Abstract
The mitotic spindle determines the cleavage furrow site during metazoan cell division1,2, but whether other mechanisms exist remains unknown. Here we identify a spindle-independent mechanism for cleavage furrow positioning in Drosophila neuroblasts. We show that early and late furrow proteins (Pavarotti, Anillin, and Myosin) are localized to the neuroblast basal cortex at anaphase onset by a Pins cortical polarity pathway, and can induce a basally-displaced furrow even in the complete absence of a mitotic spindle. Rotation or displacement of the spindle results in two furrows: an early polarity-induced basal furrow and a later spindle-induced furrow. This spindle-independent cleavage furrow mechanism may be relevant to other highly polarized mitotic cells, such as mammalian neural progenitors.
Elegant physical or genetic manipulations of the mitotic spindle have shown that the spindle determines the position of the cleavage furrow in a wide range of cells1,2. Although this is a common mechanism for furrow formation, it may not be the only one, as cleavage furrow position during the highly asymmetric mammalian meiotic divisions can be specified by a spindle-independent chromosomal cue3. The spindle pathway for furrow positioning is initiated at the overlapping microtubules of the central spindle, where the “centralspindlin” protein complex is assembled. Centralspindlin components include the kinesin Pavarotti (Zen-4 in C. elegans), the RACGAP50 Tumbleweed (Cyk-4 in C. elegans), and the RhoGEF Pebble (Ect-2 in C. elegans)1,4. After assembly, the centralspindlin complex moves to the cell cortex, possibly via a special population of stable microtubules5, to form a cortical ring at the site of the central spindle. The centralspindlin ring subsequently recruits actomyosin and initiates cleavage furrow constriction. In contrast, astral microtubules typically inhibit furrow formation4 (Figure 1a, left).
Figure 1. Polarized cortical localization of Pav/Myosin furrow markers.

(a) Summary of cortical Pav/Myosin (green) localization during a representative symmetric cell division (left) or a neuroblast asymmetric cell division (right). Central spindle microtubules, black; astral microtubules, gray.
(b) Basal cortical localization of endogenous Pav/Myosin proteins in mitotic neuroblasts.
(c,d) Localization of Pav:GFP and Sqh:GFP (Myosin) from movies 1–3. Overlay is shown below single channel image sequence Bottom rows show cortical pixel intensity plots for each protein around one half of the neuroblast cortex: from apical center (top) to basal center (bottom) of cortex. Apical up, basal down. Myo: Myosin, MTs: Microtubules. Scale bars: 10μm. Time in min:sec from anaphase onset.
Here we test whether the spindle-induced furrow model is sufficient to account for cleavage furrow positioning during asymmetric cell division of Drosophila neuroblasts. Neuroblasts establish molecular asymmetry during early prophase with the apical cortical localization of the Par complex (Bazooka; Par-6; atypical protein kinase C, aPKC) and the Pins complex (Partner of Inscuteable, Pins; Gαi; Discs large, Dlg)6. Subsequently, the scaffolding protein Miranda (Mira) and its cargo proteins Prospero (Pros), Brain tumor (Brat) and Staufen are localized to the basal cortex6. The mitotic spindle aligns along the apical/basal axis at metaphase and becomes asymmetric during anaphase, with the apical half forming longer astral and central spindle microtubules7,8. The cleavage furrow is displaced basally, generating a larger apical daughter cell and a smaller basal daughter cell. It has been assumed that the centralspindlin complex is the only mechanism for furrow positioning, because the furrow is always positioned adjacent to the central spindle, even in mutants that disrupt spindle asymmetry8–13. One model is that the basal spindle pole is anchored at the basal cortex, resulting in a basal displacement of the central spindle and subsequent cleavage furrow11 (Figure 1a, right). However, in neuroblasts, experiments such as spindle rotation, spindle displacement, or spindle ablation have never been performed to directly test whether the centralspindlin pathway is the sole mechanism for furrow positioning.
