Summary
Background
Histamine plays important biological roles in cell-to-cell communication; it is a mediator in allergic responses, a regulator of gastric acid secretion, a messenger in bronchial asthma, and a neurotransmitter in the central nervous system. Histamine acts by binding to histamine receptors, and its local action is terminated primarily by methylation. Human histamine N-methyltransferase (HNMT) has a common polymorphism at residue 105 that correlates with the high- (Thr) and low- (Ile) activity phenotypes.
Results
Two ternary structures of human HNMT have been determined: the Thr105 variant complexed with its substrate histamine and reaction product AdoHcy and the Ile105 variant complexed with an inhibitor (quinacrine) and AdoHcy. Our steady-state kinetic data indicate that the recombinant Ile105 variant shows 1.8- and 1.3-fold increases in the apparent KM for AdoMet and histamine, respectively, and slightly (16%) but consistently lower specific activity as compared to that of the Thr105 variant. These differences hold over a temperature range of 25°C–45°C in vitro. Only at a temperature of 50°C or higher is the Ile105 variant more thermolabile than the Thr105 enzyme.
Conclusions
HNMT has a 2 domain structure including a consensus AdoMet binding domain, where the residue 105 is located on the surface, consistent with the kinetic data that the polymorphism does not affect overall protein stability at physiological temperatures but lowers KM values for AdoMet and histamine. The interactions between HNMT and quinacrine provide the first structural insights into a large group of pharmacologic HNMT inhibitors and their mechanisms of inhibition.
Keywords: histamine, methylation, polymorphism, thermal stability, antimalarial drug quinacrine
Introduction
Histamine is a messenger in cell-to-cell communication. This biogenic amine is considered one of the most important mediators of allergy and inflammation [1]. Histamine also plays a key role in the regulation of gastric acid secretion [2], is involved in bronchial asthma [3, 4], and is a neurotransmitter that affects several (patho)-physiological processes such as sleep/wakefulness, hormonal cardiovascular control, thermoregulation, food intake, and memory formation [5, 6, 7].
Histamine is produced and stored in airway mast cells, in basophils (enterochromaffin-like cells in the stomach), and in the synaptic vesicles of histaminergic neurons. Upon release from storage granules (for example, in response to specific allergens through immunoglobulin E [IgE] receptor triggering [8, 9]), histamine rapidly diffuses into surrounding tissues, and changes of its plasma concentration are detectable within minutes [10]. Histamine acts primarily through pharmacologically distinct subtypes of G protein-coupled receptors [11]. Released histamine is rapidly inactivated (see below) and disappears from the bloodstream within minutes [12].
A primary messenger such as histamine requires mechanism(s) for the termination of its action. In mammals, histamine N-methyltransferase (HNMT) (reviewed in [13]) and diamine oxidase metabolize histamine. However, the relative contributions of these two enzymes to histamine metabolism differ among tissues and organisms. HNMT plays the dominant role in histamine biotransformation in bronchial epithelial and endothelial cells of the human airways [3] and gut [2] and is the only enzyme responsible for termination of the neurotransmitter action of histamine in the mammalian brain [5]. The level of histamine in brain and the activation of histaminergic neurons have been implicated in Alzheimer’s disease and other human attentional (such as attention-deficit hyperactivity disorder) and aging disorders [6, 7, 14, 15, 16].
HNMT inactivates histamine by transferring a methyl group from S-adenosyl-L-methionine (AdoMet) to the Nε2 atom of the imidazole ring, yielding methylhistamine and S-adenosyl-L-homocysteine (AdoHcy) [17]. Methylhistamine, excreted in the urine [12], does not have the pharmacologic activity of its precursor amine and is inactive at histamine receptors [18, 19].
The HNMT gene, located on chromosome 2q22.1, has six exons spanning 50 kb (http://genome.ucsc.edu/cgi-bin/hgTracks?position=U08092&db=hg6). HNMT activity is high in the trachea and bronchi, and the contractile response of isolated human bronchi to histamine is strongly augmented by an HNMT inhibitor, which suggests that HNMT plays an important role in degrading histamine and in regulating the airway response [4]. Preuss et al. [20] found HNMT mRNA in most human tissues. The highest mRNA levels were found in kidney and liver cells, with substantial expression in spleen, prostate, ovary, colon, and spinal cord cells and the lower-level expression in heart, brain, placenta, lung, stomach, thyroid, and small intestine cells. While HNMT occurs ubiquitously in vertebrate species, it does not appear to be present in invertebrates, plants, or microorganisms.
Biochemical and genetic studies of HNMT from human red blood cells demonstrated that genetic variation among individuals can result in up to 5-fold differences in HNMT activity [21, 22]. A common C-to-T genetic polymorphism at position 314 in the HNMT gene changes the specified amino acid at position 105 from threonine to isoleucine, with the frequencies of the Thr105 and Ile105 alleles being approximately 90% and 10%, respectively [20]. The Ile105 allele is associated with a decrease in both HNMT enzymatic activity and immunoreactive protein [20], which could result in reduced histamine inactivation and increased sensitivity to this amine. The relatively high allele frequency for this HNMT polymorphism suggests that the less-frequent Ile variant may provide some selective advantage. One possibility is that prolonging elevated histamine levels provides protection against infectious agents such as nematodes, for which IgE, mast cells, and histamine play major defensive roles [23].
Recently, Yan et al. [24] demonstrated an increase in the frequency of the Ile105 allele in asthmatic patients compared to controls (0.14 versus 0.08, odds ratio = 1.9, p < 0.01). This association between a functionally significant HNMT genetic polymorphism and asthma suggests individual variation in histamine metabolism might contribute to the pathophysiology and/or response to therapy for this disease. These individuals might also be more prone to scombrotoxicosis, which results from ingestion of seafood contaminated by histamine-producing bacteria [25, 26].
We have solved two ternary structures of human HNMT: a recombinant Thr105 variant in complex with histamine (substrate) and AdoHcy (reaction product), and the Ile105 variant in complex with the antimalarial drug quinacrine (an HNMT inhibitor) and AdoHcy. In addition, we have compared the biochemical, thermal, and kinetic properties of these two recombinant enzymes.
Results and Discussion
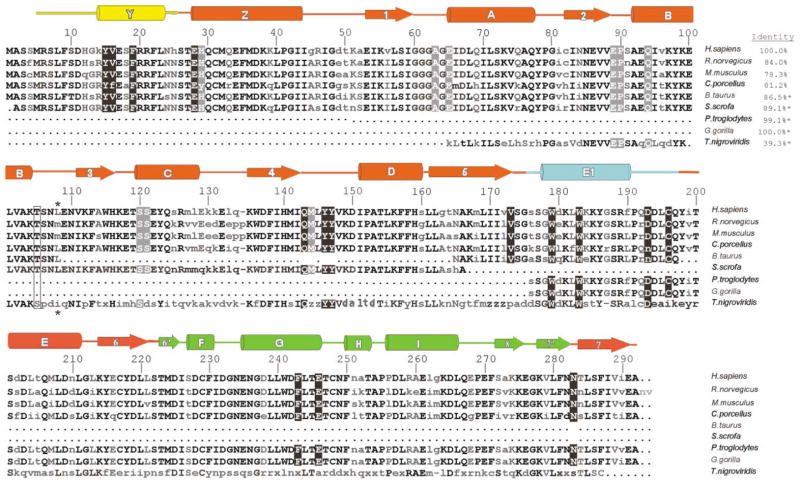
The 292 amino acid sequence of human HNMT (E.C. 2.1.1.8) is 84% identical to HNMT from rat [27] and 80% identity to that of guinea pig [28] (Figure 1). In addition, expressed sequence tags with strong similarities to human HNMT can be found in Bos taurus, Sus scrofa, Pan troglodytes, and Gorilla gorilla, and in genome survey sequences for Tetradon nigroviridis, indicating that HNMT is a highly conserved protein in vertebrates.
Figure 1. Sequence Alignment of HNMT Homologs.
