Abstract
Correlations among low-frequency spontaneous fluctuations in the blood oxygen level-dependent (BOLD) signal reflect the connectivity of intrinsic large-scale networks in the brain. These correlations have typically been characterized over the entire timecourse (mean connectivity), but the mean correlations between regions vary dynamically. By focusing on the linear relationship between activity in network nodes within the default mode network (DMN), dorsal attention network (DAN), and fronto-parietal task control network (FPTC) captured by their inter-correlations, we demonstrate that this dynamic pattern of fluctuations reveals a detailed substructure, that this substructure is robust across individuals, and that the expression of specific factors is correlated with age. To do this, we conducted a chained P-technique factor analysis of the correlations in nonoverlapping temporal windows across N=145 normal aging subjects (age 56–89). The expression of factors within the DMN, FPTC, and DAN was highly correlated with age: Decreased intercorrelations within nodes in each factor were correlated with advanced age. Although these findings converge with those from stationary analysis, the ability to quantify age-related changes in the coherence of fluctuating connectivity may yield more insights into age-related cognitive decline.
Key words: : aging, default mode network, dynamic connectivity, fMRI, functional connectivity, resting state
Introduction
Higher function (language, attention, consciousness, etc.) is neither the result of activity that is strictly localized in specific neural structures, nor the result of the brain as a whole, but it emerges from the dynamics of distributed cortical regions, with each relatively specialized for one or more aspects of the function. The composition of such systems is fundamentally enabled by patterns of anatomical connectivity, but it shifts dynamically and individual cortical fields are involved in multiple distributed systems (Chang and Glover, 2010; Smith et al., 2012). Thus, the constructs with which we describe higher function cannot be assigned directly to anatomic structures in the brain. It is at the systems level that anatomy, physiology, and function have a direct correspondence, and it is at this level that the phenomena of neuropsychology and cognitive psychology can presumably be explained.
A key insight of recent studies is that spontaneous brain activity (i.e., in absence of task engagement) measured using functional magnetic resonance imaging (fMRI) can be used to map large-scale systems. The connectivity of cortical fields is reflected through mean correlations in the low-frequency oscillations in blood oxygen level dependent (BOLD) MRI time courses of the regions, which are conveniently obtained during a resting-state scan. Connectivity maps may be obtained by correlating the timecourse in a region of interest with other voxels, or by using the data-driven independent components analysis (ICA) approach (Beckmann and Smith, 2004; Beckmann et al., 2005). The strength of correlation between regions is thought to be related to the strength and efficiency of communication. Evidence from a variety of subject populations supports this: A disruption of “normal” patterns of mean correlations obtained during functional scans (mean connectivity) has been related to aging (Andrews-Hanna et al., 2007; Grady et al., 2009), Alzheimer's disease (Grady et al., 2001; Greicius et al., 2004; Wang et al., 2007), and a variety of neuropsychiatric disorders (Hutchison et al., 2013). Functional connectivity is a new imaging tool that provides unique information about systems-level brain function which is not obtainable through structural connectivity or metabolic imaging.
Although intrinsic networks can be detected reliably in groups and in healthy individuals (Damoiseaux et al., 2006), mean network activity is only a summary of a more complicated pattern of dynamic network configuration (Smith et al., 2012) that is not yet well characterized or understood. The strength of functional connections varies not only between scans but also in the order of seconds to minutes (Allen et al., 2012; Hutchison et al., 2012).
The importance of dynamic intrinsic brain activity is only recently being appreciated through correlation with behavior. For example, Fox and Raichle (2007) demonstrate that intrinsic activity in the sensorimotor cortex is related to the force of finger tapping. It is widely supposed that dynamic aspects of connectivity reflect physiologic shifts in network configuration. For example, Bassett et al. (2011) demonstrate that flexibility of network nodes while learning a motor task in one session is predictive of learning in a subsequent session. A competitive relationship between a task-positive and a task-negative brain network is related to intraindividual variability (Kelly et al., 2008). The ability of the default mode network (DMN) to dynamically reconfigure itself to facilitate integration with an attentional network is related to recollection performance (Fornito et al., 2012).
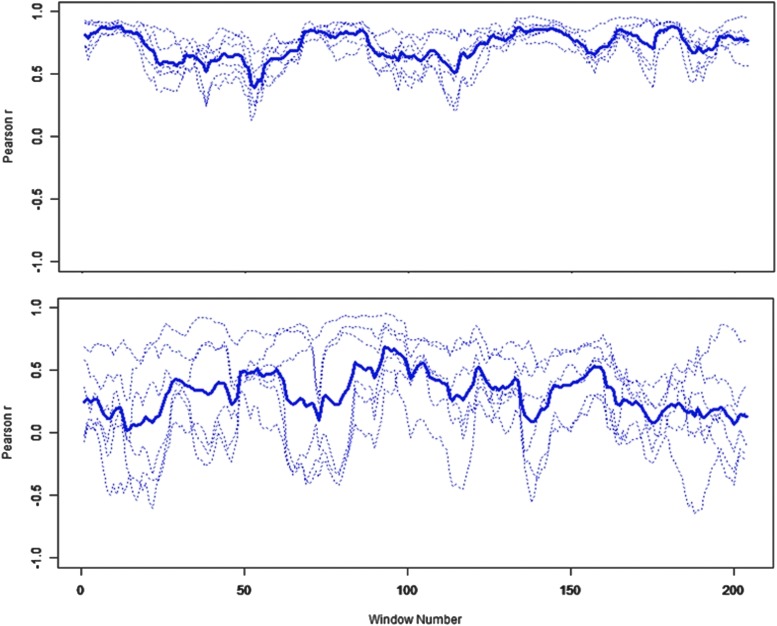
Network dynamics will vary not only with task modulation, but also with the underlying structural network, regional cortical noise (e.g., caused by alterations in dopaminergic tone, age-related sensory decline, and vascular insufficiency), and delay (caused by white matter microstructure damage and demyelinization). Figure 1, which we will describe more completely in our results, shows the pattern of time-varying pairwise correlations within a subset of nodes in the DMN for a middle-aged subject and an older subject. The mean correlation for the middle-aged subject, shown by the heavy blue line, is obviously higher than that of the older subject, and any analysis of the mean connectivity throughout the scan would detect such reliable age-related differences. Dynamic fluctuations of connectivity and mean measures of connectivity obtained from longer intervals are clearly highly correlated and closely related. However, we suggest that the more striking feature of this figure is that the connectivity among the nodes for the middle-aged subject fluctuates more tightly than the pattern of connectivity among the nodes for the older subject. This is an abstraction that describes the changing behavior of systems and subsystems through time. It may be a more useful abstraction for quantifying the effect of specific age-related processes.
FIG. 1.
Time-varying pairwise correlations among medial prefrontal cortex (mPFC), left angular gyrus (LAG), right angular gyrus (RAG), and posterior cingulate cortex (PCC) [default mode network (DMN) factor 3] at rest in sliding window of 40 sec (slide width=2 sec). Bold line is mean of correlations. At the bottom is an older subject (88 years). On top is a middle-aged subject (64 years). The older subject has more dynamic variability among correlations in this factor of the DMN.
Selective reduction of inter-correlations within reproducible subgraphs of intrinsic networks may be considered a reduction in synchronization that occurs with advanced age, due to the age-related processes described earlier. Some of this desynchronization may be related to neuronal activity: Evidence from computational modeling and electrocorticography suggests that the oscillations observable as mean correlations in the BOLD signal and low-frequency resting-state networks stem from neuronal oscillations in the gamma band in structurally connected cortical fields (Cabral et al., 2011; Deco et al., 2009; Grady et al., 2001; He et al., 2008; Ko et al., 2011). It has also been demonstrated that although resting-state networks are dominated by low frequencies in the raw BOLD signal, they are actually broadband processes which show temporal coherence over a wide frequency spectrum (Niazy et al., 2011), suggesting that higher frequencies visible within smaller timeframes may contribute to resting-state connectivity.