We began our investigation of neuroblast cleavage furrow positioning by assaying the timing and localization of three furrow components: the early furrow marker Pavarotti (Pav), an essential centralspindlin component4; Anillin, an early furrow component14; and Myosin regulatory light chain (called Myosin hereafter, encoded by the sqh gene), which is an essential component of the contractile ring. In symmetrically dividing cells, Pav/Anillin/Myosin are uniformly cortical at metaphase, and become progressively restricted to a cortical ring adjacent to the central spindle15 (Figure 1a, left). In neuroblasts, Pav/Anillin/Myosin proteins were uniform cortical at metaphase and enriched at the furrow during anaphase-telophase; in addition, we saw asymmetric localization of Pav/Anillin/Myosin to the basal cortex of the neuroblast during early anaphase (Figure 1b; Supplemental Figure 1; data not shown). The same localization was also observed by live imaging with Pav:GFP16, Anillin:GFP17, or Sqh:GFP18 (Myosin) reporter proteins (Figure 1c,d; Movies 1–3; summarized in Figure 1a, right). Pixel intensity measurements further revealed that the basal enrichment of Pav:GFP, Anillin:GFP and Sqh:GFP (Myosin) is not uniform; all markers clear from the apical cortex first, followed by partial depletion from the basal tip, prior to accumulation in a basally-shifted lateral position (Figure 1c, d; data not shown). Our data differ slightly from previous work showing apical Sqh:GFP localization in prophase neuroblasts19; our Sqh:GFP live imaging showed fluctuating weak apical or basal cortical localization during prophase (n=10; data not shown). Asymmetric basal enrichment of Pav/Myosin proteins was detectable 10–20s prior to astral microtubule asymmetry, and over 40s prior to central spindle asymmetry (Supplemental Figure 2). Pav/Anillin/Myosin asymmetric cortical localization precedes spindle asymmetry, and thus is not easily explained by a spindle-induced furrow positioning model.
We next tested the role of the mitotic spindle in generating Pav/Myosin basal cortical localization and basal furrow positioning. First, we tested whether spindle astral microtubules were required to generate Myosin cortical asymmetry. Sas-4 mutant neuroblasts lack centrioles, centrosomes, and all astral microtubules and were reported to undergo essentially normal asymmetric cell division10, as do other mutants that lack spindle pole asymmetry11,13,20,21. However, the localization of furrow proteins and the nature of the furrow positioning cue in these mutants has not been addressed. We found that Sas-4 mutant neuroblasts established normal basal cortical localization of Myosin and basal furrow formation (Figure 2a), and thus astral microtubules are not required for Myosin basal cortical localization or basal cleavage furrow positioning.
Figure 2. Spindle-independent cleavage furrow positioning.
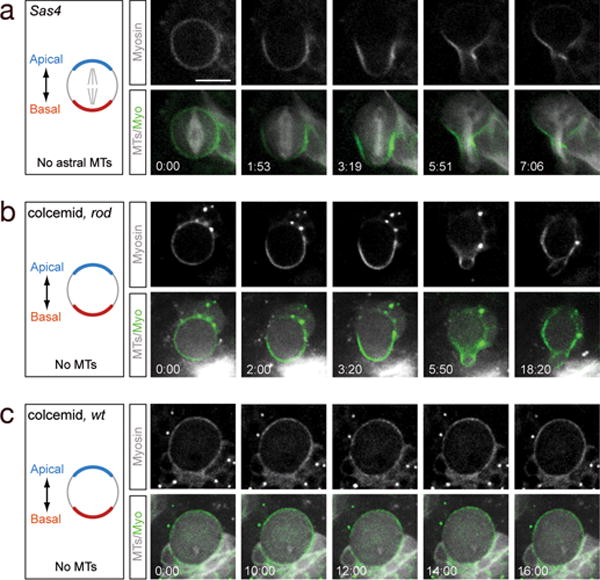
(a) Sas-4 mutant neuroblast lacks astral microtubules, yet still establishes basal Myosin localization and basal furrow position (100%, n=158).
