Abstract
Alzheimer’s disease (AD) is the most common type of senile dementia affecting elderly people. The processing of amyloid precursor protein (APP) leading to the generation of β-amyloid (Aβ) peptide contributes to neurodegeneration and development of AD pathology. The endocytic trafficking pathway, which comprises of the endosomes and lysosomes, acts as an important site for Aβ generation, and endocytic dysfunction has been linked to increased Aβ production and loss of neurons in AD brains. Since insulin-like growth factor-II (IGF-II) receptor plays a critical role in the transport of lysosomal enzymes from the trans-Golgi network to endosomes, it is likely that the receptor may have a role in regulating Aβ metabolism in AD pathology. However, very little is known on how altered levels of the IGF-II receptor can influence the expression/function of various molecules involved in AD pathology. To address this issue, we evaluated the expression profiles of 87 selected genes related to AD pathology in mouse fibroblast MS cells that are deficient in murine IGF-II receptor and corresponding MS9II cells overexpressing ∼500 times the human IGF-II receptors. Our results reveal that an elevation in IGF-II receptor levels alters the expression profiles of a number of genes including APP as well as enzymes regulating Aβ production, degradation and clearance mechanisms. Additionally, it influences the expression of various lysosomal enzymes and protein kinases that are involved in Aβ toxicity. IGF-II receptor overexpression also alters expression of several genes involved in intracellular signalling as well as cholesterol metabolism, which play a critical role in AD pathology. The altered gene profiles observed in this study closely match with the corresponding protein levels, with a few exceptions. These results, taken together, suggest that an elevation in IGF-II receptor levels can influence the expression profiles of transcripts as well as proteins that are involved in AD pathogenesis.
Introduction
Alzheimer’s disease (AD), the most common type of senile dementia affecting elderly people, is characterized neuropathologically by extracellular β-amyloid (Aβ) peptide-containing neuritic plaques, intracellular tau-positive neurofibrillary tangles and the loss of neurons in selected regions of the brain. Although most AD cases occur sporadically after 65 years of age, a small proportion of cases correspond to the early-onset (<60 years) autosomal dominant form of the disease. To date, mutations in three genes - the β-amyloid precursor protein (APP) gene on chromosome 21, the presenilin 1 (PSEN1) gene on chromosome 14 and the presenilin 2 (PSEN2) gene on chromosome 1 - have been identified as the cause of a large proportion of early-onset familial AD. Additionally, inheritance of the ε4 allele of the apolipoprotein E (APOE) gene on chromosome 19 has been shown to increase the risk of late-onset and sporadic AD [1]–[5]. At present, the underlying cause for the AD pathogenesis remains unclear, but several lines of experimental evidence suggest that cerebral accumulation of Aβ peptides may initiate and/or contribute to the development of AD pathology [1], [6], [7]. These Aβ peptides are generated from their precursor APP, which is proteolytically processed by two alternative pathways; non-amyloidogenic α-secretase and amyloidogenic β-secretase pathways [8]–[11]. While the α-secretase pathway precludes the formation of intact Aβ peptides by cleaving APP within the Aβ domain, the β-secretase pathway yields the full-length Aβ1–40/Aβ1–42 peptides. The endosomal-lysosomal system, which comprises of tubulo-vesicular endocytic organelles and the lysosomes, has been shown to play a critical role in the production of Aβ peptides as well as to certain extent, intracellular degradation of nascent Aβ peptides. There is evidence that alternative processing of APP can be regulated by multiple factors that can influence not only the generation of Aβ but also the development/progression of AD pathology [10].
The insulin-like growth factor-II/mannose-6-phosphate (IGF-II/M6P or IGF-II) receptor, is a single transmembrane domain glycoprotein widely expressed in brain and peripheral tissues. The receptor binds two different classes of ligands: i) M6P-bearing molecules such as lysosomal enzymes, and ii) IGF-II - a mitogenic polypeptide with structural homology to IGF-I and insulin [12]–[14]. At the cellular level, a subset of the receptor is located at the plasma membrane, where it regulates internalization followed by activation/clearance of various ligands or activation of intracellular signalling cascades. The majority of the receptor, however, is present within the trans-Golgi network (TGN) and endosomal organelles where they transport newly synthesized lysosomal enzymes for subsequent delivery to lysosomes [15], [16]. Since the endosomal-lysosomal system plays a critical role in the generation of Aβ-related peptides [9]–[11], it is likely that the receptor may also be involved in regulating AD pathology. This is partly supported by the evidence that i) IGF-II receptor is expressed in a subset of Aβ-containing neuritic plaques and tau-positive neurofibrillary tangles in the AD brain [17] and ii) the receptor levels are altered in affected regions of the AD brain in individuals with PSEN1 mutations or carrying APOE ε4 alleles [17], [18] and iii) the levels of the IGF-II receptor are increased along with lysosomal enzymes in mutant APP transgenic mice overproducing Aβ peptides [19]. Additionally, it has recently been shown that IGF-II receptor is a substrate for β-secretase [β-APP cleaving enzyme (BACE1)], which is involved in the generation of Aβ peptides from APP [20]. Notwithstanding these results, very little is known on how altered levels of the IGF-II receptor can influence the expression and/or function of various molecules involved in AD pathology. To address this issue, we evaluated, as a first step, the expression profiles of 87 selected genes associated AD pathology in well characterized mouse fibroblast MS cells that are deficient in murine IGF-II receptor and corresponding MS9II cells that overexpress the human IGF-II receptor [21], [22]. We use these cell lines as they have been studied extensively to characterize the role of IGF-II receptor on cell signalling and intracellular trafficking of lysosomal enzymes [22]–[25]. Additionally, no neuronal cell line that stably overexpresses IGF-II receptor is currently available. The alterations in gene expression profiles observed in MS9II cells vs MS cells were validated using Western blotting. Our results clearly show that IGF-II receptor overexpression enhances APP mRNA/protein levels and some of the enzymes involved in Aβ metabolism. Additionally, it influences the expression profiles of various lysosomal enzymes and molecules regulating Aβ toxicity as well as cholesterol metabolism that have been shown to be involved in AD pathology.
Materials and Methods
Materials
NuPAGE 4–12% Bis-Tris gels were purchased from Life technologies, Corp. (Burlington, ON, Canada). DNA isolation kit, RNeasy mini kit, SABiosciences’ RT2 First Strand Kit, RT2 SYBR Green/Fluorescein qPCR master mix and the 96-well Mouse Alzheimer’s disease RT2 Profile PCR Array were all from Qiagen Inc. (Mississauga, ON, Canada). The bicinchoninic acid (BCA) protein assay kit and enhanced chemiluminescence (ECL) kit were from ThermoFisher Scientific Inc. (Nepean, ON, Canada). Sources of all primary antibodies used in the study are listed in Table 1. All horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Paso Robles, CA, USA). All other chemicals were from Sigma-Aldrich or Thermo Fisher Scientific.
Table 1. Details of the primary antibodies used in this study.
| Antibody Type | Type | Immunogen | Dilution | Source |
| A disintegrin and metalloprotease 9 (ADAM9) | Polyclonal | H | 1∶1000 | EMD Millipore, Co. |
| Amyloid precursor protein (APP, clone 22C11) | Monoclonal | RC | 1∶2000 | Abcam |
| Anterior pharynx defective -1 (APH-1) | Polyclonal | S | 1∶500 | EMD Millipore, Co. |
| Apolipoprotein E (APOE) | Polyclonal | R | 1∶1000 | Gift from Dr. J.E. Vance |
| ATP-binding cassette, sub-family A, member 1 (ABCA1) | Polyclonal | H | 1∶1000 | Novus Biologicals, LLC |
| Cathepsin B | Polyclonal | H | 1∶400 | Santa Cruz Biotechnology, Inc. |
| Cathepsin D | Polyclonal | H | 1∶200 | Santa Cruz Biotechnology, Inc. |
| Cyclin-dependent kinase 5 (CDK5) | Polyclonal | H | 1∶1000 | Cell Signaling Technology |
| Glycogen synthase kinase (GSK) 3 beta | Monoclonal | H | 1∶3000 | Abcam |
| Insulin degrading enzyme (IDE) | Polyclonal | H | 1∶1000 | Abcam |
| Insulin-like growth factor-II (IGF-II) | Polyclonal | H | 1∶500 | Santa Cruz Biotechnology, Inc. |
| Insulin-like growth factor-II receptor | Polyclonal | H | 1∶3000 | Gift from Dr. Carolyn Scott |
| Low density lipoprotein receptor-related protein (LRP) 1 | Polyclonal | H | 1∶4000 | Gift from Dr. J.E. Vance |
| Low density lipoprotein receptor-related protein (LRP) 6 | Polyclonal | M | 1∶1000 | Gift from Dr. J.E. Vance |
| Presenilin 1 (PS1) | Monoclonal | H | 1∶1000 | EMD Millipore, Co. |
| Urokinase-type plasminogen activator (uPA) | Polyclonal | H | 1∶200 | Santa Cruz Biotechnology, Inc. |
| β-actin | Monoclonal | S | 1∶5000 | Sigma-Aldrich, Inc. |
| β-glucuronidase | Polyclonal | RC | 1∶500 | Novus Biologicals |
| β-site APP cleaving enzyme 1 (BACE1) | Monoclonal | H | 1∶2000 | R&D Systems |
M: mouse peptide; H: human peptide; R: rat peptide; RC: recombinant peptide; S: synthetic peptide.