Sequences were obtained with GenPept and GenBank identifier numbers: Homo sapiens (1236891), Rattus norvegicus (348436), Cavia porcellus guinea pig (10336511), Mus musculus (primarily dbESTs 5336012, 8079428), Bos taurus (dbESTs 9604685, 11707209), Sus scrofa European wild boar (dbEST 10874736), Pan troglodytes (7592968), Gorilla gorilla (7592970), and Tetraodon nigroviridis green-spotted puffer fish (9555631, 9555148). Percentage amino acid identities with human HNMT are given in the upper right. Asterisks designate sequences where only partial information is available. The 105Thr-to-Ile variation in humans is boxed. Residues interacting with AdoHcy have white lettering on a gray background, and residues interacting with histamine or quinacrine have white lettering on a black background.
We produced recombinant human HNMT in Escherichia coli, in methionine, and in selenomethionine-containing forms (see Materials and Methods). We calculated initial electron density maps in the space group P6 by selenium data at 3.3 Å resolution by using 4 wavelength anomalous diffraction (Table 1), and we refined the models for the Thr105 and Ile105 variants to higher resolutions of 2.3 Å and 1.9 Å, respectively (Table 2).
Table 1.
Summary of Selenium Multiwavelength Anomalous Diffraction Experiment
| λ1 | λ2 | λ3 | λ4 | |
|---|---|---|---|---|
| Wavelength (Å) | 0.97911 | 0.97977 | 0.9850 | 1.0721 |
| Energy (eV) | 12,633.03 | 12,654.50 | 12,587.31 | 11,564.69 |
| Completeness (%) | 96.4 | 92.8 | 93.7 | 96.0 |
| Rmerge = Σ(I − <I>)/Σ<I> | 0.091 | 0.082 | 0.062 | 0.075 |
| <I/σ> | 17.7 | 19.0 | 20.8 | 17.4 |
| Measured reflections | 40,019 | 35,800 | 35,952 | 40,371 |
| Unique reflections | 9,376 | 9,066 | 9,187 | 9,425 |
| Anomalous pairs | 7,715 | 7,266 | 7,398 | 7,690 |
| Overall figure of merit | 0.59 | |||
| X-ray beamline | X8C at NSLS | |||
| Crystallization conditions | 24% PEG 6000, 100 mM Tris (pH 7.5), 5% ethylene glycol | |||
| Cell dimensions (Å) | a = b = 130.5, and c = 64.1 | |||
| Resolution range (Å) | 40–3.27 | |||
Table 2.
Summary of Model Refinements (Highest Resolution Shell in Parentheses)
| Thr105-Histamine-AdoHcy | Ile105-Quinacrine-AdoHcy | |
|---|---|---|
| X-ray beamline | IMCA at APS | X12C at NSLS |
| Resolution range (Å) | 18–2.28 (2.36–2.28) | 25–1.91 (1.94–1.91) |
| Crystallization conditions | 19% PEG 6000, 19% isopropanol, 95 mM sodium citrate (pH 5.6), 5% glycerol | 20% PEG 8000, 100 mM phosphate citrate (pH 4.2), 200 mM NaCl |
| Cell dimensions (Å) | a = b = 131.14, c = 63.57 | a = b = 130.34, c = 63.01 |
| Measured reflections | 215,317 (21,749) | 202,140 (4,447) |
| Unique reflections | 27,999 (2,582) | 44,743 (946) |
| <I/σ> | 25.2 | 38.1 |
| Completeness (%) | 98.9 (89.8) | 94.2 (43.8) |
| Rmerge = Σ (I − <I>)/Σ<I> | 0.072 (0.266) | 0.033 (0.213) |
| R factor = Σ (Fo − Fc)/ΣFc | 0.205 (0.234) | 0.195 (0.235) |
| Rfree (10% of data) | 0.257 (0.284) | 0.239 (0.269) |
|
| ||
| Nonhydrogen Atoms | ||
|
| ||
| Protein | 4347 | 4202 |
| Hetrogen | 68 (AdoHcy and histamine) | 80 (AdoHcy and quinacrine) |
| Water | 304 | 384 |
|
| ||
| Rms Deviation from Ideality | ||
|
| ||
| Bond lengths (Å) | 0.007 | 0.008 |
| Bond angles (°) | 1.3 | 1.4 |
| Dihedral (°) | 22.9 | 22.6 |
| Improper (°) | 0.88 | 0.79 |
|
| ||
| Ramachandran Plot | ||
|
| ||
| Most favored (%) | 92.7 | 91.5 |
| Additionally allowed (%) | 7.3 | 8.5 |
2 Domain Structure of HNMT
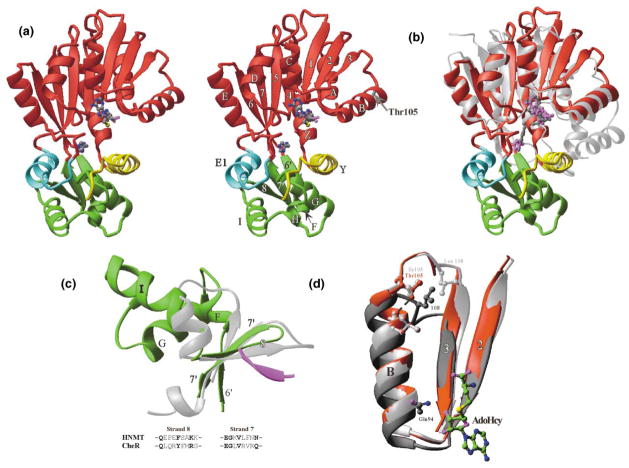
HNMT is a 2 domain protein (Figure 2a). The large domain is a classic MTase fold, as defined by 20 structurally characterized MTases to date [29]. The MTase domain contains a seven-stranded β sheet (6↑ 7↓ 5↑ 4↑ 1↑ 2↑ 3↑) flanked on each side by three helices (αZ, αA, αB and αC, αD, αE) [30]. As expected from comparison to other MTases [31], AdoHcy is bound at the carboxyl ends of the parallel strands β1-β2-β3. The histamine is at the carboxyl ends of the parallel strands β4-β5 (see below for detailed interactions). The MTase domain is structurally most similar to catechol-O-MTase (COMT) [32], a single-domain MTase; the root-mean-square (rms) deviation between the two proteins is 2.9 Å over 156 Cα atoms, and the superimposition yields coincidence of the cofactor (AdoMet or AdoHcy) and the substrate (catechol or histamine) (Figure 2b).
Figure 2. Ribbon Diagrams of HNMT-Histamine-AdoHcy.
(a) Stereo diagram of HNMT (Thr105) with bound AdoHcy and histamine (at pH 5.6). The large MTase domain is colored red (with Thr105 in gray), the N-terminal helix αY is in yellow, the helix αE1 is in cyan, and the subdomain is in green. All ribbon-type diagrams were made by using the programs RIBBONS [61]. HNMT is labeled as the helices in letters and strands in numbers, according to the nomenclature of the MTase fold defined by Schluckebier et al. [30].
(b) Superimposition of the HNMT MTase domain (red) and rat catechol-O-MTase (silver). The respective cofactors (gray AdoMet in COMT and magenta AdoHcy in HNMT) overlap well, while the substrate histamine (magenta) in HNMT (at pH 5.6) shifts a little away from the cofactor relative to 3,5-dinitrocatechol (gray) in COMT.
(c) Superimposition of the three strands from the HNMT subdomain (green) and the β subdomain of protein MTase CheR (silver). The subdomain of HNMT (green) is rotated 90° from the view in Figure 2a, looking down onto the edge of the strand β8. The β subdomain of CheR (silver) consists of a three-stranded antiparallel β sheet and a crossover α helix. The chemotaxis receptor Tar pentapeptide (magenta) binds to CheR MTase as the fourth strand of the antiparallel β sheet. Amino acids of the strands of β8 and β7′ from the two MTases are aligned.
(d) Superimposition of the β2-αB-β3 fragments from HNMT Thr105 (red), HNMT Ile105 (silver), and COMT (dark gray). In both HNMT and COMT, the amino end of helix αB and its associated loop of β2-αB is involved in AdoMet/AdoHcy binding. The human HNMT polymorphism at position 105 is located at the carboxyl end of the helix, while the human COMT polymorphism at position 108 is located in the loop of αB-β3.
Three additional segments are inserted into the HNMT MTase domain: the N-terminal helix αY (yellow in Figure 1), the helix αE1 (cyan) preceding helix αE, and a subdomain between strands β6 and β7 (green). The three non-contiguous regions interact with each other to form a separate domain, which we term the substrate binding domain (S domain). The main chain atoms of β6′ (residues 223–225) and the loop between αY and αZ (residues 25–27) interact like antiparallel β strands. The side chains of helices αE1 (residues 185–189) and αI (residues 261–265) pack together.
