Abstract
Objective
We investigated associations of early pregnancy maternal vitamin D concentrations with differential gene expression and post transcription regulation.
Method
Plasma 25-hydroxyvitamin D (25[OH]D) was measured among participants of a nested case-control study. Participants with low (<25.5 ng/ml) and high (≥31.7 ng/ml) 25[OH]D were identified among controls. Peripheral blood messenger RNA (mRNA) (N=21) and microRNA (miRNA) (N=13) expression studies were conducted among participants with low and high 25[OH]D concentrations. Differential expression between low/high groups were evaluated using Student’s T-test, fold change and SAM comparisons. We further investigated functions and functional relationships of differentially expressed mRNAs and targets of differentially expressed miRNAs.
Results
305 genes (299 up-regulated and 6 down-regulated) and 11 miRNAs (10 down-regulated and 1 up-regulated) were differentially expressed among participants with low 25[OH]D compared with those who had high 25[OH]D. Genes that participate in a wide range of cellular functions, including organ and system development (e.g. angiogenesis), inflammation and metabolic processes (e.g. carbohydrate/lipid metabolism), as well as miRNAs that target these genes were differentially expressed among women with low 25[OH]D compared with those with high 25[OH]D.
Conclusion
Early pregnancy plasma 25[OH]D concentrations are associated with maternal peripheral blood gene expression and post-transcription regulation.
Keywords: vitamin D, transcription, post-transcription regulation, microarray, early pregnancy, whole blood
Introduction
Vitamin D, a fat soluble steroid-like hormone, is a nuclear transcription factor that exerts its effects through vitamin D receptors present throughout the body and through vitamin D response elements found on hundreds of human genes [1–3]. Vitamin D influences growth, differentiation and optimal function of cells such as vascular endothelial and pancreatic beta cells, potentially playing major roles in angiogenesis, inflammation and glucose homeostasis [2]. These pleiotropic effects of vitamin D may account for associations of vitamin D insufficiency and deficiency with a number of health complications, including pregnancy related maternal, perinatal, neonatal, and child health complications.
Maternal vitamin D deficiency or insufficiency has been associated with pregnancy complications such as gestational diabetes, preeclampsia and bacterial vaginosis [3–4]. Similarly maternal vitamin D deficiency/insufficiency has also been associated with developmental origins of metabolic disorders, osteoporosis and neuropsychiatric diseases in the offspring [3, 5–7]. Vitamin D deficiency or insufficiency is highly prevalent in pregnancy, as high as 69% by some accounts [8]. Despite the wide spread prevalence of vitamin D insufficiency or deficiency in pregnancy and the strong evidence supporting its adverse consequences, research in this area is sparse. In particular, mechanistic studies (e.g. gene expression) that may enhance understanding of associations of low vitamin D with pregnancy complications are lacking.
Previous vitamin D related gene expression and post-transcription regulation studies were mostly conducted under experimental conditions [9–14]. Most of these studies evaluated expressions of few candidate genes or microRNAs (miRNAs) (e.g. PDGFA, Sorcin, IGF, TGFB, miR-125b) selected based on a priori available evidence. Extrapolations to human populations and physiologic conditions (including pregnancy) are not straightforward or at times possible. In particular, investigations did not address vitamin D related gene expression variation and/or post-transcription regulation during the critical time frame of early gestation when initial events of most pregnancy complications and fetal programming occur.
In two independent pilot microarray-based studies that allow for whole genome evaluations, we investigated associations of early pregnancy maternal plasma 25-hydroxyvitamin D (25[OH]D) concentrations with differential gene (messenger RNA, mRNA) expression and post-transcriptional regulation (miRNA). We further investigated functions and functional relationships of differentially expressed mRNAs and targets of differentially expressed miRNAs.
Materials and Methods
Study subjects were selected from participants of a nested case-control study designed to investigate associations of gestational diabetes and vitamin D concentrations. Study population and data collection procedures, described before, were briefly as follows [4]. Participants were women who attended prenatal care clinics affiliated with Swedish Medical Center, Seattle, WA and were enrolled in the Omega Study, a prospective cohort study, between September 2002 and October 2004. Women who initiate prenatal care prior to 20 weeks gestation were eligible to participate. Ineligibility criteria included younger than 18 years of age, not speaking or reading English, not planning to carry the pregnancy to term, and/or not planning to deliver at the Swedish Medical Center. Exclusion criteria of the nested case control study included history of diabetes, hypertension or other chronic diseases. Cases and controls were frequency matched for season of blood collection. Potential participants for the microarray studies were selected among controls (i.e., women who did not develop gestational diabetes or preeclampsia during the index pregnancy) of the nested case control study. The Institutional Review Board of the Swedish Medical Center approved study protocols. All participants provided written informed consent.
Information on risk factors was collected using in-person interviews, blood collection and medical records abstraction. Following enrollment, in-person interviews were conducted to collect data on risk factors including socio-demographic characteristics, reproductive and medical histories, and family histories of medical conditions. After delivery, trained personnel abstracted data from participants’ maternal and infant medical records to ascertain pregnancy outcomes. Maternal non-fasting blood samples, collected in 10-mL Vacutainer tubes containing ethylenediaminetetraacetic acid at 16 weeks gestation, were frozen at −80°C until analysis.
Samples of maternal whole blood were also collected in 2.5-mL PAXgene™ Blood RNA tubes (Qiagen Inc., Valencia, CA) for mRNA and miRNA expression studies. These samples were immediately stored at −80ºC until RNA extraction and analysis.
Plasma 25[OH]D concentrations were measured using DiaSorin enzyme immunoassay reagents and procedures (Metametrix, Norcross, GA). Intra-assay and inter-assay coefficients of variation for this method are both ≤ 12%. Participants with low 25[OH]D (<25.5 ng/ml) (10 for the mRNA study and 7 for the miRNA study) and high 25[OH]D (≥31.7 ng/ml) (11 for the mRNA study and 6 for the miRNA study) were identified.
The PAXgene Blood RNA Kit (Qiagen Inc, Valencia, CA) was used for extraction and purification of total RNA from whole blood, according to manufacturers’ protocols. The procedure, briefly, was as follows. Initial equilibration of samples to room temperature for two hours was followed by centrifugation of cell lysates. The pellets were washed and resuspended in optimized buffers and incubated with proteinase K to bring about protein digestion. Between wash steps, the silica-membrane is treated with DNase I to remove any residual DNA contaminants. After wash steps, pure RNA samples were eluted in a buffer, heat denatured at 65°C and immediately chilled on ice. The Ambion GLOBINclear™ Kit (patent pending) was used to rapidly deplete alpha and beta globin mRNA from total RNA preparations. The kit employs a novel hybridization technology that takes advantage of the strength of biotin/streptavidin binding, the specificity of nucleic acid hybridization, and the convenience of magnetic bead separations to deplete globin mRNA from blood total RNA. Extracted RNA was stored at −80° Total RNA concentration was determined by ultraviolet absorbance at 260 nm (A260) by direct measurement (i.e non-diluted) on a NanoDrop ND1000 spectrophotometer (ThermoFisher Scientific, Wilmington, DE). Purity of RNA was assessed by evaluating readings at 260 nm and 280 nm (A260/ A260). Samples with A260/280 values >1.8 were deemed acceptable. Agarose electrophoresis was also used to assess DNA contamination. Samples were then kept in frozen storage at −80°C. All RNA samples, including reference RNAs, underwent quality control checks and were labeled using the same standardized protocols.
RNA sample amplification, fragmentation and labeling were conducted using Ambion MessageAmp aRNA and Ambion amino allyl cDNA kits (Applied Biosystems, Foster City, CA). Briefly, the procedures were as follows. Total RNA was reverse transcribed using a primer containing both oligo(dT) and a T7 RNA polymerase promoter sequence. After first-strand synthesis, the reaction was treated with RNase H to cleave the RNA into small fragments. These small RNA fragments serve as primers during a second-strand synthesis reaction that produces a double-stranded cDNA template for transcription. The cDNA template was then used in in-vitro transcription reaction to produce linearly amplified aRNA. For the mRNA expression experiment, the reactive amino group of 5-(3-aminoallyl)-UTP/5-(3-aminoallyl)-dUTP was used to conjugate the purified aRNA/cDNA with the NHS-CyDye. For the miRNA experiment, Kreatech ULS™ miRNA Labeling Kit (Kreatech Diagnostics, Amsterdam, the Netherlands) was used. Labeling efficiency was calculated by the concentration of CyDye and aRNA/cDNA measured using NanoDrop ND-1000. Labeling efficiency >10 was considered adequate for further processing.
