Background: How faulty Okazaki fragments are repaired remains unclear.
Results: The DNA annealing activity of Rad52 and sister chromatid cohesion are important in the repair of faulty Okazaki fragments.
Conclusion: Rad52/Rad59-mediated sister chromatid recombination is a major means of repairing faulty Okazaki fragments.
Significance: The faithful repair of faulty Okazaki fragments is critical for genome stability.
Keywords: DNA Helicase, DNA Recombination, DNA Repair, DNA Replication, Homologous Recombination, Dna2, Rad52, Rad59, Genome Integrity, Lagging Strand Synthesis
Abstract
The correct removal of 5′-flap structures by Rad27 and Dna2 during Okazaki fragment maturation is crucial for the stable maintenance of genetic materials and cell viability. In this study, we identified RAD52, a key recombination protein, as a multicopy suppressor of dna2-K1080E, a lethal helicase-negative mutant allele of DNA2 in yeasts. In contrast, the overexpression of Rad51, which works conjointly with Rad52 in canonical homologous recombination, failed to suppress the growth defect of the dna2-K1080E mutation, indicating that Rad52 plays a unique and distinct role in Okazaki fragment metabolism. We found that the recombination-defective Rad52-QDDD/AAAA mutant did not rescue dna2-K1080E, suggesting that Rad52-mediated recombination is important for suppression. The Rad52-mediated enzymatic stimulation of Dna2 or Rad27 is not a direct cause of suppression observed in vivo, as both Rad52 and Rad52-QDDD/AAAA proteins stimulated the endonuclease activities of both Dna2 and Rad27 to a similar extent. The recombination mediator activity of Rad52 was dispensable for the suppression, whereas both the DNA annealing activity and its ability to interact with Rad59 were essential. In addition, we found that several cohesion establishment factors, including Rsc2 and Elg1, were required for the Rad52-dependent suppression of dna2-K1080E. Our findings suggest a novel Rad52/Rad59-dependent, but Rad51-independent recombination pathway that could ultimately lead to the removal of faulty flaps in conjunction with cohesion establishment factors.
Introduction
DNA replication, repair, and recombination are intricately networked to coordinate their functions for the stable maintenance of a vast amount of genetic materials. Most factors involved in these processes display a large number of physical and genetic interactions in a complicatedly interwoven manner (1–7). Lagging strand DNA synthesis is one such notable example that depends heavily on the collaborative actions of a number of proteins involved in all three processes because it is associated with a great risk of formation of a variety of aberrant DNA structures (8–11). Unlike leading strand DNA synthesis, lagging strand DNA synthesis proceeds discontinuously via a series of linked events that include the generation of Okazaki fragments, the removal of primer RNAs, and ligation as the final step (8, 12–14). In eukaryotes, the synthesis of an Okazaki fragment is initiated by DNA polymerase (Pol)2 α-primase, which synthesizes ∼10 nt of RNA followed by the addition of short stretches of DNA. The second DNA polymerase, Pol δ, elongates the nascent primer RNA-DNA to form a new Okazaki fragment with the aid of proliferating cell nuclear antigen (PCNA) and replication factor C (15). As Pol δ, a lagging strand DNA polymerase, encounters the previously synthesized downstream Okazaki fragment, it displaces the 5′-end region of the downstream fragment, creating a flap. The flaps are then removed by the combined action of Dna2 and Rad27, the two critical 5′-flap endonucleases, to create nicks that are sealed by Cdc9, a DNA ligase, to complete Okazaki fragment maturation (16–20). Considering that an extraordinary number of Okazaki fragments (for example, 2 × 107 in humans) are produced and processed per cell cycle, it would be highly deleterious to the cell if faulty processing of Okazaki fragments was not efficiently repaired.
