide A: ref 14. Papuamides: Ford PW, Gustafson KR, McKee TC, Shigematsu N, Maurizi LK, Pannell LK, Williams DE, Dilip de Silva SE, Lassota P, Allen TM, van Soest R, Andersen RJ, Boyd MR. J Am Chem Soc. 1999;121:5899–5909.. The barangamides: Roy MC, Ohtanii II, Ichiba T, Tanaka J, Satari R, Higa T. Tetrahedron. 2000;56:9079–9092..
- 10.Two different glycopeptides, theonegramide: Bewley CA, Faulkner DJ. J Org Chem. 1994;59:4849–4852.. Theopalauamide: Schmidt EW, Bewley CA, Faulkner DJ. J Org Chem. 1998;63:1254–1258..
- 11.Pederin derivatives, theopederins: Tsukamato S, Matsunaga S, Fusetani N, Toh-E A. Tetrahedron. 1999;55:13697–13702.. The onnamides: Fusetani N, Sugawara T, Matsunaga S. J Am Chem Soc. 1991;113:7811–7812..
- 12.Unusual mixed biogenetic glycosides such as the lysoplasmanylinositols: Matsunaga S, Nishimura S, Fusetani N. J Nat Prod. 2001;64:816–818. doi: 10.1021/np0100439.. The aurantosides: see ref 13c.
- 13.See Figure 1 as follows: van Soest RWM. Neth J Sea Res. 1998;23:223–230. Type I (previously identified by R. W. M. van Soest). Hamada T, Matsunaga S, Fusetani N. J Am Chem Soc. 2005;127:110–118. doi: 10.1021/ja045749e. Type II (previously identified by R. W. M. van Soest). Ratnayake AS, Davis RA, Harper MK, Veltri CA, Andjelic CD, Barrows LR, Ireland CM. J Nat Prod. 2005;68:104–107. doi: 10.1021/np049721s. Type III (previously identified by M. K. Haper).
- 14.Clark DP, Carroll J, Naylor S, Crews P. J Org Chem. 1998;63:8757–8764. [Google Scholar]
- 15.(a) Bewley CA, Holland ND, Faulkner DJ. Experientia. 1996;52:716–722. doi: 10.1007/BF01925581. [DOI] [PubMed] [Google Scholar]; (b) Magnino G, Sara A, Lancioni T, Gaino E. Invert Biol. 1999;118:213–220. [Google Scholar]; (c) Schmidt EW, Obraztsova AY, Davidson SK, Faulkner DJ, Haygood MG. Mar Biol. 2000;136:969–977. [Google Scholar]; (d) Olson JB, McCarthy PJ. Aquat Microb Ecol. 2005;39:47–55. [Google Scholar]
- 16.(a) Kaebernick M, Neilan BA. FEMS Micro Ecol. 2001;35:1–9. doi: 10.1111/j.1574-6941.2001.tb00782.x. [DOI] [PubMed] [Google Scholar]; (b) Andrianasolo EH, Gross H, Goeger D, Musafija-Girt M, McPhail KP, Leal RM, Mooberry SL, Gerwick WH. Org Lett. 2005;7:1375–1378. doi: 10.1021/ol050188x. [DOI] [PubMed] [Google Scholar]
- 17.(a) Tsukamoto S, Ishibashi M, Sasaki T, Kobayashi J. J Chem Soc, Perkin Trans 1. 1991;12:3185–3188. [Google Scholar]; (b) Kobayashi M, Tanaka J, Katori T, Kitagawa I. Chem Pharm Bull. 1990;38:2960–2966. doi: 10.1248/cpb.38.2960. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura H, Kobayashi J, Nakamura Y, Ohizumi Y, Kondo T, Hirata Y. Tetrahedron Lett. 1986;27:4319–4322. [Google Scholar]
- 19.Matsunaga S, Fusetani N. J Org Chem. 1995;60:1177–1181. [Google Scholar]
- 20.Sakai R, Higa T, Kashman Y. Chem Lett. 1986;9:1499–1502. [Google Scholar]
- 21.Kobayashi J, Kondo K, Shigemori H, Ishibashi M, Sasaki T, Mikami Y. J Org Chem. 1992;57:6680–6682. [Google Scholar]
- 22.Valentekovich RJ, Schreiber SL. J Am Chem Soc. 1995;117:9069–9070. [Google Scholar]
- 23.Marfey P. Carlsberg Res Commun. 1984;49:591–596. [Google Scholar]
- 24.Our attempts at a side-by-side comparison of motuporin (1) and isomotuporin (2) were unsuccessful, as the sample previously isolated in our lab had decomposed and no sample was available from the Andersen (see ref 3) or Schreiber labs (see ref 22)
- 25.(a) Fujii K, Ikai Y, Mayumi T, Oka H, Suzuki M, Harada K. Anal Chem. 1997;69:3346–3352. [Google Scholar]; (b) Fujii K, Ikai Y, Oka H, Suzuki M, Harada K. Anal Chem. 1997;69:5146–5151. [Google Scholar]
- 26.Wegerski CJ, Sonnenschein RN, Cabriales F, Valeriote FA, Matainaho T, Crews P. Tetrahedron. 2006;62:10393–10399. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


