Abstract
Background
Cattle babesiosis is a tick-borne disease of cattle with the most severe form of the disease caused by the apicomplexan, Babesia bovis. Babesiosis is transmitted to cattle through the bite of infected cattle ticks of the genus Rhipicephalus. The most prevalent species is Rhipicephalus (Boophilus) microplus, which is distributed throughout the tropical and subtropical countries of the world. The transmission of B. bovis is transovarian and a previous study of the R. microplus ovarian proteome identified several R. microplus proteins that were differentially expressed in response to infection. Through various approaches, we studied the reaction of the R. microplus ovarian transcriptome in response to infection by B. bovis.
Methods
A group of ticks were allowed to feed on a B. bovis-infected splenectomized calf while a second group fed on an uninfected splenectomized control calf. RNA was purified from dissected adult female ovaries of both infected and uninfected ticks and a subtracted B. bovis-infected cDNA library was synthesized, subtracting with the uninfected ovarian RNA. Four thousand ESTs were sequenced from the ovary subtracted library and annotated.
Results
The subtracted library dataset assembled into 727 unique contigs and 2,161 singletons for a total of 2,888 unigenes, Microarray experiments designed to detect B. bovis-induced gene expression changes indicated at least 15 transcripts were expressed at a higher level in ovaries from ticks feeding upon the B. bovis-infected calf as compared with ovaries from ticks feeding on an uninfected calf. We did not detect any transcripts from these microarray experiments that were expressed at a lower level in the infected ovaries compared with the uninfected ovaries. Using the technique called serial analysis of gene expression, 41 ovarian transcripts from infected ticks were differentially expressed when compared with transcripts of controls.
Conclusion
Collectively, our experimental approaches provide the first comprehensive profile of the R. microplus ovarian transcriptome responding to infection by B. bovis. This dataset should prove useful in molecular studies of host-pathogen interactions between this tick and its apicomplexan parasite.
Keywords: Cattle tick, Rhipicephalus microplus, Babesia bovis, Ovary, Transcriptome, EST
Background
The cattle tick, Rhipicephalus (Boophilus) microplus, is distributed worldwide and is detrimental to animal agriculture. Cattle producers incur substantial financial losses due to R. microplus infestations with Brazil alone experiencing losses of over $2 billion annually [1]. A substantial portion of these losses is attributable to pathogens and their associated diseases transmitted by the tick’s bite. The tick transmits two apicomplexan pathogenic agents, Babesia bovis and Babesia bigemina[2]. B. bovis is generally responsible for the more serious cases of bovine babesiosis, and frequently results in fatal infections of immunologically naive hosts. These pathogens infect the bovine erythrocyte, which is ingested by R. microplus during feeding upon an infected bovine host [2].
Following ingestion by the tick, merozoite stage apicomplexa undergo developmental changes until they are released from the bovine erythrocytes within the tick’s gut. After release, the apicomplexa complete their development to the zygote stage at which time they enter the digestive cells and begin multiplication and development until the kinete stage is reached. The apicomplexa then migrate from the digestive cells to the hemolymph and eventually spread to other tissues. Transmission of Babesia from adult tick to progeny is always trans-ovarian. After entering the developing oocytes, Babesia parasites undergo further development during the tick larval stage, and eventually occupy the tick’s salivary glands where they become infective to the vertebrate host [2].
Targeting ovarian proteins could adversely affect tick populations by causing a decrease in oogenesis and embryogenesis, thereby reducing reproduction rates, and by disrupting development and reproduction of disease causing Babesia parasites. Rachinsky et al.[3] showed that a number of R. microplus ovarian proteins are differentially expressed in response to B. bovis infection, including serine protease inhibitors, calreticulin, and peptidyl-prolyl cis-trans isomerases. These findings prompted the present study, as we wished to begin a characterization of the ovarian transcriptome, with emphasis on genes differentially expressed in response to ingestion of B. bovis-infected bovine blood. As our source of RNA, we used tissues that were used in the ovarian proteome study of Rachinsky et al.[3] to allow a direct comparison between the proteomic and transcriptomic response.
In the Rachinsky study, R. microplus fed upon a splenectomized calf suffering from bovine babesiosis due to infection with B. bovis. For the current study, we obtained dissected R. microplus ovaries archived from their study and compared gene expression in ovary dissected from adult female ticks which had fed on the infected calf with gene expression in corresponding tissue from ticks at a similar developmental stage that fed on an uninfected control calf. Our approaches included sequencing a subtracted library synthesized from infected ovarian mRNA, microarrays, serial analysis of gene expression (SAGE), and quantitative real-time polymerase chain reaction (qRT-PCR) to identify ovarian transcripts differentially expressed in association with B. bovis infection of R. microplus.
Methods
Animal use protocol
All animal use was conducted at ADRU facilities at the University of Idaho Holm Research Center (Moscow, ID) while following protocols approved by the University of Idaho Institutional Animal Care and Use Committee.
Tick strain
The ticks were taken from the f20 generation of the La Minita strain of R. microplus, which has been maintained as a Babesia-free laboratory colony at The University of Idaho Holm Research Center since 1999. La Minita was originally collected from an outbreak in Starr County, Texas in 1996 and propagated at the USDA Cattle Fever Tick Research Laboratory at Moore Field, Texas. All calves used in this study were splenectomized Holstein breed and 5–6 months of age.
Sample collection
Tissues used in the transcriptome studies were the same as those obtained and dissected for the ovarian proteome study of Rachinsky et al.[3]. During the tissue dissection stage of that study, samples had been randomly assigned for either a proteome study or a transcriptomic study. The procedures for obtaining B. bovis-infected and uninfected adult stage engorged females were reported by Rachinsky et al.[3]. Briefly, for the uninfected ticks, larvae from 1 g of R. microplus eggs were placed on a calf on study day 1 and replete female ticks began dropping on study day 22 and continued dropping until study day 31 when the animal was euthanized. The ovaries for the uninfected sample were obtained from female ticks that dropped on day 22 and were maintained for 4 days at 23°C at which time oviposition began and the ticks dissected within 24 hr. During collections of the B. bovis-infected ticks (described below), it was noted the period from when the replete females dropped from their bovine host to when ovipositioning began was twice as long compared to the uninfected ticks. Thus, we utilized the onset of oviposition as our reference point to biologically synchronize the dissection timings between the two samples.
To obtain B. bovis-infected ticks, two splenectomized calves were infested with tick larvae as above and the calves were infected on day 14 with B. bovis. This was done by intravenous inoculation with 1.8 ml of blood stabilate culture originating from the T2Bo strain of B. bovis, stored in liquid nitrogen and routinely verified as highly infective to bovines. The calf infections were monitored by daily measurement of rectal temperature, which peaked on day 22, and both animals were required to be euthanized on study day 24 due to the progression of babesiosis. All collected female ticks were incubated at 23°C for nine days, at which time oviposition and dissections commenced.
Riek [4] reported that 4–5 days after females have fed to repletion on a B. bovis-infected splenectomized calf, B. bovis vermicules can be observed in mature ova. Thus, a 9 day incubation period of the females collected from the B. bovis-infected calves should have produced infected ovaries. We confirmed ticks were infected with B. bovis by examining hemolymph smears from 66 randomly selected ticks that dropped on study day 24 from the two infected calves. Forty-six ticks had at least 5 B. bovis kinetes per high power microscopic field and the other 20 ticks had 3–5 kinetes per high power field.
SAGE was used as a third transcriptomic protocol to complement the subtracted library and microarray analyses of infection-induced differential gene expression. In the SAGE experiment on B. bovis-infected ovaries, dissections were performed using engorged female ticks that had dropped after feeding to repletion on the B. bovis-infected calves as described above. After dropping, the engorged ticks were incubated at 23°C for either 0 or 6 days prior to dissection.
Because of the status of R. microplus as an arthropod requiring adherence to strict USDA quarantine and handling restrictions for B. bovis-infected experimental calves requiring the need to sacrifice the calves at the end of each experiment, the ideal of using independent biological replicates was not met. Animal experiments were approved by the Institutional Animal Care and Use Committee at Washington State University, USA, in accordance with institutional guidelines based on the U.S. National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
RNA protocols
The Totally RNA Kit (Ambion Inc., Austin, TX, USA) was used to purify RNA from tick ovaries obtained from 20 ticks with a final lithium chloride precipitation step added per kit protocol booklet. The ovary RNA was obtained by dissecting individual engorged females into RNAlater (Ambion Inc.) and the dissected materials were pooled prior to isolating total RNA. Total RNA was treated with Turbo DNAse as per Turbo DNA-free kit protocols (Ambion Inc.). RNA integrity was verified by formaldehyde gel electrophoresis and staining in GelStar Nucleic Acid Gel Stain (Lonza, Rockland, ME, USA).
Subtracted and SAGE library synthesis
Two 250 μg samples, of B. bovis-infected or uninfected ovary total RNA, were sent to Express Genomics Inc. (Frederick, MD, USA) for subtracted library synthesis. Primary libraries were amplified from the uninfected and B. bovis-infected material, following directional cloning into the pExpress-1 vector digested with NotI and EcoRV. Subsequently, a subtracted library was produced by subtracting the B. bovis-infected material with the uninfected material, enriching for expressed genes in the B. bovis-infected material. Express Genomics quality control checks found a 100-fold reduction in the number of clones that hybridized to actin in the subtracted library compared with the primary library, verifying the subtraction process was successful. The SAGE libraries were synthesized from 50 μg of total RNA from engorged female tick ovaries using the I-SAGE Long Kit as per manufacturer’s protocols (Invitrogen Inc., Carlsbad, CA).