Expression of mRNA and miRNA were profiled using OneArray™ Human Whole Genome and OneArray™ miRNA microarray platforms, respectively (Phalanx Biotech, Palo Alto, CA). The OneArray™ Human Whole Genome array platform is comprised of 60-mer oligonucleotide probes (totaling 30,968 human genome and 1082 experimental control probes). The human gene probes are designed to hybridize to a specific target gene described in the latest public domain contents (e.g Unigene, Cancer genome Anatomy Project, BioCarta, Kyoto Encyclopedia of Gene and Genome) and validated by the Human Genome Sequencing Project. OneArray™ miRNA microarray platform is comprised of 1,040 unique miRNA probes and 108 experimental control probes. The unique miRNA probes include all human miRNAs represented in the Sanger miRBase database as well as miRNAs identified in 17 other primates. The experimental probes in both platforms are designed to monitor sample quality, hybridization and labeling reactions.
Following hybridization, arrays were scanned using Molecular Devices™ GenePix™ Scanner (Molecular Devices, Sunnyvale, CA) and data from each array was quantified using GenePix™ software (Molecular Devices, Sunnyvale, CA). Intensity values were pre-processed for analysis using Rosetta Resolver Error Models (Rosetta, Seattle, WA) [15], quality control filters and median-scale based normalization.
Analysis was conducted on normalized and log2-transformed data. Differences in mRNA expressions were evaluated using Student’s T-test, fold change (comparing low/high vitamin D groups) analysis and Significance Analysis of Microarrays (SAM) [16]. A volcano plot of fold changes and Students’ T-test p-values was used to assess differential expression of gene transcripts in the mRNA array study. At a conservative false discovery rate (FDR) of 1 per 30,968, only one gene was expected to have more extreme Students’ T-test p-value of 0.000032 (1/30,968). Genes (mRNA) that met the following three criteria in case-control comparisons constituted the final set of differentially expressed genes; Student’s T-test p-value < 0.05, absolute fold change differences ≥ 1.5, and, FDR in SAM analysis ≤ 5%. A phylogenetic tree (i.e., heat map) of differentially expressed genes was constructed using dChip software [17].
In the miRNA experiment, miRNAs that met the following two criteria in case-control comparisons constituted the final set of differentially expressed miRNA; Student’s T-test p-value < 0.05 and absolute fold change differences ≥ 1.5. Due to the relatively smaller number of comparisons in this experiment, we were not able to apply SAM comparisons. TargetScan (www.targetscan.org), a database that employs both conserved and non-conserved seed pairing algorithms, was used to identify putative miRNA targets of differentially expressed miRNA.
Functions and functional relationships of (1) differentially expressed genes and (2) genes targeted by three or more differentially expressed miRNAs were investigated, separately, using Ingenuity Pathway Analysis (IP A) (Ingenuity, Redwood City, CA). In IP A, significance of gene-enrichment of networks were determined using Fisher Exact tests comparing proportion of genes that fall under the network to the human genome. The corresponding network score (−log 10 [p-value) was then used to rank the biological significance of gene networks. We used an enrichment/network score of 3 (p-value = 0.001) as the cutoff for significance in our study.
Results
As can be seen in Table I, study participants, in low and high 25[OH]D exposure groups were similar with regards to maternal age, race/ethnicity, marital status, and parity. As expected, women with low plasma 25[OH]D concentrations, as compared with those who has higher values, had higher mean pre-pregnancy body mass index values.
Table I.
Study population characteristics
| Gene (mRNA) expression study
|
MicroRNA expression study
|
|||
|---|---|---|---|---|
| Characteristics | High Vitamin D (≥31.7 ng/ml)* |
Low Vitamin D (<25.5 ng/ml) |
High Vitamin D (≥31.7 ng/ml) |
Low Vitamin D (<25.5 ng/ml) |
| Number (N) | 11 | 10 | 6 | 7 |
| Age, years** | 34.18 (3.9) | 33.2 (5.5) | 34.33 (3.1) | 33.43 (2.4) |
| Gestational Age, weeks** | 15.91 (2.2) | 16.3 (2.5) | 15.14 (1.9) | 16.84 (2.1) |
| Pre-pregnancy BMI (kg/m2)** | 21.58 (3.1) | 23.39 (2.8) | 22.87 (3.7) | 26.40 (5.6) |
| Overweight (≥24kg/m2) | 1 (10%) | 3 (30%) | 2 (29%) | 1 (14%) |
| Non-Hispanic Whites | 11 (100%) | 10 (100%) | 6 (100%) | 7 (100%) |
| Married | 11 (100%) | 9 (90%) | 6 (100%) | 7 (100%) |
| Nulliparous | 5 (45%) | 4 (40%) | 4 (66.7%) | 3 (42.9%) |
| Gravidity** | 2.64 (1.62) | 2.10 (1.60) | 2.17 (2.4) | 2.00 (0.6) |
| Season of blood draw | ||||
| Spring/Summer | 5 (46.5) | 5 (50%) | 2 (33.3%) | 3 (42.9%) |
| Fall/Winter | 6 (64.5) | 5 (50%) | 4 (66.7%) | 4 (57.1%) |
| Plasma 25-hydroxyvitamin D (25-[OH] D) concentrations, ng/ml** | 38.84 (7.0) | 22.09 (2.6) | 39.22 (6.1) | 22.84 (2.0) |
mean (standard deviation), otherwise number (%)
Assessed by measuring maternal early pregnancy plasma 25-hydroxyvitamin D (25-[OH] D) concentrations
Abbreviations: GA=gestational age at blood draw, BMI=body mass index
The only statistically significant difference (p-value < 0.05) between the two groups in each experiment is the difference between vitamin D levels.
mRNA expression study
We initially used volcano plots to show relationships between fold change and Students’ T-test p-values. As can be seen in Figure 1, 70 genes (highlighted in green) with p-values <3.2 × 10−5 (−Log10[p-value]=4.49) reached extreme levels of statistical significance (1 FDR per 30,968). We observed both up and down regulation of gene expression according to maternal plasma 25[OH]D status. A total of 8,532 genes (27.6% of all genes) were differentially expressed (4,696 up-regulated and 3,836 down-regulated) according to the raw p-value criteria from Student’s T-test comparisons (p-value < 0.05) (Figure 2). A total of 882 genes (669 up-regulated and 213 down-regulated) had absolute fold change values ≥ 1.5 across the two study groups. In SAM analysis, FDR corresponding to expression differences of 311 genes (305 up-regulated and 6 down-regulated) was < 5%. As shown in the Venn diagram (Figure 2) a total of 305 genes (299 up-regulated and 6 down-regulated) that met all three criteria constituted our list of differentially expressed genes (Supplemental Table I). Gene expression fold change differences (comparing low/high vitamin D groups) in this set ranged from 2.63 (TRAC) to −1.81 (FKBP1B). This gene list includes genes known to be up-regulated by vitamin D (such as PDGFA) [20] in previous reports as well as novel ones (such as SLC27A4). We next evaluated the hierarchal clustering of selected differentially expressed genes and participants (Figure 3). As can be seen in the figure, based on similarity of expression profiles for selected differentially expressed genes, all but two participants with high vitamin D and all but one participant with low vitamin D clustered into the two main cluster groups. In IPA of functions and functional relationships of differentially expressed genes, 15 networks representing diverse sets of cellular functions were identified (Table II and Supplemental Figure 1). These networks are involved in functioning and development of various physiologic systems (hematologic, musculoskeletal, connective tissue, digestive, hepatic, neurological, cardiovascular and reproductive), cell-to-cell signaling, energy production and lipid/carbohydrate/amino acid metabolism. Abnormalities in these processes are associated with cancer, diabetes, inflammation/immune related disturbances and other metabolic disorders. Besides identified differentially expressed genes in our microarray study, some well described genes and proteins (such as NFKB, Estrogen receptor, VEGF, ERK1, TNF and Interferon alpha) play central roles in these over represented networks.
Figure 1. Volcano Plot of gene expression differences.
