Abstract
Induced pluripotent stem cells (iPSCs) and nuclear transfer (NT) are two of the primary routes to reprogram differentiated cells back to the pluripotent state. However, it is still unknown whether there is any correlation between the reprogramming efficiency of iPSCs and NT if the same donor cells are employed. In this study, six porcine embryonic fibroblast (PEF) lines from Landrace (L1, L6, L9) or Congjiang local pigs (C4, C5, C6) were used for iPSC induction and NT. Furthermore, the resultant iPSCs from four PEF lines (L1, L6, C4, and C5) were used for NT (iPSC-NT), and the expression of exogenous genes was detected in iPSC-NT embryos by real-time PCR. The results showed that the efficiency of iPSC lines established from different PEF lines were significantly different. When the same PEF lines were used as donor cells for NT, the blastocysts rates were also different among different PEF lines and positively related with iPSCs induction efficiency. When the iPSCs were used as donor cells for NT, compared with the source PEFs, the blastocysts rates were significantly decreased. Real-time PCR results indicated that exogenous genes (Oct4, c-Myc) continued to be expressed in iPSC-NT embryos. In summary, our results demonstrate that there was a positive correlation between iPSCs and NT reprogramming efficiency, although the mechanism of these two routes is different. This may provide a new method to select the appropriate donor cells for inducing iPSCs.
Introduction
Recently, induced pluripotent stem cells (iPSCs) have become the first potential choice for cell replacement therapy and drug screening because they can demonstrate the same characteristics of embryonic stem cells (ESCs) but do not have the ethical issues and immune rejection problems associated with ESCs (Takahashi et al., 2007). However, the efficiency of iPSC induction varies in different research laboratories due to differences in the induction systems (Huangfu et al., 2008; Nakagawa et al., 2008; Okita et al., 2007). Furthermore, the donor cells used for induction are one of the primary differences for the variation of induction efficiency. For example, the inducing efficiency could be up to 0.03% if multilineage-differentiating stress-enduring (Muse) cells are used as donor cells, compared with the efficiency of human skin fibroblasts, which was 0.0001% (Wakao et al., 2011). Some researchers have reported that hematopoietic, muscle, and liver stem cells could be more efficiently reprogrammed to iPSCs than differentiated blood, muscle, and liver cells (Eminli et al., 2009; Kleger et al., 2012; Tan et al., 2011). Donor cells with low proliferation capability showed higher iPSC induction efficiency (Xu et al., 2013). Fibroblasts are the main source of the iPSC induction because of ease of collection, but there are few reports about whether there are differences in iPSC induction efficiency among fibroblasts lines derived from different individuals.
Somatic cell nuclear transfer (SCNT) is another method for reprogramming differentiated cells to the pluripotent state. Research has indicated that donor cells have an important influence on the development of SCNT embryos. Previous studies have provided a significant amount of data on the development rates of SCNT embryos derived from donor cells of different species, tissue sources, and degree of differentiation, and these data show that donor cells could affect the cloned embryo development significantly (Inoue et al., 2003; Oback and Wells, 2007; Wakayama and Yanagimachi, 2001). Wakayama and his colleagues reported that hybrid mice (B6D2F1 and B6C3F1) were easier to clone than inbred mice (C57BL/6 and C3H/He) (Wakayama and Yanagimachi, 2001). Inoue and colleagues found that the birth rate of SCNT embryos derived from immature Sertoli cells was higher than that from cumulus cells (Inoue et al., 2003). Nuclear transfer efficiency of iPSCs with totipotent cells was significantly higher than its source mouse embryonic fibroblast (MEF) cells (Zhou et al., 2010). Meanwhile, some reports have indicated that there are differences between the blastocyst rates of SCNT embryos constructed with different cell lines derived from the same tissue source of different individuals (Heyman et al., 2002; Miyazaki et al., 2005). So, we asked the question whether there are also differences in iPSC induction efficiency between cell lines derived from the same tissue source but from different individuals.
Generally, the reprogramming mechanism of iPSCs induction and NT is considered to be different. iPSCs are induced only by several defined exogenous factors, and it is a slow process with small-probability events (Hochedlinger and Plath, 2009). In contrast, SCNT is a more efficient process that involves a wide range of largely unknown ooplasm factors. After donor cells are injected into eggs, nuclear remodeling occurs rapidly and thoroughly through cell cycle synchronization with eggs (Gurdon, 1962; Wakayama et al., 2001; Whitworth and Prather, 2010; Wilmut et al., 1997). However, there are some common characteristics of the two approaches of reprogramming, such as the alteration of the nuclear structure, removing inhibitory modifications of chromosomes, silencing of differentiation-specific genes and activating pluripotency genes, and changing the cell proliferation along with the reprogramming (Polo et al., 2012a; Whitworth and Prather, 2010). Thus, we can hypothesize that there might be a positive correlation of reprogramming efficiency between iPSCs and NT reprogramming when using donor cells from different individuals.
In this study, six porcine embryonic fibroblast (PEF) lines derived from different individuals of two breeds were used for iPSC induction and SCNT, and the efficiency of iPSC induction and SCNT embryo development was assessed. The results should test our hypothesis and deepen the understanding of the reprogramming mechanism of iPSC induction and SCNT.
Materials and Methods
The reagents and media used in our study were purchased from Life Technologies, R&D, Millipore, and Bioind unless otherwise stated.