Transcriptome sequencing
EST and SAGE library sequencing was performed at the J. Craig Venter Institute (Rockville, MD). Bacterial colonies were picked for template preparation using colony-picking robots (Genetix, Boston, MA), inoculated into 384 well plates containing liquid medium and incubated overnight at 37°C. A robotic workstation was used to prepare sequencing grade plasmid DNA via an alkaline lysis method modified for high throughput processing [5]. Beckman Multimek 96 or Biomek FX automated pipetting robot work stations (Beckman Coulter, Fullerton, CA) were used to combine pre-aliquoted templates and sequencing reaction mixes. Linear amplification steps were performed on MJ Research Tetrads PTC-225 (MJ Research, Inc., Watertown, MA) and sequencing reaction products purified by ethanol precipitation and analyzed on ABI 3730xl sequencing machines (Applied Biosystems, Foster City, CA). The unassembled ovary subtracted library EST sequences were submitted to GenBank dbEST (GenBank: FG301341-FG305398). SAGE library sequences were extracted and analyzed by a set of custom-written perl scripts.
Bioinformatics analysis
Sequence assembly and annotation were performed as described in Heekin et al.[6]. Briefly, several screening steps were applied to eliminate contaminated or low quality sequences from the subtracted library prior to assembly. A de novo transcript assembly was performed on the subtracted library using cap3 [7]. All resulting contigs and unassembled singletons (collectively referred to as unigenes) were used in subsequent analyses (Additional file 1). Annotations were initially assigned to unigenes using similarity search methods of the Uniref100 database using BLASTX with an e-value cutoff of 1e-07 [8]. Sequences with no BLASTX high-scoring pairs (HSPs) were submitted to the prot4EST application to predict the correct open reading frame (ORF) for each sequence [9]. After the ORFs were predicted, the sequences were submitted to annot8r for assignment of Gene Ontology (GO) terms [10,11].
Microarray design
Twenty μg of each DNA-free total RNA was sent to NimbleGen Systems Inc. (Madison, WI, USA) for use in microarray hybridization. A custom high-density single channel oligonucleotide array was designed by NimbleGen Systems Inc. using 13,601 of the 13,642 members of BmiGI Version 2 and these arrays were described in detail by Saldivar et al.[12].
Our array experimental design consisted of four replicates, two replicate microarrays for each of the infected and uninfected samples. Also, in the array design, each transcript represented on the array has 14 different 50-mer probes, establishing an additional layer of replication. Each probe is spotted twice on each array as technical replicates. Sample labeling, hybridization, array scanning, and image analysis was performed at NimbleGen Systems Inc. as described by Saldivar et al.[12]. Quality control measures and pre-processing were performed using the computing language R [13] and Bioconductor [14]. The quality of the arrays was assessed through standard quality control measures: pseudo-images of the arrays to detect spatial effects, scatter plots of the arrays versus a pseudo-median reference chip and summary statistics including histograms and boxplots of raw and normalized log intensities. All microarray quality control measurements were within recommended limits as established and implemented by Nimblegen.
Gene calls were generated and normalized as described [6]. The microarray data have been submitted to the GEO database (http://www.ncbi.nlm.nih.gov/geo/; GEO accession number GSE10816). Significance Analysis of Microarrays (SAM) [15,16] was performed in the Microarray Experiment Viewer (MeV Version 4.0, Dana-Farber Cancer Institute, Boston, MA, USA) to select statistically significant differentially expressed genes. The design used by SAM is a two-class unpaired design, where samples fall in either the infected or uninfected group. The cutoff for significance is determined by a tuning parameter delta and a minimum fold change threshold to ensure that called genes change at least a specified amount. The threshold value delta was set to 0.53 and fold change set to ≥ 2.0. Because of the unavailability of biological replicates, the p-values and d statistics related to the microarray data should not be interpreted as statistical probabilities. Nevertheless, these statistics remain useful for prioritizing candidates for comparison with the SAGE and subtracted library results.
Verification by real-time PCR
Array results were verified for three target genes based on their level of differential expression and the amount of annotation available for their corresponding BmiGI sequence. The same total RNA samples used for the microarrays were also used for quantitative real-time PCRs. The RETROscript Kit Reverse Transcription for RT-PCR (Ambion) was used as per manufacturer’s recommendations to produce cDNA from four micrograms of DNA-free total RNA for each sample. Primers and TaqMan probes were designed using Beacon Designer 7.5 (PREMIER BioSoft International, Palo Alto, CA; Additional file 2) and synthesized by Sigma-Aldrich Inc. (Atlanta, GA) for each gene selected and for the R. microplus 18S rRNA gene, which was the reference gene for normalization [12]. Validation experiments were run on each gene and the reference gene to determine PCR efficiencies and optimal concentrations.
All real-time reactions were carried out in clear low-profile 96 well plates (no. MLL9601, BioRad, Hercules, CA). The 25 μL total reaction volumes included primers, 250nM TaqMan probe, TaqMan Universal Master Mix No AmpErase UNG (Applied Biosystems Inc., Foster City, CA) and corresponding RETROscript cDNA. The final primer concentration for the 18S rRNA reference gene and the targeted genes was 900nM for both the forward and reverse primers. The BioRad CFX96 Real-Time System was used with a cycling protocol of 95°C for 10 min, and 50 cycles of 95°C for 15 sec, 60°C for 1 min plus plate read. The fluorescence emission data analysis was done using baseline subtracted curve fit mode with CFX Manager Software version 1.0 (BioRad). All primer and probe sequences are listed in Additional file 2.
Results and discussion
Subtracted library results
Over 4,100 bacterial colonies from the subtracted library were prepared for sequencing and this resulted in 4,045 high quality tick EST sequences. This dataset assembled into 727 unique contigs (clusters of related transcripts) and 2,161 singletons (transcripts that did not cluster) in two separate passes for a total of 2,888 unigenes (Additional file 1). Contigs that clustered during the first pass received the prefix contigA. Contigs from the first pass that clustered with additional sequences during the second pass of the assembler received the prefix contigB. Singleton sequences retained their original labels assigned during sequencing. The mean unigene length was 851.7 nucleotides. Out of 2,888 unigenes, approximately one-third received significant BLASTX HSPs (e-value < 1e-07) from the Uniref100 database and are listed in Additional file 3. The ORFs for all unigenes predicted by prot4EST are listed in Additional file 4. BLAST results reported in this study are from BLASTX searches of the Uniref100 database unless otherwise noted.
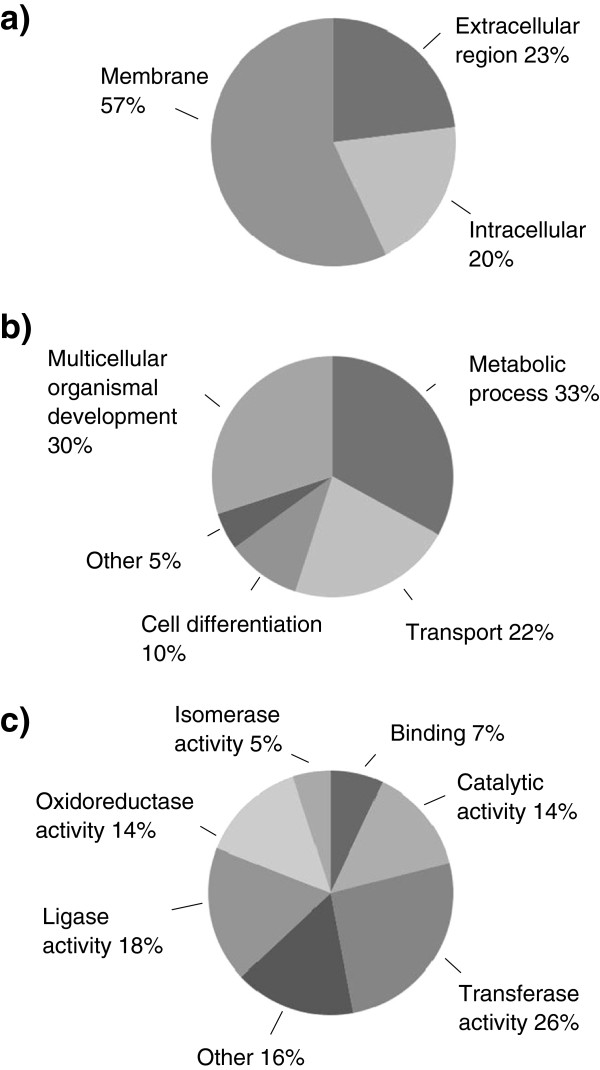
Figure 1 lists a summary of the GO annotation by the annot8r application. The unigenes are categorized by three ontology domains consisting of 29 high-level GO terms. In the cellular component domain (C), most of the differentially expressed transcripts (57%) were classified as membrane. A number of unigenes, however, were classified as extracellular (23%) or intracellular (20%). The majority of annotations in the molecular function domain (F) were assigned the GO terms: transferase activity (26%), ligase activity (18%), oxidoreductase activity (14), and catalytic activity (14%). In the biological process domain (P), metabolic process (33%), multicellular organismal development (30%), and transport (22%) were predominant. The complete GO annotation set is listed in Additional file 5.
Figure 1.
High level GO classification of unigenes from the subtracted library. a: Cellular Component Ontology, b: Biological Process Ontology, c: Molecular Function Ontology.
Genes related to stress, detoxification, and immune response
The subtracted library BLASTX analysis identified a number of unigenes with the GO term “stress response” (Table 1; Additional file 3). There were 12 unigenes with sequence similarity to cytochrome P450s, which are a group of enzymes that catalyze metabolism of organic molecules including toxins and xenobiotics. Transcripts encoding superoxide dismutase were found in the subtracted library (ContigA418 and MPOA822TR). Additional detoxification proteins related to glutathione metabolism were also observed, including ContigA295, which is a glutathione S-transferase (GST). GST expression has been previously induced upon blood feeding in R. microplus, and is postulated to be an adaptive response to reactive oxygen species created during the blood meal [17]. GSTs may also facilitate digestion of the meal by reducing proteins and lipids [18]. Additional unigenes were similar to other protein-reducing antioxidants including glutaredoxin (ContigA561), peroxiredoxin (MPOAE24TR), and several peroxinectins (ContigA694, MPOA737TR, MPOA895TR, MPOA475TR, MPOAC37TR and MPOAK85TR). The library also contained 5 stress-induced heat shock protein transcripts, ContigA105, ContigA652, MPOAC82TR, MPOAF39TR. The subtraction library contained two cytochrome c oxidase ESTs (MPOAA34TF and MPOAN31TR), and these proteins have been indirectly linked to hemocyte modifications occurring during Borrelia burgdorferi infection of Ixodes ricinus[19].