(b) Colcemid-treated rod mutant neuroblast lacks all spindle microtubules, yet still establishes basal Myosin localization and basal furrow position (100%, n=7).
(c) Colcemid-treated wild type neuroblast lacks all spindle microtubules, remains arrested at metaphase, and does not establish basal Myosin localization or initiate furrow formation (100%, n=7).
A schematic of each experiment is shown to the left (apical/basal polarity, blue/red; microtubules, gray). All genotypes imaged in late second/early third larval instar brains.
Scale bars: 10μm. Time in min:sec.
We next tested whether central spindle microtubules were required to generate Myosin cortical asymmetry and basal furrow formation. We performed live imaging of neuroblasts in which all microtubules were ablated by colcemid treatment, and a mutation in rough deal (rod) was used to bypass the metaphase-arrest checkpoint22. Surprisingly, all colcemid-treated rod mutant neuroblasts showed robust basal localization of Myosin and generated a basally-displaced cleavage furrow, despite lack of any detectable microtubules (Figure 2b, Movie 4). Thus, complete loss of microtubules does not affect basal furrow positioning. This is not a non-specific effect of microtubule loss, because most wild type neuroblasts treated with colcemid are metaphase-arrested, maintain uniform cortical Myosin, and have no furrows (Figure 2c). Furthermore, rod single mutants localize Myosin in an asymmetric fashion like wild type neuroblasts (Supplemental Figure 3). We conclude that neuroblasts have a spindle-independent mechanism for basal cleavage furrow positioning, and that activating this mechanism requires anaphase onset. We call this the “polarity-induced” pathway because it is generated by neuroblast cortical polarity cues (see below).
Wild type neuroblasts may use both spindle-induced and polarity-induced furrow positioning pathways, or just one of these pathways. To test whether both pathways are active in neuroblasts, we rotated or displaced the mitotic spindle within the neuroblast, and assayed for the ability of each pathway to specify furrow position. We performed spindle displacement experiments by examining the minority of colcemid-treated rod mutant neuroblasts where one or more tiny spindles form near the apical cortex. In these neuroblasts, we observed normal asymmetric basal localization of Myosin and basal furrow formation; slightly later we detected a second furrow adjacent to the small apical mitotic spindle (Figure 3a, b; Movie 5). Next, we performed spindle rotation experiments using the mushroom body defective (mud) mutant. In mud mutants ~15% of the spindles are orthogonal to the normal apical/basal polarity axis23–25, thereby mimicking the physical spindle rotation experiments possible in larger cells1,2. We observed that mud mutant neuroblasts with the spindle orthogonal to the apical/basal polarity axis showed basal cortical localization of Pav/Anillin/Myosin and initiated a basal furrow (Figure 3c; Movies 6, 7 and data not shown) that often pinched off an anucleate basal “polar lobe” (Figure 3d, Supplemental Figure 4a; Movie 8). Interestingly, basal furrow initiation always preceded the spindle-induced furrow initiation (Figure 3e, Movie 9). Identical findings were observed in three other mutants that show neuroblast spindle rotation (asterless, centrosomin, and Sas-4; Supplemental Figure 4b). In both spindle displacement and spindle rotation experiments, the position of the mitotic spindle is uncoupled from the cortical polarity axis, and this allows us to observe cleavage furrows formed in response to each pathway. These experiments show that neuroblasts have two distinct furrow positioning pathways: a polarity-induced pathway and a spindle-induced pathway. In wild type neuroblasts, both pathways promote basal furrow positioning, but spindle rotation/displacement experiments allow us to spatially and temporally separate each pathway (discussed below).
Figure 3. Neuroblasts use both spindle-induced and polarity-induced furrow positioning pathways.

(a) Schematic of experimental design. Apical/basal polarity, blue/red; spindle, grey.
