Abstract
Studies of human germ cell development are limited in large part by inaccessibility of germ cells during development. Moreover, although several studies have reported differentiation of mouse and human germ cells from pluripotent stem cells (PSCs) in vitro, differentiation of human germ cells from PSCs in vivo has not been reported. Here, we tested whether mRNA reprogramming in combination with xeno-transplantation may provide a viable system to probe the genetics of human germ cell development via use of induced pluripotent stem cells (iPSCs). For this purpose, we derived integration-free iPSCs via mRNA-based reprogramming with OCT3/4, SOX2, KLF4 and cMYC alone (OSKM) or in combination with the germ cell-specific mRNA, VASA (OSKMV). All iPSC lines met classic criteria of pluripotency. Moreover, global gene expression profiling did not distinguish large differences between undifferentiated OSKM and OSKMV iPSCs; however, some differences were observed in expression of pluripotency factors and germ cell-specific genes, and in epigenetic profiles and in vitro differentiation studies. In contrast, transplantation of undifferentiated iPSCs directly into the seminiferous tubules of germ cell-depleted immunodeficient mice revealed divergent fates of iPSCs produced with different factors. Transplantation resulted in morphologically and immunohistochemically recognizable germ cells in vivo, particularly in the case of OSKMV cells. Significantly, OSKMV cells also did not form tumors while OSKM cells that remained outside the seminiferous tubule proliferated extensively and formed tumors. Results indicate that mRNA reprogramming in combination with transplantation may contribute to tools for genetic analysis of human germ cell development.
INTRODUCTION
A significant challenge in elucidating genetic requirements for human germ cell formation, maintenance and differentiation is to recapitulate germ cell specification and differentiation both in vitro and in vivo. Studies in the mouse suggest that complete reconstitution of mammalian germline development from pluripotent stem cells (PSCs) is possible (1–4). In spite of successes in the mouse, in vitro differentiation of human PSCs to germ cells that progress through meiosis and form functional gametes remains a significant challenge. Notably, previous efforts, including our own, have used a variety of in vitro methodologies that advanced studies of human germ cell differentiation but consistently yielded low numbers of germ cells, inconsistency across line derivations and genotypes and incomplete imprint erasure and re-establishment in a sex-specific manner. Here, we sought to differentiate human germ cells by directly transplanting undifferentiated human induced pluripotent stem cells (iPSCs) into murine seminiferous tubules in order to make use of the germ cell niche to promote human germline formation in vivo.
Insights gained from mouse studies, combined with knowledge of genetic control of PGC fate in diverse organisms and our previous studies of human germ cell differentiation in vitro, prompted us to consider whether developmental ‘priming’ by induced expression of germ cell-specific genes in combination with xenotransplantation might overcome current roadblocks to efficient human germ cell development from PSCs. In murine studies, a critical advance was made by the induction of a transient epiblast-like cell state from mouse embryonic stem cells (mESCs) and iPSCs by addition of cytokines or via overexpression of PRDM14, DPPA3 and TFAP2C transcription factors alone (2–4). Resulting cells were developmentally competent to form sperm and oocytes following transplantation to produce live, healthy offspring after fertilization. In our own efforts, we have used overexpression of key germ cell regulators including DAZ gene family members or DAZL and VASA together to drive germ cell differentiation and meiotic progression from human ESCs and iPSCs in vitro (5–7). However, our studies and those of others using in vitro mediated differentiation have been confounded by low yields of germ cells, inefficient meiotic progression and an incomplete imprinting status (8,9). Owing to inherent differences between human and mouse PSC and, based on previous studies, we predicted that induced expression of translational regulators such as VASA, DAZ, DAZL and BOULE might promote human germ cell formation. Thus, we included VASA, a translational regulator, to the mix of factors used in mRNA reprogramming to iPSCs in hopes of alleviating hurdles that we and others have encountered with human germ cell derivation in vitro (5–9). The VASA gene encodes a highly conserved germ cell-specific RNA-binding protein whose role in germ cell development may include acting as a chaperone to enable correct folding of different target RNAs in germ cells (10). Furthermore, we note that similarities between pluripotent human ESCs and iPSCs to mouse epiblast cells lends support to our rationale that we might produce primed iPSCs for germ cell development (11–14). We then transplanted the undifferentiated iPSCs directly into the seminiferous tubules of germ cell-depleted immunodeficient mice, in order to evaluate the contribution of VASA-primed and non-primed cells to germline development in vivo.
RESULTS
Addition of VASA to OSKM-reprogramming mix does not affect reprogramming efficiencies or kinetics
Synthesized modified mRNAs, including VASA mRNA, were produced and validated prior to use as shown (Supplementary Material, Fig. S1A and B); protein expression and localization was confirmed by immunocytochemistry (Fig. 1). We added synthesized VASA mRNA to OSKM factors in molar ratios of 3:0.5:1:0.5:1 (OSKMV) and examined reprogramming efficiency and kinetics of different somatic lines [BJ (XY), HUF1 (XY), HUF3 (XX) and HUF9 (XX)] with either the OSKM or OSKMV cocktail in parallel. We adapted a previously reported protocol (15) in which feeder-free iPSC derivation was accomplished with six mRNA factors (OSKM + NANOG + LIN28A) and modified OCT3/4. We successfully derived feeder- and xeno-free iPSCs with OSKM alone and with OSKMV (Supplementary Material, Fig. S2A–F). We observed colony formation with both OSKM and OSKMV reprogramming with no significant differences in colony number (∼0.5% efficiency) or timing of colony appearance (6–12 days post first-transfection). We then further analyzed the XY lines, HUF1 and BJ, and compared OSKM- and OSKMV-derived lines across a series of functional and molecular assays beginning with a comparison of morphology, alkaline phosphatase (AP) staining and embryoid body (EB) formation. We did not observe notable differences, regardless of reprogramming cocktail used (Supplementary Material, Fig. S3A). Colonies reprogrammed with OSKMV were characterized by a high nucleus/cytoplasm ratio, prominent nucleoli, well defined borders and a distinguishing chromatin structure and nuclear architecture (speckles and heterochromatin domains), all features that are very similar to OSKM-reprogrammed colonies and human embryonic stem cells (16).
Figure 1.
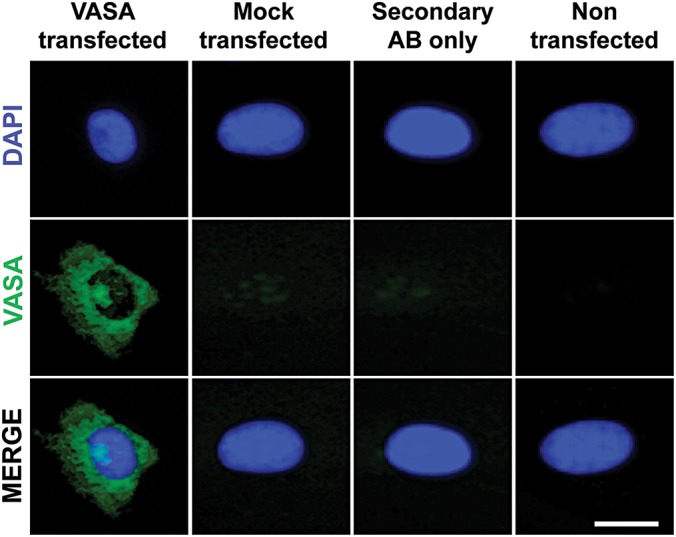
Functional validation of mRNA expression encoding for VASA. Immunostaining of VASA in mRNA-transfected fibroblasts 24 h after transfection. VASA protein localized correctly in the cytoplasm. Mock transfected, secondary antibody stained only and non-transfected samples served as negative controls. Scale bar, 10 µm.
