Abstract
The increasing availability of DNA sequence data offers an opportunity for identifying new assembly-line polyketide synthases (PKSs) that produce biologically active natural products. We developed an automated method to extract and consolidate all multimodular PKS sequences (including hybrid PKS/non-ribosomal peptide synthetases) in the National Center for Biotechnology Information (NCBI) database, generating a non-redundant catalog of 885 distinct assembly-line PKSs, the majority of which were orphans associated with no known polyketide product. Two in silico experiments highlight the value of this search method and resulting catalog. First, we identified an orphan that could be engineered to produce an analog of albocycline, an interesting antibiotic whose gene cluster has not yet been sequenced. Second, we identified and analyzed a hitherto overlooked family of metazoan multimodular PKSs, including one from Caenorhabditis elegans. We also developed a comparative analysis method that identified sequence relationships among known and orphan PKSs. As expected, PKS sequences clustered according to structural similarities between their polyketide products. The utility of this method was illustrated by highlighting an interesting orphan from the genus Burkholderia that has no close relatives. Our search method and catalog provide a community resource for the discovery of new families of assembly-line PKSs and their antibiotic products.
Keywords: assembly line, orphan antibiotics, polyketide synthase
INTRODUCTION
Multimodular polyketide synthases (PKSs) are enzymatic assembly lines responsible for the biosynthesis of many structurally and pharmacologically diverse antibiotics.1 They are typically found in bacteria, most notably in the actinomycetes, and are encoded by unusually large, clustered gene sets.2,3 Each PKS module minimally consists of a ketosynthase (KS) and an acyl carrier protein domain. The 6-deoxyerythronolide B synthase (DEBS), which catalyzes the formation of the macrocyclic core of the antibiotic erythromycin, is a prototypical example of an assembly-line PKS.4 It consists of an initiation module, six elongation modules and a thioesterase domain responsible for chain termination.
Historically, most polyketide antibiotics were discovered through activity-guided isolation, long before their PKS gene clusters were sequenced. For example, erythromycin was first isolated in 1949, but the DEBS gene cluster was not sequenced until ca. 1990.5,6 If one defines an assembly-line PKS as harboring at least three distinct elongation modules, then we estimate that the gene clusters encoding multimodular PKSs corresponding to ~200 structurally characterized polyketides have been sequenced (see below). At the same time, as genome sequencing has become easier, the number of cryptic assembly-line PKS gene clusters in the NCBI database has far surpassed the number of clusters whose product is known. These cryptic sequences have been dubbed ‘orphan PKS’ gene clusters.7-9
Several databases have been developed to catalog known PKSs.10-13 Recently developed active PKS databases include DoBISCUIT, which curates secondary metabolite gene clusters from the literature (and currently contains 86 characterized gene clusters), and ClusterMine360, which allows community users to deposit and curate gene clusters (and currently contains 254 user-deposited gene clusters). In contrast, the present work does not represent an active database of manually curated gene clusters, but rather a snapshot catalog of all automatically mined non-redundant PKS gene clusters (known and orphan) from NCBI sequence data as of June 2013, as well as the underlying method to automatically generate the catalog. As such, it is complementary to the above databases.
We restricted this study to Type I assembly-line PKSs, which consist of large multimodular polypeptides that elongate the polyketide chain by serial propagation through each of the modules. These stand in contrast with iterative PKSs, which consist of a single module that iteratively elongates and functionalizes a polyketide chain; iterative PKS classes include Type I iterative PKSs (in which the catalytic domains are fused into a single protein) and Type II PKSs (in which the catalytic domains comprise several stand-alone enzymes). Owing to their inherent modularity, Type I assembly-line PKSs have evolved to encode a greater diversity of polyketide natural products, and have commensurately greater potential for the biosynthesis of engineered polyketides.14 In this work, we sought to catalog all existing assembly-line PKS sequences to guide future studies of natural and engineered PKSs.
To generate the catalog of all assembly-line PKSs, we aimed to analyze all publicly available DNA sequences in NCBI in an unbiased manner, regardless of their previous annotation or biological source, and identify sequences containing orphan PKSs and hybrid PKS–non-ribosomal peptide synthetases (PKS–NRPSs). Our method combines the complementary capabilities of the fast BLAST algorithm15 with a recently developed tool, antiSMASH2,16 which scans a given DNA sequence for secondary metabolite domains. This high fidelity automated approach is tailored to the task at hand – discovery and comparative analysis of all assembly-line PKSs and hybrid PKS–NRPSs in publicly available sequence databases. Our analysis has not only revealed unexpected insights into PKS function and evolution, but has also set the stage for fundamentally new avenues for experimental investigation into this remarkable family of mega-synthases. The catalog of orphan PKSs is available for download and visualization at http://sequence.stanford.edu/OrphanPKS.
MATERIALS AND METHODS
Identification of assembly-line PKSs
Our approach for automated computational analysis of assembly-line PKSs is summarized in Figure 1. As of May 2013, the National Center for Biotechnology Information (NCBI) RefSeq database contained 24 656 non-redundant, annotated genomes. Ordinarily, when a genome is sequenced, it is annotated using automated gene-finding software, which identifies open reading frames and assigns putative function according to sequence similarity with proteins of known function.17-22 However, these methods only consider one open reading frame at a time and do not analyze relationships between spatially clustered genes, an approach that yields crucial insights into the enzymology of assembly-line PKSs. Moreover, there are 112 488 036 (as of June 2013) unannotated whole-genome shotgun (WGS) draft contig sequences in the NCBI database with no corresponding gene predictions. To our knowledge, there has been no attempt at large-scale characterization of assembly-line PKS clusters across the entire NCBI database.
Figure 1.
