Abstract
Genome wide association studies (GWAS) of ten different cancers have identified pleiotropic cancer predisposition loci across a region of chromosome 5p15.33 that includes the TERT and CLPTM1L genes. Of these, susceptibility alleles for pancreatic cancer have mapped to the CLPTM1L gene, thus prompting an investigation of the function of CLPTM1L in the pancreas. Immunofluorescence analysis indicated that CLPTM1L localized to the endoplasmic reticulum (ER) where it is likely embedded in the membrane, in accord with multiple predicted trans-membrane domains. Overexpression of CLPTM1L enhanced growth of pancreatic cancer cells in vitro (1.3–1.5 fold, PDAY7<0.003) and in vivo (3.46 fold, PDAY68=0.039), suggesting a role in tumor growth; this effect was abrogated by deletion of two hydrophilic domains. Affinity purification followed by mass-spectrometry identified an interaction between CLPTM1L and non-muscle myosin II (NMM-II), a protein involved in maintaining cell shape, migration, and cytokinesis. The two proteins co-localized in the cytoplasm and, after treatment with a DNA damaging agent, at the centrosomes. Overexpression of CLPTM1L and depletion of NMM-II induced aneuploidy, indicating that CLPTM1L may interfere with normal NMM-II function in regulating cytokinesis. Immunohistochemical analysis revealed enhanced staining of CLPTM1L in human pancreatic ductal adenocarcinoma (n=378) as compared to normal pancreatic tissue samples (n=17) (P=1.7×10−4). Our results suggest that CLPTM1L functions as a growth promoting gene in the pancreas and that overexpression may lead to an abrogation of normal cytokinesis, indicating that it should be considered as a plausible candidate gene that could explain the effect of pancreatic cancer susceptibility alleles on chr5p15.33.
Introduction
Risk variants in the TERT-CLPTM1L gene region on chromosome 5p15.33 have been reported in genome wide association studies (GWAS) for ten cancer types including bladder, breast, glioma, lung, melanoma, non-melanoma skin cancer, ovarian, pancreas, prostate and testicular germ cell cancer (1–13). The TERT gene encodes the catalytic subunit of the telomerase reverse transcriptase complex known for its role in maintaining telomere ends and the increased telomerase activity often seen in human cancers (14). The CLPTM1L gene encodes the cleft lip and palate associated transmembrane 1-like protein (CLPTM1L) and was originally identified in a screen for genes conferring resistance to cisplatin in ovarian cancer cells (15). When overexpressed in ovarian cancer cell lines, CLPTM1L induced apoptosis in cisplatin sensitive cells, giving rise to its original name: Cisplatin resistance-related protein (CRR9) (15). CLPTM1L was later shown to protect lung cancer cells from apoptosis after treatment with DNA damaging agents via Bcl-xL (16).
Gain of chromosome 5p is one of the most recurrent chromosomal abnormalities in human cancers (17). Although most commonly seen in thyroid, lung and cervical cancer, 5p gain is also frequent in other cancers including gastric, ovarian, colorectal, hepatocellular, esophageal, bladder, and pancreatic adenocarcinoma (17–19). The most common event in early stages of non-small cell lung cancer (NSCLC) is gain at 5p15.33 involving both TERT (78%) and CLPTM1L (53%) (20). However, a recent study of cervical cancer noted that CLPTM1L, but not TERT, was among the multiple genes on 5p (33%) that were both amplified and overexpressed (21, 22).
The most significant GWAS risk variants on 5p15.33 for pancreatic cancer lie in intron 13 of the CLPTM1L gene and are located ~27 kb from the transcriptional start of TERT (11). Although this does not exclude TERT as a plausible candidate gene explaining this pancreatic cancer risk allele, CLPTM1L should be considered a potential target gene. Thus, to explore a possible function for CLPTM1L in pancreatic cancer, we examined its role in growth control in vitro and in vivo, and searched for interacting proteins that could provide clues to its function.
Material and Methods
Cell lines and antibodies
The human embryonic kidney cell line HEK293T, human pancreatic cancer cell line PANC-1, and mouse kidney cell line IMCD3 (all from ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Mediatech Inc, Herndon, VA) supplemented with 10% fetal bovine serum (Life Technologies, Grand Island, NY).
Commercial antibodies used included those for endogenous CLPTM1L (HPA014791, Sigma, St. Louis, MO), FLAG-tagged CLPTM1L (M2 F1804, Sigma), the endoplasmic reticulum (ER) marker Calnexin, (C4731, Sigma), a mitochondrial marker (MTC02, ab3298, Abcam Cambridge, MA), the centrosome marker gamma-tubulin (T5192, Sigma), the Golgi marker GM130 (G7295, Sigma), alpha-tubulin (ab7291, Abcam) and Myosin-9/MYH10 (sc-33729, Santa Cruz Biotechnology, Santa Cruz, California). Secondary antibodies were Alexa Fluor 594® or 488® donkey anti-mouse or anti-rabbit IgG (H+L) (A21202, A21203, A21206 and A21207, Life Technologies).
Generation of CLPTM1L expression plasmids and creation of stable cell lines
Full-length human CLPTM1L cDNA (Invitrogen, Ultimate™ ORF Clone ID: IOH13343) was cloned into pDest-737 (a Gateway adapted version of Sigma’s p3xFLAG-CMV10) using the Gateway system (Invitrogen). Three constructs were generated from the full-length human CLPTM1L cDNA (RefSeq NM_030782.3) and verified by Sanger sequencing: WT CLPTM1L (full-length CLPTM1L), CLPTM1L-ΔLoop (missing amino acids (aa) 36–280 between transmembrane domains 1 and 2) and CLPTM1L-ΔCterm (lacking aa 455–538). The designed constructs include 3 tandem FLAG tags at the amino terminus of CLPTM1L. A fourth construct contained a FLAG tag at the C-terminus of CLPTM1L, to compare the effect of FLAG tags on the N- or C-termini on growth rates in vitro.
Stable cell lines were generated by transfecting PANC-1 cells (Lipofectamine 2000, 11668-019, Life technologies, Grand Island, NY) with selection in G418 (Mediatech, 30–234-CI, Manassas, VA). The constructs above, expressing WT or mutant CLPTM1L from the CMV promoter, were used to generate the following stable lines: PANC1-vo (empty vector), PANC1-CLPTM1L (full-length CLPTM1L), PANC1-CLPTM1L-ΔLoop (loop deletion) or PANC1-CLPTM1L-ΔCterm (C-terminal deletion). Transient transfections for HEK293T and mIMCD3 cells were performed with the same constructs.
Prediction of the topology of WT CLPTM1L was assessed using: TMHMM v.2.0, TMPred and TopPred2 (23–26).
In vitro and in vivo growth assays
Cell proliferation was measured in vitro by seeding PANC-1 stably expressing CLPTM1L (full-length or deletion mutants) at 3×103 cells per well in 96-well plates. Time points were taken every two days (days 1, 3, 5 and 7) and cell growth assessed using the WST-1 reagent (Roche Applied Science, Indianapolis, IN) for 30 min. The optical density (OD) change created by the metabolizing of the reagent was evaluated in a spectrophotometer (TECAN) at 450 nm. Absorbance at the reference wavelength of 600 nm was subtracted from the A450 values.