Cell Culture
IGF-II receptor deficient mouse fibroblasts MS and corresponding MS cells stably transfected with human IGF-II receptor known as MS9II cells (originally developed by Dr. W.S. Sly, Saint Louis University, MO, USA) [21], [22] used in this study were obtained as generous gifts from Dr. R. C. Bleackley (University of Alberta, AB, Canada). The cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 0.1 g/L sodium pyruvate, 2.2 g/L sodium bicarbonate, Pen/Strep 25U, 3.2 mM methotrexate and 5% dialyzed fetal bovine serum. The culture media did not contain any IGF-II or IGF-I, but the ingredients of dialyzed fetal bovine serum remain unknown. MS and MS9II cells between passages 5 and 14 were used in all of our experiments. Cells were seeded at 1×104 cells/cm2 and the medium was replaced every 3–4 days as described earlier [24]. Cultured MS and MS9II cells were harvested under basal conditions at 90% confluency as per the requirement of the specific protocol or stored at −80°C until further processing.
RNA Extraction for PCR Array
Total RNA was isolated from MS and MS9II cells using RNeasy mini kit following manufacturer’s instructions and stored at −80°C. RNA concentrations were determined using a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific) and 260/230 nm and 260/280 nm absorbance ratio were analyzed to determine RNA purity.
Real-time RT-PCR Array
At first 1 µg of total RNA was treated with genomic DNA elimination buffer at 42°C for 5 min to remove possible genomic DNA contamination. Following the elimination step, reverse transcription was carried out using the real-time RT-PCR First Strand Kit in accordance with the manufacturer’s protocol (SuperArray Biosciences Corp., MD). The resulting complementary DNA (cDNA) was diluted and combined with real-time RT-PCR SYBR Green/Fluorescein qPCR master mix and loaded onto a Mouse Alzheimer’s Disease RT2 Profiler PCR Array designed to profile the expression of 87 genes representative of biological pathways involved in APP/Aβ metabolism, cell signalling, intracellular trafficking cholesterol metabolism and cell death. All real-time PCR reactions were performed in a final volume of 25 µL using a MyiQ Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Canada) using a two-step cycling program: 10 min at 95°C (one cycle), 15 s at 95°C, followed by 1 min at 60°C (40 cycles). Data collection was performed during the annealing step (58°C) of each cycle and data were PCR-baseline subtracted and curve fitted. Threshold cycle (Ct) values were calculated using the instrument’s MyiQ optical software (Bio-Rad Laboratories, Inc.).
PCR Data Normalization and Analysis
The data were analyzed using the SABiosciences PCR Array Data analysis software based on the comparative Ct method and expressed as relative fold differences in MS9II cells compared to MS cells as described earlier [26]. All Ct values ≥35 were considered a negative call. Quality control tests for PCR reproducibility, reverse transcription efficiency and level of genomic DNA contamination were included in each plate and monitored as per the supplier’s instructions. The expression levels of two housekeeping genes included in the PCR array: Gapdh and Actb were used for normalization. The ΔCt for each gene in each plate was first calculated by subtracting the Ct value of the gene of interest from the average Ct value of the two housekeeping genes. Then, the average ΔCt value of each gene was calculated across the four replicate arrays for each cell line and ΔΔCt values were obtained by subtracting the ΔCt values of MS cells from the respective ΔCt values of MS9II cells. The fold-change for each gene from MS cells to MS9II cells was calculated as 2∧ (−ΔΔCt). Finally the fold-change for each gene was converted to fold-regulation as follows. For fold-change values >1, which indicated a positive or an up-regulation, the fold-regulation was equal to the fold-change. For fold-change values <1 indicating a negative or down-regulation, the fold-regulation was calculated as the negative inverse of the fold-change. P-values were calculated using Student’s t-test. A fold difference of ≥1.2 with a p-value <0.05 was considered as significant differential gene expression.
Immunocytochemistry
MS and MS9II cells seeded at 1×104 cells/cm2 on coverslips were fixed with 4% paraformaldehyde for 15 min, washed with phosphate-buffered saline (PBS) and then incubated overnight at 4 C with anti-IGF-II receptor antibody. The coverslips were then exposed to appropriate Alexa Fluor 594-conjugated secondary antibodies (1∶1000) for 2 h. The cell nucleus was stained with 1 µg/mL Hoechst 33258 for 5 min. The coverslips were washed with PBS and mounted with ProLong Gold antifade medium as described earlier [27], [28]. Immunostained cells were visualized using a Zeiss LSM 510 confocal microscope and the images were analyzed with ZEN 2010 (Carl Zeiss, Germany).
Western Blotting
Western blotting on cultured cell lysates was performed as described earlier [27], [28]. In brief, cultured cells were homogenized with radioimmunoprecipitation lysis buffer containing protease inhibitor cocktail and then proteins were quantified using a BCA kit. Denatured samples were resolved on 7–17% gradient sodium polyacrylamide or NuPAGE 4–12% Bis-Tris gels, transferred to polyvinylidene fluoride membranes, blocked with 5% skim milk and then incubated overnight at 4°C with different primary antibodies at proper dilutions as indicated in the Table 1. On the following day, membranes were incubated with appropriate HRP-conjugated secondary antibodies (1∶5000) and immunoreactive proteins were visualized using an ECL detection kit. All blots were re-probed with anti-β-actin antibody and quantified using a Microcomputer Imaging Device (MCID) image analysis system (Imaging Research, Inc., St Catherines, ON, Canada) as described earlier [27], [28].
Statistical Analysis
All data expressed as means ± SEM were obtained from four separate batches of cultures. Comparisons between two groups were performed using Student’s t-test. A p value of less than 0.05 was accepted as statistically significant. All statistical analyses were performed using GraphPad Prism (GraphPad software, Inc., CA, USA).
Results
Real-time RT-PCR Array Analysis of Gene Expression
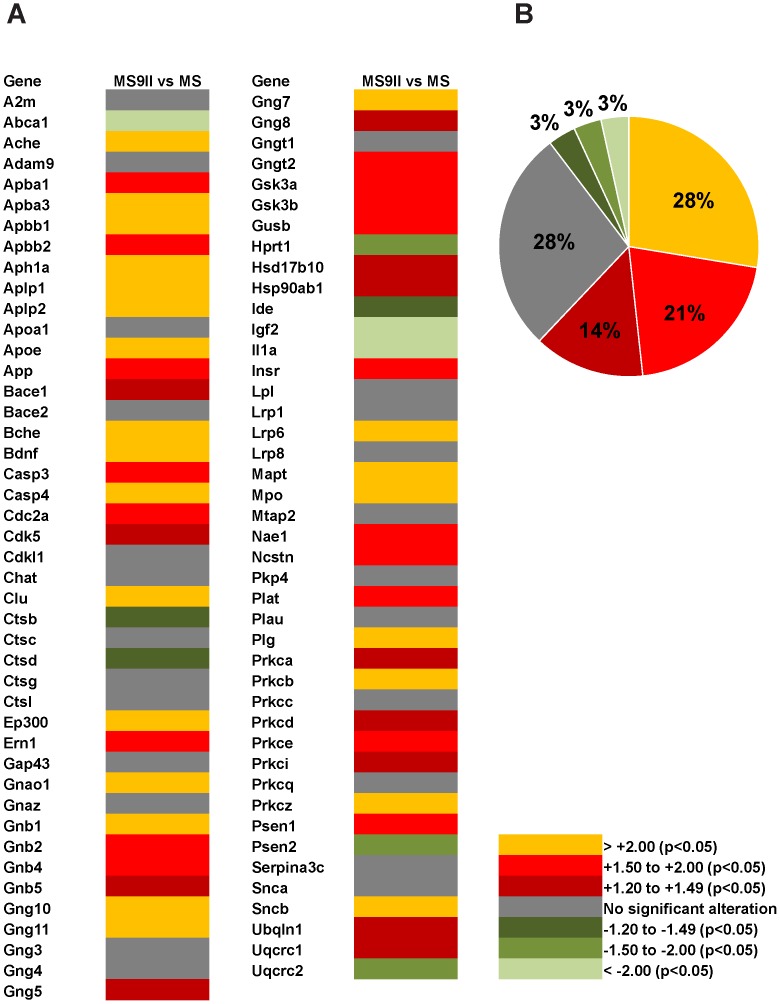
In order to gain molecular insights on the influence of the IGF-II receptor overexpression on AD pathology, we used well-characterized IGF-II receptor deficient MS cells and the corresponding MS9II cells that stably overexpresses the human IGF-II receptor ∼500 times compared to MS cells [21], [22]. These cells have been used extensively not only to define the role of the receptor in intracellular trafficking of the lysosomal enzymes but also in establishing its implication in cell signalling [23]–[25]. In our study we analyzed the expression profiles of 87 selected genes (Table 2) involved in APP/Aβ metabolism, cell signalling, cholesterol metabolism and cell death mechanism in MS and MS9II cells using real-time RT-PCR array. Our results revealed marked alterations in the relative expression of a wide-spectrum of transcripts in MS9II cells compared to MS cells (Fig. 1A). Complete list of differentially regulated genes with the respective fold-change in MS9II cells vs MS cells are provided in Table S1. Of the 87 genes evaluated, 54 genes (e.g. App, Aph1a, Apoe, Aplp1, Aplp2, Bace1, Cdk5, Clu, Gsk3a, Gsk3b, Gusb, Psen1 and Ncstn etc.) were significantly (p<0.05) up-regulated and 9 genes (e.g., Abca1, Ctsb, Ctsd, Igf2 and Ide) were significantly (p<0.05) down-regulated, while remaining 24 genes (e.g., A2m, Adam9, Bace2, Gap43, Ctsg, Ctsl and Plau) were unaltered in MS9II cells compared to MS cells (Fig. 1B). The majority of the differentially expressed genes showed 1.2- to 2-fold changes, whereas only few genes such as Apoe, Aph1a, Aplp1, Aplp2, Clu, Igf2 and Abca1 displayed more than 2-fold changes.