Analyzing time-varying correlations, as opposed to the level of signal, restricts analysis to the linear relationship between nodes, and might be more sensitive to changes in the relationship between specific brain regions. This approach has been used to develop a data-driven method for determining “eigenconnectivities” using principal components analysis (PCA) of fluctuating correlations in the brain, showing that specific eigenconnectivities differ in patients with multiple sclerosis (Leonardi et al., 2013).
Our hypothesis is that there should be a similarity in time-varying correlations within subsets of network nodes across individuals, which we call “structured variability.” This has not previously been shown. If structured variability within intrinsic networks reflects an aspect of normal systems-level physiology, we should expect that age-related desynchronization would be reflected in an alteration of the structured variability. Older subjects should exhibit greater desynchronization, or “unstructured variability,” within specific subgroups of these nodes (which are more vulnerable to age, or that have connections which are more vulnerable to age).
Methods
Participants
Data were obtained from participants in the Seattle Longitudinal Study (SLS), a cohort-sequential longitudinal study of the relationship between aging, health, cognition, and lifestyle (Schaie, 2005). The SLS members at recruitment represent a stratified-by-age and gender random sample of the membership of the Group Health Cooperative of Puget Sound, a large health maintenance organization in western Washington State. All subjects are cognitively normal. The group is ethnically homogenous: 97.2% of this sample is Caucasian, reflecting the demographics of Seattle at the time of recruitment. Cognitive and behavioral assessments have been conducted every 7 years starting in 1956 on a mixed age cohort (age 20–80) with follow-up and recruitment of new subjects every 7 years (1956 through 2005). This study has been approved by the Group Health Cooperative of Puget Sound Institutional Review Board.
The sample included 145 participants, Mage=69 (age range 56–89); Nmales=64, Nfemales=81 in the SLS at time of the third MRI (Table 1). The SLS neuroimaging group was selected from a larger group of SLS participants (n=572) who (i) had two to three assessments of episodic memory over a 14-year interval during midlife (ages ranged from 43 to 63 years); (ii) participated in the 2005 SLS data collection; and (iii) if in old age, had at least one assessment in old age (>64 years). Subjects were excluded for conditions that would contraindicate scanning (e.g., metal implants, pacemakers, and claustrophobia). Structural and functional image data were obtained on all subjects in 2010–2011. Table 1 shows the demographic information for the sample.
Table 1.
Demographics of Sample
| N | 145 |
| Age at first scan | 69 (7.8) |
| Sex (number males) | 64 (44%) |
| Education (years) | 16 (2.5) |
SD or percentage is indicated in parentheses.
MRI acquisition and processing
MRI was performed on a Philips 3.0 T Achieva scanner using an 8-channel head coil. A high-resolution MPRAGE was obtained using the following parameters: inversion time (TI)=850 msec, turbo-field echo (TFE) factor=214, repetition time (TR)=7 msec, echo time (TE)=3.20 msec, flip angle=8°, shot interval=3000 msec, acquisition matrix size 224×214, reconstructed matrix size 256×256 (field of view=220×220 mm), 160 sagittal slices, and a slice thickness of 1 mm. Functional images were obtained using the following parameters: 43 axial slices, slice thickness 3.5 mm, repeat time (TR) 2000 msec, TE=21 msec, acquisition matrix=64×64, voxel size=3.50 mm isotropic, FOV=220×220 mm, and volumes=225 (7.5 min). A B0 field map was acquired immediately after functional imaging.
All subjects underwent a resting-state scan, for which they were instructed to keep their eyes open and focus on a visual fixation cross. Cardiac and respiratory processes were monitored using the scanner's built-in photoplethysmograph that was placed on the index finger of the right hand and a pneumatic belt which was strapped around the upper abdomen. Data were sampled at 50 Hz, and files containing cardiac and respiratory waveform data were generated for each scan.
Functional images were processed using software from FSL (Jenkinson et al., 2011), FreeSurfer (Fischl and Dale, 2000), and AFNI (Cox, 1996). Data were corrected for magnetic field inhomogenities using the B0 map, and corrected for motion using FSL MCFLIRT (Jenkinson et al., 2002). Mean relative and absolute displacement were calculated for each subject. The resting-state pipeline regressed out noise from cardiac and respiratory signals (Glover et al., 2000), removed spikes using AFNI, performed slice timing correction using FSL, and regressed out time series motion parameters and the mean signal for eroded (1 mm in 3D) masks of the lateral ventricles and white matter (derived from FreeSurfer). Three-dimensional spatial smoothing was performed using a Gaussian kernel with a full-width half maximum of sigma=3 mm. Data were not normalized as a part of the pipeline. We did not perform bandpass filtering to avoid artificially inflating correlations or inducing structure that was not actually present in the data, and because resting-state networks exhibit different levels of phase synchrony at different frequencies (Handwerker et al., 2012; Niazy et al., 2011).
Statistical analysis
We selected Montreal Neurological Institute (MNI) coordinates that have been identified as nodes in the DMN, dorsal attention network (DAN), and fronto-parietal task control network (FPTC), republished by Power et al. (2011) and derived from Raichle (2001) and Dosenbach et al. (2007) (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/brain). For each coordinate, we created a 10-mm diameter mask in standard space and transformed that to subjects' native space to calculate mean subject-specific timecourses for each ROI.
Dynamic measures
Correlations change dynamically throughout the timecourse. We calculated pairwise correlations between nodes in each network in nonoverlapping windows of lengths 10 frames, 20 frames, 40 frames, and the entire scan (20, 40, 80, and 450 sec). There is no physiological motivation for these window lengths; the choice of window length is based on the desire to obtain reliable correlations while quantifying the individual variability throughout the scan. Earlier work has suggested that window lengths of ∼30 sec satisfy these criteria (Jones et al., 2012). The use of a single window per scan does not permit modeling of any individual variability throughout the scan, but enables us to test the stability of the structure obtained in smaller windows. These correlations were transformed by Fisher's z-transform to convert them to a normally distributed variable for subsequent factor analysis using Revolution R Enterprise (www.revolutionanalytics.com). We used all complete windows starting from the beginning of the scan. The first five dummy volumes in our fMRI acquisition are automatically discarded to achieve steady-state imaging. For each 7.5 min scan, this yielded 11 data points per subject for 40 sec nonoverlapping windows (22 data points per subject using windows of 10 frames, 5 data points per subject using windows of 40 frames, and 1 data point per subject using the entire scan).
Factor analysis
Factor analysis is used to identify the structure underlying groups of correlated variables and to estimate scores to measure latent factors. Factor analysis is conceptually similar to PCA except that the factor analysis model does not extract all the variance among the observed variables; it extracts only the variance which is shared by the observed variables and is due to the common factors (Gorsuch, 1983). We use factor analysis, because we hypothesize a latent structure to fluctuating correlations, which we are treating as measurements of connectivity, and because we may, ultimately, be able to leverage methods drawn from confirmatory factor analysis to verify factor structure. We conducted an exploratory factor analysis using chained P-technique factor analysis on the pairwise correlations between nodes on all individuals for each network to obtain a factor structure. P-technique factor analysis (Cattell, 1963) applies the common factor model to multivariate repeated measures of one individual obtained over many occasions. Here, we concatenate subject data, a method called chained P technique (Cattell, 1966), and apply varimax rotation to the factor-loading matrix. Varimax rotation is an orthogonal rotation that tries to create a loading matrix with a simple structure (a pattern of loadings where items load most strongly on one factor, and more weakly on the other factors) (Kaiser, 1958). The resulting factors represent subgroups of correlations that covary in time across all individuals. The factors represent the structure of the fluctuating correlations (e.g., subnetworks). A factor score is obtained for each window for each factor; we average these scores for each factor for each individual to obtain individual-level mean factor scores. Scores represent the expression of each factor (e.g., how tightly coupled the correlations are within the subnetwork). When testing the relationship between age and factor scores, we report uncorrected significance values, applying Bonferroni correction for multiple comparisons over all comparisons in all networks.