Animals
The PEF lines were derived from Landrace and Congjiang local pigs from Northeast Agricultural University Embryo Engineering Laboratory Experimental Pig Base. ICR mice for MEFs were purchased from Beijing Vital River Company. The porcine ovaries were obtained from DaZhongRouLian Slaughterhouse Company.
All studies adhered to procedures consistent with the Northeast Agriculture University Biological Sciences Guide for the care and use of laboratory animals.
PEF isolation and culture
PEFs were isolated from a 33-day-old fetus. Briefly, the fetuses were recovered and rinsed five times with Dulbecco's phosphate-buffered saline (DPBS). After removal of the head, internal organs, and limbs, the remaining tissues were finely minced into pieces. The minced tissues were digested with 1.6 mg/mL collagenase IV and 25 units of DNase I at 37°C for 15 min, followed by dispersal in PEF medium [86% high-glucose Dulbecco's modified Eagle medium (DMEM), 10% fetal bovine serum (FBS), 1% nonessential amino acids (NEAA), 2 mmol/L L-glutamine, 1% penicillin-streptomycin]. The dispersed cells were centrifuged and resuspended, and the cells were cultured in PEF medium at 37°C in 5% CO2 atmosphere and saturated humidity. At confluence, cells were dissociated, centrifuged, resuspended in FBS containing 10% dimethyl sulfoxide (DMSO), and stored in liquid nitrogen until use. A total of eight and nine PEF lines of Landrace and Congjiang local pig were successfully established from different piglets. Sex identification by PCR determined that 1#, 6#, and 9# PEFs of Landrace (L1, L6, L9) and 4#, 5#, and 6# PEFs of Congjiang local pig (C4, C5, C6) were female and were used for further study. Prior to SCNT, PEF lines were thawed, cultured, and subsequently used between passages four and five.
iPSC generation
Inducible iPSCs were generated as described previously (Wang et al., 2013). Briefly, GP293T cells were transfected with the pMX plasmids, which contained mouse Oct4, Sox2, Klf4, c-Myc, Nr5a2, and Tbx3 together with packaging plasmids vesicular stomatitis virus G protein (VSV-G). The medium was replaced 12 h after transfection, and the virus supernatants were harvested after subsequent 24 h. L1, L6, L9, and C4, C5, C6 at passage three or four in 24-well plates at a density of 104 cells/dish were incubated with filtered viral supernatants containing 8 μg/mL Polybrene. The infection medium was replaced after 24 h with PEF medium. When the PEF lines were at 90% confluence, we passaged them into 12-well plates at 1:8, and the medium was replaced with MX medium (developed in our laboratory for porcine iPSCs culture) every day until the mouse ESC-like colonies appeared at approximately 7 days after infection (Wang et al., 2013). The cells were cultured for another 10 days with MX medium, and the colonies were picked when they reached the size of 60–70 μm. Ten clones were mechanically passaged into one well of a 24-well plate, and the line was passaged with triple digestion. iPSCs were used as donor cells in SCNT within 20 passages.
Alkaline phosphatase staining
iPSCs were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 1 min and washed three times with PBS. Alkaline phosphatase (AP) staining was performed with a BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime) following the manufacturer's instructions.
Karyotype analysis
When the iPSCs reached 70–80% confluence, we added a 0.4 μg/mL concentration of colchicine for 2.5 h. We harvested the cells into a dish that had been coated with gelatin for 10 min and a hypotonic solution at 37°C was added. After 15 min, 1 mL of fixative (3 parts methanol to 1 part glacial acetic acid) was added to a 15-mL tube at room temperature for 5 min. Cultures were centrifuged at 800 rpm for 10 min followed by adding 6 mL of cold fixative at 4°C for 20 min, and then repeated once again. Then, 2 mL of cold fixative was added and centrifuged. Finally, spreads were made by “huffing” on slides, i.e., holding the cultures at 45 cm height and dropping one drop of cells onto the top of the slide. After drying for 5 min, the slides were observed using a Nikon 80i microscope. Thirty samples were selected to observe and count the chromosome number.
SCNT and in vitro fertilization
The procedure for porcine SCNT was the same as we described previously (Liu et al., 2008). After 42 h of maturation culture, the oocytes were treated with 1 mg/mL hyaluronidase to remove the surrounding cumulus cells. Oocytes with a clearly extruded first polar body were selected as recipient cytoplasts. Cumulus-free oocytes were enucleated by aspirating the first polar body and adjacent cytoplasm with a glass pipette 25 μm in diameter in tissue culture medium-199 (TCM199)-HEPES plus 0.3% bovine serum albumin (BSA) and 7.5 mg/mL cytochalasin B. A single donor cell was injected into the perivitelline space and electrically fused using two direct pulses of 120 V/mm for 30 msec in fusion medium. Fused eggs were cultured in porcine zygote medium-3 (PZM-3) medium for 7 days in an atmosphere of 5% CO2 and 95% air at 39°C. The cleavage and blastocyst rates were assessed at 48 h and 168 h after activation, and the number of blastocyst cells was examined by nuclear staining with 5 μg/mL Hoechst 33342.