Table 2. List of selected genes for Mouse Alzheimer’s disease real-time RT-PCR array.
| NCBI Ref Seq# | Gene Symbol | Official Gene Name |
| NM_175628 | A2m | Alpha-2-macroglobulin |
| NM_013454 | Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 |
| NM_009599 | Ache | Acetylcholinesterase |
| NM_007404 | Adam9 | A disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| NM_177034 | Apba1 | Amyloid beta (A4) precursor protein binding, family A, member 1 |
| NM_018758 | Apba3 | Amyloid beta (A4) precursor protein-binding, family A, member 3 |
| NM_009685 | Apbb1 | Amyloid beta (A4) precursor protein-binding, family B, member 1 |
| NM_009686 | Apbb2 | Amyloid beta (A4) precursor protein-binding, family B, member 2 |
| NM_146104 | Aph1a | Anterior pharynx defective 1a homolog (C. elegans) |
| NM_007467 | Aplp1 | Amyloid beta (A4) precursor-like protein 1 |
| NM_009691 | Aplp2 | Amyloid beta (A4) precursor-like protein 2 |
| NM_009692 | Apoa1 | Apolipoprotein A-I |
| NM_009696 | Apoe | Apolipoprotein E |
| NM_007471 | App | Amyloid beta (A4) precursor protein |
| NM_011792 | Bace1 | Beta-site APP cleaving enzyme 1 |
| NM_019517 | Bace2 | Beta-site APP-cleaving enzyme 2 |
| NM_009738 | Bche | Butyrylcholinesterase |
| NM_007540 | Bdnf | Brain derived neurotrophic factor |
| NM_009810 | Casp3 | Caspase 3 |
| NM_007609 | Casp4 | Caspase 4, apoptosis-related cysteine peptidase |
| NM_007659 | Cdc2a | Cell division cycle 2 homolog A (S. pombe) |
| NM_007668 | Cdk5 | Cyclin-dependent kinase 5 |
| NM_183294 | Cdkl1 | Cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| NM_009891 | Chat | Choline acetyltransferase |
| NM_013492 | Clu | Clusterin |
| NM_007798 | Ctsb | Cathepsin B |
| NM_009982 | Ctsc | Cathepsin C |
| NM_009983 | Ctsd | Cathepsin D |
| NM_007800 | Ctsg | Cathepsin G |
| NM_009984 | Ctsl | Cathepsin L |
| NM_177821 | Ep300 | E1A binding protein p300 |
| NM_023913 | Ern1 | Endoplasmic reticulum (ER) to nucleus signalling 1 |
| NM_008083 | Gap43 | Growth associated protein 43 |
| NM_010308 | Gnao1 | Guanine nucleotide binding protein, alpha o |
| NM_010311 | Gnaz | Guanine nucleotide binding protein, alpha z subunit |
| NM_008142 | Gnb1 | Guanine nucleotide binding protein (G protein), beta 1 |
| NM_010312 | Gnb2 | Guanine nucleotide binding protein (G protein), beta 2 |
| NM_013531 | Gnb4 | Guanine nucleotide binding protein (G protein), beta 4 |
| NM_010313 | Gnb5 | Guanine nucleotide binding protein (G protein), beta 5 |
| NM_025277 | Gng10 | Guanine nucleotide binding protein (G protein), gamma 10 |
| NM_025331 | Gng11 | Guanine nucleotide binding protein (G protein), gamma 11 |
| NM_010316 | Gng3 | Guanine nucleotide binding protein (G protein), gamma 3 |
| NM_010317 | Gng4 | Guanine nucleotide binding protein (G protein), gamma 4 |
| NM_010318 | Gng5 | Guanine nucleotide binding protein (G protein), gamma 5 |
| NM_010319 | Gng7 | Guanine nucleotide binding protein (G protein), gamma 7 |
| NM_010320 | Gng8 | Guanine nucleotide binding protein (G protein), gamma 8 |
| NM_010314 | Gngt1 | Guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| NM_023121 | Gngt2 | Guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| NM_001031667 | Gsk3a | Glycogen synthase kinase 3 alpha |
| NM_019827 | Gsk3b | Glycogen synthase kinase 3 beta |
| NM_010368 | Gusb | Glucuronidase, beta |
| NM_016763 | Hsd17b10 | Hydroxysteroid (17-beta) dehydrogenase 10 |
| NM_031156 | Ide | Insulin degrading enzyme |
| NM_010514 | Igf2 | Insulin-like growth factor 2 |
| NM_010554 | Il1a | Interleukin 1 alpha |
| NM_010568 | Insr | Insulin receptor |
| NM_008509 | Lpl | Lipoprotein lipase |
| NM_008512 | Lrp1 | Low density lipoprotein receptor-related protein 1 |
| NM_008514 | Lrp6 | Low density lipoprotein receptor-related protein 6 |
| NM_053073 | Lrp8 | Low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| NM_010838 | Mapt | Microtubule-associated protein tau |
| NM_010824 | Mpo | Myeloperoxidase |
| NM_001039934 | Mtap2 | Microtubule-associated protein 2 |
| NM_144931 | Nae1 | NEDD8 activating enzyme E1 subunit 1 |
| NM_021607 | Ncstn | Nicastrin |
| NM_026361 | Pkp4 | Plakophilin 4 |
| NM_008872 | Plat | Plasminogen activator, tissue |
| NM_008873 | Plau | Plasminogen activator, urokinase |
| NM_008877 | Plg | Plasminogen |
| NM_011101 | Prkca | Protein kinase C, alpha |
| NM_008855 | Prkcb | Protein kinase C, beta |
| NM_011102 | Prkcc | Protein kinase C, gamma |
| NM_011103 | Prkcd | Protein kinase C, delta |
| NM_011104 | Prkce | Protein kinase C, epsilon |
| NM_008857 | Prkci | Protein kinase C, iota |
| NM_008859 | Prkcq | Protein kinase C, theta |
| NM_008860 | Prkcz | Protein kinase C, zeta |
| NM_008943 | Psen1 | Presenilin 1 |
| NM_011183 | Psen2 | Presenilin 2 |
| NM_008458 | Serpina3c | Serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| NM_009221 | Snca | Synuclein, alpha |
| NM_033610 | Sncb | Synuclein, beta |
| NM_026842 | Ubqln1 | Ubiquilin 1 |
| NM_025407 | Uqcrc1 | Ubiquinol-cytochrome c reductase core protein 1 |
| NM_025899 | Uqcrc2 | Ubiquinol cytochrome c reductase core protein 2 |
Figure 1. Heat-map diagram showing gene expression profiles in MS and MS9II cells.
The figure represents data obtained using mouse AD-PCR-Array of 87 selected genes involved in APP/Aβ metabolism, cholesterol metabolism, lysosomal enzyme and cell signalling. Transcriptional levels are colored yellow and different shades of red for significant up-regulations, different shades of green for significant down-regulations and grey for no alteration in MS9II cells compared to MS cells. Of the 87 genes evaluated, 54 transcripts are up-regulated and 9 genes are down-regulated, while remaining 24 genes remained unaltered in MS9II cells compared to MS cells (A). Pie-chart showing percentage of up- and down-regulated genes in MS9II cells compared to MS cells. Gene expression levels are colored yellow and shades of red for significant up-regulation, various shades of green for significant down-regulations and grey for no alteration. As evident from the pie-charts, several genes are differentially altered following overexpression of the human IGF-II receptor in MS9II cells (B). The data included in the heat-map diagram were obtained from four different experiments.
