Abstract
Mammalian APeg3 is an antisense gene that is localized within the 3′-untranslated region of the imprinted gene, Peg3. APeg3 is expressed only in the vasopressinergic neurons of the hypothalamus, thus is predicted to play significant roles in this specific area of the brain. In the current study, we investigate the functions of APeg3 with comparative genomics and cell line-based functional approaches. The transcribed region of APeg3 displays high levels of sequence conservation among placental mammals, but without any obvious open reading frame, suggesting that APeg3 may have been selected as a ncRNA gene during eutherian evolution. This has been further supported by the detection of a conserved local RNA secondary structure within APeg3. RNA secondary structure analyses indicate a single conserved hairpin-loop structure towards the 5′ end of the transcript. The results from cell line-based transfection experiments demonstrate that APeg3 has the potential to down-regulate the transcription and protein levels of Peg3. The observed down-regulation by APeg3 is also somewhat orientation-independent. Overall, these results suggest that APeg3 has evolved as a ncRNA gene and controls the function of its sense gene Peg3 within specific neuronal cells.
Keywords: Peg3, APeg3, ncRNA, Vasopressin neuron, Genomic imprinting
1. Introduction
APeg3 is an antisense gene that is located within the 3′-untranslated region of an imprinted gene, Peg3 (paternally expressed gene 3). The expression of APeg3 is detected only in vasopressinergic neurons of the hypothalamus, suggesting a very specialized role in the neuronal cells controlling blood pressure and the volume of bodily fluid in mammals (Glasgow et al., 2005). In fact, APeg3 was initially identified as a gene that is highly up-regulated in response to osmotic challenges in rat brains (Glasgow et al., 2005). Studies on Peg3 also demonstrate up-regulation against osmotic stress in this cell type, suggesting that both APeg3 and Peg3 may play important roles in the functions of vasopressin-expressing neurons (Yamashita et al., 2002). Earlier studies from rat brain suggest that APeg3 might code for a small-sized Open Reading Frame (ORF), but were not substantiated by later studies as the observed ORF is not conserved in other mammals, such as humans and mice. Interestingly, APeg3 is also maternally imprinted as seen in Peg3: only the paternal allele is expressed while the maternal allele is repressed (Choo et al., 2008). Nevertheless, the functional impetus for APeg3 imprinting is currently not well understood.
APeg3 is one of several ncRNA genes that have been identified from mammalian imprinted domains. The list of the imprinted ncRNA genes includes H19, IPW, Kcnq1ot1, Nespas, Airn, Copg2, and Gtl2 (Koerner et al., 2009). APeg3 is very unique compared to the other imprinted ncRNA genes in the following aspects. First and foremost, APeg3 is the only imprinted antisense transcript expressed from the same (paternal) allele as its sense counterpart. Similar antisense ncRNA counterparts to imprinted genes, such as anti-Rtl1 and Copg2-AS, are selectively expressed from the opposite allele as the sense genes (Ferguson-Smith, 2011; Yamasaki et al., 2000). Secondly, the size of APeg3 is relatively small, 1.5 kb in length, whereas the size of the other imprinted non-coding genes range up to several hundreds of kb in length (Glasgow et al., 2005; Koerner et al., 2009). H19 is the only other imprinted ncRNA of similar length at ∼1.9 kb (Brannan et al., 1990). Thirdly, APeg3 lies antisense to the protein-coding gene, Peg3. Furthermore, the transcribed region overlaps with the 3′UTR of Peg3. This is quite different from other imprinted ncRNA genes, such as Gtl2, which tend to overlap with the entire imprinted region. Finally, the transcript of APeg3 is detected as an intronless mRNA with poly-A tails in vivo, suggesting that APeg3's mRNA goes through normal processes as Pol II transcripts (Choo et al., 2008). By contrast, other long ncRNA genes, such as Airn, Kcnqot1, and Rian reside inside the nucleus and do not go through normal processes as Pol II transcripts. Despite these unique features, the biological roles of APeg3 are currently unknown. However, given the antisense/sense relationship between APeg3 and Peg3, it is reasonable to predict that the main function of APeg3 may be controlling the transcription and protein levels of Peg3. To investigate this possibility, we performed a series of comparative genomics and cell-line based functional assays in the current study. The results suggest that APeg3 may have evolved as a ncRNA gene controlling Peg3 mRNA and protein levels.