CLPTM1L knock-down was performed using the Dharmacon DharmaFECT siRNA transfection reagent (Thermo Scientific Dharmacon #T-2001-01, Waltham, MA) according to the manufacturer’s instructions. Dharmacon ON-TARGET plus SMARTpool siRNA specifically targeting CLPTM1L (L-015661-02-0005) or a control non-target siRNA (D-001810-02-05) were purchased from Thermo Scientific Dharmacon. Cell proliferation experiments were performed 48 hrs after transfection with 100 nM siRNA. The efficiency of CLPTM1L knock-down was assessed by isolating RNA from PANC-1 cells, using the mirVana RNA kit (ABI). Briefly, 1 μg RNA (RIN scores >9.0) was reverse transcribed using Superscript III reverse transcriptase (Invitrogen). RT-qPCR was performed on a 7900HT system (ABI) using TaqMan gene expression assays for CLPTM1L (Hs00363947_m1) and B2M (Hs00187842_m1) from Life Technologies. Each reaction was run in quadruplicate and analyzed according to the ΔΔCt method using B2M as the housekeeping gene.
Tumor growth was measured in vivo using a xenograft mouse model. Female nude mice (8–10 weeks old) were purchased from the Animal Production Area, NCI, Frederick, MD, and housed in a pathogen free environment. Briefly, 106 PANC-1 cells, stably transfected with different CLPTM1L constructs or the vector control, were injected subcutaneously into the flank of each mouse. Tumor size was measured by a caliper three times a week for up to 77 days using the formula of length × width × width/2 to estimate tumor volumes in mm3, or when protocol experimental end points were reached (tumor diameter reached 2 cm). For each group, 5 mice were injected per stable cell line per experiment. After seeing similar results for two independent constructs expressing WT CLPTM1L with FLAG tags at either end, the CLPTM1L constructs tagged on the N-terminus were chosen for further work. Final results were pooled from 3 independent experiments: two that were performed with PANC1-vo (empty vector) and PANC1-CLPTM1L cells, and a third experiment that used PANC1-vo, PANC1-CLPTM1L, PANC1-CLPTM1L-ΔLoop and PANC1-CLPTM1L-ΔCterm cells. The difference in growth rates was analyzed by comparing tumor volumes at day 68 using the Mann-Whitney U test. Animal care and experimental procedures were approved by the NIH Animal Care and User Committees (PB-047 M1 to Dr. Javed Khan).
Affinity purification of protein complexes, tryptic digestion and fractionation
HEK293T cells were grown to ~60% confluency and transfected with 3xFLAG-tagged WT CLPTM1L (experimental analysis) or vector only (control) in a complex of polymer PEI (Polysciences Inc, Warrington, PA) and DNA at a ratio of 5:2. After 48 hrs, cells were harvested on ice in RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% SDS, 0.5% Sodium Deoxycholate, 1.0% NP-40) with 1x protease inhibitor cocktail (Roche, Nutley, NJ). Lysates were incubated with 25μL of anti-FLAG M2 agarose (Sigma) for 2 hours followed by washing and elution via a 3x-FLAG peptide as previously described (Das PMID: 20968308). The eluates were subjected to overnight digestion with trypsin (1 μg) at 37°C followed by lyophilization, reconstitution and fractionation using strong cation exchange (SCX) liquid chromatography (LC) and mass spectrometry analysis as previously described (27). Proteomics Data Analysis was performed as previously described (27).
Validation of CLPTM1L-MYH9 interaction by co-IP
HEK293T cells were transfected with 3xFLAG-tagged WT CLPTM1L expression plasmids or the vector control only, and collected 48 hours after transfection. PANC-1 cells stably expressing WT CLPTM1L, CLPTM1L-ΔLoop or CLPTM1L-ΔCterm were cultured to a confluence of ~80%. Cells were washed with ice cold PBS and harvested in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.5mM EDTA, 1.0% NP-40, with 1x protease inhibitor cocktail). Immunoprecipitation (IP) of FLAG-tagged CLPTM1L was performed by incubating the supernatant with anti-FLAG M2 agarose for 2 hours at 4°C. Affinity complexes were washed and interacting complexes released from FLAG beads by boiling in 2xLDS sample buffer (Invitrogen) and resolved on a 3–8% Tris-Acetate gel (Invitrogen). Samples were subjected to western analysis using antibodies to FLAG M2, Myosin 9/10 and beta-actin followed by appropriate secondary mouse or rabbit antibodies (Thermo Scientific, Rockford, MO) before detection of the signal with ECL (Thermo Scientific).
Cell cycle and DNA content analysis
The effect of CLPTM1L and MYH9 on cell cycle progression was studied by flow cytometric analysis of DNA content and BrdU incorporation using the FITC BrdU Flow kit (557891, BD Pharmingen BioSciences, San Jose, CA) according to the manufacturer’s recommendation. Briefly, logarithmically growing cells (transiently transfected HEK293T or stably transfected PANC-1 cells) were labeled with 10 μM BrdU for 30 min at 37°C, fixed, permeabilized and treated with DNase. A FITC conjugated anti-BrdU antibody was used for staining DNA with incorporated BrdU, and 7-amino-actinomycin D (7-AAD) for total DNA staining. Fluorescence of 30,000 cells was acquired on a FACSCalibur instrument (Becton Dickinson). The resulting DNA histograms were quantified by using the Cell Quest Pro software (Becton Dickinson) and the percentage of G0/G1, S, G2/M and aneuploidy (>4N) cells determined.
For knock-down experiments, cells were plated in six-well plates 24 hours before transfection with siRNA so they reached 80% confluency at the time of transfection. The Dharmacon DharmaFECT siRNA transfection reagent (Thermo Scientific Dharmacon #T-2001-01, Waltham, MA) was used for transfection according to the manufacturer’s instructions. Dharmacon ON-TARGET plus SMARTpool siRNA specifically targeting MYH9 (L-007668-00-0005) or a control non-target siRNA (D-001810-02-05) were purchased from Thermo Scientific Dharmacon. Twenty-four hours after transfection with 100 nM siRNA, the media was replaced with fresh complete media. Assays were performed 48 hours after transfection.
Efficiency of MYH9 knock-down was assessed by isolating RNA from PANC-1 cells, using the mirVana RNA kit (ABI). Briefly, 1 μg RNA (RIN scores >9.0) from cell lines was reverse transcribed using Superscript III reverse transcriptase (Invitrogen). RT-qPCR was performed on a 7900HT system (ABI) using TaqMan gene expression assays for MYH9 (Hs00159522_ml) and B2M (Hs00187842_m1) from Life Technologies. Each reaction was run in quadruplicate and analyzed according to the ΔΔCt method using B2M as the housekeeping gene.
Immunofluorescence (IF) and in situ Proximity Ligation Assay (DPLA)
To visualize the subcellular localization of the endogenous or FLAG-tagged CLPTM1L proteins, cells were grown on glass coverslips, fixed with 4% paraformaldehyde (PFA) or 100% methanol and stained with antibodies against endogenous CLPTM1L (1:500), FLAG-tagged CLPTM1L (1:500), Calnexin (1:500), mitochondrial marker (1:500), GM130 (1:500), alpha-tubulin (1:1000) and gamma-tubulin (1:1000) followed by visualization by confocal microscopy (Zeiss LSM 510 Meta). Assessment of co-localization with MYH9 was performed in the same manner using antibodies against MYH9 (1:500). Secondary antibodies used are Alexa Fluor 594® or 488® donkey anti-mouse or anti-rabbit IgG (H+L) (Invitrogen). Cells were counterstained with 4′6-diamidino-2-phenylindole (DAPI).
The Duolink in situ Proximity Ligation Assay (DPLA, in situ red starter kit, #92101 from Olink Bioscience, Uppsala, Sweden) was used according to the manufacturer’s protocol to evaluate interactions between CLPTM1L and other proteins. Briefly, cells were grown on coverslips and fixed with 4% PFA or 100% methanol for 10 minutes (min) at room temperature and air-dried, followed by blocking and labeling with antibodies to endogenous CLPTM1L (Sigma HPA014791) or FLAG-tagged CLPTM1L (Sigma, F1804), MYH9/10 (Santa Cruz, SC-33729), at a dilution of 1:500 in a pre-heated humidity chamber at 37°C for 30 min. After washing, slides were incubated with anti-rabbit and anti-mouse secondary antibodies with attached DPLA probes (dilution 1:5) for 30 min in a pre-heated humidity chamber at 37°C. Finally, cells were counterstained by DAPI and signals observed by confocal microscopy (Zeiss LSM 510 Meta); a red fluorescence signal indicates that two proteins are separated by less than 40 nm (28).