Table 1.
Unigenes from subtracted library annotated by BLASTX and with GO term of stress response
| Unigene a | Protein annotation | Species | Acc. no. | e -value b |
|---|---|---|---|---|
| ContigA171 |
Cytochrome P450 |
Tribolium castaneum |
D6W6R3 |
1e-68 |
| ContigA591 |
Cytochrome P450 |
Ixodes scapularis |
B7PT10 |
6e-12 |
| MPOAJ93TR |
Cytochrome P450 |
Ixodes scapularis |
B7PN37 |
2e-45 |
| MPOAN40TR |
Cytochrome P450 |
Ixodes scapularis |
B7PJW2 |
5e-24 |
| MPOAH45TF |
Cytochrome P450 |
Ixodes scapularis |
B7QJP3 |
5e-80 |
| MPOAG88TR |
Cytochrome P450 |
Ixodes scapularis |
B7P5V0 |
1e-47 |
| MPOAN40TF |
Cytochrome P450 |
Ixodes scapularis |
B7PTT2 |
4e-43 |
| MPOAG88TF |
Cytochrome P450 |
Ixodes scapularis |
B7PME9 |
6e-32 |
| MPOAG70TR, MPOAH45TR |
Cytochrome P450 |
Ixodes scapularis |
B7QJP2 |
6e-78 |
| ContigA57, ContigA143 |
Cytochrome P450 |
Ixodes scapularis |
B7P5I8 |
1e-124 |
| ContigA418 |
Superoxide dismutase |
Amblyomma maculatum |
G3MQI9 |
9e-89 |
| MPOA822TR |
Superoxide dismutase |
Amblyomma maculatum |
G3MQ28 |
2e-108 |
| ContigA652 |
Heat shock protein |
Aedes aegypti |
Q17PR3 |
7e-135 |
| MPOAC82TR, MPOAC82TF |
Heat shock protein 70 |
Moina mongolica |
D2E4A4 |
4e-103 |
| ContigA105 |
Heat shock protein |
Ixodes scapularis |
B7QJZ5 |
3e-32 |
| MPOAF39TR |
Heat shock protein |
Ixodes scapularis |
B7PAR6 |
3e-144 |
| ContigA295 |
Glutathione S-transferase |
Ixodes scapularis |
B7Q9K1 |
3e-143 |
| ContigA561 |
Glutaredoxin |
Ictalurus furcatus |
E3TDC4 |
1e-38 |
| MPOAE24TR |
Peroxiredoxin |
Ixodes scapularis |
B7Q7K9 |
6e-44 |
| MPOAA34TF |
Cytochrome c oxidase assemby |
Aedes aegypti |
Q16M46 |
4e-40 |
| MPOAN31TR |
Cytochrome c oxidase assembly |
Ixodes scapularis |
B7P8T9 |
9e-111 |
| ContigA694, MPOA737TR, MPOA895TR |
Peroxinectin |
Ixodes scapularis |
B7P9B9 |
2e-12 |
| MPOA475TR |
Peroxinectin |
Ixodes scapularis |
B7PUM7 |
2e-116 |
| MPOAK85TR, MPOAC37TR | Peroxinectin | Ixodes scapularis | B7PQ34 | 1e-84 |
a Unigene identification number as listed in Additional file 1.
b In the case of multiple e-values obtained from multiple unigenes, the highest e-value is listed.
Table 2 lists unigenes annotated with several additional stress- or immune-related GO terms including four unigenes with the GO term “response to DNA damage stimulus” (ContigA564, MPOAC91TR, MPOAF62TR, MPOAG90TR). Unigenes MPOAF05TR and ContigA290 were annotated with the GO term “carboxylesterase activity”. Carboxylesterases were induced in the midgut of the silkworm in response to insecticides [20] and are involved in the acaricide resistance mechanisms found in R. microplus[21]. Unigenes ContigA711, MPOAC37TR, and MPOAK85TR received the GO term “response to oxidative stress”. The unigene MPOA256TR was annotated with the GO term “defense response to bacterium” but did not have a useful hit in the UniRef100 database. Unigenes MPOAL11TR and ContigA709 received the GO term “immune response” but were annotated only as uncharacterized proteins.
Table 2.
Unigenes from subtracted library annotated with stress and immune-related GO ontology terms
| Unigene a | GO accession no. | Category b | GO annotation |
|---|---|---|---|
| ContigA290 |
GO:0004091 |
F |
Carboxylesterase activity |
| MPOAF05TR |
GO:0004091 |
F |
Carboxylesterase activity |
| ContigA711 |
GO:0006979 |
P |
Response to oxidative stress |
| MPOAL11TR |
GO:0006955 |
P |
Immune response |
| ContigA709 |
GO:0006955 |
P |
Immune response |
| ContigA564 |
GO:0006974 |
P |
Response to DNA damage stimulus |
| MPOAC91TR |
GO:0006974 |
P |
Response to DNA damage stimulus |
| MPOAF62TR |
GO:0006974 |
P |
Response to DNA damage stimulus |
| MPOAG90TR |
GO:0006974 |
P |
Response to DNA damage stimulus |
| MPOA256TR | GO:0042742 | P | Defense response to bacterium |
a Unigene identification number as listed in Additional file 1.
b GO category F = function, P = process.
Proteases and protease inhibitors were markedly induced in the ovary and possibly contribute to the ovarian immune response. Many of these catabolic enzymes included defensin-like molecules, including serpins, cathepsins, legumain, and microplusin (Additional files 1 and 3). Defensins are small catabolic peptides with specific antimicrobial activity. Unigene ContigA321 was orthologous to a putative defensin in I. scapularis. Defensin was induced in the gut of D. variabilis when challenged with Borrelia burgdorferi, but was not induced in the same tick when challenged with two different species of Gram-positive bacteria [22]. ContigA321 did not show significant sequence similarity to any of the ovary up-regulated transcripts reported by Stutzer et al.[23] nor to ESTs reported from other tick species. A 687 bp EST from an embryonic cell line of R. microplus, GenBank Accession EW679737, was identical to the 1188 bp ContigA321 (data not shown). Thus ContigA321 may represent a defensin unique to ovarian or embryonic tissue. Unigenes MPOA768TF and TR showed high similarity to a putative legumain-like protease from two species of ticks. Legumain is an asparaginyl endopeptidase that processes microbial antigens in lysosomes [24]. Microplusin is a unique cysteine-rich secreted antimicrobial peptide (AMP), which is active against bacteria and fungi [25]. In addition to being found in the subtracted library ContigB21, the transcript encoding microplusin was also one of the most abundant seen in the SAGE experiment (described below). Microplusin transcript levels in the ovaries of R. microplus gradually rise before peaking at the beginning of oviposition [26]. Microplusin was also among the up-regulated transcripts in a transcriptome experiment that examined gene expression in B. bovis-infected larvae of R. microplus[6].
The subtraction library contained several transcripts that may participate in the tick immune response (Additional files 1 and 3). The unigene MPOAB56TF was annotated as tumor necrosis factor receptor-associated factor (TRAF), which is an important regulator of inflammation, apoptosis, and antiviral responses (Additional file 3). ContigA62 was an ortholog to a putative alpha-2-macroglobulin in I. scapularis, which was up-regulated in Dermacentor variabilis when exposed to Anaplasma marginale[27]. Unigene MPOAA26TF showed significant similarity to fucosyltransferase, which has been demonstrated to increase microbial pathogenesis in I. scapularis[28]. Putative ixoderins were also identified (MPOAH63TR, ContigA104, and ContigA667); ixoderin is a lectin-like molecule with a possible role in innate immunity in ticks [29].
Serpins were the predominant protease inhibitor in the subtracted library. Serpins regulate blood coagulation cascades, transport of hormones and are components of the immune system of many invertebrates [30]. Serpins have significant roles in antimicrobial and antifungal responses in insects [31]. ContigA484 had sequence similarity to a cysteine peptidase inhibitor in M. musculus. A recent review of tick cysteine protease inhibitors (cystatins) characterized their roles in detoxification, innate immunity regulation, pathogen transmission and immunosuppression [32].
Cathepsins, which comprise cysteine, aspartic, and serine proteases, were the dominant protease family in the subtracted library. Cysteine proteases are important constituents of the immune response of R. microplus and participate in vitellin degradation [33,34]. The unigene MPOAH54TR had high similarity to a cysteine protease, longipain. This enzyme was recently characterized in the babesial parasite vector tick Haemaphysalis longicornis. Longipain was specifically localized to lysosomal vacuoles and was shown to be a potent parasiticide [35]. Aspartic proteases have been linked to digestion and vitellin degradation in ticks [36]. Three of the induced proteases, ContigA110, MPOA037TF, and MPOAM53TR, were serine-type proteases. Three serine protease transcripts were up-regulated in ovary tissues of R. microplus[23]. However, there was no significant sequence similarity between these ovarian serine proteases and those reported here (data not shown). Serine-type proteases may be involved in vitellin degradation, which was inferred from a study of a trypsin-like serine peptidase expressed in lice embryos [37]. Serine proteases from the gut of H. longicornis were also up-regulated during the blood-feeding process [38].
Microarray results
Our microarray approach compared the ovary transcriptome from engorged ovipositing females that fed on a B. bovis-infected bovine host to the ovary transcriptome from ticks fed on an uninfected host. The microarray experiments identified 15 transcripts that were expressed at a higher level in ticks feeding upon a B. bovis-infected calf compared with ticks feeding on an uninfected calf (Table 3; Additional file 6). No transcripts were found to be statistically expressed at a lower level in ovaries from adult female ticks feeding upon a B. bovis-infected calf compared with ovaries from ticks feeding on an uninfected calf. As similarly reported by Saldivar et al.[12] and Stutzer et al.[23], a number of the differentially expressed tick genes had no useful annotation; six of the fifteen transcripts in Table 3 did not have significant (e < 0.001) BlastX hits.
