Abstract
The respiratory tract of cystic fibrosis (CF) patients harbor persistent microbial communities (CF airway microbiome) with Pseudomonas aeruginosa emerging as a dominant pathogen. Within a polymicrobial infection, interactions between co-habitant microbes can be important for pathogenesis, but even when considered, these interactions are not well understood. Here, we show with in vitro experiments that, compared with glucose, common fermentation products from co-habitant bacteria significantly increase virulence factor production, antimicrobial activity and biofilm formation of P. aeruginosa. The maximum stimulating effect was produced with the fermentation product 2,3-butanediol, which is a substrate for P. aeruginosa, resulting in a metabolic relationship between fermenters and this pathogen. The global transcription regulator LasI LasR, which controls quorum sensing, was upregulated threefold with 2,3-butanediol, resulting in higher phenazine and exotoxin concentrations and improved biofilm formation. This indicates that the success of P. aeruginosa in CF airway microbiomes could be governed by the location within the food web with fermenting bacteria. Our findings suggest that interbacterial metabolite transfer in polymicrobial infections stimulates virulence of P. aeruginosa and could have a considerable impact on disease progression.
Keywords: Pseudomonas aeruginosa; cystic fibrosis; 2,3-butanediol; fermentation products; phenazines
Introduction
Cystic fibrosis (CF) in Caucasians is caused by inherited mutations in the CF transmembrane conductance regulator, which leads to abnormal transport of ions across membranes, resulting in excessive water adsorption and viscous secretions from epithelial surfaces and ultimately to a thicker mucus layer. This secretion is one of the causes for a chronic polymicrobial infection of the CF airway that, in turn, triggers an immunological response of the host to cause irreversible damage and eventually pulmonary failure (Boucher, 2002; Lyczak et al., 2002; Ratjen and Döring, 2003). Throughout the disease progression in conjunction with antibiotic treatments, CF airway microbiomes become less rich in community members and a dominant Pseudomonas aeruginosa population emerges (Hunter et al., 2012). P. aeruginosa plays a central role in CF through many different and complex interactions with the host and the co-habitant microbes in the CF airway microbiome (Sibley et al., 2008a; Hunter et al., 2012).
Some of these interactions occur due to the important functions of redox-active small molecules – phenazines – that are produced by P. aeruginosa within the CF airway microbiome (Wilson et al., 1988). These phenazines consist of four different chemical species with phenazine-1-carboxylic acid (PCA; a precursor for pyocyanin) and pyocyanin (PYO) as important forms with different redox potentials, resulting in different redox-active functions. In general, phenazines have been observed to increase virulence by: (1) distressing airway epithelial cell function via oxidative stress (Wilson et al., 1988; Denning et al., 1998), (2) inhibiting growth of other microbes via oxidative stress (Hassett et al., 1992) and (3) playing physiologically important roles for P. aeruginosa itself (Hunter et al., 2012). For the latter, research has shown specifically for pure cultures of P. aeruginosa that: (i) PCA increases iron availability by reducing Fe(III) to Fe(II) even when bound to human-produced proteins (Wang et al., 2011), (ii) PCA reduces swarming motility and PYO increases biofilm thickness (Ramos et al., 2010), resulting in changes in biofilm formation, (iii) PYO regulates the intracellular redox state under anaerobic conditions by extracellular electron transfer, promoting survival through redox homeostasis and thicker biofilms (Dietrich et al., 2013) and (iv) PYO enables extracellular DNA binding to the surface of the cell to change aggregation characteristics, and thus biofilm formation (Das et al., 2013). Research had already indicated that reduced PCA and PYO were considerably more reactive with Fe(III) and O2, respectively, than vice versa (Wang and Newman, 2008). Importantly, the quorum-sensing cascade regulates phenazine production by P. aeruginosa (Whiteley et al., 1999), and thus has an effect on the redox activity of the produced phenazines (Venkataraman et al., 2010).
However, much less is known about how phenazines from P. aeruginosa affect microbe–host and microbe–microbe interactions within the CF airway microbiome. Hunter et al. (2012) have observed an inverse correlation between phenazine concentrations in sputum of CF patients and lung function during disease progression with similar maximum concentrations for PCA and PYO between 75–100 μM at the highest stage of disease severity. In addition, these authors also observed a strong inverse correlation between phenazine concentration in sputum and microbiome richness. Finally, they found an important paradoxical relationship because the P. aeruginosa isolates (in laboratory cultures) from the patient group with the most severe stage produced lower rates of PYO compared with the healthy group (Hunter et al., 2012). Could this paradoxical relationship be due to a change in the behavior of P. aeruginosa when grown in a pure culture vs in a mixed culture? It is already known that avirulent sputum isolates from CF patients when co-grown with P. aeruginosa enhanced the severity of lung damage compared with pure cultures of P. aeruginosa in rats and Drosophila (Duan et al., 2003; Sibley et al., 2008a). Sibley et al. (2008a) observed with a Drosophila model and a chronic P. aeruginosa infection that co-grown avirulent microbial genera such as Staphylococcus and Streptococcus can: (1) reduce fly survival and (2) upregulate several genes that encode virulence for P. aeruginosa, including quorum sensing, and thus phenazine production. However, a mechanistic understanding was absent. We found in a bioreactor study with a co-culture of Enterobacter aerogenes and P. aeruginosa that phenazine production was greatly enhanced compared with a pure culture of P. aeruginosa with selective production toward PYO rather than the other phenazines (Venkataraman et al., 2011). Our study also showed a positive microbe–microbe interaction through phenazines, which had not been studied in detail before. We observed a beneficial physiological role for the co-habitant bacterium when grown with P. aeruginosa, resulting in some extracellular respiration instead of exclusive fermentation.