Optimal numbers of factors for this analysis were determined by establishing factor reliability through a split-half validity analysis. Our goal was to identify a subset of factors that are robust across individuals. Given the complexity of the factor structure, we used an exploratory procedure described by McCrae et al. (1996) both to select an appropriate number of factors to extract and to conduct a split-half reliability analysis. This procedure enables the exploration of factor reliability in an exploratory framework, rather than a confirmatory structural equation modeling framework, and may be more appropriate for replication of complex factor structures. Beginning by factors with eigenvalues greater than one (the Kaiser criterion); we conducted a factor analysis as described earlier for each half of a random group split. We then rotated the factor loadings of the second half to match the first using orthogonal Procrustes rotation. This procedure maximizes the size of the total congruence coefficient by optimally aligning real factors. We then computed the congruence of the factors and the significance of the Procrustes statistic. If the factor congruence was not greater than 0.8 in all but one factor, we reduced the number of extracted factors. A typical rule of thumb is that a factor congruence of 0.8–0.95 is a practical lower bound to define a factor, although there is no firm rule (Barrett, 1986). We repeated this reliability analysis on successively smaller numbers of factors to identify a set of reproducible factors within our sample for subsequent analysis.
To determine whether the temporal fluctuations that we observe in our factor structure could occur by chance, we randomly shuffled the nonoverlapping windows for each ROI and compared the congruence of the factor structure with the original solution after Procrustes rotation. This approach retains the local time series information within each window while destroying the long-range dynamic relationships.
R scripts for these analyses are available from the corresponding author on request.
Results
Inter-regional connectivity is correlated with age and fluctuates in time
Table 2 shows minimum, maximum, and mean correlations, computed using nonoverlapping windows of 40 sec, for each pair of ROIs across individuals. A window size of 40 sec reveals a large range in average correlation strength. We tested for interactions between window length and age, or window position within the scan and age, but did not find any significant interactions that would make correlations with age systematically different in different window sizes or observations. Table 2 also shows the correlation of the connectivity strength (correlation over the entire scan) with age. The strength of connectivity between many regions is significantly negatively correlated with age. Our subsequent factor analysis explores whether connectivity between these regions fluctuates independently or in distinct sub-networks, and whether all sub-networks are equally vulnerable to age.
Table 2.
Dynamic Fluctuation of Inter-Regional Connectivity and Relationship to Age
| Edge | r (age) | p | Min | Max | Mean |
|---|---|---|---|---|---|
| DAN | |||||
| LaIPS.LFEF | −0.16 | 0.055 | 0.03 | 0.77 | 0.46 |
| LaIPS.LpIPS | −0.12 | 0.162 | 0.19 | 0.84 | 0.58 |
| LaIPS.RaIPS | −0.28 | 0.001a | 0.13 | 0.80 | 0.52 |
| LaIPS.RFEF | −0.25 | 0.003 | −0.04 | 0.75 | 0.41 |
| LaIPS.RpIPS | −0.09 | 0.259 | 0.08 | 0.81 | 0.51 |
| LFEF.LpIPS | −0.15 | 0.065 | −0.05 | 0.72 | 0.38 |
| RaIPS.LFEF | −0.26 | 0.001 | −0.09 | 0.70 | 0.36 |
| RaIPS.LpIPS | −0.22 | 0.009 | −0.04 | 0.76 | 0.41 |
| RaIPS.RFEF | −0.17 | 0.044 | −0.01 | 0.75 | 0.42 |
| RaIPS.RpIPS | −0.08 | 0.367 | 0.07 | 0.80 | 0.49 |
| RFEF.LFEF | −0.21 | 0.012 | 0.09 | 0.77 | 0.48 |
| RFEF.LpIPS | −0.26 | 0.002 | −0.10 | 0.70 | 0.35 |
| RFEF.RpIPS | −0.14 | 0.089 | −0.06 | 0.71 | 0.38 |
| RpIPS.LFEF | −0.07 | 0.376 | −0.11 | 0.71 | 0.36 |
| RpIPS.LpIPS | −0.24 | 0.003 | 0.32 | 0.88 | 0.66 |
| DMN | |||||
| LAG.mPFC | −0.30 | 0.000a | −0.07 | 0.73 | 0.39 |
| LAG.PCC | −0.17 | 0.040 | 0.16 | 0.83 | 0.56 |
| Llattemp.LAG | −0.01 | 0.909 | −0.26 | 0.60 | 0.20 |
| Llattemp.mPFC | −0.28 | 0.001a | −0.27 | 0.59 | 0.19 |
| Llattemp.PCC | −0.09 | 0.262 | −0.31 | 0.58 | 0.17 |
| Llattemp.RAG | 0.01 | 0.897 | −0.32 | 0.57 | 0.13 |
| mPFC.PCC | −0.40 | 0.000a | −0.05 | 0.72 | 0.39 |
| RAG.LAG | −0.19 | 0.019 | 0.17 | 0.84 | 0.57 |
| RAG.mPFC | −0.30 | 0.000a | −0.12 | 0.69 | 0.33 |
| RAG.PCC | −0.10 | 0.249 | 0.10 | 0.82 | 0.54 |
| Rlattemp.LAG | −0.05 | 0.567 | −0.33 | 0.57 | 0.14 |
| Rlattemp.Llattemp | −0.13 | 0.120 | 0.10 | 0.79 | 0.50 |
| Rlattemp.mPFC | −0.17 | 0.040 | −0.28 | 0.57 | 0.18 |
| Rlattemp.PCC | −0.19 | 0.024 | −0.33 | 0.57 | 0.14 |
| Rlattemp.RAG | −0.05 | 0.543 | −0.32 | 0.58 | 0.15 |
| FPTC | |||||
| LdlPFC.RdlPFC | −0.13 | 0.119 | −0.07 | 0.74 | 0.40 |
| Lfrontal.LdlPFC | −0.15 | 0.071 | 0.11 | 0.81 | 0.53 |
| Lfrontal.RdlPFC | −0.18 | 0.032 | −0.11 | 0.72 | 0.36 |
| Lfrontal.Rfrontal | −0.19 | 0.019 | −0.13 | 0.72 | 0.34 |
| LIPL.LdlPFC | −0.06 | 0.492 | −0.06 | 0.73 | 0.39 |
| LIPL.Lfrontal | −0.09 | 0.292 | −0.13 | 0.72 | 0.35 |
| LIPL.RdlPFC | −0.12 | 0.155 | −0.11 | 0.71 | 0.35 |
| LIPL.Rfrontal | −0.01 | 0.915 | −0.18 | 0.68 | 0.29 |
| LIPL.RIPL | −0.09 | 0.259 | −0.03 | 0.76 | 0.42 |
| LIPS.LdlPFC | −0.26 | 0.002 | −0.01 | 0.77 | 0.45 |
| LIPS.Lfrontal | −0.26 | 0.002 | 0.00 | 0.77 | 0.46 |
| LIPS.LIPL | 0.05 | 0.570 | 0.11 | 0.81 | 0.52 |
| LIPS.RdlPFC | −0.29 | 0.000a | −0.06 | 0.74 | 0.39 |
| LIPS.Rfrontal | −0.14 | 0.086 | −0.10 | 0.73 | 0.37 |
| LIPS.RIPL | −0.15 | 0.081 | −0.10 | 0.73 | 0.38 |
| LIPS.RIPS | −0.28 | 0.001a | 0.15 | 0.83 | 0.55 |
| Rfrontal.LdlPFC | −0.10 | 0.242 | −0.19 | 0.68 | 0.29 |
| Rfrontal.RdlPFC | −0.01 | 0.865 | 0.06 | 0.81 | 0.50 |
| RIPL.LdlPFC | −0.14 | 0.097 | −0.19 | 0.68 | 0.29 |
| RIPL.Lfrontal | −0.20 | 0.014 | −0.21 | 0.65 | 0.26 |
| RIPL.RdlPFC | −0.31 | 0.000a | −0.17 | 0.69 | 0.32 |
| RIPL.Rfrontal | −0.20 | 0.015 | −0.24 | 0.63 | 0.23 |
| RIPS.LdlPFC | −0.17 | 0.039 | −0.18 | 0.68 | 0.29 |
| RIPS.Lfrontal | −0.22 | 0.007 | −0.13 | 0.70 | 0.35 |
| RIPS.LIPL | 0.13 | 0.106 | −0.16 | 0.69 | 0.31 |
| RIPS.RdlPFC | −0.32 | 0.000a | −0.12 | 0.72 | 0.35 |
| RIPS.Rfrontal | −0.15 | 0.078 | −0.14 | 0.70 | 0.32 |
| RIPS.RIPL | 0.11 | 0.207 | 0.00 | 0.76 | 0.45 |
Min, max, and mean correlation strength is computed on 40 sec windows and averaged across individuals. Correlation with age is computed using mean correlation from entire scan. Values in bold represent significant correlations with age at p<0.05.
