Abstract
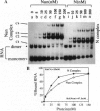
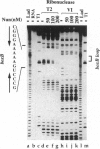
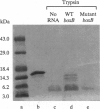
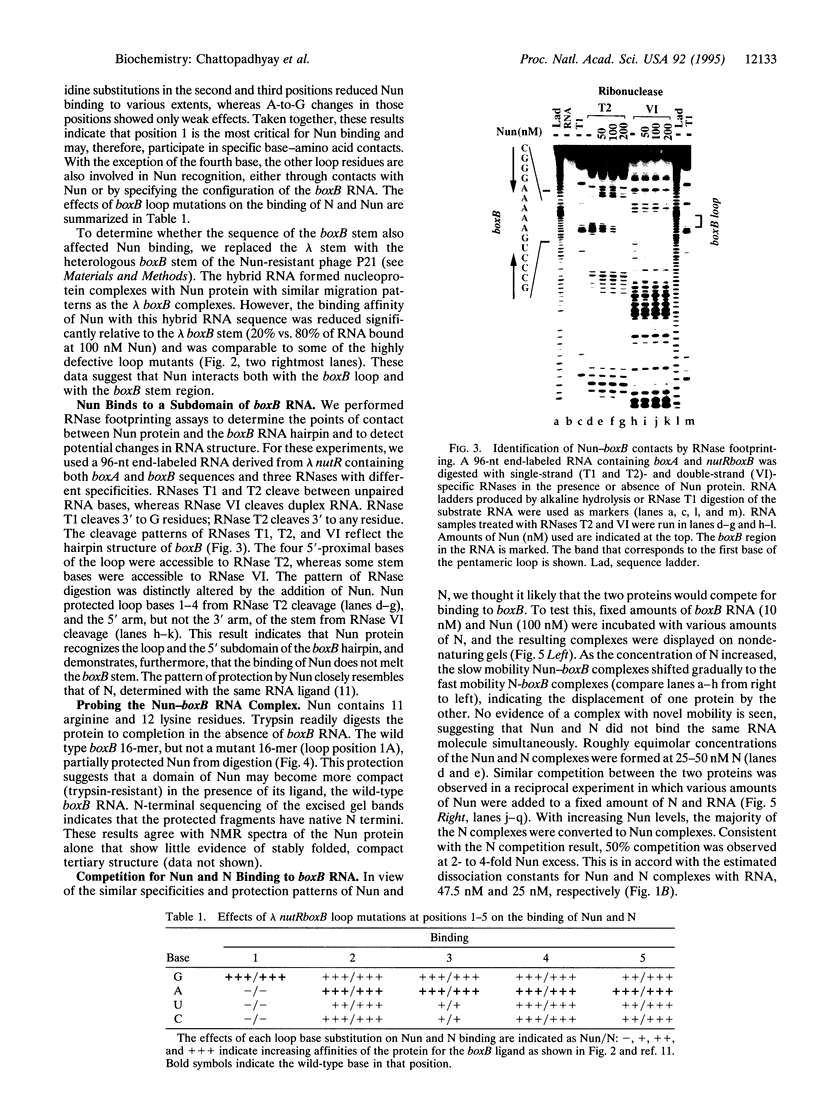
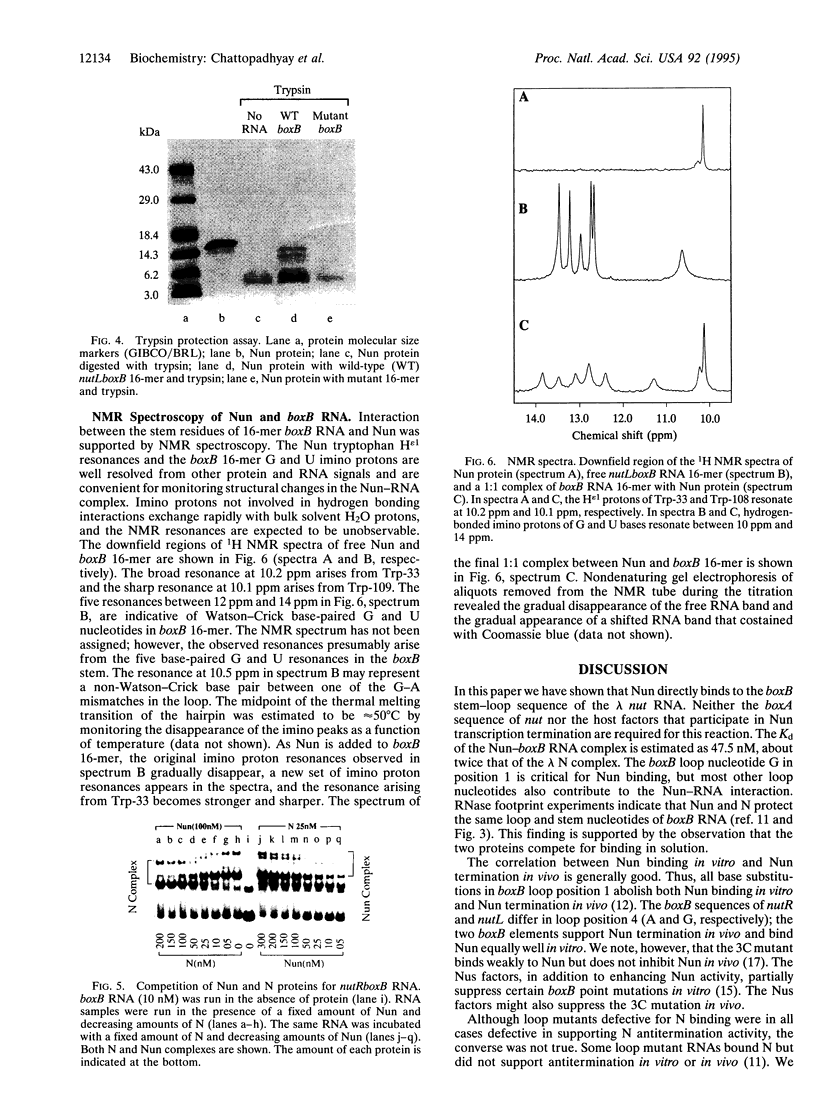
The nun gene product of prophage HK022 excludes phage lambda infection by blocking the expression of genes downstream from the lambda nut sequence. The Nun protein functions both by competing with lambda N transcription-antitermination protein and by actively inducing transcription termination on the lambda chromosome. We demonstrate that Nun binds directly to a stem-loop structure within nut RNA, boxB, which is also the target for the N antiterminator. The two proteins show comparable affinities for boxB and they compete with each other. Their interactions with boxB are similar, as shown by RNase protection experiments, NMR spectroscopy, and analysis of boxB mutants. Each protein binds the 5' strand of the boxB stem and the adjacent loop. The stem does not melt upon the binding of Nun or N, as the 3' strand remains sensitive to a double-strand-specific RNase. The binding of RNA partially protects Nun from proteolysis and changes its NMR spectra. Evidently, although Nun and N bind to the same surface of boxB RNA, their respective complexes interact differently with RNA polymerase, inducing transcription termination or antitermination, respectively.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barik S., Ghosh B., Whalen W., Lazinski D., Das A. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell. 1987 Sep 11;50(6):885–899. doi: 10.1016/0092-8674(87)90515-0. [DOI] [PubMed] [Google Scholar]
- Baron J., Weisberg R. A. Mutations of the phage lambda nutL region that prevent the action of Nun, a site-specific transcription termination factor. J Bacteriol. 1992 Mar;174(6):1983–1989. doi: 10.1128/jb.174.6.1983-1989.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994 Jul 29;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S., Garcia-Mena J., DeVito J., Wolska K., Das A. Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage lambda. Proc Natl Acad Sci U S A. 1995 Apr 25;92(9):4061–4065. doi: 10.1073/pnas.92.9.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. Control of transcription termination by RNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- Das A. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J Bacteriol. 1992 Nov;174(21):6711–6716. doi: 10.1128/jb.174.21.6711-6716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin N. C. Clustered arginine residues of bacteriophage lambda N protein are essential to antitermination of transcription, but their locale cannot compensate for boxB loop defects. J Mol Biol. 1993 May 20;231(2):343–360. doi: 10.1006/jmbi.1993.1287. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Interaction between bacteriophage lambda and its Escherichia coli host. Curr Opin Genet Dev. 1992 Oct;2(5):727–738. doi: 10.1016/s0959-437x(05)80133-9. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Nodwell J. R., Mason S. W. Transcriptional antitermination. Nature. 1993 Jul 29;364(6436):401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- Horwitz R. J., Li J., Greenblatt J. An elongation control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell. 1987 Nov 20;51(4):631–641. doi: 10.1016/0092-8674(87)90132-2. [DOI] [PubMed] [Google Scholar]
- Hung S. C., Gottesman M. E. Phage HK022 Nun protein arrests transcription on phage lambda DNA in vitro and competes with the phage lambda N antitermination protein. J Mol Biol. 1995 Mar 31;247(3):428–442. doi: 10.1006/jmbi.1994.0151. [DOI] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodwell J. R., Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993 Jan 29;72(2):261–268. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- Oberto J., Weisberg R. A., Gottesman M. E. Structure and function of the nun gene and the immunity region of the lambdoid phage HK022. J Mol Biol. 1989 Jun 20;207(4):675–693. doi: 10.1016/0022-2836(89)90237-4. [DOI] [PubMed] [Google Scholar]
- Olson E. R., Flamm E. L., Friedman D. I. Analysis of nutR: a region of phage lambda required for antitermination of transcription. Cell. 1982 Nov;31(1):61–70. doi: 10.1016/0092-8674(82)90405-6. [DOI] [PubMed] [Google Scholar]
- Robert J., Sloan S. B., Weisberg R. A., Gottesman M. E., Robledo R., Harbrecht D. The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell. 1987 Nov 6;51(3):483–492. doi: 10.1016/0092-8674(87)90644-1. [DOI] [PubMed] [Google Scholar]
- Robledo R., Atkinson B. L., Gottesman M. E. Escherichia coli mutations that block transcription termination by phage HK022 Nun protein. J Mol Biol. 1991 Aug 5;220(3):613–619. doi: 10.1016/0022-2836(91)90104-e. [DOI] [PubMed] [Google Scholar]
- Robledo R., Gottesman M. E., Weisberg R. A. Lambda nutR mutations convert HK022 Nun protein from a transcription termination factor to a suppressor of termination. J Mol Biol. 1990 Apr 20;212(4):635–643. doi: 10.1016/0022-2836(90)90226-c. [DOI] [PubMed] [Google Scholar]
- Sloan S. B., Weisberg R. A. Use of a gene encoding a suppressor tRNA as a reporter of transcription: analyzing the action of the Nun protein of bacteriophage HK022. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):9842–9846. doi: 10.1073/pnas.90.21.9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen W. A., Das A. Action of an RNA site at a distance: role of the nut genetic signal in transcription antitermination by phage-lambda N gene product. New Biol. 1990 Nov;2(11):975–991. [PubMed] [Google Scholar]