The green subdomain (amino acids 223–282) consists of an antiparallel β sheet containing three short strands (↓β8, ↑β7′, ↓β6′), two α helices (G and I), and two single-turn 310 helices (F and H) on one side of the sheet making a crossover connection between strands β6′ and β8. Strands β6 and β6′ and strands β7 and β7′ are each linked by single amino acids (Ser222 and Asn283, respectively) as if each pair was a single twisted strand. The antiparallel hydrogen bonding pattern between strands β6 and β7 (and between strands β6′ and β7′) is interrupted between the main chain carbonyl group of Asn283 and the amino group of Ser222. This interruption is probably due to the fact that the side chain of Asn283 is part of the active site and interacts with histamine (see below).
The subdomain shown in green contributes only one residue (Phe243) directly to histamine binding but may play a role in protein-protein interactions. The Vector Alignment Search Tool [33] revealed that the three short strands (β8, β7′, β6′) resemble the β motif found in the CheR protein MTase [34]. The rms deviation is 1.8 Å when structurally aligned Cα atoms, mostly strands β8 and β7′, are compared (Figure 2c). The CheR MTase interacts with cytoplasmic portions of the chemotaxis receptor by extending its antiparallel β sheet with an additional strand from the receptor [34], thus recruiting this MTase to the bacterial membrane. By analogy, the green subdomain of HNMT may interact with other proteins for its in vivo function.
Thr105Ile Polymorphic Variant
There are few significant changes between the two variant structures, particularly in the MTase domain where Thr105 is located. The overall rms deviation between two variant enzymes is 0.35 Å over the MTase domain (colored red in Figure 2a). The largest rms deviation (~1–2 Å) lies in the loop region (amino acids 104–110) between αB and β3 following Thr/Ile105. However, the S domain moves toward the MTase domain in the quinacrine bound Ile105 variant. This change is most likely due to the quinacrine binding (see below) and not to the polymorphism.
Thr105 is located at the carboxyl end of helix αB (Figure 2a). The amino end of helix αB (Gln94) and the loop prior to the helix (Glu89 and Pro90) bind AdoHcy. The hydroxyl group of Thr105 forms a hydrogen bond to the backbone carbonyl of Leu101 (Figure 2d). The surface location of residue 105 suggests that this polymorphism is unlikely to affect overall structural stability.
In the variant Ile105 structure, the hydrogen bond with the main chain of Leu101 is lost, and the loop between αB and β3 following Ile105 is more flexible (indicated by the multiple conformations of Leu108 and the higher crystallographic thermal factors in the loop). Both could slightly destabilize the carboxyl end of helix αB and affect the stability of the AdoMet binding by residues at the amino end of that helix. Surprisingly, human COMT has a common polymorphism (Met and Val) located in the corresponding loop between helix αB and strand β3 (Figure 2d), which also correlates with high- (Val) and low- (Met) activity phenotypes of COMT [35].
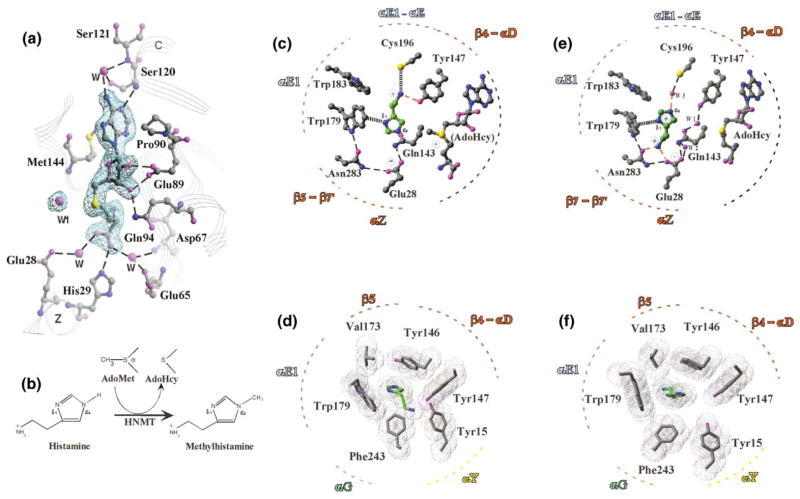
AdoHcy Binding
Figure 3a shows a network of polar and hydrophobic interactions with AdoHcy; these interactions are very similar in both HNMT variants. The interactions can be grouped according to the three moieties of AdoHcy. The adenine ring is sandwiched between Pro90 (after strand β2) and Met144 (after stand β4). The adenine ring N1 atom forms a hydrogen bond to the main chain amide of Ser120, and the N6 (NH2) forms water-mediated hydrogen bonds to the side chain of Ser120 and the main chain amide of Ser121 (after strand β3).
Figure 3. Detailed Plots of Interactions.
(a) AdoHcy is shown in stick form and is superimposed on a Fo − Fc omit electron density map at 1.91 Å resolution (Ile105 variant at pH 4.2; see Table 2). A water molecule (labeled as w1) is 3.7 Å away from the sulfur atom of AdoHcy and is held by Tyr147. The corresponding water molecule is also observed at pH 5.6 (e), but not at pH 7.5 (c).
(b) Schematic methyl transfer reaction of histamine by HNMT. The methylation only occurs at the imidazole ring Nε2 position. In aqueous solution, 80% of histamine monocation is in the Nε2-H tautomer and 20% in the Nδ1-H tautomer [18].
(c) Polar interactions involved in histamine binding at pH 7.5 (SeMet-containing Thr105 variant). The transferable methyl group was modeled onto the sulfur atom of AdoHcy.
(d) Hydrophobic interactions involved in histamine binding at pH 7.5 (SeMet-containing Thr105 variant).
(e) Polar interactions involved in histamine binding at pH 5.6 (Thr105 variant).
(f) Hydrophobic interactions involved in histamine binding at pH 5.6 (Thr105 variant).
For the adenosine ribose moiety, the ribose hydroxyl oxygen atoms (O2* and O3*) form hydrogen bonds with the carboxylic oxygen atoms of Glu89 (after strand β2) and the Nε2 atom of Gln94 of helix αB. The Glu89-ribose interaction is the most highly conserved AdoMet interaction among MTases [31].
The homocysteine portion of AdoHcy extends along the curved glycine-rich loop (60-GGGAG-64) between strand β1 and helix αA; this corresponds to motif I among MTases [36]. The terminal carboxyl oxygen atoms form hydrogen bonds to His29 (αZ) and water-mediated hydrogen bonds to Glu28 (αZ) and Glu65 (αA). The amine group (NH3) forms hydrogen bonds to the backbone oxygen atoms of Gly60 (after β1) and Ile142 (β4), and it forms water-mediated contacts to the side chains of Asp67 (αA) and His140 (β4). (For clarity, these interactions are not shown in Figure 3a.)
Two Histamine Binding Modes that Differ in pH
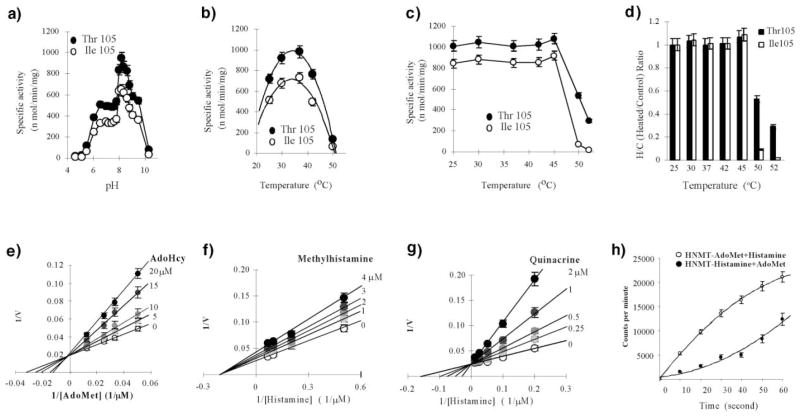
We note that the crystal of HNMT (Thr105)-histamine-AdoHcy ternary complex was formed at pH 5.6 and 16°C, where the recombinant HNMT is nearly inactive (see Figures 7a and 7b). The inactivation is probably due to the fact that histamine ring is nearly fully protonated (–Nδ1H and –Nε2H) at this pH, which is lower than the ring pKa of 6.23 at 15°C or 5.80 at 37°C [18]. However, the SeMet-containing HNMT (Thr105 variant) was crystallized in a very narrow pH range near pH 7.5 in complex with histamine and AdoHcy. At pH 7.5, the histamine ring is in the uncharged neutral form, and the SeMet-containing HNMT is active (data not shown). We observed two different modes of interactions of histamine with HNMT at the different pH. It appears likely that the protonation status of the histamine ring is essential for the methylation reaction. However, since the crystal at low pH diffracted X-rays to a higher resolution of 2.3 Å, the low pH structure is well ordered even though it is apparently not in the catalytic active mode.