(b) Spindle displacement experiment: colcemid-treated neuroblast with tiny apical spindles form two spatiotemporally-distinct furrows. Early basal furrow (arrow); later spindle-associated furrow (arrowhead).
(c–f) Spindle rotation experiment: mud mutant neuroblasts with spindles orthogonal to the apical-basal polarity axis form two spatiotemporally-distinct furrows. (c) Neuroblast forms an early basal furrow (arrow), followed by an orthogonal spindle-associated furrow (arrowhead). (d) Neuroblast forms an early basal furrow that pinches off an anucleate “polar lobe” (arrow). Cherry:Miranda marks the basal cortex. (e) Still pictures from movie 9 showing the basal contractile ring “en face” to document the progressive constriction of the contractile ring. Yellow arrows, basal furrow; white arrows, orthogonal spindle-associated contractile ring. (f) Summary of spindle rotation experiment. Myosin, green; spindle, black; midbody remnant, green dot. Time scale m:ss. Scale bar: 10μm.
What is the molecular mechanism of the polarity-induced furrow pathway? We tested the centralspindlin core component Pav, as well as each of the three major cortical polarity protein complexes (apical Par/aPKC complex, basal Miranda complex, and apical Pins complex). We used inducible pav RNAi transgene to strongly reduce Pav protein levels specifically in neuroblasts; this resulted in phenotypes matching that of a pav null mutation: the neuroblasts were enlarged and polyploid due to failure of cytokinesis, and Pav protein was undetectable by antibody staining (data not shown). Surprisingly, these Pav-depleted neuroblasts showed normal basal localization of Myosin at early anaphase, and initiated a transient basal furrow (Figure 4a). We conclude that the canonical centralspindlin pathway is not required for basal furrow formation. The apical Par complex member aPKC is essential for proper localization of all known basal proteins6, but it is not required for basal localization of Myosin (Figure 4a). Similarly, the basal scaffolding protein Miranda is not required for Myosin basal localization (Figure 4a).
Figure 4. Mechanism of polarity-induced furrow formation.
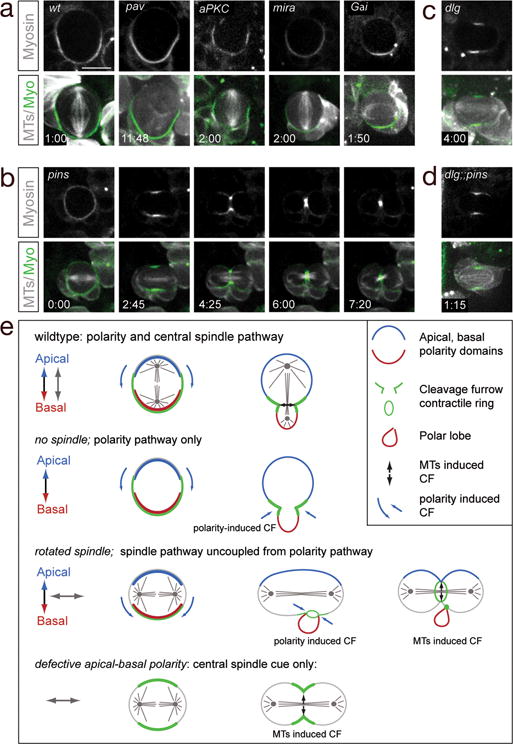
(a) Basal Myosin localization in anaphase neuroblasts is normal in neuroblasts strongly depleted for Pavarotti (Pav), aPKC, Miranda (Mira; MARCM clones) or zygotic null Gαi mutants. In all panels, time stamp: m:ss. Scale bar: 10μm.
(b) Zygotic pins single mutant larval neuroblast undergoing a symmetric division and showing symmetric Myosin (Sqh:GFP) localization.
(c) Zygotic dlg single mutant anaphase larval neuroblast undergoing a symmetric division and showing symmetric Myosin (Sqh:GFP) localization.
(d) Zygotic dlg pins double mutant anaphase neuroblast in early third larval instar undergoing a symmetric division and showing symmetric Myosin (Sqh:GFP) localization.