Cells were treated with 50 μM of the DNA damaging agent cisplatin (ALX-400-040-M250, Enzo Life Sciences, Farmingdale, NY) for 0, 24 or 48 hours for IF and Duolink experiments as indicated.
Immunohistochemistry
Tissue microarrays (TMAs) with normal- and tumor-derived pancreatic tissue samples were obtained from the Mayo Clinic and from the Surveillance, Epidemiology, and End Results (SEER) registries (29, 30). The TMAs from the Mayo Clinic (four TMAs were used: PDAC (pancreatic ductal adenocarcinoma) TMA1 (n=111), PDAC GEM (gemcitabine) (n=156), PanIN (pancreatic intraepithelial neoplasia) TMA (n=13) and pancreatic islet cell tumors (Islet cell TMAs) (n=41) along with 3 cores of tissues from normal pancreas that could be evaluated for CLPTM1L expression. The SEER TMA contained 14 cores from normal pancreas (adjacent to tumor) in addition to PDAC (n=111) and islet cell tumors (n=7).
Immunohistochemistry for CLPTM1L was performed with a rabbit polyclonal antibody (HPA014791, Sigma, St. Louis, MO) at 1:1600 dilution. Briefly, slides were de-paraffinized in xylene and graded alcohols, and subject to antigen retrieval in a pressure cooker with citrate buffer (pH6) for 20 min. Endogenous enzyme activity was blocked with 3% hydrogen peroxide in methanol with an additional block in 2% non-fat milk used to reduce nonspecific reactions. Subsequently, slides were incubated with primary antibody for 60 min at room temperature, detected antigen-antibody reaction with Dako Envision+ Dual Link system-HRP (DAKO, Carpinteria, CA), visualized in 3,3′-diaminobenzidine (DAB), counterstained with hematoxylin, dehydrated and mounted for visualization.
CLPTM1L staining intensity was scored on a scale from 0–4 as follows: 0, negative; 1, background; 2, weak; 3, positive; 4, strong. Staining extent of cells was also scored on a scale of 0–4: 0, negative; 1, <10%; 2, 10–25%; 3, 25–75% and 4, >75%. Combined scores (histoscore) were calculated by multiplying scores for intensity and extent (individual range 0–4, combined range 0–16). For statistical analysis, CLPTM1L staining was classified as negative/weak (combined scores 0–4 and intermediate/strong from 6–16). A 2×2 chi square test was used to assess the difference in staining between normal- and tumor-derived samples (df=1). A Pearson uncorrected P value was reported.
Results
Subcellular localization of CLPTM1L
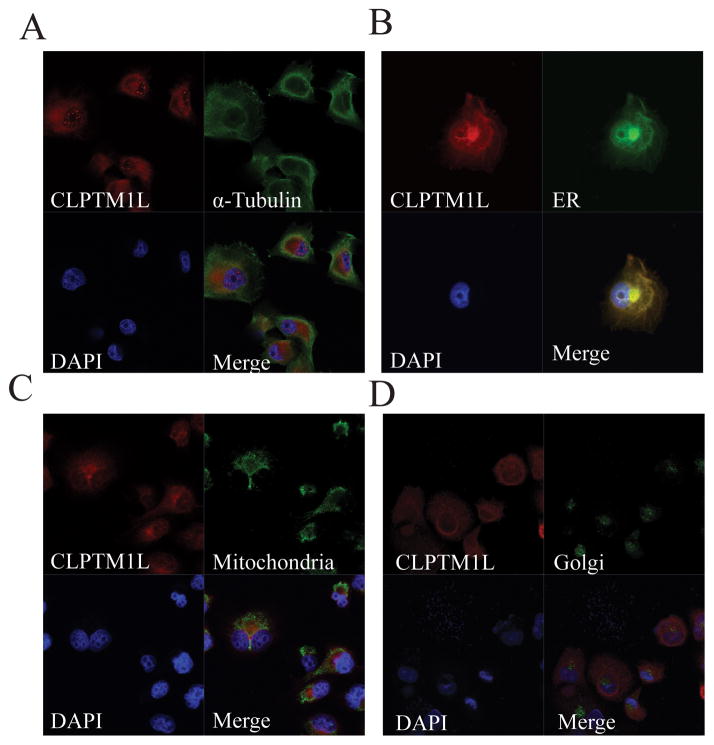
The protein product of the CLPTM1L gene is predicted to contain 6 transmembrane (TM) domains and two large hydrophilic domains: a loop between the first and second TM domains, and a C-terminal tail. It is highly conserved in primates and relatively well conserved in flies and nematodes (31). We assessed the subcellular localization of endogenous CLPTM1L in pancreatic cancer cell lines by immunofluorescence (IF) analysis and demonstrated perinuclear cytoplasmic staining and a punctate pattern of CLPTM1L over the nucleus, indicating possible nuclear or nuclear membrane staining (Figure 1a). Co-localization with markers for cytoplasmic organelles showed that CLPTM1L localized mainly to the endoplasmic reticulum (ER) (Figure 1b) in accord with its 6 predicted TM domains. No staining was detected in mitochondria (Figure 1c) despite careful assessment of co-localization with a mitochondrial marker in 200 nm optical sections in XY, XZ and YZ projections, in apparent contradiction to recent work showing mitochondrial localization in 95-D lung cancer cells (32). No staining was detected in the Golgi apparatus (Figure 1d). To confirm that the ER localization of overexpressed WT FLAG-tagged CLPTM1L observed was not due to defects in polarized sorting, we transiently expressed CLPTM1L in mouse kidney IMCD3 epithelial cells and observed co-localization with the ER-marker calnexin (Supplemental Figure 1). Two of the three programs used to predict the topology of CLPTM1L indicated that the N- and C-termini of the protein protrude into the lumen of the ER and other organelles it resides in (TMPred and TMHMM) but one (TopPred) predicted an opposite topology with both ends on the cytoplasmic face of the ER (23). The hydrophilic loop structure between TM1 and TM2 is expected to be on the opposite side of the membrane as compared to the N- and C-termini.
Figure 1. Localization of endogenous CLPTM1L in pancreatic cells and co-localization of CLPTM1L with cytoplasmic organelles.
(A) Localization of endogenous CLPTM1L in PANC-1 pancreatic cancer cells. Cytoplasmic and punctate nuclear, or nuclear membrane, staining is seen (red). Counter staining was performed for α-tubulin (green). (B) Co-localization of WT CLPTM1L (red) with the Endoplasmic Reticulum (ER) marker Calnexin (green) in PANC-1 cells stably transfected with WT CLPTM1L. Co-localization of WT CLPTM1L (red) with a mitochondrial marker (green) (C), or a Golgi marker (GM130 in green) (D), was not seen in PANC-1 cells stably transfected with WT CLPTM1L. Cells in all panels were counterstained with DAPI (blue).