2. Materials and methods
2.1. Database search and gene prediction
The intronless human antisense transcript, PEG3-AS1 (NR_023847.2), was used as a reference sequence in the BLAST program offered through the UCSC genome browser (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Using this sequence, homologous nucleotide sequences were obtained from the available genome sequences of multiple mammalian species. After our initial inspection of the retrieved sequences, one representative sequence from each order of placental mammals was used for sequence alignment with the ClustalW multiple sequence alignment (http://www.genome.jp/tools/clustalw/) as well as CLC Bio Workbench. The 1.5-kb genomic region for each species' APeg3 gene is as follows: Mus musculus for Rodentia (GRCm38/mm10 chr7: 6,706,295–6,707,624), Homo sapiens for Primates (GRCh37/hg19 chr19: 57,323,893–57,325,161), Equus caballus for Perissodactyla (Broad/EquCab2 chr10: 25,780,355–25,781,617), Oryctolagus cuniculis for Lagomorpha (Broad/oryCun2 chrUn0113: 622,308–623,587), Loxodonta africana for Proboscidea (Broad/loxAfr3 scaffold_4: 18,505,516–18,506,810), Tursiops truncatus for Cetacea (Baylor Ttru_1.4/turTru2 JH478484: 13,610–14,893), Myotis lucifugus for Chiroptera (Broad Institute Myoluc2.0/myoluc2 GL430552: 92,812– 94,034), Dasypus novemcinctus for Cingulata (Baylor/dasNov3 JH562679: 85,036–86,329) and Trichechus manatus for Sirenia (Broad v1.0/triMan1 JH594782: 4,734,304–4,735,606). These sequences were further analyzed to identify several sequence features for the transcribed region of APeg3. Transcription Start Site (TSS) and Poly-A site were identified based on switchgear analyses. A potential TATA box was identified 25-bp upstream of the TSS site as an evolutionarily conserved element. A target site for miR-124 was also identified based on TargetScan (http://www.targetscan.org/) data. The results of these analyses are readily available on the UCSC genome browser. All the sequences used for the current study are available (Supplementary materials 2–3).
2.2. Secondary structure prediction of APeg3
The primary sequence alignment of APeg3 was used as an input for covariant and minimal fold energy (MFE) analysis using Vienna Suite software (Washietl et al., 2005). RNA folding was predicted using the following two programs: RNA AliFold (http://rna.tbi.univie.ac.at/cgi-bin/RNAalifold.cgi) to predict the macro-secondary structures (global folds) and RNAz (http://www.tbi.univie.ac.at/∼wash/RNAz/) to predict the micro-secondary structures (local folds) of APeg3 (Washietl et al., 2005; Gruber et al., 2007). The programs used the primary sequence alignments to generate a consensus fold structure for APeg3. This series of analyses used two controls. The sequence of the H19 gene was included as a positive control, which has been previously shown to have significantly conserved secondary structures important for its function as a lncRNA (Juan et al., 2000), and a shuffled sequence of APeg3 was also included as a negative control.
2.3. Construction and transfection of APeg3 overexpression vectors
To construct the Short- and Full-length APeg3 expression vectors, the cDNA prepared from adult mouse brains was amplified with artificial BamHI and NotI sites using the following primers:
APeg3 Full-length-APeg3Full_BamHI CGCGGATCCGGGAATTAA GTCTGGAGACACAAAGATCTAAGG and APeg3Short_NotI ATAA GAATGCGGCCGCGCACCAGTGCAGGTGGTGCGGA. APeg3 Short-APeg3Short_BamHI CGCGGATCCCAATCAGTCTCAAGGGGTCTGGGT and APeg3Short_NotI ATAAGAATGCGGCCGCGCACCAGTGCAGGTGG TGCGGA.
(−)APeg3 Full-length-(−)APeg3Short_BamHI CGCGGATCCGCACC AGTGCAGGTGGTGCGGA and (−)APeg3Full_NotI ATAAGAATGCGGCCGCGGGAATTAAGTCTGGAGACACAAAGATCTAAGG (−)APeg3 Short-length-(−)APeg3Short_NotI ATAAGAATGCGGCCGCCAATCA GTCTCAAGGGGTCTGGGT (−)APeg3Short_BamHI CGCGGATCCGCA CCAGTGCAGGTGGTGCGGA
The amplified products were digested with BamHI and NotI and cloned into pCDNA3.1Hygro(−) vector (Invitrogen). In this vector system, the expression of APeg3 was driven by the CMV promoter and terminated by the Poly-A signal of BGH. The constructed vectors were transfected into Neuro2A cells with each well containing 0.15 × 106 cells using 4 μL lipofectamine 2000 (Invitrogen). Transfections were carried out in triplicate with 3 wells designated for subsequent RNA isolation and 3 wells for protein extraction. The transfected cells were selected using 500 μg/mL hygromycin B 24 h post-transfection. The cells were harvested for RNA isolation using TRIzol (Invitrogen) 48 h post-transfection, and also for protein extraction in lysis buffer (0.25 M Tris–HCl, pH 7.8, plus 0.1% NP-40) supplemented with fresh β-mercaptoethanol and the proteinase inhibitor cocktail (Calbiotech).