Figure 7. Kinetics and Thermal Stability of Recombinant Human HNMT.
Top panel: Specific activity and thermostability of HNMT.
(a) pH titration. We note that the sharp difference in specific activity between pH 7.8 and pH 8.0 is probably due to the different buffer salts (Hepes versus Bicine) (see Experimental Procedures).
(b) Temperature titration.
(c) Thermal stability as a function of preincubation temperature.
(d) Thermal stability expressed as heated/control ratios.
Bottom panel: Lineweaver-Burke plots of Thr105-HNMT.
(e) AdoMet-competitive AdoHcy.
(f) Histamine-noncompetitive methylhistamine.
(g) Histamine-competitive quinacrine.
(h) Pre-steady-state kinetics.
In both modes, the histamine molecule is buried at the interface of the two domains, and is therefore not visible in the molecular surface representation shown in Figure 4a. The bound histamine is in an almost-closed acidic pocket (Figure 4b). This is in sharp contrast to COMT, where catechol can diffuse into and bind at an open surface pocket (Figure 4c).
Figure 4. Solvent-Accessible Molecular Surface.
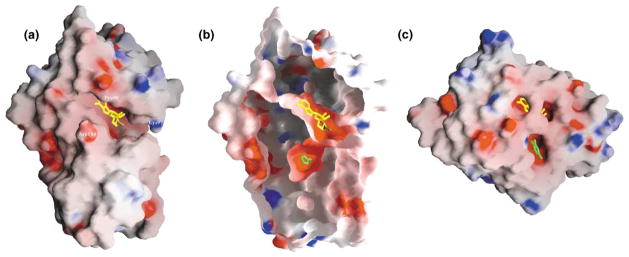
The GRASP surface [62] is colored blue for positive electrostatic potential, red for negative, and white for neutral.
(a) HNMT-AdoHcy (yellow stick)-histamine, (b) sliced cutaway of HNMT surface displaying histamine (green stick) bound in the interior, and (c) COMT-AdoMet (yellow)-3,5-dinitrocatechol (green).
Catalysis of Methyl Transfer at pH 7.5
The histamine-HNMT interactions can be grouped according to the two moieties of histamine (Figure 3b). In the pH 7.5 SeMet-containing structure, the amino group of histamine interacts with the hydroxyl group of Tyr147 via hydrogen bonding and with the sulfur atom of Cys196 via van der Waals contact. It has been shown that purified rat kidney HNMT activity is inhibited by parahydroxymercuric benzoate or iodoacetamide, but not dithiothreitol [37]. This suggested, consistent with our structure, that HNMT had a sulfhydryl near the active site.
The AdoHcy sulfur atom, where the transferable methyl group was attached, defines where catalysis must take place (Figure 3c). To determine the rotamer of the imidazole ring of histamine, we reasoned that the target Nε2 atom, not the Nδ1 atom, should be closer to the donor methyl group to become the nucleophile attacking the methyl group of AdoMet. It is ~4 Å away from the methyl group modeled onto the sulfur atom of AdoHcy and almost in line with the methyl group and sulfur atom of AdoMet. Such a linear arrangement of the nucleophile, the methyl group, and the leaving thioester group is required in the transition state by a classic SN2 reaction mechanism used by most (if not all) other MTases [38].
However, we do not know which ring nitrogen atom (Nδ1 or Nε2) is protonated. There are two possibilities, because in aqueous solution 80% of histamine monocation is in the Nε2-H tautomer, and 20% is in the Nδ1-H tautomer [18]. First, if the Nδ1 is protonated, it will likely interact with the π-electron cloud of Trp179. This leaves a lone pair of electrons on the nucleophilic Nε2 to attack the nearby methyl group of AdoMet. However, the structure reveals no obvious candidate for the enzymic base that is required to abstract the proton at Nδ1, unless Trp179 needs to be moved out of the way for a base such as a water molecule that diffuses into the active site. Second, if the Nδ2 is protonated, the proton elimination step and methyl transfer could jointly occur at Nε2. It seems reasonable to propose that the enzymic base that is required to abstract the proton at Nδ2 is the neighboring glutamic acid Glu28.
Histamine-HNMT Interactions at pH 5.6
In the pH 5.6 structure, the histamine imidazole ring (positively charged) and the terminal amino group rotate and switch positions. Two water molecules w1 and w2 are trapped between histamine and AdoHcy; each is held in position by Tyr147 and Glu28, respectively (Figure 3e). The terminal aliphatic amino group (NH3+) is anchored by ionic bonding to Glu28 (the first residue of helix αZ) and by hydrogen bonding to Asn283 (the amino acid linking strands β7 and β7′).
The imidazole ring of histamine is packed edge-to-face against to the ring of Trp179. A water molecule (w3) is located between the edge of the imidazole ring and the sulfur atom of Cys196, within hydrogen bonding distance of each. To maximize the hydrogen bonding capacity, we suggest the protonated Nε2 atom of histamine is hydrogen bonded to the water molecule w3, while the protonated ring nitrogen atom (Nδ1) points to the negatively charged π-electron cloud of Trp179 (Figure 3e). In this configuration, the target histamine Nε2 atom is located more than 8 Å away from the AdoHcy sulfur atom or ~6 Å away from the transferable methyl group modeled onto the sulfur atom of AdoHcy. This distance would be too large for methyl transfer to occur directly between the donor AdoMet methyl group and the acceptor histamine Nε2 atom (Figure 3b).
Comparison with Other Histamine Binding Proteins
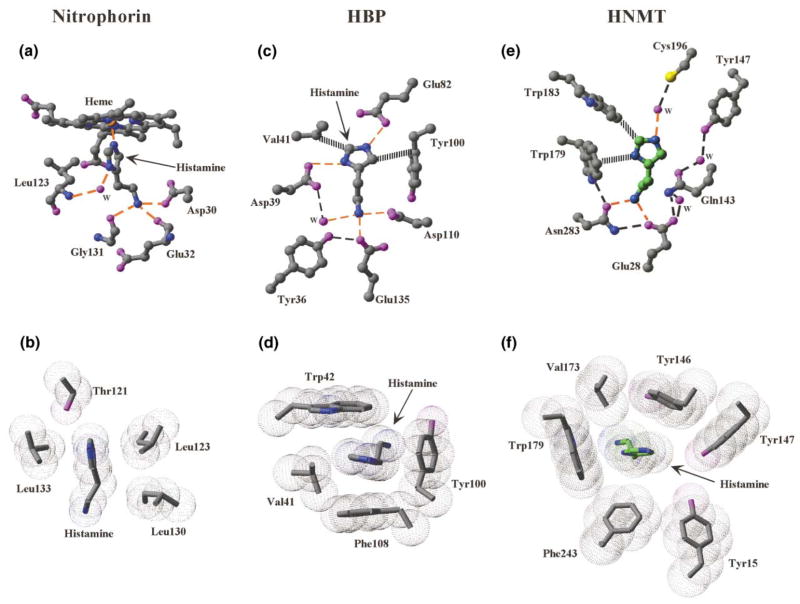
The structures of other histamine binding proteins have been described: the nitrophorins [39] and histamine binding proteins (HBPs) [40] are secreted into their hosts during feeding by blood-sucking insects and ticks, respectively. These proteins sequester histamine at the wound site to overcome the host’s inflammatory and immune responses by outcompeting binding to the host’s histamine receptors.