Owing to the oxygen-limited conditions within the thick biofilms in CF airways (Hoffman et al., 2010; Guss et al., 2011), fermentation products, which are either from bacterial metabolism or host-generated via neutrophil metabolism, are found in the sputum of CF patients (Palmer et al., 2007; Bensel et al., 2010). Many bacteria in the CF airway microbiome, such as E. aerogenes, Klebsiella pneumoniae, Staphylococcus aureus, Serratia marcescens, Streptococcus sp. and Rothia sp. (Burns et al., 1998; Guss et al., 2011; Hunter et al., 2012; Twomey et al., 2012; Zhao et al., 2012), are either obligate or facultative fermenters. On the other hand, P. aeruginosa has three high-affinity terminal oxidases and has a preference to respire with oxygen and nitrate rather than to ferment (Madigan et al., 2002; Alvarez-Ortega and Harwood, 2007), albeit it can ferment with arginine (Vander Wauven et al., 1984). Even when oxygen is not present in the deeper layers of the biofilm and respiration becomes unlikely, P. aeruginosa may be able to survive by producing reduced PYO that eventually can react extracellularly with oxygen at the boundaries of the biofilm (Dietrich et al., 2013). Regardless, bacterial fermenters have a much higher substrate turnover rate than respirers because of inefficient carbon usage, allowing fermenters to predominate when fermentable substrates (including the blood sugar glucose or extracellular polysaccharides) are present (Madigan et al., 2002). Therefore, in the lungs of CF patients, a metabolic relationship may exist in which P. aeruginosa uses the fermentation products from fermenters as its energy and carbon source. Palmer et al. (2007), indeed, found that P. aeruginosa preferentially used lactate compared with glucose in a synthetic CF medium (Palmer et al., 2007).
Microbiomes represent the summation of multiple niches for which a niche is defined as the space occupied by one organism type through adaptation to the environmental conditions and other biological variables such as position in the food chain (Hutchinson, 1957). Microbe–microbe interactions are one of the major factors dictating the success of an organism in establishing and colonizing a niche. Recent culture-independent surveys of the CF airway microbiome have hinted at the role of community dynamics in disease progression (LiPuma, 2010; Hunter et al., 2012; VanDevanter and LiPuma, 2012; Zhao et al., 2012). An almost universally observed pattern is the reduction in bacterial richness, potentially driven by antibiotic therapy, and concomitant increase in the relative abundance of P. aeruginosa throughout the disease progression (Stressmann et al., 2012). Prior to the availability of potent antibiotic therapy, few CF patients survived to adulthood (VanDevanter and LiPuma, 2012). A large fraction of lung damage for CF patients is thought to occur during periods of pulmonary exacerbation, where acute infection and inflammation lead to short periods of dramatic respiratory symptoms that can often be treated effectively with antibiotic therapy. Diagnosis of exacerbations is conducted entirely through observation of worsening clinical symptoms, and treatment is not specifically tailored to the CF airway microbiome. Therefore, insight into microbe–microbe and microbe–host interactions promote the relative success of P. aeruginosa (that is, affect its niche) can lead to treatment strategies (Sibley et al., 2008a).
We hypothesized that co-habitant fermenters in the CF airway microbiome contribute to the success of P. aeruginosa through their fermentation products, including 2,3-butanediol, and thereby influence the role this microbe plays in causing lung damage in CF patients. We tested with in vitro experiments whether different fermentation products from possible co-habitant bacteria would stimulate P. aeruginosa to exhibit higher virulence and antimicrobial activity compared with glucose. Our experimental results with bioreactors agree with our hypothesis and again establish the relevance of including ecological considerations in clinical treatment of polymicrobial infections.
Material and methods
Strains and growth conditions
We used P. aeruginosa PA14 (PA14 transposon insertion mutant library (Liberati et al., 2006)), E. aerogenes strain NRRL B-115, K. pneumoniae ATCC 9590 (Dr Martin Wiedmann, Cornell University, NY, USA), S. aureus Newman (Dr Eric Skaar, Vanderbilt University, Nashville, TN, USA) and S. marcescens NRRL B-23389. Biofilm and virulence factor quantification was performed in minimal AB medium (Clark and Maaløe, 1967) supplemented with the appropriate carbon source (30 mM each) for each experiment. AB medium includes phosphate buffer (21 mM Na2HPO4 and 11 mM KH2PO4). Because of this buffer capacity the starting pH with different carbon substrates was similar, resulting in a pH of 7.0 for glucose and 6.9 for 2,3-butanediol. In an experiment with P. aeruginosa PA 14 with 30 mM glucose in AB medium and under micro-aerobic conditions, we observed that the buffer capacity had to be lowered considerably (<50% from the standard) to decrease the pH to <6.6 (Supplementary Figure S1). This indicates that the pH remained close to the starting pH of 6.9 even after several days of growth with pure cultures of P. aeruginosa PA14 in AB medium with a standard buffer capacity, which was used in the rest of the study. We purchased 2,3-butanediol and other chemicals from Sigma-Aldrich (St Louis, MO, USA). Motility experiments were performed in NB medium as previously described (Rashid and Kornberg, 2000) with varying agar concentrations and 30 mM carbon source at aerobic conditions. Dry cell weights were determined by weighing the cell pellet after overnight drying at 105 °C. OD600 was measured with a 96-well plate reader (BioTek, Winooski, VT, USA). Statistical tests were performed using SYSTAT 12 (Systat, Chicago, IL, USA).
Bioreactor operation and biofilm experiments
For all microaerobic experiments that included: (i) the comparison between five substrates (Figures 1a and b), (ii) biofilm measurements (Figures 1a and c), (iii) antimicrobial activity and quorum sensing experiments (Figures 1e–g), (iv) supernatant production for antimicrobial screening (Figure 2a) and (v) co-culture experiments with S. marcescens (Figures 2b and c), P. aeruginosa was grown in bioreactors consisting of glass vessels (liquid volume of 400 ml) with magnetic stir-bar mixing. These bioreactors were batch fed. The headspace of the vessel was connected to the laboratory atmosphere with two 0.22 μm gas filters to allow passive diffusion of oxygen. The temperature was maintained using a jacket with 35 °C water circulation. A suspended glass cover slip was utilized as the solid substratum for biofilm development experiments. For all aerobic experiments (Figures 1e–g) and the proteomic experiments (Figure 3), P. aeruginosa was grown in 500-ml conical flasks (liquid volume of 100 ml) with vigorous shaking in an incubator maintained at 35 °C. In both microaerobic and aerobic conditions, oxygen was the terminal electron acceptor. Gram staining was performed with a gram staining kit (Difco, Becton, Dickinson and Company, Franklin Lakes, NJ, USA). Biofilm protein was extracted from the cover slip in 0.1N NaOH at 96 °C for 40 min (Marsili et al., 2008). Protein quantification was performed using the BCA assay (Thermo Scientific, Waltham, MA, USA).