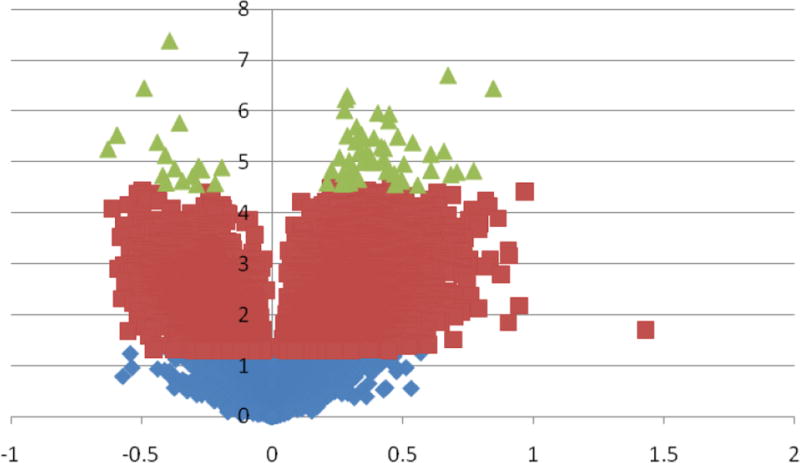
Distribution of Students’ T-test p-value (Y-axis: −Log10 [p-value]) and fold change (X-axis: Log2 [absolute fold change]) results comparing early pregnancy whole blood gene expressions of participants with low vitamin D concentrations (<25ng/ml) compared with those with high vitamin D concentrations (≥31.7ng/ml). The volcano plot indicates signal in the data as there were more genes with extreme p-values than would have been expected by chance. For example, there were 70 genes (green) with p-values more extreme than 0.000032 (−Log10 [p-value]=4.49) that corresponds to a conservative false discovery rate of 1/30968 (−Log10 [1/30968]=4.49). Purple genes are genes with t-test p-values > 0.000032 but less than < 0.05 and Blue genes are genes with t-test p-values >0.05.
Figure 2. Venn diagram summary of distribution of differentially expressed genes.
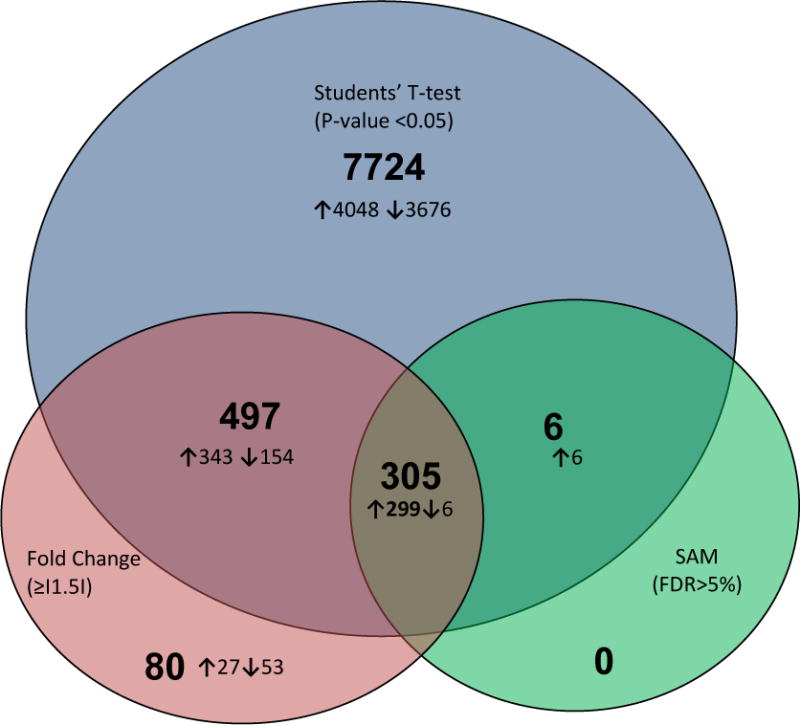
Circles represent numbers of differentially expressed genes in whole blood among pregnant women with low vitamin D concentrations (<25ng/ml) compared with pregnant women with high vitamin D concentrations (≥31.7ng/ml) in Students’ t-test (p-value < 0.05) (blue), fold change (absolute fold change ≥ 1.5) (purple) and Significance Analysis of Microarray (SAM, false discovery rate < 5%) (green) comparisons. Numbers within circles represent total number of genes and numbers of either up-regulated (↑) or down-regulated (↓) genes. The intersections of the circles represent numbers of genes differentially expressed using two or greater than two criteria as defined above.
Figure 3. Hierarchical clustering of participants and gene (mRNA) expressions.
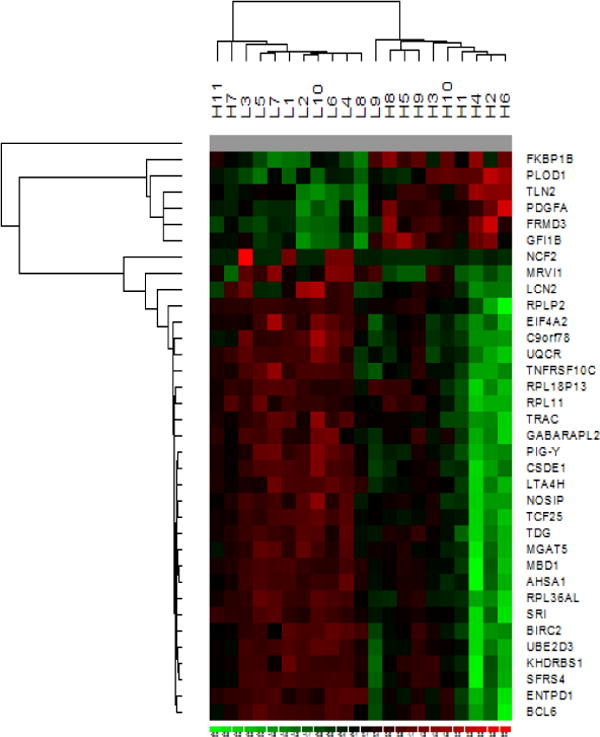
Hierarchical clustering of participants (rows) (L with <25.5ng/ml and H with ≥31.7ng/ml 25[OH]D concentrations) and selected differentially expressed genes (columns) (down-regulated genes and ≥2-fold up-regulated genes) based on similarity of expression profiles. The analysis was conducted using dChip software. The color code for signal strength in the classification scheme is as follows; induced genes are indicated by shades of red while repressed genes are indicated by shades of green. Gray represents absent data.
Table II.