The nitrophorins have a Fe3+-containing heme group carrying nitric oxide; when the nitric oxide is released, host histamine binds to the vacated binding site in a way similar to the His-Fe in myoglobin (Figure 5a). In HBPs, the carboxylate oxygen atoms of Asp39 and Glu82 form hydrogen bonds with two nitrogen atoms of the imidazole ring—indicating that the imidazole ring carries two protons or that one of the carboxylate groups is protonated (Figure 5c). Examination of the three histamine binding proteins (HNMT at pH 5.6, nitrophorins, and HBP) together reveals (1) at least one negatively changed Glu or Asp interacts with the terminal aliphatic amino group of histamine, (2) the imidazole ring is surrounded by hydrophobic residues (Figures 5b, 5d, and 5f), and (3) the two ring nitrogen atoms have very different hydrogen bond partners.
Figure 5. Histamine Binding in Known Protein Structures.
Histamine binding in (a and b) nitrophorins (pdb 1NP1), (c and d) histamine binding proteins (pdb 1QFT), and (e and f) HNMT at pH 5.6. Bonding networks are shown in (a), (c), and (e), while (b), (d), and (f) show hydrophobic/van der Waals interactions.
As mentioned, histamine functions through its binding to histamine receptors. Site-directed mutagenesis studies of H1 and H2 receptors indicated that Trp103 and Asp107 of the H1-receptors and Tyr182 and Asp186 of H2-receptors are required for histamine binding [41, 42, 43]. Histamine binding sites in HNMT and the histamine receptors may be similar as both are inhibited by many H1-antagonists, particularly those containing dimethyl-aminoalkyl groups [44]. However, the interaction with the imidazole ring Nε2 atom (where the methyl group attached) in HNMT at pH 5.6 should be unique, because methylhistamine is a potent product inhibitor of HNMT (see below) but has no effect on histamine receptors [18]. The methyl group attached onto the Nε2 atom could replace the water (w3) between the Nε2 and the sulfhydryl of Cys196 (Figure 5e).
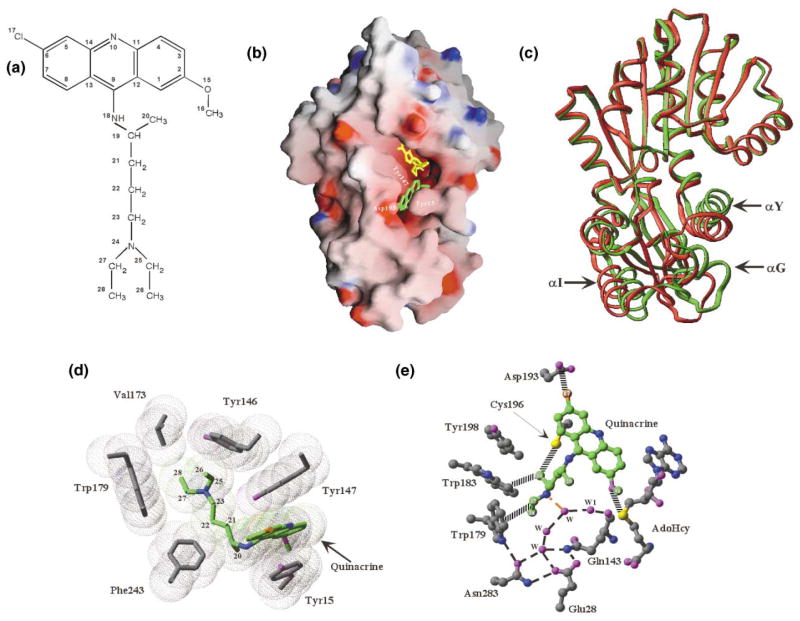
Binding of the Inhibitor Quinacrine
Inhibitors of HNMT are diverse in terms of chemical structure and clinical pharmacology. Antimalarial drugs are potent inhibitors of HNMT [45]. One of these, quinacrine, is an acridine derivative that possesses a long alkylamino tail (Figure 6a). The large tricyclic moiety blocks the entrance to the histamine pocket (Figure 6b) while its mostly hydrophobic tail fills most of the pocket. Upon inhibitor binding, the S domain undergoes a small concerted movement toward the MTase domain (Figure 6c). As described below, quinacrine is a competitive inhibitor with respective to histamine.
Figure 6. HNMT-Quinacrine Interactions.
(a) Chemical structure of quinacrine.
(b) Grasp surface of Ile105 HNMT-AdoHcy (yellow)-quinacrine (green).
(c) Superimposition of HNMT (Thr105)-AdoHcy-histamine (red), and HNMT (Ile105)-AdoHcy-quinacrine (green).
(d and e) Detailed plots of HNMT-quinacrine interactions.
The HNMT-quinacrine interactions are predominantly hydrophobic. The acridine ring is sandwiched between Tyr147 and Cys196 on one side and Tyr15 on the other (Figure 6d). The Tyr15 phenolic ring undergoes a rotamer conformational change (comparing Figures 5f and 6d) and opens the gate to the histamine pocket (Figure 6b). The chlorine atom at position 17 (Figure 6a) points perpendicularly to the plane of the carboxyl group of Asp193 and makes a van der Waals contact with the Cγ atom (Figure 6e). The O15 and C16 (CH3) are in van der Waals contact with the S of AdoHcy and Cα of Val16, respectively. The nitrogen atom at ring position 10 is exposed to the solvent and does not make any specific contact.
The quinacrine alkylamino tail fills the pocket except for the four water molecules occupying the space next to the AdoHcy (Figure 6e). These water molecules mediate hydrogen bonds between the tertiary amino group (N24) and the polar side chains of Glu28, Asn283, and Gln143. The hydrocarbon CH2 and terminal CH3 groups of the chain are able to make van der Waals contacts with Trp179 (via C27 and C28), Tyr146 (C25 and C26), Val173 (C28), Trp147 (C21 and C23), Phe243 (C22 and C20), and Trp183 and Cys196 (C26).
Kinetic Properties of HNMT
We compared the biochemical properties of the two variants. The optimum methylation conditions are similar for both forms—enzyme concentration 15 nM (within a linear range of 3–20 nM), reaction time 20 min (linear between 0 and 30 min), pH 8.2, and temperature 37°C (where maximum specific activity was observed, Figures 7a and 7b). Apparent KM values for AdoMet and histamine are similar, but those for the Ile105 variant are slightly but consistently higher (1.8-fold and 1.3-fold, respectively) than the Thr105 enzyme (Table 3).
Table 3.
Apparent KM (μM) for Histamine and AdoMet
| Enzyme | KMAdoMet (μM) | KMHistamine (μM) | Vmax (μmol/min/mg) | Reference |
|---|---|---|---|---|
| Thr105 (E. coli) | 13.1 | 3.7 | 1.4–2.0 | This study |
| Ile105 (E. coli) | 23.6 | 4.7 | 1.2–1.4 | |
| Thr105 (COS-1) | 8.2–10.0 | 23.4–25.3 | — | [20] |
| Ile105 (COS-1) | 11.7–13.0 | 35.0–35.5 | — | |
| Human (COS-1) | 6.2 | 14 | — | [63] |
| Human (kidney) | 2.0–3.0 | 13–20 | — | |
| Rat (kidney) | 13.0 | 7.3 | 1.6 | [27] |
| Rat (E. coli) | 6.3 | 7.1 | 1.6 at pH 8.5 |
We measured the Ki (dissociation constant of enzyme-inhibitor complex) of three inhibitors: AdoHcy, methylhistamine, and quinacrine (Table 4). Both reaction products, AdoHcy and methylhistamine, are potent inhibitors, and their Ki values (in μM) are similar. However, the Ki value for methylhistamine is more than 10 times lower than those reported for the rat enzyme (Table 4).
Table 4.
Dissociation Constant (Ki) of Enzyme-Inhibitor Complex
| Enzyme | KiAdoHcy (μM) | KiMethylhistamine (μM) | KiQuinacrine (μM) | Reference |
|---|---|---|---|---|
| Human | ||||
|
| ||||
| Thr105 (E. coli) | 6.9 | 6.5 | 0.45 | This study |
| Ile105 (E. coli) | 9.0 | 6.1 | 0.36 | |
|
| ||||
| Rat | ||||
|
| ||||
| E. coli expression | 9.9 | 101 | — | [27] |
| Natural (kidney) | 8.1 | 82 | — | |
The products display different modes of inhibition, diagnosed from the Lineweaver-Burke plots (Figures 7e–7g). As expected, AdoHcy is a competitive inhibitor with respect to AdoMet (Figure 7e), consistent with AdoHcy competing for the same binding site as AdoMet. However, methylhistamine is noncompetitive with histamine (Figure 7f).
