Background: Merlin controls organ size by binding to target proteins both in cytoplasm and nucleus.
Results: The structure of Merlin FERM domain in complex with its binding domain of DCAF1 is determined.
Conclusion: DCAF1 folds into a β-hairpin structure and binds to the F3 lobe of Merlin FERM domain.
Significance: The structure of the Merlin·DCAF1 complex provides a template for understanding the interactions of Merlin with its binding partners.
Keywords: Cell Growth, Cell Polarity, Ezrin, Structural Biology, Tumor Suppressor Gene
Abstract
The tumor suppressor gene Nf2 product, Merlin, plays vital roles in controlling proper development of organ sizes by specifically binding to a large number of target proteins localized both in cytoplasm and nuclei. The FERM domain of Merlin is chiefly responsible for its binding to target proteins, although the molecular basis governing these interactions are poorly understood due to lack of structural information. Here, we report the crystal structure of the Merlin FERM domain in complex with its binding domain derived from the E3 ubiquitin ligase substrate adaptor DCAF1 (also known as VPRBP). Unlike target binding modes found in ERM proteins, the Merlin-FERM binding domain of DCAF1 folds as a β-hairpin and binds to the α1/β5-groove of the F3 lobe of Merlin-FERM via extensive hydrophobic interactions. In addition to providing the first structural glimpse of a Merlin-FERM·target complex, the structure of the Merlin·DCAF1 complex is likely to be valuable for understanding the interactions of Merlin with its binding partners other than DCAF1.
Introduction
Cell-to-cell contact triggers inhibition signals for cell growth, which is crucial for living systems to maintain proper organ sizes by balancing the rate of cell proliferation and apoptosis. Disruption of the contact inhibition gives rise to cell overgrowth and often induces tumor formation (1). The tumor suppressor gene Nf2, which encodes the FERM (protein 4.1, Ezrin, Radixin, Moesin) domain-containing protein Merlin, is an important determinant in contact-mediated cell growth inhibition (2). Consistent with its critical role in growth inhibition, loss-of-function mutations of Merlin are known to cause a series of tumor formations, including the familial cancer syndrome neurofibromatosis type 2 and several other carcinomas (3–6). Mechanistically, Merlin is known for its central roles in the Hippo pathway in controlling organ sizes (7, 8). Recent investigation indicated that Merlin is also involved in a distinct cell growth regulation mechanism by accumulating in nucleus and inhibiting the activity of nuclear E3 ubiquitin ligase via binding to its substrate adaptor DCAF1 (also known as viral protein R-binding protein, VPRBP) (9). The interaction between Merlin and DCAF1 is mediated by the FERM domain of Merlin (Merlin-FERM) and the C-terminal tail of DCAF1 (DCAF1-CT) (9). However, the molecular basis for the FERM-mediated Merlin/DCAF1 interaction remains unknown.
FERM domain is a well known protein binding module that is composed of three subunits, called F1, F2, and F3 lobes (10). Numerous FERM domain-containing proteins have been identified and classified into different subfamilies (10). As one of the best studied subfamilies of FERM superfamily, ERM (ezrin, radixin, and moesin) is the most closely related to Merlin. Similar to ERM proteins, Merlin has ∼600 residues in length and contains the N-terminal FERM domain, a central helical region, and a C-terminal regulatory domain (see Fig. 1A). Despite high sequence similarity with ERM proteins, especially with their FERM domains (sequence identity of ∼60%), Merlin was shown to have distinct tissue localization and functions (11). This functional specificity indicates that the target binding mechanism of Merlin is different from ERM proteins. Extensive studies have implicated a number of potential Merlin targets (8, 9, 12, 13), most of which (including DCAF1) were found to interact with its FERM domain. Structural studies confirmed that Merlin-FERM share high structural similarity with the FERM domains of ERM proteins (14, 15). However, due to lacking of target-bound structures of Merlin-FERM, the mechanisms underlying target recognitions of Merlin were modeled largely based on the structures of ERM proteins (especially radixin) in complex with their targets.
FIGURE 1.