Gene networks overrepresented by differentially expressed genes
| # | Genes in Network* | Score | Focus genes | Functions |
|---|---|---|---|---|
| 1 | 26s Proteasome, BCR, BIRC2, CASP5, CASP6, Caspase, CD79B, COX5B, DNAJC3, DYNC1I2, DYNLL1, FKBP5, GLA, Hsp70, Hsp90, HSPA8, Igm, Ikb, IL18RAP, LRRK2, LTA4H, NFkB (complex), NFKBIA, NFKBIZ, NGLY1, PCYT1A, peptidase, PI3, RNF38, ST13, TNFRSF10C, TPP1, TXNRD1, UBE2D3 (includes EG:7323), Ubiquitin | 44 | 25 | Cellular Function and Maintenance, Hematological System Development and Function, Hematopoiesis |
| 2 | ACTR2, ACTR3, AIM1 (includes EG:202), Arp2/3, ARPC2, CASC3, CSDE1, CTSH, Cyclin A, Cyclin E, E2f, E2F3, EAPP, EGLN2, ELF4, Estrogen Receptor, EXOSC8, F Actin, GK, HIRA, HISTONE, Histone h3, Histone h4, IFI16, MBD1, MEF2C, NCOA1, P38 MAPK, PLEKHO1, Rb, RNA polymerase II, SMARCA4, TAF5L, TMEM126A, Vegf | 35 | 22 | Cellular Development, Skeletal and Muscular System Development and Function, Cell-To-Cell Signaling and Interaction |
| 3 | AHSA1, ATP5I, Calcineurin protein(s), Calpain, Cbp/p300, CD3, CD52, CEBPB, CLNS1A, CREB1, EIF4A2, Eif4g, ENTPD1, ERK1/2, GC-GCR dimer, HNRNPA3, LAT2, MKNK1, Nfat (family), NFATC3, NMI, PDGFA, Pias, PRKCH, RAB1A, RPS6, Rsk, RUNX3, SLC2A3, SNRPG, STAT5a/b, TCF7, TCR, TDG, VitaminD3-VDR-RXR | 32 | 22 | Digestive System Development and Function, Gene Expression, Energy Production |
| 4 | AQP3, ATP9A, CXORF26, DHRS3, EBNA1BP2, FNG, FSH, GDE1, GFI1B, GK7P, GRB2, GTPBP4, H1F0, hCG, Lh, LOC81691, LYAR, NOTCH1, PAPSS1 (includes EG:9061), PFKFB3, Pka, PP1 protein complex group, PPP1R16A, QRFP, RASSF2, RB1, RBM33, RFNG, SRP9, SRP14, SULF2, SUV420H1, TP53I11, VEGFA, ZNF143 | 23 | 16 | Cellular Development, Cellular Growth and Proliferation, Cancer |
| 5 | Alp, Ap1, ASCC2, ATL3, BCL6, BMP2K (includes EG:55589), Calmodulin, CYP1B1, ERK, GTPBP8, IFN Beta, IgG, IL1, IL12 (complex), IL1RN, IL4R, Immunoglobulin, Insulin, Interferon alpha, Jnk, KHDRBS1, KLHL2, LDL, Mapk, MDFIC, MIR124, Mmp, MMP25, Pdgf, PDGF BB, PI3K, Pkc(s), PTBP1, SEMA4D, VAMP3 | 22 | 15 | Cellular Development, Genetic Disorder, Inflammatory Disease |
| 6 | BZW2, C19ORF60, CEP63, COX11, COX15, COX10 (includes EG:1352), COX4I2, COX6A2, COX6B1, COX6B2, COX7A1, COX7A2, COX7A2L, COX7B, COX7B2 (includes EG:170712), COX8A, COX8B, COX8C, CTNNB1, Cytochrome c oxidase, FOXP1, HTT, ILF3, MAST3, MBTPS1, MDFI (includes EG:4188), MGAT5, MUT, NDUFB10, NPTN, PTCHD2, RASGRP2, TOMM7, TOMM20, WAC | 21 | 15 | Genetic Disorder, Neurological Disease, Skeletal and Muscular Disorders |
| 7 | ANP32B, CCDC88B, CD48, CD53, CDH1, CHTF18, CMKLR1, DDX11, DDX18, EEF1G, FAM160B1, GABARAPL2 (includes EG:11345), GBAS, HIVEP2, HNRNPAB, HPRT1, IL15, L-triiodothyronine, MGST3, MRPS22, MYC, NIPSNAP1, NOSIP, PDCD5, PFKP, Rfc, RFC2, RFX1, RFX2, RPL22, SCD2, SRM, TP53I3, TRIP12, TSPAN7 | 20 | 14 | DNA Replication, Recombination, and Repair, Gene Expression, Cell Death |
| 8 | AHCYL1, AIP, BTF3, CD93, Ck2, CLCN7, CYC1, CYTB, Cytochrome bc1, FRG1, FXR1, GPR56, HCG 25371, HMGN2, HOXA9, MEGF11, NUTF2, PTPRCAP, RPL37, RPL37A (includes EG:6168), SSB, SUGT1, TAL1, ubiquinol-cytochrome-c reductase, UQCR10, UQCR11, UQCRB, UQCRC1, UQCRC2, UQCRFS1, UQCRFSL1, UQCRH, UQCRQ, VHL | 18 | 13 | Cellular Function and Maintenance, Genetic Disorder, Metabolic Disease |
| 9 | ABHD6, APEH, BCCIP, C11ORF58, C9ORF78, CDK2, DPM1, DPM2, EIF5, GPX2, GPX7, GSTZ1 (includes EG:2954), HNF4A, KAP, KIAA0174, KIAA1012, LAPTM4A, LCMT2, MGST2, MGST3, MIPEP, MPP1, MRPL22, NAA25, NLN, PNO1, REXO2, RNGTT, SAP30BP, SLC35A1, SPCS3, SUPT5H, UBA5, USMG5, VPS29 | 18 | 13 | Genetic Disorder, Hepatic System Disease, Liver Cholestasis |
| 10 | ACSM3, ADAMDEC1, ADAMTS3, ANXA8, ART3, B3GNT1, beta-estradiol, C11ORF10, CNDP2, EGFBP2, FAM105B, FLOT2, GRAMD1C, HSF2BP, ITM2A, KDELR3, KRAS, LEPROT, METTL7A, MIR192 (includes EG:387187), MIR292 (includes EG:100049711), MME, MRVI1, MSGN1, MTMR4, MTRR, NPL, PNRC2 (includes EG:55629), progesterone, SOLH, SRI, TGFB1, TMEM164, WFDC2, ZEB2 | 18 | 13 | Cancer, Cardiovascular System Development and Function, Cell Morphology |
| 11 | ACAA1, ACSBG1, ACSL4, ATP6V1B2, C14ORF166, C6ORF48, CAPRIN1, CEP170, CFTR, CLTA, EMG1, ESYT1, GLG1, IDH2, MAN2B1, palmitic acid, PLA2G4C, PLOD1, PNPLA3, PSMB7, RPLP2, RPLP0 (includes EG:11837), RPS8, RPS18, SFXN3, SLC25A38, SLC27A2, SLC27A4, SLC2A4, TLN2, TMED2, TMED9, TMEM43, TMLHE, YWHAZ | 16 | 12 | Lipid Metabolism, Small Molecule Biochemistry, Carbohydrate Metabolism |
| 12 | AIFM3, amino acids, AMY1A, AMY2A, AMY2B (includes EG:280), ascorbic acid, B3GNT2, BHMT, CASP3, CASP8AP2, CDK11A, CUL1, DAPK2 (includes EG:23604), DNASE1L3, DPP7, DUSP22, EBAG9, EIF2AK1, EMILIN2, FBXL3, GMFG, IP6K3, MIRN101B, NR3C1, OOEP, PRKAR1B, RAB11A, RABGGTB, SEC62, SPPL2B, SRP72, SSH2, STK17B, TAOK1, WDR26 | 16 | 12 | Amino Acid Metabolism, Post-Translational Modification, Small Molecule Biochemistry |
| 13 | ABCB8, C14ORF106, CABC1, COX7A2, CTSW, DOK3, FAM86C, FKBP1B, HMGB2, HNF4A, IL2, INS1, LOC340571, MICAL1, NDUFB1, NDUFB5, POLE3, PPARGC1A, PPIL1, SAT1, SAT2, SDCCAG3 (includes EG:10807), SFRS4, SH3BGRL2, TACO1, TCF25, TMEM66, TP53, TP53INP1, TP53RK (includes EG:112858), TPRKB, VAMP5, VIM, WBP2, ZNF175 | 15 | 12 | Carbohydrate Metabolism, Hepatic System Development and Function, Organismal Functions |
| 14 | 5430435G22RIK, BTN3A3, CHST2, CIITA, CST7, CXCL10, DPP8, ERBB2, FNBP1, GBP6, GBP1 (includes EG:14468), GBP4 (includes EG:17472), IFI203, IFIT5, IFNB1, IRF6, MAP3K14, NNMT, OAS3 (includes EG:4940), PUM1, RHOV, RNF149, RNU1-1, RPL10L, RPL27A, RPL36AL, RPS15A (includes EG:6210), SEMA4C, SERPINB1, ST3GAL6, TNF, WFS1, ZNF330, ZXDA, ZXDC | 14 | 11 | Inflammatory Disease, Inflammatory Response, Connective Tissue Disorders |
| 15 | ANKRD13A, ARL4D, C20ORF30, CDS2, DUB, EML4, EP300, LAPTM5, NFYB, RBM14, SNX6, USP6, USP17, USP26, USP29, USP30, USP32, USP34, USP35, USP36, USP38, USP40, USP41, USP42, USP43, USP44, USP45, USP47, USP50, USP13 (includes EG:8975), USP27X, USP37 (includes EG:57695), USP51 (includes EG:158880), YIPF4, ZFP36 | 14 | 11 | Genetic Disorder, Reproductive System Disease, Amino Acid Metabolism |
The networks were generated using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). These genes were overlaid onto a global molecular network developed from information contained in the IPKB. Network enrichment is then assessed using a network score (negative log of p-values of Fisher tests). Focus genes (in bold) are genes identified in our list of differentially expressed genes. Networks shown here are those with network scores > 3.0.