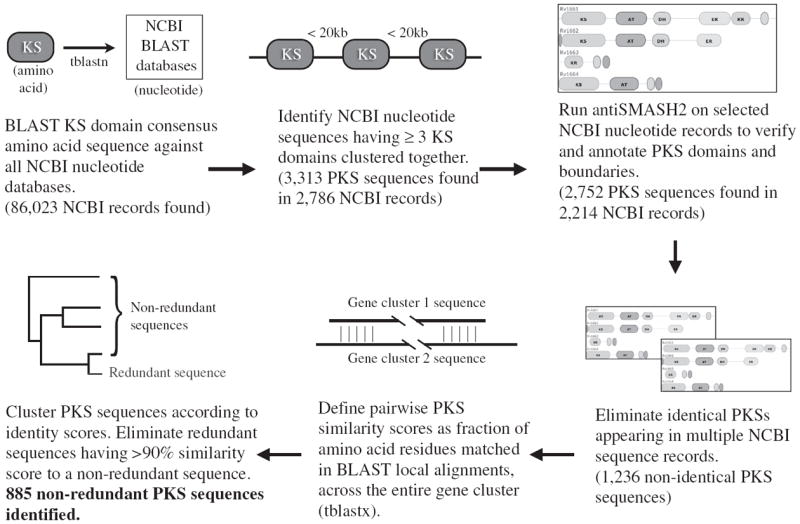
Summary of workflow. A full color version of this figure is available at The Journal of Antibiotics journal online.
A number of promising methods have been developed over the past decade for PKS protein domain annotation,10,23-29 but most of these methods are not suitable for parallel analysis of a large number of DNA sequences. A recently released program, antiSMASH2 (‘antibiotics and secondary metabolites analysis shell’), is noteworthy in this regard.16 It first performs automated gene finding on unannotated DNA sequences. Then, for assembly-line PKSs, it detects domains, analyzes enzyme specificity and predicts product structure based on previously developed algorithms. The open-source nature of this software facilitates automated analysis; however, the run-time is prohibitively slow for analysis on all sequence data in the NCBI, which houses >400 billion base pairs of information as of June 2013. On our local servers, the run-time was ~0.5 min per WGS contig record (typically ~100 kb). Given the >100 million WGS records, we estimated that >100 CPU-years would be required to mine this single data set for assembly-line PKSs, which was prohibitive. Our goal was to search all major NCBI sequence databases in an unbiased manner. We therefore first sought to narrow the list of sequences containing potential PKSs using a fast BLAST-based scan; for this, we searched for KS domains, as these are a requirement of PKS assembly lines, and their sequences are generally well-conserved.
A consensus KS domain sequence was defined by aligning KS sequences from the 56 annotated multimodular PKS protein sequences in the SBSPKS database (516 KS protein sequences in total).10 We aligned this consensus KS sequence, using tblastn, with 10 major BLAST nucleotide databases: nt, wgs, refseq_genomic, other_genomic, htgs, env_nt, est_others, gss, patnt, tsa_nt and sts. KS BLAST hits were defined as discrete KS domains if they were >3 kb apart from another KS domain (to eliminate fatty acid synthases and iterative PKSs, and to avoid multiple hits against the same KS domain). Multimodular PKSs were defined by the presence of three or more clustered KS domains, where clustering was defined as one KS existing within 20 kb of another. Sequence records meeting these criteria were then analyzed and annotated with antiSMASH2.
Notably, many of the multimodular PKSs that we identified were redundant; that is, they comprised identical sequences or subsequences of another identified PKS. The most common reasons for redundancy were: existence of the same PKS in NCBI with multiple accession numbers; a PKS cluster having been identified as both a gene sequence record and within a whole-genome sequence record; and the same PKS cluster existing in multiple unassembled whole-genome sequencing contigs. Identical gene clusters were identified and eliminated from our catalog of multimodular PKSs by identifying PKSs having either (a) identical sequence (including if one sequence was an exact subsequence of the other) or (b) identical domain architecture within a species. We noted upon manual inspection of sequence similarities (see below) that some apparently redundant sequences were not eliminated in this manner due to minor sequence variation (for example, if a genome was sequenced multiple times).
Comparative analysis of assembly-line PKSs
We next sought to examine sequence similarities between pairs of gene clusters. For PKSs, this has historically been achieved through alignment of conserved domains, such as KSs or acyltransferases (ATs).30 Because this study involved a large number of sequences, we desired a score that would summarize similarities across entire assembly lines rather than individual domains. The antiSMASH software employs a BLAST-based empirical gene cluster similarity score that counts, for each pair of clusters, the number of proteins that share a significant BLAST hit, and assigns higher scores to cluster pairs with matching ‘core’ genes.23 We instead desired a score that (1) would not rely on gene annotation, because we found that these annotations were often inaccurate or missing, (2) would compare clusters at the amino acid level (despite ignoring gene annotation), (3) would employ local alignments, given the nature of the repeating domains and modules, (4) would retain fine-grained sequence identity information rather than coarse-grained counts of similar genes, and (5) would be relatively fast to compute. These desiderata can be met by using the tblastx algorithm, where each gene cluster sequence is translated in six frames and compared with the second sequence also translated in six frames. We combined the tblastx-identified local percent identities into a heuristic gene-cluster similarity score S for gene clusters a and b as:
where k is the number of BLAST local alignments (corrected to be non-overlapping; see below), mx is the number of matched residues in local alignment x and Na is the number of total residues in cluster a. Thus, the overall percent identity represents a simple sum of the matched residues across all of the BLAST local alignments divided by the total length of the gene cluster. Because BLAST identifies multiple, often overlapping regions of local sequence similarity, we eliminated overlaps by ensuring that each residue was counted only once.