For in vitro fertilization (IVF), freshly ejaculated sperm-rich fractions were collected from fertile boars, and following a short incubation at 39°C, the semen was resuspended and washed three times in DPBS supplemented with 0.1% (wt/vol) BSA by centrifugation at 1,500° g for 4 min. The spermatozoa concentration was measured using a hemocytometer, and the proportion of motile sperm was determined. The spermatozoa were diluted with modified Tris-buffered medium (mTBM) to an optimal concentration. Cumulus-free oocytes were washed three times in mTBM. Approximately 30 oocytes were inseminated in 50-mL drops of mTBM at a final sperm concentration of 3×105/mL for 6 h. Then embryos were washed and cultured in PZM-3 medium for 7 days in an atmosphere of 5% CO2 and 95% air at 39°C.
SCNT and IVF embryos at one-cell (1C), two-cell (2C), four-cell (4C), morula (M), and blastocyst (B) stages were collected at 1, 24, 48, 96, and 168 h respectively for further assessment.
Quantitative real-time PCR
Total RNA was extracted from collected samples using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. Reverse transcription was performed using PrimeScript® RT Reagent Kit (Perfect Real Time, TaKaRa) with the parameters 37°C for 15 min and 85°C for 5 sec, and the cDNA was stored at −20°C until use. For quantitative real-time PCR, reactions were performed using SYBR® Premix ExTaqTM II (Perfect Real Time, TaKaRa) and 7500 Real-Time PCR System (Applied Biosystems). Reactions were performed in 96-well optical reaction plates (Applied Biosystems) with the following conditions: 95°C for 30 sec, followed by 40 two-step cycles at 95°C for 5 sec, and at 60°C for 34 sec, and finally a dissociation stage consisting of 95°C for 15 sec, 60°C for 1 min, and 95°C for 15 sec. For each sample, the cycle threshold (CT) values were obtained from three replicates. The primers used for amplification of target and internal reference genes were presented in Table S1 (Supplementary Data are available at www.liebertpub.com/cell/). The relative expression levels of target genes were analyzed using the 2−δδCT method.
Statistical analysis
Differences of data [mean±standard error of the mean (SEM)] were analyzed by SPSS statistical software. Statistical analysis of data regarding embryo development was performed using the general linear models (GLM) procedure. The data of cell number, blastocyst cell number, and gene expression were analyzed using one-way analysis of variance (ANOVA). The relationship of the efficiency of iPSCs and the development rate of NT embryos was analyzed by Pearson correlation. For all analyses, differences were considered to be statistically significant when p<0.05.
Results
The reprogramming efficiency of donor cells from different individuals was different during iPSC induction
iPSCs were induced by Oct4, Sox2, Klf4, c-Myc, Nr5a2, and Tbx3 (m-pMX-OSKMNT) from six PEF lines, which were derived from Landrace (L1, L6, L9) and Congjiang local pigs (C4, C5, C6). On the 7th day after infection, the numbers of AP-positive colonies were detected. There were no significant differences within L1, L6, L9 or C4, C5, C6 (Fig. 1A–B). However, the numbers of iPSC clones from L1, L6, and L9 that could still be stably grown after three passages were 17, 4, and 8, and the numbers from C4, C5, and C6 were 2, 24, and 3, respectively (Fig. 1C). These results indicated that iPSC induction efficiencies were different among different PEF lines.
FIG. 1.
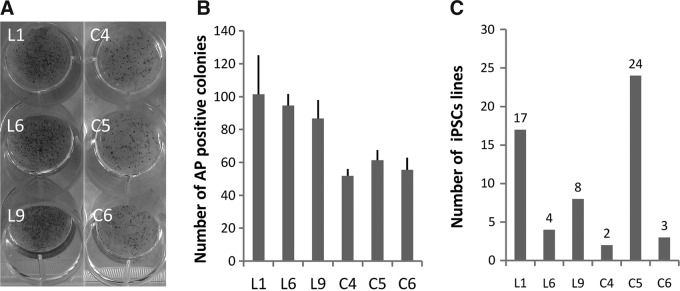
iPSC induction efficiency is different between different PEF cell lines. (A) AP-positive colonies from different PEF lines of Landrace and Congjiang local pigs on the 7th day after infection. (B) The numbers of AP-positive colonies were similar among different PEF lines of Landrace or Congjiang local pigs. (C) The numbers of stable iPSC lines were different between different PEF lines of Landrace and Congjiang local pigs.