2.4. cDNA synthesis and quantitative Reverse-Transcription PCR (qRT-PCR)
Total RNA was isolated from transfected Neuro2A cells using a commercial kit (Trizol, Invitrogen). The isolated RNA was first reverse-transcribed using the MLV First-Strand Synthesis System (Invitrogen) with random and oligo dT primers. Subsequent cDNA was used as a template for qRT-PCR. This analysis was performed with the iQ SYBR green supermix (Bio-Rad) using the iCycler iQTM multicolor real-time detection system (Bio-Rad). All qRT-PCR reactions were carried out for 40 cycles under standard PCR conditions. The expression levels of Peg3 were first normalized to those of β-Actin, then compared to those of the empty vector (pCDNA) controls. The primers for Peg3 and β-Actin were as follows: Peg3-1a (5′-GGTTCAGTGTGGGTGCACTAGA CT-3′), Peg3-1b.1 (5′-GCTCACACCCAAGGGCTTGAGCGT-3′), Peg3-1b.3 (5′-TCCCTAGTGTGCATGATCTGGT-3′), bactin-1a (5′-GAGCACCCTGTGCT GCTCACCGA-3′) and bactin-1b (5′-CTCTTTGATGTCACGCACGATTTC-3′).
2.5. Protein isolations and Western blots
The transfected Neuro2A cells were first rinsed with 1× PBS, and lysed with the lysis buffer (0.25 M Tris–HCl, pH 7.8, plus 0.1% NP-40). The subsequent crude lysates were collected and snap-frozen in liquid nitrogen. Later, the lysate was then thawed at 4 °C to prevent protein degradation. The cellular debris was removed by centrifugation for 10 min at 4 °C. Protein concentrations were determined by the Bradford assay kit (Pierce), using diluted BSA as protein standards. Sixty micrograms of each lysate was separated on 10% SDS-PAGE gels and transferred to PVDF membranes (Hybond-P, Amersham) using a Mini Trans-Blot wet transfer cell (Bio-Rad). Membranes were blocked for 1 h in the Tris-buffered saline containing 1% skim milk and 0.05% Tween100, and incubated overnight at 4 °C with the custom-made anti-PEG3 antibody (Kim et al., 2012). These blots were incubated for an additional 1 h with the secondary antibody linked to horseradish peroxidase (Sigma). The blots were developed using the Western blot detection system according to the manufacturer's protocol (Intron Biotech). Densitometry was measured using Jmol: an open-source Java viewer for chemical structures in 3D.
3. Results
3.1. APeg3 as an evolutionarily conserved ncRNA in placental mammals
To analyze the evolutionary conservation of APeg3, human PEG3-AS1 sequence (NR 023847.2) was used as a probe to search the UCSC genome browser, yielding genomic sequences for each of the 9 eutherian orders (Supplementary material 1). Using these sequences, pairwise sequence comparison analyses were conducted, and the results indicated high levels of sequence conservation between different mammals: 76 to 84% nucleotide sequence identity in the 1.5-kb transcribed region of APeg3 (Table 1). Multiple sequence alignments were also performed using the 9 representative sequences, and the results were visualized as a graph in the bottom panel of Fig. 1. The UCSC PhyloP-derived placental mammal sequences used for the zoomed-out conservation include multiple species corresponding mainly to primate lineages as these are the most complete sequenced genomes and do not account for gaps in sequences. In this way, the conservation analysis could be biased. However, our custom zoomed-in conservation was derived from only one representative species of each order within the class eutheria, offering an unbiased characterization of the region. Differences in conservation level between the zoomed-out view and the zoomed-in view are the result of hand-picking genomic regions of APeg3 to enhance the accuracy of the initial PhyloP conservation. High levels of sequence conservation are again observed throughout the 1.5-kb region with many small-size insertions/deletions specific to individual species, which can be seen as sudden drop-offs in the graph. Several sequence motifs, important for Pol II transcription, are also found within or around this 1.5-kb genomic region, including a TATA box, Transcription Start Site (TSS) and two Poly-Adenylation sites. However, there are no ORFs or Kozak sequences that are conserved among placental mammals. These results suggest that the 1.5-kb genomic region of APeg3 is a gene template for a poly-adenylated non-coding RNA (ncRNA). The high levels of sequence conservation detected within the 1.5-kb region are also unusual, and can be contrasted with the 3′-UTR of an adjacent gene, Zim1, which shows no sequence conservation (Mammalian Cons Plot in Fig. 1). Overall, the high level of primary sequence conservation further supports that the region has not evolved simply as an UTR for the sense gene, Peg3. Instead, this region demonstrates signs of selection as a ncRNA gene during mammalian evolution.
Table 1.