The above approach yielded a similarity score for every pair of gene clusters, with similarities ranging from 0 to 1, corresponding roughly to 0–100% identity. We selected a score of 0.9 (roughly 90% identity) to define redundancy (Figure 1). This threshold was selected by manual inspection of clusters that we deemed to be redundant (for example, multiple sequences of the erythromycin gene cluster).
To visualize the PKS similarity scores as a dendrogram, the scores were converted to distances between 0 and 1, and the pairwise distance matrix was made symmetric by choosing the larger of the two scores. For example, in cases where one PKS was shorter than the other, the pairwise score derived from the shorter sequence was higher due to a smaller denominator in the above formula. The distance matrix was visualized as a dendrogram using the R software package (hclust) and the McQuitty method of linkage;31 we selected this linkage method owing to its ease of visualization and because it maintained the percent identity distances on the visualized dendrogram branches (as opposed to the neighbor-joining method, for example). This clustering and visualization approach was first applied to known PKS gene clusters (Figure 2), and subsequently to the entire set of discovered PKS clusters (Supplementary Figure 1).
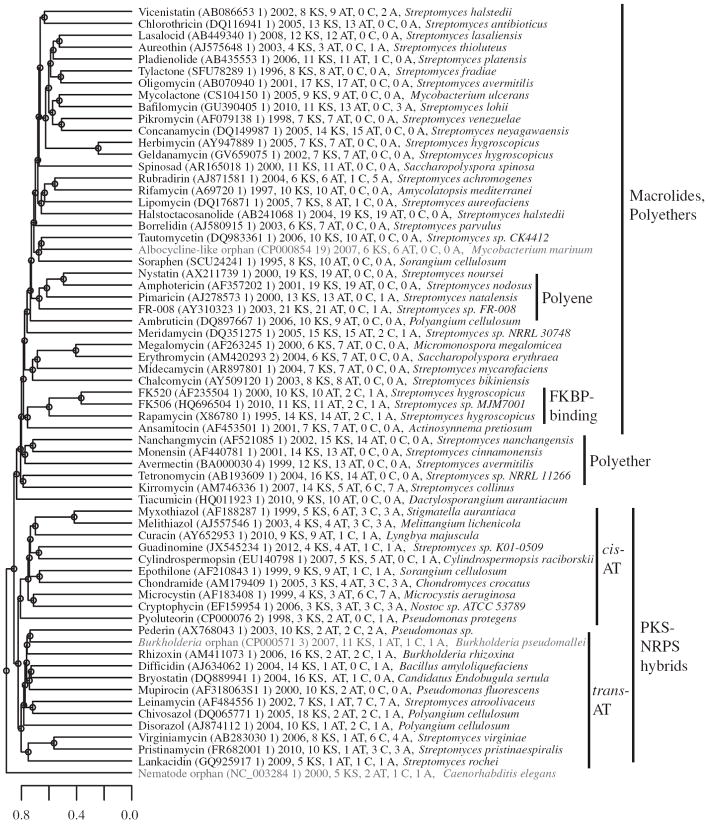
Figure 2.
Sequence similarity relationships among PKSs involved in the biosynthesis of known polyketide natural products. The amino-acid sequences of 62 representative assembly-line PKSs and PKS–NRPSs, plus the three orphan PKSs highlighted in this report, were compared in a pairwise manner using a gene-cluster similarity score. Scale bar displays the distance between gene clusters (between 0 and 1). The label for each PKS lists the Genbank ID and cluster number, date the sequence was deposited in NCBI, number of KS, AT, C and A domains, and sequence origin as listed in NCBI. The three orphans highlighted in the text are displayed in red. A full color version of this figure is available at The Journal of Antibiotics journal online.
For characterization of individual orphan PKSs, we began by manually analyzing the automatically generated antiSMASH annotation, verifying and sometimes correcting the annotation. We manually predicted chemical structures based on the predicted domains and PKS colinearity rules.
RESULTS
Identification of PKS sequences using BLAST and antiSMASH2
A total of 3313 putative PKS sequences (spanning 2786 NCBI sequence records) were identified using the BLAST-based scan for ≥3 clustered KS domains (Figure 1, Supplementary Table 1). These NCBI records were analyzed and annotated with antiSMASH2, which verified the identity of 2752 (spanning 2214 NCBI records) of these as containing PKSs. We manually investigated the discrepancy (Supplementary Table 2) and found that those putative PKSs identified by the BLAST scan but not antiSMASH2 included (1) eukaryotic fatty acid synthases in which the KS was separated into ≥3 exons, leading to false positives by the BLAST approach, (2) eukaryotic PKSs (in particular, algal) that appeared to be true assembly lines with multiple KSs and (3) a small number of prokaryotic PKSs that appeared to be true assembly lines.
We defined identical PKSs within the set of 2752 as those having (a) identical sequence or (b) identical domain architectures within a species. Eliminating these duplicates resulted in a catalog of 1236 non-identical PKSs (Figure 1, Supplementary Table 3).
Survey of assembly-line PKS characteristics
The 1236 non-identical assembly-line PKS gene clusters spanned 536 species. Of these, 172 corresponded to PKSs annotated as gene clusters involved in the biosynthesis of known natural products; most of the remainder appeared to be orphan PKSs. Approximately one-half of the PKSs were encoded within unfinished whole-genome sequences, with an additional one-quarter derived from complete genome sequences. One quarter of the PKSs were trans-AT systems,32,33 and nearly one-half included one or more NRPS modules. The GC content of gene clusters ranged from 22% to 77%; the distribution was bimodal, with one mode ~70% and the other ~45% (Supplementary Figure 2).