miRNA expression study
Eleven miRNAs (10 down-regulated and 1 up-regulated) were differentially expressed in Students’ T-test (p-value < 0.05) and fold change (absolute fold change > 1.5) comparisons (Table III). MiRNA expression fold change differences (comparing low/high vitamin D groups) ranged from 1.79 (miR-574-5p) to −2.32 (miR-589). The list of differentially expressed miRNAs includes well characterized miRNAs (e.g. miR-93 and miR-573). We identified 73 genes targeted by ≥ 3 differentially expressed miRNAs. Of these, one (BCL11A) is targeted by six miRNAs; four are targeted by five miRNAs; 14 are targeted by four miRNAs; and, 54 are targeted by three miRNAs. Notably, several of these target genes and/or their gene families, including the BCL11A gene, were differentially expressed in our mRNA microarray experiment (Table IV). In IPA of functions and functional relationships of these target genes, networks involved in cellular morphology, function and maintenance, system development (including the nervous system and skeletal systems), cell-to-cell signaling, cell death and carbohydrate/lipid metabolism were over represented (Table V). Other genes and proteins with strong associations to metabolic and inflammatory disorders (e.g., NFKB, Estrogen receptor, VEGF and Insulin) were featured prominently in these networks (Supplemental Figure 2).
Table III.
List of differentially expressed microRNAs
| MicroRNA | Fold change | Students’ T test P-value | Chromosomal location | Gene targets (number)* |
|---|---|---|---|---|
| hsa-miR-589 | −2.32 | 0.0024 | 7q22.1 | 114 |
| hsa-miR-601 | −2.00 | 0.0017 | 9q33.3 | 61 |
| hsa-miR-573 | −1.89 | 0.0006 | 4p15.2 | 117 |
| hsa-miR-138 | −1.86 | 0.0052 | 3p21.32 | 388 |
| hsa-miR-320d | −1.81 | 0.0465 | 13q14.11 | 539 |
| hsa-miR-196a* | −1.79 | 0.0039 | 12q13.13 | 210 |
| hsa-miR-92b | −1.73 | 0.0440 | 1q22 | 692 |
| hsa-miR-423-3p | −1.72 | 0.0102 | 17q11.2 | 3 |
| hsa-miR-484 | −1.66 | 0.0485 | 16p13.11 | 175 |
| hsa-miR-93 | −1.65 | 0.0009 | 7q22.1 | 991 |
| hsa-miR-574-5p | 1.79 | 0.0090 | 4p14 | 90 |
Putative gene targets (targeted by 3 or more differentially expressed microRNAs) were identified using (http://www.targetscan.org/).
Table IV.
List of genes targeted by three or more differentially expressed microRNAs
| Gene Symbol | Gene Name | MicroRNAs targeting the gene | |
|---|---|---|---|
| # | List of microRNAs | ||
| ABL2 | v-abl Abelson murine leukemia viral oncogene homolog 2 | 4 | miR-196a/miR-589/miR-93/miR-92b |
| ALS2CR13 | Amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 13 | 3 | miR-320d/miR-92b/miR-93 |
| ANKFY1** | Ankyrin repeat and FYVE domain containing 1 | 3 | miR-138/miR-320d/miR-93 |
| ANKRD52** | Ankyrin repeat domain 52 | 4 | miR-320d/miR-484/miR-574-5p/miR-93 |
| ARF1 | ADP-ribosylation factor 1 | 3 | miR-320d/miR-574-5p/miR-92b |
| ATP11A** | ATPase, class VI, type 11A | 3 | miR-320d/miR-92b/miR-93 |
| BCL11A** | B-cell CLL/lymphoma 11A (zinc finger protein) | 6 | miR-138/miR-196a/miR-573/miR-574-5p/miR-589/miR-92b |
| C17orf39** | Chromosome 17 open reading frame 39 | 4 | miR-138/miR-320d/miR-92b/miR-93 |
| CALM3 | Calmodulin 3 | 3 | miR-196a/miR-320d/miR-92b |
| CALN1 | Calneuron 1 | 3 | miR-138/miR-320d/miR-92b |
| CCNL2 | Cyclin L2 | 3 | miR-138/miR-320d/miR-574-5p |
| CDK6 | Cyclin-dependent kinase 6 | 3 | miR-138/miR-320d/miR-92b |
| CNOT6L | CCR4-NOT transcription complex, subunit 6-like | 3 | miR-138/miR-320d/miR-93 |
| CPEB3 | Cytoplasmic polyadenylation element binding protein 3 | 3 | miR-320d/miR-92b/miR-93 |
| CUGBP2 | CUG triplet repeat, RNA binding protein 2 | 4 | miR-196a/miR-589/miR-92b/miR-93 |
| DCBLD2 | Discoidin, CUB and LCCL domain containing 2 | 4 | miR-320d/miR-484/miR-573/miR-93 |
| DCP1A | DCP1 decapping enzyme homolog A | 4 | miR-138/miR-320d/miR-487/miR-92b |
| DLGAP2 | Discs, large (Drosophila) homolog-associated protein 2 | 4 | miR-196a/miR-589/miR-92b/miR-93 |
| E2F3** | E2F transcription factor 3 | 5 | miR-138/miR-320d/miR-573/miR-92b/miR-93 |
| EIF5A2** | Eukaryotic translation initiation factor 5A2 | 3 | miR-320d/miR-589/miR-93 |
| FAM160B1** | Family with sequence similarity 160, member B1 | 3 | miR-320d/miR-92b/miR-93 |
| FOSL2** | FOS-like antigen 2 | 3 | miR-138/miR-484/miR-92b |
| FXR1** | Fragile X mental retardation, autosomal homolog 1 | 3 | miR-320d/miR-92b/miR-93 |
| GCT8D3 | Glycosyl transferase 8 domain containing 3 | 3 | miR-320d/miR-92b/miR-93 |
| GNS | Glucosamine (N-acetyl)-6-sulfatase | 3 | miR-320d/miR-92b/miR-93 |
| GOLGA1 | Golgin A1 | 3 | miR-320d/miR-92b/miR-93 |
| H3F3B | H3 histone, family 3B (H3.3B) | 3 | miR-138/miR-320d/miR-92b |
| HDAC4 | Histone deacetylase 4 | 3 | miR-138/miR-320d/miR-93 |
| IGF1 | Insulin-like growth factor 1 (somatomedin C) | 3 | miR-196a/miR-320d/miR-573 |
| JARID1A | Jumonji, AT rick interactive domain A | 3 | miR-196a/miR-320d/miR-93 |
| KIAA0831 | KIAA0831 | 3 | miR-320d/miR-92b/miR-93 |
| KIAA1549 | KIAA1549 | 3 | miR-320d/miR-484/miR-573 |
| LIMCH1 | LIM and calponin homology domains 1 | 3 | miR-138/miR-320d/miR-92b |
| LOC16207 | 3 | miR-320d/miR-92b/miR-93 | |
| MEX3D | Mex-3 homolog D (C. elegans) | 3 | miR-320d/miR-573/miR-93 |
| MYT1L | Myelin transcription factor 1-like | 3 | miR-320d/miR-92b/miR-93 |
| NEGR1 | Neuronal growth regulator 1 | 3 | miR-574-5p/miR-320d/miR-92b |
| NELF | Nasal embryonic LHRH factor | 3 | miR-138/miR-320d/miR-92b |
| NFAT5** | Nuclear factor of activated T-cells 5, tonicity-responsive | 5 | miR-138/miR-574-5p/miR-601/miR-92b/miR-93 |
| NR2C2 | Nuclear receptor subfamily 2, group C, member 2 | 3 | miR-196a/miR-320d/miR-93 |
| NRK | Nik related kinase | 3 | miR-196a/miR-320d/miR-92b |
| NUFIP2 | Nuclear fragile X mental retardation protein interacting protein 2 | 4 | miR-320d/miR-574-5p/miR-92b/miR-93 |