CLPTM1L overexpression enhances cell proliferation in vitro and in vivo
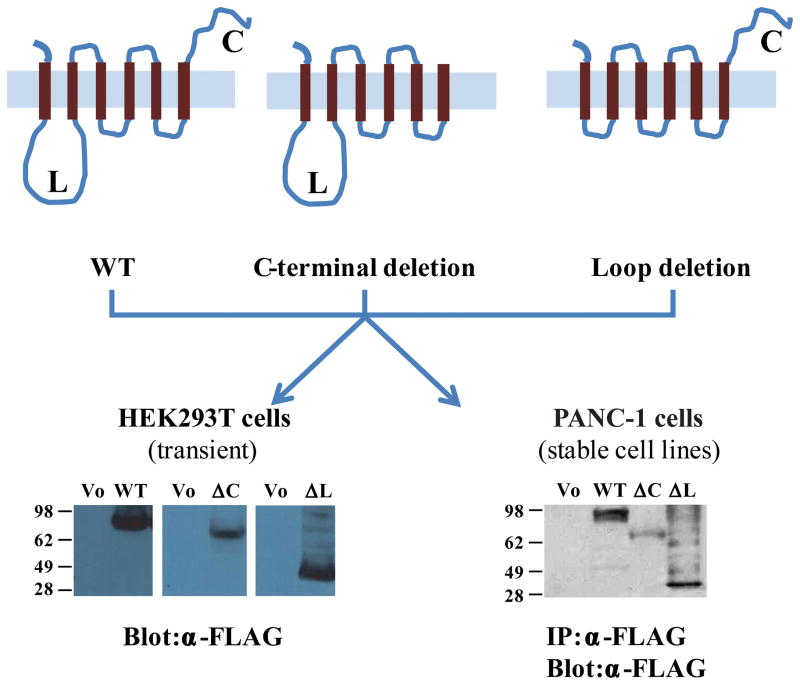
To investigate if CLPTM1L influenced growth, we overexpressed full-length or mutant CLPTM1L in HEK293T (transient transfection) and PANC-1 (stable transfection) cells. These included four FLAG epitope-tagged cDNA expression constructs: wild type (WT) full-length CLPTM1L (tagged on either C- or N-termini), and two deletion mutants tagged at the N-termini: CLPTM1L-ΔCterm (hydrophilic C-terminal domain after the last transmembrane domain deleted) and CLPTM1L-ΔLoop (hydrophilic loop between the first and second trans-membrane domains deleted). Figure 2 shows a schematic figure of the resulting proteins (upper panels) as well as protein expression of the three different forms of CLPTM1L (WT and 2 mutant) in transiently transfected HEK293T cells (by western blotting in lower left panel) and in PANC-1 cells stably expressing the proteins (by FLAG immunoprecipitation and western blotting in lower right panel).
Figure 2. Wild type and mutant CLPTM1L proteins.
Top panel shows schematic figures of WT and mutant CLPTM1L proteins. From left to right: WT CLPTM1L (full-length CLPTM1L) with its 6 predicted transmembrane (TM) domains, CLPTM1L-ΔCterm (lacking the C-terminal (C) end of the protein or aa 455–538) and CLPTM1L-ΔLoop (missing aa 36–280 that form the transmembrane loop (L) between TM1 and TM2). Bottom panel shows protein expression of WT and mutant (ΔC and ΔL) N-FLAG-tagged CLPTM1L in transiently transfected HEK293T cells (left) and stably transfected PANC-1 cells (right) by western blot and immunoprecipitation analysis. Empty vector is V0.
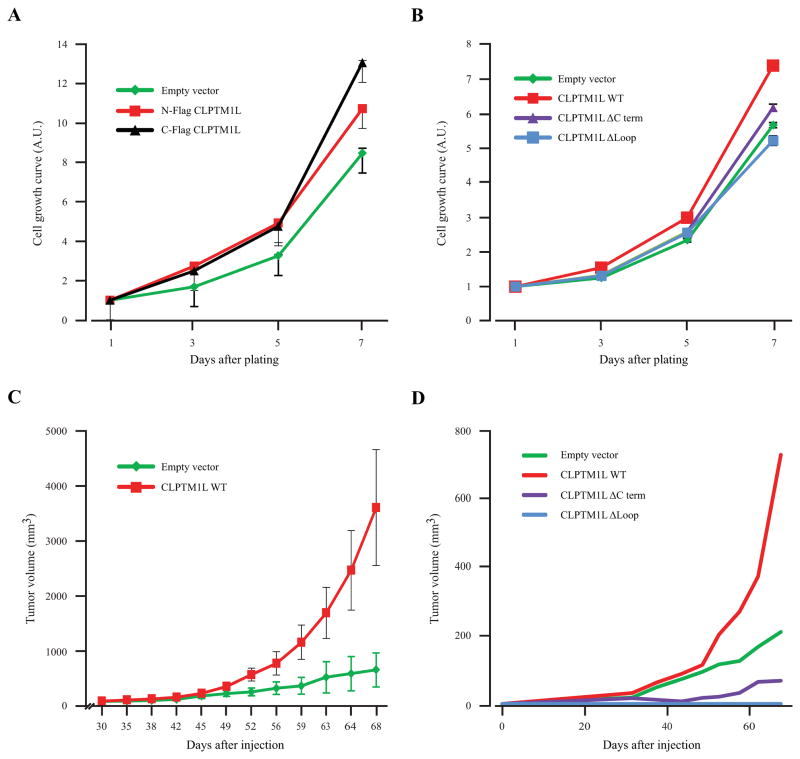
We assessed cell proliferation in vitro and observed that cells overexpressing full-length WT CLPTM1L grew faster than cells containing empty vector (Figure 3a). Two sets of PANC-1 cells stably transfected with CLPTM1L (with N- or C-terminal FLAG epitope tags) showed similar results (1.3–1.5 fold increase at day 7, P=0.0019 and P=0.0027, respectively) indicating that the FLAG tag does not influence the growth promoting function of the protein. This effect was abolished by the two mutants, CLPTM1L-ΔLoop and CLPTM1L-ΔCterm (Figure 3b) and inhibited by siRNA targeting CLPTM1L (Supplemental Figure 2). These results prompted an investigation of whether the same growth effect was mediated by CLPTM1L in vivo. PANC-1 cells stably overexpressing WT CLPTM1L generated larger tumors in nude mice in vivo than those containing empty vector (average tumor volume is shown for one representative experiment in Figure 3c; median tumor volume is shown for all three experiments combined in Figure 3d). A significantly larger tumor size was observed in vivo for cells overexpressing CLPTM1L as compared to the empty vector (3.46 fold at day 68, P=0.039). The two deletion mutants did not induce growth as compared to the empty vector indicating that these two domains are critical for the growth promoting function of the CLPTM1L protein. In fact, PANC-1 cells expressing CLPTM1LΔLoop and CLPTM1LΔCterm grew slower than controls indicating a possible dominant negative effect (Figure 3d). This effect was significant for the CLPTM1L construct lacking the Loop structure (P=0.012) but not for the construct lacking the C terminus (P=0.69). To rule out the possibility that the lack of growth promotion by the mutant CLPTM1L proteins is caused by abrogated subcellular localization, we evaluated the localization of the two mutants in PANC-1 cells. Both mutants localized to the ER (Supplemental Figure 3), indicating that mislocalization does not explain the reduced biological effect. However, a defect in folding of the mutants within the membranes cannot be ruled out.
Figure 3. CLPTM1L overexpression induces growth in vitro and in vivo.
(A) Growth rates in vitro measured over 7 days in PANC-1 cells stably expressing N-terminal FLAG-tagged WT CLPTM1L (red) or C-terminal FLAG-tagged WT CLPTM1L (black) as compared to cells containing empty vector (green). (B) Growth rates in vitro for PANC-1 cells expressing WT N-FLAG CLPTM1L (red), N-FLAG CLPTM1L-ΔCterm (purple) or N-FLAG CLPTM1L-ΔLoop (blue) as compared to cells containing empty vector (green). (C) Effects of overexpressing WT N-FLAG CLPTM1L on in vivo growth in nude mice (subcutaneous growth). Results for one representative experiment using PANC-1 cells stably expressing WT CLPTM1L or empty vector are shown in in this panel. Each point shows the average volume and SE of tumors in 5 mice. (D) A compilation of in vivo tumor growth for three separate experiments showing median tumor volumes of PANC-1 cells expressing WT N-FLAG CLPTM1L (red), N-FLAG CLPTM1L-ΔCterm (purple) and N-FLAG CLPTM1L-ΔLoop (blue) as compared to cells containing the empty vector (green). The lines represent median tumor volume over 68 days after injection (5 mice per construct per experiment).