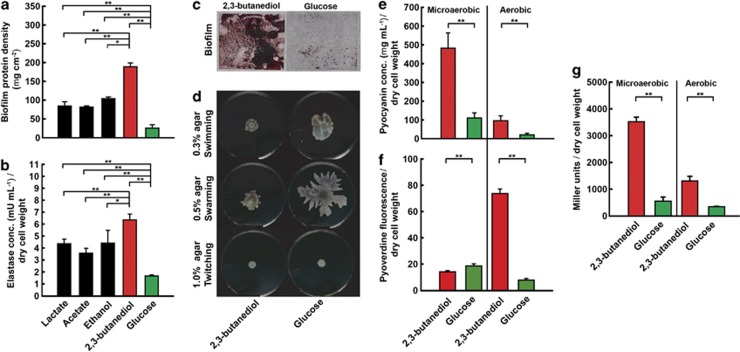
Figure 1.
Biofilm formation, virulence, motility, antimicrobial activity and quorum sensing for P. aeruginosa PA14 with fermentation products and glucose: (a) biofilm protein density (as mass per unit area); (b) elastase concentrations (as milliunits per ml normalized for dry cell weight; including LasA and LasB activities); (c) biofilm images on glass cover slips (view is 20 mm × 20 mm); (d) motilities for culture plates; (e) PYO concentration (as micrograms per milliliter normalized for dry cell weight); (f) pyoverdine fluorescence normalized for dry cell weight; and (g) concentration of 3O-C12-HSL (expressed as β-galactosidase activity of E. coli indicator strain in Miller units normalized for dry cell weight) in supernatant. Experiments for panels a and b were performed with four fermentation products and glucose, while experiments for panels c–g were performed with only 2,3-butanediol as the fermentation product and glucose as the control. Microaerobic conditions for all panels were juxtaposed with aerobic conditions for only panels e–g. Error bars indicate standard deviation among triplicates. A two-way ANOVA was performed (*P-value <0.05; **P-value <0.01).
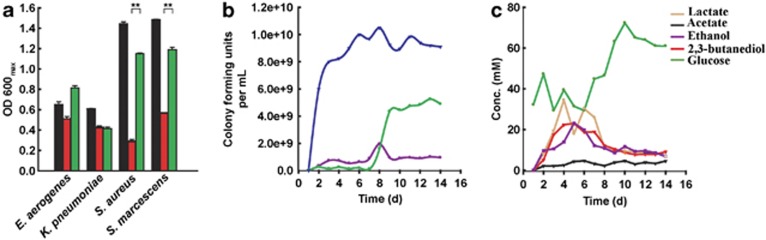
Figure 2.
Antimicrobial activity of P. aeruginosa grown with 2,3-butanediol and glucose: (a) maximum optical density at 600 nm attained by each microorganism in unsupplemented AB medium (black), in AB medium supplemented with cell-free supernatant of P. aeruginosa grown with 2,3-butanediol (red) and in AB medium supplemented with cell-free supernatant of P. aeruginosa grown with glucose (green), the initial glucose concentration for all three media conditions was adjusted to 30 mM; (b) tracking the populations of S. marcescens (purple) and P. aeruginosa (green) in a co-culture of a batch-fed bioreactor over the operating period (glucose was added each day), and the population of a pure culture of S. marcescens (blue) for similar operating conditions; and (c) metabolite profile with glucose as a substrate for the same co-culture experiment with S. marcescens and P. aeruginosa in b. For b and c: the result of one trial is shown; the biological duplicate is shown in Supplementary Figure S2. Error bars in a indicate standard deviation among triplicates. A two-way ANOVA was performed (*P-value <0.05; **P-value <0.01).
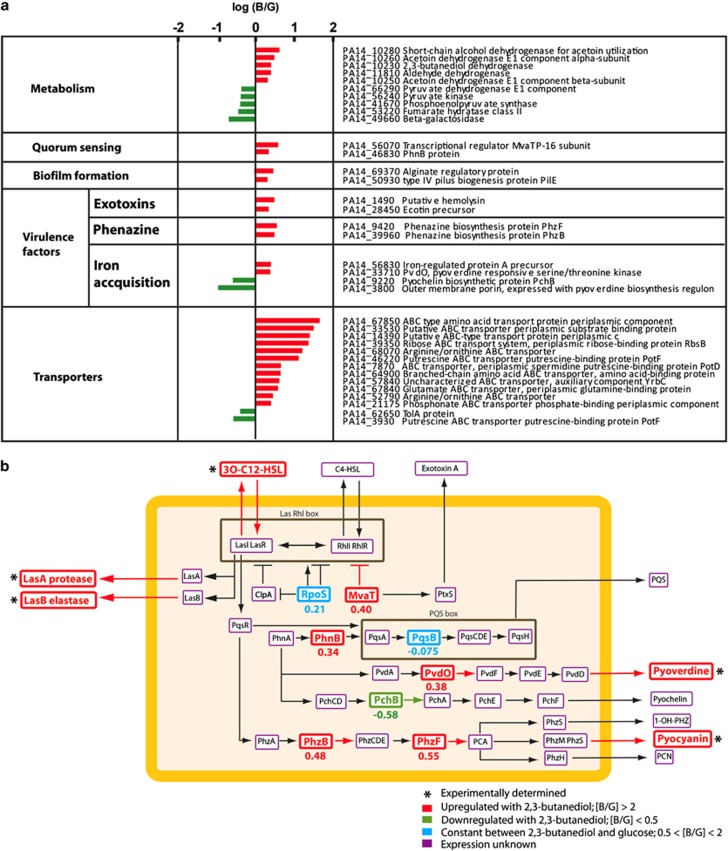
Figure 3.