Pairwise sequence comparison of APeg3.
| Organism | E. caballus | T. truncatus | M. lucifugus | L. africana | T. manatus | D. novemcinctus | M. musculus | O. cuniculis | H. Sapiens |
|---|---|---|---|---|---|---|---|---|---|
| Equus caballus | 100 | 84.73 | 82.41 | 77.93 | 79.82 | 78.56 | 75.5 | 80.18 | 83.2 |
| Tursiops truncatus | 84.73 | 100 | 79.78 | 75.12 | 76.47 | 75.23 | 71.66 | 77.13 | 80.99 |
| Myotis lucifugus | 82.41 | 79.78 | 100 | 74.03 | 74.92 | 73.96 | 72.64 | 77.18 | 79.22 |
| Loxodonta africana | 77.93 | 75.12 | 74.03 | 100 | 89.01 | 77.34 | 68.61 | 73.15 | 75.53 |
| Trichechus manatus | 79.82 | 76.47 | 74.92 | 89.01 | 100 | 79.1 | 70.19 | 74.41 | 77.78 |
| Dasypus novemcinctus | 78.56 | 75.23 | 73.96 | 77.34 | 79.1 | 100 | 69.5 | 74.17 | 76.42 |
| Mus musculus | 75.5 | 71.66 | 72.64 | 68.61 | 70.19 | 69.5 | 100 | 79.18 | 77.33 |
| Oryctolagus cuniculis | 80.18 | 77.13 | 77.18 | 73.15 | 74.41 | 74.17 | 79.18 | 100 | 83.84 |
| Homo sapiens | 83.2 | 80.99 | 79.22 | 75.53 | 77.78 | 76.42 | 77.33 | 83.84 | 100 |
Fig. 1.
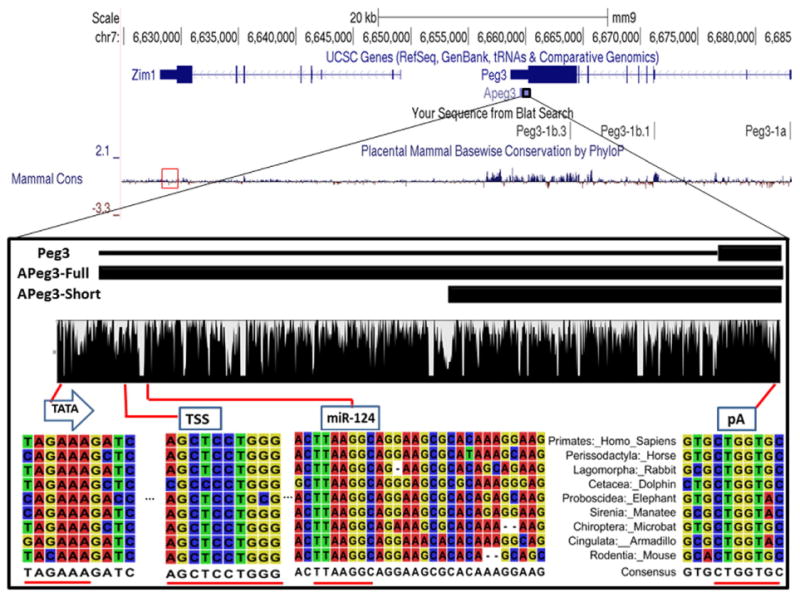
Evolutionary conservation of APeg3 among placental mammals. UCSC Genome Browser Mus musculus assembly 9 (mm9) displays the 100-kb genomic region surrounding mouse APeg3. The transcribed and exon regions of Peg3 and Zim1 are also presented along with the transcribed region of APeg3. Evolutionary conservation of this genomic interval among placental mammals is shown with a graphical output of the PhyloP analysis. PhyloP conservation analysis of Zim1 3′ UTR is designated in a red box. In a zoom-in view, two black boxes represent the different-length known cDNAs for APeg3. APeg3-Full is based on the deposited cDNA sequences from human and rat, while APeg3-Short is based on the cDNA sequences from mouse. The evolutionary conservation of APeg3 was further analyzed using the genomic sequences obtained from 9 representative placental mammals (Supplementary Fig. 1). This analysis identified a highly conserved TATA box (TATA), transcriptional start site (TSS) and a poly-adenylation signal (pA1). This analysis also confirmed the conservation of a potential target site for miRNA (miRNA-124). Areas of high conservation within these regions are underscored in red.
3.2. Global and local secondary structure of APeg3 with evolutionary conservation
To determine potential functions of APeg3 as a structural ncRNA, secondary structure predictions were performed using RNAalifold and RNAz programs. First, RNAalifold uses sequence alignments and determines folding free energies of relatively large RNA sequences (>200 bp) (Bernhart et al., 2008; Ge and Zhang, 2013). This program produces a consensus fold structure based on the lowest folding energy modeled and weights the conservation of folded structures among individual sequences. Both APeg3 and the corresponding reverse complement (−)APeg3 alignment were analyzed using RNAalifold to detect potentially large-scale secondary structures in both orientations. This series of analyses also used two controls: H19 as a positive control, an imprinted ncRNA previously seen to harbor functional characteristics of similar length to APeg3, and a shuffled APeg3 alignment as a negative control. According to the outcomes of the prediction (Fig. 2), the sense and antisense APeg3 exhibit similar conservation levels of secondary structure with different overall structures.
Fig. 2.