(e) Summary.
The final known polarity complex we tested was the apical Pins complex. We scored pins zygotic mutant neuroblasts at late second-third larval instar; the majority formed an asymmetric spindle and divided asymmetrically (89%; n=147), and thus could not be assayed for polarity-induced furrow positioning due to the presence of the canonical spindle-induced furrow pathway. More informative were the ~11% of pins mutant neuroblasts that had a symmetrical spindle and divided symmetrically; all of these neuroblasts lacked Pav/Myosin basal cortical enrichment, lacked basal furrows, and never formed “polar lobes” (100%, n=19, Figure 4b, Movie 10 and data not shown). To increase the percentage of symmetrically dividing pins mutant neuroblasts, we combined pins with a mutation in dlg, which is required for normal spindle asymmetry9. We found that 100% of the dlg pins double mutant neuroblasts showed symmetrical spindles, and they all lacked Myosin basal cortical enrichment and basally-displaced furrows (100%, n=20, Figure 4d). The lack of asymmetric Myosin localization in the pins and dlg pins mutant neuroblasts is due to the loss of Pins, not the symmetric spindle, because mud and Gαi mutant neuroblasts have symmetric spindles and still show basal Myosin localization and basal “polar lobe” formation (Figure 3c, 4a; Movie 11; data not shown). Thus, Pins is an essential component of the polarity-induced cleavage furrow pathway. To test if Dlg has a role in the polarity induced furrow pathway, we examined dlg single mutants. Only a small fraction had a symmetrical spindle (6%, n=65), and of these neuroblasts, two exhibited basally-enriched Myosin (data not show) and two showed symmetric cortical Myosin (Figure 4c). This partial phenotype suggests that Dlg plays a role in furrow positioning, but that Pins is likely to act through at least one other protein to regulate cleavage furrow position. We conclude that Pins/Dlg are components of the spindle-independent cortical polarity-induced cleavage positioning mechanism.
We have shown that neuroblasts use two pathways for specifying the site of cleavage furrow position: the well-studied centralspindlin pathway, and a new cortical polarity pathway. In neuroblasts these pathways appear to work partially redundantly: the polarity-induced pathway alone can give a basal furrow (e.g. in colcemid-treated rod mutant neuroblasts), whereas the spindle alone can induce an equatorial furrow (e.g. in dlg pins mutant neuroblasts) (summarized in Figure 4e). Although neuroblasts normally use both pathways redundantly, other cell types may uncouple the polarity-induced and spindle-induced pathways. For example, molluscan embryos often create determinant-filled “polar lobes” which form earlier and orthogonal to the spindle-induced furrow26. Mammalian embryonic neuroepithelial cells are highly elongated along their apical/basal axis and can initiate cleavage furrowing at their basal endfoot, far from the site of the apical mitotic spindle27. It will be interesting to see if a polarity-induced furrow pathway exists in mammalian neuroepithelial cells, as well as other polarized cell types.
Methods summary
We used these mutant alleles (aPKCK06403, mirazz178, pinsP89, dlgm52 mud4, gai8, cnnhk21, Sas-4M, asl2, rodH4.8); the UAS-PavRNAi line 46137 from VDRC; and the above-mentioned Gal4, UAS, and FRT stocks (see online methods for full stock references). Previously described methods were used for drug treatment28, live imaging29, and antibody staining29. Detailed methods are available in the supplemental material. All neuroblasts were imaged were from second or third larval instar central brains.