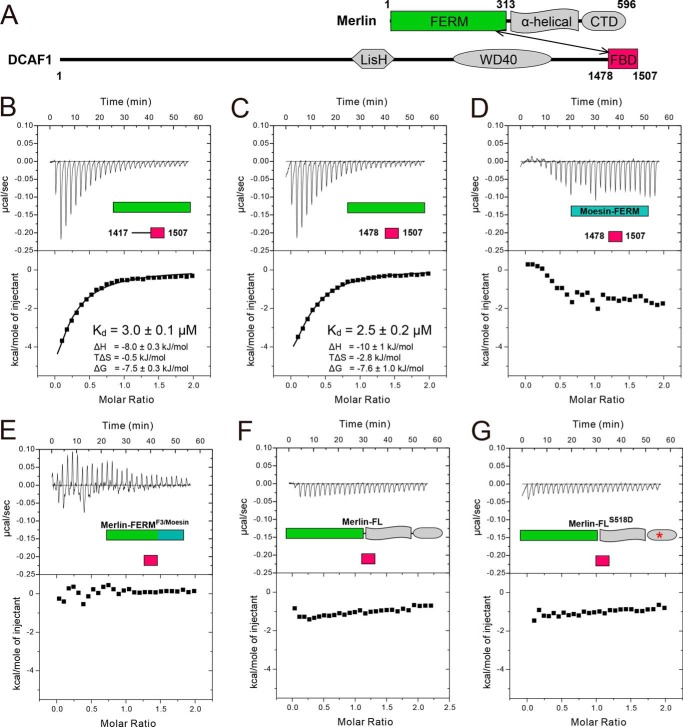
Characterization of the Merlin-FERM/DCAF1 interaction. A, domain organizations of Merlin and DCAF1 are shown as schemes. The Merlin-FERM/DCAF1-FBD interaction is indicated by a double-headed arrow. B--G, ITC-based measurements showing the binding affinities between DCAF1-CT and Merlin-FERM (B), and between DCAF1-FBD and Merlin-FERM (C), Moesin-FERM (D), Merlin-FERMF3/Moesin (E), Merlin-FL (F), and Merlin-FLS518D (G).
Here, we show that Merlin-FERM directly binds to a short fragment of DCAF1 located at the extreme C-terminal end (termed FERM-binding domain or FBD,3 see Fig. 1A). We determined the crystal structure of Merlin-FERM·DCAF1-FBD complex at 2.6 Å resolution. In addition to uncovering the molecular basis governing the specific Merlin-FERM/DCAF1 interaction, the complex structure also reveals a distinct FERM-mediated targeting mechanism in general. Unlike target binding modes found in ERM proteins, DCAF1-FBD folds as a β-hairpin to augment the β-sheet of the F3 lobe of Merlin-FERM via extensive hydrophobic interactions. Additionally, the highly conserved, negatively charged DCAF1 β-hairpin loop was found to contribute to the DCAF1/Merlin interaction, likely via binding to a positively charged cleft between the F1 and F3 lobes of Merlin-FERM.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The Merlin constructs (Merlin-FL, residues 1–596; Merlin-FERM, residues 1–313), and the DCAF1 constructs (DCAF1-CT, residues 1417–1507; DCAF1-FBD, residues 1478–1507) were amplified by PCR using the mouse and human cDNA libraries as the template, respectively, and individually cloned into the modified pET32a vector. Various mutants were created using standard two-step PCR-based methods and confirmed by DNA sequencing. Recombinant proteins with N-terminal thioredoxin- and His6-tagged were transformed to Escherichia coli BL21(DE3) cells, cultured at 37 °C to an OD ∼ 0.6 and induced with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside at 16 °C overnight. The expressed proteins were purified by a Ni2+-nitrilotriacetic acid agarose affinity chromatography followed by a size-exclusion chromatography. During purification, all protein samples were detected and analyzed by SDS-PAGE coupled with Coomassie Blue staining.
Isothermal Titration Calorimetry (ITC) Assay
ITC was carried out on a MicroCal VP-ITC at 25 °C. All proteins were dissolved in a buffer containing 50 mm Tris, pH 7.5, 250 mm NaCl, 1 mm DTT, and 1 mm EDTA. The titration processes were performed by injecting 5–10-μl aliquots of protein samples in syringe (concentration of 100 μm) into protein samples in cell (concentration, 10 μm) 27 times and at time intervals of 120 s to ensure the titration peak returned to the baseline. The data were analyzed using the Origin (version 7.0) and fitted by the one-site binding model.
Crystallization
For crystallization, the tagged proteins were treated by a small amount of human rhinovirus 3C protease at 4 °C overnight to cleave the fusion tags and further purified by size-exclusion chromatography. Crystals of the Merlin-FERM·DCAF1-FBD complex were obtained by hanging drop vapor diffusion method at 16 °C within 5 days. To set up a hanging drop, 1 μl of concentrated protein mixture (∼20 mg/ml) at 1:1 stoichiometric ratio was mixed with 1 μl of crystallization solution with 20% isopropyl alcohol and 5% PEG 8000, pH 8.0. Before diffraction experiments, crystals were soaked in crystallization solution containing 30% glycerol for cryoprotection. The diffraction data were collected at Shanghai Synchrotron Radiation Facility and were processed and scaled using HKL2000 (Table 1) (16).
TABLE 1.