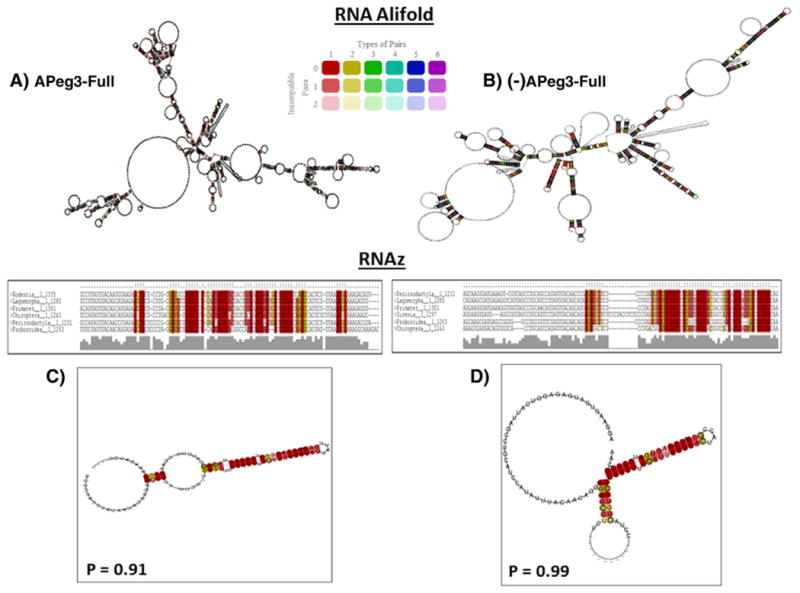
Predicted secondary structures of APeg3 mRNA. The 1.5-kb nucleotide sequences from 9 representative mammals were used to predict the global secondary structures of APeg3 using the Alifold prediction program: sense (A) and antisense (B) strands of APeg3. Conserved bases are color-coded based on their conservation level (legend inset). Local secondary structures were also modeled using the lowest fold energy as well as covariant analysis on small (100 bp) windows within APeg3. The RNAz program predicted a single conserved hairpin-loop structure within the sense (P = 0.91) and antisense (P = 0.99) APeg3 transcripts (C, D) with the 98.93% folding accuracy. The predicted hairpin-loop structure is located in the 5′ side of the APeg3 transcript (nucleotide positions 145–185 in mouse APeg3).
Second, the RNAz program was used to detect any conserved micro-secondary structures within APeg3. The program analyzes multiple sequence alignments and recognizes conserved fold structures in small (200 bp) sliding-windows (Gruber et al., 2007). These folds are based on the lowest MFE of each short sequence. Covariant analysis was then performed on each fold and the resulting folds were scored by the program. In this program, a class probability (P) is used to measure the likelihood of a fold. The program uses a cutoff of P = 0.9, which can accurately predict 75% of known ncRNAs and classify folds with 99% specificity (Washietl et al., 2005). According to the outcomes, a single fold was predicted for APeg3 in the 1.5-kb interval (Fig. 2C). The fold was conserved at the 5′-side of the transcript (nucleotide positions 145–185 in NR 023846.1) (P = 0.99). Interestingly, the antisense of APeg3 also displayed one conserved stem-loop structure, located at the 3′-side of the transcript (P = 0.91) (Fig. 2D). Overall, these secondary structure fold predictions indicate the presence of a conserved hairpin loop structure in both sense and antisense orientations of APeg3. A series of predictions were also conducted using two controls, H19 and the shuffled APeg3. This series of analyses predicted a known small fold, miR675, within H19 (P = 1.0), and only one insignificant fold within the shuffled APeg3 (P = 0.76); confirming the sensitivity of the RNAz program.
3.3. APeg3 down-regulates Peg3 mRNA
As seen in many antisense transcripts (Osato et al., 2007; Katayama et al., 2005), the potential function of APeg3 may be to regulate the expression of the sense gene, Peg3. To determine whether APeg3 has the potential to regulate Peg3, we performed a series of in vitro experiments as described below. We have constructed 4 expression vectors containing the different portions of mouse APeg3: APeg3-Full and -Short in both sense and antisense directions (the extent of APeg3-Short is indicated in Fig. 1). After transfections of these vectors to Neuro2A cells, we measured Peg3 mRNA levels using qRT-PCR (Fig. 3). This series of expression analyses used an empty vector as a control (pCDNA). Overexpression of APeg3-Short resulted in down-regulation of Peg3 by 57.60 ± 0.4% (P = 0.0034), whereas overexpression of (−) APeg3-Short resulted in a much smaller change in Peg3 expression (27.80 ±C 0.16%)(P = 0.0536). On the other hand, overexpression of APeg3-Full caused a much greater down-regulation of Peg3 (81.32 ± 0.03%) (P = 0.0008), which was an ∼20% lower expression than those from APeg3-Short. Interestingly, however, overexpression of (−) APeg3-Full also caused down-regulation of Peg3 (49.30 ±0.054%) (P = 0.0271). Given the results from both directions of APeg3-Full, the observed down-regulation by APeg3-Full may be orientation-independent. Furthermore, it is possible that the potential region causing this down-regulation might be located within the 5′-side of the 1.5-kb region of APeg3 since this down-regulation is much more obvious in the Full-length constructs than in the Short-length constructs lacking the 5′-side of APeg3. To account for any minor transcriptional variants of Peg3, we also performed the previous qRT-PCR with primers amplifying the entire Peg3 transcript (Peg3-1a, Peg3-1b.3) and observed a similar trend (Supplementary Fig. 2). Peg3 mRNA levels were decreased in transfections of APeg3-Short (71.43 ± 0.056%) (P = 0.0118), APeg3-Full (11.13 ± 0.0635%) (P = 0.0229) and (−)APeg3-Full (24.61 ± 0.237%) (P = 0.0332), while changes in Peg3 mRNA were not statistically significant in the (−) APeg3-Short (54.18 ± 0.425%) (P = 0.2135) transfections. However, these alternative forms of Peg3 have minimal contribution as compared to the major Peg3 transcript (Kim et al., 1997). Taken together, these results demonstrate that APeg3 has the potential to down-regulate the level of Peg3 mRNA in vitro.