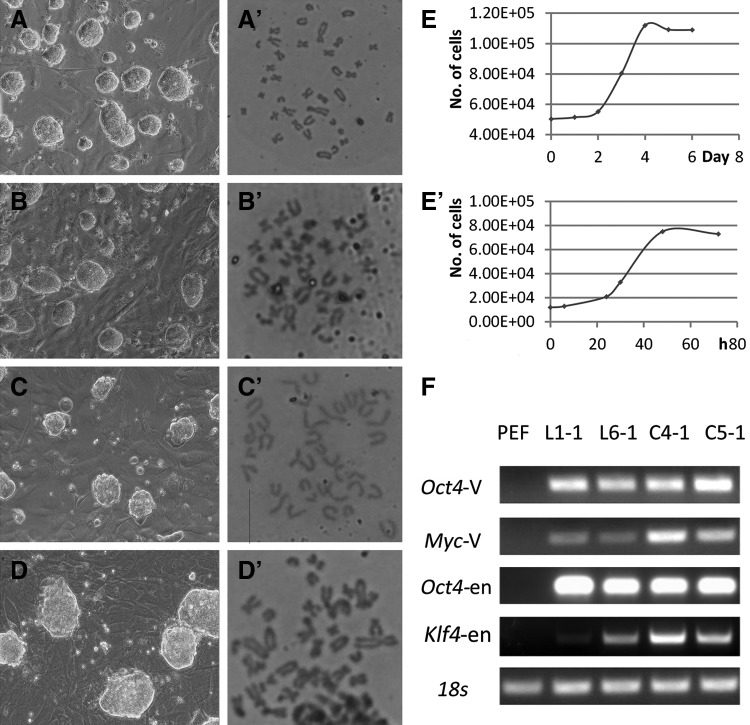
The stable iPSC lines in this study were dome shaped and were similar in appearance to mouse ESC colonies (Fig. 2A–D). They also had a normal karyotype of 38 chromosomes (Fig. 2A′–D′). Analysis indicated that all the six PEF lines had a similar doubling time (43.5 h) (Fig. 2E), whereas the doubling time of each iPSC line was approximately 16 h (Fig. 2E′). The results of RT-PCR showed that both exogenes (Oct4-V, Myc-V) and endogenes (Oct4-en, Klf-en) were expressed in iPSCs (Fig. 2F). Characterization of the pluripotency of the porcine iPSCs showed that they could express OCT4 and SOX2 and could differentiate into cells from all three germ layers both in vitro and in vivo (Fig. S1).
FIG. 2.
Characterization of porcine iPSCs. (A–D′) iPSC lines derived from all PEF lines were dome shaped and similar in morphology to mouse ESCs colonies (A–D, L1, L6, C4, C5), and had a normal karyotype of 38 chromosomes (A′–D′). Scale bar, 100 μm. (E–E′) Proliferation characteristics of the PEF and iPSC lines. (F) RT-PCR assessment showed that both the exogenous (Oct4-V, Myc-V) and the endogenous pluripotency genes (Oct4-en, Klf-en) were expressed in iPSCs. The number of each type cells for RT-PCR was ∼103 and the number of cycles was 30.
The reprogramming efficiency of donor cells from different individuals were different during SCNT and positively related to the efficiency of iPSC induction
To determine whether there are also differences in NT reprogramming efficiency among such PEF lines, and what the relationship is between the efficiency of NT embryonic development and iPSC induction, we performed SCNT using six PEF lines as donor cells. The NT blastocyst rates of L1, L6, L9 were 10.73%, 4.63%, 8.72% (Table 1), and that of C4, C5, C6 were 9.20%, 14.99%, 8.40% respectively (Table 2). There were significant differences (p<0.05) between the blastocyst rates of L1, L9, and L6, and between those of C4, C6, and C5. Interestingly, the PEF lines that had a higher NT blastocyst rate also had a higher iPSC induction efficiency. Statistical analysis indicated that the efficiency of NT embryonic development is positively correlated with the efficiency of iPSC induction (Pearson correlation was 0.837, p<0.05) (Fig. S2).
Table 1.
PEF-SCNT and iPSC-SCNT Development of Landrace Pigs
| Cell line | Duplicate number | Reconstructed embryo (%) | Cleavage (%) | Blastocyst (%) | No. of blastocyst cells |
|---|---|---|---|---|---|
| L1 | 3 | 157 | 131 (83.67±8.82) | 17 (10.73±3.45)a | 31.47±12.43 |
| L1-1 | 3 | 142 | 96 (67.49±3.87) | 17 (11.77±1.36)a | 50.88±24.26 |
| L1-2 | 3 | 134 | 56 (49.29±4.03) | 13 (9.68±0.84)a | 36.62±13.87 |
| L1-3 | 4 | 140 | 89 (63.66±5.60) | 16 (11.43±0.27)a | 39.15±14.93 |
| L6 | 3 | 108 | 82 (75.93±9.75) | 5 (4.63±1.61)b | 31.40±17.91 |
| L6-1 | 4 | 127 | 71 (55.82±4.27) | 11 (8.36±3.22)a | 34.67±17.24 |
| L9 | 3 | 104 | 89 (85.37±13.22) | 9 (8.72±3.22)a | 41.33±9.19 |
Different superscripts represent a significant difference, p<0.05.
PEF, porcine embryonic fibroblasts; SCNT, somatic cell nuclear transfer; iPSC, induced pluripotent stem cells.
Table 2.
PEF-SCNT and iPSC-SCNT Development of Congjiang Local Pigs
| Cell line | Duplicate number | Reconstructed embryo (%) | Cleavage (%) | Blastocyst (%) | No. of blastocyst cells |
|---|---|---|---|---|---|
| C4 | 3 | 121 | 95 (79.12±8.50) | 11 (9.20±1.66)a | 31.60±11.64 |
| C4-1 | 3 | 113 | 74 (68.32±13.30) | 6 (5.68±1.75)a | 33.50±19.09 |
| C5 | 3 | 119 | 92 (76.54±4.43) | 17 (14.99±3.34)b | 34.47±10.72 |
| C5-1 | 3 | 156 | 113 (70.71±8.23) | 8 (5.06±1.70)a | 38.00±29.70 |
| C6 | 3 | 157 | 124 (79.27±5.68) | 11 (8.40±3.20)a | 32.83±11.71 |
Different superscripts represent a significant difference, p<0.05.