Shift in proteome with 2,3-butanediol (three biological replicates) compared with glucose (three biological replicates): (a) protein abundance ratios between 2,3-butanediol and glucose for selected proteins with statistically significant differences (P <0.05) (ratios for all proteins are shown in Supplementary Table S2); and (b) simplified cellular network linking quorum sensing and virulence factors for P. aeruginosa PA14. The orange rectangle denotes the cell wall. Upregulated virulence factors (determined experimentally and marked with *) and protein expression (detected through proteomics; [B/G] >2) are shown in red. Downregulated protein expression (detected through proteomics; [B/G] <0.5) is shown in green. Unchanged protein expression (detected through proteomics; 0.5 <[B/G] <2) is shown in blue. Undetected proteins are framed in purple. The numbers below the proteins are the average log ratio of the protein abundances (2,3-butanediol:glucose [B/G]).
Virulence factor, autoinducer measurement, and proteomics
PYO was quantified via a LC/MS protocol adapted from the study by Dekimpe and Deziel (2009). Elastase measurement was performed via elastin congo red hydrolysis activity (Suh et al., 1999) with elastase from porcine pancreas (Sigma-Aldrich) as a reference. Pyoverdine was measured fluorometrically by excitation at 400 nm and emission at 460 nm (Suh et al., 1999). 3O-C12-HSL was measured with Escherichia coli indicator strain MG4 with plasmid pkdT17 (kindly provided by Dr Peter E. Greenberg, University of Washington, Seattle, WA, USA) according to Pearson et al. (1994). For comparative shotgun proteomics, samples were prepared and processed as previously described (Werner et al., 2009); tryptic peptides were labeled with isobaric tags (Thermo TMT, Thermo Scientific) for relative quantification, and ten peptide fractions (separated by high-pH RP HPLC) were analyzed using a LTQ Orbitrap Velos nLC-MS/MS.
Antimicrobial activity, co-culture experiments and metabolite analyses
To quantify the antimicrobial activity (Figure 2a), we grew P. aeruginosa in microaerobic bioreactors with 2,3-butanediol or glucose for 48 h. Next, we obtained cell-free supernatant by centrifuging at 10 000 g for 10 min. Glucose concentrations were measured using HPLC, and appropriate volumes of sterile stock glucose solutions were added to the supernatants such that the final concentration was 30 mM for all supernatants. On a 96-well plate, these supernatants were inoculated with the appropriate organism (E. aerogenes/K. pneumoniae/S. aureus/S. marcescens) (200 ul final volume), covered with a sterile film to prevent dehydration after which the OD600 was monitored at 35 °C every 60 min for 16 h in a plate reader (BioTek) with constant shaking. Controls (Fresh AB medium with glucose) were also included for each fermenter. To monitor co-culturing of S. marcescens and P. aeruginosa (Figures 2b and c), we operated bioreactors at microaerobic conditions. To prevent substrate depletion and pH-based inhibition of S. marcescens, we added glucose and adjusted the pH daily. Serial dilutions of daily bioreactor samples with the co-culture were plated on LB agar plates using an autoplater (Autoplate 4000, Spiral Biotech, Advanced Instruments, Norwood, MA, USA) and incubated for 18 h at 32 °C at aerobic conditions. We quantified the colonies of S. marcescens, which turned pink (prodigiosin production), and of P. aeruginosa, which turned green (PYO (blue) production with some PCA (yellow)). Metabolites were detected and quantified with a HPLC (Waters, Milford, MA, USA; Venkataraman et al., 2011).
Results and discussion
We evaluated virulence of P. aeruginosa cultures that were fed with the fermentation products lactate, acetate, ethanol and 2,3-butanediol, which are produced by common fermenters (Figure 1) (Burns et al., 1998; Ji et al., 2011). We quantified: (i) biofilm formation and (ii) elastase activity, which in P. aeruginosa includes the protease LasA and elastase LasB activities. Biofilm formation by P. aeruginosa is relevant to CF patient treatment because of the increased persistence and antibiotic resistance of the microbe in this state (Singh et al., 2002), while elastase causes pulmonary hemorrhages and inhibition of immune defense mechanisms (Kuang et al., 2011). We maintained microaerobic conditions by passive O2 diffusion into bioreactors (dissolved oxygen <9 μM), mimicking the oxygen-limited state of the lung biofilm of CF patients that is observed for the CF airway through the presence of: (1) both strict aerobic and strict anaerobic bacteria (Sibley et al., 2011) and (2) both PCA and PYO (Hunter et al., 2012), because oxygen is required to convert the precursor PCA into PYO (Parsons et al., 2007). We found that both biofilm protein density and elastase concentrations were considerably higher with fermentation products compared with glucose (Figures 1a and b). For both biofilm formation and elastase production, 2,3-butanediol had the maximum stimulating effect. Specifically, P. aeruginosa biofilm protein density was 7.4-fold higher with 2,3-butanediol-grown cells than with glucose-grown cells (Figure 1a; 188.69±10.35 μg cm−2 vs 25.57±8.92 μg cm−2).