Fig. 3.
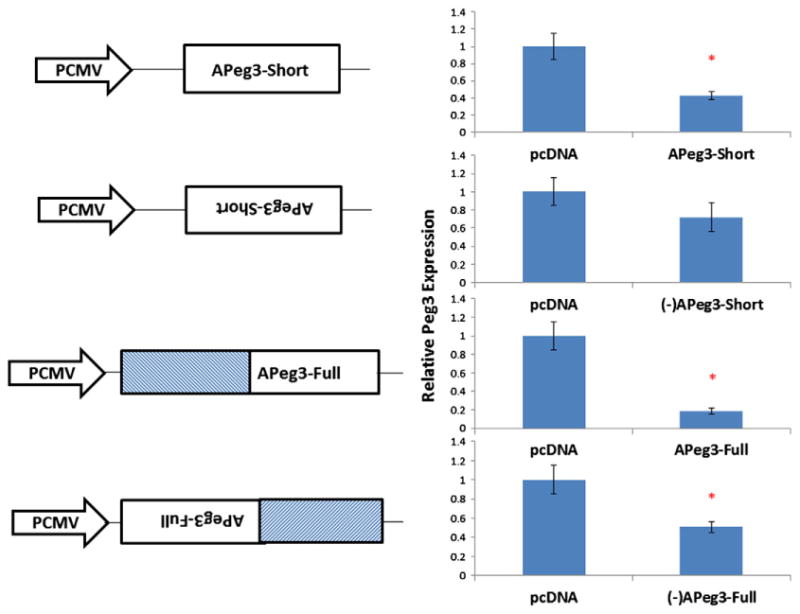
Effects of APeg3 overexpression on the transcription levels of Peg3. A series of constructs transcribing APeg3 were transfected into Neuro2A cells. APeg3 transcription was driven by the Cytomegalovirus (CMV) promoter, and terminated and polyadenylated by a Bovine Growth Hormone (BGH) terminator. Transfected cells were selected with hygromycin antibiotic (500 mg/mL). Potential effects of APeg3 overexpression were measured by determining the transcription levels of Peg3 using the total RNA isolated from the transfected cells with qRT-PCR. The transcription levels of Peg3 were first normalized to β-Actin, then compared to a control (pCDNA, empty vector). The results derived from multiple transfections (n = 6) with each trial representing three individual wells were summarized and presented as graphs with standard deviations. Asterisks represent statistically significant changes in the transcription levels of Peg3 between two constructs.
3.4. APeg3 down-regulates the protein levels of PEG3
To confirm whether the observed down-regulation of Peg3 transcript resulted in reduced levels of the PEG3 protein, we performed Western blots with protein extracts prepared from the transfected Neuro2A cells (Fig. 4). The protein extracts from the two samples transfected with the sense and antisense APeg3-Full, respectively, were analyzed and compared with those from the sample transfected with the empty vector (pCDNA). The protein levels of PEG3 in each sample were first quantified using densitometric assays through the Jmol application http://www.jmol.org, and later normalized with those of β-Actin. The normalized values were finally compared with serial dilutions (100–40%)of the control sample transfected with pCDNA. According to the results, the protein levels of PEG3 were lowered by 85% in the transfectants with APeg3-Full, and by 74% in the transfectant with (−)APeg3-Full (lanes 2 and 3 in Fig. 4). The reduced protein levels observed from these transfections with APeg3-Full are consistent with the down-regulation observed in Peg3 mRNA (Fig. 3). In conclusion, the results from both qRT-PCR and Western blotting demonstrate that the overexpression of APeg3 indeed results in reduction in the mRNA and protein levels of Peg3.
Fig. 4.