WT CLPTM1L interacts with non-muscle myosin II (NMM-II) in pancreatic cancer cells
To investigate further the molecular function of the CLPTM1L protein, we searched for interacting proteins using affinity purification, followed by liquid chromatography and mass spectrometry (LC MS/MS) analysis. Since CLPTM1L is predicted to be highly hydrophobic, we performed the screen in RIPA buffer to facilitate solubilization of membrane-associated proteins. The most frequently observed peptides (Supplemental Table 1) belonged to Myosin-9 (MYH9) and Myosin-10 (MYH10), two heavy chain non-muscle myosin type II (NMM-II) proteins involved in maintaining cell shape, migration, secretion and cytokinesis. PANC-1 cells express ~25 fold higher amounts of MYH9 compared to MYH10 at the RNA level (Supplemental Figure 4) suggesting that this interaction may be primarily between CLPTM1L and MYH9. Interestingly, the ER marker calnexin was identified as a potential CLPTM1L interacting protein (Supplemental Table 1), consistent with the observed ER localization shown above using immunofluorescence analysis.
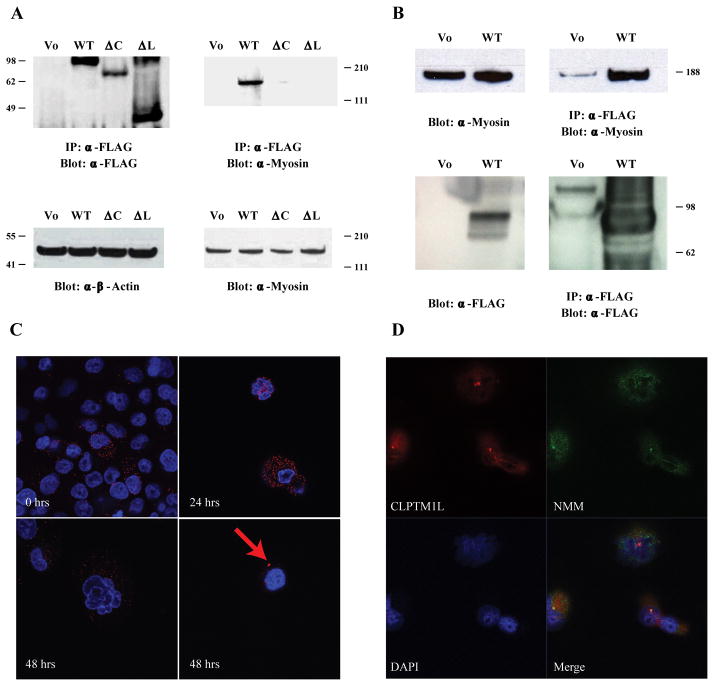
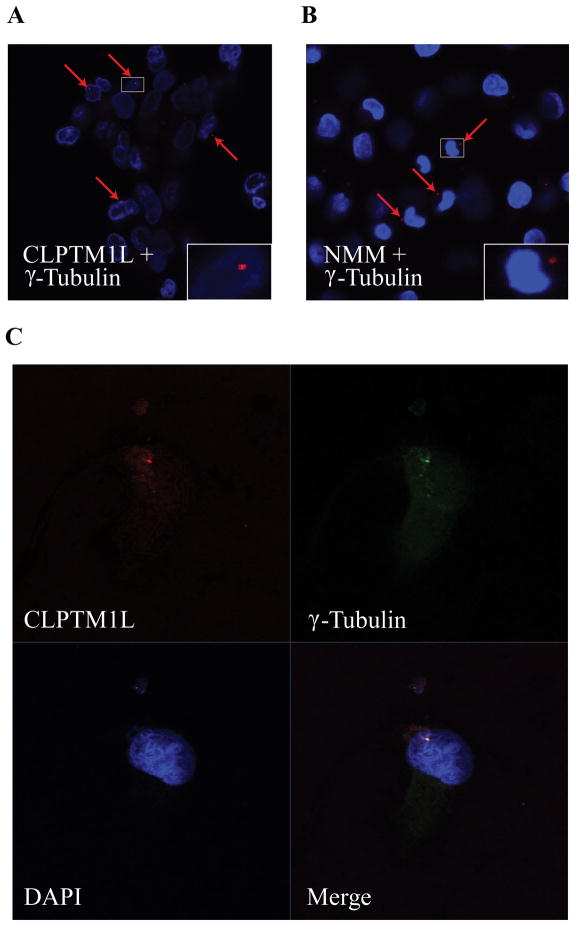
The interaction between CLPTM1L and non-muscle myosin II (NMM-II) was confirmed by co-immunoprecipitation in both PANC-1 (Figure 4a) and 293T cells (Figure 4b). An interaction between the two CLPTM1L deletion mutants and non-muscle myosin II was absent or very low (Figure 4a), indicating that the loop and C-terminal domains are required for the interaction between the two proteins. To assess where in the cells CLPTM1L and non-muscle myosin II interact, we performed co-localization experiments by in situ Proximity Ligation Assay (DPLA) (Duolink) (Figure 4c) and immunofluorescence (IF) (Figure 4d) and confirmed the interaction. Since CLPTM1L has been shown to protect cells from cisplatin induced apoptosis (16, 32) we treated PANC-1 stably transfected cells with 50 μM cisplatin for 0, 24 or 48 hours and observed a shift in the extent of co-localization after 48 hours of cisplatin treatment (Figure 4c). Instead of widespread cytoplasmic co-localization for CLPTM1L and NMM-II, fewer areas of co-localization were seen after cisplatin treatment, especially after 48 hrs (lower panels in Figure 4c). These were often as few as one or two prominent dots per cell, located close to the nucleus (lower right panel in Figure 4c). This pattern was confirmed using IF, showing prominent co-localization of CLPTM1L and non-muscle myosin II after cisplatin treatment in dot-like structures with tubular projections located at the nuclear periphery (Figure 4d, right panels). We hypothesized that these structures were centrosomes, and confirmed that both CLPTM1L and non-muscle myosin II co-localized with the centrosome marker γ-tubulin, as shown in Figure 5a for the former and in Figure 5b for the latter, indicating that the two proteins probably interact specifically at centrosomes after DNA damage induction by cisplatin. Co-localization of CLPTM1L and γ-tubulin was confirmed using IF (Figure 5c) indicating co-localization in dot-like structures close to the nucleus consistent with a single centrosome per cell.
Figure 4. CLPTM1L and non-muscle Myosin II interact.