Next, in batch-fed bioreactors, we tested the antimicrobial activity of P. aeruginosa cultures grown with 2,3-butanediol or glucose toward possible co-habitant fermenters. Antimicrobial activity is important because it may result in the dominance of P. aeruginosa in a microbial community (Hunter et al., 2012), and consequently accentuates the virulence of this microbe. First, we obtained growth curves with four fermenters that are capable of 2,3-butanediol production – E. aerogenes, K. pneumoniae, S. aureus and S. marcescens in: (i) unsupplemented AB medium, (ii) AB medium supplemented with the cell-free supernatant of P. aeruginosa grown with 2,3-butanediol and (iii) AB medium supplemented with the cell-free supernatant of P. aeruginosa grown with glucose. Microaerobic, pure culture studies of S. aureus and S. marcescens showed that cell-free supernatant from a P. aeruginosa PA14 culture grown with 2,3-butanediol was considerably more inhibitory to these fermenters than supernatant from P. aeruginosa grown on glucose (Figure 2a). However, there was no significant inhibition of E. aerogenes and K. pneumoniae and it is known that, for example, E. aerogenes can co-exist with P. aeruginosa (Venkataraman et al., 2011), whereas S. aureus showed growth retardation (Pal'chykovs'ka et al., 2008).
Because 2,3-butanediol had the most pronounced effect on the virulence of P. aeruginosa (Figures 1a and b) and because 2,3-butanedione, which is a volatile precursor of 2,3-butanediol, has specifically been detected in airways of CF patients (Whiteson et al., 2014), we studied the interaction of this fermentation product with P. aeruginosa in more detail. Finally, 2,3-butanediol is pH neutral, which is advantageous for microbiota, because it prevents lethal acidification unlike other fermentation products such as acetic acid and lactic acid. Biofilm density increased after growth with 2,3-butanediol compared with glucose (Figure 1c). Since decreased motility positively affects biofilm formation by encouraging cell clustering and aggregation in P. aeruginosa (Singh et al., 2002; Ramos et al., 2010), we tested swimming motility (bacterial locomotion in liquids), swarming motility (biosurfactant induced locomotion), and twitching motility (surface locomotion by type IV pili). We observed a notable reduction in swimming and swarming motility with 2,3-butanediol compared with glucose (Figure 1d), which is likely involved in the increased biofilm formation with 2,3-butanediol. However, we did not detect a difference in twitching motility (Figure 1d).
We further quantified changes in antimicrobial activity of P. aeruginosa grown with 2,3-butanediol by assaying the concentrations of PYO to ascertain oxidative stress to microbes (Hassett et al., 1992), and pyoverdine, which is a siderophore essential for P. aeruginosa in iron-limiting situations, to ascertain iron sequestration from other bacteria (Kim et al., 2005). Oxygen concentrations influence the production of these antimicrobials by P. aeruginosa through quorum sensing (Schuster and Greenberg, 2008), and therefore we maintained two oxygen tensions: (i) microaerobic by passive oxygen diffusion and (ii) aerobic by vigorously shaken cultures. Similar to our earlier observation (Venkataraman et al., 2011), the PYO concentrations in P. aeruginosa cultures were considerably higher with 2,3-butanediol compared with glucose for both microaerobic and aerobic conditions (Figure 1e). The outcome was more nuanced for pyoverdine, which had lower concentrations with 2,3-butanediol for microaerobic conditions, but higher concentrations for aerobic conditions compared with glucose (Figure 1f). This is likely because iron was only limiting to the aerobically grown cells, which had a much higher dry cell weight than the microaerobic cells (Supplementary Table S1). Thus, the fermentation product 2,3-butanediol may increase the antimicrobials PYO and pyoverdine for conditions in which P. aeruginosa could have a competitive advantage.
In addition to being an antimicrobial, PYO is also utilized by P. aeruginosa as an electron mediator (that is, a temporary electron acceptor or electron shuttle) for extracellular electron transfer with oxygen as the terminal electron acceptor at the periphery of the biofilm (Wang and Newman, 2008). The advantage for P. aeruginosa is to maintain redox homeostasis, and thus to survive, under anaerobic conditions within the deeper layers of the biofilm when PYO is present (Dietrich et al., 2013). This extracellular electron transfer with oxygen over a distance can be measured quantitatively and in real time by electric current production through the oxidation of reduced PYO at an electrode as the solid-state electron acceptor in a bioelectrochemical system (Rabaey et al., 2005; Venkataraman et al., 2011). In our earlier work with bioelectrochemical systems under microaerobic conditions (Venkataraman et al., 2011), we observed that the electric current doubled for P. aeruginosa with 2,3-butanediol vs glucose due to an increase in total phenazine concentration and a shift to its production of mainly PYO. In addition, in the presence of oxidized PYO, E. aerogenes (the producer of 2,3-butanediol with glucose as a substrate) respired at least partially with the electrode, resulting in enhancement of its growth. When growing a binary culture of E. aerogenes and P. aeruginosa in a bioelectrochemical system after glucose addition as the only substrate, we observed a 14-fold increase in electric current production compared with the individual strains under similar conditions (Venkataraman et al., 2011). We found that glucose was quickly converted to 2,3-butanediol by E. aerogenes' fermentation, and that it became the substrate for P. aeruginosa, resulting in a high concentration of reduced PYO that was subsequently converted into oxidized PYO by the electrode. This resulted in the ability for E. aerogenes to become a respirer with oxidized PYO even though oxygen supply was limited (Venkataraman et al., 2011).
Here, we did not utilize an electrode in our bioreactors and of course no electrode is present in vivo. Regardless, in the lungs of CF patients both P. aeruginosa and fermenters, such as E. aerogenes, could have a fitness benefit via PYO trough extracellular electron transfer with oxygen. Other bacteria within the human host have already shown a beneficial interaction with electron mediators under anaerobic conditions. For example, Faecalibacterium prausnitzii interacted with another electron mediator – riboflavin – in an experiment that mimicked the human gut environment (Khan et al., 2012) and Salmonella typhimurium has been shown to outcompete other microbes because of its ability to use ethanolamine as a reduced substrate for respiration with tetrathionate as the electron mediator (Thiennimitr et al., 2011). Considering this advantage, it is not surprising that we observed considerably higher PYO production for microaerobic vs aerobic conditions with P. aeruginosa because with high oxygen concentrations no mediators are needed for survival (Figure 1e). Thus, both the antimicrobial and electron mediator effects of this compound may improve the fitness of P. aeruginosa in the microbial communities of CF patients.