Transient ectopic VASA expression alters gene expression signatures of derived iPSC lines
As part of the assessment of pluripotency, we examined endogenous gene and protein expression of various markers associated with pluripotency including POU5F1, NANOG, SALL4 and DNMT3B in both OSKM and OSKMV colonies (Supplementary Material, Fig. S3C). Notably, expression of a subset of markers associated with pluripotency was lower (P < 0.05) in lines reprogrammed with OSKMV relative to their OSKM counterpart, with PRMT5, SALL4 and DPPA4 being the most significantly different (P < 0.001). We also confirmed a similar reduction in expression of a subset of genes in lines that were derived with OSKM or OSKMV via a lentiviral reprogramming strategy to exclude reprogramming strategy related events (Supplementary Material, Fig. S3C). We then examined effects of transient ectopic expression of VASA during reprogramming on expression of genes associated with early germ cell development. We observed that the majority of markers showed gene expression levels similar to the lines reprogrammed with OSKM alone and/or the parental fibroblast line, indicating no gene activation (exemplified by PRDM1). However, a subset (PRDM14, DPPA3 [STELLA] and VASA) was expressed at significantly higher levels (P < 0.001) in iPSC lines reprogrammed with OSKMV relative to OSKM-derived colonies, indicated for iPSC.HUF1 cells (Fig. 2A). Results were partially mirrored by the lentiviral-derived HUF1 iPSC line and mRNA-derived iPSC.BJ cells. We detected a smaller subset of germ cell markers at a higher expression level in OSKMV-derived lines of which only PRDM14 (for iPSC.BJ) was significantly upregulated compared with the OSKM counterpart (Supplementary Material, Fig. S3D). We note that gene expression was measured at two different passages (passage 4 and 14) to eliminate the possibility of expression from exogenous mRNA and to demonstrate stability of the distinct endogenous gene expression profile. We further confirmed endogenous VASA gene expression in OSKMV cells with immunocytochemistry (Supplementary Material, Fig. S3B).
Figure 2.
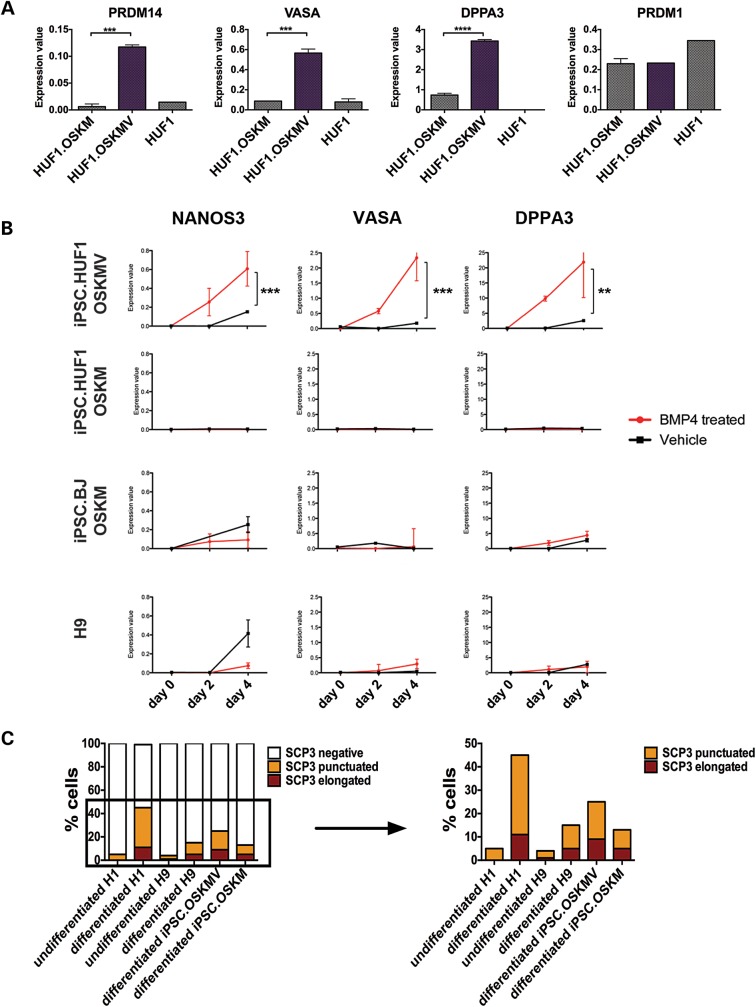
Functional and molecular studies of iPSC.HUF1 derived with OSKM and OSKMV. (A) Gene expression analysis of markers associated with the germline lineage in an undifferentiated state. Three genes (PRDM14, VASA, DPPA3) showed significantly higher expression in OSKMV-derived clones (purple bar) compared with OSKM (Student's t-test, mean ± SD; n ≥ 16 for each gene and sample *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (B) Gene expression analysis of clones derived with OSKM and OSKMV, iPSC.BJ line and H9 during in vitro differentiation into primordial germ cells with BMP4. Samples were isolated at day 0, 2 and 4 post-BMP4/vehicle treatment. Three key markers (NANOS3, VASA, DPPA3) were upregulated in OSKMV-derived clones (Student's t-test, mean ± SD; n ≥ 16 for each gene and sample *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (C) Quantification of meiotic spreads. Approximately 200–300 cells were counted for each line. Staining patterns were classified as negative, punctuated or elongated.
OSKMV-derived lines, similar to OSKM-derived lines, are fully pluripotent
To further assess whether mRNA-reprogrammed OSKM and OSKMV lines are both fully pluripotent, we performed a variety of molecular and functional assays. Spontaneous differentiation revealed that all lines form all three germ layers in vitro (Supplementary Material, Fig. S3E). Epigenetic analysis revealed similar methylation status of OCT3/4 and NANOG promoters of both OSKM- and OSKMV-derived HUF1 lines comparable to hESCs, which were substantially different relative to the parental fibroblast line (Supplementary Material, Fig. S3F). Moreover, we observed that all clones were karyotypically normal and differentiated to all three germ layers in vivo in teratoma assays, following kidney capsule injection into SCID mice (Supplementary Material, Fig. S3G and H). Taken together, these results indicate that mRNA-derived iPSC clones from multiple independent fibroblast lines, with and without addition of VASA are fully reprogrammed by the most stringent assays available to date for human pluripotent cells (17).
OSKMV- and OSKM-derived clones respond differently to BMP4 treatment
VASA overexpression from lentiviral vectors has been reported to drive differentiation into primordial germ cells (PGCs) and more mature germ cells when overexpressed in both human ESCs and iPSCs (6). In mice, PGC specification is characterized by localized BLIMP1 (PRDM1) expression in a small number of cells in the posterior, proximal mouse epiblast at E6.25 followed by upregulation of DPPA3 around E7.25 (18). Several groups have demonstrated in vitro differentiation of PGCs from both mouse ESCs and hESCs via media supplementation with bone morphogenetic proteins (BMPs) (3,5,7,19). Thus, we hypothesized that lines derived with OSKMV might reveal a different potential in their ability to respond to BMP4 relative to OSKM cells. We supplemented culture media of both OSKM- and OSKMV-derived lines with BMP4 over the course of 4 days and assessed activation of gene expression of early germline markers. We included two controls—an mRNA-derived (OSKM) BJ fibroblast line and a hESC line (H9). Although a subset of genes, including NANOS2, RET and Y chromosome DAZ2 among others, did not show significant changes in gene expression in either the OSKM- or OSKMV-derived lines, expression of NANOS3, VASA and DPPA3 was significantly upregulated (P < 0.01) post-BMP4 treatment in the line derived with OSKMV after 4 days of BMP4 treatment (Fig. 2B). iPSCs derived with OSKM as well as controls did not demonstrate differential expression of these germ cell markers in response to BMP4 treatment, indicating that observed differences are confined to different reprogramming factor cocktails used during iPSC derivation.
We also examined gene expression changes of pluripotency-associated markers during BMP4 supplementation (Supplementary Material, Fig. S4A and B). DNMT3B, POUF51 and SALL4, all genes that are strongly expressed in hESCs and iPSCs, showed a rapid decrease in gene expression after 4 days of BMP4 treatment in both OSKM- and OSKMV-derived lines as well as both control lines, as expected (Supplementary Material, Fig. S4B).