(A) Co-immunoprecipitation and western blot analysis showing robust interaction between WT N-FLAG CLPTM1L and non-muscle myosin II (Myosin) in stably transfected PANC-1 cells (upper right panel). Little or no interaction was seen with the two deletion mutants of CLPTM1L (upper left panel). β-Actin protein levels in cell lysates are shown as a loading control (lower left panel) and non-muscle myosin II (Myosin) protein levels are shown by western blot (lower right panel). (B) Co-immunoprecipitation and western blot analysis showing the interaction between WT N-FLAG CLPTM1L and non-muscle myosin II (Myosin) in transiently transfected HEK293T cells. Non-muscle myosin levels are shown in the upper left panel, and WT CLPTM1L levels are shown in the lower left panel. Robust pull-down of non-muscle myosin is seen with CLPTM1L in the cell line stably expressing WT CLPTM1L (WT, upper right panel) whereas little is seen in the cell line containing the empty vector (Vo). (C) The interaction between WT N-FLAG CLPTM1L and non-muscle myosin II (NMM-II) in stably transfected PANC-1 was confirmed by in situ Proximity Ligation Assay with or without treatment with 50 μM cisplatin for 24 or 48 hrs. The interaction is seen as red dots where the two proteins are in close proximity. Two representative figures are shown for the 48 hours’ timepoint. A red arrow points to a prominent area of co-localization located close to the nucleus. (D) Immunofluorescence analysis showing co-localization of WT N-FLAG CLPTM1L and non-muscle myosin II (NMM-II) as orange to yellow staining after treatment with cisplatin for 48 hrs.
Figure 5. CLPTM1L and non-muscle Myosin II localize to centrosomes.
Co-localization of CLPTM1L (A) and non-muscle myosin II (B) with the centrosome marker γ-tubulin by in situ Proximity Ligation Assay (DPLA). (C) Co-localization of WT CLPTM1L and γ-tubulin by immunofluorescence analysis. PANC-1 cells stably expressing N-FLAG CLPTM1L were treated for 2 days with 50 μM cisplatin in all panels. The yellow signal in the lower right panel shows co-localization of CLPTM1L and γ-tubulin.
Overexpression of WT CLPTM1L results in increased aneuploidy
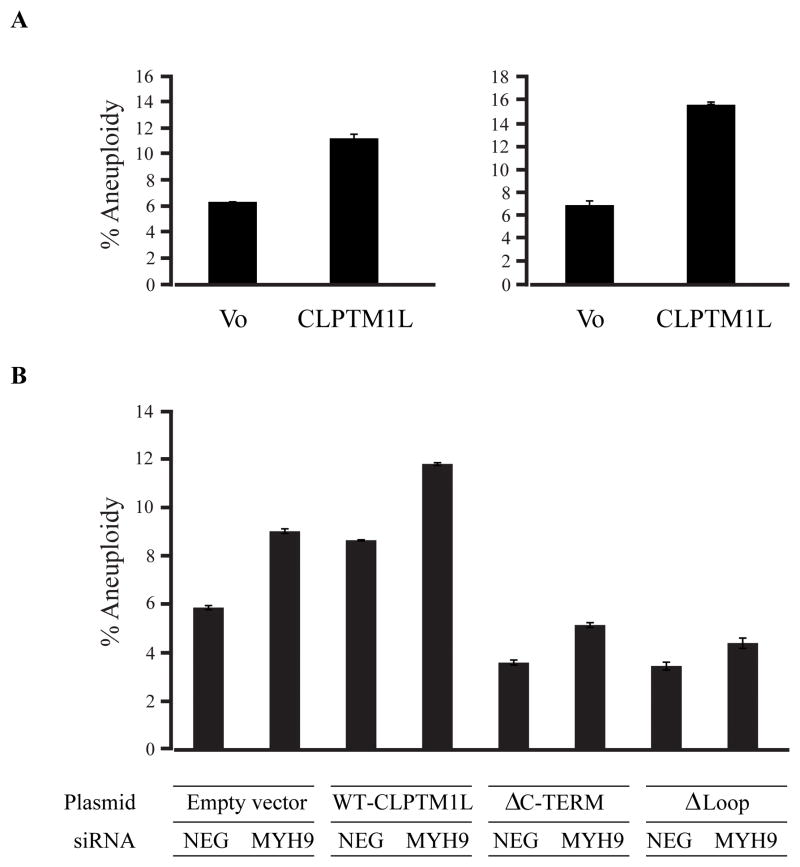
Based on the interaction with non-muscle myosin II and co-localization with γ-tubulin at the centrosome, we assessed DNA content and cell cycle progression by FACS analysis after transiently or stably overexpressing WT CLPTM1L in HEK293T cells and PANC-1 cells, respectively. This analysis did not reveal a significant difference in the fraction of cells in G0/G1, S or G2/M phase (data not shown) but indicated that the percentage of aneuploid cells with DNA content double than that of the majority of cells, was enhanced in cells overexpressing CLPTM1L; a 1.8 fold increase was seen in PANC-1 cells (left panel) and a 2.3 fold increase was seen in HEK293T cells (right panel) as compared to empty vector (Figure 6a). It has previously been shown that inhibiting the function of non-muscle myosin II with a specific chemical inhibitor (blebbistatin), or with specific siRNA, results in abnormal cytokinesis and increased rates of aneuploidy (33). To address this, we assessed aneuploidy in PANC-1 cells stably expressing WT and mutant CLPTM1L. After knock-down of MYH9 expression with a specific siRNA (knock-down efficiency 77.8–86.0%, Supplemental Figure 5), we noted an increase in the fraction of aneuploid cells in PANC-1 cells containing empty vector (1.54 fold) and in cells that overexpressed WT CLPTM1L (1.36 fold) as compared to negative siRNA (Figure 6b, bars 1–4). Simultaneous overexpression of WT CLPTM1L and knock-down of MYH9 gave the highest fraction of aneuploid cells (2.01 fold), suggesting an additive effect. On the other hand, PANC-1 cells expressing the two deletion mutants showed lower levels of aneuploid cells than cells containing empty vector or WT-CLPTM1L. Knock-down of MYH9 in these cells increased the fraction of aneuploid cells, but not to the levels seen in cells containing empty vector or expressing WT-CLPTM1L (Figure 6b, bars 5–8).
Figure 6. CLPTM1L overexpression increases the fraction of aneuploid cells.
(A) PANC-1 cells stably expressing WT N-FLAG CLPTM1L (right) and HEK293T cells transiently expressing N-FLAG CLPTM1L (left) and were labeled with BrDU and analyzed for cell cycle content. The percentage of aneuploid cells (DNA content >G2M peak) is shown in the graphs. (B) DNA content was also measured in PANC-1 cells stably expressing WT N-FLAG CLPTM1L or mutant N-FLAG CLPTM1L (CLPTM1L-ΔCterm or CLPTM1L-ΔLoop) with transient siRNA knock-down of non-muscle myosin II MYH9 or a non-target (NEG) siRNA. The percentage of aneuploid cells was compared between cells expressing WT and mutant CLPTM1L and cells containing empty vector. The average and SE of n=3 replicates is shown in the graphs.
CLPTM1L protein levels were increased in human pancreatic adenocarcinoma
To assess CLPTM1L protein expression in normal- and tumor-derived pancreatic samples, we performed immunohistochemical staining of formalin fixed paraffin embedded tumor- and normal-derived pancreatic samples. Initially, we stained tissue microarrays (TMAs) obtained from the Mayo Clinic, containing normal (n=3) and pancreatic adenocarcinoma (PDAC TMA1, n=111 and PDAC GEM; n=156) pancreatic samples. To increase the sample size for normal pancreatic samples, we stained a TMA from the Surveillance, Epidemiology, and End Results (SEER) registries with normal (n=14) and tumor (SEER PDAC, n=111) tissue samples. All normal samples scored as negative or weak (combined “intensity*extent” or histoscores from 0–2) whereas PDAC-derived samples scored in all four categories (histoscores from 0–16). In fact, the normal-derived samples on both TMAs never scored above a histoscore of 2. The distribution of average histoscores across all PDAC samples combined (PDAC TMA1, PDAC GEM and PDAC SEER) were: 24.9% negative, 28.8% weak, 22.2% intermediate and 24.1% strong (see scores and percentages for individual TMAs in Table 1 and representative staining in Supplemental Figure 6). When score categories were collapsed into two groups with lower (negative and weak; histoscore 0–4) vs. higher (intermediate and strong; histoscore 6–16) scores, a statistically significant difference was seen for CLPTM1L staining between the normal- and tumor-derived samples (P=1.7×10−4) indicating elevated expression levels of CLPTM1L in pancreatic tumors of PDAC histology. Significant correlations between CLPTM1L protein levels (histoscore) with survival, gender, age, stage and grade were not seen (data not shown). PanINs (n=13) stained mostly in the negative (46.2%) and weak (46.2%) categories but with a small fraction (7.7%) of samples falling within the intermediate group. The average percentages of staining scores for islet cell tumors (n=48) on both TMA sets was 6.3% negative, 2.1% weak, 8.3% intermediate and 83.3% strong. Negative or weak staining was seen in islets in the normal samples indicating that CLPTM1L levels may also be increased in pancreatic endocrine tumors.
