Abstract
Diffuse and unstoppable infiltration of brain and spinal cord tissue by neoplastic glial cells is the single most important therapeutic problem posed by the common glioma group of tumors: astrocytoma, oligoastrocytoma, oligodendroglioma, their malignant variants and glioblastoma. These neoplasms account for more than two thirds of all malignant central nervous system tumors. However, most glioma research focuses on an examination of the tumor cells rather than on host-specific, tumor micro-environmental cells and factors. This can explain why existing diffuse glioma therapies fail and why these tumors have remained incurable. Thus, there is a great need for innovation. We describe a novel strategy for the development of a more effective treatment of diffuse glioma. Our approach centers on gaining control over the behavior of the microglia, the defense cells of the CNS, which are manipulated by malignant glioma and support its growth. Armoring microglia against the influences from glioma is one of our research goals. We further discuss how microglia precursors may be genetically enhanced to track down infiltrating glioma cells.
Keywords: Glioblastoma, M2 polarization, microglia, pathway analysis, systems biology, zinc finger nucleases
INTRODUCTION
Gliomas are tumors derived from glial cells, the non-neuronal cells of the brain and spinal cord. They are the most common tumors of the central nervous tissue and many are fatal comprising more than two thirds of all malignant CNS neoplasms. Gliomas occur in adults as well as children, and their prognosis depends on subtype and grade (www.pubcan.org). A tentative diagnosis can be made by means of modern neuroimaging techniques but the gold standard for glioma diagnosis is the microscopic examination of tumor tissue following a surgical biopsy. Currently, the microscopic examination can be complemented but not replaced by molecular tests. Therefore, only a trained neuropathologist should make the diagnosis by morphologic criteria. This is important because other CNS diseases can present with tumor-like signs. An accurate diagnosis is of key importance for the decision on treatment strategies. Surgery as well as radio- and chemotherapy may be applied.
The vast majority of contemporary glioma research focuses on the tumor as such rather than on what the central nervous system (CNS) does or does not do to the infiltrating neoplastic glial cells. This can explain why all existing diffuse glioma therapies fail and why the prognosis of these tumors has changed insignificantly in more than a century and they have remained incurable. Consequently, there is a great need for innovation. Recent advances in neuroscience knowledge and technological progress in molecular biology raise hope that such innovation is within reach.
In this article we outline a novel strategy that focuses on making use of the presence of microglia within diffuse glioma for therapeutic purposes. There is increasing evidence that high-grade gliomas very effectively attract microglia/macrophages and subsequently control their activity eliciting mainly tumor-supportive functions that facilitate glioma growth [1]. We are interested in the question of whether this fatal attraction can be used against the tumor by employing bone-marrow transplantation of genetically enhanced [2] microglia precursors. We further discuss the need for the development of an in silico model of the microglia, which is expected to yield a blueprint of the molecular controls that are required to modify the behavior of glioma associated microglia. In addition, our vision for engineering microglia that are capable of tracking down individual, deeply infiltrating glioma cells is outlined. Lastly, we describe a technology known as zinc finger nucleases (ZFNs) that may be employed to implement the required genetic modifications.
THE CURRENT GOLD STANDARD OF GLIOMA DIAGNOSIS
The microscopic examination of a tumor tissue reveals the histological tumor type. Gliomas are named after the normal glial cell types with which each tumor variant shares morphological similarities. The two main neuroglial cell types of the CNS are astrocytes and oligodendrocytes. The third common glial cell type, the microglia, populate the CNS during embryonic and early postnatal development and cause tumors so rarely that there is no official classification entry [3]. After the type of brain tumor has been determined based on morphological criteria, a WHO grade is assigned. The WHO classification of tumors (www.pubcan.org) currently distinguishes the main subtypes of common glioma shown in Table 1.
Table 1.
Main Types of Human Glioma
| Astrocytic tumors |
| Pilocytic astrocytoma (I) |
| Pilomyxoid astrocytoma (II) |
| Subependymal giant cell astrocytoma (I) |
| Pleomorphic xanthoastrocytoma (II) |
| Diffuse astrocytoma (II) |
| Fibrillary astrocytoma |
| Gemistocytic astrocytoma |
| Protoplasmic astrocytoma |
| Anaplastic astrocytoma (III) |
| Glioblastoma (IV) |
| Giant cell glioblastoma |
| Gliosarcoma |
| Gliomatosis cerebri (III) |
| Oligodendroglial tumors |
| Oligodendroglioma (II) |
| Anaplastic oligodendroglioma (III) |
| Oligoastrocytic tumors |
| Oligoastrocytoma (II) |
| Anaplastic oligoastrocytoma (III) |
| Ependymal tumors |
| Subependymoma (I) |
| Myxopapillary ependymoma (I) |
| Ependymoma (II) |
| Cellular |
| Papillary |
| Clear cell |
| Tanycytic |
| Anaplastic ependymoma (III) |
Grades of the different glioma variants are indicated by Roman numerals shown in brackets following the respective glioma subtype.
In general terms, a tumor is referred to as grade I if the biopsy shows only very few dividing cells and the chances of the patient is being cured by the surgical resection of the tumor alone are high. A grade II tumor in contrast is likely to recur and may even worsen over time, i.e. become anaplastic. The latter is called tumor progression and is regularly the case for diffuse astrocytoma, oligoastrocytoma and oligodendroglioma. Histological signs of a malignant or WHO grade III glioma include so-called atypical nuclei and the presence of dividing cells (mitosis). Patients with grade III tumors are treated by adjuvant radiation and/or chemotherapy. Glioblastoma is an example of a WHO grade IV tumor. In addition to cells that appear malignant and variable numbers of mitotic cells, such tumors tend to show large areas of cell death (necrosis) because they grow so rapidly that the blood supply cannot keep up with their growth rate. However, grade IV tumors also stimulate the formation of new blood vessels (neoangiogenesis) and the presence of the latter is another and perhaps even more significant histological sign of their malignancy.
In summary, the WHO classification of CNS tumors (www.pubcan.org) represents a malignancy scale aimed at aiding the clinician to choose the right treatment. It is not always strictly logical. For instance, there is no pilocytic astrocytoma grade II and there is no diffuse astrocytoma grade I. These two entities are very different biologically but share the common family name astrocytoma. Patients with a WHO grade II glioma usually survive more than five years whereas survival of 2–3 years are typical for an individual with a WHO grade III tumor. The outcome is much worse for WHO grade IV glioblastoma where less than half the patients survive more than one year.
THE PRESENCE OF MICROGLIA AND RELATED CELLS WITHIN DIFFUSE GLIOMA
The occurrence of microglial cells in glioma is not a new finding [4, 5]. However, their role remained unclear for many decades. In 1998 we reported that microglia support glioma growth [6]. This finding has been widely reproduced in the meantime and microglia research has yielded much information on the molecular characteristics of these cells. In 1998 we also demonstrated that bone marrow-derived precursors can give rise to typical ramified parenchymal microglia in the adult [7, 8], a concept now adopted by others [9]. These observations are of great relevance in the context of our plan to send genetically enhanced microglia as therapeutic agents into diseased brains. It is likely that many if not most of the glioma-associated macrophages and microglia (ramified cells of typical morphology with perpendicularly branching cell processes) have an extra-cerebral source. The phenotypic and functional differences between these largely “M2 polarized” cells and classically activated (inflammatory) “M1 macrophages” [10] are intriguing and point to a strategy gliomas employ to manipulate microglial behavior in favor of tumor survival and growth.
Analyses of the signaling networks by which microglia interact with the glioma suggests the following view [1]: Glioma-microglia synergies drive a self-amplifying cascade of events that spirals out of control as the tumor progresses. Glioma and microglial cells not only appear to have a symbiotic relationship but one that becomes highly skewed in favor of the glioma [11]. Specifically, the immunosuppressive microenvironment in a glioma created by molecules such as TGFB1, CSF1, and IL10 polarizes glioma-infiltrating microglia towards the M2 phenotype. Gliomas also produce chemotactic factors, such as MCP-1, resulting in the recruitment of large numbers of additional microglia and macrophages. Gliomas further promote the proliferation of microglial cells. In turn, microglia support glioma angiogenesis as well as glioma cell invasion. This cross talk between glioma and microglia is governed by multiple paracrine loops formed by glioma- and microgliareleased molecules and their receptors. Some of the molecules involved also act in an autocrine manner regulating glioma and microglia behavior, respectively (for details see [1]). It therefore appears that immune cells, which are a major source of angiogenic and growth factors as well as matrix-remodeling enzymes that have an entirely normal and necessary function in wound healing, are recruited and subverted to support neoplastic progression [12] in glioma.
There is an additional and particularly interesting aspect of microglia-tumor cell interactions that has attracted attention only very recently. Fusion of tumor cells with bone marrow-derived cells has been proposed as a mechanism underlying invasion and metastasis in human cancer [13]. Accordingly, cellular fusion of microglia with glioma cells is being considered as a possible explanation for the surprising finding that isocitrate dehydrogenase mutations can be observed in microglia/macrophages associated with glioma [14]. We have suggested earlier that some macrophages in glioblastoma may derive from tumor stem cells [15], which would be in line with the view of a significant role of microglia/macrophages in glial tumorigenesis [14]. The fusion hypothesis is of particularly great interest as macrophages are highly migratory cells and gliomas are the most diffusely growing tumors of all. One way to test this hypothesis experimentally will be to use gender-mismatched microglia and glioma cells in combination with FISH to detect Y-chromosomal sequences in cells exhibiting a macrophage phenotype (the tumor cell line, e.g. CNS-1, being derived from male animals).
Taken together, the microglial contributions to glioma growth appear significant and justify serious efforts to study microglia/macrophage-glioma interactions in the greatest detail possible and not only to bring genetically modified microglia into the glioma-affected brain to exert an inhibitory influence on glioma growth but to reduce the vulnerability of the microglia towards glioma influences as the first step.
NON-INVASIVE ACCESS TO THE CNS
Using bone-marrow chimeras carrying a non-expressed marker gene and a combined model of facial nerve axotomy and transfer experimental autoimmune encephalitis, we demonstrated that cells from the macrophage precursor cell pool of the bone marrow have the ability to become typical ramified microglia in the adult [8]. Thus, if recently bone marrow-derived parenchymal microglia fully integrate into a regenerating brain nucleus’ architecture, entirely new approaches for delivering genes into the adult CNS become a possibility [8]. The validity of this hypothesis has been dramatically confirmed by the recent finding that pathological grooming in Hoxb8 mutant mice can be cured through a bone marrow transplant [16]. Furthermore, transplantation of wild-type bone marrow into irradiation-conditioned Mecp2-null hosts resulted in engraftment of brain parenchyma by bone-marrow-derived myeloid cells of microglial phenotype, arresting the development of disease [17]. Importantly, only the use of a conditioning regimen capable of ablating functionally defined brain-resident myeloid precursors allows turnover of microglia that is mediated by local proliferation of early immigrants rather than entrance of mature cells from the circulation [18].
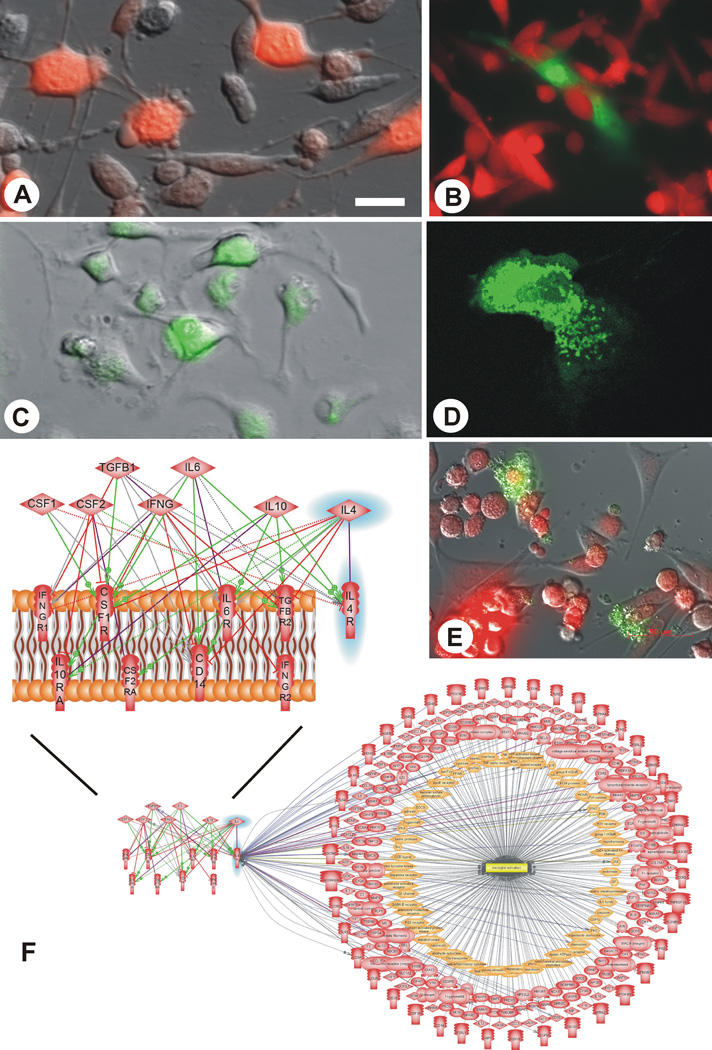
We are using a syngeneic glioma model that employs GFP-transgenic Lewis rats and mCherry-transduced CNS-1 [19, 20] glioma cells (Fig. 1). Our goal is to achieve non-invasive access to the experimental glioma through autologous bone marrow-transplants. As a first step, we are replicating earlier results from a C6 glioma model, which based on use of a non-expressed marker gene strongly suggested that a significant portion of the microglia/macrophages in glioma are bone marrow-derived [21].