Next, we tested whether quorum sensing, which controls virulence, antimicrobial activity, and biofilm formation for P. aeruginosa (Whiteley et al., 1999; Wagner et al., 2003), was upregulated with 2,3-butanediol compared with glucose. Quorum sensing in P. aeruginosa is mediated by diffusible acylhomoserine lactones that are produced by LasI, which synthesizes 3-oxo-dodecadonyl homoserine lactone (3O-C12-HSL), and RhlI, which synthesizes n-butyryl homoserine lactone (C4-HSL). We quantified 3O-C12-HSL via a bioassay (Pearson et al., 1994) and observed that with 2,3-butanediol, P. aeruginosa produced nearly threefold higher concentrations for both microaerobic and aerobic conditions (Figure 1g). Quorum sensing is an integral part of P. aeruginosa physiology in CF patients (Singh et al., 2000) and our findings show that 2,3-butanediol affects quorum sensing signaling, thereby contributing to the success of P. aeruginosa in polymicrobial infections. We realize that for all these results on the effect of 2,3-butanediol on biofilm formation, host virulence, antimicrobial activity and quorum sensing (Figures 1a–c and e–g), further verification with in vivo experiments will be pertinent.
We conducted a comparative proteome analysis with aerobic, planktonic pure cultures of P. aeruginosa grown with 2,3-butanediol or glucose (each had three biological replicates). Cells were harvested for protein extraction in the late log phase. We did not perform a biofilm experiment and/or used microaerobic conditions to prevent stratification of cells, and therefore to ensure that all bacterial cells were universally grown under similar conditions. We used aerobic conditions since we had observed that microaerobic and aerobic conditions resulted in similar functions that were important for this study (Figures 1e and g). We detected 268 significant protein abundance differences (out of a total of 509 detected proteins, two-way ANOVA, n=3, P-value<0.05) of which 109 were upregulated with 2,3-butanediol (average protein abundance ratio, 2,3-butanediol [B]:glucose [G] [B/G] >2 (or >100.30 for the logarithmic scale in Figure 3)) and 102 were downregulated ([B/G] <0.5). We classified these detected proteins into functional categories (Figure 3a; Supplementary Table S2).
We found that most of the quantified proteins for quorum sensing, biofilm formation and virulence factors (that is, both host virulence and antimicrobial activity) were upregulated for 2,3-butanediol compared with glucose (10 out of 12 in Figure 3a). In accordance with our quorum sensing bioassay (Figure 1g), we observed PhnB at nearly 2.2-fold greater abundance (Figures 3a and b). PhnB is involved in the production of Pseudomonas quinolone signal, which is a part of the quorum sensing cascade (McKnight et al., 2000). Two additional quorum-sensing regulatory proteins were not significantly different (RpoS and PqsB; Figure 3b), but as regulatory proteins their lack of induction may not be surprising. Expression for MvaT was higher with 2,3-butanediol (Figures 3a and b), and this protein regulates LasI LasR and RhlI RhlR during stationary growth phase (Diggle et al., 2002). It also influences Exotoxin A (Figure 3b); two exotoxin-related proteins were upregulated with 2,3-butanediol (Figure 3a). Our comparative proteome analysis verified and expanded on the link between quorum sensing and 2,3-butanediol metabolism for P. aeruginosa (2,3-butanediol dehydrogenase was upregulated; Figure 3a). It remains unclear, however, how this metabolic activity specifically connects to the quorum sensing signaling cascade.
In regards to biofilm formation, PilE and an alginate regulatory protein AlgP – alginate overproduction is characteristic of P. aeruginosa in CF (Nivens et al., 2001) – were upregulated 2.0 and 3.1-fold with 2,3-butanediol, respectively (Figure 3a). For the virulence factors, the higher measured PYO concentration with 2,3-butanediol (Figure 1e) corresponded with higher ratios of PhzB and PhzF (Figures 3a and b), which are two proteins in the phenazine synthesis pathway with PYO as one of the final possible products (Figure 3b). The outcome was more nuanced for iron acquisition with upregulated PvdO (Figure 3a), verifying the measured higher pyoverdine concentrations for aerobic conditions (Figure 1f), but with lower PchB abundance (Figures 3a and b). We also found a significant increase in ABC transporter proteins with 2,3-butanediol (12 out of 14 proteins in Figure 3a). In summary, our comparative proteome analysis validates the link between 2,3-butanediol metabolism and biofilm formation/virulence factors through quorum sensing regulation.
Finally, we verified that fermenters would first convert glucose into fermentation products, such as 2,3-butanediol, which P. aeruginosa then uses as its substrate. S. marcescens (unlike E. aerogenes, K. pneumoniae and S. aureus) produces all four fermentation products – lactate, acetate, ethanol and 2,3-butanediol – from glucose, and was inhibited by the antimicrobial activity of P. aeruginosa (Figure 2a). Therefore, we tracked the population of S. marcescens in co-culture with P. aeruginosa over the operating period (Figure 2b and Supplementary Figure S1A). We found that during days 1–6 of the bioreactor operation, S. marcescens dominated the co-culture, and glucose was fermented to lactate, acetate, ethanol and 2,3-butanediol (Figures 2b and c and Supplementary Figure S1A and B). Subsequently, P. aeruginosa consumed these fermentation products, and became the dominant organism in the bioreactor during days 8–14 of the experiment. After day 8, S. marcescens was inhibited (the colony-forming units of S. marcescens decreased by 40.6%) by the antimicrobial activity of P. aeruginosa despite the presence of sufficient concentrations of glucose (at least 30 mM). A control experiment with a pure culture of S. marcescens indicated inhibition in the co-culture from the start of the operating period (Figure 2b and Supplementary Figure S1A). Even though inhibition occurred in a microaerobic co-culture with P. aeruginosa PA14, S. marcescens initially dominated by fermenting glucose to products that subsequently became substrates for P. aeruginosa. Regardless, P. aeruginosa displayed antimicrobial activity and emerged as the final dominant species (Figure 2b; Supplementary Figure S1A).