Values marked with (a) survive Bonferonni correction for multiple comparisons.
DMN, default mode network; FPTC, fronto-parietal task control network; DAN, dorsal attention network; PCC, posterior cingulate cortex; LAG, left angular gyrus; RAG, right angular gyrus; dlPFC, dorsal lateral prefrontal cortex; mPFC, medial prefrontal cortex; LFEF, left frontal eye field; RFEF, right frontal eye field; LaIPS, left anterior intraparietal sulcus; RaIPS, right anterior intraparietal sulcus; LpIPS, left posterior intraparietal sulcus; RpIPS, right posterior intraparietal sulcus.
Factor structure of intrinsic networks is robust
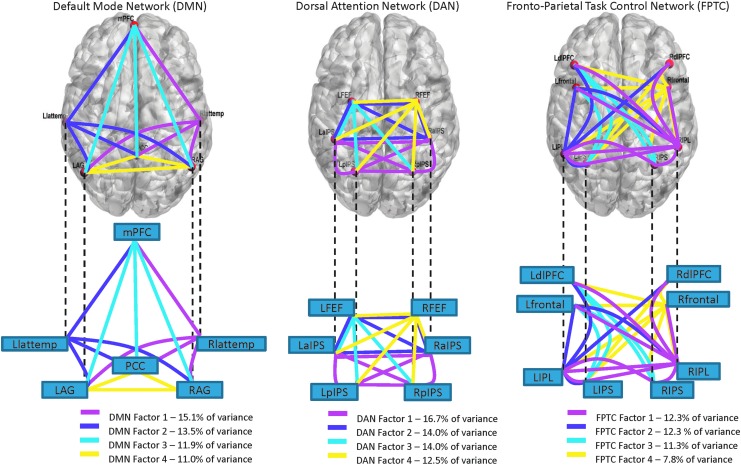
For the DMN and DAN, both eigenvalues and our procedure using Procrustes rotation yielded four factors. The structure of the FPTC is more variable: There are seven eigenvalues greater than one, but, ultimately, only four reliable factors could be extracted. The remaining three factors vary significantly by population (i.e., factor correspondence was low). Figure 2 shows the factor structure for time-varying intercorrelations in these three networks, where lines indicate loadings greater than 0.4 for the entire sample. Factor loadings for the entire sample, using a window size of 40 sec, are shown in Supplementary Tables S2–S4.
FIG. 2.
Factor structure (showing factors with loadings >0.4) of the DMN, dorsal attention network (DAN), and fronto-parietal task control network (FPTC).
Factor analysis results for window sizes of 20 and 80 sec and the entire scan are similar to the solution obtained at 40 sec. We quantify the similarity by rotating the solutions at different window sizes to maximum congruence with the 40 sec solution and calculating the Procrustes correlations: DMN20=0.99, DMN80=0.99, DMN450=0.98, DAN20=0.99, DAN80=0.98, DAN450=0.92, FPTC20=0.99, FPTC80=0.95, and FPTC450=0.83. The structure is virtually identical, as indicated by the extremely high correlations, at window sizes of 20 and 80 sec; so, we report results from window sizes of 40 sec. We note that the congruence of the DMN factor structure derived from the full scan (450 sec) is almost identical to the DMN structure derived from smaller timescales. The task-positive network factor structures derived from the full scan are quite similar to those derived from the shorter windows in the task-positive networks, although slightly less so than the DMN. This suggests that there is more individual variability in task-positive networks, so that long windows might obscure dynamic patterns which are visible at smaller time windows.
In the DMN, the factor accounting for the most variance (15.1%) consists of inter-correlations of the posterior cingulate cortex (PCC), the left and right angular gyrus (LAG and RAG), and the medial prefrontal cortex (mPFC) with the right lateral temporal lobe (Rlattemp). The second largest factor (13.5% of the variance) is the symmetric opposite. The third largest factor (11.9% of variance) connects the mPFC to the PCC, the LAG, and the RAG (the frontal part of the DMN). The last factor (11% of variance) connects the PCC with the LAG and RAG, and it connects the RAG with the LAG (the posterior part of the DMN).
In the DAN, the factor accounting for the most variance (16.7%) consists of intercorrelations among the left and right anterior intraparietal sulcus (LaIPS and RaIPS) and the left and right posterior intraparietal sulcus (LpIPS and RpIPS). The second largest factor (14.0%) consists of correlations between the left frontal eye field (LFEF) and the LaIPS, LpIPS, and RpIPS. The third factor (14.0% of variance) consists of correlations between the right frontal eye field (RFEF) and the RpIPS, LpIPS, and LaIPS. The fourth factor (12.5% of variance) loads on all intercorrelations between the left and right FEFs and the left and right aIPS.
In the FPTC, the first factor (12.3% of variance) connects the right IPL and the right and left dorsal lateral prefrontal cortex (dlPFC), RIPS, LIPS, LIPL, and left and right frontal. It also loads on intercorrelations between the RIPS and the LIPL and the LIPL and the LIPS. The second factor (12.3% of variance) is dominated by connections between the right frontal and all other nodes. In addition, this factor loads on the correlation between the right dlPFC and the LIPS. The third factor (11.3%) is nearly a left-handed subset of factor two, including connections between the left frontal and LIPS, RIPS, and left dlPFC, with a link between the left dLPFC and the LIPS. The fourth factor (7.8% of variance) includes correlations between the LIPL and the left and right dlPFC, left frontal, and LIPS (a left-handed subset of factor 1).
The factor structures obtained from an analysis of the whole sample were reproducible in a split-half analysis with high congruence (Procrustes correlations for the DMN=0.89, DAN=0.90, and FPTC=0.83). Tables 3–5 give the factor loadings for the split-half analysis.
Table 3.