| OLA1 | Obg-like ATPase 1 | 3 | miR-320d/miR-484/miR-574-5p |
| PALM2 | Paralemmin 2 | 3 | miR-320d/miR-484/miR-92b |
| PAPD5 | PAP associated domain containing 5 | 3 | miR-138/miR-320d/miR-92b |
| PARD6B | Par-6 partitioning defective 6 homolog beta (C. elegans) | 3 | miR-196a/miR-320d/miR-93 |
| PBX3 | Pre-B-cell leukemia homeobox 3 | 3 | miR-196a/miR-320d/miR-93 |
| PCDH19 | Protocadherin 19 | 3 | miR-196a/miR-320d/miR-484 |
| PCGF3 | Polycomb group ring finger 3 | 4 | miR-138/miR-320d/miR-573/miR-92b |
| PHOX2B | Paired-like homeobox 2b | 3 | miR-138/miR-320d/miR-589 |
| PHTF2 | Putative homeodomain transcription factor 2 | 3 | miR-320d/miR-92b/miR-93 |
| PRRG1 | Proline rich Gla (G-carboxyglutamic acid) 1 | 3 | miR-320d/miR-573/miR-93 |
| PTEN | Phosphatase and tensin homolog | 3 | miR-320d/miR-92b/miR-93 |
| PURB | Purine-rich element binding protein B | 4 | miR-320d/miR-573/miR-601/miR-93 |
| RAD23B | RAD23 homolog B (S. cerevisiae) | 4 | miR-196a/miR-320d/miR-573/miR-93 |
| SEMA3A** | Sema, immunoglobulin, (semaphorin) 3A | 3 | miR-196a/miR-320d/miR-92b |
| SH2B3 | SH2B adaptor protein 3 | 3 | miR-196a/miR-320d/miR-601 |
| SIN3A | SIN3 homolog A, transcription regulator (yeast) | 3 | miR-196a/miR-320d/miR-589 |
| SLITRK3 | SLIT and NTRK-like family, member 3 | 3 | miR-320d/miR-589/miR-93 |
| SNTB2 | Syntrophin, beta 2 (dystrophin-associated protein A1) | 3 | miR-196a/miR-320d/miR-93 |
| SNX16** | Sorting nexin 16 | 3 | miR-196a/miR-320d/miR-93 |
| SOBP | Sine oculis binding protein homolog (Drosophila) | 4 | miR-138/miR-320d/miR-92b/miR-93 |
| SOX4 | SRY (sex determining region Y)-box 4 | 5 | miR-138/miR-320d/miR-573/miR-92b/miR-93 |
| SPOPL | Speckle-type POZ protein-like | 3 | miR-320d/miR-573/miR-93 |
| TMCC1 | Transmembrane and coiled-coil domain family 1 | 3 | miR-320d/miR-92b/miR-93 |
| TNRC6B | Trinucleotide repeat containing 6B | 5 | miR-138/miR-320d/miR-573/miR-92b/miR-93 |
| TRIM36 | Tripartite motif-containing 36 | 3 | miR-320d/miR-92b/miR-93 |
| TSC1 | Tuberous sclerosis 1 | 3 | miR-196a/miR-320d/miR-92b |
| TWF1 | Twinfilin, actin-binding protein, homolog 1 (Drosophila) | 3 | miR-320d/miR-92b/miR-93 |
| XIAP | X-linked inhibitor of apoptosis | 3 | miR-320d/ miR-601/miR-93 |
| ZC3H7B** | Zinc finger CCCH-type containing 7B | 4 | miR-138/miR-320d/miR-484/miR-93 |
| ZFHX4 | Zinc finger homeobox 4 | 3 | miR-320d/miR-601/miR-92b |
| ZNF148** | Zinc finger protein 148 | 4 | miR-138/miR573/miR-92b/miR-93 |
Putative gene targets (targeted by 3 or more differentially expressed miRNAs) were identified using (http://www.targetscan.org/).
Gene or gene families that were differentially expressed in our mRNA microarray study.
Table V.
MicroRNA target gene enrichment network analysis results
| # | Genes in Network | Score | Focus genes | Functions |
|---|---|---|---|---|
| 1 | 26s Proteasome, Akt, Ap1, ARF1, CDK6, Cyclin A, Cyclin E, DCBLD2, E2F3, ERK, Estrogen Receptor, FOSL2, H3F3B, hCG, Hdac, HDAC4, IGF1, Insulin, KDM5A, MIR1, N-cor, NFkB (complex), NR2C2, PDGF BB, Pi3-kinase, PTEN, RAD23B, SH2B3, SIN3A, SOX4, TSC1, TWF1, Ubiquitin, Vegf, XIAP | 37 | 18 | Cell Morphology, Nervous System Development and Function, Organismal Development |
| 2 | AKIRIN1, ATXN1, ATXN2L, C1ORF144, C9ORF25, CALN1, CELF2, CNOT6L, DBC1, DMPK, FXR1, HIVEP1, ING3, KLF7, KLHL34, MIR199A1, MIR9-1 (includes EG:407046), MIRN101B, MLL4, NOVA1, NXPH1, OLA1, PALM2, PAPD5, PAPOLA, PHF12, PHTF2, POLE4, RBM9, SELS, SGK1, SLITRK3, SNX12, ZC3H7B, ZFHX4 | 20 | 11 | RNA Post-Transcriptional Modification, Neurological Disease, Genetic Disorder |
| 3 | AMMECR1L, ARID4B, ARRDC3, ATP11A, BASP1, BHLHE41, C12ORF53, C17ORF39, C5ORF41, CALM3, E2F3, EMG1, FAM117B, FBN2 (includes EG:2201), GALNT1, GXYLT1, KIAA1274, LIMCH1, LRP4, LYPLA2, MIR122 (includes EG:406906), MIR181B2, MIR28 (includes EG:407020), MIRLET7E (includes EG:406887), NUFIP2, OFD1, PCDH19, PREB, PURB, RIOK3, TBX18, TMCC1, WDR44, ZIC3, ZNF503 | 20 | 11 | Genetic Disorder, Skeletal and Muscular Disorders, Cellular Function and Maintenance |
| 4 | ABL2, Akt-Calmodulin-Hsp90-Nos3, ARHGEF19, CCL27, CDC42BPG, CDC42SE1, CES2 (includes EG:234671), CPEB3, CSRNP1, CYP2D12, DCP1A, DEFA5 (includes EG:1670), EIF2C2, ENPP1, HRAS, Hsp90, IL2, IL1B, ITPA, MIR146A (includes EG:406938), NCR3, NFAT5, PARD6B, PHOX2B, PROK2, RAC1, SLC22A4 (includes EG:6583), SLC39A8, SNTB2, SP1, SPOPL, TGFB1, TNRC6B, TYMP, ZNF148 | 18 | 10 | Cell-To-Cell Signaling and Interaction, Gene Expression, Cellular Movement |
| 5 | ANKRD52, AQP3, BCL11A, CAP2, CCNL2, CSDC2, DLGAP2, DUSP14, EIF5A2, ERK1/2, FAM160B1, FSH, GPRC5A, IL15, Il15r, IL17RD, IL1F8, IL22R1-IL10R2, Jnk, Jun-GABP, KSR2, MICA, MIR373, MKRN1, MT1H, NELF, PBX3, PEBP4, PI3K, RNA polymerase II, SEMA3A, SLC12A4, SLC12A7, TERT, TFF2 | 16 | 9 | Cellular Compromise, Cell Death, Cellular Growth and Proliferation |
| 6 | ACOT2, Aldolase, ANKFY1, ARHGAP33, DYNC1I2, GNS, HTT, INPP4A, INPP5E, JPH2, KIAA1549, MAPK8, MDH1, MTM1, MTMR9, MYT1L, NDUFV1, NEGR1, NRK, PCGF3, phosphatidylinositol-3-phosphate, RAB5A, RNF2, RNF34, SFXN3, SLC25A11, SLC2A4, SNX13, SNX16, TPP1, TRIM36, UBE2H (includes EG:7328), UBE2S, ZBTB16, ZNF675 | 16 | 9 | Carbohydrate Metabolism, Lipid Metabolism, Small Molecule Biochemistry |
The networks were generated using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). Each gene (targeted by 3 or more differentially expressed microRNA identified using http://www.targetscan.org) identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). These genes were overlaid onto a global molecular network developed from information contained in the IPKB. Network enrichment is then assessed using a network score (negative log of p-values of Fisher tests). Focus genes (in bold) are genes identified from our list of target genes. Networks shown here are those with network scores > 3.0.