Sequence similarities between assembly-line PKSs
In order to better understand the relationships among the identified assembly-line PKSs, we performed pairwise comparisons of the amino-acid sequences of the 1236 non-identical PKSs identified above. It should be noted that standard phylogenetic methods are not applicable for comparisons of PKS genes because the sequences are not strictly homologous; rather, they evolved through numerous events of horizontal transfer and module duplication.34,35 Owing to this manner of evolution, many PKSs share multiple local regions of sequence similarity. We therefore developed a strategy to facilitate cluster-wide sequence comparisons and visualizations, as detailed in the Methods section. In brief, we used a heuristic BLAST-based pairwise similarity score whose value ranges from 0 to 100, which corresponds roughly to the percent identity across the entire length of the gene cluster (including tailoring enzymes outside the core assembly-line genes) (Supplementary Figure 3). Although our method has inherent biases, it provides a reasonable basis for establishing sequence relationships.
As a preliminary test of the gene cluster similarity score, we aligned 62 PKSs corresponding to well-known polyketide antibiotics and visualized their relationships as a dendrogram (Figure 2). The dendrogram is not a phylogenetic tree, because the sequences are not homologous; rather, the distances are based on our heuristic gene cluster similarity score. PKSs with known close relationships were found to cluster together (for example, the clusters for erythromycin and megalomicin, FK506 and FK520, amphotericin and nystatin). Higher order relationships were also evident, such as clusters of macrolides, polyethers, trans-AT PKSs and PKS–NRPS hybrids. There is a single trans-AT clade, consistent with previous phylogenetic analyses of individual KS domains from cis- and trans-AT PKSs, which also suggest distinct lineages of these two classes.36 Interesting outliers are apparent in the tree, such as kirromycin and tetronomycin, both of which are encoded by gene clusters that contain both cis-AT and trans-AT genes. Their placement in the cis-AT clade suggests a greater degree of overall similarity to cis-AT clusters than the trans-AT clusters, though their peripheral position in the cis-AT clade reveals that they are relatively distant from the rest.
Interestingly, the trans-AT clade is entirely contained within a larger PKS–NRPS clade, suggesting that the evolution of trans-AT PKSs may have involved a PKS–NRPS hybrid ancestor. We further investigated this possibility by performing phylogenetic analysis of the KS domains in the 62 gene clusters (Supplementary Figure 4). The KS domain phylogeny parallels the trends observed in Figure 2: KS domains from PKS–NRPS hybrids constitute a separate clade from the PKS-only sequences, and within this PKS–NRPS hybrid clade, there are distinct cis-AT and trans-AT system clades.
Having established the heuristic gene-cluster similarity score on known PKS clusters, we next applied the same method to the entire set of 1236 known and orphan PKSs. This resulted in a dendrogram analogous to that in Figure 2, but with 1236 leaves (Supplementary Figure 1). PKSs from the same species sometimes appeared together in clades, but often PKSs from the same species were spread diversely across clades. Instead of clustering by host species, the PKSs clustered according to the trends observed in Figure 2: trans-AT clusters, PKS–NRPS hybrids, and by characteristics of the encoded chemical. We noted that some clades in the dendrogram contained both orphan and known gene clusters, suggesting clues about the origin and possibly the encoded chemistries of the orphans, whereas many clades in the large tree contained only orphan clusters with no known relative. To quantify this observation, we ‘cut’ the tree of 1236 gene clusters into clades at varying tree heights (Table 1). At each cut threshold, we counted the total number of clades and the number of clades containing a characterized PKS. We denoted those clades containing no characterized PKS as ‘orphan clades’. At varying thresholds, the fraction of clades that were orphan clades was consistently near 80%. These results suggest a large degree of unexplored diversity in orphan PKS gene clusters.
Table 1.
Characterization of orphan PKS clades
| Tree cut height | Total number of clades | Number of clades containing a characterized PKS | % of clades containing a characterized PKS | % of clades that are orphan clades |
|---|---|---|---|---|
| 1 | 1 | 1 | 100 | 0 |
| 0.9 | 7 | 2 | 29 | 71 |
| 0.8 | 129 | 27 | 21 | 79 |
| 0.7 | 378 | 77 | 20 | 80 |
| 0.6 | 531 | 104 | 20 | 80 |
| 0.5 | 636 | 125 | 20 | 80 |
| 0.4 | 709 | 137 | 19 | 81 |
| 0.3 | 762 | 143 | 19 | 81 |
| 0.2 | 833 | 152 | 18 | 82 |
| 0.1 | 922 | 162 | 18 | 82 |
| 0 | 1236 | 172 | 14 | 86 |
The similarity scores among 1236 gene clusters were visualized as a dendrogram (Supplementary Figure 1), which was ‘cut’ at various heights. Decreasing the cut height increased the number of clades. At each threshold, we quantified the number of clades that contained a characterized PKS versus orphan clades that contained no characterized PKS.
Redundancy defined by sequence similarity
We noted upon manual browsing of the dendrogram of PKS sequence similarities (Supplementary Figure 1) that many sequences were extremely similar, though not identical, and therefore not eliminated by the above redundancy criteria. We used our gene cluster similarity score to eliminate any remaining redundancy in our set of 1236 PKS clusters, defining a redundance threshold at a similarity score >0.9 (that is, sequences that shared roughly 90% sequence identity, see Methods). We found that 351 of the PKS clusters were redundant by this definition, leading to a final count of 885 non-redundant PKS clusters (Supplementary Table 4).
Timeline of PKS sequencing
Using the date that each PKS gene cluster was first deposited in NCBI, we calculated the rate of assembly-line PKS sequence discovery (Figure 3). For gene clusters identified within a larger sequence (for example, whole-genome sequences or contigs), this date represents the date that the sequence was first deposited in NCBI, regardless of its annotation at that time. PKS gene clusters with assigned early dates (pre-2000) were usually deposited in NCBI with specific annotation as biosynthetic gene clusters corresponding to known natural products, and were often in the patent database. Subsequent PKSs tended to be derived from genome sequences or unfinished WGS contigs (the latter of which contained no gene annotation).