Full methods
Fly strains and genetics
All mutant chromosomes were balanced over Cyo, actin:GFP, TM3 actin:GFP, Ser, e or TM6B, Tb. We used Oregon R as wild type, and the following mutant chromosomes and fly strains:
aPKCK06403 (reference [1])
FRT82B mirazz178 (reference [2])
pinsP89 (flybase = rapsP89 ; reference [3])
dlgm52 (flybase = dlg14; reference [4])
mud4 (reference [5])
Gαi8 (reference [6])
cnnhk21 (reference [7])
FRT82B Sas-4M (reference [8])
asl2 (reference [9])
rodH4.8 (reference [10])
worGal4 (reference [11])
worGal4, UAS-Cherry:Jupiter (reference [12])
worGal4, UAS-Cherry:Mira (reference [12])
anillin:GFP (reference [13])
baz:GFP (reference [14])
baz:GFP, mud4 (reference [12])
pUAST-GFP:PavNLS5 (reference [15])
Sqh:Cherry (reference [16])
Sqh:GFP (reference [17])
worGal4, UAS-GFP:Mira, UAS-cherry:Jupiter (reference [12])
UAS-PavRNAi46137 (reference [18])
Recombinant chromosomes
The following recombinant chromosomes were generated using standard genetic procedures:
worGal4, UAS-Cherry:Jupiter, Sqh:GFP (this work)
worGal4, pUAST-GFP:PavNLS5 (this work)
MARCM analysis
For generating Mira MARCM clones [19], we crossed the analysis line hsFLP70/hsFLP70; worGal4, UAS-Cherry:Jupiter, Sqh:GFP, tubGal80 FRT82B/TM6C, Sb (this work) to Mira FRT82B mirazz178 and heat-shocked the progeny 24–48h after larval hatching for 1h at 37°C. mira mutant clones of third instar larvae were used for live imaging.
Pavarotti RNAi experiment
Pavarotti knock-down was achieved by crossing worGal4 (reference [11]) driver line to UAS-PavRNAi46137 [18]. Loss of Pavarotti was confirmed using the anti-Pav antibody [14].
Colcemid experiments
For colcemid experiments, the following strains were used +; worGal4, UAS-Cherry:Jupiter, Sqh:GFP (this work)) or +; worGal4, UAS-Cherry:Jupiter, Sqh:GFP; rodH4.8 (this work)). Wild type or rodH4.8 mutant neuroblasts were incubated with colcemid in live imaging medium [12] at a final concentration of 0.1um/ml. Live imaging was started without delay. Mild spindle phenotypes became apparent immediately after colcemid exposure, whereas complete spindle depolymerization was seen ~30–60min after colcemid addition.
Immunohistochemistry
The following antibodies were used for this study: guinea pig anti-Miranda (1:1000), rabbit anti-Zipper (1:500; this work), rabbit anti-Pavarotti (1:500)[15]. Mouse anti-Tubulin DM1A (Sigma, 1:1500), Rat anti-Pins (1:300)[20], Rabbit anti-Gαi (1:500)[21]. Secondary antibodies were from Invitrogen/Molecular Probes (Eugene, OR).
Imaging, post imaging procedures and measurements
Live imaging methods were previously described [12]. Fixed preparations were imaged on a Leica SP2, and for Supplemental Figure 1a on a Leica SP5 confocal microscope. Live samples were imaged on a McBain spinning disc confocal microscope equipped with a Hamamatsu EM-CCD camera, using a 63× 1.4NA oil-immersion objective. Pixel intensity measurements (Figure 1 c,d) were performed using ImageJ. Only one half of the neuroblasts cortex was measured starting at the apical cortex and ending at the basal cortex. Post-imaging processing and measurements were performed in ImageJ or Imaris 6.2–7.0 (Bitplane).
Larval staging
For all experiments, late second/third instar larvae were used for analysis.
Supplementary Material
Acknowledgments
We thank A. Brand, D. Glover, C-Y. Lee, M. Peifer and J. Raff, E. Wieschaus, A. Wilde the Bloomington and VDRC stock centers for fly stocks and/or antibody reagents; R. Andersen, B. Bowerman, M. Goulding, and B. Nolan for comments on the manuscript; and Taryn Gillies and Keiko Hirono for technical support. This work was supported by the NIH (GM068032; K.P.), the American Heart Association (C.C, K.P.), the Swiss National Science Foundation (C.C), and HHMI (C.Q.D.).