OSKMV- and OSKM-derived lines display distinct SCP3 staining patterns
Synaptonemal complex protein 3 (SCP3) encodes a meiosis-specific protein that is essential for formation of meiotic synaptonemal complexes of the maternal and paternal homologous chromosomes (20). Its localization and distribution along the chromosomes indicates meiotic progression in PSC differentiation to germ cells (5,7,8). Ectopic overexpression of VASA in pluripotent human iPSCs has recently been shown to promote meiotic progression as judged by positive SCP3 staining patterns (6). Thus, we compared expression and localization patterns of SCP3 protein in OSKM- and OSKMV-differentiated cells. Our analysis revealed that the majority of cells regardless of VASA mRNA addition during reprogramming stained negative for SCP3 protein indicating no meiotic activity (Fig. 2C and Supplementary Material, Fig. S4C). Nonetheless, clusters of differentiated cells showed a punctuated staining pattern, indicative of early premeiotic events (leptotene stage); moreover, cells reprogrammed with OSKMV displayed slightly higher percentages (P < 0.05) of cells positive for SCP3 staining. In addition, we noticed a distinct staining pattern that was most prominent in H1 hESCs and in iPSCs reprogrammed with OSKMV consisting of elevated SCP3 clusters with elongated structures indicative of late, albeit disorganized, assembly of meiotic chromosomes in zygotene, pachytene or diplotene meiotic prophase I stages (5,7). This staining pattern was observed in ∼6% of cells, a percentage that is significantly higher (P < 0.05) than in the OSKM-reprogrammed cells or the female hESC control (8 and 10%, respectively) (Fig. 2C). Few cells in the undifferentiated negative female control (H9 hESC) displayed SCP3 staining patterns as expected. Interestingly, the second differentiated male control (H1 hESC) line displayed the greatest number of cells with elongated SCP3 staining pattern comparable with differentiated OSKMV cells.
Analysis of epigenetic status and global transcription profiles
We next examined the epigenetic status and global transcriptional profiles of derived lines (Supplementary Material, Fig. S4D and E). We analyzed four imprinted genes via bisulfite sequencing: KCNQ1OT1- and PEG1/MEST-linked differentially methylated regions (DMRs), both maternally imprinted (21,22) as well as the H19 DMR and H19 promoter loci, both paternally imprinted (23). Our analysis revealed slight but significant differences (P < 0.0001) in methylation status of both maternally and paternally imprinted loci, in particular, the maternally imprinted KCQ1OT1 and the paternally imprinted H19 promoter region (Supplementary Material, Fig. S4E). For both loci, the OSKMV line showed a significantly lower percentage of methylated sequences compared with the OSKM counterpart and the parental HUF1 fibroblast line. To further test our findings, we also included an additional control of three independent pooled male sperm samples. We observed little if any methylation at the KCNQ1OT1 and PEG1/MEST loci as expected for maternal-specific germline methylated loci. Also, as expected, the paternally imprinted H19 DMR region demonstrated near-complete methylation. In contrast, to the H19 DMR, which acquires methylation during gametogenesis, the H19 promoter is further methylated following implantation (24) and was characterized by few methylated sequences (18%). Equal proportions of fully methylated and fully unmethylated DNA was detected for PEG1/MEST in both, the OSKM and OSKMV line (P > 0.05, χ2 test). We note that methylation patterns of imprinted genes are diagnostic of germ cells and that cells were analyzed in their undifferentiated state.
To examine global gene expression, we performed RNAseq followed by differential gene expression analysis on the OSKM- and OSKMV-derived HUF1 line together with controls (parental HUF1 and H9). As expected, gene expression profiles between HUF1 fibroblasts and the three pluripotent populations (OSKM, OSKMV, H9 hESCs) were significantly different (P < 0.05) (Supplementary Material, Fig. S5). Interestingly, HUF1 fibroblasts and H9 hESCs expressed more genes at significantly different levels (P < 0.05) than HUF1 fibroblasts compared with their iPSC derivatives (Supplementary Material, Fig. S5C). Pairwise volcano and scatter plots emphasize the gene expression differences between each sample (Supplementary Material, Fig. S5A and B). We further explored the relationship between all four lines and visualized the Jensen–Shannon (JS) distances in a heat map and a significant genes (P < 0.05) overview matrix across all genes (Supplementary Material, Fig. S5C and D). Principal component analysis (PCA) and multidimensional scaling (MDS) revealed a very similar global gene expression in the OSKM- and OSKMV-derived lines that were significantly different (P < 0.05) from the parental fibroblasts (Supplementary Material, Fig. S5E and F). Derived iPSC lines also were distinct from the H9 hESC cell control but revealed a similar global gene expression phenotype (all three pluripotent lines with a positive second PC in PCA analysis). Hierarchical clustering analysis (Supplementary Material, Fig. S5G) and a heat map of differentially expressed genes (Supplementary Material, Fig. S5H) highlighted the relationship between all PSC lines. When we compared both derived iPSC lines with hESCs, we observed only a few genes, isoforms and transcription start sites (TSSs) with statistically significant differential expression, consistent with previous reports (25,26).
Xenotransplantation directs germ cell formation in OSKM- and OSKMV-reprogrammed cells
There are no previous reports of transplantation of human PSCs to the seminiferous environment in vivo. The use of testicular transplantation is common for teratoma analysis; however, as commonly used, teratoma formation assays rely on transplantation under the testis capsule. In contrast, here we made use of a xenotransplant assay in which cells from reprogrammed lines were directly injected into the seminiferous tubules of busulfan-treated immunodeficient nude mice as previously described (27,28). We used this assay to investigate OSKMV and OSKM donor cell differentiation into germ cells in vivo with injection directly into mouse seminiferous tubules. Specifically, we hypothesized that the inclusion of VASA during reprogramming would confer a unique germ cell competent state. As previously reported, transplanted human spermatogonial stem cells migrate to the seminiferous tubule basement membrane and proliferate to form chains and patches of spermatogonia that persist long term (28). Note that complete spermatogenesis is a function of evolutionary distance such that rat transplantation into mouse tubules or human transplantation into non-human primates is expected to yield complete spermatogenesis whereas human transplantation of germ cells into mouse or rat tubules will be limited to formation of prospermatogonia, spermatogonia and/or possibly early meiotic derivatives (28). As a positive control, we transplanted human fetal testicular cells (22-week-old tissue) into busulfan-depleted spermatogenic tubules of immunodeficient mice and observed clusters and chains of spermatogonia 2-month post-transplantation (Fig. 3A, left panel). Transplantation of H1 hESCs (XY karyotype) and H9 hESCs (XX karyotype) served as controls; we hypothesized that H9 hESCs would result in formation of few, if any, germ cells in seminiferous tubules relative to the XY hESCs. Two standard methods to examine the potential to form germ cells in transplants were pursued: Immunohistochemistry of serial sections of fixed tissue and whole-mount staining of testis to assess potential differentiation to chains of spermatogonia, in vivo. Whole mounting is only possible when engraftment occurs in the absence of large masses post-transplantation. For immunohistochemical (IHC) analysis of serial cross sections of transplanted tissues, we used a panel of very well-characterized germ cell markers (2,5,29–32).
Figure 3.