Table 1.
CLPTM1L protein levels are enhanced in pancreatic cancer
|
|
|
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Score
|
Mayo Clinic TMAs
|
SEER TMA
|
|||||||||||||||
| Group | Value | Normal TMA1 | PanIN TMA1 | PDAC TMA1 | PDAC GEM | Islet Cell TMA | Normal | PDAC | Islet Cell | ||||||||
|
|
|
|
|
|
|
|
|
|
|||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | ||
|
|
|
|
|
|
|
|
|
||||||||||
| Negative | 0–1 | 2 | 66.7 | 6 | 46.2 | 25 | 22.5 | 10 | 6.4 | 0 | 0.0 | 8 | 57.1 | 59 | 53.2 | 3 | 42.9 |
| Weak | 2–4 | 1 | 33.3 | 6 | 46.2 | 42 | 37.8 | 20 | 12.8 | 0 | 0.0 | 6 | 42.9 | 47 | 42.3 | 1 | 14.3 |
| Intermediate | 5–8 | 0 | 0 | 1 | 7.7 | 31 | 27.9 | 49 | 31.4 | 2 | 4.9 | 0 | 0 | 4 | 3.6 | 2 | 28.6 |
| Strong | 9–16 | 0 | 0 | 0 | 0.0 | 13 | 11.7 | 77 | 49.4 | 39 | 95.1 | 0 | 0 | 1 | 0.9 | 1 | 14.3 |
|
|
|
|
|
|
|
|
|
||||||||||
| Total | 3 | 100 | 13 | 100 | 111 | 100 | 156 | 100 | 41 | 100 | 14 | 100 | 111 | 100 | 7 | 100 | |
|
|
|
||||||||||||||||
Combined staining intensity and extent for CLPTM1L by immunohistochemistry in normal-derived pancreas, PanIN, PDAC and pancreatic islet cell tumors. CLPTM1L immunohistochemical staining extent (area) and intensity was scored from 0–4 and the combined extent*intensity calculated and shown above (histoscore). Acronyms are as follows for the Mayo Clinic and SEER TMAs used: PanIN: pancreatic intraepithelial neoplasia; PDAC: pancreatic ductal adenocarcinoma, Islet cell: Islet cell tumors, GEM: gemcitabine.
Discussion
Chromosome 5p15.33 contains a pleiotropic cancer predisposition locus identified in GWAS of at least ten cancer types (34). Multiple independent loci, in the TERT gene or the neighboring CLPTM1L gene, have been identified in this region (1–13, 35, 36). Some, but not all, risk loci have been associated with TERT expression or with telomere length (35, 36). To further define the underlying biology of the susceptibility alleles on 5p15.33, we investigated the function of a plausible candidate gene, CLPTM1L, and its encoded protein, cleft lip and palate transmembrane protein 1-like protein (CLPTM1L). We observed a positive effect on tumor cell growth both in vitro and in vivo, after overexpression of full-length CLPTM1L. Deletion mutants of CLPTM1L lacking either one of the two large hydrophilic domains of this predominantly hydrophobic and presumably membrane bound protein did not induce growth in vitro or in vivo, indicating that these domains may be crucial for the growth promoting function of the protein.
A search for interacting proteins identified non-muscle myosin heavy chain, types IIA and IIB (NMM-II, also named myosin heavy chain 9/10, MYH9 or MYH10) as protein partners. The two hydrophilic domains of CLPTM1L were required for this interaction. MYH9 and MYH10 are heavy chain subunits of Myosin, a hexameric protein containing 2 heavy chains, 2 light chains and 2 regulatory chain subunits, and are important for maintaining cell shape, migration and secretion (37–39). In addition, NMM-II plays a role in cytokinesis where it, in concert with actin and furrow components, drives the contraction needed for cytokinesis, the final step in cell division (33). A well-known effect of blocking the expression or function of NMM-II in model organisms is a defect in cytokinesis that leads to the formation of multinucleate cells (40–42). In HeLa cells, treatment with blebbistatin, a chemical inhibitor of NMM-II, inhibits contraction of the cell cleavage furrow resulting in an increased number of binucleate cells (33). NMM-II is also important for the migration of centrosomes to the opposite sides of chromosomes during assembly of the mitotic spindle, indicating an important role during multiple steps of cell division (43).
In our study, under normal growth conditions, CLPTM1L and NMM-II co-localized to the ER. After treatment with a DNA damaging agent, both CLPTM1L and NMM-II localized to centrosomes, where they appear to interact. As seen in other cell types (33), knocking down the levels of NMM-II resulted in an increase in aneuploidy. The opposite was true for CLPTM1L, as increased levels resulted in enhanced rates of aneuploidy. This suggests that the two proteins could have antagonizing effects on cells proceeding through cell division, which may involve a disruption of normal NMM-II function by CLPTM1L.
The interaction between MYH9 and CLPTM1L appears to be mediated through the loop domain and C-terminal tail of CLPTM1L. Since the ER membrane is continuous with the nuclear membrane, which is absorbed into the ER during mitosis (44), CLPTM1L may reside in the outer nuclear membrane (as per punctate staining pattern in Figure 1a) and interact with centrosome proteins during cell division. It is also possible that the interaction between the two proteins in the ER and other subcellular organelles has additional functional consequences that may influence cell growth, apoptosis or other cellular activities. CLPTM1L has recently been shown to be overexpressed in lung cancer where it protects cells from apoptosis after treatment with DNA damaging agents via Bcl-xL, indicating a negative role on mitochondrial apoptotic pathways (16, 32).
In a recent proteomics screen of the human autophagy network, CLPTM1L was shown to bind PIK3C3 (Phosphatidylinositol 3-kinase catalytic subunit type 3), indicating a possible role in autophagy (45). Although PIK3C3 plays a central role in autophagy, it is also important for vesicle trafficking, receptor signaling and cytokinesis. Depletion of PIK3C3 in mammalian cells leads to an arrest of cytokinesis and an increased number of multinucleate cells (46). Interestingly, in the autophagy screen, both CLPTM1L and MYH9 were also listed as interacting proteins for CLN3 (45), a transmembrane vesicle trafficking protein that regulates cytokinesis in yeast (47) and is mutated in Batten disease (48).
Immunohistochemical analysis showed that a substantial number of pancreatic cancers have higher levels of the CLPTM1L protein than normal pancreatic tissues, approximately 46% (range 5%–81%) of pancreatic adenocarcinomas and 92% (range 43%–100%) of islet cell tumors, indicating a possible role in carcinogenesis in both the exocrine and endocrine pancreas. A possible limitation of our study is that we have a substantially larger number of pancreatic cancers (PDAC, n=378) compared to normal samples (n=17). Thus, our findings would benefit from additional validation studies.