Culture-independent techniques have recently correlated a decrease in bacterial diversity with the relative abundance of P. aeruginosa, duration of antibiotic therapy, and disease severity in CF patients (Stressmann et al., 2012). One question that has been raised is whether increasing the microbial diversity would ‘remediate' CF microbiota (VanDevanter and LiPuma, 2012). But this in turn raises more questions, including which bacterial species should increase the diversity to specifically challenge P. aeruginosa? In addition, measures of taxonomic diversity do not specifically account for the encoded functions and metabolic potential of a microbial community. Therefore, a link between diversity loss and pulmonary exacerbation may not be meaningful for disease progression. Our finding that interbacterial interactions help P. aeruginosa to create and occupy a dominant niche in the CF habitat by stimulating virulence and antimicrobial activity can guide efforts for strategic management of the lung microbiota in CF patients. In fact, efforts to prevent P. aeruginosa virulence by targeting other members of the CF polymicrobial community, specifically members of the Streptococcus milleri group, have been successful (Sibley et al., 2008b). The authors of the study did not understand, however, by which mechanisms Streptococcus spp. triggered the virulence of P. aeruginosa.
Our results indicate that solving pulmonary exacerbation by enhancing fermenters to increase the diversity of the microbial community would, in fact, contribute to the virulence of P. aeruginosa, causing damage to both the human host and co-habitant microbes, rather than relieving this. On the other hand, blocking access to fermentation products could be a viable strategy for altering the niche in which P. aeruginosa now succeeds in the lung of CF patients. Elucidation of chemical signaling between microbes that trigger pulmonary exacerbations may enable early diagnosis before severe inflammatory damage occurs. Subsequent blocking of these signals could reduce virulence of P. aeruginosa, avoiding pulmonary exacerbations and possibly slowing disease progression. In summary, our results point toward indirectly targeting the niche of P. aeruginosa by reducing access to virulence-provoking fermentations products. However, further studies are now necessary to verify this with model systems.
Interbacterial metabolite transfer fueling pathogenesis of polymicrobial infections is an important clinical consideration. Our data suggest that for infections involving cohabitation of fermenters and P. aeruginosa, fermenters alter P. aeruginosa physiology by producing compounds that enhance its virulence. Therefore, the presence of fermentation products, such as 2,3-butanediol or 2,3-butanedione in a polymicrobial infection can be a warning sign for virulence of P. aeruginosa (Whiteson et al., 2014). Finally, our work suggests that P. aeruginosa can utilize fermentation products as a substrate, leading to increased expression of virulence factors. Further insight into the mechanism of this food-web interaction may lead to more sophisticated diagnosis and treatment for patients with CF and other polymicrobial infections involving P. aeruginosa.
Acknowledgments
This research was supported through NSF CAREER grant #0939883 to LTA. We thank Dr Katrine Whiteson (San Diego State University) for helpful comments.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alvarez-Ortega C, Harwood CS. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol. 2007;65:153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensel T, Stotz M, Borneff-Lipp M, Wollschläger B, Wienke A, Taccetti G, et al. Lactate in cystic fibrosis sputum. J Cyst Fibros. 2010;10:37–44. doi: 10.1016/j.jcf.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Boucher RC. An overview of the pathogenesis of cystic fibrosis lung disease. Adv Drug Deliver Rev. 2002;54:1359–1371. doi: 10.1016/s0169-409x(02)00144-8. [DOI] [PubMed] [Google Scholar]
- Burns JL, Emerson J, Stapp JR, Yim DL, Krzewinski J, Louden L, et al. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis. 1998;27:158–163. doi: 10.1086/514631. [DOI] [PubMed] [Google Scholar]
- Clark DJ, Maaløe O. DNA replication and division cycle in Escherichia coli. J Mol Biol. 1967;23:99–103. [Google Scholar]
- Das T, Kutty SK, Kumar N, Manefield M. Pyocyanin facilitates extracellular DNA binding to Pseudomonas aeruginosa influencing cell surface properties and aggregation. PLoS One. 2013;8:e58299. doi: 10.1371/journal.pone.0058299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekimpe V, Deziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiol-SGM. 2009;155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- Denning GM, Railsback MA, Rasmussen GT, Cox CD, Britigan BE. Pseudomonas pyocyanine alters calcium signaling in human airway epithelial cells. Am J Physiol—Lung C. 1998;274:L893–L900. doi: 10.1152/ajplung.1998.274.6.L893. [DOI] [PubMed] [Google Scholar]
- Dietrich LEP, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC, Newman DK. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J Bacteriol. 2013;195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Winzer K, Lazdunski A, Williams P, Camara M. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of n-acylhomoserine lactone production and virulence gene expression. J Bacteriol. 2002;184:2576–2586. doi: 10.1128/JB.184.10.2576-2586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Dammel C, Stein J, Rabin H, Surette MG. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- Guss AM, Roeselers G, Newton ILG, Young CR, Klepac-Ceraj V, Lory S, et al. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J. 2011;5:20–29. doi: 10.1038/ismej.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett DJ, Charniga L, Bean K, Ohman DE, Cohen MS. Response of Pseudomonas aeruginosa to pyocyanin—mechanisms of resistance, antioxidant defenses, and demonstration of a manganese-cofactored superoxide-dismutase. Infect Immun. 1992;60:328–336. doi: 10.1128/iai.60.2.328-336.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LR, Richardson AR, Houston LS, Kulasekara HD, Martens-Habbena W, Klausen M, et al. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog. 2010;6:e1000712. doi: 10.1371/journal.ppat.1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RC, Klepac-Ceraj V, Lorenzi MM, Grotzinger H, Martin TR, Newman DK. Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am J Respir Cell Mol Biol. 2012;47:738–745. doi: 10.1165/rcmb.2012-0088OC. [DOI] [PubMed] [Google Scholar]