Split-Half Validity Results for DMN
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 1 | Factor 2 | Factor 3 | Factor 4 | |
|---|---|---|---|---|---|---|---|---|
| DMNPCC.DMNRlattemp | 0.827 | 0.158 | 0.060 | 0.080 | 0.691 | 0.381 | 0.141 | 0.075 |
| DMNLAG.DMNRlattemp | 0.626 | 0.405 | 0.086 | 0.086 | 0.800 | 0.103 | 0.062 | 0.124 |
| DMNRAG.DMNRlattemp | 0.711 | 0.192 | 0.054 | 0.153 | 0.690 | 0.211 | −0.022 | 0.105 |
| DMNPCC.DMNLlattemp | 0.408 | 0.540 | 0.121 | 0.070 | 0.269 | 0.992 | 0.210 | 0.100 |
| DMNLAG.DMNLlattemp | 0.193 | 0.966 | 0.125 | 0.099 | 0.372 | 0.547 | 0.094 | 0.110 |
| DMNPCC.DMNmPFC | 0.114 | 0.103 | 0.720 | 0.200 | 0.111 | 0.114 | 0.754 | 0.189 |
| DMNmPFC.DMNLAG | 0.106 | 0.118 | 0.780 | 0.174 | 0.128 | 0.027 | 0.757 | 0.223 |
| DMNmPFC.DMNRAG | 0.161 | 0.021 | 0.623 | 0.302 | 0.148 | 0.018 | 0.650 | 0.312 |
| DMNPCC.DMNLAG | 0.059 | 0.181 | 0.271 | 0.570 | 0.015 | 0.210 | 0.261 | 0.530 |
| DMNPCC.DMNRAG | 0.096 | 0.018 | 0.090 | 0.822 | 0.083 | 0.087 | 0.166 | 0.668 |
| DMNLAG.DMNRAG | 0.030 | 0.083 | 0.189 | 0.581 | 0.097 | 0.070 | 0.156 | 0.651 |
| DMNRAG.DMNLlattemp | 0.341 | 0.537 | 0.090 | 0.227 | 0.320 | 0.498 | 0.054 | 0.216 |
| DMNmPFC.DMNLlattemp | 0.285 | 0.322 | 0.359 | 0.013 | 0.244 | 0.386 | 0.272 | 0.001 |
| DMNmPFC.DMNRlattemp | 0.503 | 0.119 | 0.246 | −0.073 | 0.536 | 0.124 | 0.235 | −0.066 |
| DMNLlattemp.DMNRlattemp | 0.224 | 0.069 | 0.161 | 0.043 | 0.180 | 0.063 | 0.185 | 0.109 |
Congruence of factors is 0.94, 0.72, 0.98, and 0.98 (correlation=0.89, p=0.001).
Loadings >0.4 are shaded.
Table 4.
Split-Half Validity Results for DAN
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 1 | Factor 2 | Factor 3 | Factor 4 | |
|---|---|---|---|---|---|---|---|---|
| DANLpIPS.DANRaIPS | 0.190 | 0.519 | 0.622 | 0.038 | 0.107 | 0.613 | 0.517 | 0.071 |
| DANLpIPS.DANLaIPS | 0.214 | 0.581 | 0.179 | 0.131 | 0.208 | 0.585 | 0.247 | 0.232 |
| DANRpIPS.DANRaIPS | 0.087 | 0.374 | 0.533 | 0.205 | 0.133 | 0.452 | 0.466 | 0.046 |
| DANRpIPS.DANLaIPS | 0.142 | 0.575 | 0.201 | 0.264 | 0.237 | 0.444 | 0.396 | 0.187 |
| DANRaIPS.DANLaIPS | 0.291 | 0.219 | 0.696 | 0.019 | 0.251 | 0.223 | 0.651 | 0.016 |
| DANLpIPS.DANLFEF | 0.353 | 0.375 | 0.077 | 0.550 | 0.318 | 0.450 | −0.036 | 0.777 |
| DANLFEF.DANRpIPS | 0.249 | 0.315 | 0.132 | 0.871 | 0.276 | 0.302 | 0.120 | 0.668 |
| DANLFEF.DANLaIPS | 0.451 | 0.130 | 0.256 | 0.434 | 0.383 | 0.070 | 0.243 | 0.670 |
| DANRFEF.DANRaIPS | 0.489 | 0.085 | 0.327 | 0.239 | 0.555 | 0.094 | 0.385 | 0.056 |
| DANRFEF.DANLaIPS | 0.717 | 0.176 | 0.213 | 0.117 | 0.782 | 0.171 | 0.248 | 0.095 |
| DANLpIPS.DANRFEF | 0.733 | 0.473 | 0.093 | 0.130 | 0.669 | 0.533 | −0.002 | 0.120 |
| DANLpIPS.DANRpIPS | 0.114 | 0.586 | 0.122 | 0.115 | 0.181 | 0.463 | 0.263 | 0.195 |
| DANLFEF.DANRFEF | 0.491 | 0.058 | 0.305 | 0.226 | 0.499 | 0.083 | 0.234 | 0.335 |
| DANLFEF.DANRaIPS | 0.418 | 0.006 | 0.556 | 0.388 | 0.295 | −0.034 | 0.618 | 0.455 |
| DANRpIPS.DANRFEF | 0.616 | 0.365 | 0.105 | 0.307 | 0.664 | 0.387 | 0.067 | 0.112 |
Congruence of factors is 0.95, 0.95, 0.90, 0.83 (correlation=0.90, p=0.001).
Loadings >0.4 are shaded.
Table 5.
Split-Half Validity Results for Fronto-FPTC
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 1 | Factor 2 | Factor 3 | Factor 4 | |
|---|---|---|---|---|---|---|---|---|
| FPTCLdlPFC.FPTCRIPL | 0.404 | 0.075 | 0.461 | 0.108 | 0.304 | 0.044 | 0.384 | 0.285 |
| FPTCLfrontal.FPTCRIPL | 0.460 | 0.042 | 0.428 | 0.054 | 0.367 | 0.039 | 0.443 | 0.240 |
| FPTCRIPL.FPTCLIPL | 0.046 | 0.054 | 0.640 | 0.147 | 0.168 | 0.043 | 0.378 | 0.228 |
| FPTCRIPL.FPTCRIPS | 0.049 | 0.124 | 0.558 | 0.092 | 0.142 | 0.143 | 0.627 | 0.106 |
| FPTCRIPL.FPTCLIPS | 0.094 | 0.119 | 0.800 | 0.047 | 0.244 | 0.126 | 0.597 | 0.225 |
| FPTCLfrontal.FPTCLIPL | 0.475 | 0.075 | 0.121 | 0.386 | 0.249 | 0.043 | 0.195 | 0.469 |
| FPTCLfrontal.FPTCRIPS | 0.667 | 0.223 | 0.199 | 0.023 | 0.579 | 0.279 | 0.363 | −0.071 |
| FPTCLfrontal.FPTCLIPS | 0.694 | 0.198 | 0.096 | 0.113 | 0.502 | 0.243 | 0.125 | 0.206 |
| FPTCLdlPFC.FPTCRfrontal | 0.224 | 0.543 | 0.053 | 0.168 | 0.285 | 0.560 | −0.067 | 0.303 |
| FPTCRfrontal.FPTCLfrontal | 0.282 | 0.523 | 0.109 | 0.080 | 0.234 | 0.612 | −0.089 | 0.255 |
| FPTCRfrontal.FPTCLIPL | −0.014 | 0.492 | 0.168 | 0.388 | −0.099 | 0.530 | 0.257 | 0.409 |
| FPTCRfrontal.FPTCRIPS | 0.249 | 0.638 | 0.252 | −0.025 | 0.156 | 0.638 | 0.311 | −0.066 |
| FPTCRfrontal.FPTCLIPS | 0.150 | 0.718 | 0.171 | 0.159 | 0.029 | 0.701 | 0.223 | 0.192 |
| FPTCRdlPFC.FPTCLdlPFC | 0.266 | 0.264 | 0.060 | 0.405 | 0.506 | 0.277 | −0.033 | 0.258 |
| FPTCRdlPFC.FPTCLfrontal | 0.350 | 0.274 | 0.123 | 0.343 | 0.482 | 0.338 | 0.015 | 0.250 |
| FPTCRdlPFC.FPTCRIPS | 0.282 | 0.429 | 0.265 | 0.156 | 0.401 | 0.389 | 0.388 | −0.087 |
| FPTCRdlPFC.FPTCRfrontal | 0.065 | 0.507 | 0.007 | 0.196 | 0.076 | 0.488 | −0.012 | 0.206 |
| FPTCRdlPFC.FPTCRIPL | 0.139 | 0.284 | 0.479 | 0.150 | 0.322 | 0.170 | 0.346 | 0.228 |
| FPTCRdlPFC.FPTCLIPL | 0.098 | 0.237 | 0.161 | 0.677 | 0.171 | 0.240 | 0.247 | 0.478 |