In summary, our results show that CLPTM1L promotes growth of pancreatic cancer cells in vitro and in vivo and that its function, at least in part, may be through abrogation of cell division. We have shown that CLPTM1L interacts with NMM-II, and that the two proteins have opposing effects on aneuploidy. Since NMM-II is required for cytokinesis, it is possible that CLPTM1L blocks this process, perhaps by interfering with processing, localization or function of NMM-II. Furthermore we show that CLPTM1L is overexpressed in a substantial fraction of pancreatic tumors. Although further studies are needed to characterize CLPTM1L function in greater detail, our results indicate that CLPTM1L influences processes that are important for cancer growth and thus represents a plausible candidate gene for investigating the biological basis of cancer susceptibility alleles on chr5p15.33, particularly for pancreatic cancer.
Supplementary Material
Acknowledgments
We thank Dr. Javed Khan, Pediatric Oncology Branch, National Cancer Institute, National Institutes of Health, for guidance with mouse experiments.
Footnotes
Conflicts of Interest statement: the authors declare that there are no potential conflicts of interest
References
- 1.Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42:978–84. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet. 2011;43:1210–4. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–9. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–6. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–41. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–7. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stacey SN, Sulem P, Masson G, Gudjonsson SA, Thorleifsson G, Jakobsdottir M, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41:909–14. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, Yang B, Li B, Liu Y. Association between TERT-CLPTM1L rs401681[C] allele and NMSC cancer risk: a meta-analysis including 45,184 subjects. Archives of dermatological research. 2012 doi: 10.1007/s00403-012-1275-8. [DOI] [PubMed] [Google Scholar]
- 11.Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–8. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43:785–91. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–7. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–5. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto K, Okamoto A, Isonishi S, Ochiai K, Ohtake Y. A novel gene, CRR9, which was up-regulated in CDDP-resistant ovarian tumor cell line, was associated with apoptosis. Biochem Biophys Res Commun. 2001;280:1148–54. doi: 10.1006/bbrc.2001.4250. [DOI] [PubMed] [Google Scholar]
- 16.James MA, Wen W, Wang Y, Byers LA, Heymach JV, Coombes KR, et al. Functional characterization of CLPTM1L as a lung cancer risk candidate gene in the 5p15.33 locus. PloS one. 2012;7:e36116. doi: 10.1371/journal.pone.0036116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baudis M. Genomic imbalances in 5918 malignant epithelial tumors: an explorative meta-analysis of chromosomal CGH data. BMC cancer. 2007;7:226. doi: 10.1186/1471-2407-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoglund M, Gisselsson D, Hansen GB, Mitelman F. Statistical dissection of cytogenetic patterns in lung cancer reveals multiple modes of karyotypic evolution independent of histological classification. Cancer genetics and cytogenetics. 2004;154:99–109. doi: 10.1016/j.cancergencyto.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 19.Baudis M, Cleary ML. Progenetix.net: an online repository for molecular cytogenetic aberration data. Bioinformatics. 2001;17:1228–9. doi: 10.1093/bioinformatics/17.12.1228. [DOI] [PubMed] [Google Scholar]
- 20.Kang JU, Koo SH, Kwon KC, Park JW, Kim JM. Gain at chromosomal region 5p15.33, containing TERT, is the most frequent genetic event in early stages of non-small cell lung cancer. Cancer genetics and cytogenetics. 2008;182:1–11. doi: 10.1016/j.cancergencyto.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Vazquez-Mena O, Medina-Martinez I, Juarez-Torres E, Barron V, Espinosa A, Villegas-Sepulveda N, et al. Amplified genes may be overexpressed, unchanged, or downregulated in cervical cancer cell lines. PloS one. 2012;7:e32667. doi: 10.1371/journal.pone.0032667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lando M, Holden M, Bergersen LC, Svendsrud DH, Stokke T, Sundfor K, et al. Gene dosage, expression, and ontology analysis identifies driver genes in the carcinogenesis and chemoradioresistance of cervical cancer. PLoS Genet. 2009;5:e1000719. doi: 10.1371/journal.pgen.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Punta M, Forrest LR, Bigelow H, Kernytsky A, Liu J, Rost B. Membrane protein prediction methods. Methods. 2007;41:460–74. doi: 10.1016/j.ymeth.2006.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.v.2.0 T. TMHMM v.2.0. http://wwwcbsdtudk/services/TMHMM/
- 25.TMPred TMPred. http://wwwchembnetorg/software/TMPRED_formhtml.
- 26.TopPred2. TopPred2. http://wwwsbcsuse/~erikw/toppred2/
- 27.Das S, Bosley AD, Ye X, Chan KC, Chu I, Green JE, et al. Comparison of strong cation exchange and SDS-PAGE fractionation for analysis of multiprotein complexes. Journal of proteome research. 2010;9:6696–704. doi: 10.1021/pr100843x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–7. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 29.Takikita M, Altekruse S, Lynch CF, Goodman MT, Hernandez BY, Green M, et al. Associations between selected biomarkers and prognosis in a population-based pancreatic cancer tissue microarray. Cancer Res. 2009;69:2950–5. doi: 10.1158/0008-5472.CAN-08-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herreros-Villanueva M, Zhang JS, Koenig A, Abel EV, Smyrk TC, Bamlet WR, et al. SOX2 promotes dedifferentiation and imparts stem cell-like features to pancreatic cancer cells. Oncogenesis. 2013;2:e61. doi: 10.1038/oncsis.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.HomoloGene N. http://www.ncbi.nlm.nih.gov/homologene.
- 32.Ni Z, Tao K, Chen G, Chen Q, Tang J, Luo X, et al. CLPTM1L Is Overexpressed in Lung Cancer and Associated with Apoptosis. PloS one. 2012;7:e52598. doi: 10.1371/journal.pone.0052598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–7. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 34.Mocellin S, Verdi D, Pooley KA, Landi MT, Egan KM, Baird DM, et al. Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J Natl Cancer Inst. 2012;104:840–54. doi: 10.1093/jnci/djs222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kote-Jarai Z, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Dadaev T, Jugurnauth-Little S, et al. Fine-mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with TERT expression. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–84. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nature reviews Molecular cell biology. 2009;10:778–90. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bond LM, Brandstaetter H, Sellers JR, Kendrick-Jones J, Buss F. Myosin motor proteins are involved in the final stages of the secretory pathways. Biochemical Society transactions. 2011;39:1115–9. doi: 10.1042/BST0391115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang A, Ma X, Conti MA, Adelstein RS. Distinct and redundant roles of the non-muscle myosin II isoforms and functional domains. Biochemical Society transactions. 2011;39:1131–5. doi: 10.1042/BST0391131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mabuchi I, Okuno M. The effect of myosin antibody on the division of starfish blastomeres. The Journal of cell biology. 1977;74:251–63. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–91. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 42.Urven LE, Yabe T, Pelegri F. A role for non-muscle myosin II function in furrow maturation in the early zebrafish embryo. Journal of cell science. 2006;119:4342–52. doi: 10.1242/jcs.03197. [DOI] [PubMed] [Google Scholar]
- 43.Rosenblatt J, Cramer LP, Baum B, McGee KM. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell. 2004;117:361–72. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- 44.English AR, Voeltz GK. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harbor perspectives in biology. 2013:5. doi: 10.1101/cshperspect.a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sagona AP, Nezis IP, Pedersen NM, Liestol K, Poulton J, Rusten TE, et al. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nature cell biology. 2010;12:362–71. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- 47.Codlin S, Haines RL, Burden JJ, Mole SE. Btn1 affects cytokinesis and cell-wall deposition by independent mechanisms, one of which is linked to dysregulation of vacuole pH. Journal of cell science. 2008;121:2860–70. doi: 10.1242/jcs.030122. [DOI] [PubMed] [Google Scholar]
- 48.Consortium TIBD. Isolation of a novel gene underlying Batten disease, CLN3. Cell. 1995;82:949–57. doi: 10.1016/0092-8674(95)90274-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.