Figure 3.
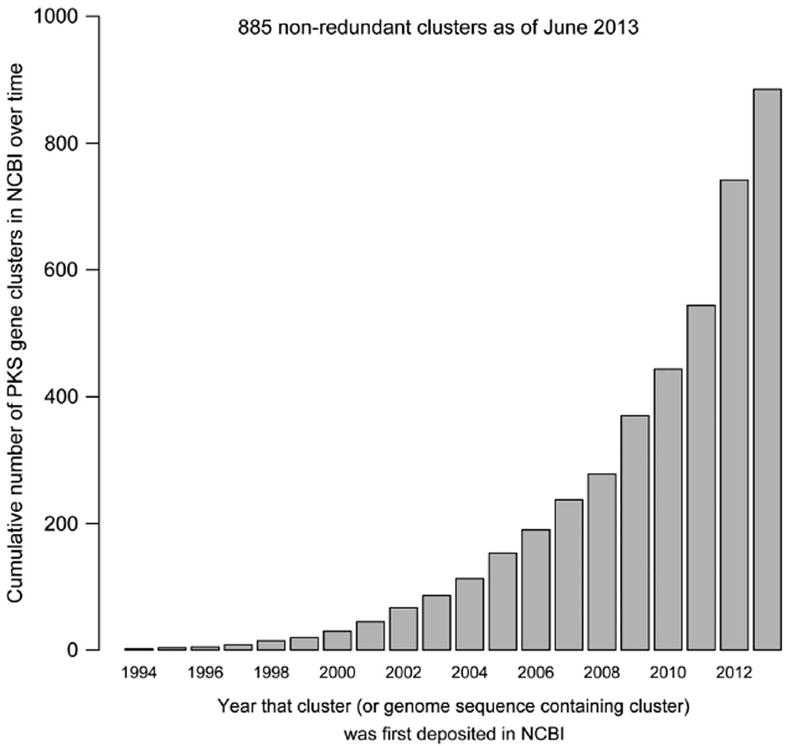
Cumulative number of non-redundant PKS gene clusters in NCBI over time: data were collected through the first half of 2013. A full color version of this figure is available at The Journal of Antibiotics journal online.
Below we highlight three orphan PKSs: one that putatively encodes a polyketide with a similar structure to a known natural product, and two that putatively encode polyketides with little similarity to any known natural product. For reference, we included these three orphan PKSs in the analysis of known PKSs (Figure 2).
An orphan PKS with the potential of producing an albocycline analog
Numerous polyketide natural products have been isolated from organisms whose genomes have yet to be sequenced: for example, the antibiotic albocycline (1, Figure 4) was isolated from the bacterium Streptomyces sp. 6–31 and the cluster responsible for its biosynthesis remains unknown.37,38 We asked whether any of the orphans identified in our survey might produce a polyketide of similar structure to albocycline. Because albocycline is predicted to be produced by a PKS comprised of six elongation modules, we filtered the list of orphan clusters based on those that possessed exactly six KS domains and a single enoyl reductase domain (albocycline biosynthesis is expected to require only a single fully reducing module). Five PKSs met these criteria. Of these, an orphan assembly line found in the genome of Mycobacterium marinum had the most plausible sequence and AT domain specificity (Figure 4). Interestingly, this PKS has sequence similarity to the soraphen PKS from Sorangium cellulosum So ce26.39 In order to cyclize into a 14-membered macrolide, the dehydratase domain of module 1 would need to be inactive to provide the requisite hydroxyl group at C2. It is not unusual for certain domains to be inactive in PKSs; for example, in the rapamycin PKS/NRPS, certain dehydratase, ketoreductase and enoyl reductase domains do not act on the elongating polyketide chain.40 A second difference between albocycline and the predicted orphan polyketide from M. marinum is the C10–C11 double bond. Following the activity of module 5, this would have to undergo isomerization to form a skipped diene; such an isomerization could, in principle, occur through the action of either the dehydratase domain of module 5 or module 6, in analogy with the trans- to cis-isomerization seen in epothilone biosynthesis.41 Finally, the double bond between C4–C5 would either have to isomerize or become reduced in order to accommodate the required conformation for macrolactonization. Although there are differences between albocycline itself and the predicted product of the M. marinum orphan cluster, this example highlights the presence of orphan clusters within the NCBI genome database that could, in principle, produce analogs of known polyketide natural products. Such orphan clusters could thus serve as a platform for producing polyketides with unknown biosynthetic clusters; in such cases, the orphan could be expressed and subsequently engineered to produce the desired compounds. As the database grows, this may be a particularly effective strategy for accessing polyketides derived from unculturable sources such as marine antibiotics.
Figure 4.
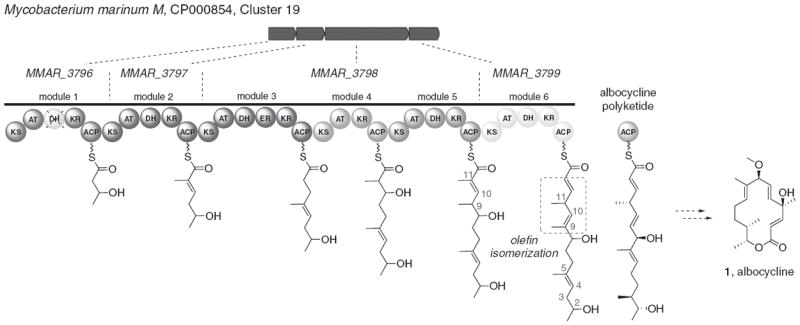
An orphan cluster that may produce a natural product that is structurally analogous to albocycline, a potent antibiotic derived from Streptomyces sp. 6–31, whose encoding gene cluster is unknown. This cluster could be engineered to produce albocycline analogs. A full color version of this figure is available at The Journal of Antibiotics journal online.