Statistics of data collection and model refinement
Numbers in parentheses represent the value for the highest resolution shell.
| Data collection | |
| Space group | P3121 |
| Unit cell parameters (Å) | a = 97.1, c = 224.3 |
| Resolution range (Å) | 50–2.6 (2.64–2.6) |
| No. of unique reflections | 37,815 |
| Redundancy | 4.6 (4.8) |
| I/σ | 20.11 (2.22) |
| Completeness (%) | 98.16 (99.05) |
| Rmerge (%)a | 7.09 (92.5) |
| Structure refinement | |
| Rcryst/Rfree (%)b | 22.4 (31.9)/25.6 (35.1) |
| r.m.s.d. bonds (Å)/angles | 0.003/0.7° |
| Average B factor | 96.2 |
| No. of atoms | |
| Protein atoms | 5070 |
| Water molecules | 31 |
| Other molecules | 6 |
| No. of reflections | |
| Working set | 53,440 |
| Test set | 1994 |
| Ramachandran plotc | |
| Favored regions (%) | 97.5 |
| Allowed regions (%) | 2.2 |
| Outlier (%) | 0.3 |
a Rmerge = Σ|Ii − Im /ΣIi, where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry-related reflections.
b Rcryst = Σ||Fobs| − |Fcalc||/Σ|Fobs|, where Fobs and Fcalc are observed and calculated structure factors. Rfree = ΣT||Fobs| − |Fcalc||/ΣT|Fobs|, where T is a test data set of ∼5% of the total reflections randomly chosen and set aside prior to refinement.
c Values were defined by MolProbity.
Structure Determination
The initial phase was determined by molecular replacement using the apo form of Merlin-FERM (Protein Data Bank code 1ISN) as the search model. The model was refined in Phenix (17) against the 2.6 Å data set. The DCAF1-FBD peptide was built subsequently in COOT (18). In the final stage, an additional TLS refinement was performed in Phenix. The final model was further validated by using MolProbity (19). The refinement statistics are listed in Table 1. The structural model of DCAF1-FBD was well assigned except for the connecting loop for the β-hairpin structure. All structure figures were prepared using PyMOL. The sequence alignments were prepared and presented using ClustalW (20) and ESPript (21), respectively.
RESULTS
The FERM Domain of Merlin Specifically Binds to a Short, C-terminal Tail Fragment of DCAF1
Merlin-FERM was shown to interact with the DCAF1 C-terminal region recently (9). We first tried to verify this interaction using purified recombinant proteins. Quantitative binding assays showed that the C-terminal region of DCAF1 (DCAF1-CT, residues 1417–1507) binds to Merlin-FERM with a dissociation constant (Kd) of ∼3 μm (Fig. 1B). Further boundary mapping revealed that the N-terminal part of DCAF1-CT (residues 1417–1477) showed no detectable binding to Merlin-FERM (data not shown), and the remaining C-terminal 30 residues (residues 1478–1507) displayed the same binding affinity as the entire DCAF1-CT does (Fig. 1C), indicating that the last 30 residues in DCAF1 contains the complete FERM-binding domain (DCAF1-FBD). The binding of Merlin-FERM to DCAF1-FBD is mainly driven by enthalpy (Fig. 1, B and C). Interestingly, although sharing a high amino acid sequence identity with Merlin, the FERM domain of moesin had no detectable binding to DCAF1-FBD (Fig. 1D), indicating that the FERM domain of Merlin encodes its intrinsic target binding specificities. A Merlin-FERM chimera (termed Merlin-FERMF3/Moesin), in which the F3 lobe was replaced by the corresponding F3 lobe of moesin, failed to bind DCAF1-FBD (Fig. 1E), indicating that the F3 lobe is chiefly responsible for Merlin-FERM to bind to DCAF1.
Overall Structure of the Merlin-FERM·DCAF1-FBD Complex
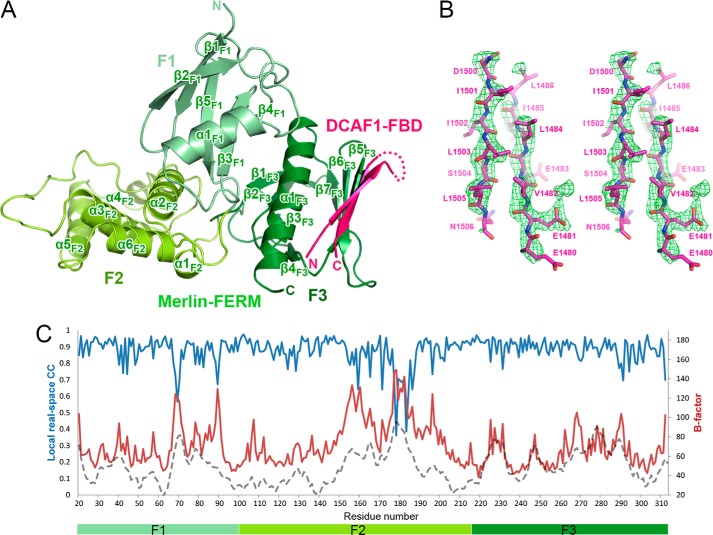
To understand the molecular basis governing the Merlin/DCAF1 interaction, we determined the crystal structure of Merlin-FERM in complex with DCAF1-FBD at 2.6 Å resolution using the molecular replacement method (Table 1). In the crystal structure, Merlin-FERM and DCAF1-FBD forms a 1:1 ratio complex with two complexes per asymmetric unit (Fig. 2A). In the complex, Merlin-FERM adopts a typical FERM architecture, comprised of three lobes, F1, F2, and F3. Consistent with the biochemistry data shown in Fig. 1, the DCAF1-FBD peptide folds as a β-hairpin to bind to the α1/β5-groove of the F3 lobe of Merlin-FERM (Fig. 2A). The two β-strands of DCAF1-FBD are well defined, whereas the loop connecting the hairpin is completely disordered (Fig. 2, A and B). The overall fold of Merlin-FERM in the DCAF1·FBD complex is almost identical to the apo form structure (14) and the FERM domain of radixin in complex with CD44 (22) (overall root mean square deviation of 0.7 Å with 285 aligned residues and of 0.9 Å with 294 aligned residues, respectively), indicating that the DCAF1 binding does not induce obvious conformational changes to the F3 lobe as well as the entire FERM domain. By analyzing the B factor distribution of Merlin-FERM in complex and apo forms (Fig. 2C), we found that the F3 lobe in the DCAF1-bound structure shows similar B-factors with that in the apo form structure, although the overall B-factor of the DACF1-bound structure is much higher than that of the apo structure, suggesting that the binding to DCAF1 stabilizes the F3 lobe of the Merlin-FERM domain.