Discussion
In pilot microarray-based expression profile studies of maternal peripheral blood, we show that early pregnancy low 25[OH]D concentrations are associated with maternal peripheral blood gene expression and post-transcription regulation. Genes that participate in a wide range of cellular functions, including organ and system development (e.g. angiogenesis), inflammation and metabolic processes (e.g. carbohydrate/lipid metabolism), as well as miRNAs that target these genes were differentially expressed among women with low 25[OH]D compared with those with high 25[OH]D concentrations.
Previous candidate expression profiling studies, mostly conducted using specific cell lines or tissue under laboratory settings, have reported vitamin D related gene expression and post-transcription regulation [9–14]. Our genome-wide microarray investigations replicated most of their findings either directly (up-regulation or down-regulation of genes in our microarray experiment) or indirectly (up-regulation or down-regulation of miRNAs that target these genes). In addition, our investigations expand the literature to include study of pregnant women as few previous studies evaluated vitamin D related gene expressions and post-transcription regulation during pregnancy [18–19]. In a recent candidate gene expression study by Rochat et al., maternal vitamin D supplementation was associated with up regulation of ILT3 and ILT4 genes in cord blood [18]. While we did not find differential regulation of these specific genes in maternal blood in our study, other genes related to antigen presentation like CD79B and CD48 were differentially expressed in mothers enrolled in our study.
Our findings support the thesis that vitamin D is involved in physiological functions and low concentrations of vitamin D may be associated with pathophysiologic processes throughout the body [1]. A physiologic 2–3 fold increase in vitamin D concentrations is observed during the first trimester of pregnancy possibly related to active synthesis in placental decidual cells [20]. The ubiquity of vitamin D receptors and easy placental passage of Vitamin D to the fetus serve to underscore potentially important physiologic effects of maternal vitamin D status during gestation and parturition [3–4, 20]. Pregnancy complications such as preeclampsia and gestational diabetes share similar pathophysiologic processes (such as inflammation, cell signaling and abnormal lipid metabolism) with deregulation associated with low vitamin D concentrations that are demonstrated in our findings [21–22]. Thus, vitamin D may influence risk for pregnancy complications through these pathomechanisms.
Since early pregnancy maternal characteristics influence the intrauterine environment and subsequently fetal programming, vitamin D may also have substantial influence on the developmental origins of childhood and adult disorders [5–7]. Our findings of across the board deregulation of genes involved in organ/system development are particularly significant. Similarly, these pathophysiologic mechanisms may account for associations of vitamin D with gestational metabolic disorders (e.g., gestational diabetes and preeclampsia) and chronic disorders (e.g., Type 2 diabetes, hypertension and cardiovascular disease) across the life course [4].
We identified differential expressions of both candidate and novel mRNAs (e.g. PDGFA) and miRNAs (e.g. miR-93). PDGFA is part of the platelet-derived growth factor family, potent mitogens whose expressions are highly regulated and impact development of the cardiovascular and nervous system, as well as the kidney and the lung [13]. Importantly, platelet derived growth factors have been implicated in glucose homeostasis [23] and preeclampsia [24]. Pedigo et al previously demonstrated that vitamin D induces expression of PDGFA, a role through which it influences pathogenesis of gestational diabetes and preeclampsia, similar to atherosclerosis, Type 2 diabetes, fibrotic diseases and cancer [13]. We and others have previously shown that VEGF, whose functions involve angiogenesis and vascular homeostasis, is associated with a number of disease states including preeclampsia, gestational diabetes and diabetes complications [21–22, 25–26]. The regulatory mechanisms for expression of this important gene are not fully described. Recently, Long et al demonstrated that miR-93, one of the differentially expressed miRNAs in our experiment, regulates VEGF expression in experimental models of diabetes both in vitro and in vivo [26]. This novel association which may have implications in pregnancy and pregnancy complications may provide future opportunities for better understanding of pregnancy complications and potential preventative targets.
Vitamin D deficiency can follow lack of exposure to sunlight, medication/supplement intake (e.g. glucocorticoids and antiseizure medications), malabsorption, hepatic and renal failure [1]. Major sources of vitamin D are endogenous production following exposure to sunlight and dietary intake (and supplementation) [1,4]. There is no consensus regarding the optimal levels of vitamin D concentrations in pregnancy due to variations in the state of vitamin D sufficiency throughout the life cycle, inherent differences across study populations investigated and since most information on current recommendations are based on observations related to bone-related diseases in non-pregnant populations [20]. Recent reports indicate widespread vitamin D deficiency among adults and children. For instance, in the NHANES III report, prevalence of vitamin D deficiency in the US, among adults, was estimated to be between 25–57% [27]. Given the importance of vitamin D in pregnancy and the widespread prevalence of vitamin D insufficiency in the general population, further research is needed to better understand optimum levels of vitamin D in pregnancy.
Our study has several strengths. It was conducted using samples that were collected in early pregnancy, a critical period related to pregnancy complications and fetal development. The use of peripheral blood, a relatively accessible tissue for research investigations provides a proof-of-concept for future investigations and clinical applications. Further, peripheral blood evaluations may provide a good sample of evaluating systemic pathophysiological processes that may account for observed epidemiological associations of vitamin D insufficiency or deficiency with gestational diabetes and preeclampsia risk. We used microarrays to investigate both global mRNA and miRNA expressions (transcription and post-transcription regulation). Concordance of findings of networks over represented by differentially expressed genes or target genes of differentially expressed miRNAs provide validations for the importance of the networks related to vitamin D concentrations. Finally, we used multiple statistical analyses tools including the SAM based false discovery rate to minimize false positives.
Several limitations of our study merit discussion. The cut-off vitamin D concentrations we used to define groups of low/high vitamin D concentrations is based on available evidence that may not reflect optimum levels in pregnancy, as described earlier [28]. In addition, single measurements of vitamin D may not adequately reflect vitamin D levels throughout pregnancy. While there are advantages to the use of peripheral blood for these kinds of investigations, the caveat that peripheral blood changes may not reflect changes at target tissue (e.g. pancreatic cells of beta cells) is a potential limitation of our study. However, collection of such samples is prohibitive in epidemiological studies. There was limited overlap between participants (three with low and three with high vitamin D concentrations) of the two studies on mRNA and miRNA expressions. We were unable to conduct both profiles in the complete study population due to sample unavailability. As a result, we could not directly compare gene expressions and post-transcription regulation in the whole study population. Sample shortages, also prohibited us from conducting confirmatory quantitative real time polymerase chain reaction (qRT-PCR) experiments of microarray-based expression measures. However, similarities between genes and gene networks that are either differentially expressed in our experiment or are targeted by differentially expressed miRNAs mitigate this concern. Finally, our modest sample size as well as unaccounted variations in the study populations may have limited the statistical power of our pilot study to identify gene expression and post-transcription regulation differences. Larger studies will be needed to confirm and expand upon these initial observations.
In summary, early pregnancy plasma 25[OH]D concentrations are associated with maternal peripheral blood gene expressions and post-transcription regulation. Findings from this pilot and feasibility study enhance understanding of pathophysiologic processes influenced by vitamin D and shed light on potential mechanisms of associations of vitamin D concentrations with pregnancy complications and outcomes. Given the prevalence and public health importance of low vitamin D concentrations, further research in this area may provide opportunities for preventative and therapeutic applications.
Supplementary Material
Supplemental Figure 1 Top networks overrepresented by differentially expressed genes
The networks were generated using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). These genes were overlaid onto a global molecular network developed from information contained in the IPKB. Network enrichment is then assessed using a network score (negative log of p-values of Fisher tests). Focus genes (shaded) are genes identified in our list of differentially expressed genes.
Supplemental Figure 2 Top network overrepresented by gene targets of differentially expressed microRNAs
The network was generated using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). Each target gene (targeted by three or more differentially expressed microRNAs identified using http://www.targetscan.org) identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). These genes were overlaid onto a global molecular network developed from information contained in the IPKB. Network enrichment is then assessed using a network score (negative log of p-values of Fisher tests). Focus genes (shaded) are genes identified in our list of differentially expressed microRNA targeted genes.
Supplemental Table I Full list of differentially expressed genes
Acknowledgments
The authors are indebted to the participants of the Omega study for their cooperation. They are also grateful for the technical expertise of staff of the Center for Perinatal Studies, Swedish Medical Center. This work was supported by grants from the National Institute of Child Health and Human Development, National Institutes of Health (HD/HL R01-32562) and the March of Dimes (#1 FY08-425).