PEF, porcine embryonic fibroblasts; SCNT, somatic cell nuclear transfer; iPSC, induced pluripotent stem cells.
iPSCs from different individuals had a similar SCNT embryonic development rate
To explore the effects of iPSC induction on NT reprogramming, selected iPSC lines derived from different PEF lines were used for NT. Because there was more than one iPSC line from each PEF line, it was necessary to evaluate whether different iPSC lines from the same PEF line have a similar SCNT development rate. We randomly selected three iPSC lines (L1-1, L1-2, L1-3) from L1 to examine SCNT development. The SCNT blastocyst rates of L1-1, L1-2, and L1-3 were 11.77%, 9.68%, and 11.43%, respectively, (p>0.05, Table 1). The results indicated that different iPSC lines from the same PEF line had similar SCNT development rates. So we chose one iPSC line from each of L1, L6 (L1-1, L6-1) and C4, C5 (C4-1, C5-1) for further studies because there were significant differences between the SCNT blastocyst rates between PEF lines of L1 and L6 and between C4 and C5. The blastocyst rates of L1-1 and L6-1 iPSC-SCNT were 11.77% and 8.36% (p>0.05, Table 1), and the blastocysts rate of C4-1 and C5-1 iPSC-SCNT were 5.68% and 5.06% (p>0.05, Table 2). iPSC-SCNT development rates decreased significantly (p<0.05) compared with its source PEF-SCNT development rate in Congjiang local pig group (Table 2).
Continued exogenous gene expression in iPSC-SCNT embryos
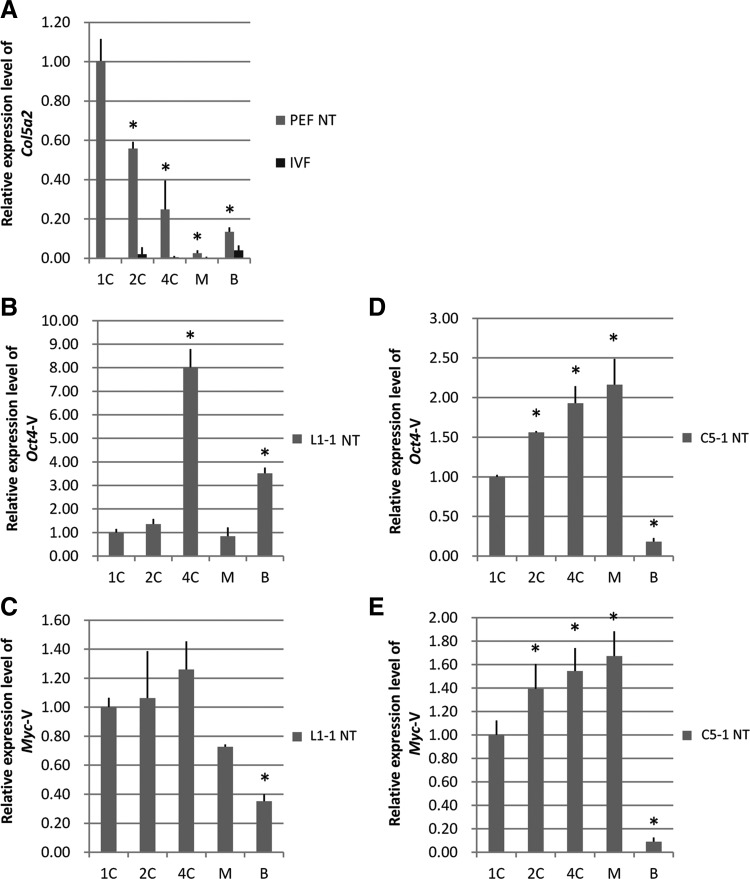
The key early event of reprogramming that occurs in SCNT is the rapid silencing of somatic-specific gene expression and the activation of embryonic gene expression. Theoretically, exogenous genes should be silenced in iPSC-SCNT embryos. We detected the expression of a PEF-specific gene (Col5a2) in PEF-SCNT embryos and exogenous genes (Oct4-V, Myc-V) in iPSC-SCNT embryos by real-time PCR. The results showed that Col5a2 expression declined quickly after SCNT (Fig. 3A), but exogenous genes (Oct4-V, Myc-V) in iPSC-NT embryos continued to be expressed until the blastocyst stage (Fig. 3B–E).
FIG. 3.
Exogenous gene continued to be expressed in iPSC-SCNT embryos. (A) Col5a2, a somatic cell-specific gene, was quickly silenced during PEF-SCNT development. (B–E) Exogenous genes (Oct4-V, Myc-V) continued to express in iPSC-SCNT embryos until the blastocyst stage. There were significant differences between data with different numbers of asterisks, p<0.05.
Discussion
iPSC induction and SCNT are the two main reprogramming approaches. However, it is unknown whether there is correlation in the reprogramming efficiency of these two approaches. Here, for the first time, we made a systematic analysis of the reprogramming efficiency of the two reprogramming approaches. We found that there is a positive correlation between iPSC and SCNT reprogramming efficiency among different PEF lines, which will increase our understanding of the reprogramming mechanisms of both iPSCs and SCNT.