Fig. (1).
Experimental tools used in the project. A. mCherry labeled CNS-1 glioma cells in culture; B. transgenic astrocytes (green, obtained from GFP-transgenic Lewis rats) being overrun in culture by syngeneic mCherry labeled CNS-1 glioma cells (red) mirroring the in vivo situation; C. bone marrow cells isolated from GFP-transgenic Lewis rats in culture; D. a GFP-transgenic microglial cell/macrophage in culture; E. Co-culture of mCherry labeled CNS-1 glioma cells and microglia isolated from GFP transgenic rats; F. schematic representation of intercellular microglia “polarization signaling”: CSF2 (GM-CSF) and IFNG are the molecules that drive microglia towards the M1 phenotype, whereas TGFB1, CSF1, IL10, IL4 and IL6 are molecules that polarize microglia towards the M2 phenotype when binding to their respective receptors located on the surface of microglia (purple lines); however, these signals may also act on other receptors on the surface of the microglia; stimulatory regulation is represented by green lines, inhibitory regulation by red lines, and ambiguous effects are shown as grey lines; IL-4 and its receptor, IL-4R are highlighted in blue; the biological associations of the latter with molecules that are regulated during microglial activation are illustrated by the lower panel of this figure and referenced in detail in Table 2. Scale bar: 40 µm (appx. 80 in E).
TOWARDS AN IN SILICO MODEL OF MICROGLIAL CELLS
There is currently no complete systems biological definition of microglial cells (or of any other cell type). Starting out from a first partial transcriptome signature, which we obtained previously [22], we are currently complementing our database by mining publicly available microarray datasets. Pathway analysis software [23, 24] is used to extract information that can assist with the design of an in silico microglia pathway model, which will serve as a blueprint for the controls of microglial behavior in vivo. Reducing microglial susceptibilities to the influence of glioma (e.g. by knocking out receptors such as IL4R, Table 2, Figs. 1, 2) while strengthening or introducing other properties, e.g. the ability to track down glioma cells similar to what stem cells can do [25–32], represent key areas where this in silico knowledge will be applied.
Table 2.
Biological Associations Relating to the IL4R Receptor
| Relation | Type | Reference |
|---|---|---|
| CD4 ---> IL4R | Regulation | [53, 54] |
| CD40 ---> IL4R | Expression | [55–58] |
| CD40LG --+> IL4R | Expression | [59, 60] |
| CD86 --+> IL4R | Regulation | [61] |
| IFNB1 ---> IL4R | Expression | [62] |
| IL12 --+> IL4R | Expression | [63–66] |
| IL13 --+> IL4R | Direct Regulation | [67–165] |
| IL1B ---> IL4R | Expression | [166–168] |
| IL4R --+> CD8A | Regulation | [169–171] |
| IL4R --+> CNR1 | Expression | [172] |
| IL4R --+> NGF | Molecular Transport | [173] |
| IL4R --+> STAT3 | Regulation | [174, 175] |
| IL4R ---- PTPN6 | Binding | [176–181] |
| IL4R ---- SOCS3 | Binding | [182] |
| IL4R ---- TP53 | Binding | [183] |
| IL4R ---> ADAM8 | Expression | [184] |
| IL4R ---> AKT1 | Regulation | [185, 186] |
| IL4R ---> CCL2 | Expression | [187] |
| IL4R ---> CD4 | Regulation | [188–195] |
| IL4R ---> CD86 | Expression | [57, 61, 196] |
| IL4R ---> EGFR | Regulation | [197] |
| IL4R ---> IL12 | Expression | [95, 198, 199] |
| IL4R ---> IL1B | Regulation | [83] |
| IL4R ---> MAPK14 | Regulation | [200] |
| IL4R ---> MAPK3 | Regulation | [156, 201] |
| IL4R ---> MAPK8 | Regulation | [156] |
| IL4R ---> STAT1 | Regulation | [202, 203] |
| IL4R ---| FAS | Regulation | [204, 205] |
| IL4R ---| IFNGR1 | Regulation | [202] |
| IL4R ---| MAPK1 | Regulation | [206] |
| IL4R ---| NOS2 | Regulation | [90] |
| IL4R ---| TLR4 | Regulation | [207] |
| MAPK1 --+> IL4R | Regulation | [208] |
| MAPK14 --+> IL4R | Expression | [209] |
| MIF --+> IL4R | Regulation | [210] |
| RAF1 ---> IL4R | Regulation | [211] |
| REL ---| IL4R | Expression | [212] |
| SOCS1 ---| IL4R | Regulation | [213] |
| STAT1 ---> IL4R | Expression | [214] |
| TNF --+> IL4R | Expression | [215–219] |
| TNFRSF1A ---> IL4R | Regulation | [217] |
--+>Positive influence.
---|Negative influence.
(Pathway Studio, Elsevier).
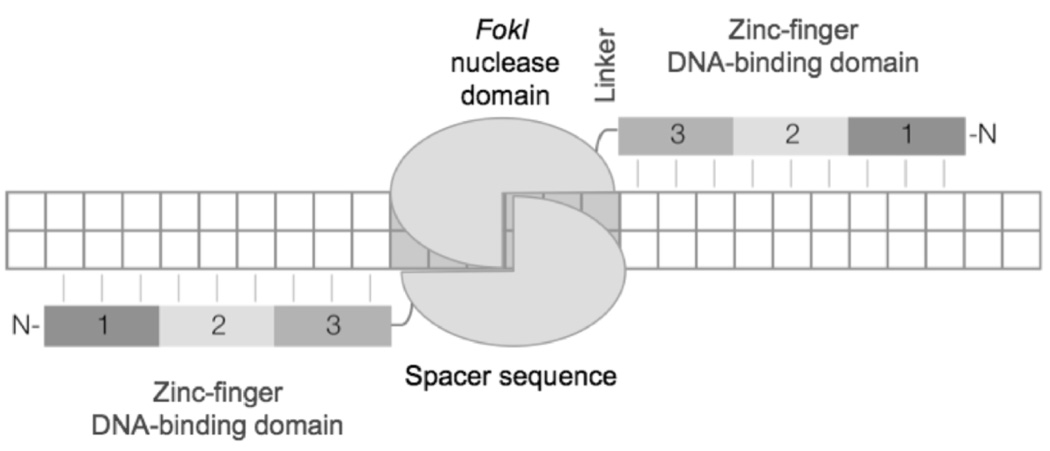
Fig. (2).
Zinc finger nuclease (ZFN)-mediated DNA double-strand break. A ZFN designed to create a DNA double-strand break (DSB) in the target consists of two monomers. Each monomer encompasses three zinc-fingers (1, 2, 3), which recognize 9 base pairs within the target and a FokI nuclease domain. A short “linker” sequence connects the two domains. The FokI nuclease only functions as a dimer and therefore, following dimerization the nuclease is activated and cleaves the DNA within the spacer sequence.
NEW MOLECULAR GENETIC METHODS FOR REPROGRAMMING CELLS
Technologies with the potential for editing the genome hold great promise for cell-based therapies. Termed zinc finger nucleases (ZFN), one such technology, which has matured significantly in recent years, combines the most abundant DNA binding motif, zinc fingers and the power of restriction endonucleases to provide sequence-specific modification of the genome. Zinc finger proteins (ZFPs), discovered in Xenopus the mid-1980’s [33], are the largest class of DNA-binding proteins found in eukaryotic cells. They serve diverse roles in most cell processes including DNA replication and repair, transcription, translation, metabolism and cell signaling among others [34]. Each zinc finger motif consists of approximately 30 amino acids folded into a ββα structure. A ZFP recognizes three bases in a DNA sequence via the single α-helical structure in the C-terminal region of the protein and binds by inserting the α-helix into the major groove of the DNA double helix [35]. The stability of the entire protein complex is afforded by the anti-parallel β-β hairpin structure present at the N-terminal region. The hairpin is created by the binding of a Zn2+ ion to two canonical cysteine residues that are generally 2–4 amino acids apart followed by the zinc ion interaction with two histidine residues, commonly referred to as the C2H2 zinc finger [36]. The discovery that several ZFPs linked in tandem are capable of recognizing a broad spectrum of DNA sequences with high specificity opened a “toolbox” capable of tinkering with the molecular machinery of a cell.
In 1996, a report on the generation of a fusion construct between zinc finger proteins and the nuclease domain of the non-discriminative, type IIS restriction enzyme FokI heralded a new era in DNA manipulation [37]. The uniqueness of this design was many-fold. The first among these was the ability to engineer tandem ZFPs to target specific DNA sequences. This implied that by linking engineered ZFPs in tandem, it was technically possible to target any DNA sequence in the human genome – a highly sought after molecular tool with the ability to manipulate the human genome at desired sites. Secondly, the use of the FokI restriction nuclease implied that enzymatic activity would only be present when the cleavage domain was present as a dimer, an intrinsic characteristic of the enzyme [38]. This empowers the ZFP-FokI hybrid, commonly referred to as a zinc finger nuclease (ZFN), with further specificity as each half of the nuclease dimer is fused to ZFPs flanking the desired cleavage site in DNA. Additionally, since the ZFPs bind to opposite strands of the DNA, the ZFN creates a highly desirable double-strand break (DSB) at the target locus in the genome. It is well established that cells employ the universal process of homologous recombination (HR) to mediate site-specific recombination following DSB in DNA in order to maintain genomic stability and integrity. This phenomenon offers another advantage to the ZFN technology whereby “correction” of the cleaved DNA helix can be afforded by introducing a targeting DNA sequence homologous to the cleaved segment but bearing the “corrected” or “edited” gene sequence. DSB repair of damaged DNA by HR is the most accurate form of cellular repair that usually employs the undamaged sister-chromatid as a template.
ZFN technology is gaining more wide use. Investigations in mammalian as well as other systems have revealed the key parameters that offer maximum efficacy of targeting. Continuous minor modifications are honing the technology. A plethora of studies have demonstrated the potential of the technology and clinical trials are underway [39–46]. We are planning to apply this technology to the genetic modification of microglial cells and their precursors.
SUMMARY
The clinical consequences of diffuse glioma are serious and their prognosis is dire. Symptoms range from neurological and other somatic deficits to cognitive and psychological problems. As a result, brain tumors cause the fourth highest loss of potential life years of all cancers. This justifies an intense research effort. Importantly, after decades of failure there is a clear case for more interdisciplinary research and specifically studies into the question of what CNS constituents do and do not do in support of glioma cell growth. By combining experimental neuropathological and immunological with some of the latest molecular genetics techniques, the approach outlined here will contribute to an improved understanding of bone marrow-derived microglia and their suitability for the treatment of CNS disorders. It will be tested using glioma as a first target. There is reason for optimism because successful cell-based treatments for nervous system disorders employing bone marrow-transplantation are already beginning to emerge in other areas [47]. If successful the results of the work proposed here are likely to be of relevance also for other cancers that are characterized by the presence of macrophages [48]. ZFNs and new methods such as TALENs that are currently being developed [49] are expected to facilitate the synthetic biological engineering of microglia precursors and will assist in making use of the microglia as a novel and powerful vehicle for treating glioma. Ultimately such engineered cells could also be made to carry a payload [50, 51] that may be of additional diagnostic as well as therapeutic utility through interactions with hadrons for instance [52] (Fig. 3).