References
- 1.Holick MF, Chen TC. Vitamin D deficiency: a worldwide problem with health consequences. Am J Clin Nutr. 2008;87:1080S–1086S. doi: 10.1093/ajcn/87.4.1080S. [DOI] [PubMed] [Google Scholar]
- 2.Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357:266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 3.Mulligan ML, Felton SK, Riek AE, Bernal-Mizrachi C. Implications of vitamin D deficiency in pregnancy and lactation. Am J Obstet Gynecol. 2010;202:429.e1–9. doi: 10.1016/j.ajog.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang C, Qiu C, Hu FB, David RM, van Dam RM, Bralley A, Williams MA. Maternal plasma 25-hydroxyvitamin D concentrations and the risk for gestational diabetes mellitus. PLoS One. 2008;3:e3753. doi: 10.1371/journal.pone.0003753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper C, Harvey N, Cole Z, Hanson M, Dennison E. Developmental origins of osteoporosis: the role of maternal nutrition. Adv Exp Med Biol. 2009;646:31–39. doi: 10.1007/978-1-4020-9173-5_3. [DOI] [PubMed] [Google Scholar]
- 6.Lanham SA, Roberts C, Cooper C, Oreffo RO. Intrauterine programming of bone. Part 1: alteration of the osteogenic environment. Osteoporos Int. 2008;19:147–156. doi: 10.1007/s00198-007-0443-8. [DOI] [PubMed] [Google Scholar]
- 7.Eyles DW, Feron F, Cui X, Kesby JP, Harms LH, Ko P, McGrath JJ, Burne TH. Developmental vitamin D deficiency causes abnormal brain development. Psychoneuroendocrinology. 2009;34:S247–S257. doi: 10.1016/j.psyneuen.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Ginde AA, Sullivan AF, Mansbach JM, Camargo CA., Jr Vitamin D insufficiency in pregnant and nonpregnant women of childbearing age in the United States. Am J Obstet Gynecol. 2010;202:436.e1–8. doi: 10.1016/j.ajog.2009.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Towsend K, Trevino V, Falciani F, Stewart PM, Hewison M, Campbell MJ. Identification of VDR-responsive gene signatures in breast cancer cells. Oncology. 2006;71:111–123. doi: 10.1159/000100989. [DOI] [PubMed] [Google Scholar]
- 10.Mohri T, Nakajima M, Takagi S, Komagata S, Yokoi T. MicroRNA regulates human vitamin D receptor. Int J Cancer. 2009;125:1328–1333. doi: 10.1002/ijc.24459. [DOI] [PubMed] [Google Scholar]
- 11.Wu-Wong JR, Nakane M, Ma J, Ruan X, Kroeger PE. VDR-mediated gene expression patterns in resting human coronary artery smooth muscle cells. J Cell Biochem. 2007;100:1395–1405. doi: 10.1002/jcb.21133. [DOI] [PubMed] [Google Scholar]
- 12.Wood RJ, Tchack L, Angelo G, Pratt RE, Sonna LA. DNA microarray analysis of vitamin D-induced gene expression in a human colon carcinoma cell line. Physiol Genomics. 2004;17:122–129. doi: 10.1152/physiolgenomics.00002.2003. [DOI] [PubMed] [Google Scholar]
- 13.Pedigo N, Zhang H, Koszewski NJ, Kaetzel DM. A 5′-distal element mediates vitamin D-inducibility of PDGF-A gene transcription. Growth Factors. 2003;21:151–160. doi: 10.1080/08977190310001636595. [DOI] [PubMed] [Google Scholar]
- 14.Pike JW, Meyer MB. The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D(3) Endocrinol Metab Clin North Am. 2010;39:255–269. doi: 10.1016/j.ecl.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weng L, Dai H, Zhan Y, He Y, Stepaniants SB, Bassett DE. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- 16.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. PNAS. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rochat MK, Ege MJ, Plabst D, Steinle J, Bitter S, Braun-Fahrländer C, Dalphin JC, Riedler J, Roponen M, Hirvonen MR, Büchele G, Renz H, Lauener R, Krauss-Etschmann S, von Mutius E, PASTURE Study group Maternal vitamin D intake during pregnancy increases gene expression of ILT3 and ILT4 in cord blood. Clin Exp Allergy. 2010;40:786–794. doi: 10.1111/j.1365-2222.2009.03428.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Y, Goff JP, Reinhardt TA, Horst RL. Pregnancy and lactation increase vitamin D-dependent intestinal membrane calcium adenosine triphosphatase and calcium binding protein messenger ribonucleic acid expression. Endocrinology. 1998;139:3520–3524. doi: 10.1210/endo.139.8.6141. [DOI] [PubMed] [Google Scholar]
- 20.Lapillonne A. Vitamin D deficiency during pregnancy may impair maternal and fetal outcomes. Med Hypotheses. 2010;74:71–75. doi: 10.1016/j.mehy.2009.07.054. [DOI] [PubMed] [Google Scholar]
- 21.Enquobahrie DA, Meller M, Rice KM, Psaty BM, Siscovick DS, Williams MA. Differential placental gene expression in preeclampsia. Am J Obstet Gynecol. 2008;199:566.e1–11. doi: 10.1016/j.ajog.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enquobahrie DA, Williams MA, Qiu C, Meller M, Sorenson TK. Global placental gene expression in gestational diabetes mellitus. Am J Obstet Gynecol. 2009;200:206.e1–13. doi: 10.1016/j.ajog.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 23.Whiteman EL, Chen JJ, Birnbaum MJ. Platelet-derived growth factor (PDGF) stimulates glucose transport in 3T3-L1 adipocytes overexpressing PDGF receptor by a pathway independent of insulin receptor substrates. Endocrinology. 2003;144:3811–3820. doi: 10.1210/en.2003-0480. [DOI] [PubMed] [Google Scholar]
- 24.Jurcovicová J, Krueger KS, Nandy I, Lewis DF, Brooks GG, Brown EG. Expression of platelet-derived growth factor-A mRNA in human placenta: effect of magnesium infusion in pre-eclampsia. Placenta. 1998;19:423–427. doi: 10.1016/s0143-4004(98)90083-2. [DOI] [PubMed] [Google Scholar]
- 25.Muy-Rivera M, Vadachkoria S, Woelk GB, Qiu C, Mahomed K, Williams MA. Maternal plasma VEGF, sVEGF-R1, and PlGF concentrations in preeclamptic and normotensive pregnant Zimbabwean women. Physiol Res. 2005;54:611–622. [PubMed] [Google Scholar]
- 26.Long J, Wang Y, Wang W, Chang BH, Danesh FR. Identification of MicroRNA-93 as a novel regulator of vascular endothelial growth factor (VEGF) In hyperglycemic conditions. J Biol Chem. 2010 May 25; doi: 10.1074/jbc.M110.136168. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Looker AC, Dawson-Hughes B, Calvo MS, Gunter EW, Sahyoun NR. Serum 25-hydroxyvitamin D status of adolescents and adults in two seasonal subpopulations from NHANES III. Bone. 2002;30:771–777. doi: 10.1016/s8756-3282(02)00692-0. [DOI] [PubMed] [Google Scholar]
- 28.Dietary Supplement Fact Sheet: Vitamin D. Bethesda (MD): Office of Dietary Supplements, National Institutes of Health; 2009. Nov, [cited 2010 July 17]. Available from http://ods.od.nih.gov/factsheets/vitamind.asp#en4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 Top networks overrepresented by differentially expressed genes
The networks were generated using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). These genes were overlaid onto a global molecular network developed from information contained in the IPKB. Network enrichment is then assessed using a network score (negative log of p-values of Fisher tests). Focus genes (shaded) are genes identified in our list of differentially expressed genes.
Supplemental Figure 2 Top network overrepresented by gene targets of differentially expressed microRNAs
The network was generated using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). Each target gene (targeted by three or more differentially expressed microRNAs identified using http://www.targetscan.org) identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). These genes were overlaid onto a global molecular network developed from information contained in the IPKB. Network enrichment is then assessed using a network score (negative log of p-values of Fisher tests). Focus genes (shaded) are genes identified in our list of differentially expressed microRNA targeted genes.
Supplemental Table I Full list of differentially expressed genes