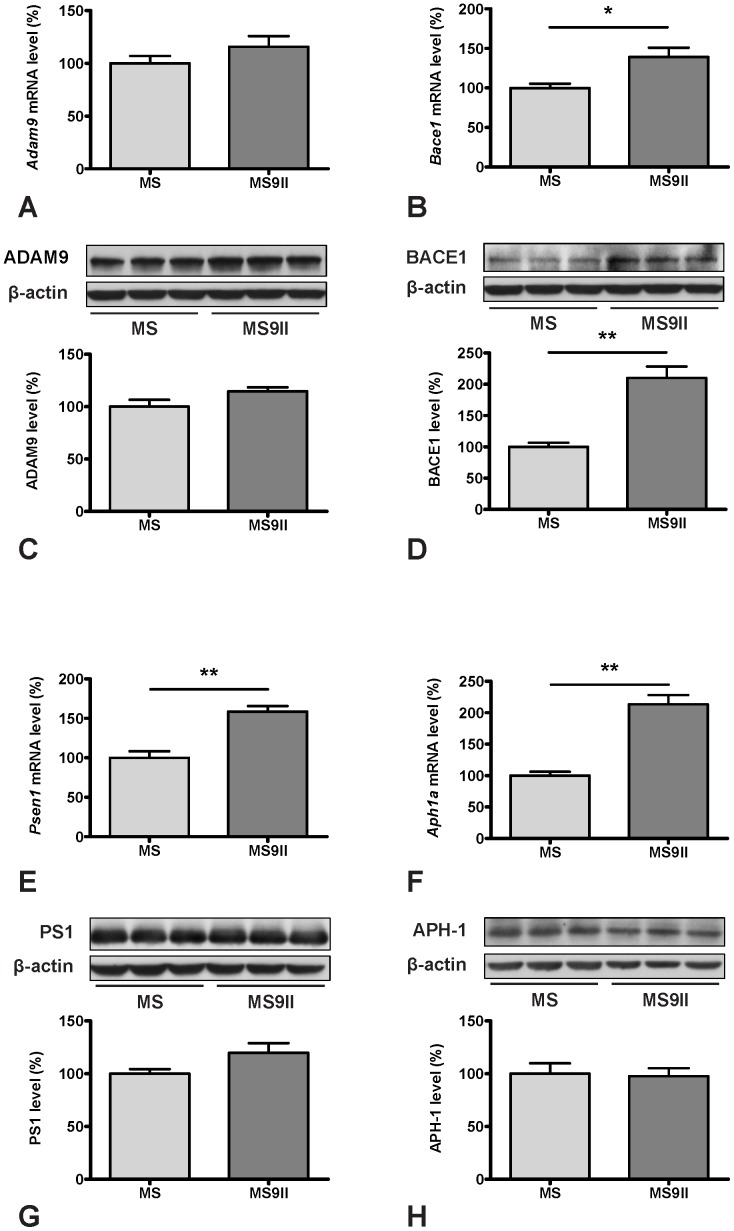
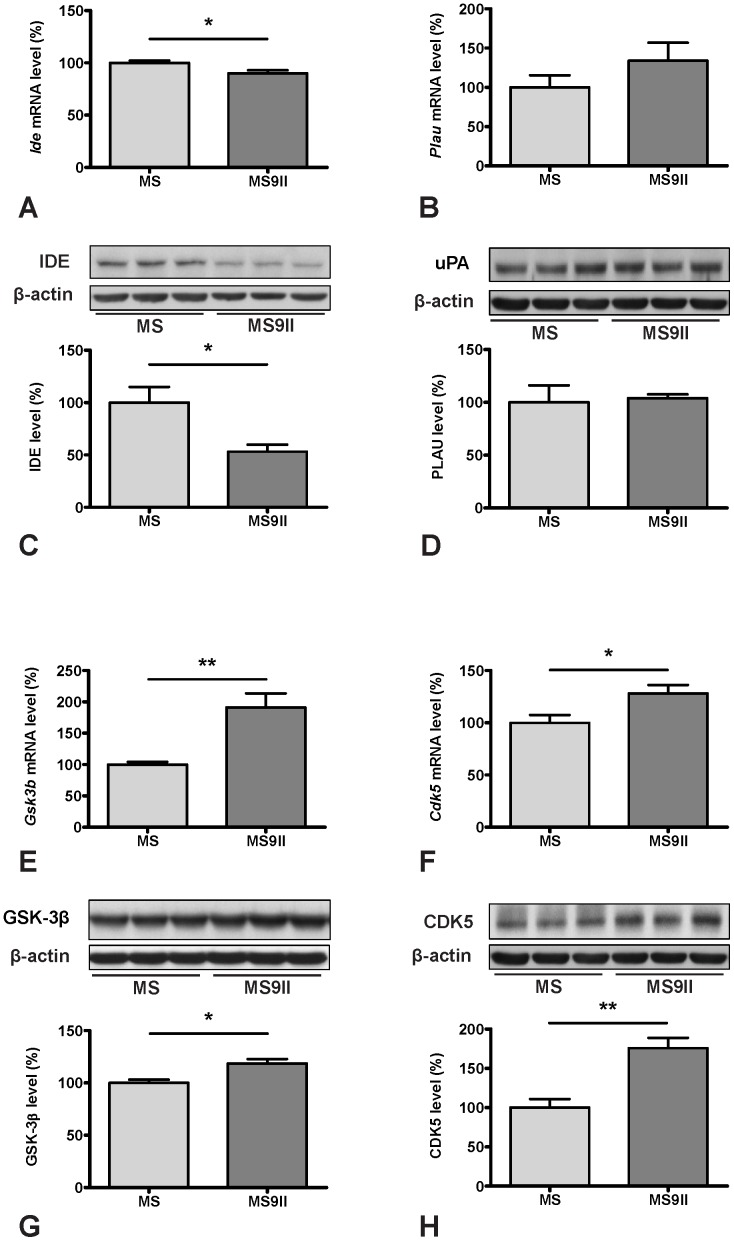
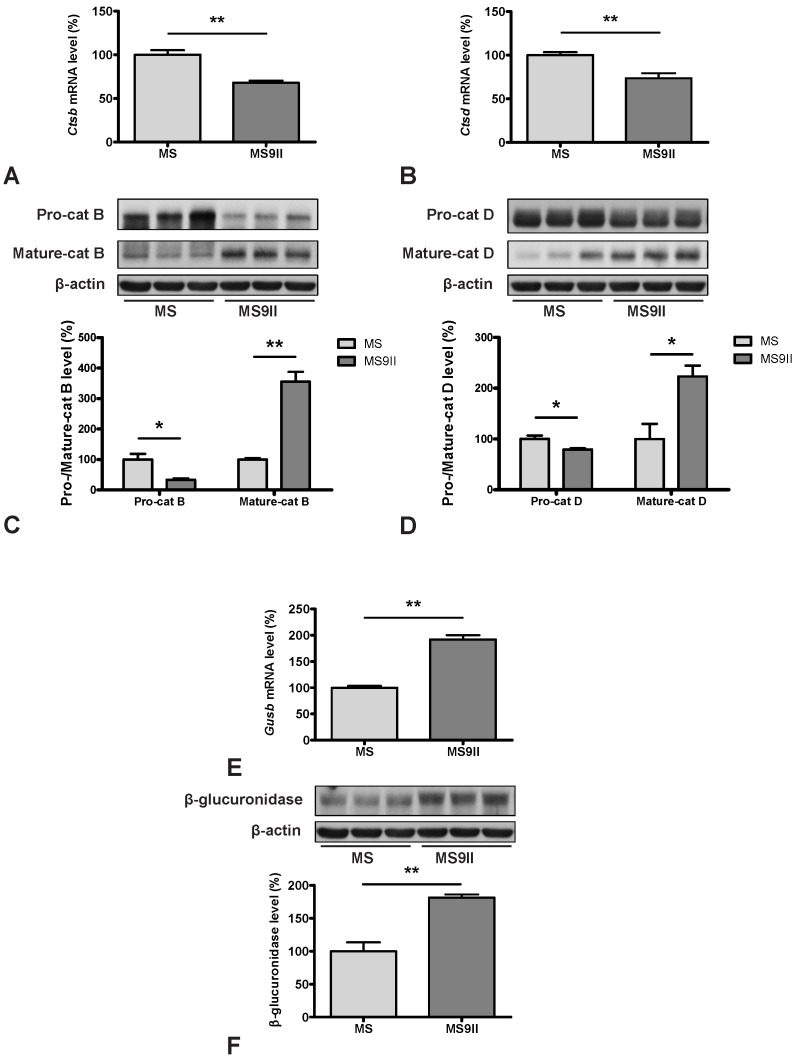
AD-PCR-Array data revealed that expression of genes directly involved in Aβ production such as App, Bace1, Psen1, Ncstn and Aph1a but not Adam9 were significantly increased in MS9II cells compared to MS cells (Figs. 1A; 2D; 3A, B, E and F). In contrast, expression of some of the genes involved in Aβ degradation such as Ide was decreased, while the others such as A2m and Plau encoding urokinase-type plasminogen activator (uPA), which activates an Aβ degrading enzyme plasminogen, remains unaltered (Figs. 1A; 4A and B). A number of transcripts that may have a role in regulating Aβ-mediated toxicity [7], [8] were either increased (such as GSk3a, Gsk3b, Prkca, Casp3, Casp4, Cdk5) or showed no alterations (i.e. Cdk11, Prkcc) in MS9II vs MS cells (Figs. 1A; 4E and F). Given the significance of cholesterol in AD pathology [29]–[32], it is of relevance that some of the transcripts involved in cholesterol metabolism were either markedly increased (i.e. Apoe, Clu and Lrp6), decreased (i.e. Abca1) or remained unaltered (i.e. Apoa1, Lpl, Lrp1 and Lrp8) in MS9II cells compared to MS cells (Figs. 1A; 5A, B, E and F). Transcripts corresponding to various lysosomal enzymes that are transported by the IGF-II receptor and are known to be involved in AD pathology [15], [16], [33]–[35] were also found to be differentially expressed. While the expression of Ctsb and Ctsd were decreased and Gusb was increased (Figs. 1A; 6A, B and E), Ctsg and Ctsl did not exhibit any significant alterations between the two cell lines. Apart from its well established trafficking role, IGF-II receptor is known to mediate certain biological effects of IGF-II by triggering specific cellular signalling pathways [36]–[40]. In fact some of these effects including regulation of acetylcholine release from adult rat brain [39], [40] as well as hypertrophy of myocardial cells [41] are known to be mediated by G protein linked protein kinase C (PKC) pathways. In keeping with these results, we observed that various subunits/isoforms of G proteins as well as PKC were differentially expressed in MS9II cells compared to MS cells. Some of the transcripts such as Gnao1, Gnb1, Gnb2, Gnb4, Gnb5, Gng5, Gng8, Gngt2, Prkca, Prkcb, Prkcd, Prkce, Prkci and Prkcz were found to be significantly increased while others (i.e. Gnaz, Gng3, Gng4, Gngt1 and Prkcc) did not exhibit any marked changes following overexpression of IGF-II receptor (Fig. 1A).