FIGURE 2.
Overall structure of the Merlin-FERM·DCAF1-FBD complex. A, the crystal structure is presented as a ribbon diagram with the three lobes of Merlin-FERM, F1 (light green), F2 (green), F3 (dark green), and DCAF1-FBD (pink) drawn in their specific colors. The same color code is used throughout the rest of the figures except as otherwise indicated. The disordered loop connecting two β-strands is indicated by a dotted line. B, the simulated annealing omit map (green mesh) for the DCAF1-FBD hairpin structure. The map was contoured at 3σ with the structural model superimposed. C, the B-factor (red curve) and real-space (blue curve) correlation plot of Merlin-FERM in our crystal structure. The B-factor distribution (gray dashed curve) of the apo Merlin-FERM structure (Protein Data Bank code 1ISN) was shown for comparison.
An Atypical Target Binding Mode for FERM Domains Revealed by the Merlin-FERM·DCAF1-FBD Complex
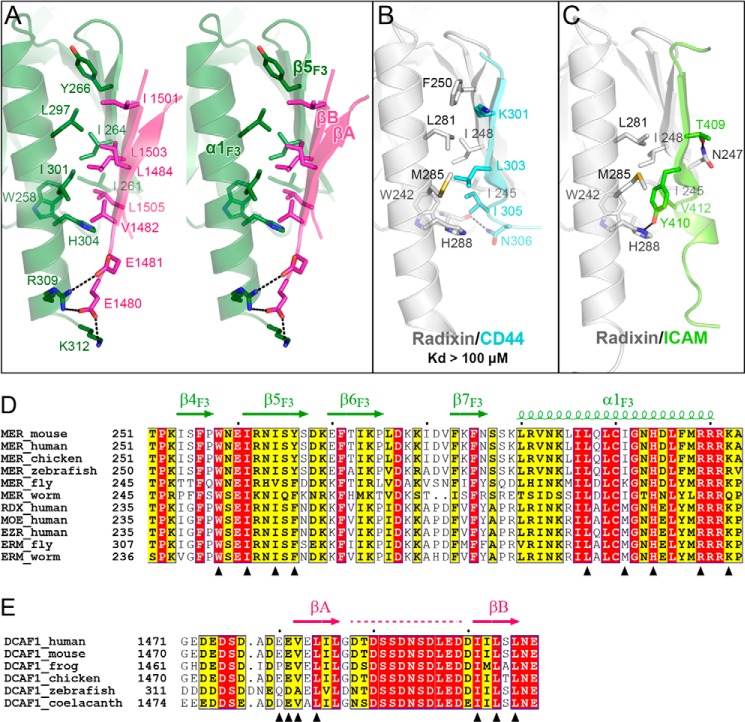
Although other lobes are also known to participate in binding to target proteins, the F3 lobes of FERM domains are the major target binding sites (22–31). The F3 lobes of Merlin and ERM proteins adopt a PTB-like fold, consisting of a seven-stranded β-sandwich and a C-terminal α-helix (Fig. 2A) (14, 15, 24). A groove (termed αβ-groove) mainly formed by β5F3 and α1F3 is a well characterized binding region in FERM domains, including those of Moesin (24), Radixin (22, 25–28), Talin (29, 32, 33), Myosin-X (30, 34), and Sorting Nexin-17 (31). Most of these targets, especially those of ERM-binding proteins, bind to the αβ-groove via a β-strand or a β-strand-like extended structures (Fig. 3, B and C), and thus, the contact between the target and the F3 lobe is not extensive (and their interactions are often very weak; Fig. 3B). DCAF1-FBD also binds to the αβ-groove of Merlin-FERM (Fig. 3A). However, instead of using a single β-strand, DCAF1-FBD adopts a β-hairpin structure and augments one of the anti-parallel β-sheets of Merlin-FERM formed by β5F3, β6F3, and β7F3. Residues from both DCAF1-FBD β-hairpin strands participate in the binding to Merlin-FERM, mainly via hydrophobic interactions (Fig. 3A). The highly conserved hydrophobic residues (Val-1482 and Leu-1484 in βA and Ile-1501, Leu-1503, and Leu-1505 in βB) from DCAF1-FBD contact with the hydrophobic αβ-groove to form a hydrophobic core (Fig. 3, A and D), presumably stabilizing the β-hairpin folding of DCAF1-FBD as well as the Merlin·DCAF1 complex. Consistent with this analysis, the ITC-based measurements showed that the mutant with the deletion of either βA or βB in DCAF1-FBD failed to bind to Merlin-FERM (Table 2). Similarly, disruption of the hydrophobic interaction by substituting Leu-1503, which is central to the hydrophobic core formed by DCAF1-FBD and the αβ-groove of the F3 lobe, with lysine also abolished the binding of DCAF1 to Merlin (Table 2). In addition, several conserved salt bridges formed between negatively charged residues (Glu-1480 and Glu-1481) in DCAF1-FBD and positively charged residues (Arg-309 and Lys-312) in the F3 lobe also contribute to the FBD/F3 interaction (Fig. 3A). Fitting with this analysis, a charge-reversal mutation of DCAF1-FBD (E1480K/E1481K) largely impaired its binding to Merlin-FERM (Table 2).
FIGURE 3.