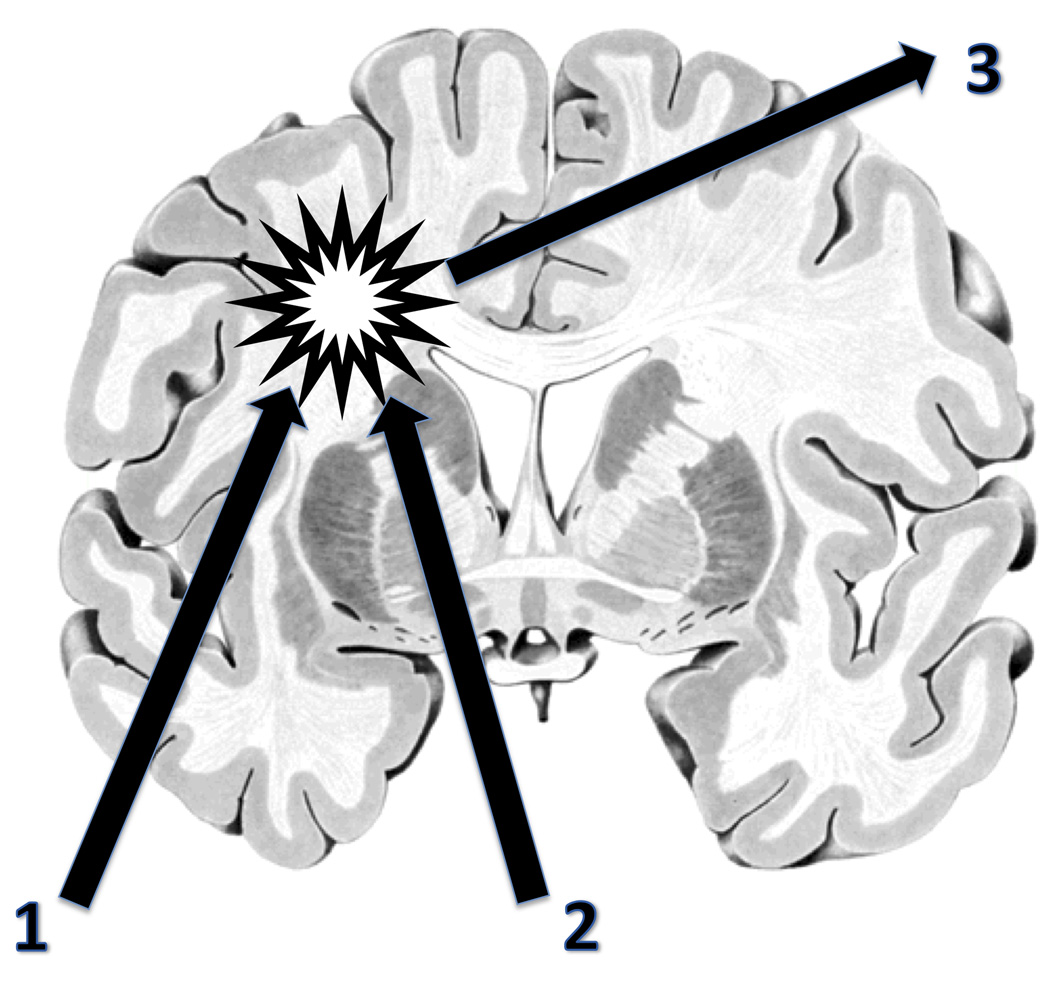
Fig. (3).
Vision for a future glioma treatment utilizing the potential of genetically enhanced microglia: two non-invasive strategies converging on the tumor. 1. Once the trafficking (entry into the CNS) and tracking (following glioma cells) behavior of the microglia can be controlled satisfactorily, these cells like other macrophages could also be employed to carry a payload [50, 51]. 2. Very precise and effective and therefore perhaps even curative tissue destruction/stimulation could then be achieved by means of a hadron beam [52]. 3. This approach may also allow real-time monitoring of the treatment if appropriate radiation sensors are implanted stereotactically (initially, for calibration purposes) and suitable radiochemicals for visualization become available. The glioma and its diffusely infiltrating cells is represented by the star. Coronal brain slice taken from inter BRAIN 1.1 for Windows, Springer 1998.
ACKNOWLEDGEMENTS
RMD Holsinger is the recipient of a NHMRC project grant (570398).
ABBREVIATIONS
- ADAM8
ADAM metallopeptidase domain 8
- AKT1
v-akt murine thymoma viral oncogene homolog 1
- CCL2
Chemokine (C-C motif) ligand 2
- CD4
CD4 molecule
- CD14
CD14 molecule
- CD40
CD40 molecule, TNF receptor super family member 5
- CD40LG
CD40 ligand
- CD86
CD86 molecule
- CD8A
CD8a molecule
- CNR1
Cannabinoid receptor 1
- CNS-1
Glioma cell line syngeneic to Lewis rats
- CNS
Central nervous system
- CSF1
Colony stimulating factor 1 (macrophage)
- CSF1R
Colony stimulating factor 1 receptor
- CSF2
Colony stimulating factor 2 (granulocyte-macrophage)
- CSF2RA
Colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
- DNA
Deoxyribonucleic acid
- DSB
Double-strand break
- EGFR
Epidermal growth factor receptor
- FAS
Fas (TNF receptor super family, member 6)
- FISH
Fluorescent in situ hybridization
- FOKI
A restriction endonuclease
- IFNB1
Interferon, beta 1
- IFNG
Interferon, gamma
- IFNGR2
Interferon gamma receptor 2 (interferon gamma transducer 1)
- IFNGR1
Interferon gamma receptor 1
- IL1B
Interleukin-1, beta
- IL4
Interleukin-4
- IL4R
Interleukin-4 receptor
- IL6
Interleukin-6 (interferon, beta 2)
- IL6R
Interleukin-6 receptor
- IL10
Interleukin-10
- IL10RA
Interleukin-10 receptor, alpha
- IL12
Interleukin-12
- IL13
Interleukin-13
- MAPK1
Mitogen-activated protein kinase 1
- MAPK14
Mitogen-activated protein kinase 14
- MAPK3
Mitogen-activated protein kinase 3
- MAPK8
Mitogen-activated protein kinase 8
- MIF
Macrophage migration inhibitory factor (glycosylation-inhibiting factor)
- NGF
Nerve growth factor (beta polypeptide)
- NOS2
Nitric oxide synthase 2, inducible
- PTPN6
Protein tyrosine phosphatase, non-receptor type 6
- RAF1
v-raf-1 murine leukemia viral oncogene homolog 1
- REL
v-rel reticuloendotheliosis viral oncogene homolog
- SOCS1
Suppressor of cytokine signaling 1
- SOCS3
Suppressor of cytokine signaling 3
- STAT1
Signal transducer and activator of transcription 1
- STAT3
Signal transducer and activator of transcription 3 (acute-phase response factor)
- TGFB1
Transforming growth factor, beta 1
- TGFBR2
Transforming growth factor, beta receptor II (70/80kDa)
- TLR4
Toll-like receptor 4
- TNF
Tumor necrosis factor
- TNFRSF1A
Tumor necrosis factor receptor super family, member 1A
- TP53
Tumor protein p53
- ZFN
Zinc finger nucleases
- ZFP
Zinc finger protein
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Li W, Graeber MB. The molecular profile of microglia under the influence of glioma. Neurooncology. 2012;14(8):958–978. doi: 10.1093/neuonc/nos116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graeber MB. Changing face of microglia. Science. 2010;330(6005):783–788. doi: 10.1126/science.1190929. [DOI] [PubMed] [Google Scholar]
- 3.Hulette CM. Microglioma, a histiocytic neoplasm of the central nervous system. Mod. Pathol. 1996;9(3):316–319. [PubMed] [Google Scholar]
- 4.Imamura S. Microglia in gliomas. Folia Psych. Neurol. Jpn. 1954;8(2):99–126. doi: 10.1111/j.1440-1819.1954.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 5.Penfield W. Microglia and the process of phagocytosis in gliomas. Am. J. Pathol. 1925;1(1):77–90. [PMC free article] [PubMed] [Google Scholar]
- 6.Grasbon-Frodl EM, Flügel A, Wolz P, Klinkert WE, Kreutzberg GW, Graeber MB. Jahrestagung der Neuroonkologischen Arbeitsgemeinschaft der Deutschen Gesellschaft für Neurochirurgie. Germany: Dresden; 1998. Nov 6–7, Untersuchungen zur Funktion von Mikroglia in Hirntumoren: Förderung des Wachstums von C6-Gliomzellen in vitro. (Abstract). [Google Scholar]
- 7.Flügel A, Kreutzberg GW, Graeber MB. Transformation of bone marrow-derived macrophages into ramified microglia during autoimmune inflammation of the axotomized rat facial nucleus. J. Neuroimmunol; 5th Congress of the International Society of Neuroimmunology; Montreal, Canada. August 23–27, 1998; 1998. p. 27. [Google Scholar]
- 8.Flügel A, Bradl M, Kreutzberg GW, Graeber MB. Transformation of donor-derived bone marrow precursors into host microglia during autoimmune CNS inflammation and during the retrograde response to axotomy. J. Neurosci. Res. 2001;66(1):74–82. doi: 10.1002/jnr.1198. [DOI] [PubMed] [Google Scholar]
- 9.Priller J, Flügel A, Wehner T, Boentert M, Haas CA, Prinz M, Fernández-Klett F, Prass K, Bechmann I, de Boer BA, Frotscher M, Kreutzberg GW, Persons DA, Dirnagl U. Targeting gene-modified hematopoietic cells to the central nervous system: use of green fluorescent protein uncovers microglial engraftment. Nat. Med. 2001;7(12):1356–1361. doi: 10.1038/nm1201-1356. [DOI] [PubMed] [Google Scholar]
- 10.Sica A, Larghi P, Mancino A, Rubino L, Porta C, Totaro MG, Rimoldi M, Biswas SK, Allavena P, Mantovani A. Macrophage polarization in tumour progression. Semin. Cancer Biol. 2008;18(5):349–355. doi: 10.1016/j.semcancer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Zhai H, Heppner FL, Tsirka SE. Microglia/macrophages promote glioma progression. Glia. 2010;59(3):472–485. doi: 10.1002/glia.21117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Pawelek JM, Chakraborty AK. Fusion of tumour cells with bone marrow-derived cells: a unifying explanation for metastasis. Nat. Rev. Cancer. 2008;8(5):377–386. doi: 10.1038/nrc2371. [DOI] [PubMed] [Google Scholar]
- 14.Zheng P-P, van der Weiden M, van der Spek PJ, Vincent AJPE, Kros JM. Isocitrate dehydrogenase 1R132H mutation in microglia/macrophages in gliomas: Indication of a significant role of microglia/macrophages in glial tumorigenesis. Cancer Biol. Ther. 2012;13(10) doi: 10.4161/cbt.20836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graeber MB, Flügel A, Moran LB, Scheithauer BW. In: Immune Biology of Brain Tumours. Stavrou D, Hagel C, editors. München-Orlando: Dustri-Verlag Dr. Karl Feistle; 2010. pp. 69–80. [Google Scholar]
- 16.Chen S-K, Tvrdik P, Peden E, Cho S, Wu S, Spangrude G, Capecchi MR. Hematopoietic origin of pathological grooming in hoxb8 mutant mice. Cell. 2010;141(5):775–785. doi: 10.1016/j.cell.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derecki NC, Cronk JC, Lu Z, Xu E, Abbott SBG, Guyenet PG, Kipnis J. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature. 2012;484(7392):105–109. doi: 10.1038/nature10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capotondo A, Milazzo R, Politi LS, Quattrini A, Palini A, Plati T, Merella S, Nonis A, di Serio C, Montini E, Naldini L, Biffi A. Brain conditioning is instrumental for successful microglia reconstitution following hematopoietic stem cell transplantation. Proc. Natl. Acad. Sci. USA. 2012;109(37):15018–15023. doi: 10.1073/pnas.1205858109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruse CA, Molleston MC, Parks EP, Schiltz PM, Kleinschmidt-DeMasters BK, Hickey WF. A rat glioma model, CNS-1, with invasive characteristics similar to those of human gliomas: a comparison to 9L gliosarcoma. J. Neurooncol. 1994;22(3):191–200. doi: 10.1007/BF01052919. [DOI] [PubMed] [Google Scholar]
- 20.Owens GC, Orr EA, De Masters BK, Muschel RJ, Berens ME, Kruse CA. Overexpression of a transmembrane isoform of neural cell adhesion molecule alters the invasiveness of rat CNS-1 glioma. Cancer Res. 1998;58(9):2020–2028. [PubMed] [Google Scholar]
- 21.Graeber MB, Flügel A. Bone marrow-derived microglia in experimental glioma. 40th Annual Meeting of the Society for Neuroscience; San Diego, USA. 13–17 November 2010; Poster 262.21.2010. [Google Scholar]
- 22.Moran LB, Duke DC, Turkheimer FE, Banati RB, Graeber MB. Towards a transcriptome definition of microglial cells. Neurogenetics. 2004;5(2):95–108. doi: 10.1007/s10048-004-0172-5. [DOI] [PubMed] [Google Scholar]
- 23.Moran LB, Duke DC, Graeber MB. The microglial gene regulatory network activated by interferon-gamma. J. Neuroimmunol. 2007;183(1–2):1–6. doi: 10.1016/j.jneuroim.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Moran LB, Graeber MB. Towards a pathway definition of Parkinson’s disease: a complex disorder with links to cancer, diabetes and inflammation. Neurogenetics. 2008;9(1):1–13. doi: 10.1007/s10048-007-0116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SA, Ryu CH, Kim SM, Lim JY, Park SI, Jeong CH, Jun JA, Oh JH, Park SH, Oh W, Jeun SS. CXCR4-transfected human umbilical cord blood-derived mesenchymal stem cells exhibit enhanced migratory capacity toward gliomas. Intl. J. Oncol. 2010;38(1) [PubMed] [Google Scholar]
- 26.Zhao Y, Wang S. Human NT2 neural precursor-derived tumor-infiltrating cells as delivery vehicles for treatment of glioblastoma. Hum. Gene Ther. 2010;21(6):683–694. doi: 10.1089/hum.2009.196. [DOI] [PubMed] [Google Scholar]
- 27.Lee DH, Ahn Y, Kim SU, Wang KC, Cho BK, Phi JH, Park IH, Black PM, Carroll RS, Lee J, Kim SK. Targeting rat brainstem glioma using human neural stem cells and human mesenchymal stem cells. Clin. Cancer Res. 2009;15(15):4925–4934. doi: 10.1158/1078-0432.CCR-08-3076. [DOI] [PubMed] [Google Scholar]
- 28.An JH, Lee SY, Jeon JY, Cho KG, Kim SU, Lee MA. Identification of gliotropic factors that induce human stem cell migration to malignant tumor. J. Prot. Res. 2009;8(6):2873–2881. doi: 10.1021/pr900020q. [DOI] [PubMed] [Google Scholar]
- 29.Magge SN, Malik SZ, Royo NC, Chen HI, Yu L, Snyder EY, O’Rourke DM, Watson DJ. Role of monocyte chemoattractant protein-1 (MCP-1/CCL2) in migration of neural progenitor cells toward glial tumors. J. Neurosci. Res. 2009;87(7):1547–1555. doi: 10.1002/jnr.21983. [DOI] [PubMed] [Google Scholar]
- 30.Kim D, Lee J, Choi S, Kim J, Jeun S, Oh W, Yang Y, Chang J. Overexpression of CXC chemokine receptors is required for the superior glioma-tracking property of umbilical cord blood-derived mesenchymal stem cells. Stem Cells Devel. 2008 doi: 10.1089/scd.2008.0050. [DOI] [PubMed] [Google Scholar]