An orphan PKS in Burkholderia with little similarity to known PKSs
Burkholderia mallei and Burkholderia pseudomallei are human and animal pathogens whose genome sequences recently became available.42 A polyketide metabolite (called malleilactone or burkholderic acid) has been identified by two different groups, and the hybrid PKS/NRPS cluster responsible for its biosynthesis has been characterized.43,44 In addition, Biggins and co-workers have noted that numerous other PKS/NRPS clusters exist in the genomes of B. mallei, B. pseudomallei and Burkholderia thailandensis.44 Some of these PKSs share close similarity to PKSs in other Burkholderia species, whereas others are relatively unique. Among the latter, an orphan PKS that is found in both B. mallei and B. pseduomallei (but not B. thailandensis) appears to make an unusual natural product (Figure 5). It is important to note that Nguyen et al.33 have found that some trans-AT containing PKSs in a variety of organisms (including a Burkholderia species) produce polyketides with structures that do not follow the canonical rules of enzyme domain colinearity; the structure shown is what is predicted based on colinearity rules. This orphan is grouped within the trans-AT containing PKS/NRPS hybrid clusters in the dendrogram shown in Figure 2, and its closest relative there is the pederin synthase.45 The PKS contains two C-methyl transferase domains and an aminotransferase and has an NRPS module in the middle of the cluster. It is noteworthy that the malleilactone/burkholderic acid PKS also harbors a partial NRPS module in the middle of the cluster, even though no amino acid is incorporated into the observed natural product. Instead, the C domain serves to unite the PKS fragments required to form the natural product. The aminotransferase domain in the orphan PKS shown in Figure 5 is particularly striking, given their rarity in known PKSs (one also exists in the malleilactone/burkholderic acid PKS, but it is silent).46,47 Notably, this Burkholderia orphan and the malleilactone/burkholderic acid cluster appear in clades that are quite distant from each other in the comparative sequence analysis, suggesting substantial evolutionary distance and encoded product between the two gene clusters (Supplementary Figure 1). Given the significant threat posed by Burkholderia species to human and animal health, the existence of a hitherto unexplored polyketide natural product putatively produced by both B. mallei and B. pseudomallei may warrant investigation to provide insight into their biology and pathogenesis.
Figure 5.
Predicted PKS assembly line for a cluster from Burkholderia spp. that does not appear related to any known biosynthetic clusters A full color version of this figure is available at The Journal of Antibiotics journal online.
Eukaryotic PKSs
To our knowledge, no assembly-line PKS has been functionally characterized in eukaryotes. In Dictyostelium discoideum (a slime mold amoeba), the Dif-1 (differentiation factor 1, also called ‘steely’) polyketide has been identified as a product of a unimodular iterative PKS.48,49 In fact, a few protozoan parasites do harbor orphan assembly-line PKSs, including a conserved PKS in Toxoplasma gondii and Neospora caninum and another in Cryptosporidium.50,51 However, assembly-line PKSs are not thought to exist in metazoans.
We were therefore surprised that the above analysis revealed the existence of an orphan clade that spanned a range of nematode species. Specifically, a hybrid PKS–NRPS encoded by a single open reading frame was found in these species; the homolog from Caenorhabditis elegans is shown in Figure 6. It is not known whether the system acts in an assembly-line fashion or in an iterative fashion (or both). By our heuristic gene cluster similarity score, the closest relatives are three orphan PKS assembly lines from Clostridium spp., although the similarity was weak and the architectures of the two PKSs were quite different. RNAi data in the WormBase database offered no information on knockdown phenotypes.52 Recent RNA-seq data generated by modENCODE suggest that the gene is transcribed during embryo, L4 larvae and young adult life stages.53
Figure 6.
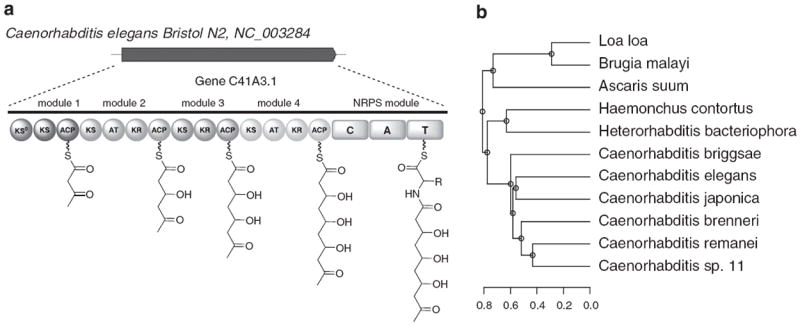
(a) Predicted structure of a hybrid PKS/NRPS assembly-line gene in C. elegans. (b) Clade of nematode homologs of the hybrid PKS/NRPS C. elegans gene in part A, clustered according to our heuristic PKS sequence similarity score (see also Supplementary Figure 5). A full color version of this figure is available at The Journal of Antibiotics journal online.