Structural and sequence analysis of the Merlin-FERM/DCAF1-FBD interaction. A, molecular details of the Merlin-FERM/DCAF1-FBD interaction. The residues involved in forming the FERM/FBD interface are shown as sticks. Hydrogen bonds and salt bridges are indicated by dashed lines. B and C, two examples showing that CD44 (Protein Data Bank code 2ZPY) and ICAM-2 (Protein Data Bank code 1J19) bind to the αβ-groove in the F3 lobe of Radixin (an ERM protein) as the single β-strand structures. We also measured the binding between the CD44 peptide and Moesin-FERM by ITC and found that their interaction is very weak (Kd >100 μm). D, sequence alignment of the αβ-groove region in the FERM domains from Merlin and ERM family members. E, sequence alignment of the FBD regions from DCAF1 proteins across different species. In these two alignments, residues that are absolutely conserved and highly conserved are highlighted in red and yellow, respectively. The secondary structural elements are indicated above the alignments. The residues shown in Fig. 3A are indicated by triangles in D and E, respectively. The disordered region in DCAF1-FBD is indicated by a dashed line in E.
TABLE 2.
ITC-based analysis of the interactions between Merlin-FERM and various DCAF1-FBD mutants
| DCAF1-FBD | Kd |
|---|---|
| μm | |
| WT | 2.5 ± 0.2 |
| Δ1478–1487 | Undetectable |
| Δ1501–1507 | Undetectable |
| Δ1490–1499 | 7.0 ± 2.1 |
| D1490A/D1493A/D1496A/E1498A/D1499A | 9.0 ± 1.8 |
| E1480K/E1481K | 12 ± 2 |
| L1484K | 22 ± 2 |
| L1503K | Undetectable |
The Negatively Charged Connecting Loop of the DCAF1-FBD Hairpin Is Likely to Be Involved in the Binding to Merlin-FERM
We were surprised to find that the most conserved region in DCAF1-FBD is the structurally disordered, β-hairpin connecting loop (Fig. 3E). This loop is rich in negatively charged residues as well as four serine/threonine residues, some of which are predicted to be potential phosphorylation sites of protein kinases such as CK2. Interestingly, a cleft nearby the αβ-groove and at the inter-face of the F1 and F3 lobes of Merlin-FERM is highly positively charged (Fig. 4A). A number of conserved residues from both the F1 and F3 lobes form this positively charged cleft (Figs. 3D and 4A). The binding of the DCAF1-FBD hairpin to the αβ-groove of the F3 lobe juxtaposes the negatively charged β-hairpin loop to the positively charged F1/F3 cleft of the FERM domain (Fig. 4A). It is rational to hypothesize that the charge-charge attraction between the DCAF1-FBD hairpin loop and Merlin-FERM F1/F3 cleft may further enhance the Merlin/DCAF1 interaction. To test this hypothesis, we deleted the connecting loop or mutated five negatively charged residues in the connecting loop to non-charged Ala (D1490A/D1493A/D1496A/E1498A/D1499A) and measured their binding affinity with Merlin-FERM and found that both of the mutants indeed showed decreased binding (albeit rather modestly) to Merlin-FERM (Table 2). It is possible that addition of negative charges by phosphorylation(s) of Ser/Thr within the loop may further enhance the interaction between DCAF1 and Merlin-FERM. We note with interest that the F1/F3 cleft of the ERM proteins (e.g. the Radixin FERM domain shown in Fig. 4B) is also positively charged. Importantly, the positively charged F1/F3 cleft of Radixin-FERM is known to bind to negatively charged phosphoinositol phosphates such as InsP3, which is involved in the activation of ERM proteins (35).