- 31.Jurvansuu J, Zhao Y, Leung DS, Boulaire J, Yu YH, Ahmed S, Wang S. Transmembrane protein 18 enhances the tropism of neural stem cells for glioma cells. Cancer Res. 2008;68(12):4614–4622. doi: 10.1158/0008-5472.CAN-07-5291. [DOI] [PubMed] [Google Scholar]
- 32.Xu F, Zhu J-H. Stem cells tropism for malignant gliomas. Neurosci. Bull. 2007;23(6):363–369. doi: 10.1007/s12264-007-0054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31(2):532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 36.Diakun GP, Fairall L, Klug A. EXAFS study of the zinc-binding sites in the protein transcription factor IIIA. Nature. 1986;324(6098):698–699. doi: 10.1038/324698a0. [DOI] [PubMed] [Google Scholar]
- 37.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wah DA, Bitinaite J, Schildkraut I, Aggarwal AK. Structure of FokI has implications for DNA cleavage. Proc. Natl. Acad. Sci. USA. 1998;95(18):10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wayengera M. Zinc finger nucleases for targeted mutagenesis and repair of the sickle-cell disease mutation: An insilico study. BMC Blood Disord. 2012;12(1):5. doi: 10.1186/1471-2326-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat. Biotechnol. 2010:1–5. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 41.Liu P-Q, Chan EM, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Holmes MC, Collingwood TN, Gregory PD. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol. Bioengin. 2010 doi: 10.1002/bit.22654. [DOI] [PubMed] [Google Scholar]
- 42.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325(5939):433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santiago Y, Liu P-Q, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA. 2008;105(15):5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moehle EA, Moehle EA, Rock JM, Rock JM, Lee Y-L, Lee YL, Jouvenot Y, Jouvenot Y, DeKelver RC, Dekelver RC, Gregory PD, Gregory PD, Urnov FD, Urnov FD, Holmes MC, Holmes MC. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl. Acad. Sci. USA. 2007;104(9):3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urnov FD, Miller JC, Lee Y-L, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 46.Cannon P, June C. Chemokine receptor 5 knockout strategies. Curr. Opin. HIV AIDS. 2011;6(1):74–79. doi: 10.1097/COH.0b013e32834122d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brenneman M, Sharma S, Harting M, Strong R, Cox CS, Aronowski J, Grotta JC, Savitz SI. Autologous bone marrow mononuclear cells enhance recovery after acute ischemic stroke in young and middle-aged rats. J. Cereb. Blood Flow Metab. 2009;30(1):140–149. doi: 10.1038/jcbfm.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pollard JW. Macrophages define the invasive microenvironment in breast cancer. J. Leukocyte Biol. 2008;84(3):623–630. doi: 10.1189/jlb.1107762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, II, Tan W, Penheiter SG, Ma AC, Leung AYH, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a highefficiency TALEN system. Nature. 2012;491(7422):114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madsen SJ, Baek S-K, Makkouk AR, Krasieva T, Hirschberg H. Macrophages as cell-based delivery systems for nanoshells in photothermal therapy. Ann. Biomed. Engin. 2011;40(2):507–515. doi: 10.1007/s10439-011-0415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi J, Kim H-Y, Ju EJ, Jung J, Park J, Chung H-K, Lee JS, Lee JS, Park HJ, Song SY, Jeong S-Y, Choi EK. Use of macrophages to deliver therapeutic and imaging contrast agents to tumors. Biomaterials. 2012;33(16):4195–4203. doi: 10.1016/j.biomaterials.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 52.Boldeman JW, Banati R. Options for a light ion facility for hadron therapy and research. J. Proc. R. Soc. NSW. 2011;144:58–65. [Google Scholar]
- 53.Tanaka Y, Hamano S, Gotoh K, Murata Y, Kunisaki Y, Nishikimi A, Takii R, Kawaguchi M, Inayoshi A, Masuko S, Himeno K, Sasazuki T, Fukui Y. T helper type 2 differentiation and intracellular trafficking of the interleukin 4 receptor-alpha subunit controlled by the Rac activator Dock2. Nat. Immunol. 2007;8(10):1067–1075. doi: 10.1038/ni1506. [DOI] [PubMed] [Google Scholar]
- 54.Perona-Wright G, Mohrs K, Mayer KD, Mohrs M. Differential regulation of IL-4Ralpha expression by antigen versus cytokine stimulation characterizes Th2 progression in vivo. J. Immunol. 2010;184(2):615–623. doi: 10.4049/jimmunol.0902408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haas KM, Estes DM. Activation of bovine B cells via surface immunoglobulin M cross-linking or CD40 ligation results in different B-cell phenotypes. Immunology. 2000;99(2):272–278. doi: 10.1046/j.1365-2567.2000.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Atamas SP, Luzina IG, Dai H, Wilt SG, White B. Synergy between CD40 ligation and IL-4 on fibroblast proliferation involves IL-4 receptor signaling. J. Immunol. 2002;168(3):1139–1145. doi: 10.4049/jimmunol.168.3.1139. [DOI] [PubMed] [Google Scholar]
- 57.Jirapongsananuruk O, Hofer MF, Trumble AE, Norris DA, Leung DY. Enhanced expression of B7.2 (CD86) in patients with atopic dermatitis: a potential role in the modulation of IgE synthesis. J. Immunol. 1998;160(9):4622–4627. [PubMed] [Google Scholar]
- 58.Wagner EF, Hanna N, Fast LD, Kouttab N, Shank PR, Vazquez A, Sharma S. Novel diversity in IL-4-mediated responses in resting human naive B cells versus germinal center/memory B cells. J. Immunol. 2000;165(10):5573–5579. doi: 10.4049/jimmunol.165.10.5573. [DOI] [PubMed] [Google Scholar]
- 59.Avery DT, Ma CS, Bryant VL, Santner-Nanan B, Nanan R, Wong M, Fulcher DA, Cook MC, Tangye SG. STAT3 is required for IL-21-induced secretion of IgE from human naive B cells. Blood. 2008;112(5):1784–1793. doi: 10.1182/blood-2008-02-142745. [DOI] [PubMed] [Google Scholar]
- 60.Tian C, Kron GK, Dischert KM, Higginbotham JN, Crowe JE. Low expression of the interleukin (IL)-4 receptor alpha chain and reduced signalling via the IL-4 receptor complex in human neonatal B cells. Immunology. 2006;119(1):54–62. doi: 10.1111/j.1365-2567.2006.02405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Podojil JR, Sanders VM. Selective regulation of mature IgG1 transcription by CD86 and beta 2-adrenergic receptor stimulation. J. Immunol. 2003;170(10):5143–5151. doi: 10.4049/jimmunol.170.10.5143. [DOI] [PubMed] [Google Scholar]
- 62.Shirey KA, Pletneva LM, Puche AC, Keegan AD, Prince GA, Blanco JCG, Vogel SN. Control of RSV-induced lung injury by alternatively activated macrophages is IL-4R alpha-, TLR4-, and IFN-beta-dependent. Muc. Immunol. 2010;3(3):291–300. doi: 10.1038/mi.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naume B, Gately MK, Desai BB, Sundan A, Espevik T. Synergistic effects of interleukin 4 and interleukin 12 on NK cell proliferation. Cytokine. 1993;5(1):38–46. doi: 10.1016/1043-4666(93)90022-w. [DOI] [PubMed] [Google Scholar]
- 64.Oriss TB, McCarthy SA, Campana MA, Morel PA. Evidence of positive cross-regulation on Th1 by Th2 and antigen-presenting cells: effects on Th1 induced by IL-4 and IL-12. J. Immunol. 1999;162(4):1999–2007. [PubMed] [Google Scholar]
- 65.Moll H, Scharner A, Kämpgen E. Increased interleukin 4 (IL-4) receptor expression and IL-4-induced decrease in IL-12 production by Langerhans cells infected with Leishmania major. Infect. Immun. 2002;70(3):1627–1630. doi: 10.1128/IAI.70.3.1627-1630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jankovic D, Kullberg MC, Noben-Trauth N, Caspar P, Ward JM, Cheever AW, Paul WE, Sher A. Schistosome-infected IL-4 receptor knockout (KO) mice, in contrast to IL-4 KO mice, fail to develop granulomatous pathology while maintaining the same lymphokine expression profile. J. Immunol. 1999;163(1):337–342. [PubMed] [Google Scholar]
- 67.Yang X, Lee J, Brooks J, Wilhelm J, Myszka D, Kasaian MT, Goldman S, Wolf S, Fitz LJ. Binding characterization of the interleukin-13 signaling complex and development of a ternary time-resolved fluorescence resonance energy transfer assay. Anal. Biochem. 2008;376(2):206–212. doi: 10.1016/j.ab.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 68.Mueller TD, Zhang JL, Sebald W, Duschl A. Structure, binding, and antagonists in the IL-4/IL-13 receptor system. Biochim. Biophys. Acta. 2002;1592(3):237–250. doi: 10.1016/s0167-4889(02)00318-x. [DOI] [PubMed] [Google Scholar]
- 69.Berry LM, Adams R, Airey M, Bracher MG, Bourne T, Carrington B, Cross AS, Davies GC, Finney HM, Foulkes R, Gozzard N, Griffin RA, Hailu H, Lamour SD, Lawson AD, Lightwood DJ, McKnight AJ, O'Dowd VL, Oxbrow AK, Popplewell AG, Shaw S, Stephens PE, Sweeney B, Tomlinson KL, Uhe C, Palframan RT. In vitro and in vivo characterisation of anti-murine IL-13 antibodies recognising distinct functional epitopes. Int. Immunopharmacol. 2009;9(2):201–206. doi: 10.1016/j.intimp.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 70.Madala SK, Dolan MA, Sharma D, Ramalingam TR, Wilson MS, Mentink-Kane MM, Masison DC, Wynn TA. Mapping mouse IL-13 binding regions using structure modeling, molecular docking, and high-density peptide microarray analysis. Proteins. 2011;79(1):282–293. doi: 10.1002/prot.22881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kioi M, Kawakami K, Puri RK. Mechanism of action of interleukin-13 antagonist (IL-13E13K) in cells expressing various types of IL-4R. Cell Immunol. 2004;229(1):41–51. doi: 10.1016/j.cellimm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 72.Obiri NI, Debinski W, Leonard WJ, Puri RK. Receptor for interleukin 13. Interaction with interleukin 4 by a mechanism that does not involve the common gamma chain shared by receptors for interleukins 2, 4, 7, 9, and 15. J. Biol. Chem. 1995;270(15):8797–8804. doi: 10.1074/jbc.270.15.8797. [DOI] [PubMed] [Google Scholar]
- 73.Terada N, Hamano N, Nomura T, Numata T, Hirai K, Nakajima T, Yamada H, Yoshie O, Ikeda-Ito T, Konno A. Interleukin-13 and tumour necrosis factor-alpha synergistically induce eotaxin production in human nasal fibroblasts. Clin. Exp. Allergy. 2000;30(3):348–355. doi: 10.1046/j.1365-2222.2000.00750.x. [DOI] [PubMed] [Google Scholar]
- 74.Honjo E, Shoyama Y, Tamada T, Shigematsu H, Hatanaka T, Kanaji S, Arima K, Ito Y, Izuhara K, Kuroki R. Expression of the extracellular region of the human interleukin-4 receptor alpha chain and interleukin-13 receptor alpha1 chain by a silkworm-baculovirus system. Prot. Expr. Purif. 2008;60(1):25–30. doi: 10.1016/j.pep.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 75.Raia V, Schilling M, Böhm M, Hahn B, Kowarsch A, Raue A, Sticht C, Bohl S, Saile M, Möller P, Gretz N, Timmer J, Theis F, Lehmann W-D, Lichter P, Klingmüller U. Dynamic mathematical modeling of IL13-induced signaling in Hodgkin and primary mediastinal B-cell lymphoma allows prediction of therapeutic targets. Cancer Res. 2011;71(3):693–704. doi: 10.1158/0008-5472.CAN-10-2987. [DOI] [PubMed] [Google Scholar]