Quinacrine is an extremely potent inhibitor (Ki = 450nM) that is competitive to histamine (Figure 7g), suggesting that quinacrine competes for the same binding site as histamine. The determined Ki value is about four times higher than those previous reported (10–100 nM) [44, 49–51].
The pre-steady-state kinetics were carried out in different mixing orders of AdoMet and histamine with HNMT. Whereas the reactions preincubated with Ado-Met and started with histamine show an immediate burst of methylation, a delay was observed for the preincubation with histamine (Figure 7h). Such delay suggests that the HNMT-histamine complex is nonproductive and must dissociate to allow AdoMet binding to the free enzyme. We conclude that the methyl-group donor Ado-Met is the first substrate that adds to the enzyme and that histamine must be the second substrate. Methylhistamine is the first product to leave the enzyme and AdoHcy is the second. These results are consistent with the proposed ordered sequential bi-bi reaction mechanism [47, 48]. From a structural point of view, the AdoMet binding site is open to the solvent, while the histamine binding site is much less accessible (Figure 4a).
Thermal Stability of HNMT Variants
As shown in Figure 7b, the Ile105 variant of HNMT has slightly but consistently lower specific activity measured at temperatures between 25°C and 50°C. We compared the thermal properties of the two variants by preincubating the enzymes at different temperatures and then measuring the specific activity at 37°C (Figure 7c). Preincubation for 30 min between 25°C and 45°C did not affect the activity of either variant; the Ile105 variant constantly had slightly lower (~84%) activity than the Thr105 enzyme. Only at a higher temperature of 50°C or 52°C, was Ile105-HNMT more thermolabile than the Thr105 enzyme (Figure 7d). Our data are consistent with the trend observed by Preuss et al. [20].
In addition to the differences in thermal properties of the two variants, Preuss et al. [20] suggested that differences in protein quantity (due to expression or degradation) might contribute to the larger differences in activity observed in vivo—HNMT immunoreactive protein levels are higher with the alleles that encoded Thr105 than with those that encoded Ile105. In addition, other factors such as protein-protein interactions might be involved in vivo. However, our data suggest that both variants of HNMT are much more stable over a wide range of temperatures than COMT, despite both MTases having a polymorphism at corresponding loops. Met-COMT had significantly higher thermolability than Val-COMT at the physiological temperature of 37°C [46], enough to account for the lower activity of Met-COMT.
Biological Implications
We have described the crystallographic structure of human HNMT. The results reveal that HNMT has a two-domain structure—the MTase domain and the S domain. The MTase domain primarily carries out cofactor binding and probably catalysis, while both domains contribute to histamine binding. The known polymorphic amino acid 105 resides on the outer surface of the MTase domain, away from the AdoMet binding site, and thus does not contribute directly to AdoMet or histamine binding. The Thr/Ile variation at position 105 does not affect overall protein stability at 37°C, but may alter the AdoMet binding site locally in the free enzyme (reflected in increased KM for AdoMet).
It is well documented that HNMT is inhibited by structurally diverse groups of pharmacologic agents and naturally occurring compounds [17]. As seen from our HNMT structures, the S domain and the interface between the two domains are very accommodating and provide a unique target for designing inhibitors selective for HNMT. A large number of aromatic residues can form hydrophobic contacts with structurally diverse molecules. A combination of three polar residues, Glu28, Gln143, and Asn283, may provide selectivity through direct or water-mediated interactions for inhibitors carrying an amino group.
Experimental Procedures
Cloning, Expression, and Purification
An expressed sequence tag (EST) clone (ATCC 948206) was obtained from American Type Culture Collection, supposedly as a cDNA clone with complete coding sequences of human HNMT. However, we found that this clone lacks the first 17 base pairs of the HNMT coding sequence. The missing sequence was built into the 5′ oligonucleotide primer to allow PCR of the intact human HMNT gene, as reported in GenBank (accession U08092). The PCR fragment was cloned into appropriate sites of pGEX-4T-3 (Pharmacia Biotech) to allow the production of HNMT fused to glutathione S-transferase (GST). The GST tag can be cleaved off by thrombin to leave three amino acids (Gly-Ser-His) prior to the first methionine of HNMT. The Ile105 variant was engineered by PCR-based site-directed mutagenesis and verified by DNA sequencing.
E. coli DH5α cells containing HNMT were induced by 0.1 mM IPTG at an OD600 of 0.4–0.7 in LB medium at 37°C. Three columns were used for purification: glutathione sepharose, MonoQ (20 mM Tris [pH 7.4]), and a gel-filtration Superdex 200HR column. Either MonoS (10 mM MES [pH 5.5]) or an inhibitor (amodiaquine-sepharose 6B)-based affinity column replaced the MonoQ step in some preparations. The protease digestion by thrombin (Calbiochem) allowed the cutting of the bound GST-HNMT on a Glutathione Sepharose 4B column. Approximately 3–5 mg of pure protein was obtained from 1 liter of induced culture. The selenomethionine (SeMet) derivative of GST-HNMT was expressed in methionine auxotrophs strain DL41(DE3) with a defined medium supplemented with 50 μg/ml SeMet [52].
For binding of histamine and AdoHcy, HNMT (~1 mg/ml or 30 μM) was mixed with 50 mM histamine and 0.6 mM AdoHcy in 15 mM Tris-HCl (pH 7.4), 1 mM DTT, and 1% polyethylene glycol (PEG) 400. The mixture was further concentrated to 15–20 mg/ml HNMT (0.45–0.6 mM), based on OD280 absorption and a calculated extinction coefficient of 41,610 M−1cm−1 [53]. We kept the ratio of enzyme to AdoHcy at approximately 1:1 and saturated the binary complex with histamine or quinacrine (~80-fold molar excess) for crystallization by vapor diffusion using the hanging-drop method. For the conditions of crystallization, see Tables 1 and 2.
Crystallography
All crystals had space group P6, with two molecules in the asymmetric unit. Four wavelength 3.27 Å SeMet anomalous diffraction data were collected and processed by using HKL [54] (Table 1). SOLVE [55] found 15 Se sites while CNS [56] found 16 sites out of 20 in the asymmetric unit. SOLOMON solvent-flipping [57] was used for density modification. The SeMet residues served as markers in the primary sequence during tracing. Model building with O [58] and refinement with CNS [56] continued by using a 2.3 Å data set from a crystal of the Thr105 HNMT-histamine-AdoHcy complex and a 1.9 Å data set from a crystal of the Ile105 HNMT-quinacrine-AdoHcy (Table 2).
Methylation Assay
A radiometric procedure was utilized to measure HNMT activity [59, 60, 17]. We used [methyl-3H]-AdoMet to form radiolabeled methylhistamine. The product was isolated from unincorporated [methyl-3H]-AdoMet by organic extraction by using toluene and isopentanol and quantified by liquid scintillation spectrometry. Typically, a 100 μl reaction mixture was incubated at 37°C for 20 min and contained the following components: 15 nM HNMT, 40 μM histamine, 40 μM unlabeled AdoMet, 0.125 μCi labeled AdoMet, 125 mM Bicine (pH 8.2), 1 mM EDTA, and 0.025% bovine serum albumin. The reaction was terminated with 75 μl of 2.5 M sodium borate (pH 11.0) and 1.25 ml of toluene:isopentanol (1:1). After the layers had separated, 1 ml of the organic phase was transferred to another tube containing 250 μl of 0.5 M HCl. Again, the aqueous and organic layers were allowed to separate, and then 200 μl of the aqueous phase was mixed with 10 ml of ICN-CytoScint scintillation fluid and quantified by liquid scintillation counting. Each point shown was the mean of two determinations.
The pH optimization (Figure 7a) was carried out in 125 mM buffers of varying pH (sodium acetate, pH 4–5; sodium citrate, pH 5.5–6.5; HEPES, pH 7–7.8; Bicine, pH 8–9; glycine, pH 9.5–10). Running the reaction at different temperatures for 20 min allowed temperature optimization (Figure 7b).