In this study, we found that the efficiencies of iPSC induction were different among different PEF donors. Generally, iPSC induction efficiency is determined by the percentage of the marker gene expression in cells if there is a reporter system in the donor cells, such as the Oct4 promoter driving the enhanced green fluorescent protein (EGFP) system, or determined by the numbers of stable iPSC lines derived from the donor cells (Su et al., 2013; Xue et al., 2013; Zhao et al., 2009). The number of AP-positive colonies is not considered an ideal index for the efficiency of iPSC induction, because AP-positive colonies cannot represent the full reprogramming status; it is only an early reprogramming marker (Huang et al., 2011).
In this study, where there was no reporter system in the porcine PEF donors, we checked both of the number of AP-positive colonies and the numbers of iPSC lines that could be sustained for at least three passages with triple digestion. There was a significant difference among the number of iPSCs lines from different PEF lines derived from different individuals, both in the two breeds of Landrace and Congjiang local pigs, although there were no differences in the number of AP-positive colonies on the 7th day after infection. These results indicate that it might be the genetic or epigenetic background of different PEF lines that affects the iPSC induction efficiency.
Evidence is emerging that early epigenetic priming events in the reprogramming process may be critical for subsequent induction of pluripotency. For example, cells that are unable to silence Thy1 expression early enough upon Oct4, Sox2, Klf4, and c-Myc induction become refractory to the action of the reprogramming factors and yield iPSCs with dramatic delay and at much lower efficiency (Polo et al., 2012b). Also, the original epigenetic states of source cells have a significant impact on iPSC induction. Researchers have reported that hematopoietic, muscle, and liver stem cells can be more efficiently reprogrammed to iPSCs than differentiated blood, muscle, and liver cells (Eminli et al., 2009; Kleger et al., 2012; Tan et al., 2011). The differences of epigenetic background in these two groups of cells are considered to be the main reason for the differences in induction efficiencies. Comparison of the global methylation level of different PEF sublines has shown that there were significant differences between them (Bonk et al., 2007). The six PEF lines examined in this study were derived from the same litter but were from different fetuses of the same sex. The cells were used after the same number of passages in culture, and their growth rate and cell cycle synchrony were also similar, suggesting that differences in their iPSC induction efficiency may be mainly caused by the differences of their epigenetic state.
The PEF lines that were used for iPSC induction in this experiment were also used for SCNT, and the results showed that the SCNT embryonic development rates of different PEF lines were also different. This suggests that the difference in the epigenetic state of these donor cells also has a significant impact on SCNT reprogramming, a result that is consistent with previous research. Bonk et al. had found that there is an inverse correlation between the methylation status of the donor cell and the SCNT blastocyst rates when fibroblasts and blastomeres were used as donor cells in swine. (Bonk et al., 2007). Liu et al. and Mason et al. indicated that chromosome organization of donor cells is also important for SCNT reprogramming (Liu et al., 2012; Mason et al., 2012). In mice, when both iPSCs and their parental cells are used as donor cells for SCNT, iPSCs show better constitutive heterochromatin remodeling and developmental potential (Liu et al., 2012). Theoretically, nuclear reprogramming in SCNT needs to drive the donor cell nuclei from a differentiated state back to the embryonic state, during which dramatic epigenetic changes take place. So the original epigenetic status of donor cells should have an effect on the reprogramming efficiency.
Interestingly, we found the reprogramming efficiency of iPSC induction and SCNT was positively related in this study. Previously, some similarities of reprogramming mechanism have been suggested between the two programming methods, such as an alteration of nuclear structure, modification of the chromosomes, silencing of differentiation-specific genes, activating of pluripotency genes, and changing cell proliferation (Polo et al., 2012a; Whitworth and Prather, 2010). In addition, methods that can enhance reprogramming efficiency in SCNT also work in iPSC induction. For example, addition of small molecules (such as the deacetylase inhibitor, 5-azacytidine) that can artificially modulate the epigenetic status of the genome can improve the efficiency of both SCNT embryonic development and iPSC induction (Ding et al., 2008; Kishigami et al., 2006; Kong et al., 2011; Mali et al., 2010; Mikkelsen et al., 2008). Starvation of donor cells (Chen et al., 2012; Zakhartchenko et al., 1999) or inhibition of the P53 pathway (Hong et al., 2009) can also improve the efficiency both of iPSC induction and SCNT embryo development. All of these studies suggested that although there might be some differences in the reprogramming mechanism of iPSC induction and SCNT, similarities do exist between the two reprogramming methods. No matter which of the reprogramming methods is employed, the epigenetic modifications of differentiated cells are the main “obstacle,” which should be overcome before achieving the pluripotent state, and this could explain why there is positive correlation between iPSCs and SCNT reprogramming efficiency.
When the induced porcine iPSCs were used for SCNT, iPSC-SCNT development tended to be low, which is consistent with the recent reports (Fan et al., 2013). When tracked, the expression of exogenous genes in iPSC-SCNT embryos, Oct4-V and Myc-V continued to be expressed in iPSC-SCNT embryos. We deduced that the expression of exogenous genes may affect porcine iPSC-SCNT embryo development.