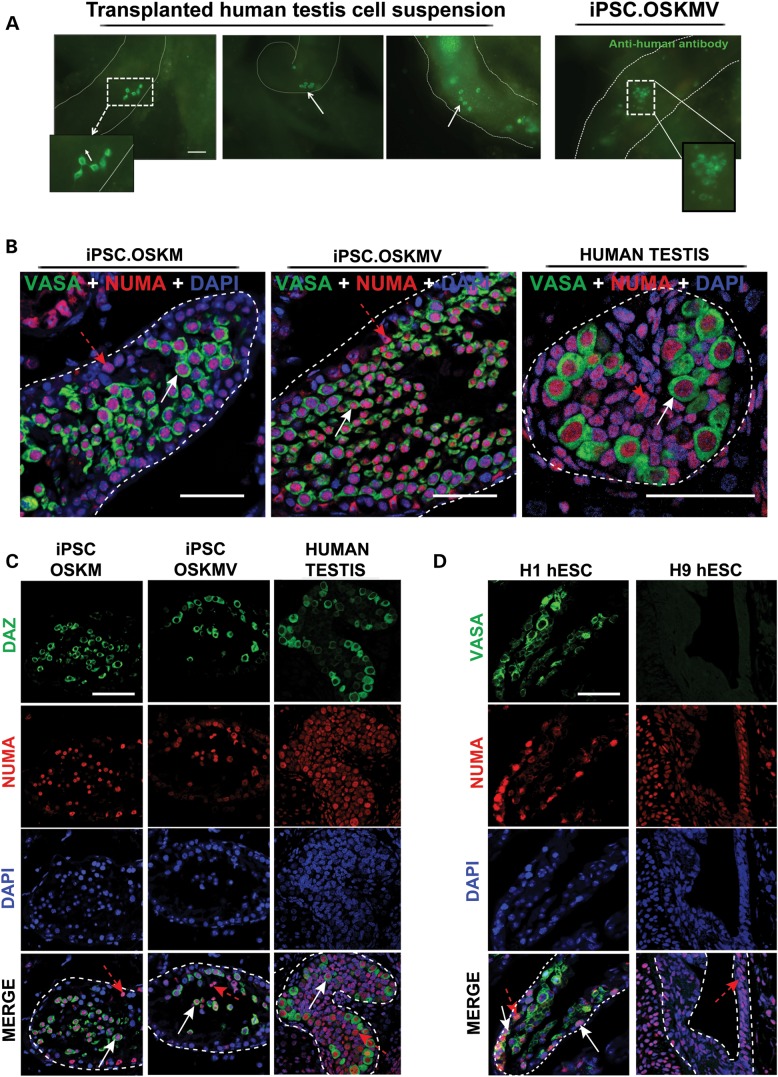
Transplantation of OSKM and OSKMV cells into busulfan-treated mouse testes. (A) Whole-mount analysis on transplanted human fetal testis cells and OSKMV cells into mouse testes. Chain (white dashed rectangle) and cluster formation (white arrows) visible in human fetal testis control cells. Transplanted OSKMV cells gave rise to cluster formation indicated by white dashed rectangle. Scale bar, 50 µm. (B) Histology cross sections of tubules inside mouse testes stained with co-localizing human-specific NUMA (red) and VASA (green). White arrows indicate transplanted cells positive for VASA germ cell marker; red dashed arrows indicate VASA-negative transplanted donor cells. Scale bar, 50 µm. (C) Histology cross sections of tubules of mouse testes stained with additional germ cell-specific marker DAZ demonstrating NUMA co-localization. Scale bar, 50 µm. (D) H1 and H9 hESC controls. H1 cells are positive for VASA/NUMA co-staining; H9 cells are negative for germ cell marker selection. White arrows indicate transplanted cells positive for VASA germ cell marker; red dashed arrows indicate VASA-negative donor cells. Scale bar, 50 µm.
We injected undifferentiated OSKM and OSKMV iPSCs into seminiferous tubules of eight testes each to evaluate their potential to form PGCs, gonocytes or spermatogonia in vivo in a transplant assay. Depending on which factors were used during reprogramming, we obtained strikingly different results. Testes injected with OSKMV cells maintained their naïve tissue structure 2-month post injection and one of eight testes was positive for a cluster of human cells that persisted long term (Fig. 3A, right panel and Supplementary Material, Table S3). In contrast, testes transplanted with OSKM cells developed large internal proliferating cell masses with no signs of teratoma formation in all eight of eight transplanted testes—transforming the testis into an enlarged tissue (Fig. 4A and B and Supplementary Material, Fig. S6A). To examine outcomes more extensively, we then repeated the transplantation and used the same method of direct comparison of germ cell activity across all transplantations. Our replicate transplantations consisted of OSKMV cell injections into the seminiferous tubules of an additional six testes with subsequent IHC analysis of serial cross sections regardless of tissue structure. We confirmed previous observations that indicated that OSKMV-derived iPSCs do not form large masses of cells but instead leave the mouse testis structure intact. In direct contrast, testes transplanted with undifferentiated H9 (Fig. 4Av) and H1 (Fig. 4Aiii) hESCs, developed enlarged tissues indicative of tumors similar to OSKM cells (Fig. 4Aii) and in addition formed teratoma-like structures (in H9 cells only, Fig. 4Avi) indicative of multiple germ layer tissue formation (see also Supplementary Material, Table S3). We observed that H1 cells that were not localized to the basement membrane did not demonstrate clear differentiation to either germ cells or somatic cells. Instead, based on the histology of multiple xenografts, H1 and OSKM cells outside the tubules resembled the histology of embryonal carcinoma (EC) cells and yolk sac tumors (Fig. 4Avii and viii) (33).
Figure 4.
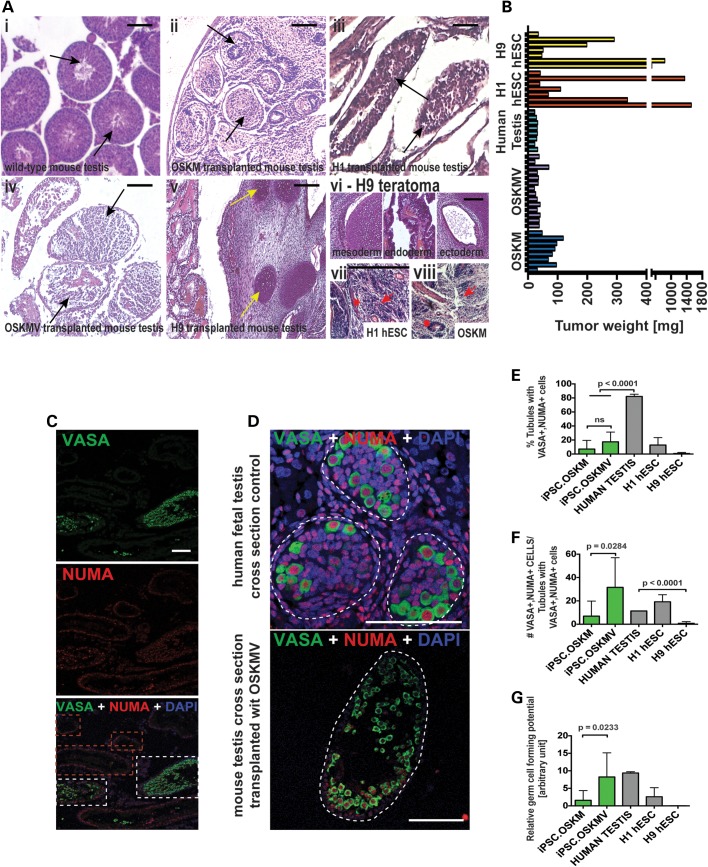
Transplantation of OSKM and OSKMV followed by quantification analysis. (A) Hematoxylin and eosin staining of histology cross sections of tubules inside mouse testes. (i–v) Black arrows indicate individual tubules. Yellow arrows indicate cartilage formation in H9 transplanted cells as example for teratoma formation. Scale bar, 100 µm. (vi) H9 transplanted cells formed teratomas indicated by three germ layers. (vii and viii) H1 and OSKM cells formed fibrotic structures that resembled embryonal carcinoma (EC) cells and yolk sac tumors. (B) Weight of testis 2 months after transplantation of cells (OSKM and OSKMV cells with three controls: human testis, H1 hESC and H9 hESC line). (C) Representative low-magnification image of histology cross section of multiple tubules stained with NUMA and VASA indicating that only a fraction of tubules formed VASA-positive cells. Red dashed rectangle indicates tubules with NUMA only cells along the basement membrane. White rectangle indicates NUMA/VASA-positive tubules. Scale bar, 50 µm. (D) Representative image of histology cross section of one single tubule. Single NUMA/VASA-positive cells were counted for quantification. Scale bar, 50 µm. (E–G) Quantification of immunohistochemistry results of cross sections from OSKM and OSKMV cells compared with both controls. All serial sections were subject to counting (see Materials and Methods) Scale bar, 80 µm. (E) Percentage of tubules with positive VASA/NUMA co-staining calculated (against total number of counted tubules). iPSC.OSKMV is also significantly different from H9 control (P = 0.0432) (F) For each positive tubule, VASA-positive cells that co-stained with NUMA were counted and calculated against positively stained tubules. iPSC.OSKMV is also significantly different from H9 control (P = 0.0397) (G) Relative germ cell forming potential calculated by multiplying fraction of positively stained tubules with number of VASA/NUMA co-stained cells for each sample. iPSC.OSKMV is also significantly different from H9 control (P = 0.0417).