We hypothesized that the gene may have arisen via one of two evolutionary mechanisms: (a) horizontal gene transfer from bacteria or (b) divergence from a nematode fatty acid synthase gene. To explore these possibilities, individual domains of the C. elegans PKS gene were aligned with several bacterial PKS domains or alternatively domains from the C. elegans fatty acid synthase gene FASN-1 (Supplementary Figure 5). The worm AT and ketoreductase domains were most similar to the FASN-1 AT and ketoreductase domains, respectively, suggesting a possible origin from the worm FAS. In contrast, the worm KS domains were different from all other homologs but related to each other, suggesting a possible duplication event. Of the five KS domains in the gene, the first was most similar to the fourth, and the second was most similar to the fifth; this pattern is also suggestive of duplication. Alignment of the domains of the C. elegans gene with those in homologous genes from nematodes Loa loa, Brugia malayi, Ascaris suum, Haemonchus contortus and Heterorhabditis bacteriophora, as well as several other Caenorhabditis species suggested a possible point of duplication (Supplementary Figure 5). Taken together, these results suggest an evolutionary history in which a fatty acid synthase gene was duplicated and diverged to form this gene.
The function of this putative PKS gene is unknown. It is conceivable that it is involved in biosynthesis of signaling molecules similar to the ascarosides, which are widely distributed among nematodes.54 These molecules are assembled in a modular fashion from chemical building blocks of sugars, fatty acid-like side chains and other tailoring groups.55
DISCUSSION
Our automated search for all assembly-line PKSs in NCBI sequence records revealed a total of 885 non-redundant PKSs and hybrid PKS–NRPSs. Of these, approximately 20% synthesize known natural products; the rest are orphan assembly lines with no associated polyketide compound. Many orphan PKSs are similar to other orphans but not to known PKSs; these clades of orphans may encode new classes of polyketide natural products with unique structures and biological activities. Characterizing these sequences and encoded chemical structures will be an important next step in the natural product discovery. The catalog and analysis presented here can be expected to aid the refactoring of these biosynthetic pathways in heterologous hosts, thereby expanding the accessible repertoire of bioactive complex polyketides.
We explored several potential applications of this data set of assembly-line PKSs. Natural products of unknown biosynthetic origin can be used to search for candidate clusters that may be involved in the biosynthesis of a close structural analog; these analogous orphans could thus serve as a platform to engineer the biosynthesis of the known natural product. Furthermore, this method allowed the identification of an orphan cluster that occurs across two different species of pathogenic Burkholderia bacteria that contains an unusual aminotransferase domain and appears quite distinct (by sequence and putatively encoded chemical) from previously characterized PKS clusters. Finally, surprising examples of PKSs were found in metazoans, a finding that was made possible by the unbiased approach used in our search method.
The abundance of gene clusters now available in public sequence databases offers an opportunity to study their relationships and evolution. Historically, such gene cluster sequence comparisons have been carried out at the level of individual proteins, modules or domains, but as genomic data continues to expand, such approaches become cumbersome. The cluster-wide similarity score used here is an attempt to simplify comparisons among gene clusters. This comparative analysis recapitulated known relationships among known PKSs (for example, that PKS sequences cluster according encoded chemical product), and predicted new relationships (for example, that trans-AT clusters arose from a PKS/NRPS hybrid cluster). However, this one-dimensional score is limited, in that it offers no information about relationships among individual modules or domains. Richer sequence models and methods of comparison should yield insights into the evolution of natural product gene clusters, as well as the chemical diversity and bioactivities of their encoded natural products.
Supplementary Material
Acknowledgments
We thank Colin Harvey, Caleb Chan and Gergana Vandova for helpful discussions. This research was supported by grants from the National Institutes of Health (R01 GM087936 to CK, and P01 HG000205 to RWD) and the Stanford Institute for Immunity, Transplantation, and Infection (to MEH). RVO is a recipient of a National Institute of General Medical Sciences Postdoctoral Fellowship (GM103165-01A1) and is a fellow of the Center for Molecular Analysis and Design (CMAD) at Stanford University. MEH is a recipient of a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
Supplementary Information accompanies the paper on The Journal of Antibiotics website (http://www.nature.com/ja)
In honor of Professor Christopher T. Walsh’s extraordinary contributions to antibiotic research.
References
- 1.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 2.Hopwood DA. Genetic contributions to understanding polyketide synthases. Chem Rev. 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 3.Katz L. Manipulation of modular polyketide synthases. Chem Rev. 1997;97:2557–2575. doi: 10.1021/cr960025+. [DOI] [PubMed] [Google Scholar]
- 4.Khosla C, Tang Y, Chen AY, Schnarr NA, Cane DE. Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu Rev Biochem. 2007;76:195–221. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- 5.Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 6.Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature. 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 7.Gross H. Strategies to unravel the function of orphan biosynthesis pathways: recent examples and future prospects. Appl Microbiol Biotechnol. 2007;75:267–277. doi: 10.1007/s00253-007-0900-5. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, et al. A proteomic survey of nonribosomal peptide and polyketide biosynthesis in actinobacteria. J Proteome Res. 2012;11:85–94. doi: 10.1021/pr2009115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang Y-M, Chang S-L, Oakley BR, Wang CCC. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol. 2011;15:137–143. doi: 10.1016/j.cbpa.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anand S, et al. SBSPKS: Structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 2010;38:W487–W496. doi: 10.1093/nar/gkq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tae H, Sohng JK, Park K. MapsiDB: an integrated web database for type I polyketide synthases. Bioprocess Biosyst Eng. 2009;32:723–727. doi: 10.1007/s00449-008-0296-3. [DOI] [PubMed] [Google Scholar]