| FPTCRdlPFC.FPTCLIPS | 0.253 | 0.398 | 0.175 | 0.414 | 0.383 | 0.401 | 0.225 | 0.234 |
| FPTCLdlPFC.FPTCLfrontal | 0.464 | 0.182 | 0.016 | 0.150 | 0.479 | 0.107 | −0.016 | 0.220 |
| FPTCLdlPFC.FPTCLIPL | 0.444 | 0.064 | 0.057 | 0.491 | 0.278 | −0.055 | 0.164 | 0.528 |
| FPTCLdlPFC.FPTCRIPS | 0.605 | 0.238 | 0.248 | 0.103 | 0.628 | 0.286 | 0.343 | −0.026 |
| FPTCLdlPFC.FPTCLIPS | 0.637 | 0.135 | 0.070 | 0.248 | 0.590 | 0.167 | 0.073 | 0.252 |
| FPTCRfrontal.FPTCRIPL | 0.084 | 0.545 | 0.481 | −0.059 | 0.115 | 0.382 | 0.342 | 0.211 |
| FPTCLIPL.FPTCRIPS | 0.161 | 0.160 | 0.521 | 0.375 | 0.113 | 0.195 | 0.643 | 0.129 |
| FPTCLIPL.FPTCLIPS | 0.168 | 0.037 | 0.348 | 0.465 | 0.041 | 0.086 | 0.409 | 0.355 |
| FPTCRIPS.FPTCLIPS | 0.265 | 0.243 | 0.407 | 0.079 | 0.316 | 0.353 | 0.594 | −0.082 |
Split half validity results for FPTC. Congruence of factors is 0.81, 0.95, 0.84, and 0.66 (correlation=0.83, p=0.001).
Loadings >0.4 are shaded.
Congruence-of-factor structures derived from shuffled windows to the entire sample were much lower than for the unshuffled windows (Procrustes correlation for DMN=0.56, FPTC=0.61, DAN=0.56). In addition, the factor structure obtained from an analysis of shuffled windows showed little temporal co-fluctuation of correlations and accounted for significantly less of the total variance than the original solutions (Supplementary Tables S5–S7). Thus, the unshuffled relationship is special, suggesting that temporal dynamics are meaningful.
Specific factors show increased variability among correlations with age
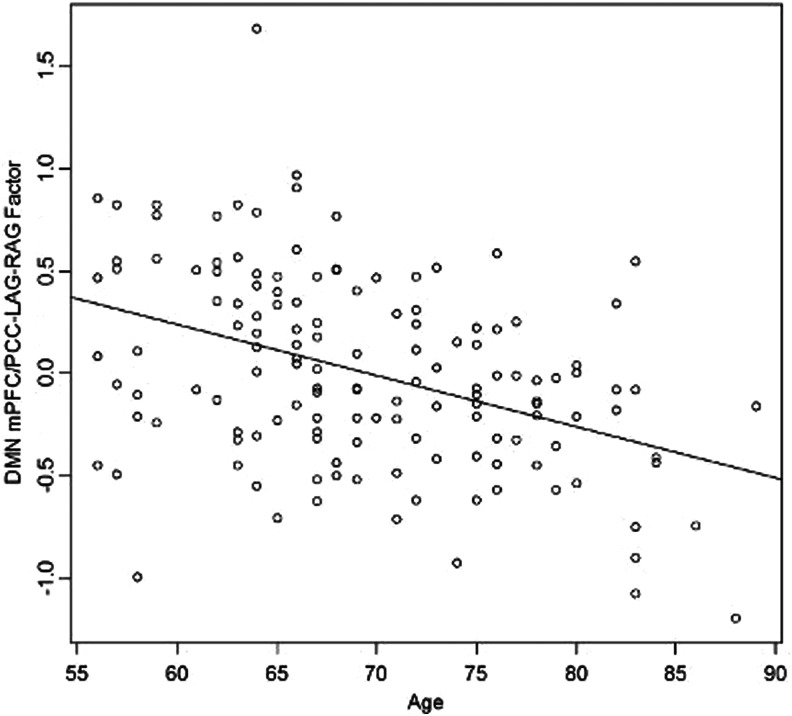
We examined the correlations between the mean factor score for each subject and age. In the DMN, the mean individual scores for the factor that loads most highly on connections between the mPFC to the PCC, LAG, and RAG are significantly negatively correlated with age [r(143)=−0.41, p<0.001] (Fig. 3). On average, subjects with higher values of this factor score have more similar correlations in that these networks link through time. Figure 1 shows the intercorrelations of nodes loading most highly on this factor for subjects with extreme values of this factor score. The younger subject has much more tightly coupled intercorrelations through time than the older subject.
FIG. 3.
Correlation of DMN factor 3 and age (r=−0.41).
In the DAN, scores for factors 1, 3, and 4 were negatively correlated with age: [factor 1: r(143)=−0.18, p=0.031, factor 3: r(143)=−0.22, p=0.008, factor 4: r(143)=−0.24, p=0.004]. In all cases, subjects with higher factor scores have more similar correlations in network links in each factor throughout time.
In the FPTC, scores for factor 3 (connecting the left dlPFC with the RIPS, LIPS, Lfrontal, and Lfrontal with the RIPS and LIPS) are negatively correlated with age [r(143)=−0.30, p<0.001]. Higher factor scores are associated with similar correlations in network links throughout time.
All correlations with age except for the DAN factor 1 and 3 survive Bonferroni correction for multiple comparisons (12 factors tested, Bonferroni threshold p<0.004).
Motion does not account for the relationship between factor scores and age
Power et al. (2011) have recently demonstrated that motion systematically causes artificially reduced or exaggerated patterns of correlations in resting-state data, and they recommend the scrubbing of frames surrounding motion artifacts as a preprocessing step to prevent this. Since we are looking at patterns of correlations through time, we cannot scrub our data without changing the number of observations available for each subject. We examined the potential impact of motion on our findings as follows. For each subject, the absolute and relative mean displacement during the resting state scan was computed. Age was uncorrelated with absolute displacement [r(143)=0.047, p=0.570] but was slightly correlated with mean relative displacement [r(143)=0.183, p=0.028]. This correlation was due to five older subjects with mean absolute displacement >0.23 mm. Removing these five subjects and repeating the factor analysis did not qualitatively change the factors obtained or our split-half validity analysis.
We re-examined the relationship between the factors in each network that were significantly related to age, controlling for relative displacement. All correlations decreased slightly but remained significant [DMN factor 3 r(143)=−0.38, p<0.001, DAN factor 1 r(143)=−0.17, p=0.045, DAN factor 3 r(143)=−0.20, p=0.015, DAN factor 4 r(143)=−0.23, p=0.005, FPTC factor 3 r(143)=−0.26, p=0.001].
Discussion
The contribution of this work is to demonstrate that fluctuating connectivity within large-scale networks of the brain exhibits a factor structure that can be replicated, is stable across multiple timescales, and that the expression of specific factors within networks is related to age. In the introduction, we emphasize that this abstraction may reveal more about age-related processes in functional networks than mean correlations.