Validation of Altered Gene Expression Profiles by Western Blotting
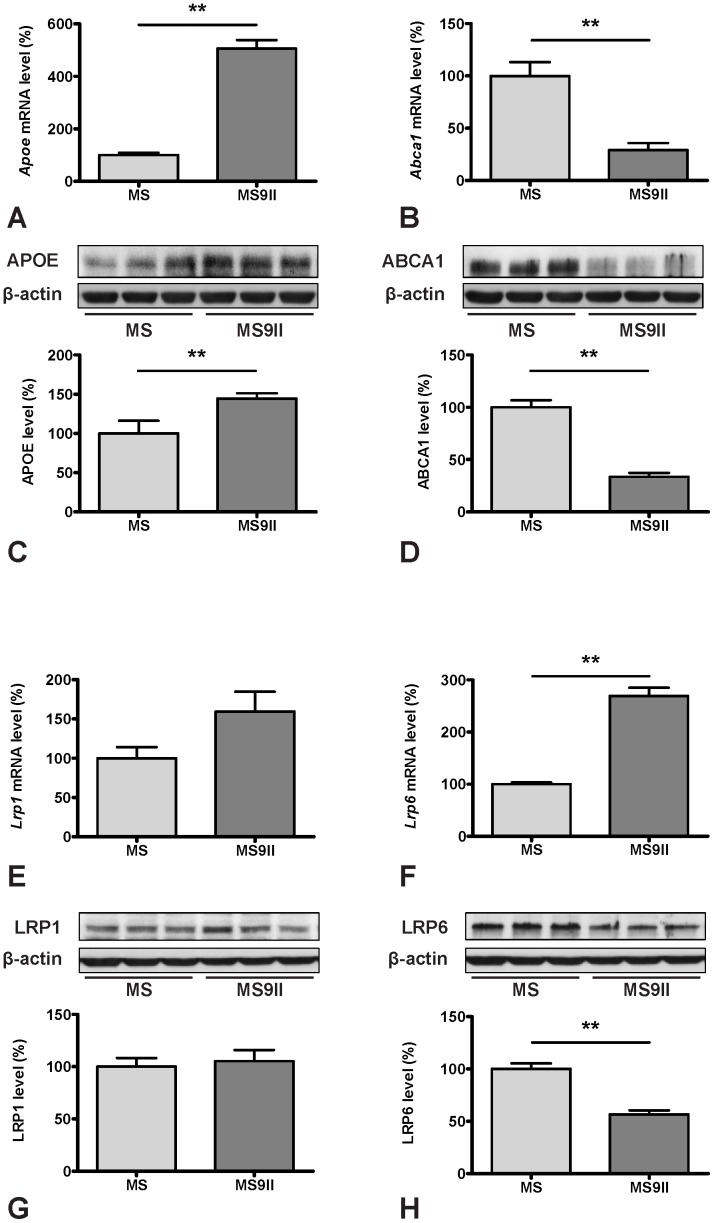
To validate the changes observed in the expression profiles of the genes involved in AD pathology, we evaluated steady-state protein levels of selected transcripts in MS and MS9II cells by immunoblot analysis. Consistent with our AD-PCR-Array data, we observed significant increase in the levels of APP (Fig. 2F) and its processing enzyme BACE1 (Fig. 3D), while the levels of ADAM9 remained unaltered in MS9II cells as compared with MS cells (Fig. 3C). Interestingly, the levels of certain proteins with long half-lives such as PS1 and APH1, in contrast to their respective transcripts, were found to be unaltered, consistent with the reports that stable formation of γ-secreatse complex determines the steady-state protein levels of these proteins (Figs. 3G and H) [42]–[44]. In keeping with the transcript levels, we observed significant decrease in the levels of IDE and ABCA1 (Figs. 4C; 5D), marked increase in GSK3β, CDK5 and APOE levels (Figs. 4G and H; 5C) and no alteration in uPA and LRP1 levels (Figs. 4D; 5G) in MS9II cells vs MS cells. The levels of LRP6, in contrast to its transcript, were found to be decreased (Fig. 5H) suggesting post-translational regulatory mechanism likely contributes to the steady-state protein levels. With regards to the lysosomal enzymes, the pro-forms of cathepsins B and D correlated rather well with the decreased levels of both transcripts observed in MS9II cells compared to MS cells (Figs. 6C and D). However, the mature forms of these enzymes were found to be markedly increased suggesting an intriguing post-translation mechanism that may have a role in regulating the levels/activity of these enzymes within the cells (Figs. 6C and D). On the other hand, both the Gusb transcript levels and the steady-state β-glucuronidase protein levels were higher in MS9II cells as compared with MS cells (Fig. 6F).
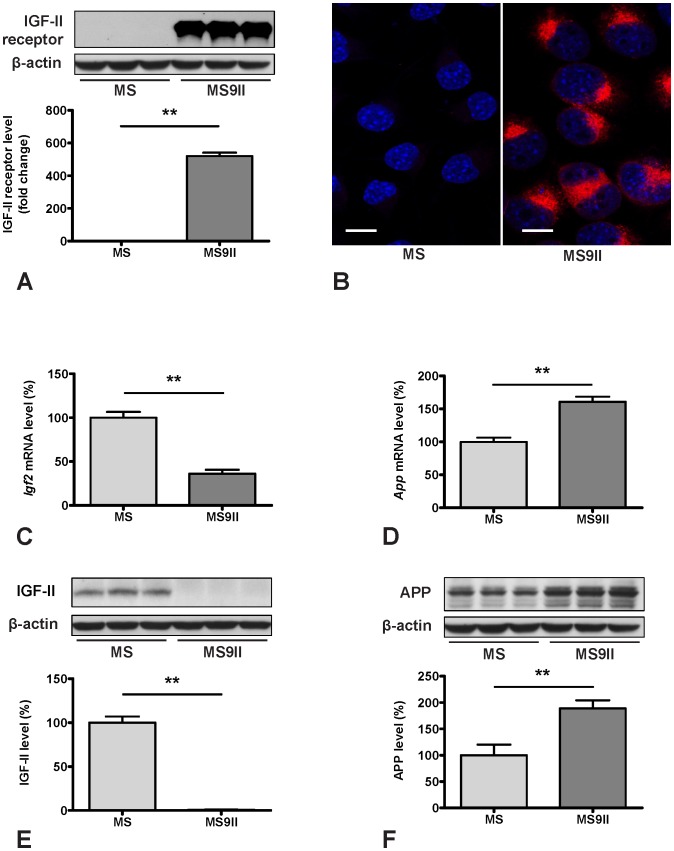
Figure 2. Transcript and protein expression levels of IGF-II receptor (A, B), IGF-II (C, E) and APP (D, F) in MS and MS9II cells.
Increased levels and expression of IGF-II receptor in MS9II vs MS cells are validated using Western blotting and immunofluorescence staining respectively (A, B). Histograms showing decreased level of Igf2 mRNA (C) and increased level of App mRNA in MS9II cells compared to MS cells as obtained using AD-PCR-Array. Immunoblots and respective histograms validating decreased levels of IGF-II and increased levels of APP in MS9II cells. The protein levels were normalized to the β-actin and the values from four different experiment are expressed as means ± SEM, **p<0.01. Scale bar = 10 µm.
Figure 3. Transcript and protein expression levels of ADAM9 (A, C), BACE1 (B, D), PS1 (E, G) and APH-1 (F, H) in MS and MS9II cells.
Histograms showing unaltered levels of Adam9 mRNA (A) and increased levels of Bace1 mRNA (B) in MS9II cells compared to MS cells. Immunoblots and respective histograms validating unchanged ADAM9 (C) and increased BACE1 (D) levels in MS9II cells. Histograms showing increased mRNA levels for Psen1 (E) and Aph1a (F) in MS9II cells compared to MS cells. Immunoblots and respective histograms showing unaltered protein levels of PS1 (G) and APH-1 (H) in MS9II cells. The protein levels were normalized to the β-actin and the values from four different experiments are expressed as means ± SEM, *p<0.05, **p<0.01.
Figure 4. Transcript and protein expression levels of IDE (A, C), PLAU (B, D), GSK-3β (E, G) and CDK5 (F, H) in MS and MS9II cells.
Histograms showing decreased levels of Ide mRNA (A) and unaltered levels of Plau mRNA (B) in MS9II cells compared to MS cells. Immunoblots and respective histograms validating decreased IDE (C) and unchanged uPA (D) levels in MS9II cells. Histograms showing increased mRNA levels for Gsk3b (E) and Cdk5 (F) in MS9II cells compared to MS cells. Immunoblots and respective histograms showing marked increase in protein levels of GSK-3β (G) and CDK5 (H) in MS9II cells. The protein levels were normalized to the β-actin and the values from four different experiments are expressed as means ± SEM, *p<0.05, **p<0.01.
Figure 5. Transcript and protein expression levels of APOE (A, C), ABCA1 (B, D), LRP1 (E, G) and LRP6 (F, H) in MS and MS9II cells.
Histograms showing increased levels of Apoe mRNA (A) and decreased levels of Abca1 mRNA (B) in MS9II cells compared to MS cells. Immunoblots and respective histograms validating increased APOE (C) and decreased ABCA1 (D) levels in MS9II cells. Histograms showing unaltered levels of Lrp1 mRNA (E) and increased levels of Lrp6 mRNA (F) in MS9II cells compared to MS cells. Immunoblots and respective histograms showing unchanged LRP1 but decreased levels of LRP6 in MS9II cells. The protein levels were normalized to the β-actin and the values from four different experiments are expressed as means ± SEM, *p<0.05, **p<0.01.
Figure 6. Transcript and protein expression levels of cathepsin B (A, C), cathepsin D (B, D) and β-glucorinidase (E, F) in MS and MS9II cells.