Next, we focused on directly comparing germ cell activity and extensively screened serial cross sections for a panel of well-established germ cell markers in all transplanted mouse testis (OSKM, OSKMV, H1, H9). We compared our results with cross sections from a 22-week-old human fetal testis sample which contain abundant prospermatogonia and spermatogonia. Our analysis depended on the identification of human donor cells and the ability to discriminate them from mouse cells. For this purpose, we used a human-specific antibody, NUMA, that stains the cell nucleus and validated its specificity by staining various tissues of human and mouse origin with latter staining negative for NUMA (Supplementary Material, Fig. S6B and C). Our analysis revealed germ cells inside tubules of OSKM- and OSKMV-transplanted testes that are of human origin (nuclear NUMA) and co-localize with several germ cell-specific markers including VASA and DAZ (Fig. 3C and D and Supplementary Material, Movies S1 and S2). In contrast, transplanted H9 (XX female) cells that were detected inside tubules did not stain positive for any germ cell markers (Fig. 3D). Note, busulfan-depleted seminiferous tubules bear the risk of incomplete germ cell depletion resulting in infrequent mouse germ cells present; thus, we carefully discriminated positively stained germ cells that are of human or mouse origin (Fig. 5A) in all of our analysis and subsequent quantification efforts.
Figure 5.
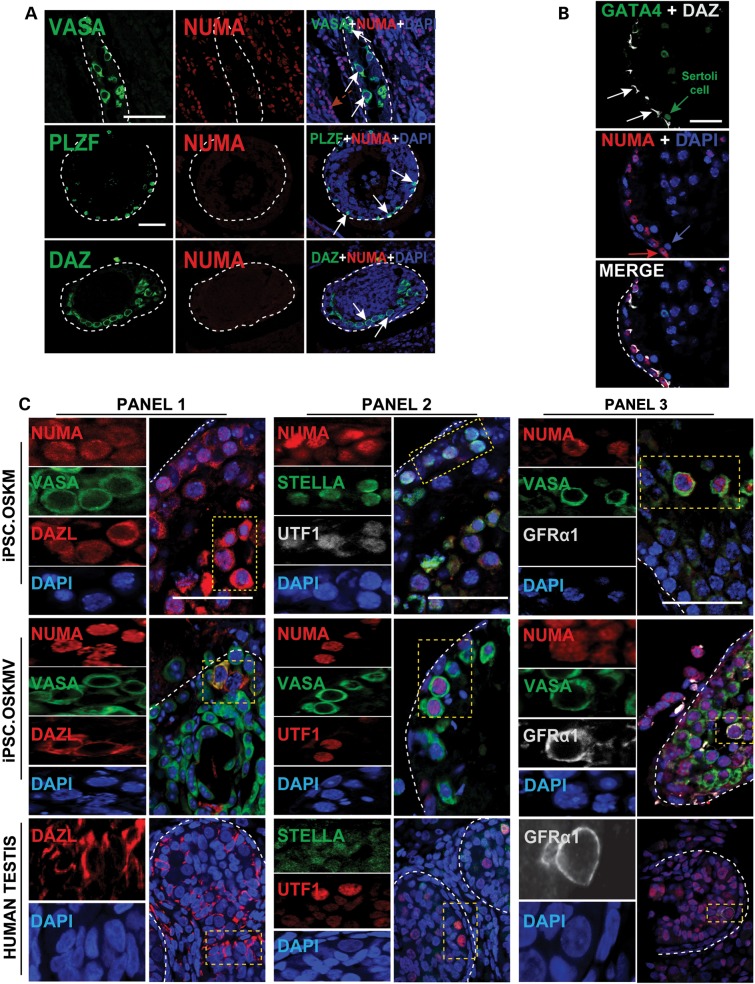
Germ cell formation of OSKM- and OSKMV-transplanted cells in vivo. (A) Observed endogenous germ cell activity inside the basement membrane of transplanted mouse testes. Cells stained positive for VASA, PLZF and DAZ but do not co-localize with NUMA indicating residual germ cells after busulfan treatment. Scale bar, 50 µm. (B) Detection of Sertoli cells indicated by GATA4 staining that is of mouse origin and near OSKMV-transplanted cells at the basement membrane of mouse testes. Scale bar, 50 µm. (C) Additional germ cell markers stain positive with NUMA in OSKM- and OSKMV-transplanted cells in mouse testes and co-localize with each other. GFRα1 was only detected in OSKMV-transplanted cells and human fetal testis control sections. Yellow dashed rectangles indicate magnified snapshots in each panel and for each sample. Each panel shows single individual protein and DAPI (left) and a zoomed out merged image (right). Scale bar, 80 µm.
OSKMV in contrast to OSKM reveals greater germ cell forming potential
In order to quantify our immunohistochemical analysis, we counted tubules that stained positive for all VASA/NUMA cells to determine the average fraction of tubules that had residing human germ cells present (Fig. 4C). We observed 7 and 18% of tubules positive for both OSKM- and OSKMV-transplanted cells, respectively (Fig. 4D and E). In contrast, we observed over 80% of tubules filled with NUMA/VASA double-positive cells in the human fetal testis cross sections (notably, these are non-transplanted cells and naturally existing germ cells). We then considered only those tubules positive for NUMA/VASA activity and counted individual cells across all sections; results indicate a more than 4-fold difference between OSKM and OSKMV with the latter being superior (Fig. 4F). To compare germ cell production between OSKM and OSKMV derivations and our human fetal testis control, we calculated relative germ cell numbers by multiplying the percent occupied tubules by the number of cells per tubule and compared them to our positive control. This indicated a 5-fold difference between OSKM compared with OSKMV and the human fetal testis, which was significant (Fig. 4G). Analysis of H1 donor-derived cells revealed results that were similar to OSKM-transplanted cells. As highlighted from the IHC analysis, mouse testes transplanted with H9 cells had lower germ cell activity in our quantitative analysis, which, however, was not significant (Fig. 4E–G).
In order to further verify that germ cells were induced from iPSCs in vivo, we extended our analysis of previously characterized germ cell markers (2,5,29–32) and demonstrated co-localization of three sets of PGC and premeiotic germ cell markers (VASA/DAZL, DPPA3/UTF1 and VASA/UTF1) with NUMA (Fig. 5C, Panels 1 and 2) for OSKM- and OSKMV-transplanted cells in conjunction with the positive control (Fig. 5C). In addition, we observed, in rare instances, cells that stained positive for GFRα1 cells, suggesting that OSKMV iPSCs are not only superior in germ cell formation to OSKM iPSCs in vitro, but also in vivo.
To evaluate whether transplanted cells differentiated to other lineages (such as Sertoli cells) or remained in their undifferentiated state, we co-stained for NUMA and germ cell markers with GATA4, a Sertoli cell marker. We observed GATA4-positive cells (Fig. 5B) at the basement membrane of tubules that did not co-stain with NUMA, indicating their murine origin; we note that the GATA4 cells resided in the vicinity of NUMA/DAZ double-positive cells highlighting their supportive phenotype in germ cell differentiation. Tubules that were entirely filled with large numbers of cells both intratubular and extratubular appeared to be unorganized, of human origin and OCT3/4 positive, suggesting an excess of transplanted cells that retained their undifferentiated state (Supplementary Material, Fig. S6D). This is in contrast to OCT3/4 positive cells that did not co-localize with NUMA but instead revealed an organized staining pattern along the basement membrane indicating endogenous cell of mouse origin. We observed that a small number of the transplanted iPSCs might escape during injection particularly in H1 and H9 hESC-containing testes. Specifically, H1 and H9 donor cells appeared to exit the seminiferous tubules and entered the extratubular space within the mouse testis as illustrated by positive NUMA staining outside mouse tubules (Supplementary Material, Fig. S6D, middle panel). Since the cells are undifferentiated iPSCs; however, they may subsequently proliferate.