Thermal stability (Figure 7c) was assayed by incubation of the enzymes at a given temperature for 30 min, quenching on ice, and then running the reaction at 37°C. In addition, the thermal stability was expressed as heated/control (H/C) ratios (Figure 7d) using the 25°C (room temperature) as the control temperature.
The KM and Vmax values (Table 3) were determined from Line-weaver-Burke plots. HNMT (15 nM) was preincubated with histamine (40 μM) or AdoMet (40 μM). Addition of AdoMet (5, 10, 15, 20, 30, 40, 80, and 100 μM) or histamine (1, 2, 4, 5, and 10 μM) started the reaction.
For each inhibitor (AdoHcy, methylhistamine, or quinacrine), inhibition was examined as a function of concentration of AdoMet or histamine for both HNMT variants. The inhibitor (for each indicated concentration in Figures 7e–7g) was preincubated with HNMT (15 nM) and histamine (40 μM) or AdoMet (40 μM). The addition of AdoMet (20, 30, 40, and 80 μM) or histamine (2, 5, 10, and 15 μM or 5, 10, 20, 40, and 100 μM) started the reaction. The slope of each linear fit was plotted against the concentration of inhibitor and the intercept on the x axis gave an estimate of Ki (shown in Table 4).
Pre-steady-state kinetics (Figure 7h) were studied under the conditions of HNMT (2 μM), histamine (40 μM), unlabeled AdoMet (40 μM), and 0.5 μCi labeled AdoMet. HNMT was preincubated with histamine or AdoMet, and the addition of AdoMet or histamine started the reaction.
Acknowledgments
We thank Paul Kearney (Emory University) for cloning and expression and Michael Becker (Brookhaven National Laboratory) for FedEx remote X-ray data collection at beamline X12C in the National Synchrotron Light Source. We thank Dieter Schneider (Brookhaven National Laboratory), Joel Berendzen and Leon Flaks (Los Alamos National Laboratory), and Lisa Keefe (Illinois Institute of Technology), respectively, for help with X-ray data collection at beamlines X26C (NSLS), X8C (NSLS), and IMCA (Advanced Photon Source). We thank Robert M. Blumenthal (Medical College of Ohio) and two referees for critical comments on the manuscript. These studies were supported in part by the Emory University School of Medicine Department of Biochemistry start-up funds, the Georgia Research Alliance, the National Science Foundation INT-0003815, and the Japan Society for the Promotion of Science.
Footnotes
Accession Numbers
The atomic coordinates of the two ternary complexes of HNMT(Thr105)/Histamine/AdoHcy (pH 5.6) and HNMT(Ile105)/Quinacrine/AdoHcy have been deposited in the Protein Data Bank under accession numbers 1JQD and 1JQE, respectively.
References
- 1.Moss RB. Allergic etiology and immunology of asthma. Ann Allergy. 1989;63:566–577. [PubMed] [Google Scholar]
- 2.Rangachari PK. Histamine: mercurial messenger in the gut. Am J Physiol. 1992;262:G1–13. doi: 10.1152/ajpgi.1992.262.1.G1. [DOI] [PubMed] [Google Scholar]
- 3.Okinaga S, Sasaki H, et al. The role of HMT (histamine N-methyltransferase) in airways: a review. Methods Find Exp Clin Pharmacol. 1995;17(Suppl C):16–20. [PubMed] [Google Scholar]
- 4.Yamauchi K, Takishima T, et al. Structure and function of human histamine N-methyltransferase: critical enzyme in histamine metabolism in airway. Am J Physiol. 1994;267:L342–349. doi: 10.1152/ajplung.1994.267.3.L342. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz JC, Arrang JM, Garbarg M, Pollard H, Ruat M. Histaminergic transmission in the mammalian brain. Physiol Rev. 1991;71:1–51. doi: 10.1152/physrev.1991.71.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Passani MB, Bacciottini L, Mannaioni PF, Blandina P. Central histaminergic system and cognition. Neurosci Biobehav Rev. 2000;24:107–113. doi: 10.1016/s0149-7634(99)00053-6. [DOI] [PubMed] [Google Scholar]
- 7.Brown RE, Stevens DR, Haas HL. The physiology of brain histamine. Prog Neurobiol. 2001;63:637–672. doi: 10.1016/s0301-0082(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 8.Barnes PJ. Anti-IgE therapy in asthma: rationale and therapeutic potential. Int Arch Allergy Immunol. 2000;123:196–204. doi: 10.1159/000024444. [DOI] [PubMed] [Google Scholar]
- 9.Wood-Baker R. Histamine and its receptors. In: Busse WW, Holgate ST, editors. Asthma and Rhinitis. Oxford, England: Blackwell Science; 2000. pp. 961–67. [Google Scholar]
- 10.Kaplan AP, Beaven MA. In vivo studies of the pathogenesis of cold urticaria, cholinergic urticaria, and vibration-induced swelling. J Invest Dermatol. 1976;67:327–332. doi: 10.1111/1523-1747.ep12514352. [DOI] [PubMed] [Google Scholar]
- 11.Hill SJ, Haas HL, et al. International union of pharmacology. XIII. classification of histamine receptors. Pharmacol Rev. 1997;49:253–278. [PubMed] [Google Scholar]
- 12.Beaven MA. Histamine: its role in physiological and pathological processes. Monogr Allergy. 1978;13:1–113. [PubMed] [Google Scholar]
- 13.Weinshilboum RM, Otterness DM, Szumlanski CL. Methylation pharmacogenetics: catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu Rev Pharmacol Toxicol. 1999;39:19–52. doi: 10.1146/annurev.pharmtox.39.1.19. [DOI] [PubMed] [Google Scholar]
- 14.Ligneau X, Schwartz J-C, et al. Neurochemical and behavioral effects of ciproxifan, a potent histamine H3-receptor antagonist. J Pharmacol Exp Ther. 1998;287:658–666. [PubMed] [Google Scholar]
- 15.Panula P, et al. Relja M. Neuronal histamine deficit in Alzheimer’s disease. Neuroscience. 1998;82:993–997. doi: 10.1016/s0306-4522(97)00353-9. [DOI] [PubMed] [Google Scholar]
- 16.Morisset S, et al. Arrang JM. High constitutive activity of native H3 receptors regulates histamine neurons in brain. Nature. 2000;408:860–864. doi: 10.1038/35048583. [DOI] [PubMed] [Google Scholar]
- 17.Verburg KM, Henry DP. Histamine N-methyltransferase. enzymology and functional aspects. In: Boulton AA, Baker GB, Yu PH, editors. Neuromethods 5 Neurotransmitter Enzymes. Clifton, NJ: Humana Press; 1986. pp. 147–204. [Google Scholar]
- 18.Ganellin CR. Chemistry and structure-activity relationship of drugs acting at histamine receptors. In: Ganellin CR, Parsons ME, editors. Pharmacology of Histamine Receptors. Bristol, UK: Wright PSG; 1982. pp. 10–102. [Google Scholar]
- 19.Arrang JM, Garbarg M, Schwartz JC. Auto-inhibition of brain histamine release mediated by a novel class (H3) of histamine receptor. Nature. 1983;302:832–837. doi: 10.1038/302832a0. [DOI] [PubMed] [Google Scholar]
- 20.Preuss CV, Weinshilboum RM, et al. Human histamine N-methyltransferase pharmacogenetics: common genetic polymorphisms that alter activity. Mol Pharmacol. 1998;53:708–717. doi: 10.1124/mol.53.4.708. [DOI] [PubMed] [Google Scholar]
- 21.Scott MC, Van Loon JA, Weinshilboum RM. Pharmacogenetics of N-methylation: heritability of human erythrocyte histamine N-methyltransferase activity. Clin Pharmacol Ther. 1988;43:256–262. doi: 10.1038/clpt.1988.30. [DOI] [PubMed] [Google Scholar]
- 22.Price RA, Scott MC, Weinshilboum RM. Genetic segregation analysis of red blood cell (RBC) histamine N-methyltransferase (HNMT) activity. Genet Epidemiol. 1993;10:123–131. doi: 10.1002/gepi.1370100205. [DOI] [PubMed] [Google Scholar]
- 23.King CL, Xianli J, Malhotra I, Liu S, Mahmoud AA, Oettgen HC. Mice with a targeted deletion of the IgE gene have increased worm burdens and reduced granulomatous inflammation following primary infection with schistosoma mansoni. J Immunol. 1997;158:294–300. [PubMed] [Google Scholar]