Table 3.
R. microplus genes with highest up-regulation associated with B. bovis infection in microarrays
|
ID
a
|
d
b
|
FC
c
|
BlastX annotation |
|||
|---|---|---|---|---|---|---|
| Protein name | Species | Acc. no. | e -value | |||
|
TC12551 |
2.1 |
47.8 |
put. secreted salivary gland peptide |
Ixodes scapularis |
XP002411978.1 |
3e-04 |
|
TC9311 |
1.9 |
60.8 |
Kunitz-like protease inhibitor |
Ancylostoma caninum |
AAN10061.1 |
5e-105 |
| TC6492 |
1.9 |
32.8 |
Kunitz-like protease inhibitor 6 |
Rhipicephalus microplus |
P83606.2 |
0.0 |
|
TC13077 |
1.9 |
39.6 |
Kunitz-like protease inhibitor |
Ancylostoma caninum |
AAN10061.1 |
5e-116 |
| TC9020 |
1.8 |
30.8 |
NSSd |
- |
- |
- |
|
TC6491 |
1.5 |
16.1 |
Kunitz-like protease inhibitor 6 |
Rhipicephalus microplus |
P83606.2 |
2e-94 |
|
TC6326 |
1.5 |
16.6 |
put. secreted salivary gland peptide |
Ixodes scapularis |
XP002411978.1 |
3e-10 |
| BEAAA85TR |
1.5 |
19.9 |
NSS |
- |
- |
- |
| TC11578 |
1.4 |
16.9 |
NSS |
- |
- |
- |
|
BEABQ71TR |
1.2 |
10.5 |
GGY domain protein |
Amblyomma variegatum |
DAA34729.1 |
1e-09 |
| BEAC749TR |
1.2 |
10.1 |
NSS |
- |
- |
- |
|
TC6758 |
1.1 |
7.9 |
hypo. protein IscW |
Ixodes scapularis |
XP002411179.1 |
1e-54 |
| TC8946 |
1.1 |
7.9 |
NSS |
- |
- |
- |
| TC6671 |
1.1 |
8.9 |
NSS |
- |
- |
- |
| TC5979 | 1.0 | 7.5 | hypo. conserved protein 57 | Amblyomma variegatum | DAA34262.1 | 2e-33 |
Gene IDs in bold type are also represented in the subtracted library.
a The identification number from BmiGI Version 2 (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=b_microplus).
b d is the d statistic as performed by SAM.
c FC is the fold change ratio.
d No statistically significant similarity found in BlastX search, based on e-value < 0.001.
Four of the most up-regulated transcripts in the adult female ovary microarrays had high sequence similarity to Kunitz-like protease inhibitors (Table 3). Kunitz-type inhibitors have been studied in R. microplus and exhibit activity against bovine trypsin and human neutrophil elastase [39]. These inhibitors have also been shown to be differentially expressed in tick salivary glands in response to pathogen infection [40]. Thus, Kunitz-type inhibitors likely play a defensive role in the tick ovary. Four Kunitz-type protease inhibitors were found in the subtracted library dataset (Additional file 3), thus corroborating the microarray findings.
To verify the microarray results, we selected 3 transcripts with differential expression of varying fold-changes and performed qRT-PCR to compare transcript levels in the B. bovis-infected ovary tissue with the uninfected control (Table 4). The directional expression changes for all selected transcripts were qualitatively similar in both the microarrays and qRT-PCRs. In fact, the three transcripts showed higher differential regulation in the RT-PCR than in the microarrays, which was expected since array results were compressed towards zero during their analysis.
Table 4.
RT-PCR Verification of selected microarray results
| |
Microarray
a
|
Relative quantitative PCR
a
|
||
|---|---|---|---|---|
| EST | Uninfected | Infected | Uninfected | Infected |
| TC9020 |
1 |
30.8 |
1 |
395 |
| TC9311 |
1 |
60.8 |
1 |
206 |
| TC13077 | 1 | 39.6 | 1 | 61 |
a Normalized data to set lower value to 1 for comparison purposes.
SAGE results
SAGE was used to identify genes that are responding to the infection process whereby B. bovis enters and replicates in the tick ovary. SAGE libraries were produced from ovaries dissected from engorged female ticks the day of dropping from the bovine host (designated day 0) and 6 days after dropping. Since the subtracted library and microarray protocols assessed the ovarian transcriptome 9 days post-repletion, the time period between repletion and the onset of oviposition was investigated using SAGE. The 6 day time point was selected because Riek [4] reported that B. bovis vermicules could be found in mature ova 4–5 days after females fed to repletion. Additional files 7 and 8 contain the complete raw data sets for SAGE tag counts and associations between SAGE tags. Table 5 shows the numbers of tags in both libraries, the number of unique tags and their distribution into abundance classes. Combining both libraries, 792 of the tags had exact matches to BmiGI Version 2.0 in the forward direction (Additional file 9) and 568 of the tags had matches in the reverse complement direction (Additional file 10). Tables 6 and 7 present the 50 most abundant SAGE tags in the control and infected libraries, respectively. Each tag’s corresponding match to BmiGI Version 2 is indicated and annotation is given if available. Twenty-two tags from the Control (Day 0) library and 16 tags from the Infected (Day 6) library had useful annotation. Table 8 lists the tags with >10-fold differential expression comparing the Control and Infected SAGE libraries. Twenty-four tags were in the up-regulated category while 13 tags were in the down-regulated category. Seven of the up-regulated tags and 6 of the down-regulated tags in this table had an exact match to a member of BmiGI Version 2.0. All up-regulated and down-regulated tags are listed in Additional files 11 and 12, respectively.
Table 5.
Number of LongSAGE tags and abundance classes from each library
| % Abundance |
Overall number of tags |
Number of unique tags |
||
|---|---|---|---|---|
| Control | Infected | Control | Infected | |
| > 1.0 |
0 |
481 |
0 |
2 |
| 0.2 - 1.0 |
1032 |
903 |
39 |
32 |
| 0.05 - < 0.2 |
1429 |
1110 |
227 |
143 |
| < 0.02 |
1152 |
1907 |
516 |
789 |
| 0.02 - < 0.05 |
3151 |
4349 |
3151 |
4349 |
| Total | 6764 | 8750 | 3933 | 5315 |
Table 6.
The 50 most abundant SAGE tags in the pooled tag data from control LongSAGE library
| Rank | Tag a | Count |
Infected |
BmiGI match |
|||
|---|---|---|---|---|---|---|---|
| Library rank | Description | Species | e-value | ID | |||
| 1 |
ACGTGACTGTCGCCACC |
57 |
29 |
- |
- |
- |
- |
| 2 |
TGGTGCCCGAAACGAAG |
55 |
106 |
Uro-adherence factor A |
Taloromyces stipitatus |
1e-05 |
TC12322 |
| 3 |
TGGCTGGCTGCCCACTG |
48 |
18 |
Ribosomal protein P0 |
Haemaphysalis longicornis |
2e-35 |
TC9039 |
| 4 |
GACGGCGAGTGGGAACC |
45 |
116 |
calreticulin |
Rhipicephalus microplus |
1e-165 |
TC8950 |
| 5 |
TACGAAGCGCTGGCAGA |
43 |
49 |
Disulfide isomerase |
Haemaphysalis longicornis |
4e-48 |
TC8716 |
| 6 |
ACGCGACTGTCGCCACC |
40 |
29 |
Putative secreted protein |
Ixodes scapularis |
3e-72 |
TC5798 |
| 7 |
TCTGGACGCGGCAAGGG |
38 |
425 |
- |
- |
|
- |
| 8 |
GCCCGCAGCGGCTGAAC |
35 |
50 |
Hypothetical protein |
Ixodes scapularis |
2e-10 |
TC7866 |
| 9 |
CTCACCGACCCGTCGGC |
35 |
43 |
- |
- |
- |
- |
| 10 |
GGTCCACCCCAGCGACT |
33 |
31 |
Hypothetical protein |
Ixodes scapularis |
8e-104 |
TC12372 |
| 11 |
CGCAAGGCCCAAGGAGG |
29 |
260 |