Histograms showing decreased levels of Ctsb mRNA (A) and Ctsd mRNA (B), but increased levels of Gusb mRNA (E) in MS9II cells compared to MS cells. Immunoblots and respective histograms showed decreased levels of pro-cathepsin B and D but increased levels of mature cathepsins B and D in MS9II cells than in MS cells. Immunoblot analysis of β-glucuronidase level, consistent with mRNA, was enhanced in MS9II cells. The protein levels were normalized to the β-actin and the values from four different experiments are expressed as means ± SEM, *p<0.05, **p<0.01.
Discussion
The present study using real-time RT-PCR arrays reveals that an increase in IGF-II receptor levels can influence the expression profiles of several genes involved in AD pathology. Notably, some of the differentially expressed genes are directly associated with the production and clearance of Aβ peptides, while the others are linked to cholesterol metabolism and the endosomal-lysosomal system function, all of which are known to play critical roles in the development of AD pathology. The altered gene profiles with few exceptions correlated well with alterations in the corresponding steady-state protein levels. With reference to the quantification of differential gene expression, it is important to highlight two points in context of the results obtained in the present study. First, the absolute fold-changes in the level of a specific transcript need not be of high magnitude to have a significant effect on cell physiology. Second, post-translational modifications on proteins, apart from altered levels of the transcripts, can have an important role in regulating the function and/or development of AD-related pathology. It is also of interest to note that the present study, in the absence of any neuronal cell lines overexpressing IGF-II receptor, was carried out using mouse fibroblast MS9II cells. Thus, the results obtained in the present study may not precisely recapitulate the changes that can be seen in neurons following overexpression of the IGF-II receptor. Nevertheless, these results suggest that an alteration in IGF-II receptor levels can influence the expression profiles of a number of transcripts as well as proteins that are involved either directly and/or indirectly in the development of AD pathology.
IGF-II Receptor Overexpression and APP/Aβ Metabolism
Previous studies have shown that IGF-II receptor level and expression are increased in the hippocampus and cortex but not in the striatum of mutant APP transgenic mice compared to age-matched control mice [19]. However, the levels/expression of the receptor, unlike transgenic mice, are usually unaltered in AD brains [17], [18], but there is evidence that the receptor levels/expression can be decreased selectively in the hippocampus of AD patients carrying two copies of APOE ε4 allele [17] or increased in the cortical region of familial cases with a PSEN1 mutation [18]. More recently, a quantitative proteomics study reveals that the IGF-II receptor may be a substrate for BACE1, albeit its functional significance in relation to AD pathology remains unclear [20]. Our results clearly indicate that overexpression of the IGF-II receptor markedly increases the expression of App, Bace1, Psen1, Ncstn and Aph1a but not Adam9. Consistent with transcript levels, we observe up-regulation of APP and BACE1 and no alteration in ADAM9 between MS9II and MS cells. The protein levels of PS1 and APH1, unlike their transcripts, are not markedly altered; this is not surprising because it is known that their steady-state levels are tightly regulated by stoichiometric interaction between the four γ-secretase complex subunits [42]–[44]. Nevertheless, it remains to be determined whether IGF-II receptor overexpression can influence γ-secretase enzyme activity. Interestingly, the levels of Ide and its corresponding protein insulin degrading enzyme, which is involved in the clearance of Aβ peptides [45], [46], are significantly down-regulated in MS9II cells compared to MS cells. These results suggest that IGF-II receptor overexpression may influence the clearance of Aβ peptides. However, we did not observe an alteration in the expression of A2m or Plau, which code for two major proteins that mediate Aβ degradation [45]–[47]. Thus it remains to be established to what extent IGF-II receptor can influence Aβ clearance mechanisms, which are known to play an important role in pathogenesis of sporadic AD.
IGF-II Receptor Overexpression and Cholesterol Metabolism
A number of studies have indicated that altered cholesterol homeostasis can influence AD pathology. This is supported by the evidence that i) inheritance of ε4 isoform of the cholesterol transporter APOE is a major risk factor for late-onset AD [2], [4], ii) epidemiological data suggest statins, drugs that block cholesterol biosynthesis, reduce the prevalence of AD, albeit more recent prospective studies have produced conflicting results [48]–[50], iii) elevated cholesterol levels increase Aβ production/deposition, whereas inhibition of cholesterol synthesis lowers Aβ levels/deposition [29], [51]–[53], and iv) some genes related to cholesterol metabolism have been linked to AD including Clu (involved in the transport of cholesterol), LRP (a major receptor for ApoE in the brain) and ABCA1 (involved in the efflux of cholesterol), though their associations need to be validated in future studies [54]–[56]. Our real-time RT-PCR array data reflect a dysregulation in cholesterol metabolism following elevation of IGF-II receptor levels as we observed an up-regulation of Apoe, Clu and Lrp6 and down-regulation of Abca1 in MS9II cells compared to MS cells. Interestingly, Apoa1, Lpl, Lrp1 and Lrp8 transcripts did not exhibit marked alterations between the two cell lines. Consistent with the transcripts levels, we observed an increase in APOE, a decrease in ABCA1 and no alteration in LRP1 levels. The levels of LRP6, in contrast to its transcript, is decreased in MS9II cells than MS cells. Some recent data indicate that APOE can influence AD not only by regulating the transport of cholesterol but also the extent of Aβ fibrilization as well as clearance of Aβ peptides [57], [58]. The ABCA1, on the other hand, has been shown to modulate Aβ deposition by regulating its production as well as lipidation of APOE [59]. Thus, it is possible that IGF-II receptor can influence AD pathology by APOE and ABCA1 regulated cholesterol metabolism.
IGF-II Receptor Overexpression and Lysosomal Enzymes
IGF-II receptor plays an important role in the transport of newly synthesized lysosomal enzymes from TGN to lysosomes where they regulate the clearance of various cellular proteins [15], [16], [60], [61]. Some of the enzymes such as cathepsins B and D are also known to affect cell viability following their release into the cytosol [28], [60]–[63]. Evidence suggests that cathepsins may be involved in the generation of Aβ peptides and their levels/expressions are increased in the vulnerable neurons as well as plasma of AD patients [33]–[35], [64]–[67]. Inhibitors of cathepsin B or deletion of the gene have been shown to reduce Aβ burden in mutant APP transgenic mice [68], [69]. Interestingly, overexpression of the IGF-II receptor shows a decreased expression of Ctsb and Ctsd transcripts and pro-forms of the enzymes, while the levels of mature enzymes are increased possibly due to efficient M6P-dependent trafficking and proteolytic conversion of the pro-forms to active enzymes in endosomes and lysosomes [70]. The profile of Gusb transcript and the corresponding protein levels, however, are increased following overexpression of the IGF-II receptor levels. Although these results suggest that IGF-II receptor may differentially regulate various lysosomal enzymes, increased levels of mature cathepsins B and D as well as β-glucuronidase, apart from degradation of cellular proteins, can influence AD pathology via other pathways including APP/Aβ metabolism.
IGF-II Receptor Overexpression and Cell Signalling
In contrast to the well established trafficking role of the IGF-II receptor, its significance in triggering intracellular signalling in response to IGF-II binding remains controversial. A number of studies, however, indicate that IGF-II receptor can mediate certain biological effects of IGF-II in multiple cell types, including stimulation of calcium influx in Balb/c-3T3 fibroblasts and CHO cells [36], motility of human rhabdomyosarcoma cells [37], migration of human extravillous trophoblasts [71], stimulation of Na+/H+ exchange and inositol trisphosphate production [38] and insulin exocytosis by pancreatic β cells [72]. Some of these ligand-induced responses of the IGF-II receptor are triggered by interaction with G protein-induced PKC-dependent signalling pathways [14], [73]. In addition, we have earlier reported that IGF-II receptor activation can potentiate acetylcholine release via a G protein sensitive PKCα-dependent pathway in the brain [39], [40]. The results of the present study show that IGF-II receptor overexpression can induce marked alterations in the levels of various transcripts associated with G protein subunits and PKC isoforms. Although significance of these alterations remains to be established, it is possible that these changes may partly be linked to the signalling effects of the IGF-II receptor. Recently it has been reported that IGF-II, by activating its own receptor, can induce long-term potentiation and promote memory consolidation [74]. This is indeed relevant as IGF-II mRNA levels are decreased in AD brains with the progression of disease pathology [75]. The current study also showed a decrease in IGF-II transcript and protein levels in MS9II cells thus suggesting that overexpression of IGF-II receptor may have role not only in APP/Aβ metabolism but also in regulating AD-related cognitive functions. Additionally, we observed that enhanced expression of the IGF-II receptor can increase transcripts and protein levels of GSK and Cdk5 both of which are associated with toxicity induced by Aβ peptides [1], [7], [8]. Thus, it would be of interest to define whether overexpression of the receptor can render the cells more vulnerable to Aβ-mediated toxicity.
Conclusion
The present study reports that elevation of IGF-II receptor expression differentially alters not only the expression profiles of various transcripts including APP as well as enzymes regulating Aβ production/clearance mechanisms but also certain lysosomal enzymes and protein kinases that are involved in Aβ metabolism, clearance and toxicity. The overexpression of the IGF-II receptor is also found to alter various genes that regulate cholesterol metabolism, which may be of relevance to AD pathogenesis. Additionally, we observe profound changes in a number of G protein and PKC transcripts, which may be associated with IGF-II receptor signalling. The altered gene profiles, with some exceptions, match with the corresponding protein levels. Collectively these results suggest that elevation of IGF-II receptor levels can differentially influence the transcription and protein levels of genes that are involved either directly or indirectly with pathogenesis of AD.
Supporting Information
Gene expression profiles in MS9II cell compared to MS cells as studied using real-time RT-PCR arrays.