Our approach of analyzing the fluctuations of correlations among nodes enables us to quantify a finer-grained network structure that can be isolated by regional covariation of the BOLD signal. Shuffling the nonoverlapping windows for each ROI, thereby destroying the long range of temporal fluctuations, removed this structure. We refer to this co-fluctuation of correlations within subnetworks as “structured variability” to indicate that this type of variability is a physiological manifestation of the dynamic behavior of the brain. We hypothesized and determined that older adults exhibit additional “unstructured variability,” or lower inter-correlations within subnetworks, resulting in lower dynamic connectivity in some, but not all, factors. The correlation of specific factor scores with age makes it unlikely that meaningful factors are solely due to our sample, scanner, or method of fMRI preprocessing. The relationship of specific factor scores to age lends interpretability to the general age-related reduction of connectivity (Table 2) as affecting specific subsystems.
The analysis of multiple small windows per individual enables us to quantify dynamic (within-scan) connectivity. Our ability to replicate this structure in a split-half sample means that this dynamic structure is robust across individuals. Finally, we found that factors obtained using correlations from multiple window sizes ranging from 20 to 450 sec (the entire scan) generated similar factor structures, with task-positive networks showing slightly more variability of factor structure at larger window sizes than the DMN. This scale-free characteristic does not mean that there is no dynamic information within each scan; as correlations fluctuate throughout a scan, so do factor scores describing the expression of each factor at each window. The findings indicate we can be confident that scores computed for smaller window sizes reflect the same structure which is evident across individuals at large timescales. Determining how these scores can predict ongoing performance in dynamic analyses is a subject for future work.
We are able to subdivide each network into at least four replicable subnetworks. The patterns of loadings are determined by rotating the solution using varimax rotation, but this solution is not unique; one could imagine alternate rotations with different loading patterns. Different rotations and consequent loading patterns are formally equivalent, and they may be more interpretable neurobiologically. We present the rotation (varimax) that tries to create a pattern of loadings where items load most strongly on one factor, and weaker on other factors. The rotation criteria are analogous to spatiotemporal independence in ICA. Although in ICA the solution is unique, it is based on the assumption of maximizing spatiotemporal independence, and to the degree that the data violate these assumptions, either approach will generate results which may not be interpretable. The key observation is that we can quantify an intrinsic substructure based on the fluctuations of connectivity.
We cannot in this study attribute specific function to the identified subnetworks. There is precedent for dividing DMN into anterior and posterior components with unique patterns of connectivity (Uddin et al., 2009). These findings mirror our separate DMN factors 3 and 4. Andrews-Hanna et al. (2010) identified a midline (anterior MPFC and PCC) core and a medial temporal lobe subsystem that share some functional properties. Similarly, using accelerated imaging data, Smith et al. (2012) show that the DMN is the sum of multiple distinct temporal processes, including a lateralized semantic network. Task-positive networks are also the sum of multiple temporal processes.
Our approach differs from the approach of identifying “temporal functional modes” (Smith et al., 2012) in several ways. First, we examine the time-varying behavior of correlations of network ROIs rather than the fluctuations of the BOLD signal within spatially derived components. We believe that this enables us to examine features of the time-varying correlation graph structure of brain networks without a fast TR sequence. Instead of temporal functional modes, we obtain factor scores that describe how closely correlations among subgroups of nodes covary in each individual for each time window. Our approach does not tell us anything about what modes are present spatially, and nothing about whether the BOLD signal in correlated ROIs is increasing or decreasing. Second, we go further to relate individual factor scores to age.
Noise caused by motion artifacts likely affects the time-varying correlation structure as it does the mean correlation structure (Power et al., 2011). However, we do not believe that this alters the pattern of results. First, removing subjects with large relative motion does not qualitatively change the observed factor structure or its robustness across individuals. Second, although it is an imperfect method, controlling for relative motion does not alter the significance or relative magnitude of the relationships between factors related to age. Our interpretation is that motion introduces a small amount of noise into the individual factor scores, which is difficult to control for. Longer resting-state acquisitions that would permit removal of windows with motion, or the use of multi-echo EPI denoising (Kundu et al., 2012) may reduce the impact of noise.
The factor structure which we observe may capture important information about the dynamic structure of the brain at rest that describes its health and ability to support higher-order cognitive processes. The results converge with prior studies showing reduced mean functional connectivity with age (Andrews-Hanna et al., 2007), but provide a richer description that enables one to postulate biological mechanisms for differential variability of individual factors. We find that not all network subcomponents show decreased synchronization with age. This suggests that the cause of this desynchronization of correlations in the BOLD signal is not global, but instead reflects damage to age-vulnerable network subcomponents and their interconnections. In particular, we notice that the only age-related component in the DMN involves the connection between the medial prefrontal wall and the posterior nodes of the DMN (PCC and angular gyri). This connectivity between the anterior hub of the DMN and the parietal hubs may be particularly vulnerable to age, suggesting a closer investigation of the intrinsic functioning of the medial prefrontal wall or long-range connectivity between these association hubs.
We used Fischer z-transformed correlations between regions of interest, our measurements of connectivity, as raw input into a factor analysis. Correlations are sample statistics that partial out extraneous variance. As such, the input to our factor analyses reflects these concentrated linear relationships, excluding any systematic nonlinear relationships. These correlations are also very susceptible to noise within any time period. Although this limited focus is almost certainly incomplete, the fact that it allows us to extract a robust structure of covarying fluctuations, some of which are related to age, is important.
Although sliding windows have been used to quantify dynamics (e.g., Chang and Glover, 2010; Hutchison et al., 2012; Jones et al., 2012), there are several open issues with regard to the use of fixed window sizes [see Hutchinson et al. (2013) for a review]. Nonstationary sources of noise in fMRI time series and white noise can induce changes in functional connectivity over time (Smith, 2012). Furthermore, the window size should be large enough to provide a good signal-to-noise ratio while being short enough to reflect the underlying dynamics. Since our approach quantifies the structure of fluctuations in connectivity and models error explicitly, it is relatively insensitive to noise. We have demonstrated that factors obtained using correlations from multiple window sizes generated similar factor structures, indicating a robustness of the method to noise in small windows.
The approach that we have adopted to demonstrate robustness of factors relies on the congruence of factors derived from exploratory analysis. This approach is not state of the art in behavioral studies, where typically the replicability of factor structures is tested with confirmatory factor analysis. We chose a model-free approach for evaluating factor replicability because of the complexity of the factor structure and our lack of theoretical understanding of what these factors should resemble. Future work remains to determine to what degree this structure can be reproduced in confirmatory factor analysis, and across different populations and ages. Confirmatory factor analysis, or a framework such as exploratory structural equation modeling (ESEM) (Asparouhov and Muthén, 2009) is necessary for completely testing invariance of the factor structure across groups. However, commonly useful factor structures (e.g., the “Big Five” personality factor structure) often do not fit confirmatory factor models well, and confirmatory models may inflate correlations among factors or generate other misleading results (Browne, 2001; McCrae et al., 1996). Analogous to existing approaches for finding spatiotemporal components based on ICA that assume maximal independence of components, we assume factors are orthogonal. This assumption is probably an oversimplification, and more work will be necessary to test this hypothesis.
Our emphasis here was on within-network dynamic connectivity. An important goal, addressed by Smith et al. (2012), is to examine between-network connections. A factor analysis approach does not scale to the interconnections of the brain, because there should be more observations than variables to fit a factor model, motivating the PCA approach taken to identify eigenconnectivities (Leonardi et al., 2013). However, factor analysis within a structural equation framework enables testing of the significance of factor loadings in a hypothesis-driven analysis, to carefully examine interrelationships between dynamic connectivity of networks. This is a direction of future research.