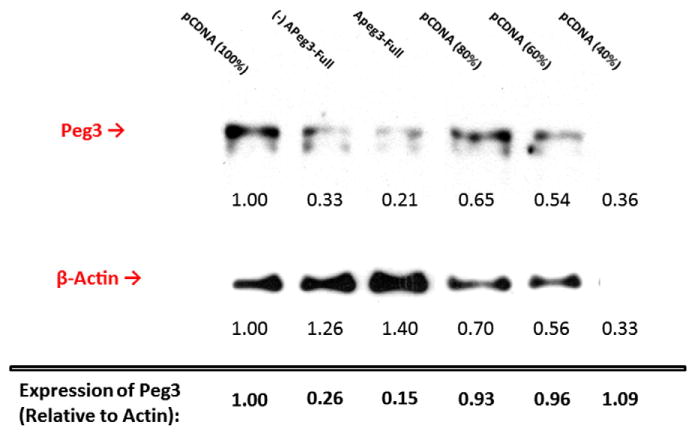
Effects of APeg3 overexpression on the protein levels of PEG3. A set of transfected Neuro2A were further analyzed using Western blotting to measure the protein levels of PEG3. The density of the detected PEG3 bands by anti-PEG3 antibodies was normalized to that of the β-Actin band, then compared with serial dilutions of pCDNA-transfected Neuro2A as standards. Numbers below the bands describe the protein levels of PEG3 and β-Actin. Numbers within the red box indicate protein levels PEG3 relative to β-Actin.
4. Discussion
In the current study, the unknown functions of APeg3 have been investigated using two different approaches, comparative genomic and cell line-based functional analyses. First, comparison of the sequences from different mammals revealed unusually high levels of conservation, but without any obvious ORF in the transcribed region of APeg3, suggesting that APeg3 may have been selected as a ncRNA gene during mammalian evolution (Fig. 1). This has been further supported by the detection of a conserved RNA secondary structure within APeg3 (Fig. 2). Second, the results from cell line-based analyses further demonstrated that APeg3 has the potential to down-regulate the transcription and protein levels of Peg3 (Figs. 3 & 4). Thus, these results suggest that APeg3 has likely evolved as a ncRNA gene controlling the function of the sense gene, Peg3.
The high level of sequence conservation within the transcribed region of APeg3 is very unusual given the following reasoning (Fig. 1): The transcribed region of APeg3 does not harbor any obvious ORF and Kozak consensus motifs that are conserved among mammals, thus suggesting that APeg3 is a ncRNA gene. This is further supported by the presence of many insertions/deletions in the transcribed region of APeg3 between different mammals. At the same time, the sense direction of APeg3 is part of the 3′UTR of Peg3. In terms of functional selection during evolution, both ncRNA and 3′UTRs are expected to have much less constraints than the protein-coding region of individual genes (Li and Graur, 1991). As a result, these non-coding regions should have much less sequence conservation than the coding regions of genes. This can be easily observed in the 3′UTR of many genes, including Zim1, which shows no conservation at all (Fig. 1). Nevertheless, the transcribed region of APeg3 has maintained high levels of sequence identity during mammalian evolution (Table 1), which is quite comparable to the conservation levels of protein-coding regions. This might be related to the fact that this region harbors sequence elements for both genes, APeg3 and Peg3 with opposite transcriptional orientations. Given this special situation, this is the most likely explanation for the unusual sequence conservation observed from APeg3, which needs further investigation in the near future.
According to the results derived from in vitro transfection experiments, APeg3 has the potential to down-regulate expression of Peg3 (Figs. 3 & 4). The actual mechanism by which APeg3 down-regulates Peg3 is currently unknown, but the following scenario can be envisioned with the limited evidence presented in the current study. The most dramatic down-regulation by APeg3 was observed with the full-length APeg3 constructs, yet this down-regulation appears to be somewhat orientation-independent. The complementarity between Peg3 and APeg3 could allow for RNA:RNA interactions at the proposed hairpin-loop which could be causing transcriptional or translational interference of Peg3 via APeg3 regardless of APeg3 orientation. However, in silico predictions do not correlate this interaction to be the most likely at the 5′ hairpin-loop structure (Mueckstein et al., 2006).
Another possibility for the orientation-independent down-regulation of Peg3 is through small RNA-mediated silencing. There are currently four known mechanisms for small RNA to down-regulate protein-coding mRNA, miRNA, snoRNA, pi-RNA and siRNA (Meister and Tuschl, 2004). Of these four, the miRNA-mediated mechanism is mainly known to be associated with imprinted domains, and functions independent of orientation (Royo and Cavaille, 2008; Meister and Tuschl, 2004). Thus, the orientation-independent down-regulation by APeg3 might be mediated through some unknown miRNA embedded within the APeg3 locus. In that regard, it is relevant to point out the fact that the 5′-side of APeg3 contains a potential target site for miR-124 (Fig. 1), further supporting this possibility.