Footnotes
Author contribution
C.C., K.E.P. and C.Q.D conceived and designed the project. C.C performed all the experiments. C.C. and C.Q.D wrote the manuscript with input from K.E.P.
References
- 1.Oliferenko S, Chew TG, Balasubramanian MK. Positioning cytokinesis. Genes Dev. 2009;23(6):660. doi: 10.1101/gad.1772009. [DOI] [PubMed] [Google Scholar]
- 2.von Dassow G. Concurrent cues for cytokinetic furrow induction in animal cells. Trends Cell Biol. 2009;19(4):165. doi: 10.1016/j.tcb.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Deng M, Suraneni P, Schultz RM, Li R. The Ran GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Dev Cell. 2007;12(2):301. doi: 10.1016/j.devcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Somers WG, Saint R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev Cell. 2003;4(1):29. doi: 10.1016/s1534-5807(02)00402-1. [DOI] [PubMed] [Google Scholar]
- 5.Foe VE, von Dassow G. Stable and dynamic microtubules coordinately shape the myosin activation zone during cytokinetic furrow formation. J Cell Biol. 2008;183(3):457. doi: 10.1083/jcb.200807128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132(4):583. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Cai Y, et al. Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell. 2003;112(1):51. doi: 10.1016/s0092-8674(02)01170-4. [DOI] [PubMed] [Google Scholar]
- 8.Fuse N, Hisata K, Katzen AL, Matsuzaki F. Heterotrimeric G proteins regulate daughter cell size asymmetry in Drosophila neuroblast divisions. Curr Biol. 2003;13(11):947. doi: 10.1016/s0960-9822(03)00334-8. [DOI] [PubMed] [Google Scholar]
- 9.Albertson R, Doe CQ. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol. 2003;5(2):166. doi: 10.1038/ncb922. [DOI] [PubMed] [Google Scholar]
- 10.Basto R, et al. Flies without centrioles. Cell. 2006;125(7):1375. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 11.Giansanti MG, Gatti M, Bonaccorsi S. The role of centrosomes and astral microtubules during asymmetric division of Drosophila neuroblasts. Development. 2001;128(7):1137. doi: 10.1242/dev.128.7.1137. [DOI] [PubMed] [Google Scholar]
- 12.Izumi Y, et al. Differential functions of G protein and Baz-aPKC signaling pathways in Drosophila neuroblast asymmetric division. J Cell Biol. 2004;164(5):729. doi: 10.1083/jcb.200309162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11(2):116. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- 14.Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131(1):165. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hickson GR, Echard A, O’Farrell PH. Rhokinase controls cell shape changes during cytokinesis. Curr Biol. 2006;16(4):359. doi: 10.1016/j.cub.2005.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minestrini G, Harley AS, Glover DM. Localization of Pavarotti-KLP in living Drosophila embryos suggests roles in reorganizing the cortical cytoskeleton during the mitotic cycle. Mol Biol Cell. 2003;14(10):4028. doi: 10.1091/mbc.E03-04-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverman-Gavrila RV, Hales KG, Wilde A. Anillin-mediated targeting of peanut to pseudocleavage furrows is regulated by the GTPase Ran. Mol Biol Cell. 2008;19(9):3735. doi: 10.1091/mbc.E08-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Royou A, Sullivan W, Karess R. Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. J Cell Biol. 2002;158(1):127. doi: 10.1083/jcb.200203148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barros CS, Phelps CB, Brand AH. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev Cell. 2003;5(6):829. doi: 10.1016/s1534-5807(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 20.Giansanti MG, Bucciarelli E, Bonaccorsi S, Gatti M. Drosophila SPD-2 Is an Essential Centriole Component Required for PCM Recruitment and Astral-Microtubule Nucleation. Curr Biol. 2008;18(4):303. doi: 10.1016/j.cub.2008.01.058. [DOI] [PubMed] [Google Scholar]
- 21.Bonaccorsi S, Giansanti MG, Gatti M. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat Cell Biol. 2000;2(1):54. doi: 10.1038/71378. [DOI] [PubMed] [Google Scholar]
- 22.Basto R, Gomes R, Karess RE. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat Cell Biol. 2000;2(12):939. doi: 10.1038/35046592. [DOI] [PubMed] [Google Scholar]