- 76.Howard TD, Koppelman GH, Xu J, Zheng SL, Postma DS, Meyers DA, Bleecker ER. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am. J. Hum. Genet. 2002;70(1):230–236. doi: 10.1086/338242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smerz-Bertling C, Duschl A. Both interleukin 4 and interleukin 13 induce tyrosine phosphorylation of the 140-kDa subunit of the interleukin 4 receptor. J. Biol. Chem. 1995;270(2):966–970. doi: 10.1074/jbc.270.2.966. [DOI] [PubMed] [Google Scholar]
- 78.Skapenko A, Kalden JR, Lipsky PE, Schulze-Koops H. The IL-4 receptor alpha-chain-binding cytokines, IL-4 and IL-13, induce forkhead box P3-expressing CD25+CD4+ regulatory T cells from CD25-CD4+ precursors. J. Immunol. 2005;175(9):6107–6116. doi: 10.4049/jimmunol.175.9.6107. [DOI] [PubMed] [Google Scholar]
- 79.Kondo M, Tamaoki J, Takeyama K, Nakata J, Nagai A. Interleukin-13 induces goblet cell differentiation in primary cell culture from Guinea pig tracheal epithelium. Am. J. Respir Cell Mol. Biol. 2002;27(5):536–541. doi: 10.1165/rcmb.4682. [DOI] [PubMed] [Google Scholar]
- 80.Mandal D, Fu P, Levine AD. REDOX regulation of IL-13 signaling in intestinal epithelial cells: usage of alternate pathways mediates distinct gene expression patterns. Cell. Signal. 2010;22(10):1485–1494. doi: 10.1016/j.cellsig.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81.Morimoto M, Morimoto M, Zhao A, Madden KB, Dawson H, Finkelman FD, Mentink-Kane M, Urban JF, Wynn TA, Shea-Donohue T. Functional importance of regional differences in localized gene expression of receptors for IL-13 in murine gut. J. Immunol. 2006;176(1):491–495. doi: 10.4049/jimmunol.176.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee SG, Kim BS, Kim JH, Lee SY, Choi SO, Shim JY, Hong TJ, Hong SJ. Gene-gene interaction between interleukin-4 and interleukin-4 receptor alpha in Korean children with asthma. Clin. Exp. Allergy. 2004;34(8):1202–1208. doi: 10.1111/j.1365-2222.2004.02015.x. [DOI] [PubMed] [Google Scholar]
- 83.Hirst SJ, Hallsworth MP, Peng Q, Lee TH. Selective induction of eotaxin release by interleukin-13 or interleukin-4 in human airway smooth muscle cells is synergistic with interleukin-1beta and is mediated by the interleukin-4 receptor alpha-chain. Am. J. Respir. Crit. Care Med. 2002;165(8):1161–1171. doi: 10.1164/ajrccm.165.8.2107158. [DOI] [PubMed] [Google Scholar]
- 84.Ostrand-Rosenberg S, Clements VK, Terabe M, Park JM, Berzofsky JA, Dissanayake SK. Resistance to metastatic disease in STAT6-deficient mice requires hemopoietic and non hemopoietic cells and is IFN-gamma dependent. J. Immunol. 2002;169(10):5796–5804. doi: 10.4049/jimmunol.169.10.5796. [DOI] [PubMed] [Google Scholar]
- 85.Yasunaga S, Yuyama N, Arima K, Tanaka H, Toda S, Maeda M, Matsui K, Goda C, Yang Q, Sugita Y, Nagai H, Izuhara K. The negative-feedback regulation of the IL-13 signal by the IL-13 receptor alpha2 chain in bronchial epithelial cells. Cytokine. 2003;24(6):293–303. doi: 10.1016/j.cyto.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 86.Andrews AL, Holloway JW, Puddicombe SM, Holgate ST, Davies DE. Kinetic analysis of the interleukin-13 receptor complex. J. Biol. Chem. 2002;277(48):46073–46078. doi: 10.1074/jbc.M209560200. [DOI] [PubMed] [Google Scholar]
- 87.Antoniu SA. Pitrakinra, a dual IL-4/IL-13 antagonist for the potential treatment of asthma and eczema. Curr. Opin. Invest. Drugs. 2010;11(11):1286–1294. [PubMed] [Google Scholar]
- 88.Faffe DS, Flynt L, Bourgeois K, Panettieri RA, Shore SA. Interleukin-13 and interleukin-4 induce vascular endothelial growth factor release from airway smooth muscle cells: role of vascular endothelial growth factor genotype. Am. J. Respir. Cell Mol. Biol. 2006;34(2):213–218. doi: 10.1165/rcmb.2005-0147OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Finkelman FD, Yang M, Perkins C, Schleifer K, Sproles A, Santeliz J, Bernstein JA, Rothenberg ME, Morris SC, Wills-Karp M. Suppressive effect of IL-4 on IL-13-induced genes in mouse lung. J. Immunol. 2005;174(8):4630–4638. doi: 10.4049/jimmunol.174.8.4630. [DOI] [PubMed] [Google Scholar]
- 90.Bian K, Zhong M, Harari Y, Lai M, Weisbrodt N, Murad F. Helminth regulation of host IL-4Ralpha/Stat6 signaling: mechanism underlying NOS-2 inhibition by Trichinella spiralis. Proc. Natl. Acad. Sci. USA. 2005;102(11):3936–3941. doi: 10.1073/pnas.0409461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kasaian MT, Tan XY, Jin M, Fitz L, Marquette K, Wood N, Cook TA, Lee J, Widom A, Agostinelli R, Bree A, Schlerman FJ, Olland S, Wadanoli M, Sypek J, Gill D, Goldman SJ, Tchistiakova L. Interleukin-13 neutralization by two distinct receptor blocking mechanisms reduces immunoglobulin E responses and lung inflam mation in cynomolgus monkeys. J. Pharmacol. Exp. Ther. 2008;325(3):882–892. doi: 10.1124/jpet.108.136515. [DOI] [PubMed] [Google Scholar]
- 92.Nakano T, Inoue H, Fukuyama S, Matsumoto K, Matsumura M, Tsuda M, Matsumoto T, Aizawa H, Nakanishi Y. Niflumic acid suppresses interleukin-13-induced asthma phenotypes. Am. J. Respir. Crit. Care Med. 2006;173(11):1216–1221. doi: 10.1164/rccm.200410-1420OC. [DOI] [PubMed] [Google Scholar]
- 93.Wong GW, Foster PS, Yasuda S, Qi JC, Mahalingam S, Mellor EA, Katsoulotos G, Li L, Boyce JA, Krilis SA, Stevens RL. Biochemical and functional characterization of human transmembrane tryptase (TMT)/tryptase gamma. TMT is an exocytosed mast cell protease that induces airway hyper responsiveness in vivo via an interleukin-13/interleukin-4 receptor alpha/signal transducer and activator of transcription (STAT) 6-dependent pathway. J. Biol. Chem. 2002;277(44):41906–41915. doi: 10.1074/jbc.M205868200. [DOI] [PubMed] [Google Scholar]
- 94.Kawakami M, Kawakami K, Takahashi S, Abe M, Puri RK. Analysis of interleukin-13 receptor alpha2 expression in human pediatric brain tumors. Clin. Cancer Res. 2004;101(5):1036–1042. doi: 10.1002/cncr.20470. [DOI] [PubMed] [Google Scholar]
- 95.Hölscher C, Arendse B, Schwegmann A, Myburgh E, Brombacher F. Impairment of alternative macrophage activation delays cutaneous leishmaniasis in non healing BALB/c mice. J. Immunol. 2006;176(2):1115–1121. doi: 10.4049/jimmunol.176.2.1115. [DOI] [PubMed] [Google Scholar]
- 96.Mochizuki M, Bartels J, Mallet AI, Christophers E, Schröder JM. IL-4 induces eotaxin: a possible mechanism of selective eosinophil recruitment in helminth infection and atopy. J. Immunol. 1998;160(1):60–68. [PubMed] [Google Scholar]
- 97.Junttila IS, Mizukami K, Dickensheets H, Meier-Schellersheim M, Yamane H, Donnelly RP, Paul WE. Tuning sensitivity to IL-4 and IL-13: differential expression of IL-4Ralpha, IL-13Ralpha1, and gammac regulates relative cytokine sensitivity. J. Exp. Med. 2008;205(11):2595–2608. doi: 10.1084/jem.20080452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zheng T, Liu W, Oh SY, Zhu Z, Hu B, Homer RJ, Cohn L, Grusby MJ, Elias JA. IL-13 receptor alpha2 selectively inhibits IL-13-induced responses in the murine lung. J. Immunol. 2008;180(1):522–529. doi: 10.4049/jimmunol.180.1.522. [DOI] [PubMed] [Google Scholar]
- 99.Malabarba MG, Rui H, Deutsch HH, Chung J, Kalthoff FS, Farrar WL, Kirken RA. Interleukin-13 is a potent activator of JAK3 and STAT6 in cells expressing interleukin-2 receptorgamma and interleukin-4 receptor-alpha. Biochem. J. 1996;319(Pt 3):865–872. doi: 10.1042/bj3190865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang JG, Hilton DJ, Willson TA, McFarlane C, Roberts BA, Moritz RL, Simpson RJ, Alexander WS, Metcalf D, Nicola NA. Identification, purification, and characterization of a soluble interleukin (IL)-13-binding protein. Evidence that it is distinct from the cloned Il-13 receptor and Il-4 receptor alphachains. J. Biol. Chem. 1997;272(14):9474–9480. doi: 10.1074/jbc.272.14.9474. [DOI] [PubMed] [Google Scholar]
- 101.Palmer-Crocker RL, Hughes CC, Pober JS. IL-4 and IL-13 activate the JAK2 tyrosine kinase and Stat6 in cultured human vascular endothelial cells through a common pathway that does not involve the gamma c chain. J. Clin. Invest. 1996;98(3):604–609. doi: 10.1172/JCI118829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen W, Sivaprasad U, Tabata Y, Gibson AM, Stier MT, Finkelman FD, Hershey GKK. IL-13R alpha 2 membrane and soluble isoforms differ in humans and mice. J. Immunol. 2009;183(12):7870–7876. doi: 10.4049/jimmunol.0901028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oshima Y, Puri RK. Characterization of a powerful high affinity antagonist that inhibits biological activities of human interleukin-13. J. Biol. Chem. 2001;276(18):15185–15191. doi: 10.1074/jbc.M010159200. [DOI] [PubMed] [Google Scholar]
- 104.Orchansky PL, Kwan R, Lee F, Schrader JW. Characterization of the cytoplasmic domain of interleukin-13 receptor-alpha. J. Biol. Chem. 1999;274(30):20818–20825. doi: 10.1074/jbc.274.30.20818. [DOI] [PubMed] [Google Scholar]
- 105.Kagami S, Saeki H, Komine M, Kakinuma T, Tsunemi Y, Nakamura K, Sasaki K, Asahina A, Tamaki K. Interleukin-4 and interleukin-13 enhance CCL26 production in a human keratinocyte cell line, HaCaT cells. Clin. Exp. Immunol. 2005;141(3):459–466. doi: 10.1111/j.1365-2249.2005.02875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Elias JA, Lee CG, Zheng T, Ma B, Homer RJ, Zhu Z. New insights into the pathogenesis of asthma. J. Clin. Invest. 2003;111(3):291–297. doi: 10.1172/JCI17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Andrews AL, Holloway JW, Holgate ST, Davies DE. IL-4 receptor alpha is an important modulator of IL-4 and IL-13 receptor binding: implications for the development of therapeutic targets. J. Immunol. 2006;176(12):7456–7461. doi: 10.4049/jimmunol.176.12.7456. [DOI] [PubMed] [Google Scholar]
- 108.Zuegg J, Webb DC, Foster PS, Casarotto MG. Structural model of human IL-13 defines the spatial interactions with the IL- 13Ralpha/IL-4Ralpha receptor. Immunol. Cell Biol. 2001;79(4):332–339. doi: 10.1046/j.1440-1711.2001.01035.x. [DOI] [PubMed] [Google Scholar]
- 109.Tachdjian R, Mathias C, Al Khatib S, Bryce PJ, Kim HS, Blaeser F, O'Connor BD, Rzymkiewicz D, Chen A, Holtzman MJ, Hershey GK, Garn H, Harb H, Renz H, Oettgen HC, Chatila TA. Pathogenicity of a disease-associated human IL-4 receptor allele in experimental asthma. J. Exp. Med. 2009;206(10):2191–2204. doi: 10.1084/jem.20091480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fukushi J, Ono M, Morikawa W, Iwamoto Y, Kuwano M. The activity of soluble VCAM-1 in angiogenesis stimulated by IL-4 and IL-13. J. Immunol. 2000;165(5):2818–2823. doi: 10.4049/jimmunol.165.5.2818. [DOI] [PubMed] [Google Scholar]