We have not yet speculated on the underlying reason for robust patterns of dynamic connectivity. It is possible that these are a result of the brain's normal operation as a nonlinear system of coupled oscillators as filtered through the BOLD response. Evidence from computational modeling and electrocorticography suggests that the oscillations observable as mean correlations in the BOLD signal and low-frequency resting-state networks stem from neuronal oscillations in the gamma band in structurally connected cortical fields (Cabral et al., 2011; Deco et al., 2009; Grady et al., 2001; He et al., 2008; Ko et al., 2011). Therefore, structural connectivity and cortical network architecture should largely determine the observed factor structure, with individual differences in structure and function causing deviations from normal operation. This opens possibilities for the use of dynamic connectivity as a physiological marker for brain functioning in normal aging.
Supplementary Material
Acknowledgments
The authors appreciate the critical reviews of earlier versions of this article provided by Sherry L. Willis, K.W. Schaie, and Brian Flaherty. This research was supported by grants from the National Institutes of Health 1RC4NS073008-01 and R37 AG024102.
Author Disclosure Statement
No competing financial interests exist.
References
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. 2012. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex. DOI: 10.1093/cercor/bhs352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews-Hanna JR, Reidler JS, Sepulcre J, Poulin R, Buckner RL. 2010. Functional-anatomic fractionation of the brain's default network. Neuron 65:550–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews-Hanna JR, Snyder AZ, Vincent JL, Lustig C, Head D, Raichle ME, Buckner RL. 2007. Disruption of Large-Scale Brain Systems in Advanced Aging. Neuron 56:924–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asparouhov T, Muthén B. 2009. Exploratory structural equation modeling. Struct Equ Modeling 16:397–438 [Google Scholar]
- Barrett P. 1986. Factor comparison: an examination of three methods. Pers Ind Diff 7:327–340 [Google Scholar]
- Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Carlson JM, Grafton ST. 2011. Dynamic reconfiguration of human brain networks during learning. Proc Natl Acad Sci USA 108:7641–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. 2005. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci 360:1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. 2004. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging 23:137–152 [DOI] [PubMed] [Google Scholar]
- Browne MW. (2001). An overview of analytic rotation in exploratory factor analysis. Multivariate Behav Res 36:111–150 [Google Scholar]
- Cabral J, Hugues E, Sporns O, Deco G. 2011. Role of local network oscillations in resting-state functional connectivity. Neuroimage 57:130–139 [DOI] [PubMed] [Google Scholar]
- Cattell RB. 1963. The structuring of change by P-technique and incremental R-technique. In: Harris CW. (ed.) Problems in Measuring Change, Madison, WI: University of Wisconsin Press; pp. 167–198 [Google Scholar]
- Cattell RB. 1966. Patterns of change: measurement in relation to state dimensions, trait change, lability and process concepts. In: Cattell RB. (ed.), Handbook of Multivariate Experimental Psychology, Chicago: Rand McNally; pp. 355–402 [Google Scholar]
- Chang C, Glover GH. 2010. Time–frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage 50:81–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW. 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29:162–173 [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. 2006. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci USA 103:13848–13853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirs V, McIntosh AR, Sporns O, Kötter R. 2009. Key role of coupling, delay, and noise in resting brain fluctuations. Proc Natl Acad Sci U S A 106:10302–10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach NUF, Fair DA, Miezin FM, Cohen AL, Wenger KK, Dosenbach RAT, et al. . 2007. Distinct brain networks for adaptive and stable task control in humans. Proc Natl Acad Sci U S A 104:11073–11078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. 2000. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A 97:11050–11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Harrison BJ, Zalesky A, Simons JS. 2012. Competitive and cooperative dynamics of large-scale brain functional networks supporting recollection. Proc Natl Acad Sci USA 109:12788–12793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. 2007. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci 8:700–711 [DOI] [PubMed] [Google Scholar]
- Glover GH, Li T-Q, Ress D. 2000. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med 44:162–167 [DOI] [PubMed] [Google Scholar]
- Gorsuch RL. 1983. Factor Analysis, 2nd ed. Hillsdale, NJ and London: Lawrence Erlbaum Associates [Google Scholar]
- Grady CL, Furey ML, Pietrini P, Horwitz B, Rapoport SI. 2001. Altered brain functional connectivity and impaired short-term memory in Alzheimer's disease. Brain 124:739–756 [DOI] [PubMed] [Google Scholar]
- Grady CL, Protzner AB, Kovacevic N, Strother SC, Afshin-Pour B, Wojtowicz M, Anderson JA, et al. . 2009. A Multivariate Analysis of Age-Related Differences in Default Mode and Task-Positive Networks across Multiple Cognitive Domains. Cereb Cortex 20:1432–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. 2004. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: Evidence from functional MRI. Proc Natl Acad Sci USA 101:4637–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker DA, Roopchansingh V, Gonzalez-Castillo J, Bandettini PA. 2012. Periodic changes in fMRI connectivity. Neuroimage 63:1712–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He BJ, Snyder AZ, Zempel JM, Smyth MD, Raichle ME. 2008. Electrophysiological correlates of the brain's intrinsic large-scale functional architecture. Proc Natl Acad Sci U S A 105:16039–16044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, et al. . 2013. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage 80:360–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Gati JS, Everling S, and Menon RS. 2012. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum Brain Mapp 34:2154–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, and Smith S. 2002. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17:825–841 [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. 2011. FSL. Neuroimage. DOI: 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Jones DT, Vemuri P, Murphy MC, Gunter JL, Senjem ML, Machulda MM, et al. . 2012. Non-stationarity in the “Resting Brain's” modular architecture. PLoS One 7:e39731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser HF. 1958. The varimax criterion for analytic rotation in factor analysis. Psychometrika 23:187–200 [Google Scholar]
- Kelly AMC, Uddin LQ, Biswal BB, Castellanos FX, Milham MP. 2008. Competition between functional brain networks mediates behavioral variability. NeuroImage 39:527–537 [DOI] [PubMed] [Google Scholar]
- Ko AL, Darvas F, Poliakov A, Ojemann J, Sorensen LB. 2011. Quasi-periodic fluctuations in default mode network electrophysiology. J Neurosci 31:11728–11732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Inati SJ, Evans JW, Luh W-M, Bandettini PA. 2012. Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. Neuroimage 60:1759–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi N, Richiardi J, Gschwind M, Simioni S, Annoni J-M, Schluep M, et al. . 2013. Principal components of functional connectivity: a new approach to study dynamic brain connectivity during rest. Neuroimage 83:937–950 [DOI] [PubMed] [Google Scholar]
- McCrae RR, Zonderman AB, Costa PT, Jr., Bond MH, Paunonen SV. 1996. Evaluating replicability of factors in the Revised NEO Personality Inventory: confirmatory factor analysis versus Procrustes rotation. J Pers Social Psychol 70:552–566 [Google Scholar]
- Niazy RK, Xie J, Miller K, Beckmann CF, Smith SM. 2011. Spectral characteristics of resting state networks. Prog Brain Res 193:259–276 [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2011. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59:2142–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. 2001. A default mode of brain function. Proc Natl Acad Sci U S A 98:676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaie KW. 2012. Developmental influences on adult intelligence: the Seattle longitudinal study (Update). New York, NY: Oxford University Press [Google Scholar]
- Smith SM. 2012. The future of FMRI connectivity. Neuroimage 62:1257–1266 [DOI] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, et al. . 2012. Temporally-independent functional modes of spontaneous brain activity. Proc Natl Acad Sci U S A 109:3131–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Kelly AMC, Biswal BB, Castellanos FX, Milham MP. 2009. Functional connectivity of default mode network components: correlation, anticorrelation, and causality. Hum Brain Mapp 30:625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Liang M, Wang L, Tian L, Zhang X, Li K, Jiang T. 2007. Altered functional connectivity in early Alzheimer's disease: a resting-state fMRI study. Hum Brain Mapp 28:967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.