(DOCX)
Acknowledgments
We would like to thank Dr. R.C. Bleackley (University of Alberta, Edmonton, AB, Canada) for providing us the MS and MS9II cell lines, Dr. J.E. Vance (University of Alberta, Edmonton, AB, Canada) for the gift of the anti-APOE, anti-LRP1 and anti-LRP6 antibodies, Dr. S. Sipione (University of Alberta, Edmonton, AB, Canada) for providing the anti-ABCA1 antibody and Dr. C. Scott (Kolling Institute of Medical Research, NSW, Australia) for the anti-IGF-II receptor antibody.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the paper.
Funding Statement
This work was supported by grants from Natural Sciences and Engineering Research Council of Canada (#203518) and Canadian Institutes of Health Research (MOP-84480) to SK. YW is a recipient of Doctoral Awards from Alzheimer Society of Canada and a Graduate Studentship Award from Alberta Innovates - Health Solutions. GT is supported by R01AG019070 grant from National Institutes on Aging. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Selkoe DJ (2008) Biochemistry and molecular biology of amyloid beta-protein and the mechanism of Alzheimer’s disease. Handb Clin Neurol 89: 245–260. [DOI] [PubMed] [Google Scholar]
- 2. Bertram L, Tanzi RE (2012) The genetics of Alzheimer’s disease. Prog Mol Biol Transl Sci 107: 79–100. [DOI] [PubMed] [Google Scholar]
- 3. Lopez OL, DeKosky ST (2003) [Neuropathology of Alzheimer’s disease and mild cognitive impairment]. Rev Neurol 37: 155–163. [PubMed] [Google Scholar]
- 4. St George-Hyslop PH, Petit A (2005) Molecular biology and genetics of Alzheimer’s disease. C R Biol 328: 119–130. [DOI] [PubMed] [Google Scholar]
- 5. Nelson PT, Braak H, Markesbery WR (2009) Neuropathology and cognitive impairment in Alzheimer disease: a complex but coherent relationship. J Neuropathol Exp Neurol 68: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hardy J (2009) The amyloid hypothesis for Alzheimer’s disease: a critical reappraisal. J Neurochem 110: 1129–1134. [DOI] [PubMed] [Google Scholar]
- 7. Tickler AK, Wade JD, Separovic F (2005) The role of Abeta peptides in Alzheimer’s disease. Protein Pept Lett 12: 513–519. [DOI] [PubMed] [Google Scholar]
- 8. Nathalie P, Jean-Noel O (2008) Processing of amyloid precursor protein and amyloid peptide neurotoxicity. Curr Alzheimer Res 5: 92–99. [DOI] [PubMed] [Google Scholar]
- 9. Thinakaran G, Koo EH (2008) Amyloid precursor protein trafficking, processing, and function. J Biol Chem 283: 29615–29619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O’Brien RJ, Wong PC (2011) Amyloid precursor protein processing and Alzheimer’s disease. Annu Rev Neurosci 34: 185–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haass C, Kaether C, Thinakaran G, Sisodia S (2012) Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med 2: a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghosh P, Dahms NM, Kornfeld S (2003) Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol 4: 202–212. [DOI] [PubMed] [Google Scholar]
- 13. Hawkes C, Kar S (2004) The insulin-like growth factor-II/mannose-6-phosphate receptor: structure, distribution and function in the central nervous system. Brain Res Brain Res Rev 44: 117–140. [DOI] [PubMed] [Google Scholar]
- 14. El-Shewy HM, Luttrell LM (2009) Insulin-like growth factor-2/mannose-6 phosphate receptors. Vitam Horm 80: 667–697. [DOI] [PubMed] [Google Scholar]
- 15. Dahms N, Hancock MK (2002) P-type lectins. Biochim Biophys Acta 1572: 317–340. [DOI] [PubMed] [Google Scholar]
- 16. Hille-Rehfeld A (1995) Mannose 6-phosphate receptors in sorting and transport of lysosomal enzymes. Biochim Biophys Acta 1241: 177–194. [DOI] [PubMed] [Google Scholar]
- 17. Kar S, Poirier J, Guevara J, Dea D, Hawkes C, et al. (2006) Cellular distribution of insulin-like growth factor-II/mannose-6-phosphate receptor in normal human brain and its alteration in Alzheimer’s disease pathology. Neurobiol Aging 27: 199–210. [DOI] [PubMed] [Google Scholar]
- 18. Cataldo AM, Peterhoff CM, Schmidt SD, Terio NB, Duff K, et al. (2004) Presenilin mutations in familial Alzheimer disease and transgenic mouse models accelerate neuronal lysosomal pathology. J Neuropathol Exp Neurol 63: 821–830. [DOI] [PubMed] [Google Scholar]
- 19. Amritraj A, Hawkes C, Phinney AL, Mount HT, Scott CD, et al. (2009) Altered levels and distribution of IGF-II/M6P receptor and lysosomal enzymes in mutant APP and APP+PS1 transgenic mouse brains. Neurobiol Aging 30: 54–70. [DOI] [PubMed] [Google Scholar]
- 20. Hemming ML, Elias JE, Gygi SP, Selkoe DJ (2009) Identification of beta-secretase (BACE1) substrates using quantitative proteomics. PLoS One 4: e8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gabel CA, Goldberg DE, Kornfeld S (1983) Identification and characterization of cells deficient in the mannose 6-phosphate receptor: evidence for an alternate pathway for lysosomal enzyme targeting. Proc Natl Acad Sci U S A 80: 775–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kyle JW, Nolan CM, Oshima A, Sly WS (1988) Expression of human cation-independent mannose 6-phosphate receptor cDNA in receptor-negative mouse P388D1 cells following gene transfer. J Biol Chem 263: 16230–16235. [PubMed] [Google Scholar]
- 23. Wood RJ, Hulett MD (2008) Cell surface-expressed cation-independent mannose 6-phosphate receptor (CD222) binds enzymatically active heparanase independently of mannose 6-phosphate to promote extracellular matrix degradation. J Biol Chem 283: 4165–4176. [DOI] [PubMed] [Google Scholar]
- 24. Motyka B, Korbutt G, Pinkoski MJ, Heibein JA, Caputo A, et al. (2000) Mannose 6-phosphate/insulin-like growth factor II receptor is a death receptor for granzyme B during cytotoxic T cell-induced apoptosis. Cell 103: 491–500. [DOI] [PubMed] [Google Scholar]
- 25. Di Bacco A, Gill G (2003) The secreted glycoprotein CREG inhibits cell growth dependent on the mannose-6-phosphate/insulin-like growth factor II receptor. Oncogene 22: 5436–5445. [DOI] [PubMed] [Google Scholar]
- 26. Maulik M, Thinakaran G, Kar S (2013) Alterations in gene expression in mutant amyloid precursor protein transgenic mice lacking Niemann-Pick type C1 protein. PLoS One 8: e54605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maulik M, Ghoshal B, Kim J, Wang Y, Yang J, et al. (2012) Mutant human APP exacerbates pathology in a mouse model of NPC and its reversal by a beta-cyclodextrin. Hum Mol Genet 21: 4857–4875. [DOI] [PubMed] [Google Scholar]
- 28. Amritraj A, Wang Y, Revett TJ, Vergote D, Westaway D, et al. (2013) Role of cathepsin D in U18666A-induced neuronal cell death: potential implication in Niemann-Pick type C disease pathogenesis. J Biol Chem 288: 3136–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maulik M, Westaway D, Jhamandas JH, Kar S (2013) Role of cholesterol in APP metabolism and its significance in Alzheimer’s disease pathogenesis. Mol Neurobiol 47: 37–63. [DOI] [PubMed] [Google Scholar]
- 30. Di Paolo G, Kim TW (2011) Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat Rev Neurosci 12: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leduc V, Jasmin-Belanger S, Poirier J (2010) APOE and cholesterol homeostasis in Alzheimer’s disease. Trends Mol Med 16: 469–477. [DOI] [PubMed] [Google Scholar]
- 32. Martins IJ, Berger T, Sharman MJ, Verdile G, Fuller SJ, et al. (2009) Cholesterol metabolism and transport in the pathogenesis of Alzheimer’s disease. J Neurochem 111: 1275–1308. [DOI] [PubMed] [Google Scholar]
- 33. Nixon RA, Cataldo AM (2006) Lysosomal system pathways: genes to neurodegeneration in Alzheimer’s disease. J Alzheimers Dis 9: 277–289. [DOI] [PubMed] [Google Scholar]
- 34. Cataldo AM, Barnett JL, Pieroni C, Nixon RA (1997) Increased neuronal endocytosis and protease delivery to early endosomes in sporadic Alzheimer’s disease: neuropathologic evidence for a mechanism of increased beta-amyloidogenesis. J Neurosci 17: 6142–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haque A, Banik NL, Ray SK (2008) New insights into the roles of endolysosomal cathepsins in the pathogenesis of Alzheimer’s disease: cathepsin inhibitors as potential therapeutics. CNS Neurol Disord Drug Targets 7: 270–277. [DOI] [PubMed] [Google Scholar]
- 36. Nishimoto I, Hata Y, Ogata E, Kojima I (1987) Insulin-like growth factor II stimulates calcium influx in competent BALB/c 3T3 cells primed with epidermal growth factor. Characteristics of calcium influx and involvement of GTP-binding protein. J Biol Chem 262: 12120–12126. [PubMed] [Google Scholar]
- 37. Minniti CP, Kohn EC, Grubb JH, Sly WS, Oh Y, et al. (1992) The insulin-like growth factor II (IGF-II)/mannose 6-phosphate receptor mediates IGF-II-induced motility in human rhabdomyosarcoma cells. J Biol Chem 267: 9000–9004. [PubMed] [Google Scholar]