- 111.Takeda N, O&apos, Dea EL, Doedens A, Kim J-w, Weidemann A, Stockmann C, Asagiri M, Simon MC, Hoffmann A, Johnson RS. Differential activation and antagonistic function of HIF-{alpha} isoforms in macrophages are essential for NO homeostasis. Genes Develop. 2010;24(5):491–501. doi: 10.1101/gad.1881410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zurawski SM, Chomarat P, Djossou O, Bidaud C, McKenzie AN, Miossec P, Banchereau J, Zurawski G. The primary binding subunit of the human interleukin-4 receptor is also a component of the interleukin-13 receptor. J. Biol. Chem. 1995;270(23):13869–13878. doi: 10.1074/jbc.270.23.13869. [DOI] [PubMed] [Google Scholar]
- 113.Reibman J, Hsu Y, Chen LC, Bleck B, Gordon T. Airway epithelial cells release MIP-3alpha/CCL20 in response to cytokines and ambient particulate matter. Am. J. Respir. Cell Mol. Biol. 2003;28(6):648–654. doi: 10.1165/rcmb.2002-0095OC. [DOI] [PubMed] [Google Scholar]
- 114.Henderson WR, Chi EY, Maliszewski CR. Soluble IL-4 receptor inhibits airway inflammation following allergen challenge in a mouse model of asthma. J. Immunol. 2000;164(2):1086–1095. doi: 10.4049/jimmunol.164.2.1086. [DOI] [PubMed] [Google Scholar]
- 115.Kelly-Welch AE, Melo ME, Smith E, Ford AQ, Haudenschild C, Noben-Trauth N, Keegan AD. Complex role of the IL-4 receptor alpha in a murine model of airway inflammation: expression of the IL-4 receptor alpha on nonlymphoid cells of bone marrow origin contributes to severity of inflammation. J. Immunol. 2004;172(7):4545–4555. doi: 10.4049/jimmunol.172.7.4545. [DOI] [PubMed] [Google Scholar]
- 116.Kozma N, Halasz M, Polgar B, Poehlmann TG, Markert UR, Palkovics T, Keszei M, Par G, Kiss K, Szeberenyi J, Grama L, Szekeres-Bartho J. Progesterone-induced blocking factor activates STAT6 via binding to a novel IL-4 receptor. J. Immunol. 2006;176(2):819–826. doi: 10.4049/jimmunol.176.2.819. [DOI] [PubMed] [Google Scholar]
- 117.Mattes J, Yang M, Siqueira A, Clark K, MacKenzie J, McKenzie AN, Webb DC, Matthaei KI, Foster PS. IL-13 induces airways hyper reactivity independently of the IL-4R alpha chain in the allergic lung. J. Immunol. 2001;167(3):1683–1692. doi: 10.4049/jimmunol.167.3.1683. [DOI] [PubMed] [Google Scholar]
- 118.Welham MJ, Learmonth L, Bone H, Schrader JW. Interleukin-13 signal transduction in lymphohemopoietic cells. Similarities and differences in signal transduction with interleukin-4 and insulin. J. Biol. Chem. 1995;270(20):12286–12296. doi: 10.1074/jbc.270.20.12286. [DOI] [PubMed] [Google Scholar]
- 119.Strait RT, Morris SC, Smiley K, Urban JF, Finkelman FD. IL-4 exacerbates anaphylaxis. J. Immunol. 2003;170(7):3835–3842. doi: 10.4049/jimmunol.170.7.3835. [DOI] [PubMed] [Google Scholar]
- 120.Ohta Y, Hayashi M, Kanemaru T, Abe K, Ito Y, Oike M. Dual modulation of airway smooth muscle contraction by Th2 cytokines via matrix metalloproteinase-1 production. J. Immunol. 2008;180(6):4191–4199. doi: 10.4049/jimmunol.180.6.4191. [DOI] [PubMed] [Google Scholar]
- 121.Yamada A, Takami M, Kawawa T, Yasuhara R, Zhao B, Mochizuki A, Miyamoto Y, Eto T, Yasuda H, Nakamichi Y, Kim N, Katagiri T, Suda T, Kamijo R. Interleukin-4 inhibition of osteoclast differentiation is stronger than that of interleukin-13 and they are equivalent for induction of osteoprotegerin production from osteoblasts. Immunology. 2007;120(4):573–579. doi: 10.1111/j.1365-2567.2006.02538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Orchansky PL, Ayres SD, Hilton DJ, Schrader JW. An interleukin (IL)-13 receptor lacking the cytoplasmic domain fails to transduce IL-13-induced signals and inhibits responses to IL-4. J. Biol. Chem. 1997;272(36):22940–22947. doi: 10.1074/jbc.272.36.22940. [DOI] [PubMed] [Google Scholar]
- 123.Xiang J, Rir-Sim-Ah J, Tesfaigzi Y. IL-9 and IL-13 induce mucous cell metaplasia that is reduced by IFN-gamma in a Bax-mediated pathway. Am. J. Respir. Cell Mol. Biol. 2008;38(3):310–317. doi: 10.1165/rcmb.2007-0078OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Madala SK, Pesce JT, Ramalingam TR, Wilson MS, Minnicozzi S, Cheever AW, Thompson RW, Mentink-Kane MM, Wynn TA. Matrix metalloproteinase 12-deficiency augments extracellular matrix degrading metalloproteinases and attenuates IL-13-dependent fibrosis. J. Immunol. 2010;184(7):3955–3963. doi: 10.4049/jimmunol.0903008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang J, Paré PD, Sandford AJ. Recent advances in asthma genetics. Respir. Res. 2008;9:4. doi: 10.1186/1465-9921-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao Y, He D, Zhao J, Wang L, Leff AR, Spannhake EW, Georas S, Natarajan V. Lysophosphatidic acid induces interleukin-13 (IL-13) receptor alpha2 expression and inhibits IL-13 signaling in primary human bronchial epithelial cells. J. Biol. Chem. 2007;282(14):10172–10179. doi: 10.1074/jbc.M611210200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Finkelman FD, Hogan SP, Hershey GKK, Rothenberg ME, Wills-Karp M. Importance of cytokines in murine allergic airway disease and human asthma. J. Immunol. 2010;184(4):1663–1674. doi: 10.4049/jimmunol.0902185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stevens DA, Moss RB, Kurup VP, Knutsen AP, Greenberger P, Judson MA, Denning DW, Crameri R, Brody AS, Light M, Skov M, Maish W, Mastella G. Allergic bronchopulmonary aspergillosis in cystic fibrosis--state of the art: Cystic Fibrosis Foundation Consensus Conference. Clin. Infect. Dis. 2003;37(Suppl 3):S225–S264. doi: 10.1086/376525. [DOI] [PubMed] [Google Scholar]
- 129.Whittaker L, Niu N, Temann UA, Stoddard A, Flavell RA, Ray A, Homer RJ, Cohn L. Interleukin-13 mediates a fundamental pathway for airway epithelial mucus induced by CD4 T cells and interleukin-9. Am. J. Respir. Cell Mol. Biol. 2002;27(5):593–602. doi: 10.1165/rcmb.4838. [DOI] [PubMed] [Google Scholar]
- 130.Vita N, Lefort S, Laurent P, Caput D, Ferrara P. Characterization and comparison of the interleukin 13 receptor with the interleukin 4 receptor on several cell types. J. Biol. Chem. 1995;270(8):3512–3517. doi: 10.1074/jbc.270.8.3512. [DOI] [PubMed] [Google Scholar]
- 131.Walker W, Healey GD, Hopkin JM. RNA interference of STAT6 rapidly attenuates ongoing interleukin-13-mediated events in lung epithelial cells. Immunology. 2009;127(2):256–266. doi: 10.1111/j.1365-2567.2008.02951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brown CR, Reiner SL. Experimental lyme arthritis in the absence of interleukin-4 or gamma interferon. Infect. Immun. 1999;67(7):3329–3333. doi: 10.1128/iai.67.7.3329-3333.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang Y, Malabarba MG, Nagy ZS, Kirken RA. Interleukin 4 regulates phosphorylation of serine 756 in the transactivation domain of Stat6. Roles for multiple phosphorylation sites and Stat6 function. J. Biol. Chem. 2004;279(24):25196–25203. doi: 10.1074/jbc.M313668200. [DOI] [PubMed] [Google Scholar]
- 134.Yang M, Hogan SP, Henry PJ, Matthaei KI, McKenzie AN, Young IG, Rothenberg ME, Foster PS. Interleukin-13 mediates airways hyper reactivity through the IL-4 receptor-alpha chain and STAT-6 independently of IL-5 and eotaxin. Am. J. Respir. Cell Mol. Biol. 2001;25(4):522–530. doi: 10.1165/ajrcmb.25.4.4620. [DOI] [PubMed] [Google Scholar]
- 135.Jung YJ, LaCourse R, Ryan L, North RJ. Evidence inconsistent with a negative influence of T helper 2 cells on protection afforded by a dominant T helper 1 response against Mycobacterium tuberculosis lung infection in mice. Infect. Immun. 2002;70(11):6436–6443. doi: 10.1128/IAI.70.11.6436-6443.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Heinzmann A, Mao XQ, Akaiwa M, Kreomer RT, Gao PS, Ohshima K, Umeshita R, Abe Y, Braun S, Yamashita T, Roberts MH, Sugimoto R, Arima K, Arinobu Y, Yu B, Kruse S, Enomoto T, Dake Y, Kawai M, Shimazu S, Sasaki S, Adra CN, Kitaichi M, Inoue H, Yamauchi K, Tomichi N, Kurimoto F, Hamasaki N, Hopkin JM, Izuhara K, Shirakawa T, Deichmann KA. Genetic variants of IL-13 signaling and human asthma and atopy. Hum. Mol. Genet. 2000;9(4):549–559. doi: 10.1093/hmg/9.4.549. [DOI] [PubMed] [Google Scholar]
- 137.Madhankumar AB, Mintz A, Debinski W. Alanine-scanning mutagenesis of alpha-helix D segment of interleukin-13 reveals new functionally important residues of the cytokine. J. Biol. Chem. 2002;277(45):43194–43205. doi: 10.1074/jbc.M205047200. [DOI] [PubMed] [Google Scholar]
- 138.Palmqvist P, Lundberg P, Persson E, Johansson A, Lundgren I, Lie A, Conaway HH, Lerner UH. Inhibition of hormone and cytokine-stimulated osteoclastogenesis and bone resorption by interleukin-4 and interleukin-13 is associated with increased osteoprotegerin and decreased RANKL and RANK in a STAT6-dependent pathway. J. Biol. Chem. 2006;281(5):2414–2429. doi: 10.1074/jbc.M510160200. [DOI] [PubMed] [Google Scholar]
- 139.Kraft P. Multiple comparisons in studies of gene x gene and gene x environment interaction. Am. J. Hum. Genet. 2004;74(3):582–584. doi: 10.1086/382051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Oshima Y, Joshi BH, Puri RK. Conversion of interleukin-13 into a high affinity agonist by a single amino acid substitution. J. Biol. Chem. 2000;275(19):14375–14380. doi: 10.1074/jbc.275.19.14375. [DOI] [PubMed] [Google Scholar]
- 141.Madden KB, Whitman L, Sullivan C, Gause WC, Urban JF, Katona IM, Finkelman FD, Shea-Donohue T. Role of STAT6 and mast cells in IL-4- and IL-13-induced alterations in murine intestinal epithelial cell function. J. Immunol. 2002;169(8):4417–4422. doi: 10.4049/jimmunol.169.8.4417. [DOI] [PubMed] [Google Scholar]
- 142.Zhao A, Morimoto M, Dawson H, Elfrey JE, Madden KB, Gause WC, Min B, Finkelman FD, Urban JF, Shea-Donohue T. Immune regulation of protease-activated receptor-1 expression in murine small intestine during Nippostrongylus brasiliensis infection. J. Immunol. 2005;175(4):2563–2569. doi: 10.4049/jimmunol.175.4.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao A, McDermott J, Urban JF, Gause W, Madden KB, Yeung KA, Morris SC, Finkelman FD, Shea-Donohue T. Dependence of IL-4, IL-13, and nematode-induced alterations in murine small intestinal smooth muscle contractility on Stat6 and enteric nerves. J. Immunol. 2003;171(2):948–954. doi: 10.4049/jimmunol.171.2.948. [DOI] [PubMed] [Google Scholar]
- 144.Arima K, Sato K, Tanaka G, Kanaji S, Terada T, Honjo E, Kuroki R, Matsuo Y, Izuhara K. Characterization of the interaction between interleukin-13 and interleukin-13 receptors. J. Biol. Chem. 2005;280(26):24915–24922. doi: 10.1074/jbc.M502571200. [DOI] [PubMed] [Google Scholar]