Hypothetical protein |
Ixodes scapularis |
1e-20 |
TC9377 |
| 12 |
CGCAAGGCCCAAGGAGG |
29 |
50 |
- |
- |
- |
- |
| 13 |
GTGGTGCACGCCAACCC |
28 |
80 |
Superoxide dismutase- |
Apis mellifera |
8e-44 |
TC12062 |
| 14 |
CCAGCGCTAAAGATGCG |
27 |
59 |
- |
- |
- |
- |
| 15 |
GAGGCGGTGCGGGAGAG |
27 |
178 |
- |
- |
- |
- |
| 16 |
GCACGGCGATGCGACGG |
26 |
39 |
- |
- |
- |
- |
| 17 |
TGTGGCTGGGGCTCCGC |
26 |
50 |
- |
- |
- |
- |
| 18 |
GACTCCAATGAAGGCCC |
25 |
25 |
Alpha tubulin |
Mus musculus |
0.0 |
TC9399 |
| 19 |
ACACGACTGTCGCCACC |
23 |
116 |
- |
- |
- |
- |
| 20 |
GCGAGGAGCTTGTCGGG |
22 |
35 |
- |
- |
- |
- |
| 21 |
GCTGTGGTTGCGCGCAC |
22 |
59 |
- |
- |
- |
- |
| 22 |
CTGCAGACGTTGACGGG |
21 |
97 |
Adipose differentiation related protein |
Ixodes scapularis |
8e-75 |
TC9098 |
| 23 |
GGCCCCCTCCCGCCCAA |
21 |
Nob |
Transaldolase |
Macaca mulatta |
5e-57 |
TC13335 |
| 24 |
TGCGCAAAGGACGCCCG |
21 |
178 |
Serine proteinase inhibitor |
Rhipicephalus microplus |
1e-26 |
BEAET94TR |
| 25 |
ATCTGAGTTTAGACCGA |
21 |
1 |
Mitochondrial DNA |
Rhipicephalus sanguineus |
0.00 |
TC5761 |
| 26 |
CTGAGGATTGCCGAGCC |
20 |
97 |
60S ribosomal protein L7 |
Argas monolakensis |
1e-104 |
TC5935 |
| 27 |
CACGTACAACCTCTGCG |
19 |
No |
- |
- |
- |
- |
| 28 |
GGGGAGTCTGACGACTG |
19 |
80 |
- |
- |
- |
- |
| 29 |
GGCGACCGCTTCACCGA |
19 |
68 |
Myosin regulatory light chain |
|
5e-83 |
TC12444 |
| 30 |
TTGTGCAGCGATCGGCA |
19 |
425 |
- |
- |
- |
- |
| 31 |
GCACCTGGCGCTGGCAA |
18 |
260 |
- |
- |
- |
- |
| 32 |
CTGGCCGCTTGGGTCCG |
18 |
116 |
- |
- |
- |
- |
| 33 |
GTAGGCCCGGTATTGGT |
17 |
18 |
- |
- |
- |
- |
| 34 |
CACCTTTGCATCGACGC |
15 |
68 |
- |
- |
- |
- |
| 35 |
GTACCAGAGGACAAGCC |
15 |
967 |
- |
- |
- |
- |
| 36 |
GGAAGCGCTAAGCGGCC |
14 |
50 |
- |
- |
- |
- |
| 37 |
GAGGCACAGGCGCCGAA |
14 |
143 |
60S ribosomal protein L13e |
Amblyomma americanum |
3e-85 |
TC12299 |
| 38 |
TCTGTGCGTGCCAAGGA |
14 |
116 |
60S ribosomal protein L10 |
Ixodes scapularis |
2e-48 |
TC8894 |
| 39 |
GTCAGCTGATGGGCAGA |
14 |
178 |
G nucleotide binding protein |
Dermacentor variabilis |
2e-167 |
TC6908 |
| 40 |
CAAATCTCTGCGTGGCA |
13 |
260 |
Translation initiation factor 2C |
Ixodes scapularis |
4e-27 |
TC6114 |
| 41 |
GCCTGCGTTTGCTGCAG |
13 |
143 |
Nucleolysin RNA binding protein |
Pediculus humanus corporis |
2e-122 |
TC12242 |
| 42 |
TTGCGGCTGCGCCGCAC |
13 |
260 |
Golgi protein involved in ER retention |
Ixodes scapularis |
2e-81 |
TC12211 |
| 43 |
GTTTGTGAGAGCGCCGC |
13 |
260 |
- |
- |
- |
- |
| 44 |
CCCGCGGTCATCACGGA |
13 |
143 |
- |
- |
- |
- |
| 45 |
AAGGCGCCAGCGGTGAT |
13 |
68 |
- |
- |
- |
- |
| 46 |
GCCGCACACTTTGACAG |
12 |
97 |
Ubiquitin/ribosomal protein S27A |
Dermacentor variabilis |
1e-61 |
TC10071 |
| 47 |
GGTTGGGCGCCGACGCG |
12 |
178 |
Ubiquitin protein ligase |
Ixodes scapularis |
1e-76 |
BEABI57TR |
| 48 |
GCGTTTGCTGGTGCCAG |
12 |
178 |
Maleate dehydrogenase |
Ixodes scapularis |
2e-164 |
TC9744 |
| 49 |
TGGTGGTAGCTGGTGCG |
12 |
14 |
- |
- |
- |
- |
| 50 | GTGGTGCCGTCGGCGCT | 12 | 260 | - | - | - | - |
a CATG trimmed from each tag’s 5′ end for clarity.
b Indicates tag not found in Infected library.
Table 7.
The 50 most abundant SAGE tags in the pooled tag data from Infected LongSAGE library
|
Rank |
Tag
a
|
Count |
Control |
BmiGI match |
|||
|---|---|---|---|---|---|---|---|
| Library rank | Description | Species | e -value | ID | |||
| 1 |
ATCTGAGTTTAGACCGA |
339 |
25 |
Mitochondrial DNA |
Rhipicephalus sanguineus |
0.00 |
TC5761 |
| 2 |
TGATTGTGTGCTATGTG |
142 |
783 |
- |
- |
- |
|
| 3 |
GGGGCAAACACTATGGA |
85 |
Nob |
- |
- |
- |
|
| 4 |
AAGATCACACTGGCATT |
57 |
783 |
- |
- |
- |
|
| 5 |
TTTTCCCCAACCCAGGA |
50 |
No |
Microplusin |
R. microplus |
7e-49 |
BEACP61TR |
| 6 |
CAGGCTGTCCCAGCAAT |
43 |
267 |
Secreted salivary gland peptide |
I. scapularis |
1e-05 |
BEAE009TR |
| 7 |
TTTTTCCCAACCCAGGA |
39 |
783 |
- |
- |
- |
|
| 8 |
GGTCAAGGGGTAATAAA |
37 |
No |
- |
- |
- |
|
| 9 |
GTGGTTACGGAGGCGGG |
36 |
No |
- |
- |
- |
|
| 10 |
GACGGCCCTTGCAAGTG |
33 |
128 |
- |
- |
- |
|
| 11 |
CAGAAGCTTCAAAGCCA |
33 |
783 |
- |
- |
- |
|
| 12 |
TCGACAGGGTCATTCCG |
31 |
No |
- |
- |
- |
|
| 13 |
GCCGTTCTTAGTTGGTG |
28 |
783 |
- |
- |
- |
|
| 14 |
TGGTGGTAGCTGGTGCG |
25 |
49 |
- |
- |
- |
|
| 15 |
GCGGTTACGGAAGCGGG |
25 |
No |
- |
- |
- |
|
| 16 |
CAGTTGTTGTTGCAGGG |
24 |
77 |
- |
- |
- |
|
| 17 |
AAGATCACGCTGGCATT |
24 |
No |
- |
- |
- |
|
| 18 |
GTAGCCGCCAGCCAAGG |
22 |
387 |
- |
- |
- |
|
| 19 |
TGGCTGGCTGCCCACTG |
22 |
3 |
Ribosomal protein P0 |
Haemaphysalis longicornis |
2e-36 |
TC9039 |
| 20 |
GTAGGCCCGGTATTGGT |
22 |
33 |
- |
- |
- |
|
| 21 |
CACATCATAGAACAGCT |
21 |
No |
- |
- |
- |
|
| 22 |
CTGTCCAATAAATGTCC |
21 |
195 |
H3 Histone - |
Canis familiaris |
1e-68 |
TC12182 |
| 23 |
GAAATAAATGCTGCCCT |
21 |
No |
- |
- |
- |
|
| 24 |
ACAAATAAAATTGAGCT |
21 |
No |
- |
- |
- |
|
| 25 |
CTTACTGCCCCAGCAAT |
20 |
387 |
Secreted salivary gland peptide |
I. scapularis |
9e-15 |
TC8005 |
| 26 |
GTGGGCTTCGGGGTCGC |
20 |
157 |
- |
- |
- |
|
| 27 |
GACTCCAATGAAGGCCC |
20 |
18 |
Alpha tubulin |
Mus musculus |
0.0 |
TC9399 |
| 28 |
TTGAGAGGTGGACAGGT |
19 |
53 |
- |
- |
- |
|
| 29 |
ACGCGACTGTCGCCACC |
18 |
6 |
Secreted protein |
I. scapularis |
3e-72 |
TC5798 |
| 30 |
ACGTGACTGTCGCCACC |
18 |
1 |
- |
- |
- |
|
| 31 |
GGTCCACCCCAGCGACT |
17 |
10 |
Ribosomal protein L10A |
I. scapularis |
8e-104 |
TC12372 |
| 32 |
TACTGTACCGAGGCCAG |
17 |
No |
- |
- |
- |
|
| 33 |
GTTGTTACGGGTAACGG |
17 |
No |
Secreted protein |
I. scapularis |
6e-20 |
BEAE880TF |
| 34 |
ATATTGACATTTCGTAG |
17 |
No |
Mitochondrial DNA |
R. sanguineus |
0.0 |
TC12054 |
| 35 |
GCGAGGAGCTTGTCGGG |
16 |
20 |
- |
- |
- |
|
| 36 |
AAAAAGGCTCAAGAAAT |
15 |
783 |
- |
- |
- |
|
| 37 |
GGACTCTGTAAGCACCG |
15 |
53 |
- |
- |
- |
|
| 38 |
CCGGTTCTTTCTTGGTG |
15 |
No |
- |
- |
- |
|
| 39 |
GGCGGAATAAAAGCGGT |
14 |
60 |
60S ribosomal protein L5 |
I. scapularis |
4e-146 |
TC8903 |
| 40 |
ATCTGAGTTTAAACCGA |
14 |
No |
- |
- |
- |
|
| 41 |
CCTCCAACGTACTCCGG |
14 |
783 |
Hypothetical protein |
I. scapularis |
8e-15 |
TC11473 |
| 42 |
GCACGGCGATGCGACGG |
14 |
16 |
- |
- |
- |
|
| 43 |
GGTCAGTCGGTCCTTAG |
13 |
783 |
10kD secreted protein |
A. monolakensis |
5e-28 |
TC12507 |
| 44 |
TAGGAATTTAAAAGTTG |
13 |
No |
Mitochondrial DNA |
R. sanguineus |
0.0 |
TC5761 |
| 45 |
CTCACCGACCCGTCGGC |
13 |
8 |
- |
- |
- |
|
| 46 |
CCGAAATAAGGCGAAAC |
13 |
No |
- |
- |
- |
|
| 47 |
ACCAGTTCAGGAGAGCC |
13 |
65 |
- |
- |
- |
|
| 48 |
CCCGGCCACAACCAGGA |
13 |
100 |
Hypothetical protein |
I. scapularis |
5e-68 |
TC10088 |
| 49 |
TACGAAGCGCTGGCAGA |
12 |
5 |
Disulfide isomerase |
Haemaphysalis longicornis |
4e-48 |
TC8716 |
| 50 | GGATTTGGTCTCTTTGA | 11 | 783 | 60S acidic ribosomal protein P1 | I. scapularis | 1e-29 | TC13709 |
a CATG trimmed from each tag’s 5′ end for clarity.
b Indicates tag not found in Control library.