Epigenetic analysis of donor-derived germ cells from H1 hESCs, OSKM and OSKMV iPSCs
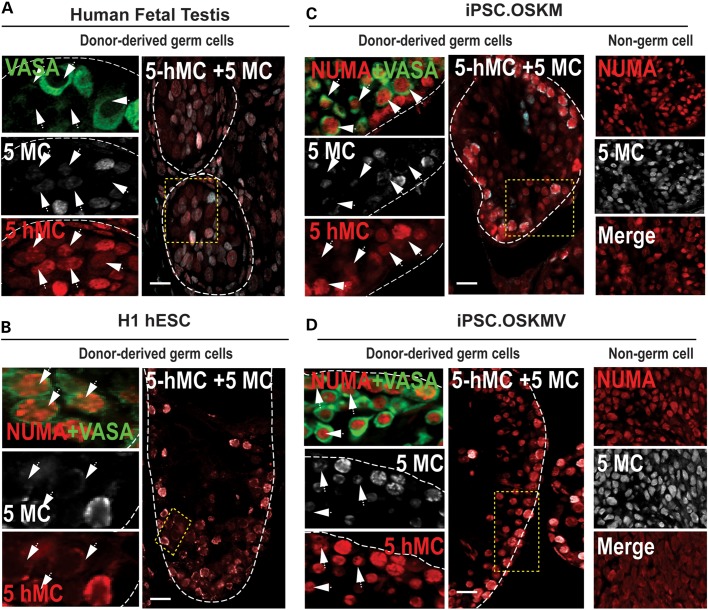
In order to evaluate if germ cells derived from donor iPSCs were specifically undergoing epigenetic remodeling events such as demethylation and conversion of 5-methylcytosine (5-MC) to 5-hydroxy-methylation (5-hMC) (34,35), we performed immunohistochemistry for 5-MC and 5-hMC in human fetal testes and in recipient mouse testes xenografts as previously demonstrated for embryo analysis (36) (Fig. 6). We observed that, in fetal testes, the levels of 5-MC appeared reduced in a large number of germ cells compared with 5-hMC levels. This was particularly evident in that a subset of VASA-positive germ cells had little to no 5-MC signal and a robust 5-hMC signal in fetal germ cells (Fig. 6A). We stained for NUMA/VASA in adjacent tissue sections and observed that OSKM, OSKMV and H1 hESC donor-derived cells exhibited a similar pattern of 5-MC and 5-hMC signals to fetal germ cells (Fig. 6, arrows). 5-MC levels were low to absent in NUMA + VASA+ donor cells indicating that global DNA demethylation was initiated but only partially complete owing to the maintenance of 5-hMC levels, which is an intermediate step in germ cell DNA demethylation. We also noted that NUMA-positive donor cells of OSKM and OSKMV xenografts that were VASA-negative and not considered germ cells expressed very high levels of both 5-MC and 5-hMC (Fig. 6C and D, right column). These observations collectively suggest an epigenetic transition from 5-MC to 5-hMC expression in fetal germ cells which is mirrored in germ cells derived in vivo from undifferentiated OSKM and OSKMV donor cells.
Figure 6.
DNA demethylation in xenotransplanted hESCs and iPSCs. Human fetal testis section (22 weeks) and testis xenografts of H1 hESCs, OSKM and OSKMV human iPSCs were stained for VASA, 5-methylcytosine (5-MC) and 5-hydroxymethlcytosine (5-hMC) in NUMA+ regions. (A) Cross section of a human fetal testis with positive immunostaining for VASA, 5-MC and 5-hMC. Areas in dotted yellow rectangles are shown in higher magnification on the left of each panel. White arrows indicate germ cells with low to none 5-MC signal relative to 5-hMC. (B–D) Cross sections of mouse testes xenografts after transplantation of undifferentiated H1 hESCs, OSKM and OSKMV cells, respectively, immunostained for NUMA, VASA, 5-MC and 5-hMC. Areas in dotted yellow rectangles are shown in higher magnification on the left of each panel. White arrows indicate NUMA + VASA-positive donor cells with low to none 5-MC signal relative to 5-hMC. Non-germ cell containing regions of the xenografts are shown in far right column for OSKM and OSKMV cells. Scale bar, 50 µm.
DISCUSSION
Patient-specific iPSCs may enable genetic analysis of naturally occurring deletions, insertions and mutations in human germ cell development. However, although there has been great progress in differentiation of mouse PGCs from PSCs via a combination of in vitro priming and in vivo use of the niche to complete spermatogenesis and oogenesis (2–4,37–39), several roadblocks must be overcome in order to achieve efficient, reproducible human germ cell differentiation amenable to developmental genetic studies. Here, we tested the hypothesis that somatic cells could be reprogrammed to a pluripotent state with an additional factor, VASA, in concert with the Yamanaka factors. We further hypothesized that VASA would endow reprogrammed cells with an enhanced ability to form human germ cells relative to cells reprogrammed with Yamanaka factors alone. Findings presented here provide the most viable strategy to date to direct human germ cell differentiation from PSCs for several reasons: (1) iPSCs were produced via mRNA reprogramming, thus insuring they are integration and xeno-free and do not require extensive culture (which introduces genetic and epigenetic mutations) to remove DNA-based reprogramming factors, (2) undifferentiated iPSCs are modified via inclusion of VASA in the reprogramming cocktail in order to enhance germ cell development and minimize tumorigenesis post-transplantation and (3) our strategy makes use of the seminiferous tubule niche in vivo in order to efficiently direct human germ cell formation from iPSCs, thereby potentially minimizing errors in erasure and establishment in sex-specific imprinted genes as have previously been observed in all human studies to date (5–8,40).
It is notable that efforts to simply translate results, in whole, from mouse to humans in diverse systems from hematopoietic, neural, cardiac, liver and others have all demonstrated a need to develop a specific protocol for differentiation of human tissues relative to the mouse. We have found this to be the case to date with differentiation of germ cells from mouse and human PSCs. Although some methods and molecules are transferrable, others are not and thus, we focused here on alterations to accommodate human germ cell differentiation. Our data reveal that reprogramming with inclusion of VASA can endow a ground state of pluripotency that is unique from OSKM and hESC lines. This is reflected at the undifferentiated state in OSKMV cells by notably lowered expression of OCT3/4 (POU5F1), TERT and SALL4. We suggest that the lowered level of these core pluripotency regulators is sufficient to maintain several phenotypic features in OSKMV cells consistent with conventially derived OSKM iPSCs. Interestingly, ectopic VASA expression does not affect the global transcriptional programs of OSKMV- and OSKM-derived iPSCs. It appears that ectopic VASA expression during reprogramming to iPSCs provides a signal that downregulates genes linked to pluripotency but does not induce in vitro differentiation under proliferative cell culture conditions. Our data also reveal that the OSKMV-reprogramming process facilitates germ cell formation from these cells. Evidence for this is provided in part by significant upregulation of premeiotic germ cell markers NANOS3, VASA and DPPA3 upon BMP4 differentiation and a greater potential to enter meiosis compared with the OSKM counterparts. It is worth nothing that PGC culture in vitro under proliferative conditions can result in derivation of pluripotent embryonic germ cell lines capable of differentiating to somatic and germline progeny cells; moreover, several studies have provided evidence of extensive similarity between ESCs and early germ cells (11,41). In contrast, the ontogeny of mouse iPSCs versus mouse PGCs is a more distant one than in humans (3). Mouse ESCs and iPSCs are considered to have a more naïve ground state than hESCs and therefore, an epiblast-like state is induced prior to PGC formation (2–4).