- 145.McCusker CT, Wang Y, Shan J, Kinyanjui MW, Villeneuve A, Michael H, Fixman ED. Inhibition of experimental allergic airways disease by local application of a cell-penetrating dominant-negative STAT-6 peptide. J. Immunol. 2007;179(4):2556–2564. doi: 10.4049/jimmunol.179.4.2556. [DOI] [PubMed] [Google Scholar]
- 146.Lim RH, Kobzik L. Transplacental passage of interleukins 4 and 13? PLoS One. 2009;4(3):e4660. doi: 10.1371/journal.pone.0004660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Desmet C, Gosset P, Henry E, Garzé V, Faisca P, Vos N, Jaspar F, Mélotte D, Lambrecht B, Desmecht D, Pajak B, Moser M, Lekeux P, Bureau F. Treatment of experimental asthma by decoy-mediated local inhibition of activator protein-1. Am. J. Respir. Crit. Care Med. 2005;172(6):671–678. doi: 10.1164/rccm.200410-1431OC. [DOI] [PubMed] [Google Scholar]
- 148.Pillemer BB, Qi Z, Melgert B, Oriss TB, Ray P, Ray A. STAT6 activation confers upon T helper cells resistance to suppression by regulatory T cells. J. Immunol. 2009;183(1):155–163. doi: 10.4049/jimmunol.0803733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kraich M, Klein M, Patiño E, Harrer H, Nickel J, Sebald W, Mueller TD. A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor. BMC Biol. 2006;4:13. doi: 10.1186/1741-7007-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pernis AB, Rothman PB. JAK-STAT signaling in asthma. J. Clin. Invest. 2002;109(10):1279–1283. doi: 10.1172/JCI15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tomlinson KL, Davies GCG, Sutton DJ, Palframan RT. Neutralisation of interleukin-13 in mice prevents airway pathology caused by chronic exposure to house dust mite. PloS One. 2010;5(10) doi: 10.1371/journal.pone.0013136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gallina G, Dolcetti L, Serafini P, De Santo C, Marigo I, Colombo MP, Basso G, Brombacher F, Borrello I, Zanovello P, Bicciato S, Bronte V. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J. Clin. Invest. 2006;116(10):2777–2790. doi: 10.1172/JCI28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Müller U, Stenzel W, Köhler G, Werner C, Polte T, Hansen G, Schütze N, Straubinger RK, Blessing M, McKenzie AN, Brombacher F, Alber G. IL-13 induces disease-promoting type 2 cytokines, alternatively activated macrophages and allergic inflammation during pulmonary infection of mice with Cryptococcus neoformans. J. Immunol. 2007;179(8):5367–5377. doi: 10.4049/jimmunol.179.8.5367. [DOI] [PubMed] [Google Scholar]
- 154.Lopez M, Clarkson MR, Albin M, Sayegh MH, Najafian N. A novel mechanism of action for anti-thymocyte globulin: induction of CD4+CD25+Foxp3+ regulatory T cells. J. Am. Soc. Nephrol. 2006;17(10):2844–2853. doi: 10.1681/ASN.2006050422. [DOI] [PubMed] [Google Scholar]
- 155.Heller NM, Matsukura S, Georas SN, Boothby MR, Rothman PB, Stellato C, Schleimer RP. Interferon-gamma inhibits STAT6 signal transduction and gene expression in human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2004;31(5):573–582. doi: 10.1165/rcmb.2004-0195OC. [DOI] [PubMed] [Google Scholar]
- 156.Tobin MJ. Asthma, airway biology, and nasal disorders in AJRCCM 2002. Am. J. Respir. Crit. Care Med. 2003;167(3):319–332. doi: 10.1164/rccm.2212007. [DOI] [PubMed] [Google Scholar]
- 157.Shimamura T, Fujisawa T, Husain SR, Joshi B, Puri RK. Interleukin 13 mediates signal transduction through interleukin 13 receptor alpha2 in pancreatic ductal adenocarcinoma: role of IL-13 Pseudomonas exotoxin in pancreatic cancer therapy. Clin. Cancer Res. 2010;16(2):577–586. doi: 10.1158/1078-0432.CCR-09-2015. [DOI] [PubMed] [Google Scholar]
- 158.Chen J, Olsen J, Ford S, Mirza S, Walker A, Murphy JM, Young IG. A new isoform of interleukin-3 receptor {alpha} with novel differentiation activity and high affinity binding mode. J. Biol. Chem. 2009;284(9):5763–5773. doi: 10.1074/jbc.M808197200. [DOI] [PubMed] [Google Scholar]
- 159.Lutz MB, Schnare M, Menges M, Rössner S, Röllinghoff M, Schuler G, Gessner A. Differential functions of IL-4 receptor types I and II for dendritic cell maturation and IL-12 production and their dependency on GM-CSF. J. Immunol. 2002;169(7):3574–3580. doi: 10.4049/jimmunol.169.7.3574. [DOI] [PubMed] [Google Scholar]
- 160.Barnes PJ. The cytokine network in asthma and chronic obstructive pulmonary disease. J. Clin. Invest. 2008;118(11):3546–3556. doi: 10.1172/JCI36130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kawakami M, Kawakami K, Kasperbauer JL, Hinkley LL, Tsukuda M, Strome SE, Puri RK. Interleukin-13 receptor alpha2 chain in human head and neck cancer serves as a unique diagnostic marker. Clin. Cancer Res. 2003;9(17):6381–6388. [PubMed] [Google Scholar]
- 162.Ahn MH, Kang CM, Park CS, Park SJ, Rhim T, Yoon PO, Chang HS, Kim SH, Kyono H, Kim KC. Titanium dioxide particle-induced goblet cell hyperplasia: association with mast cells and IL-13. Respir. Res. 2005;6:34. doi: 10.1186/1465-9921-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Suzuki T, Schirra F, Richards SM, Jensen RV, Sullivan DA. Estrogen and progesterone control of gene expression in the mouse meibomian gland. Invest. Ophthalmol. Vis. Sci. 2008;49(5):1797–1808. doi: 10.1167/iovs.07-1458. [DOI] [PubMed] [Google Scholar]
- 164.Wynes MW, Frankel SK, Riches DW. IL-4-induced macrophage-derived IGF-I protects myofibroblasts from apoptosis following growth factor withdrawal. J. Leukoc. Biol. 2004;76(5):1019–1027. doi: 10.1189/jlb.0504288. [DOI] [PubMed] [Google Scholar]
- 165.Lotz M, König T, Ménard S, Gütle D, Bogdan C, Hornef MW. Cytokine-mediated control of lipopolysaccharide-induced activation of small intestinal epithelial cells. Immunology. 2007;122(3):306–315. doi: 10.1111/j.1365-2567.2007.02639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pousset F, Cremona S, Dantzer R, Kelley K, Parnet P. Interleukin-4 and interleukin-10 regulate IL1-beta induced mouse primary astrocyte activation: a comparative study. Glia. 1999;26(1):12–21. doi: 10.1002/(sici)1098-1136(199903)26:1<12::aid-glia2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 167.van der Velden VH, Naber BA, Wierenga-Wolf AF, Debets R, Savelkoul HF, Overbeek SE, Hoogsteden HC, Versnel MA. Interleukin 4 receptors on human bronchial epithelial cells. An in vivo and in vitro analysis of expression and function. Cytokine. 1998;10(10):803–813. doi: 10.1006/cyto.1998.0365. [DOI] [PubMed] [Google Scholar]
- 168.Yamamoto S, Kobayashi I, Tsuji K, Nishi N, Muro E, Miyazaki M, Zaitsu M, Inada S, Ichimaru T, Hamasaki Y. Upregulation of interleukin-4 receptor by interferon-gamma: enhanced interleukin-4-induced eotaxin-3 production in airway epithelium. Am. J. Respir. Cell Mol. Biol. 2004;31(4):456–462. doi: 10.1165/rcmb.2004-0128OC. [DOI] [PubMed] [Google Scholar]
- 169.Morris SC, Heidorn SM, Herbert DR, Perkins C, Hildeman DA, Khodoun MV, Finkelman FD. Endogenously produced IL-4 nonredundantly stimulates CD8+ T cell proliferation. J. Immunol. 2009;182(3):1429–1438. doi: 10.4049/jimmunol.182.3.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Morrot A, Hafalla JC, Cockburn IA, Carvalho LH, Zavala F. IL-4 receptor expression on CD8+ T cells is required for the development of protective memory responses against liver stages of malaria parasites. J. Exp. Med. 2005;202(4):551–560. doi: 10.1084/jem.20042463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ueda N, Kuki H, Kamimura D, Sawa S, Seino K, Tashiro T, Fushuku K, Taniguchi M, Hirano T, Murakami M. CD1d-restricted NKT cell activation enhanced homeostatic proliferation of CD8+ T cells in a manner dependent on IL-4. Int. Immunol. 2006;18(9):1397–1404. doi: 10.1093/intimm/dxl073. [DOI] [PubMed] [Google Scholar]
- 172.Börner C, Höllt V, Sebald W, Kraus J. Transcriptional regulation of the cannabinoid receptor type 1 gene in T cells by cannabinoids. J. Leukoc. Biol. 2007;81(1):336–343. doi: 10.1189/jlb.0306224. [DOI] [PubMed] [Google Scholar]
- 173.Brodie C, Goldreich N, Haiman T, Kazimirsky G. Functional IL-4 receptors on mouse astrocytes: IL-4 inhibits astrocyte activation and induces NGF secretion. J. Neuroimmunol. 1998;81(1–2):20–30. doi: 10.1016/s0165-5728(97)00154-9. [DOI] [PubMed] [Google Scholar]
- 174.Rahaman SO, Vogelbaum MA, Haque SJ. Aberrant Stat3 signaling by interleukin-4 in malignant glioma cells: involvement of IL-13Ralpha2. Cancer Res. 2005;65(7):2956–2963. doi: 10.1158/0008-5472.CAN-04-3592. [DOI] [PubMed] [Google Scholar]
- 175.Malek TR, Yu A, Scibelli P, Lichtenheld MG, Codias EK. Broad programming by IL-2 receptor signaling for extended growth to multiple cytokines and functional maturation of antigen-activated T cells. J. Immunol. 2001;166(3):1675–1683. doi: 10.4049/jimmunol.166.3.1675. [DOI] [PubMed] [Google Scholar]
- 176.Huang H, Paul WE. Protein tyrosine phosphatase activity is required for IL-4 induction of IL-4 receptor alpha-chain. J. Immunol. 2000;164(3):1211–1215. doi: 10.4049/jimmunol.164.3.1211. [DOI] [PubMed] [Google Scholar]
- 177.Kashiwada M, Giallourakis CC, Pan PY, Rothman PB. Immunoreceptor tyrosine-based inhibitory motif of the IL-4 receptor associates with SH2-containing phosphatases and regulates IL-4-induced proliferation. J. Immunol. 2001;167(11):6382–6387. doi: 10.4049/jimmunol.167.11.6382. [DOI] [PubMed] [Google Scholar]
- 178.Haque SJ, Harbor P, Tabrizi M, Yi T, Williams BR. Protein-tyrosine phosphatase Shp-1 is a negative regulator of IL-4- and IL-13-dependent signal transduction. J. Biol. Chem. 1998;273(51):33893–33896. doi: 10.1074/jbc.273.51.33893. [DOI] [PubMed] [Google Scholar]
- 179.Wurster AL, Withers DJ, Uchida T, White MF, Grusby MJ. Stat6 and IRS-2 cooperate in interleukin 4 (IL-4)-induced proliferation and differentiation but are dispensable for IL-4- dependent rescue from apoptosis. Mol. Cell Biol. 2002;22(1):117–126. doi: 10.1128/MCB.22.1.117-126.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hanson EM, Dickensheets H, Qu CK, Donnelly RP, Keegan AD. Regulation of the dephosphorylation of Stat6. Participation of Tyr-713 in the interleukin-4 receptor alpha, the tyrosine phosphatase SHP-1, and the proteasome. J. Biol. Chem. 2003;278(6):3903–3911. doi: 10.1074/jbc.M211747200. [DOI] [PubMed] [Google Scholar]
- 181.Imani F, Rager KJ, Catipovic B, Marsh DG. Interleukin-4 (IL-4) induces phosphatidylinositol 3-kinase (p85) dephosphorylation. Implications for the role of SHP-1 in the IL-4-induced signals in human B cells. J. Biol. Chem. 1997;272(12):7927–7931. doi: 10.1074/jbc.272.12.7927. [DOI] [PubMed] [Google Scholar]