FIGURE 4.
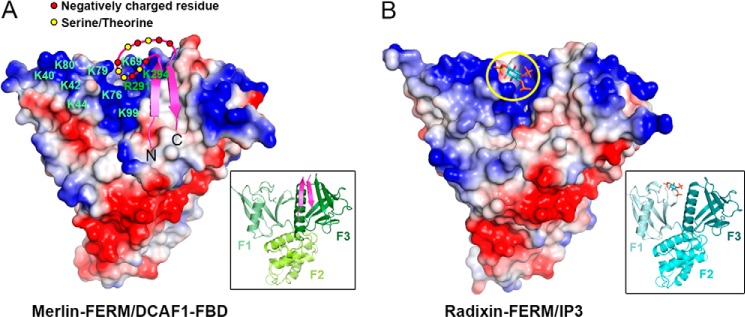
The negatively charged connecting loop of the DCAF1-FBD β-hairpin contributes to the binding to Merlin-FERM. A, the model of the connecting loop interacting with the highly positively charged F1/F3 cleft on Merlin-FERM. The figure was drawn using an electrostatic surface charge potential map with blue for the positive charge potential and red for the negative charge potential. The positively charged residues involved in forming the cleft are labeled. B, the structure of the FERM domain of Radixin in complex with the headgroup of phosphatidylinositol 4,5-bisphosphate (Protein Data Bank code 1GC6) was shown in the same presentation mode as A for comparison. The headgroup structure is highlighted by a yellow circle.
The Auto-inhibited and Structurally Closed Form of Merlin Cannot Bind to DCAF1-FBD
Similar with ERM proteins, Merlin is believed to adopt a closed conformation in solution, in which the C-terminal regulatory tail binds to the FERM domain and keeps the full-length Merlin in an auto-inhibited conformation (36). Based on the structure of auto-inhibited Moesin (24, 37) and in view of the very high amino acid sequence identity between Merlin and Moesin, it is generally accepted that the C-terminal tail of Merlin is likely to binding to its own FERM domain with a mode similar to that found in Moesin. If this model-based analysis were true, the auto-inhibitory tail binding site and the DCAF1-FBD binding site on the FERM domain partially overlap with each other. We predict that DCAF1-FBD is not expected to bind to the full-length, auto-inhibited Merlin, as the intra-molecular FERM head and inhibitory tail interaction would prevail. To test our hypothesis, we measured the binding affinity between the full-length Merlin protein (Merlin-FL) and DCAF1-FBD. Fully fitted with our structural analysis and prediction, Merlin-FL showed no detectable binding to DCAF1-FBD (Fig. 1F). These data indicate that the productive interaction between Merlin and DCAF1 will require Merlin to be activated (i.e. release of the tail inhibitory domain from the FERM domain) by certain regulatory factor(s). Our in vitro biochemical binding data obtained using highly purified recombinant proteins are in apparent odd with the conclusion drawn by Li et al. (9) (see “Discussion” for details). Curiously, the phosphorylation mimic S518D-mutant of the full-length Merlin (Merlin-FLS518D) also showed no detectable binding to DCAF1 (Fig. 1G), a finding that is consistent with that by Li et al. (9).
DISCUSSION
Although it has been studied for many years, the target binding mechanisms for Merlin remains elusive, partly due to the lack of the structural data of the full-length Merlin or Merlin-FERM in complex with its targets. Currently, much of the mechanistic interpretations of Merlin/target interactions are derived from the homology-based structural models based on the structures of ERM proteins as the templates. The crystal structure of Merlin-FERM in complex with DCAF1 provides the first atomic picture of how the FERM domain of Merlin recognizes its target. Specifically, Merlin-FERM uses the conventional αβ-groove in the F3 lobe as the binding site to interact with DCAF1. However, the binding mode between Merlin-FERM and DCAF1 is distinct from those of ERM proteins (22, 28). Merlin-FERM binds to DCAF1 with a much more extensive hydrophobic interface. In this novel binding mode, DCAF1 adopts a β-hairpin conformation with residues from both strands participating in binding to Merlin. Additionally, the binding of the DCAF1 β-hairpin to the αβ-groove of the F3 lobe positions the negatively charged β-hairpin loop closely to the positively charged FERM F1/F3 cleft and thereby enhances the Merlin/DCAF1 interaction. Together, our structural and biochemical analysis reveals a distinct target binding mode of Merlin-FERM.