Given, the close relationship between human iPSCs and PGCs and the more distant relationship to mouse ESCs, we tested whether differentiation of RNA-reprogrammed iPSCs might simply be directed to the germline via injection directly into seminiferous tubules. Our observations reveal differences in OSKMV versus OSKM cells in favor of a germ-cell fate. This interpretation is based on several observations: First, NUMA+ OSKM and OSKMV cells in testicular cross sections integrated into or proximal to the basal membrane of the mouse tubules where spermatogonia and gonocytes normally reside in early fetal stages. Second, OSKM and OSKMV donor cells expressed key premeiotic germline markers with specific expression of the early spermatogonial marker, GFRα1 limited to OSKMV cells only. Third, the efficiency of derivation of germ cells was greater with OSKMV donor cells relative to OSKM cells. Fourth, we detected partial and in some cases complete DNA demethylation of H1, OSKM and OSKMV donor-derived cells in a germ cell-specific manner, consistent with their progression through the germ cell lineage. The DNA demethylation pattern in donor-derived germ cells closely mirrored that observed in second trimester fetal gonocytes and prospermatogonia and offered us a hint that donor cells may be undergoing a germ cell-specific event. However, in order to extend our results in future studies, locus-specific imprinting and chromatin modifications will be determined and a detailed analysis of donor-derived cells and their equivalent fetal germ cells will be performed in order to pinpoint the precise stage of germ cell differentiation that donor cells are progressing through. Finally, and of particular importance if we are to probe genetics of human germ cell development in the absence of complicating factors, cells reprogrammed with OSKMV formed no teratomas, tumors or undifferentiated cell masses in all injected sites (14 testes) in contrast to OSKM cells (similar to H1 [XY] control cells).
Our studies indicate that the PSC state is under control of a transcriptional circuitry that includes the Yamanaka reprogramming factors (42). Previous studies indicate that this transcriptional program is implemented in the context of an ‘open’ chromatin state, and it has been proposed that this state allows transcriptional programs to switch rapidly upon induction of differentiation or reprogramming (16,43). We suggest that chromatin remodelers that maintain the ESC/iPSC state, may re-open somatic chromatin during reprogramming thus allowing VASA (and the Yamanaka factors) to activate the transcriptional network for pluripotency and/or PGC specification. Furthermore, we suggest that the activation of a germline differentiation pathway in OSKMV cells is mediated by a molecular ‘switch’, potentially a chromatin remodeler, which detects changes in the cell culture environment or when cells are exposed to the seminiferous tubule niche. In the case of OSKMV cells, this ‘switch’ guarantees a germline fate while, in OSKM cells, the switch may require the function of additional molecular mechanisms to favor the same fate. In support of this notion, chicken ESCs primed with ectopic VASA (Cvh) maintained pluripotency and exhibited increased germline competence when differentiated (44). Collectively, we compare differential potential of OSKMV cells over OSKM cells to form PGCs in the environment of the seminiferous tubule without the risk of tumor formation (Fig. 7). We therefore highlight the inclusion of VASA during iPSC reprogramming as a defined method to induce germ cell fate in humans. We further speculate that other factors with conserved functions in germ cell specification in mice and humans such as the transcription factor PRDM14 (3,4) or the translational regulators, NANOS3, DAZL and PUM1/2 (5,6,45) may also be employed in mRNA-based reprogramming to confer higher germ cell potential on target cells.
Figure 7.
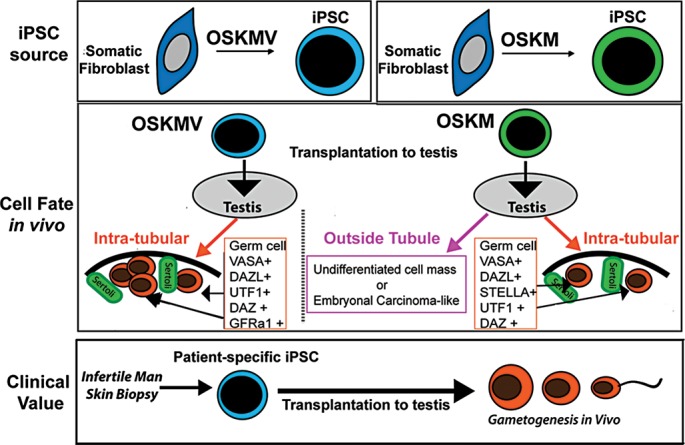
Model of transplantation of undifferentiated iPSCs into mouse testes. A schematic summarizing the major findings of this study. iPSCs reprogrammed with OSKM and OSKMV displayed differences in their phenotype after derivation. When transplanted, iPSC.OSKMV cells specifically differentiate to PGCs inside the spermatogonial tubule niche where they contact sertoli cells and were positive for a set of different stage-specific germ cell markers including GFRα1. iPSC.OSKM cells also localized inside the mouse testis niche and expressed key germ cell markers but to a lower extend and frequency. In addition, iPSC.OSKM cells developed enlarged cell masses inside mouse testes that resembled embryonal carcinoma-like tissue. This is in contrast to iPSC.OSKMV where all transplanted mouse testes kept their naïve structure. The transplantation strategies proposed here offer a potential avenue for fertility restoration for infertile men.
MATERIALS AND METHODS
Xenotransplantation assay
Human cell lines were transplanted into the testes of busulfan-treated, immunodeficient nude mice (NCr nu/nu; Taconic) as previously described for primate (27) and human (46) spermatogonia. Briefly, immunodeficient nude mice were treated with a single dose of busulfan (40 mg/kg, Sigma) at 6 weeks of age to eliminate endogenous spermatogenesis. Xenotransplantation was then performed 5 weeks after busulfan treatment by injecting cell suspensions containing 10% trypan blue (Invitrogen) into the seminiferous tubules of recipient testes via cannulation of the efferent ducts. Approximately 7 µl of cell suspension was injected per testis.
Eight weeks after transplantation, recipient mouse seminiferous tubules were dispersed gently with Collagenase IV (1 mg/ml) and DNase I (1 mg/ml) in D-PBS and fixed in 4% paraformaldehyde, as previously described (27,46). Clusters of human cells were observed on the basement membranes of recipient mouse seminiferous tubules by staining with a rabbit anti-primate testis cell primary antibody (27) and a goat anti-rabbit IgG secondary antibody conjugated with AlexaFluor488 (Invitrogen). All dehydration, rehydration and staining steps were carried out in 12-mm Transwell baskets (Corning Life Sciences) to prevent loss of seminiferous tubules.