Table 8.
LongSAGE tags with >10-fold differential expression
| Tag a |
Control |
Infected |
Difference |
BmiGI match |
|||
|---|---|---|---|---|---|---|---|
| Count b | Count b | (fold) | Description | Species | e-value | ||
| Upregulated with infection |
|
|
|
|
|
|
|
| TGATTGTGTGCTATGTG |
1 |
148 |
148 |
Noc |
|
|
|
| GGGGCAAACACTATGGA |
0 |
89 |
>89 |
No |
|
|
|
| AAGATCACACTGGCATT |
1 |
59 |
59 |
No |
|
|
|
| TTTTCCCCAACCCAGGA |
0 |
52 |
>52 |
BEACP61TR |
Microplusin |
R. microplus |
7e-49 |
| TTTTTCCCAACCCAGGA |
1 |
41 |
41 |
No |
|
|
|
| GGTCAAGGGGTAATAAA |
0 |
38 |
>38 |
No |
|
|
|
| GTGGTTACGGAGGCGGG |
0 |
38 |
>38 |
No |
|
|
|
| CAGAAGCTTCAAAGCCA |
1 |
35 |
35 |
No |
|
|
|
| TCGACAGGGTCATTCCG |
0 |
32 |
>32 |
No |
|
|
|
| GCCGTTCTTAGTTGGTG |
1 |
30 |
30 |
No |
|
|
|
| GCGGTTACGGAAGCGGG |
0 |
27 |
>27 |
No |
|
|
|
| AAGATCACGCTGGCATT |
0 |
25 |
>25 |
No |
|
|
|
| CACATCATAGAACAGCT |
0 |
23 |
>23 |
No |
|
|
|
| ACAAATAAAATTGAGCT |
0 |
23 |
>23 |
No |
|
|
|
| TACTGTACCGAGGCCAG |
0 |
18 |
>18 |
No |
|
|
|
| GTTGTTACGGGTAACGG |
0 |
18 |
>18 |
BEAE880TF |
Secreted protein |
I. scapularis |
6e-20 |
| ATATTGACATTTCGTAG |
0 |
18 |
>18 |
TC12054 |
Mitochondrial DNA |
R. sanguineus |
0.0 |
| CCGGTTCTTTCTTGGTG |
0 |
15 |
>15 |
No |
|
|
|
| AAAAAGGCTCAAGAAAT |
1 |
15 |
15 |
No |
|
|
|
| ATCTGAGTTTAGACCGA |
25 |
354 |
14.2 |
TC5761 |
Mitochondrial DNA |
R. sanguineus |
0.0 |
| GGTCAGTCGGTCCTTAG |
1 |
14 |
14 |
TC12507 |
10kD secreted protein |
A. monolakensis, |
5e-28 |
| GTAGCCGCCAGCCAAGG |
2 |
23 |
11.5 |
No |
|
|
|
| CAGGCTGTCCCAGCAAT |
4 |
45 |
11.2 |
BEAE009TR |
Secreted salivary peptide |
I. scapularis |
1e-05 |
| CTTACTGCCCCAGCAAT |
2 |
21 |
10.5 |
TC8005 |
Secreted salivary peptide |
I. scapularis |
9e-15 |
| Down-regulated with infection |
|
|
|
|
|
|
|
| TCTGGACGCGGCAAGGG |
48 |
1 |
-48.0 |
No |
|
|
|
| CACGTACAACCTCTGCG |
22 |
0 |
<-14.0 |
No |
|
|
|
| TTGTGCAGCGATCGGCA |
22 |
1 |
-22.0 |
No |
|
|
|
| GTACCAGAGGACAAGCC |
18 |
1 |
-18.0 |
No |
|
|
|
| TCGAACCCCCGGCAGTA |
14 |
0 |
<-14.0 |
No |
|
|
|
| TGGGGCACGTCCAAGCT |
14 |
1 |
-14.0 |
TC6102 |
Elongation factor beta |
Ornithodoros parkeri, |
1e-85 |
| GTGGTGCCATCGGCGCT |
14 |
1 |
-14.0 |
No |
|
|
|
| TCGAACCCCCGGCAGTA |
12 |
1 |
-12.0 |
TC14104 |
Ribosomal protein S18 |
Ornithodoros parkeri, |
1e-67 |
| GAAGAAGCCATCGGCCG |
12 |
1 |
-12.0 |
No |
|
|
|
| AACCCCGTCGAGCACCC |
12 |
1 |
-12.0 |
TC12306 |
Ribosomal protein L8 |
Glossina morsitans morsitans |
2e-116 |
| GGCCGCTACCCGGACCT |
12 |
1 |
-12.0 |
TC10648 |
Hypothetical protein |
I. scapularis |
1e-93 |
| TGGTGCCCGAAACGAAG |
68 |
6 |
-11.3 |
TC12322 |
Uro-adherence factor A |
Taloromyces stipitatus |
1e-05 |
| AAGAGCGTGTGCGGCTG | 36 | 3 | -11.0 | TC9377 | Hypothetical protein | I. scapularis | 1e-20 |
a CATG trimmed from each tag’s 5′ end for clarity.
b To allow comparisons, tag count data normalized to 8,750 total tag counts per library and individual counts adjusted accordingly. Thus, Control Library tags multiplied by 8750/6764 and Infected Library tags by 8750/8750.
c Indicates no match to BmiGI
The only host defense-related protein detected in the SAGE of the infected sample was microplusin, which exhibits antimicrobial activity during oogenesis in R. microplus[26]. A tag matching with the microplusin transcript was the fifth-most abundant tag in the B. bovis-infected library. However, a microplusin tag was not found in the non-infected library. The rest of the proteins occurring in the infected sample that were annotated had primarily house-keeping functions. Two notable proteins, calreticulin and superoxide dismutase, appeared in the SAGE uninfected library. Calreticulin was up-regulated in ovarian tissue from B. bovis-infected ticks in an earlier study [3]. Superoxide dismutase (SOD), a potent antioxidant, is often up-regulated when a cell is experiencing stress and plays a role in the virulence of pathogens [41]. A tag corresponding to SOD was not found in the SAGE infected ovary dataset, and perhaps the absence of this transcript assists in the successful B. bovis infection of the tick.
Overlap between the approaches
In the three approaches described in this study, the genes found to be up-regulated in response to B. bovis infection were compared with each other and with the up-regulated proteins found by Rachinsky et al.[3] under the same conditions (Figure 2, Table 9). No overlap was seen between genes detected in the SAGE ovary experiment and those of the microarray and Rachinsky et al.[3] proteome study. Only 4 of the 37 transcripts that were differentially expressed in the SAGE protocol were found in the subtracted library dataset. The microarray, subtracted library, and proteome studies compared transcript/protein profiles of infected and uninfected ovary tissues derived from similar conditions (i. e. incubation temperature and time point sampled). In contrast, the SAGE experiment was conducted with the control consisting of engorged females collected and dissected on the day they reached repletion and dropped from the host and the “infected” ovaries from ticks held 6 days post-repletion. Thus, some of the differential expression seen in the SAGE experiment could be strictly related to developmental events in the ovary during this preoviposition stage rather than related to B. bovis infection. Another possible confounding factor in the microarray, subtracted library, and the Rachinsky et al.[3] experiments was the tick infestations on the uninfected control and the B. bovis-infected bovine host took place one month apart due to space limitations in the quarantine facility and an accident that required the euthanization of the control calf. Additionally, the engorgement process of ticks feeding on the infected animal took longer than the engorgement of the ticks feeding on the uninfected control animal. Thus, we adjusted the dissection dates to ensure the females were in similar developmental stages for both groups. This difference in days post-drop before dissection could contribute to developmental differences unrelated to B. bovis infection and these might be reflected in the subtracted library and microarray datasets. It also must be noted that infection with Babesia alters the serum profile in the affected host as it struggles to cope with the infection [42]. These serum changes are likely causing altered transcription of genes in the tick feeding on Babesia-infected host blood. These changes induced by the altered host blood might be confounding the differential transcription study. Differentially expressed genes that we ascribe to B. bovis infection of the cattle tick might actually be responding to the altered serum components between the control and infected bovine. Seven transcripts were common to both the microarray and the subtracted library datasets. One of these seven, the Kunitz-type serpin represented by TC9311, was also described in the proteome study of Rachinsky et al.[3]. Five other transcripts in the subtracted library also overlapped with overexpressed proteins reported in the ovarian proteome study.
Figure 2.
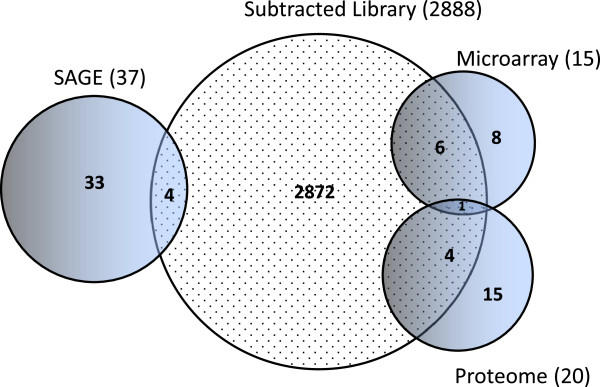
Overlaps between the differentially expressed transcript datasets. A total of 2,888, 37, 20, and 15 transcripts make up the datasets from the subtracted library, SAGE, Rachinsky et al.[3] proteome and the microarray experiments. The numbers in the intersecting regions of the circles represent the number of transcripts that are common between the represented datasets.
Table 9.