The FERM domain of Merlin interacts with its own tail, and this intra-molecular interaction is believed to keep Merlin in an auto-inhibited conformation (36). Release of the auto-inhibition (e.g. by truncation of a part of its C-terminal tail) constitutively activates Merlin (8), indicating that the FERM domain-mediated activities of Merlin requires the release of the C-terminal tail-mediated auto-inhibition. Interestingly, phosphorylation of Ser-518 or substitutions of Ser-518 with Glu/Asp in the predicted helical region between the FERM domain and the inhibitory tail domain convert Merlin into a functionally less active state (36). This functionally less active state of Merlin is often equated to a conformationally more closed state, although no direct biochemical or structural evidences exist. We show, using highly purified proteins in vitro, that the WT C-terminal tail auto-inhibited Merlin FERM cannot bind to DCAF1 (Fig. 1F), and the full-length S518D Merlin cannot bind to DCAF1 either (Fig. 1G). The data indicate that the biologically less active S518D Merlin does not necessarily correspond to the conformational, more closed form of Merlin FERM domain. We hypothesize that additional factor(s) can sense the phosphorylation status of Ser-518 and thus regulate the biological activity of Merlin. Finding such Merlin regulatory factor(s) will be an important future topic in the Hippo signaling pathway.
The unique target binding specificities of the FERM domains are likely to account for a large part of functional differences between Merlin and ERM proteins. With this in mind, we carefully analyzed the DCAF1-FBD binding site on Merlin-FERM. Despite of large affinity difference in the bindings of DCAF1-FBD to the FERM domains of Merlin and Moesin (Fig. 1, C and D), the amino acid residues of the DCAF1-FBD binding site (i.e. the αβ-groove) in Merlin, are highly conserved across Merlin and ERM proteins except for a few amino acid residues (Fig. 3D). Paradoxically, substitution of these few residues in the F3 lobe of Merlin-FERM with the corresponding residues from Moesin (Y266F, E270K, D281P, V282D, K284V, N286Y, L295R, and Q298A) did not result in a loss of DCAF1 binding (data not shown). This result indicates that the differences in other regions of the F3 lobe or even in the F1 or F2 lobes contribute to the unique target binding specificities of FERM domains from Merlin and ERM proteins. Additional work is required to decode such target binding specificities of FERM domain proteins.
Acknowledgment
We thank the Shanghai Synchrotron Radiation Facility BL17U for x-ray beam time.
This work was supported by Research Grant Council of Hong Kong Grants 663610, 663811, 663812, HKUST6/CRF/10, SEG_HKUST06, AoE/M09/12, and T13-607/12R (to M. Z.).
- FBD
- FERM-binding domain
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 2. Morrison H., Sherman L. S., Legg J., Banine F., Isacke C., Haipek C. A, Gutmann D. H., Ponta H., Herrlich P. (2001) The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 15, 968–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trofatter J. A., MacCollin M. M., Rutter J. L., Murrell J. R., Duyao M. P., Parry D. M., Eldridge R., Kley N., Menon A. G., Pulaski K. (1993) A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell 72, 791–800 [DOI] [PubMed] [Google Scholar]
- 4. Rouleau G. A., Merel P., Lutchman M., Sanson M., Zucman J., Marineau C., Hoang-Xuan K., Demczuk S., Desmaze C., Plougastel B. (1993) Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature 363, 515–521 [DOI] [PubMed] [Google Scholar]
- 5. Li W., Cooper J., Karajannis M. A., Giancotti F. G. (2012) Merlin: a tumour suppressor with functions at the cell cortex and in the nucleus. EMBO Rep. 13, 204–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McClatchey A. I., Giovannini M. (2005) Membrane organization and tumorigenesis–the NF2 tumor suppressor, Merlin. Genes Dev. 19, 2265–2277 [DOI] [PubMed] [Google Scholar]
- 7. Grusche F. A., Richardson H. E., Harvey K. F. (2010) Upstream regulation of the hippo size control pathway. Curr. Biol. 20, R574–82 [DOI] [PubMed] [Google Scholar]
- 8. Yin F., Yu J., Zheng Y., Chen Q., Zhang N., Pan D. (2013) Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154, 1342–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li W., You L., Cooper J., Schiavon G., Pepe-Caprio A., Zhou L., Ishii R., Giovannini M., Hanemann C. O., Long S. B., Erdjument-Bromage H., Zhou P., Tempst P., Giancotti F. G. (2010) Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell 140, 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bretscher A., Edwards K., Fehon R. G. (2002) ERM proteins and merlin: integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 3, 586–599 [DOI] [PubMed] [Google Scholar]