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported by a National Institutes of Health grant through the National Heart Lung and Blood Institute (U01HL100397) and the National Institute of Child Health and Human Development (NICHD U54 HD068158 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research). Funding to pay the Open Access publication charges for this article was provided by grants from the National Institutes of Health and private funds.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Reijo Pera laboratory for helpful suggestions. In particular, we are grateful to S. Briggs and V. Sebastiano for assistance with mRNA reprogramming. We are grateful to B. Behr, director of the IVF/ART laboratory for assistance in obtaining valuable samples, H. Steyer for assistance in immunohistology staining, C. Cheng for epigenetic analysis. Finally, we thank R. Lauster, director of Medical Biotechnology at Technical University Berlin for thoughtful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Grabole N., Tischler J., Hackett J.A., Kim S., Tang F., Leitch H.G., Magnusdottir E., Surani M.A. Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 2013;14:629–637. doi: 10.1038/embor.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayashi K., Ogushi S., Kurimoto K., Shimamoto S., Ohta H., Saitou M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science. 2012;338:971–975. doi: 10.1126/science.1226889. [DOI] [PubMed] [Google Scholar]
- 3.Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 4.Nakaki F., Hayashi K., Ohta H., Kurimoto K., Yabuta Y., Saitou M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- 5.Kee K., Angeles V.T., Flores M., Nguyen H.N., Reijo Pera R.A. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medrano J.V., Ramathal C., Nguyen H.N., Simon C., Reijo Pera R.A. Divergent RNA-binding proteins, DAZL and VASA, induce meiotic progression in human germ cells derived in vitro. Stem Cells. 2012;30:441–451. doi: 10.1002/stem.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panula S., Medrano J.V., Kee K., Bergstrom R., Nguyen H.N., Byers B., Wilson K.D., Wu J.C., Simon C., Hovatta O., et al. Human germ cell differentiation from fetal- and adult-derived induced pluripotent stem cells. Hum. Mol. Genet. 2011;20:752–762. doi: 10.1093/hmg/ddq520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Easley C.A.T., Phillips B.T., McGuire M.M., Barringer J.M., Valli H., Hermann B.P., Simerly C.R., Rajkovic A., Miki T., Orwig K.E., et al. Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep. 2012;2:440–446. doi: 10.1016/j.celrep.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park T.S., Galic Z., Conway A.E., Lindgren A., van Handel B.J., Magnusson M., Richter L., Teitell M.A., Mikkola H.K., Lowry W.E., et al. Derivation of primordial germ cells from human embryonic and induced pluripotent stem cells is significantly improved by coculture with human fetal gonadal cells. Stem Cells. 2009;27:783–795. doi: 10.1002/stem.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gustafson E.A., Wessel G.M. Vasa genes: emerging roles in the germ line and in multipotent cells. Bioessays. 2010;32:626–637. doi: 10.1002/bies.201000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark A.T., Bodnar M.S., Fox M., Rodriquez R.T., Abeyta M.J., Firpo M.T., Pera R.A. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum. Mol. Genet. 2004;13:727–739. doi: 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- 12.Qin H., Blaschke K., Wei G., Ohi Y., Blouin L., Qi Z., Yu J., Yeh R.F., Hebrok M., Ramalho-Santos M. Transcriptional analysis of pluripotency reveals the Hippo pathway as a barrier to reprogramming. Hum. Mol. Genet. 2012;21:2054–2067. doi: 10.1093/hmg/dds023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabour D., Arauzo-Bravo M.J., Hubner K., Ko K., Greber B., Gentile L., Stehling M., Scholer H.R. Identification of genes specific to mouse primordial germ cells through dynamic global gene expression. Hum. Mol. Genet. 2011;20:115–125. doi: 10.1093/hmg/ddq450. [DOI] [PubMed] [Google Scholar]
- 14.Zwaka T.P., Thomson J.A. A germ cell origin of embryonic stem cells? Development. 2005;132:227–233. doi: 10.1242/dev.01586. [DOI] [PubMed] [Google Scholar]
- 15.Warren L., Ni Y., Wang J., Guo X. Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Sci. Rep. 2012;2:657. doi: 10.1038/srep00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meshorer E., Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 17.Chan E.M., Ratanasirintrawoot S., Park I.H., Manos P.D., Loh Y.H., Huo H., Miller J.D., Hartung O., Rho J., Ince T.A., et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi K., de Sousa Lopes S.M., Surani M.A. Germ cell specification in mice. Science. 2007;316:394–396. doi: 10.1126/science.1137545. [DOI] [PubMed] [Google Scholar]
- 19.Kee K., Gonsalves J.M., Clark A.T., Pera R.A. Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cells Dev. 2006;15:831–837. doi: 10.1089/scd.2006.15.831. [DOI] [PubMed] [Google Scholar]
- 20.Page S.L., Hawley R.S. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- 21.Kerjean A., Dupont J.M., Vasseur C., Le Tessier D., Cuisset L., Paldi A., Jouannet P., Jeanpierre M. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum. Mol. Genet. 2000;9:2183–2187. doi: 10.1093/hmg/9.14.2183. [DOI] [PubMed] [Google Scholar]
- 22.Smilinich N.J., Day C.D., Fitzpatrick G.V., Caldwell G.M., Lossie A.C., Cooper P.R., Smallwood A.C., Joyce J.A., Schofield P.N., Reik W., et al. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc. Natl. Acad. Sci. USA. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorvaldsen J.L., Duran K.L., Bartolomei M.S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferguson-Smith A.C., Sasaki H., Cattanach B.M., Surani M.A. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- 25.Guenther M.G., Frampton G.M., Soldner F., Hockemeyer D., Mitalipova M., Jaenisch R., Young R.A. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermann B.P., Sukhwani M., Lin C.C., Sheng Y., Tomko J., Rodriguez M., Shuttleworth J.J., McFarland D., Hobbs R.M., Pandolfi P.P., et al. Characterization, cryopreservation, and ablation of spermatogonial stem cells in adult rhesus macaques. Stem Cells. 2007;25:2330–2338. doi: 10.1634/stemcells.2007-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagano M., Patrizio P., Brinster R.L. Long-term survival of human spermatogonial stem cells in mouse testes. Fertil. Steril. 2002;78:1225–1233. doi: 10.1016/s0015-0282(02)04345-5. [DOI] [PubMed] [Google Scholar]
- 29.Anderson R.A., Fulton N., Cowan G., Coutts S., Saunders P.T. Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC Dev. Biol. 2007;7:136. doi: 10.1186/1471-213X-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buaas F.W., Kirsh A.L., Sharma M., McLean D.J., Morris J.L., Griswold M.D., de Rooij D.G., Braun R.E. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- 31.Castrillon D.H., Quade B.J., Wang T.Y., Quigley C., Crum C.P. The human VASA gene is specifically expressed in the germ cell lineage. Proc. Natl. Acad. Sci. USA. 2000;97:9585–9590. doi: 10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kristensen D.M., Nielsen J.E., Skakkebaek N.E., Graem N., Jacobsen G.K., Rajpert-De Meyts E., Leffers H. Presumed pluripotency markers UTF-1 and REX-1 are expressed in human adult testes and germ cell neoplasms. Hum. Reprod. 2008;23:775–782. doi: 10.1093/humrep/den010. [DOI] [PubMed] [Google Scholar]
- 33.Chaganti R.S., Houldsworth J. Genetics and biology of adult human male germ cell tumors. Cancer Res. 2000;60:1475–1482. [PubMed] [Google Scholar]
- 34.Hackett J.A., Sengupta R., Zylicz J.J., Murakami K., Lee C., Down T.A., Surani M.A. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi S., Hong K., Liu R., Inoue A., Shen L., Zhang K., Zhang Y. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 2013;23:329–339. doi: 10.1038/cr.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wossidlo M., Nakamura T., Lepikhov K., Marques C.J., Zakhartchenko V., Boiani M., Arand J., Nakano T., Reik W., Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 37.Haston K.M., Tung J.Y., Reijo Pera R.A. Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. PLoS One. 2009;4:e5654. doi: 10.1371/journal.pone.0005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholas C.R., Chavez S.L., Baker V.L., Reijo Pera R.A. Instructing an embryonic stem cell-derived oocyte fate: lessons from endogenous oogenesis. Endocr. Rev. 2009;30:264–283. doi: 10.1210/er.2008-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholas C.R., Haston K.M., Grewall A.K., Longacre T.A., Reijo Pera R.A. Transplantation directs oocyte maturation from embryonic stem cells and provides a therapeutic strategy for female infertility. Hum. Mol. Genet. 2009;18:4376–4389. doi: 10.1093/hmg/ddp393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gkountela S., Li Z., Vincent J.J., Zhang K.X., Chen A., Pellegrini M., Clark A.T. The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat. Cell Biol. 2013;15:113–122. doi: 10.1038/ncb2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pirouz M., Klimke A., Kessel M. The reciprocal relationship between primordial germ cells and pluripotent stem cells. J. Mol. Med. (Berl.) 2012;90:753–761. doi: 10.1007/s00109-012-0912-1. [DOI] [PubMed] [Google Scholar]
- 42.Jaenisch R., Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramalho-Santos M. iPS cells: insights into basic biology. Cell. 2009;138:616–618. doi: 10.1016/j.cell.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavial F., Acloque H., Bachelard E., Nieto M.A., Samarut J., Pain B. Ectopic expression of Cvh (Chicken Vasa homologue) mediates the reprogramming of chicken embryonic stem cells to a germ cell fate. Dev. Biol. 2009;330:73–82. doi: 10.1016/j.ydbio.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 45.Gilboa L., Lehmann R. Repression of primordial germ cell differentiation parallels germ line stem cell maintenance. Curr. Biol. 2004;14:981–986. doi: 10.1016/j.cub.2004.05.049. [DOI] [PubMed] [Google Scholar]
- 46.Dovey S.L., Valli H., Hermann B.P., Sukhwani M., Donohue J., Castro C.A., Chu T., Sanfilippo J.S., Orwig K.E. Eliminating malignant contamination from therapeutic human spermatogonial stem cells. J. Clin. Invest. 2013;123:1833–1843. doi: 10.1172/JCI65822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.