If this is the case, is this potential miRNA from APeg3 mainly targeting the transcript from Peg3? Given the similar expression profiles in vasopressin-expressing neurons, we believe that Peg3 is most likely the main target of APeg3. However, we cannot rule out the possibility that other genes in those specific neuronal cells could be targeted by a potential miRNA from APeg3. In conclusion, the observed down-regulation of Peg3 by APeg3 is very intriguing given the crucial roles played by Peg3 in eutherians, thus it would be of great interest to further investigate this observed down-regulation by APeg3 in terms of molecular mechanisms as well as potential targets.
Supplementary Material
Acknowledgments
We would like to thank Corey Bretz, Arundhati Bakshi and Dr. Hana Kim for their careful reading and discussion of the manuscript. We would like to thank Dr. David Donze and Asawari Korde for providing the facilities and guidance in northern blotting for confirmation of APeg3 over-expression and Michelle Thiaville for her mentorship in the APeg3 transfection experiments. We would also like to thank Rui Lu, Ryoichi Teruyama and James Moroney for their intellectual input towards experimental designs. Finally, another acknowledgement should go out to HK for her help with data analysis and comparative genomics. We would also like to thank the reviewers for their insightful comments. This research was supported by the National Institutes of Health (J.K. R01-GM066225 and R01-GM097074).
Abbreviations
- ORF
Open Reading Frame
- MFE
minimal fold energy
- ncRNA
non-coding RNA
- TSS
transcriptional start site
- Peg3
paternally expressed gene 3
- APeg3
antisense paternally expressed gene 3
- CMV
Cytomegalovirus
- BGH
Bovine Growth Hormone
- mm9
Mus musculus assembly 9
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gene.2014.02.056.
Conflict of interest: The authors declare no conflict of interest.
References
- Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Molecular and Cellular Biology. 1990;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo JH, Kim JD, Kim J. Imprinting of an evolutionarily conserved antisense transcript gene APeg3. Gene. 2008;409(1–2):28–33. doi: 10.1016/j.gene.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith Anne C. Genomic imprinting: the emergence of an epigenetic paradigm. Nature Reviews. Genetics. 2011;12(8):565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- Ge P, Zhang S. Incorporating phylogenetic-based covarying mutations into RNAalifold for RNA consensus structure prediction. BMC Bioinformatics. 2013;14:142. doi: 10.1186/1471-2105-14-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow E, Ryu SL, Yamashita M, Zhang BJ, Mutsuga N, Gainer H. APeg3, a novel paternally expressed gene 3 antisense RNA transcript specifically expressed in vasopressinergic magnocellular neurons in the rat supraoptic nucleus. Molecular Brain Research. 2005;137(1–2):143–151. doi: 10.1016/j.molbrainres.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Neubock R, Hofacker IL, Washietl S. The RNAz web server: prediction of thermodynamically stable and evolutionarily conserved RNA structures. Nucleic Acids Research. 2007;35:W335–8. doi: 10.1093/nar/gkm222. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan V, Crain C, Wilson C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Research. 2000;28(5):1221–1227. doi: 10.1093/nar/28.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309(5740):1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kim J, Ashworth L, Branscomb E, Stubbs L. The human homolog of a mouse-imprinted gene, Peg3, maps to a zinc finger gene-rich region of human chromosome 19q13.4. Genome Research. 1997;7(5):532–540. doi: 10.1101/gr.7.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Ekram MB, Kim H, Faisal M, Frey WD, Huang JM, Tran K, Kim MM, Yu S. Imprinting control region (ICR) of the Peg3 domain. Human Molecular Genetics. 2012;21(12):2677–2687. doi: 10.1093/hmg/dds092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner MV, Pauler FM, Huang R, Barlow DP. The function of non-coding RNAs in genomic imprinting. Development. 2009;136(11):1771–1783. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Graur D. Fundamentals of Molecular Evolution. Vol. 1. Sinauer Associates; Sunderland, Mass: 1991. p. 73. [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(2006):343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Mueckstein U, Tafer H, Hackermueller J, Bernhart SH, Stadler PF, Hofacker IL. Thermodynamics of RNA–RNA binding. Bioinformatics. 2006;22(10):1177–1182. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- Osato N, Suzuki Y, Ikeo K, Gojobori T. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176(2):1299–1306. doi: 10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H, Cavaille J. Non-coding RNAs in imprinted gene clusters. Biology of the Cell. 2008;100(3):149–166. doi: 10.1042/BC20070126. [DOI] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(7):2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Hayashida S, Miura K, Masuzaki H, Ishimaru T, Niikawa N, Kishino T. The novel gene, gamma2-COP (COPG2), in the 7q32 imprinted domain escapes genomic imprinting. Genomics. 2000;68(3):330–335. doi: 10.1006/geno.2000.6265. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Glasgow E, Zhang BJ, Kusano K, Gainer H. Identification of cell-specific messenger ribonucleic acids in oxytocinergic and vasopressinergic magnocellular neurons in rat supraoptic nucleus by single-cell differential hybridization. Endocrinology. 2002;143(11):4464–4476. doi: 10.1210/en.2002-220516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