- 11. McCartney B. M., Fehon R. G. (1996) Distinct cellular and subcellular patterns of expression imply distinct functions for the Drosophila homologues of moesin and the neurofibromatosis 2 tumor suppressor, merlin. J. Cell Biol. 133, 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yi C., Troutman S., Fera D., Stemmer-Rachamimov A., Avila J. L., Christian N., Persson N. L., Shimono A., Speicher D. W., Marmorstein R., Holmgren L., Kissil J. L. (2011) A tight junction-associated Merlin-angiomotin complex mediates Merlin's regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell 19, 527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kissil J. L., Wilker E. W., Johnson K. C., Eckman M. S., Yaffe M. B., Jacks T. (2003) Merlin, the product of the Nf2 tumor suppressor gene, is an inhibitor of the p21-activated kinase, Pak1. Mol. Cell 12, 841–849 [DOI] [PubMed] [Google Scholar]
- 14. Shimizu T., Seto A., Maita N., Hamada K., Tsukita S., Tsukita S., Hakoshima T. (2002) Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain. J. Biol. Chem. 277, 10332–10336 [DOI] [PubMed] [Google Scholar]
- 15. Kang B. S., Cooper D. R., Devedjiev Y., Derewenda U., Derewenda Z. S. (2002) The structure of the FERM domain of merlin, the neurofibromatosis type 2 gene product. Acta Crystallogr. D. Biol. Crystallogr. 58, 381–391 [DOI] [PubMed] [Google Scholar]
- 16. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 17. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 18. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 19. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 22. Mori T., Kitano K., Terawaki S., Maesaki R., Fukami Y., Hakoshima T. (2008) Structural basis for CD44 recognition by ERM proteins. J. Biol. Chem. 283, 29602–29612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tepass U. (2009) FERM proteins in animal morphogenesis. Curr. Opin. Genet. Dev. 19, 357–367 [DOI] [PubMed] [Google Scholar]
- 24. Pearson M. A., Reczek D., Bretscher A., Karplus P. A. (2000) Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell 101, 259–270 [DOI] [PubMed] [Google Scholar]
- 25. Takai Y., Kitano K., Terawaki S., Maesaki R., Hakoshima T. (2008) Structural basis of the cytoplasmic tail of adhesion molecule CD43 and its binding to ERM proteins. J. Mol. Biol. 381, 634–644 [DOI] [PubMed] [Google Scholar]
- 26. Takai Y., Kitano K., Terawaki S., Maesaki R., Hakoshima T. (2007) Structural basis of PSGL-1 binding to ERM proteins. Genes Cells 12, 1329–1338 [DOI] [PubMed] [Google Scholar]
- 27. Terawaki S., Kitano K., Hakoshima T. (2007) Structural basis for type II membrane protein binding by ERM proteins revealed by the radixin-neutral endopeptidase 24.11 (NEP) complex. J. Biol. Chem. 282, 19854–19862 [DOI] [PubMed] [Google Scholar]
- 28. Hamada K., Shimizu T., Yonemura S., Tsukita S., Tsukita S., Hakoshima T. (2003) Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex. EMBO J. 22, 502–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wegener K. L., Partridge A. W., Han J., Pickford A. R., Liddington R. C., Ginsberg M. H., Campbell I. D. (2007) Structural basis of integrin activation by talin. Cell 128, 171–182 [DOI] [PubMed] [Google Scholar]
- 30. Wei Z., Yan J., Lu Q., Pan L., Zhang M. (2011) Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain. Proc. Natl. Acad. Sci. U.S.A. 108, 3572–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghai R., Bugarcic A., Liu H., Norwood S. J., Skeldal S., Coulson E. J., Li S. S., Teasdale R. D., Collins B. M. (2013) Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins. Proc. Natl. Acad. Sci. U.S.A. 110, E643–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wegener K. L., Basran J., Bagshaw C. R., Campbell I. D., Roberts G. C., Critchley D. R., Barsukov I. L. (2008) Structural basis for the interaction between the cytoplasmic domain of the hyaluronate receptor layilin and the talin F3 subdomain. J. Mol. Biol. 382, 112–126 [DOI] [PubMed] [Google Scholar]
- 33. García-Alvarez B., de Pereda J. M., Calderwood D. A., Ulmer T. S., Critchley D., Campbell I. D., Ginsberg M. H., Liddington R. C. (2003) Structural determinants of integrin recognition by talin. Mol. Cell 11, 49–58 [DOI] [PubMed] [Google Scholar]
- 34. Hirano Y., Hatano T., Takahashi A., Toriyama M., Inagaki N., Hakoshima T. (2011) Structural basis of cargo recognition by the myosin-X MyTH4-FERM domain. EMBO J. 30, 2734–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hamada K., Shimizu T., Matsui T., Tsukita S., Hakoshima T. (2000) Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 19, 4449–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sher I., Hanemann C. O., Karplus P. A., Bretscher A. (2012) The tumor suppressor merlin controls growth in its open state, and phosphorylation converts it to a less-active more-closed state. Dev. Cell 22, 703–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Q., Nance M. R., Kulikauskas R., Nyberg K., Fehon R., Karplus P. A., Bretscher A., Tesmer J. J. (2007) Self-masking in an intact ERM-merlin protein: an active role for the central α-helical domain. J. Mol. Biol. 365, 1446–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]