- 24.Yan L, Galinsky RE, Bernstein JA, Liggett SB, Weinshilboum RM. Histamine N-methyltransferase pharmacogenetics: association of a common functional polymorphism with asthma. Pharmacogenetics. 2000;10:261–266. doi: 10.1097/00008571-200004000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Morrow JD, Margolies GR, Rowland J, Roberts LJ. Evidence that histamine is the causative toxin of scombroid-fish poisoning. N Engl J Med. 1991;324:716–720. doi: 10.1056/NEJM199103143241102. [DOI] [PubMed] [Google Scholar]
- 26.Smart DR. Scombroid poisoning. a report of seven cases involving the Western Australian salmon, Arripis truttaceus. Med J Aust. 1992;157:748–751. doi: 10.5694/j.1326-5377.1992.tb141274.x. [DOI] [PubMed] [Google Scholar]
- 27.Takemura M, Wada H, et al. Histamine N-methyltransferase from rat kidney. Cloning, nucleotide sequence, and expression in Escherichia coli cells. J Biol Chem. 1992;267:15687–15691. [PubMed] [Google Scholar]
- 28.Kitanaka J, Kitanaka N, Tsujimura T, Kakihana M, Terada N, Takemura M. Guinea pig histamine N-methyltransferase: cDNA cloning and mRNA distribution. Jpn J Pharmacol. 2001;85:105–108. doi: 10.1254/jjp.85.105. [DOI] [PubMed] [Google Scholar]
- 29.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.18.3784. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schluckebier G, O’Gara M, Saenger W, Cheng X. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J Mol Biol. 1995;247:16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- 31.Fauman EC, Blumenthal RM, Cheng X. Structure and evolution of AdoMet-dependent methyltransferases. In: Cheng X, Blumenthal RM, editors. S-adenosylmethionine-dependent Methyltrasferases: structures and functions. River Edge, NJ: World Scientific Publishing; 1999. pp. 1–38. [Google Scholar]
- 32.Vidgren J, Svensson LA, Liljas A. Crystal structure of catechol O-methyltransferase. Nature. 1994;368:354–358. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- 33.Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 34.Djordjevic S, Stock AM. Chemotaxis receptor recognition by protein methyltransferase CheR. Nature Struct Biol. 1998;5:446–450. doi: 10.1038/nsb0698-446. [DOI] [PubMed] [Google Scholar]
- 35.Grossman MH, Szumlanski C, Littrell JB, Weinstein R, Weinshilboum RM. Electrophoretic analysis of low and high activity forms of catechol-O-methyltransferase in human erythrocytes. Life Sci. 1992;50:473–480. doi: 10.1016/0024-3205(92)90386-4. [DOI] [PubMed] [Google Scholar]
- 36.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 37.Harvima RJ, Kajander EO, Harvima IT, Fraki JE. Purification and partial characterization of rat kidney histamine-N-methyltransferase. Biochim Biophys Acta. 1985;841:42–49. doi: 10.1016/0304-4165(85)90272-7. [DOI] [PubMed] [Google Scholar]
- 38.Coward JK. Chemical mechanisms of methyl transfer reactions: comparison of methylases with nonenzymic “model reactions”. In: Salvatore F, Borek E, Zappia V, Williams-Ashman HG, Schlenk F, editors. The biochemistry of adenosylmethionine. New York: Columbia University Press; 1977. pp. 127–144. [Google Scholar]
- 39.Weichsel A, Andersen JF, Champagne DE, Walker FA, Montfort WR. Crystal structures of a nitric oxide transport protein from a blood-sucking insect. Nature Struct Biol. 1998;5:304–309. doi: 10.1038/nsb0498-304. [DOI] [PubMed] [Google Scholar]
- 40.Paesen GC, Adams PL, Harlos K, Nuttall PA, Stuart DI. Tick histamine-binding proteins: isolation, cloning, and three-dimensional structure. Mol Cell. 1999;3:661–671. doi: 10.1016/s1097-2765(00)80359-7. [DOI] [PubMed] [Google Scholar]
- 41.Smit MJ, Hoffmann M, Timmerman H, Leurs R. Molecular properties and signalling pathways of the histamine H1 receptor. Clin Exp Allergy. 1999;29(Suppl 3):19–28. [PubMed] [Google Scholar]
- 42.Ohta K, Hayashi H, Mizuguchi H, Kagamiyama H, Fujimoto K, Fukui H. Site-directed mutagenesis of the histamine H1 receptor: roles of aspartic acid107, asparagine198 and threonine194. Biochem Biophys Res Commun. 1994;203:1096–1101. doi: 10.1006/bbrc.1994.2295. [DOI] [PubMed] [Google Scholar]
- 43.Gantz I, Yamada T, et al. Molecular basis for the interaction of histamine with the histamine H2 receptor. J Biol Chem. 1992;267:20840–20843. [PubMed] [Google Scholar]
- 44.Taylor KM, Snyder SH. Histamine methyltransferase: inhibition and potentiation by antihistamines. Mol Pharmacol. 1972;8:300–310. [PubMed] [Google Scholar]
- 45.Cohn VH. Inhibition of histamine methylation by antimalarial drugs. Biochem Pharmacol. 1965;14:1686–1688. doi: 10.1016/0006-2952(65)90026-2. [DOI] [PubMed] [Google Scholar]
- 46.Lotta T, et al. Taskinen J. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry. 1995;34:4202–4210. doi: 10.1021/bi00013a008. [DOI] [PubMed] [Google Scholar]
- 47.Gitomer WL, Tipton KF. Purification and kinetic properties of ox brain histamine N-methyltransferase. Biochem J. 1986;233:669–676. doi: 10.1042/bj2330669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Francis DM, Thompson MF, Greaves MW. The kinetic properties and reaction mechanism of histamine methyltransferase from human skin. Biochem J. 1980;187:819–828. doi: 10.1042/bj1870819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beaven MA, Shaff RE. Inhibition of histamine methylation in vivo by the dimaprit analog, SKF Compound 91488. Agents Actions. 1979;9:455–460. doi: 10.1007/BF01968110. [DOI] [PubMed] [Google Scholar]
- 50.Thithapandha A, Cohn VH. Brain histamine N-methyltransferase purification, mechanism of action, and inhibition by drugs. Biochem Pharmacol. 1978;27:263–271. doi: 10.1016/0006-2952(78)90227-7. [DOI] [PubMed] [Google Scholar]
- 51.Tachibana T, Taniguchi S, Imamura S, Fujiwara M, Hayashi H. Effects of drugs on the activity of histamine-N-methyltransferase from guinea pig skin. Biochem Pharmacol. 1988;37:2872–2876. doi: 10.1016/0006-2952(88)90056-1. [DOI] [PubMed] [Google Scholar]
- 52.Hendrickson WA, Horton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwave-length anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 54.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.Terwilliger TC, Berendzen J. Automated MAD and MIR sructure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brünger AT, et al. Warren GL. Crystallography and NMR system: a new suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 57.Abrahams JP, Leslie AGW. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 58.Jones TA, Kjeldgaard M. Electron density interpretation. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 59.Verburg KM, Bowsher RR, Henry DP. A new radioenzymatic assay for histamine using purified histamine N-methyltransferase. Life Sci. 1983;32:2855–2867. doi: 10.1016/0024-3205(83)90322-3. [DOI] [PubMed] [Google Scholar]
- 60.Bowsher RR, Verburg KM, Henry DP. Rat histamine N-methyltransferase. Quantification, tissue distribution, purification, and immunologic properties. J Biol Chem. 1983;258:12215–12220. [PubMed] [Google Scholar]
- 61.Carson M. Ribbons. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- 62.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Protein Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 63.Girard B, Otterness DM, Wood TC, Honchel R, Wieben ED, Weinshilboum RM. Human histamine N-methyltransferase pharmacogenetics: cloning and expression of kidney cDNA. Mol Pharmacol. 1994;45:461–468. [PubMed] [Google Scholar]