Differentially expressed members of BmiGI Version 2.0 in common among subtracted library, SAGE, microarray results and up-regulated proteins from Rachinsky et al. proteome study[3]
| Proteome study | Microarray | SAGE | Subtracted library | |
|---|---|---|---|---|
| Proteome |
- |
TC9311 Kunitz-type serpin |
None |
TC8919 Pep.-prolyl cis-trans isomerase |
| |
|
|
|
TC8931 Myosin light chain |
| |
|
|
|
TC8950 Calreticulin |
| |
|
|
|
TC9311 Kunitz-type serpin |
| |
|
|
|
TC12119 Cytochrome C oxidase |
| Array |
TC9311: Kunitz-type Serpin |
- |
None |
TC6326 Salivary gland peptide |
| |
|
|
|
TC6491 Kunitz-type protease inhibitor |
| |
|
|
|
TC6758 Protein IscW |
| |
|
|
|
TC9311 Kunitz-type serpin |
| |
|
|
|
TC12551 Salivary gland peptide |
| |
|
|
|
TC13077 Kunitz-type protease inhibitor |
| |
|
|
|
BEABQ71TR GGY domain protein |
| SAGE |
None |
None |
- |
TC6102 Elongation factor beta |
| |
|
|
|
TC9377 Hypothetical protein |
| |
|
|
|
BEACP61TR Microplusin |
| |
|
|
|
BEAE009TR Putative salivary protein |
| Library |
TC8919 Pep.-prolyl cis-trans isomerase |
TC6326 Salivary gland peptide |
TC6102 Elongation factor beta |
- |
| |
TC8931 Myosin light chain |
TC6491 Kunitz-type protease inhib. |
TC9377 Hypothetical protein |
|
| |
TC8950 Calreticulin |
TC6758 Protein IscW |
BEACP61TR Microplusin |
|
| |
TC9311 Kunitz-type serpin |
TC9311 Kunitz-type serpin |
BEAE009TR Putative salivary protein |
|
| |
TC12119 Cytochrome C oxidase |
TC12551 Salivary gland peptide |
|
|
| |
|
TC13077 Kunitz-type protease inhib. |
|
|
| BEABQ71TR GGY domain protein |
Conclusion
The complementary experimental approaches in this study produced several differential gene expression datasets associated with the infection of R. microplus by B. bovis. With the caveats discussed above in mind, transcripts that were detected as differentially expressed by more than one experimental protocol are priority targets for further study of the interactions at the vector-pathogen interface between R. microplus and B. bovis. A recent report [23] profiled gene expression in R. microplus ovarian tissues during feeding, reporting 417 up-regulated ovary-specific transcripts. Their results are not strictly comparable to ours, as our study was designed to detect infection-related differential transcription while [23] was designed to look at overall ovarian transcription during feeding. However, A and G family ABC transporter-, several zinc finger protein-, microcephalin-, cysteine rich secretory protein-, and serine protease-encoding transcripts, among others, were noted in our subtracted library study and that of Stutzer et al.[23]. A range of proteases and protease inhibitors were also noted as up-regulated in both studies. It would be interesting to design and conduct a study to discern signaling and regulatory mechanisms that might be occurring on these enzyme systems in the ovary of R. microplus. Stutzer et al.[23] profiled the adult female transcriptome of R. microplus, including that of the ovary, in response to feeding. The study reported here is the first comprehensive profile of the ovarian transcriptome responding to infection. Although a large percentage of tick genes remain without functional annotation, these newly identified gene expression patterns contribute to our understanding of the R. microplus transcriptome.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AH participated in the bioinformatic analysis of the subtracted library and drafted the manuscript. FDG conceived the study, participated in the design, data collection, and analysis of the study and participated in drafting the manuscript. KGB participated in the data collection, data analysis, and designed and conducted the RT-PCR verification study. LS, SED and CG participated in analysis of the microarray data; GAS participated in the overall study design and collection of tick materials. VN, SED and AD participated in study design and coordinated the sequencing phases. KAB participated in study design and microarray experimental design. All authors read and approved the final manuscript.
Supplementary Material
Unigene dataset from subtracted library sequencing. This Excel file contains assembled contig and singleton sequences from the subtracted library synthesized from the Babesia bovis-infected female tick ovaries (using uninfected tick ovary for the subtraction).
Relative quantitative real-time PCR primers and probes. This Word document contains the sequences of the primers and TaqMan probes used in real-time PCR verifications of the microarray results.
BLASTX annotations of subtracted library unigenes. This Excel file contains information on the BLASTX analysis of the Unigenes from Additional file 1, including definition line and e-values.
ORFs for all unigenes as predicted by prot4EST. Excel file contains the prot4EST predicted open reading frame for each unigene of Additional file 1.
Unigene GO annotations. Excel file containing GO terms for Unigenes from Additional file 1 that had corresponding GO classification terms.
Microarray values for ovary transcripts that were up-regulated in ticks feeding upon a B. bovis-infected calf. FC denotes fold change and d is the SAM statistic. This table in Excel format gives BLASTX definition line and e-value information for members of BmiGI Version 2 that were statistically significantly up- or down-regulated in the microarray experiment. The threshold value delta was set to 0.53 and fold change set to ≥ 2.0 to determine significance. No transcripts were found to be down-regulated under these conditions.
Overall statistics for SAGE experiment. Excel file containing raw counts for the SAGE tag libraries, including total number of tags, number of unique tags, and number of mutually occurring tags.
SAGE tag raw counts in ovary infected tissue and in ovary control uninfected tissue. Excel file lists each tag and its corresponding number of occurrences for both SAGE libraries.
SAGE tags with matches to sequences in BmiGI Version 2 in the forward direction. Excel file lists each SAGE tag that has an exact match to a member of BmiGI Version 2.
SAGE tags that match sequences in BmiGI Version 2 when tag is reverse complemented. Excel file lists each SAGE tag that, when reverse complemented, has an exact match to a member of BmiGI Version 2.
SAGE sequence tags that show higher normalized tag counts in infected samples compared to control samples. This Excel file contains a list of SAGE tags that show higher normalized tag counts in infected samples compared to control samples. This includes the tag sequence, the normalized tag count for both the control uninfected sample library and the Babesia bovis-infected sample library, and the calculated fold-change.
SAGE tags that show lower counts in infected sample compared to control uninfected sample. This Excel file contains a list of SAGE tags that show lower normalized tag counts in infected samples compared to control samples. This includes the tag sequence, the normalized tag count for both the control uninfected sample library and the Babesia bovis-infected sample library, and the calculated fold-change.
Contributor Information
Andrew M Heekin, Email: andyheekin@gmail.com.
Felix D Guerrero, Email: Felix.Guerrero@ars.usda.gov.
Kylie G Bendele, Email: kylie.bendele@ars.usda.gov.
Leo Saldivar, Email: leo.sal@gmail.com.
Glen A Scoles, Email: scoles@vetmed.wsu.edu.
Scot E Dowd, Email: sdowd@mrdnalab.com.
Cedric Gondro, Email: cgondro2@une.edu.au.
Vishvanath Nene, Email: v.nene@cgiar.org.
Appolinaire Djikeng, Email: a.djikeng@cgiar.org.
Kelly A Brayton, Email: kbrayton@vetmed.wsu.edu.
Acknowledgements
LS was supported by the National Research Initiative of the USDA CSREES grant #2005-35604-15440 (to KAB and FDG). This article reports the results of research only. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation of endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
References
- Grisi L, Massard CL, Moya Borja GE, Pereira JB. Impacto economico das principais ectoparasitoses em bovinos no Brasil. Hora Vet. 2002;6:8–10. [Google Scholar]
- Bock R, Jackson L, de Vos A, Jorgensen W. Babesiosis of cattle. Parasitology. 2004;6(Suppl):247–269. doi: 10.1017/s0031182004005190. [DOI] [PubMed] [Google Scholar]
- Rachinsky A, Guerrero FD, Scoles GA. Differential protein expression in ovaries of uninfected and Babesia-infected southern cattle ticks, Rhipicephalus (Boophilus) microplus. Insect Biochem Mol Biol. 2007;6:1291–1308. doi: 10.1016/j.ibmb.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Riek RF. The life cycle of Babesia argentina (Lignieres, 1903) (Sporozoa: Piroplasmidae) in the tick vector Boophilus microplus (Canestrini) Aust J Agric Res. 1966;6:247–254. doi: 10.1071/AR9660247. [DOI] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Heekin AM, Guerrero FD, Bendele KG, Saldivar L, Scoles GA, Gondro C, Nene V, Djikeng A, Brayton KA. Analysis of Babesia bovis infection-induced gene expression changes in larvae from the cattle tick, Rhipicephalus (Boophilus) microplus. Parasit Vectors. 2012;6:162. doi: 10.1186/1756-3305-5-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;6:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;6:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Wasmuth JD, Blaxter ML. prot4EST: Translating expressed sequence tags from neglected genomes. BMC Bioinforma. 2004;6:187. doi: 10.1186/1471-2105-5-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Eppig JT, Harris MA, Hill DP, Issel-Traver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;6:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid R, Blaxter ML. Annot8r: GO, EC and KEGG annotation of EST datasets. BMC Bioinforma. 2008;6:180. doi: 10.1186/1471-2105-9-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldivar L, Guerrero FD, Miller RJ, Bendele KG, Gondro C, Brayton KA. Microarray analysis of acaricide-inducible gene expression in the southern cattle tick, Rhipicephalus (Boophilus) microplus. Insect Mol Biol. 2008;6:597–606. doi: 10.1111/j.1365-2583.2008.00831.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing . Vienna: R Foundation for Statistical Computing; 2009. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang JH. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;6:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci. 2001;6:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B Stat Methodol. 2002;6:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- Pereira LS, Oliveira PL, Barja-Fidalgo C, Daffre S. Production of reactive oxygen species by hemocytes from the cattle tick Boophilus microplus. Exp Parasitol. 2001;6:66–72. doi: 10.1006/expr.2001.4657. [DOI] [PubMed] [Google Scholar]