- 38. Rogers SA, Purchio AF, Hammerman MR (1990) Mannose 6-phosphate-containing peptides activate phospholipase C in proximal tubular basolateral membranes from canine kidney. J Biol Chem 265: 9722–9727. [PubMed] [Google Scholar]
- 39. Hawkes C, Jhamandas JH, Harris KH, Fu W, MacDonald RG, et al. (2006) Single transmembrane domain insulin-like growth factor-II/mannose-6-phosphate receptor regulates central cholinergic function by activating a G-protein-sensitive, protein kinase C-dependent pathway. J Neurosci 26: 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kar S, Seto D, Dore S, Hanisch U, Quirion R (1997) Insulin-like growth factors-I and -II differentially regulate endogenous acetylcholine release from the rat hippocampal formation. Proc Natl Acad Sci U S A 94: 14054–14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chu CH, Tzang BS, Chen LM, Kuo CH, Cheng YC, et al. (2008) IGF-II/mannose-6-phosphate receptor signaling induced cell hypertrophy and atrial natriuretic peptide/BNP expression via Galphaq interaction and protein kinase C-alpha/CaMKII activation in H9c2 cardiomyoblast cells. J Endocrinol 197: 381–390. [DOI] [PubMed] [Google Scholar]
- 42. Thinakaran G, Borchelt DR, Lee MK, Slunt HH, Spitzer L, et al. (1996) Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron 17: 181–190. [DOI] [PubMed] [Google Scholar]
- 43. Thinakaran G, Harris CL, Ratovitski T, Davenport F, Slunt HH, et al. (1997) Evidence that levels of presenilins (PS1 and PS2) are coordinately regulated by competition for limiting cellular factors. J Biol Chem 272: 28415–28422. [DOI] [PubMed] [Google Scholar]
- 44. Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, et al. (2003) The role of presenilin cofactors in the gamma-secretase complex. Nature 422: 438–441. [DOI] [PubMed] [Google Scholar]
- 45. Eckman EA, Eckman CB (2005) Abeta-degrading enzymes: modulators of Alzheimer’s disease pathogenesis and targets for therapeutic intervention. Biochem Soc Trans 33: 1101–1105. [DOI] [PubMed] [Google Scholar]
- 46. Miners JS, Baig S, Palmer J, Palmer LE, Kehoe PG, et al. (2008) Abeta-degrading enzymes in Alzheimer’s disease. Brain Pathol 18: 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Leissring MA (2008) The AbetaCs of Abeta-cleaving proteases. J Biol Chem 283: 29645–29649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haag MD, Hofman A, Koudstaal PJ, Stricker BH, Breteler MM (2009) Statins are associated with a reduced risk of Alzheimer disease regardless of lipophilicity. The Rotterdam Study. J Neurol Neurosurg Psychiatry 80: 13–17. [DOI] [PubMed] [Google Scholar]
- 49. Sparks DL, Kryscio RJ, Sabbagh MN, Connor DJ, Sparks LM, et al. (2008) Reduced risk of incident AD with elective statin use in a clinical trial cohort. Curr Alzheimer Res 5: 416–421. [DOI] [PubMed] [Google Scholar]
- 50. Zamrini E, McGwin G, Roseman JM (2004) Association between statin use and Alzheimer’s disease. Neuroepidemiology 23: 94–98. [DOI] [PubMed] [Google Scholar]
- 51. Simons M, Keller P, De Strooper B, Beyreuther K, Dotti CG, et al. (1998) Cholesterol depletion inhibits the generation of beta-amyloid in hippocampal neurons. Proc Natl Acad Sci U S A 95: 6460–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fassbender K, Simons M, Bergmann C, Stroick M, Lutjohann D, et al. (2001) Simvastatin strongly reduces levels of Alzheimer’s disease beta -amyloid peptides Abeta 42 and Abeta 40 in vitro and in vivo. Proc Natl Acad Sci U S A 98: 5856–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Puglielli L, Tanzi RE, Kovacs DM (2003) Alzheimer’s disease: the cholesterol connection. Nat Neurosci 6: 345–351. [DOI] [PubMed] [Google Scholar]
- 54. Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, et al. (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41: 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kolsch H, Lutjohann D, Ludwig M, Schulte A, Ptok U, et al. (2002) Polymorphism in the cholesterol 24S-hydroxylase gene is associated with Alzheimer’s disease. Mol Psychiatry 7: 899–902. [DOI] [PubMed] [Google Scholar]
- 56. Shibata N, Kawarai T, Lee JH, Lee HS, Shibata E, et al. (2006) Association studies of cholesterol metabolism genes (CH25H, ABCA1 and CH24H) in Alzheimer’s disease. Neurosci Lett 391: 142–146. [DOI] [PubMed] [Google Scholar]
- 57. Ma J, Yee A, Brewer HB Jr, Das S, Potter H (1994) Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature 372: 92–94. [DOI] [PubMed] [Google Scholar]
- 58. Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, et al. (2011) Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci Transl Med 3: 89ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, et al. (2008) Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J Clin Invest 118: 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mullins C, Bonifacino JS (2001) The molecular machinery for lysosome biogenesis. Bioessays 23: 333–343. [DOI] [PubMed] [Google Scholar]
- 61. Repnik U, Stoka V, Turk V, Turk B (2012) Lysosomes and lysosomal cathepsins in cell death. Biochim Biophys Acta 1824: 22–33. [DOI] [PubMed] [Google Scholar]
- 62. Roberg K, Ollinger K (1998) Oxidative stress causes relocation of the lysosomal enzyme cathepsin D with ensuing apoptosis in neonatal rat cardiomyocytes. Am J Pathol 152: 1151–1156. [PMC free article] [PubMed] [Google Scholar]
- 63. Amritraj A, Peake K, Kodam A, Salio C, Merighi A, et al. (2009) Increased activity and altered subcellular distribution of lysosomal enzymes determine neuronal vulnerability in Niemann-Pick type C1-deficient mice. Am J Pathol 175: 2540–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schechter I, Ziv E (2008) Kinetic properties of cathepsin D and BACE 1 indicate the need to search for additional beta-secretase candidate(s). Biol Chem 389: 313–320. [DOI] [PubMed] [Google Scholar]
- 65. Schechter I, Ziv E (2011) Cathepsins S, B and L with aminopeptidases display beta-secretase activity associated with the pathogenesis of Alzheimer’s disease. Biol Chem 392: 555–569. [DOI] [PubMed] [Google Scholar]
- 66. Sundelof J, Sundstrom J, Hansson O, Eriksdotter-Jonhagen M, Giedraitis V, et al. (2010) Higher cathepsin B levels in plasma in Alzheimer’s disease compared to healthy controls. J Alzheimers Dis 22: 1223–1230. [DOI] [PubMed] [Google Scholar]
- 67. Cataldo AM, Barnett JL, Berman SA, Li J, Quarless S, et al. (1995) Gene expression and cellular content of cathepsin D in Alzheimer’s disease brain: evidence for early up-regulation of the endosomal-lysosomal system. Neuron 14: 671–680. [DOI] [PubMed] [Google Scholar]
- 68. Hook VY, Kindy M, Hook G (2008) Inhibitors of cathepsin B improve memory and reduce beta-amyloid in transgenic Alzheimer disease mice expressing the wild-type, but not the Swedish mutant, beta-secretase site of the amyloid precursor protein. J Biol Chem 283: 7745–7753. [DOI] [PubMed] [Google Scholar]
- 69. Kindy MS, Yu J, Zhu H, El-Amouri SS, Hook V, et al. (2012) Deletion of the cathepsin B gene improves memory deficits in a transgenic ALZHeimer’s disease mouse model expressing AbetaPP containing the wild-type beta-secretase site sequence. J Alzheimers Dis 29: 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Braulke T, Bonifacino JS (2009) Sorting of lysosomal proteins. Biochim Biophys Acta 1793: 605–614. [DOI] [PubMed] [Google Scholar]
- 71. McKinnon T, Chakraborty C, Gleeson LM, Chidiac P, Lala PK (2001) Stimulation of human extravillous trophoblast migration by IGF-II is mediated by IGF type 2 receptor involving inhibitory G protein(s) and phosphorylation of MAPK. J Clin Endocrinol Metab 86: 3665–3674. [DOI] [PubMed] [Google Scholar]
- 72. Zhang Q, Tally M, Larsson O, Kennedy RT, Huang L, et al. (1997) Insulin-like growth factor II signaling through the insulin-like growth factor II/mannose-6-phosphate receptor promotes exocytosis in insulin-secreting cells. Proc Natl Acad Sci U S A 94: 6232–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hawkes C, Amritraj A, Macdonald RG, Jhamandas JH, Kar S (2007) Heterotrimeric G proteins and the single-transmembrane domain IGF-II/M6P receptor: functional interaction and relevance to cell signaling. Mol Neurobiol 35: 329–345. [DOI] [PubMed] [Google Scholar]
- 74. Chen DY, Stern SA, Garcia-Osta A, Saunier-Rebori B, Pollonini G, et al. (2011) A critical role for IGF-II in memory consolidation and enhancement. Nature 469: 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rivera EJ, Goldin A, Fulmer N, Tavares R, Wands JR, et al. (2005) Insulin and insulin-like growth factor expression and function deteriorate with progression of Alzheimer’s disease: link to brain reductions in acetylcholine. J Alzheimers Dis 8: 247–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression profiles in MS9II cell compared to MS cells as studied using real-time RT-PCR arrays.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the paper.