In summary, this study proved that there is positive correlation between the reprogramming efficiency of iPSC induction and SCNT. This result can help us to further understand the reprogramming mechanism of both iPSC induction and SCNT. Furthermore, this result also indicates that SCNT has the potential to be used as a screening method for the selection of appropriate donor cells suitable for iPSC induction, which would be beneficial considering the time-consuming task of iPSC induction.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Important Scientific Problems Program of China (973 Program, 2011CBA01006).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Bonk A.J., Cheong H.T., Li R., Lai L., Hao Y., Liu Z., Samuel M., Fergason E.A., Whitworth K.M., Murphy C.N., Antoniou E., and Prather R.S. (2007). Correlation of developmental differences of nuclear transfer embryos cells to the methylation profiles of nuclear transfer donor cells in Swine. Epigenetics 2, 179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Huang J., Yang X., Liu B., Zhang W., Huang L., Deng F., Ma J., Bai Y., Lu R., Huang B., Gao Q., Zhuo Y., and Ge J. (2012). Serum starvation induced cell cycle synchronization facilitates human somatic cells reprogramming. PloS One 7, e28203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Wang Y., Zhang D., Wang Y., Guo Z., and Zhang Y. (2008). Increased pre-implantation development of cloned bovine embryos treated with 5-aza-2′-deoxycytidine and trichostatin A. Theriogenology 70, 622–630 [DOI] [PubMed] [Google Scholar]
- Eminli S., Foudi A., Stadtfeld M., Maherali N., Ahfeldt T., Mostoslavsky G., Hock H., and Hochedlinger K. (2009). Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat. Genet. 41, 968–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan N., Chen J., Shang Z., Dou H., Ji G., Zou Q., Wu L., He L., Wang F., Liu K., Liu N., Han J., Zhou Q., Pan D., Yang D., Zhao B., Ouyang Z., Liu Z., Zhao Y., Lin L., Zhong C., Wang Q., Wang S., Xu Y., Luan J., Liang Y., Yang Z., Li J., Lu C., Vajta G., Li Z., Ouyang H., Wang H., Wang Y., Yang Y., Liu Z., Wei H., Luan Z., Esteban M.A., Deng H., Yang H., Pei D., Li N., Pei G., Liu L., Du Y., Xiao L., and Lai L. (2013). Piglets cloned from induced pluripotent stem cells. Cell Res. 23, 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon J.B. (1962). Adult frogs derived from the nuclei of single somatic cells. Dev. Biol. 4, 256–273 [DOI] [PubMed] [Google Scholar]
- Heyman Y., Zhou Q., Lebourhis D., Chavatte-Palmer P., Renard J.P., and Vignon X. (2002). Novel approaches and hurdles to somatic cloning in cattle. Cloning Stem Cells 4, 47–55 [DOI] [PubMed] [Google Scholar]
- Hochedlinger K., and Plath K. (2009). Epigenetic reprogramming and induced pluripotency. Development 136, 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., and Yamanaka S. (2009). Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Fan N., Cai J., Yang D., Zhao B., Ouyang Z., Gu W., and Lai L. (2011). Establishment of a porcine Oct-4 promoter-driven EGFP reporter system for monitoring pluripotency of porcine stem cells. Cell. Reprogram. 13, 93–98 [DOI] [PubMed] [Google Scholar]
- Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., and Melton D.A. (2008). Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 26, 1269–1275 [DOI] [PubMed] [Google Scholar]
- Inoue K., Ogonuki N., Mochida K., Yamamoto Y., Takano K., Kohda T., Ishino F., and Ogura A. (2003). Effects of donor cell type and genotype on the efficiency of mouse somatic cell cloning. Biol. Reprod. 69, 1394–1400 [DOI] [PubMed] [Google Scholar]
- Kishigami S., Mizutani E., Ohta H., Hikichi T., Thuan N.V., Wakayama S., Bui H.T., and Wakayama T. (2006). Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem. Biophys. Res. Commun. 340, 183–189 [DOI] [PubMed] [Google Scholar]
- Kleger A., Mahaddalkar P.U., Katz S.F., Lechel A., Joo J.Y., Loya K., Lin Q., Hartmann D., Liebau S., Kraus J.M., Cantz T., Kestler H.A., Zaehres H., Scholer H., and Rudolph K.L. (2012). Increased reprogramming capacity of mouse liver progenitor cells, compared with differentiated liver cells, requires the BAF complex. Gastroenterology 142, 907–917 [DOI] [PubMed] [Google Scholar]
- Kong Q.R., Zhu J., Huang B., Huan Y.J., Wang F., Shi Y.Q., Liu Z.F., Wu M.L., and Liu Z.H. (2011). [TSA improve transgenic porcine cloned embryo development and transgene expression]. Yi chuan=Hereditas / Zhongguo yi chuan xue hui bian ji 33, 749–756 [DOI] [PubMed] [Google Scholar]
- Liu Z., Wan H., Wang E., Zhao X., Ding C., Zhou S., Li T., Shuai L., Feng C., Yu Y., Zhou Q., and Beaujean N. (2012). Induced pluripotent stem-induced cells show better constitutive heterochromatin remodeling and developmental potential after nuclear transfer than their parental cells. Stem Cells Dev. 21, 3001–3009 [DOI] [PubMed] [Google Scholar]
- Liu Z.H., Song J., Wang Z.K., Tian J.T., Kong Q.R., Zheng Z., Yin Z., Gao L., Ma H.K., Sun S., Li Y.T., Wang H.B., and Prather R.S. (2008). Green fluorescent protein (GFP) transgenic pig produced by somatic cell nuclear transfer. Chinese Science Bulletin 53, 1035–1039 [Google Scholar]