- Rosa de Lima MF, Sanchez Ferreira CA, Joaquim de Freitas DR, Valenzuela JG, Masuda A. Cloning and partial characterization of a Boophilus microplus (Acari: Ixodidae) glutathione S-transferase. Insect Biochem Mol Biol. 2002;6:747–754. doi: 10.1016/S0965-1748(01)00157-6. [DOI] [PubMed] [Google Scholar]
- Rudenko N, Golovchenko M, Edwards MJ, Grubhoffer L. Differential expression of Ixodes ricinus tick genes induced by blood feeding or Borrelia burgdorferi infection. J Med Entomol. 2005;6:36–41. doi: 10.1603/0022-2585(2005)042[0036:DEOIRT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Yu QY, Lu C, Li WL, Xiang ZH, Zhang Z. Annotation and expression of carboxylesterases in the silkworm. Bombyx mori. BMC Genomics. 2009;6:553. doi: 10.1186/1471-2164-10-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero FD, Pruett JH, Li AY. Molecular and biochemical diagnosis of esterase-mediated pyrethroid resistance in a Mexican strain of Boophilus microplus (Acari: Ixodidae) Exp Appl Acarol. 2002;6:257–264. doi: 10.1023/a:1025319104529. [DOI] [PubMed] [Google Scholar]
- Sonenshine DE, Ceraul SM, Hynes WE, Macaluso KR, Azad AF. Expression of defensin-like peptides in tick hemolymph and midgut in response to challenge with Borrelia burgdorferi, Escherichia coli, and Bacillus subtilis. Exp Appl Acarol. 2002;6:127–134. doi: 10.1023/a:1025354326877. [DOI] [PubMed] [Google Scholar]
- Stutzer C, van Zyl WA, Olivier NA, Richards S, Maritz-Olivier C. Gene expression profiling of adult female tissues in feeding Rhipicephalus microplus cattle ticks. Int J Parasitol. 2013;6:541–554. doi: 10.1016/j.ijpara.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Manoury B, Hewitt EW, Morrice N, Dando PM, Barrett AJ, Watts C. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature. 1998;6:695–699. doi: 10.1038/25379. [DOI] [PubMed] [Google Scholar]
- Silva FD, Rezende CA, Rossi DC, Esteves E, Dyszy FH, Schreier S, Gueiros-Filho F, Campos CB, Pires JR, Daffre S. Structure and mode of action of microplusin, a copper II-chelating antimicrobial peptide from the cattle tick Rhipicephalus (Boophilus) microplus. J Biol Chem. 2009;6:34735–34746. doi: 10.1074/jbc.M109.016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves E, Fogaça AC, Maldonado R, Silva FD, Manso APP, Pelajo-Machado M, Valle D, Daffre S. Antimicrobial activity in the tick Rhipicephalus (Boophilus) microplus eggs: Cellular localization and temporal expression of microplusin during oogenesis and embryogenesis. Dev Comp Immunol. 2009;6:913–919. doi: 10.1016/j.dci.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Jaworski DC, Zou Z, Bowen J, Wasala NB, Madden R, Wang Y, Kocan KM, Jiang H, Dillwith JW. Pyrosequencing and characterization of immune response genes from the American dog tick, Dermacentor variabilis (L.) Insect Mol Biol. 2010;6:617–630. doi: 10.1111/j.1365-2583.2010.01037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedra JHF, Narasimhan S, Rendic D, DePonte K, Bell-Sakyi L, Wilson IBH, Fikrig E. Fucosylation enhances colonization of ticks by Anaplasma phagocytophilum. Cell Microbiol. 2010;6:1222–1234. doi: 10.1111/j.1462-5822.2010.01464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rego RO, Haidusek O, Kovar V, Kopacek P, Grubhoffer L, Hypsa V. Molecular cloning and comparative analysis of fibrinogen-related proteins from the soft tick Ornithodoros moubata and the hard tick Ixodes ricinus. Insect Biochem Mol Biol. 2005;6:991–1004. doi: 10.1016/j.ibmb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Park SH, Jiang R, Piao S, Zhang B, Kim E-H, Kwon H-M, Jin XL, Lee BL, Ha N-C. Structural and functional characterization of a highly specific serpin in the insect innate immunity. J Biol Chem. 2011;6:1567–1575. doi: 10.1074/jbc.M110.144006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA, Reichhart J-M. Drosophila innate immunity: an evolutionary perspective. Nature Immunol. 2002;6:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- Schwarz A, Valdés JJ, Kotsyfakis M. The role of cystatins in tick physiology and blood feeding. Ticks Tick Borne Dis. 2012;6:117–127. doi: 10.1016/j.ttbdis.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl PC, Sorgine MHF, Leall AT, Logullo C, Oliveira PL, da Silva Vaz I Jr, Masuda A. An extraovarian aspartic protease accumulated in tick oocytes with vitellin-degradation activity. Comp Biochem Physiol B Biochem Mol Biol. 2008;6:392–399. doi: 10.1016/j.cbpb.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Oldiges DP, Parizi LF, Zimmer KR, Lorenzini DM, Seixas A, Masuda A, de Silva Vaz I Jr. A Rhipicephalus (Boophilus) microplus cathepsin with dual peptidase and antimicrobial activity. Int J Parasitol. 2012;6:635–645. doi: 10.1016/j.ijpara.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Tsuji N, Miyoshi T, Battsetseg B, Matsuo T, Xuan X, Fujisaki K. A cysteine protease is critical for Babesia spp. transmission in haemaphysalis ticks. PLoS Pathog. 2008;6:e1000062. doi: 10.1371/journal.ppat.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seixas A, Oliveira P, Termignoni C, Logullo C, Masuda A, da Silva Vas I. , JrRhipicephalus (Boophilus) microplus embryo proteins as target for tick vaccine. Vet Immunol Immunopathol. pp. 149–156. [DOI] [PubMed]
- Skern-Mauritzen S, Frost P, Dalvin S, Kvamme BO, Sommerset I, Nilsen F. A trypsin-like protease with apparent dual function in early Lepeophtheirus salmonis (Krøyer) development. BMC Mol Biol. 2009;6:44. doi: 10.1186/1471-2199-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Tsuji N, Islama KM, Kamioa T, Fujisaki K. Cloning and molecular characterization of a cubilin-related serine proteinase from the hard tick Haemaphysalis longicornis. Insect Biochem Mol Biol. 2004;6:799–808. doi: 10.1016/j.ibmb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Lima CA, Torquato RJ, Sasaki SD, Justo GZ, Tanaka AS. Biochemical characterization of a Kunitz type inhibitor similar to dendrotoxins produced by Rhipicephalus (Boophilus) microplus (Acari: Ixodidae) hemocytes. Vet Parasitol. 2010;6:279–287. doi: 10.1016/j.vetpar.2009.09.030. [DOI] [PubMed] [Google Scholar]
- Francischetti IMB, Valenzuela JG, Andersen JF, Mather TN, Ribeiro JMC. Ixolaris, a novel recombinant tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick, Ixodes scapularis: identification of factor X and factor Xa as scaffolds for the inhibition of factor VIIa/tissue factor complex. Blood. 2002;6:2602–2612. doi: 10.1182/blood-2001-12-0237. [DOI] [PubMed] [Google Scholar]
- Champion OL, Cooper IAM, James SL, Ford D, Karlyshev A, Wren BW, Duffield M, Oyston PCF, Titball RW. Galleria mellonella as an alternative infection model for Yersinia pseudotuberculosis. Microbiology. 2009;6:1516–1522. doi: 10.1099/mic.0.026823-0. [DOI] [PubMed] [Google Scholar]
- Barrera R, Carapeto MV, Habela MA, Zaragoza C. Electrophoretic pattern of serum proteins in horses with babesiosis. Arch Med Vet. 2010;6:173–178. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Unigene dataset from subtracted library sequencing. This Excel file contains assembled contig and singleton sequences from the subtracted library synthesized from the Babesia bovis-infected female tick ovaries (using uninfected tick ovary for the subtraction).
Relative quantitative real-time PCR primers and probes. This Word document contains the sequences of the primers and TaqMan probes used in real-time PCR verifications of the microarray results.
BLASTX annotations of subtracted library unigenes. This Excel file contains information on the BLASTX analysis of the Unigenes from Additional file 1, including definition line and e-values.
ORFs for all unigenes as predicted by prot4EST. Excel file contains the prot4EST predicted open reading frame for each unigene of Additional file 1.
Unigene GO annotations. Excel file containing GO terms for Unigenes from Additional file 1 that had corresponding GO classification terms.
Microarray values for ovary transcripts that were up-regulated in ticks feeding upon a B. bovis-infected calf. FC denotes fold change and d is the SAM statistic. This table in Excel format gives BLASTX definition line and e-value information for members of BmiGI Version 2 that were statistically significantly up- or down-regulated in the microarray experiment. The threshold value delta was set to 0.53 and fold change set to ≥ 2.0 to determine significance. No transcripts were found to be down-regulated under these conditions.
Overall statistics for SAGE experiment. Excel file containing raw counts for the SAGE tag libraries, including total number of tags, number of unique tags, and number of mutually occurring tags.
SAGE tag raw counts in ovary infected tissue and in ovary control uninfected tissue. Excel file lists each tag and its corresponding number of occurrences for both SAGE libraries.
SAGE tags with matches to sequences in BmiGI Version 2 in the forward direction. Excel file lists each SAGE tag that has an exact match to a member of BmiGI Version 2.
SAGE tags that match sequences in BmiGI Version 2 when tag is reverse complemented. Excel file lists each SAGE tag that, when reverse complemented, has an exact match to a member of BmiGI Version 2.
SAGE sequence tags that show higher normalized tag counts in infected samples compared to control samples. This Excel file contains a list of SAGE tags that show higher normalized tag counts in infected samples compared to control samples. This includes the tag sequence, the normalized tag count for both the control uninfected sample library and the Babesia bovis-infected sample library, and the calculated fold-change.
SAGE tags that show lower counts in infected sample compared to control uninfected sample. This Excel file contains a list of SAGE tags that show lower normalized tag counts in infected samples compared to control samples. This includes the tag sequence, the normalized tag count for both the control uninfected sample library and the Babesia bovis-infected sample library, and the calculated fold-change.