- 182.O'Connor JC, Sherry CL, Guest CB, Freund GG. Type 2 diabetes impairs insulin receptor substrate-2-mediated phosphatidylinositol 3-kinase activity in primary macrophages to induce a state of cytokine resistance to IL-4 in association with overexpression of suppressor of cytokine signaling-3. J. Immunol. 2007;178(11):6886–6893. doi: 10.4049/jimmunol.178.11.6886. [DOI] [PubMed] [Google Scholar]
- 183.Sasaki Y, Mita H, Toyota M, Ishida S, Morimoto I, Yamashita T, Tanaka T, Imai K, Nakamura Y, Tokino T. Identification of the interleukin 4 receptor alpha gene as a direct target for p73. Cancer Res. 2003;63(23):8145–8152. [PubMed] [Google Scholar]
- 184.King NE, Zimmermann N, Pope SM, Fulkerson PC, Nikolaidis NM, Mishra A, Witte DP, Rothenberg ME. Expression and regulation of a disintegrin and metalloproteinase (ADAM) 8 in experimental asthma. Am. J. Respir. Cell Mol. Biol. 2004;31(3):257–265. doi: 10.1165/rcmb.2004-0026OC. [DOI] [PubMed] [Google Scholar]
- 185.Stephenson LM, Park DS, Mora AL, Goenka S, Boothby M. Sequence motifs in IL-4R alpha mediating cell-cycle progression of primary lymphocytes. J. Immunol. 2005;175(8):5178–5185. doi: 10.4049/jimmunol.175.8.5178. [DOI] [PubMed] [Google Scholar]
- 186.Kelly E, Won A, Refaeli Y, Van Parijs L. IL-2 and related cytokines can promote T cell survival by activating AKT. J. Immunol. 2002;168(2):597–603. doi: 10.4049/jimmunol.168.2.597. [DOI] [PubMed] [Google Scholar]
- 187.Yamaji-Kegan K, Su Q, Angelini DJ, Myers AC, Cheadle C, Johns RA. Hypoxia-induced mitogenic factor (HIMF/FIZZ1/RELMalpha) increases lung inflammation and activates pulmonary microvascular endothelial cells via an IL-4-dependent mechanism. J. Immunol. 2010;185(9):5539–5548. doi: 10.4049/jimmunol.0904021. [DOI] [PubMed] [Google Scholar]
- 188.Huang Z, Coleman JM, Su Y, Mann M, Ryan J, Shultz LD, Huang H. SHP-1 regulates STAT6 phosphorylation and IL-4-mediated function in a cell type-specific manner. Cytokine. 2005;29(3):118–124. doi: 10.1016/j.cyto.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 189.Webb DC, Cai Y, Matthaei KI, Foster PS. Comparative roles of IL-4, IL-13, and IL-4Ralpha in dendritic cell maturation and CD4+ Th2 cell function. J. Immunol. 2007;178(1):219–227. doi: 10.4049/jimmunol.178.1.219. [DOI] [PubMed] [Google Scholar]
- 190.Shiroiwa W, Tsukamoto K, Ohtsuji M, Lin Q, Ida A, Kodera S, Ohtsuji N, Nakamura K, Tsurui H, Kinoshita K, Nishimura H, Shirai T, Hirose S. IL-4Ralpha polymorphism in regulation of IL-4 synthesis by T cells: implication in susceptibility to a subset of murine lupus. Int. Immunol. 2007;19(2):175–183. doi: 10.1093/intimm/dxl134. [DOI] [PubMed] [Google Scholar]
- 191.Kubo M, Yamashita M, Abe R, Tada T, Okumura K, Ransom JT, Nakayama T. CD28 costimulation accelerates IL-4 receptor sensitivity and IL-4-mediated Th2 differentiation. J. Immunol. 1999;163(5):2432–2442. [PubMed] [Google Scholar]
- 192.Wiethe C, Debus A, Mohrs M, Steinkasserer A, Lutz M, Gessner A. Dendritic cell differentiation state and their interaction with NKT cells determine Th1/Th2 differentiation in the murine model of Leishmania major infection. J. Immunol. 2008;180(7):4371–4381. doi: 10.4049/jimmunol.180.7.4371. [DOI] [PubMed] [Google Scholar]
- 193.Tamura T, Igarashi O, Hino A, Yamane H, Aizawa S, Kato T, Nariuchi H. Impairment in the expression and activity of Fyn during differentiation of naive CD4+ T cells into the Th2 subset. J. Immunol. 2001;167(4):1962–1969. doi: 10.4049/jimmunol.167.4.1962. [DOI] [PubMed] [Google Scholar]
- 194.Webb DC, Matthaei KI, Cai Y, McKenzie AN, Foster PS. Polymorphisms in IL-4R alpha correlate with airways hyperreactivity, eosinophilia, and Ym protein expression in allergic IL-13−/− mice. J. Immunol. 2004;172(2):1092–1098. doi: 10.4049/jimmunol.172.2.1092. [DOI] [PubMed] [Google Scholar]
- 195.Bix M, Wang ZE, Thiel B, Schork NJ, Locksley RM. Genetic regulation of commitment to interleukin 4 production by a CD4(+) T cell-intrinsic mechanism. J. Exp. Med. 1998;188(12):2289–2299. doi: 10.1084/jem.188.12.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lu P, Urban JF, Zhou XD, Chen SJ, Madden K, Moorman M, Nguyen H, Morris SC, Finkelman FD, Gause WC. CD40-mediated stimulation contributes to lymphocyte proliferation, antibody production, eosinophilia, and mastocytosis during an in vivo type 2 response, but is not required for T cell IL-4 production. J. Immunol. 1996;156(9):3327–3333. [PubMed] [Google Scholar]
- 197.Liu TF, Tatter SB, Willingham MC, Yang M, Hu JJ, Frankel AE. Growth factor receptor expression varies among high-grade gliomas and normal brain: epidermal growth factor receptor has excellent properties for interstitial fusion protein therapy. Mol. Cancer Ther. 2003;2(8):783–787. [PubMed] [Google Scholar]
- 198.Dhiman N, Ovsyannikova IG, Cunningham JM, Vierkant RA, Kennedy RB, Pankratz VS, Poland GA, Jacobson RM. Associations between measles vaccine immunity and single-nucleotide polymorphisms in cytokine and cytokine receptor genes. J. Infect. Dis. 2007;195(1):21–29. doi: 10.1086/510596. [DOI] [PubMed] [Google Scholar]
- 199.Levings MK, Schrader JW. IL-4 inhibits the production of TNF-alpha and IL-12 by STAT6-dependent and -independent mechanisms. J. Immunol. 1999;162(9):5224–5229. [PubMed] [Google Scholar]
- 200.Dodeller F, Schulze-Koops H. The p38 mitogen-activated protein kinase signaling cascade in CD4 T cells. Arthritis Res. Ther. 2006;8(2):205. doi: 10.1186/ar1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Peng Q, Matsuda T, Hirst SJ. Signaling pathways regulating interleukin-13-stimulated chemokine release from airway smooth muscle. Am. J. Respir. Crit. Care Med. 2004;169(5):596–603. doi: 10.1164/rccm.200307-888OC. [DOI] [PubMed] [Google Scholar]
- 202.Maldonado RA, Soriano MA, Perdomo LC, Sigrist K, Irvine DJ, Decker T, Glimcher LH. Control of T helper cell differentiation through cytokine receptor inclusion in the immunological synapse. J. Exp. Med. 2009;206(4):877–892. doi: 10.1084/jem.20082900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hackett RH, Wang YD, Larner AC. Mapping of the cytoplasmic domain of the human growth hormone receptor required for the activation of Jak2 and Stat proteins. J. Biol. Chem. 1995;270(36):21326–21330. doi: 10.1074/jbc.270.36.21326. [DOI] [PubMed] [Google Scholar]
- 204.Rothstein TL. Inducible resistance to Fas-mediated apoptosis in B cells. Cell Res. 2000;10(4):245–266. doi: 10.1038/sj.cr.7290053. [DOI] [PubMed] [Google Scholar]
- 205.Rothstein TL, Zhong X, Schram BR, Negm RS, Donohoe TJ, Cabral DS, Foote LC, Schneider TJ. Receptor-specific regulation of B-cell susceptibility to Fas-mediated apoptosis and a novel Fas apoptosis inhibitory molecule. Immunol. Rev. 2000;176:116–133. doi: 10.1034/j.1600-065x.2000.00616.x. [DOI] [PubMed] [Google Scholar]
- 206.Lee JR, Koretzky GA. Extracellular signal-regulated kinase-2, but not c-Jun NH2-terminal kinase, activation correlates with surface IgM-mediated apoptosis in the WEHI 231 B cell line. J. Immunol. 1998;161(4):1637–1644. [PubMed] [Google Scholar]
- 207.Ganley-Leal LM, Liang Y, Jagannathan-Bogdan M, Farraye FA, Nikolajczyk BS. Differential regulation of TLR4 expression in human B cells and monocytes. Mol. Immunol. 2010;48(1–3):82–88. doi: 10.1016/j.molimm.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mora AL, Stephenson LM, Enerson B, Youn J, Keegan AD, Boothby M. New programming of IL-4 receptor signal transduction in activated T cells: Stat6 induction and Th2 differentiation mediated by IL-4Ralpha lacking cytoplasmic tyrosines. J. Immunol. 2003;171(4):1891–1900. doi: 10.4049/jimmunol.171.4.1891. [DOI] [PubMed] [Google Scholar]
- 209.Tsuji K, Yamamoto S, Ou W, Nishi N, Kobayashi I, Zaitsu M, Muro E, Sadakane Y, Ichimaru T, Hamasaki Y. dsRNA enhances eotaxin-3 production through interleukin-4 receptor upregulation in airway epithelial cells. Eur. Respir. J. 2005;26(5):795–803. doi: 10.1183/09031936.05.00010805. [DOI] [PubMed] [Google Scholar]
- 210.Prieto-Lafuente L, Gregory WF, Allen JE, Maizels RM. MIF homologues from a filarial nematode parasite synergize with IL-4 to induce alternative activation of host macrophages. J. Leukoc. Biol. 2009;85(5):844–854. doi: 10.1189/jlb.0808459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wang D, Zamorano J, Keegan AD, Boothby M. HMG-I(Y) phosphorylation status as a nuclear target regulated through insulin receptor substrate-1 and the I4R motif of the interleukin-4 receptor. J. Biol. Chem. 1997;272(40):25083–25090. doi: 10.1074/jbc.272.40.25083. [DOI] [PubMed] [Google Scholar]
- 212.Mora A, Youn J, Keegan A, Boothby M. NF-kappa B/Rel participation in the lymphokine-dependent proliferation of T lymphoid cells. J. Immunol. 2001;166(4):2218–2227. doi: 10.4049/jimmunol.166.4.2218. [DOI] [PubMed] [Google Scholar]
- 213.Haque SJ, Harbor PC, Williams BR. Identification of critical residues required for suppressor of cytokine signaling-specific regulation of interleukin-4 signaling. J. Biol. Chem. 2000;275(34):26500–26506. doi: 10.1074/jbc.275.34.26500. [DOI] [PubMed] [Google Scholar]
- 214.Kotanides H, Reich NC. Interleukin-4-induced STAT6 recognizes and activates a target site in the promoter of the interleukin-4 receptor gene. J. Biol. Chem. 1996;271(41):25555–25561. doi: 10.1074/jbc.271.41.25555. [DOI] [PubMed] [Google Scholar]
- 215.Puri RK, Leland P. Tumor necrosis factor upregulates interleukin-4 receptors on murine sarcoma cells. Biochem. Biophys. Res. Commun. 1993;197(3):1424–1430. doi: 10.1006/bbrc.1993.2636. [DOI] [PubMed] [Google Scholar]
- 216.Guizani L, Lang P, Stancou R, Bertoglio J. Regulation of interleukin-2 and interleukin-4 receptor expression in human and ape lymphoid cell lines. Lymphokine Cytokine Res. 1993;12(1):25–32. [PubMed] [Google Scholar]
- 217.Lugli SM, Feng N, Heim MH, Adam M, Schnyder B, Etter H, Yamage M, Eugster HP, Lutz RA, Zurawski G, Moser R. Tumor necrosis factor alpha enhances the expression of the interleukin (IL)-4 receptor alpha-chain on endothelial cells increasing IL-4 or IL-13-induced Stat6 activation. J. Biol. Chem. 1997;272(9):5487–5494. doi: 10.1074/jbc.272.9.5487. [DOI] [PubMed] [Google Scholar]
- 218.Blease K, Seybold J, Adcock IM, Hellewell PG, Burke-Gaffney A. Interleukin-4 and lipopolysaccharide synergize to induce vascular cell adhesion molecule-1 expression in human lung microvascular endothelial cells. Am. J. Respir. Cell Mol. Biol. 1998;18(5):620–630. doi: 10.1165/ajrcmb.18.5.3052. [DOI] [PubMed] [Google Scholar]
- 219.Nonaka M, Ogihara N, Fukumoto A, Sakanushi A, Pawankar R, Yagi T. Combined stimulation of nasal polyp fibroblasts with poly IC, interleukin 4, and tumor necrosis factor alpha potently induces production of thymus- and activation-regulated chemokine. Arch. Otolaryngol. Head Neck Surg. 2008;134(6):630–635. doi: 10.1001/archotol.134.6.630. [DOI] [PubMed] [Google Scholar]