- Mali P., Chou B.K., Yen J., Ye Z., Zou J., Dowey S., Brodsky R.A., Ohm J.E., Yu W., Baylin S.B., Yusa K., Bradley A., Meyers D.J., Mukherjee C., Cole P.A., and Cheng L. (2010). Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 28, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason K., Liu Z., Aguirre-Lavin T., and Beaujean N. (2012). Chromatin and epigenetic modifications during early mammalian development. Anim. Reprod. Sci. 134, 45–55 [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B.E., Jaenisch R., Lander E.S., and Meissner A. (2008). Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K., Tomii R., Kurome M., Ueda H., Hirakawa K., Ueno S., Hiruma K., and Nagashima H. (2005). Evaluation of the quality of porcine somatic cell nuclear transfer embryo by gene transcription profiles. J. Reprod. Dev. 51, 123–131 [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., and Yamanaka S. (2008). Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26, 101–106 [DOI] [PubMed] [Google Scholar]
- Oback B., and Wells D.N. (2007). Donor cell differentiation, reprogramming, and cloning efficiency: Elusive or illusive correlation? Mol. Reprod. Dev. 74, 646–654 [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., and Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J., Bar-Nur O., Cheloufi S., Stadtfeld M., Figueroa M.E., Robinton D., Natesan S., Melnick A., Zhu J., Ramaswamy S., and Hochedlinger K. (2012a). A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151, 1617–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J., Bar-Nur O., Cheloufi S., Stadtfeld M., Figueroa M.E., Robinton D., Natesan S., Melnick A., Zhu J.F., Ramaswamy S., and Hochedlinger K. (2012b). A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151, 1617–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R.J., Baylink D.J., Neises A., Kiroyan J.B., Meng X., Payne K.J., Tschudy-Seney B., Duan Y., Appleby N., Kearns-Jonker M., Gridley D.S., Wang J., Lau K.H., and Zhang X.B. (2013). Efficient generation of integration-free ips cells from human adult peripheral blood using BCL-XL together with Yamanaka factors. PloS One 8, e64496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- Tan K.Y., Eminli S., Hettmer S., Hochedlinger K., and Wagers A.J. (2011). Efficient generation of iPS cells from skeletal muscle stem cells. PloS One 6, e26406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakao S., Kitada M., Kuroda Y., Shigemoto T., Matsuse D., Akashi H., Tanimura Y., Tsuchiyama K., Kikuchi T., Goda M., Nakahata T., Fujiyoshi Y., and Dezawa M. (2011). Multilineage-differentiating stress-enduring (Muse) cells are a primary source of induced pluripotent stem cells in human fibroblasts. Proc. Natl. Acad. Sci. USA 108, 9875–9880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T., and Yanagimachi R. (2001). Mouse cloning with nucleus donor cells of different age and type. Mol. Reprod. Dev. 58, 376–383 [DOI] [PubMed] [Google Scholar]
- Wakayama T., Tabar V., Rodriguez I., Perry A.C., Studer L., and Mombaerts P. (2001). Differentiation of embryonic stem cell lines generated from adult somatic cells by nuclear transfer. Science 292, 740–743 [DOI] [PubMed] [Google Scholar]
- Wang J., Gu Q., Hao J., Jia Y., Xue B., Jin H., Ma J., Wei R., Hai T., Kong Q., Bou G., Xia P., Zhou Q., Wang L., and Liu Z. (2013). Tbx3 and Nr5alpha2 play important roles in pig pluripotent stem cells. Stem Cell Rev. 9, 700–708 [DOI] [PubMed] [Google Scholar]
- Whitworth K.M., and Prather R.S. (2010). Somatic cell nuclear transfer efficiency: How can it be improved through nuclear remodeling and reprogramming? Mol. Reprod. Dev. 77, 1001–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmut I., Schnieke A.E., McWhir J., Kind A.J., and Campbell K.H. (1997). Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813 [DOI] [PubMed] [Google Scholar]
- Xu Y., Wei X., Wang M., Zhang R., Fu Y., Xing M., Hua Q., and Xie X. (2013). Proliferation rate of somatic cells affects reprogramming efficiency. J. Biol. Chem. 288, 9767–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Cai X., Wang L., Liao B., Zhang H., Shan Y., Chen Q., Zhou T., Li X., Hou J., Chen S., Luo R., Qin D., Pei D., and Pan G. (2013). Generating a non-integrating human induced pluripotent stem cell bank from urine-derived cells. PloS One 8, e70573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhartchenko V., Durcova-Hills G., Stojkovic M., Schernthaner W., Prelle K., Steinborn R., Muller M., Brem G., and Wolf E. (1999). Effects of serum starvation and re-cloning on the efficiency of nuclear transfer using bovine fetal fibroblasts. J. Reprod. Fertil. 115, 325–331 [DOI] [PubMed] [Google Scholar]
- Zhao X.Y., Li W., Lv Z., Liu L., Tong M., Hai T., Hao J., Guo C.L., Ma Q.W., Wang L., Zeng F.Y., and Zhou Q. (2009). iPS cells produce viable mice through tetraploid complementation. Nature 461, 86–90 [DOI] [PubMed] [Google Scholar]
- Zhou S., Ding C., Zhao X., Wang E., Dai X., Liu L., Li W., Liu Z., Wan H., and Feng C. (2010). Successful generation of cloned mice using nuclear transfer from induced pluripotent stem cells. Cell Res. 20, 850–853 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.