- 12.Conway KR, Boddy CN. ClusterMine360: a database of microbial PKS/NRPS biosynthesis. Nucl Acids Res. 2013;41:D402–D407. doi: 10.1093/nar/gks993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichikawa N, et al. DoBISCUIT: a database of secondary metabolite biosynthetic gene clusters. Nucl Acids Res. 2013;41:D408–D414. doi: 10.1093/nar/gks1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menzella HG, et al. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat Biotechnol. 2005;23:1171–1176. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Blin K, et al. antiSMASH 2.0- a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guigó R, Knudsen S, Drake N, Smith T. Prediction of gene structure. J Mol Biol. 1992;226:141–157. doi: 10.1016/0022-2836(92)90130-c. [DOI] [PubMed] [Google Scholar]
- 18.Salamove AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukashin AV, Borodovsky M. GeneMark.hmm: New solutions for gene finding. Nucl Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucl Acids Res. 2004;32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucl Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medema MH, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li MHT, Ung PMU, Zajkowski J, Garneau-Tsodikova S, Sherman DH. Automated genome mining for natural products. BMC Bioinformatics. 2009;10:185. doi: 10.1186/1471-2105-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber T, et al. CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J Biotechnol. 2009;140:13–17. doi: 10.1016/j.jbiotec.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Starcevic S, et al. ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 2008;36:6882–6892. doi: 10.1093/nar/gkn685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minowa Y, Araki M, Kanehisa M. Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J Mol Biol. 2007;368:1500–1517. doi: 10.1016/j.jmb.2007.02.099. [DOI] [PubMed] [Google Scholar]
- 28.Rōttig M, et al. NRPSpredictor2- a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011;39:W362–W367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenke-Kodama H, Börner T, Dittmann E. Natural biocombinatorics in the polyketide synthase genes of the actinobacterium Streptomyces avermitilis. PLoS Comput Biol. 2006;2:e132. doi: 10.1371/journal.pcbi.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McQuitty LL. Similarity analysis by reciprocal pairs for discrete and continuous data. Educ Psychol Meas. 1966;26:825–831. [Google Scholar]
- 32.Yadav G, Gokhale RS, Mohanty D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J Mol Biol. 2003;328:335–363. doi: 10.1016/s0022-2836(03)00232-8. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen T, et al. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- 34.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 35.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 36.Jenke-Kodama H, Sandmann A, Muller R, Dittmann E. Evolutionary implications of bacterial polyketide synthases. Mol Biol Evol. 2005;22:2027–2039. doi: 10.1093/molbev/msi193. [DOI] [PubMed] [Google Scholar]
- 37.Koyama N, Yotsumoto M, Onaka H, Tomoda H. New structural scaffold 14-membered macrocyclic lactone ring for selective inhibitors of cell wall peptidoglycan biosynthesis in Staphylococcus aureus. J Antibiot. 2013;66:303–304. doi: 10.1038/ja.2012.122. [DOI] [PubMed] [Google Scholar]
- 38.Nagahama N, Suzuki M, Awataguchi S, Okuda T. Studies on a new antibiotic, albocycline. I. isolation, purification and properties. J Antibiot. 1967;20:261–266. [PubMed] [Google Scholar]
- 39.Ligon J, et al. Characterization of the biosynthetic gene cluster for the antifungal polyketide soraphen A from Sorangium cellulosum So ce26. Gene. 2002;285:257–267. doi: 10.1016/s0378-1119(02)00396-7. [DOI] [PubMed] [Google Scholar]
- 40.Schwecke T, et al. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang L, et al. Cloning and heterologous exoression of the epothilone gene cluster. Science. 2000;287:640–642. doi: 10.1126/science.287.5453.640. [DOI] [PubMed] [Google Scholar]
- 42.Galyov EE, Brett PJ, DeShazer D. Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu Rev Microbiol. 2010;64:495–517. doi: 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- 43.Franke J, Ishida K, Hertweck C. Genomics-driven discovery of burkholderic acid, a noncanonical cryptic polyketide from human pathogenic Burkholderia species. Angew Chem Int Ed. 2012;51:11611–11615. doi: 10.1002/anie.201205566. [DOI] [PubMed] [Google Scholar]
- 44.Biggins JB, Ternei MA, Brady SF. Malleilactone, a polyketide synthase-derived virulence factor encoded by the cryptic secondary metabolome of Burkholderia pseudomallei group pathogens. J Am Chem Soc. 2012;134:13192–13195. doi: 10.1021/ja3052156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci USA. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holmes TC, et al. Molecular insights into the biosynthesis of guadinomine: a type III secretion system inhibitor. J Am Chem Soc. 2012;134:177797–17806. doi: 10.1021/ja308622d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rounge TB, Rohrlack T, Nederbragt AJ, Kristensen T, Jakobsen KS. A genome-wide analysis of nonribosomal peptide synthetase gene clusters and their peptides in Planktothrix rubescens strain. BMC Genomics. 2009;10:396. doi: 10.1186/1471-2164-10-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neumann CS, Walsh CT, Kay RR. A flavin-dependent halogenase catalyzes the chlorination step in the biosynthesis of Dictyostelium differentiation-inducing factor 1. Proc Natl Acad Sci USA. 2010;107:5798–5803. doi: 10.1073/pnas.1001681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Austin MB, et al. Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase. Nat Chem Biol. 2006;2:494–502. doi: 10.1038/nchembio811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.John U, et al. Novel insights into evolution of protistan polyketide synthases through phylogenomic analysis. Protist. 2007;159:21–30. doi: 10.1016/j.protis.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 51.Zhu G, et al. Cryptosporidium parvum: the first protest known to encode a putative polyketide synthase. Gene. 2002;298:79–89. doi: 10.1016/s0378-1119(02)00931-9. [DOI] [PubMed] [Google Scholar]
- 52.Yook K, et al. WormBase 2012: more genomes, more data, new website. Nucl Acids Res. 2012;40:D735–D741. doi: 10.1093/nar/gkr954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choe A, et al. Ascaroside signaling is widely conserved among nematodes. Curr Biol. 2012;22:772–780. doi: 10.1016/j.cub.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Reuss SH, et al. Comparative metabolomics reveals biogenesis of ascarosides, a modular library of small-molecule signals in C. elegans. J Am Chem Soc. 2012;134:1817–1824. doi: 10.1021/ja210202y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.