- Hutchinson GE. Concluding Remarks. Cold Spring Harbor Symposia on Quantitat Biol. 1957;22:415–427. [Google Scholar]
- Ji X-J, Huang H, Ouyang P-K. Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol Adv. 2011;29:351–364. doi: 10.1016/j.biotechadv.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Khan MT, Duncan SH, Stams AJM, van Dijl JM, Flint HJ, Harmsen HJM. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 2012;6:1578–1585. doi: 10.1038/ismej.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Wang W, Deckwer WD, Zeng AP. Expression of the quorum-sensing regulatory protein LasR is strongly affected by iron and oxygen concentrations in cultures of Pseudomonas aeruginosa irrespective of cell density. Microbiol-SGM. 2005;151:1127–1138. doi: 10.1099/mic.0.27566-0. [DOI] [PubMed] [Google Scholar]
- Kuang Z, Hao Y, Walling BE, Jeffries JL, Ohman DE, Lau GW. Pseudomonas aeruginosa elastase provides an escape from phagocytosis by degrading the pulmonary surfactant protein-A. PloS One. 2011;6:e27091. doi: 10.1371/journal.pone.0027091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103:19931–19931. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LiPuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. 2010;23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan TM, Martinko MJ, Parker J.2002Brock Biology of Microorganisms10 edn.Pearson Education Inc.: New Jersey [Google Scholar]
- Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. Shewanella secretes flavins that mediate extracellular electron transfer. Proc Natl Acad Sci USA. 2008;105:3968–3973. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight SL, Iglewski BH, Pesci EC. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2000;182:2702–2708. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivens DE, Ohman DE, Williams J, Franklin MJ. Role of alginate and its o-acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol. 2001;183:1047–1057. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal'chykovs'ka LH, Aleksieieva IV, Kostina VH, Platonov MO, Nehruts'ka VV, Deriabin OM, et al. New amides of phenazine-1-carboxylic acid: antimicrobial activity and structure-activity relationship. Ukr Biokhim Zh. 2008;80:140–147. [PubMed] [Google Scholar]
- Parsons JF, Greenhagen BT, Shi K, Calabrese K, Robinson H, Ladner JE. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry. 2007;46:1821–1828. doi: 10.1021/bi6024403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaey K, Boon N, Hofte M, Verstraete W. Microbial phenazine production enhances electron transfer in biofuel cells. Environ Sci Technol. 2005;39:3401–3408. doi: 10.1021/es048563o. [DOI] [PubMed] [Google Scholar]
- Ramos I, Dietrich LEP, Price-Whelan A, Newman DK. Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res Microbiol. 2010;161:187–191. doi: 10.1016/j.resmic.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid MH, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratjen F, Döring G. Cystic fibrosis. The Lancet. 2003;361:681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- Schuster M, Greenberg EP.(eds). (2008Chemical Communications Among Bacteria ASM press: Washington DC; p12 [Google Scholar]
- Sibley CD, Duan K, Fischer C, Parkins MD, Storey DG, Rabin HR, et al. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CD, Grinwis ME, Field TR, Eshaghurshan CS, Faria MM, Dowd SE, et al. Culture enriched molecular profiling of the cystic fibrosis airway microbiome. PLoS One. 2011;6:e22702. doi: 10.1371/journal.pone.0022702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci USA. 2008;105:15070–15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Stressmann FA, Rogers GB, van der Gast CJ, Marsh P, Vermeer LS, Carroll MP, et al. Long-term cultivation-independent microbial diversity analysis demonstrates that bacterial communities infecting the adult cystic fibrosis lung show stability and resilience. Thorax. 2012;67:867–873. doi: 10.1136/thoraxjnl-2011-200932. [DOI] [PubMed] [Google Scholar]
- Suh SJ, Silo-Suh L, Woods DE, Hassett DJ, West SEH, Ohman DE. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey KB, O'Connell OJ, McCarthy Y, Dow JM, O'Toole GA, Plant BJ, et al. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J. 2012;6:939–950. doi: 10.1038/ismej.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Wauven C, Piérard A, Kley-Raymann M, Haas D. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J Bacteriol. 1984;160:928–934. doi: 10.1128/jb.160.3.928-934.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDevanter DR, LiPuma JJ. Microbial diversity in the cystic fibrosis airways: where is thy sting. Future Microbiol. 2012;7:801–803. doi: 10.2217/fmb.12.55. [DOI] [PubMed] [Google Scholar]
- Venkataraman A, Rosenbaum AM, Perkins SD, Werner JJ, Angenent LT. Metabolite-based mutualism between Pseudomonas aeruginosa PA14 and Enterobacter aerogenes enhances current generation in bioelectrochemical systems. Energy Environ Sci. 2011;4:4550–4559. [Google Scholar]
- Venkataraman A, Rosenbaum M, Arends JBA, Halitschke R, Angenent LT. Quorum sensing regulates electric current generation of Pseudomonas aeruginosa PA14 in bioelectrochemical systems. Electrochem Comm. 2010;12:459–462. [Google Scholar]
- Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Newman DK. Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ Sci Technol. 2008;42:2380–2386. doi: 10.1021/es702290a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol. 2011;193:3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JJ, Ptak AC, Rahm BG, Zhang S, Richardson RE. Absolute quantification of Dehalococcoides proteins: enzyme bioindicators of chlorinated ethene dehalorespiration. Environ Microbiol. 2009;11:2687–2697. doi: 10.1111/j.1462-2920.2009.01996.x. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Lee KM, Greenberg EP. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteson KL, Meinardi S, Lim YW, Schmieder R, Maughan H, Quinn R, et al. 2014Breath gas metabolites and bacterial metagenomes from cystic fibrosis airways indicate active pH neutral 2,3-butanedione fermentation ISMEJe-pub ahead of print 9 January 2014; doi: 10.1038/ismej.2013.229 [DOI] [PMC free article] [PubMed]
- Wilson R, Sykes DA, Watson D, Rutman A, Taylor GW, Cole PJ. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect Immun. 1988;56:2515–2517. doi: 10.1128/iai.56.9.2515-2517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci USA. 2012;109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.