- 23.Bowman SK, et al. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10(6):731. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Izumi Y, et al. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006 doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- 25.Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006 doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- 26.Conrad GW, Williams DC. Polar lobe formation and cytokinesis in fertilized eggs of Ilyanassa obsoleta. I. Ultrastructure and effects of cytochalasin B and colchicine. Dev Biol. 1974;36(2):363. doi: 10.1016/0012-1606(74)90058-x. [DOI] [PubMed] [Google Scholar]
- 27.Kosodo Y, et al. Cytokinesis of neuroepithelial cells can divide their basal process before anaphase. EMBO J. 2008;27(23):3151. doi: 10.1038/emboj.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegrist SE, Doe CQ. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development. 2006;133(3):529. doi: 10.1242/dev.02211. [DOI] [PubMed] [Google Scholar]
- 29.Cabernard C, Doe CQ. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev Cell. 2009;17(1):134. doi: 10.1016/j.devcel.2009.06.009. [DOI] [PubMed] [Google Scholar]
References
- 1.Rolls MM, et al. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol. 2003;163(5):1089–98. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat Genet. 2005;37(10):1125–9. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- 3.Yu F, et al. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100(4):399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
- 4.Woods DF, Bryant PJ. Molecular cloning of the lethal(1)discs large-1 oncogene of Drosophila. Dev Biol. 1989;134(1):222–35. doi: 10.1016/0012-1606(89)90092-4. [DOI] [PubMed] [Google Scholar]
- 5.Guan Z, et al. Mushroom body defect, a gene involved in the control of neuroblast proliferation in Drosophila, encodes a coiled-coil protein. Proc Natl Acad Sci U S A. 2000;97(14):8122–7. doi: 10.1073/pnas.97.14.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu F, et al. Distinct roles of Galphai and Gbeta13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J Cell Biol. 2003;162(4):623–33. doi: 10.1083/jcb.200303174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11(2):116–20. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- 8.Basto R, et al. Flies without centrioles. Cell. 2006;125(7):1375–86. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 9.Bonaccorsi S, Giansanti MG, Gatti M. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J Cell Biol. 1998;142(3):751–61. doi: 10.1083/jcb.142.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basto R, Gomes R, Karess RE. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat Cell Biol. 2000;2(12):939–43. doi: 10.1038/35046592. [DOI] [PubMed] [Google Scholar]
- 11.Albertson R, Doe CQ. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol. 2003;5(2):166–70. doi: 10.1038/ncb922. [DOI] [PubMed] [Google Scholar]
- 12.Cabernard C, Doe CQ. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev Cell. 2009;17(1):134–41. doi: 10.1016/j.devcel.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Silverman-Gavrila RV, Hales KG, Wilde A. Anillin-mediated targeting of peanut to pseudocleavage furrows is regulated by the GTPase Ran. Mol Biol Cell. 2008;19(9):3735–44. doi: 10.1091/mbc.E08-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buszczak M, et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175(3):1505–31. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minestrini G, Harley AS, Glover DM. Localization of Pavarotti-KLP in living Drosophila embryos suggests roles in reorganizing the cortical cytoskeleton during the mitotic cycle. Mol Biol Cell. 2003;14(10):4028–38. doi: 10.1091/mbc.E03-04-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin AC, Kaschube M, Wieschaus EF. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 2009;457(7228):495–9. doi: 10.1038/nature07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Royou A, Sullivan W, Karess R. Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. J Cell Biol. 2002;158(1):127–37. doi: 10.1083/jcb.200203148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–6. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 19.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–61. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 20.Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006 doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- 21.Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123(7):1323–35. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