Among the enzymes involved in the maturation of Okazaki fragments, Dna2 appears to link diverse DNA metabolisms by playing multiple roles in Okazaki fragment processing (11, 17, 21, 22), long-range resection of double strand break (DSB) repair (23–26), and S-phase checkpoint activation (27, 28). It has been shown that yeast Dna2 consists of three domains, each of which encodes a distinct biochemical activity: helicase, endonuclease, and secondary structure-specific DNA binding activity (28–32). The endonuclease activity of Dna2 is essential (33, 34), whereas other activities such as DNA helicase and structure-specific DNA binding activities are dispensable for cell survival under some growth conditions (31, 35). For example, the Dna2 helicase activity, although normally essential, becomes dispensable when cells are grown in the presence of poor carbon sources such as lactate or glycerol (35). Cells with the dna2Δ405N mutant allele lacking the N-terminal 405-amino acid (aa) display temperature-sensitive (ts) growth defects (31). The endonuclease activity of Dna2 is critical for both Okazaki fragment processing and DSB resection (23, 24), whereas the N-terminal 405-aa domain of Dna2 is required for the checkpoint function and also for DSB resection to some extent (27, 36). This N-terminal domain of Dna2 becomes critical in the processing of secondary structured flaps in Okazaki fragment (28). In contrast to the multiple functions of the endonuclease and N-terminal DNA binding activities of Dna2, the helicase activity of Dna2 seems to be involved exclusively in DNA replication, most likely in the processing of secondary structure or higher order structure flaps present in a subset of Okazaki fragments (23, 27, 32, 36, 37). The failure to deal with this problem could result in DNA damages, checkpoint activation, and cell death. Therefore, the coordinated action of lagging strand replication and DNA repair would be important for the maintenance of genome integrity.
Rad52 is a key recombination protein that possesses two important functions in homologous recombination (HR). First, it facilitates the formation of the Rad51 nucleoprotein filament by replacing RPA with Rad51 (38, 39), which is termed the “recombination mediator activity” of Rad52. The C-terminal 409–420-amino acid region of Rad52, which is responsible for its interaction with Rad51, is identified as crucial for the recombination mediator function of Rad52 (40, 41). Second, Rad52 has single-stranded (ss) DNA annealing activity in its N-terminal region (42–44), which is required for the second-end capture during the canonical HR event (45) or other alternative recombination events such as synthesis-dependent strand annealing (46, 47) and single strand annealing (SSA) (43, 48). Located in the central part of Rad52, the RPA-interacting domain is responsible for the displacement of RPA from ssDNA (49, 50). The RPA displacement function of Rad52 is important for both HR mediator (50) and ssDNA annealing activities (51) because the RPA-coated ssDNA is inhibitory to Rad51-ssDNA filament formation or DNA annealing between two complementary ssDNA.
It has been suggested that HR works as a backup pathway for faulty Okazaki fragment processing. Previous genetic data show that rad27Δ, which is hyper-recombinogenic by itself, exhibits synthetic lethal phenotypes in combination with deletions of the genes involved in HR, implying important roles for HR in the repair of faulty Okazaki fragments (52, 53). In addition, the genetic data showing that dna2-1 rad52Δ and dna2-2 rad52Δ double mutants are synthetic lethal (9), whereas dna2-22 and dna2Δ405N single mutants show increased rates of recombination, are also consistent with the above possibility (54, 58). In this article, we have identified the RAD52 gene as a suppressor of the dna2-K1080E mutant in Saccharomyces cerevisiae that lacks ATPase/helicase activity (30, 55). The suppression was dependent on both the strand annealing activity and the ability of Rad52 to interact with Rad59 but not its recombination mediator activity. Thus, a Rad52/Rad59-dependent, but Rad51-independent, recombination pathway appears crucial for Okazaki fragment metabolism. This pathway also requires several factors for cohesion establishment. We will discuss the interplay among these factors for faithful lagging strand DNA synthesis.
MATERIALS AND METHODS
Strains
The genotypes of the S. cerevisiae strains used in this study are summarized in Table 1. All strains are haploid and were derived from YPH499. YJA2 has the dna2Δ405N allele that lacks the N-terminal 405-aa domain, whereas YJA1B lacks a chromosomal version of DNA2 but harbors pRS316-DNA2, a low copy plasmid with URA3 as a selection marker expressing wild type Dna2. YJA1BJK65, a derivative of YJA1B, possesses pRS314-dna2-K1080E, a low copy plasmid with TRP1 as a selection marker containing helicase-negative dna2-K1080E. YMJ1 is a derivative of YJA1BJK65 that harbors pRS325-ADH1-RAD52, a multicopy plasmid with LEU2 as a selection marker expressing Rad52. The RAD50, RAD51, RAD54, RAD55, RAD57, RAD59, ELG1, RSC2, and RSC7 genes were deleted separately from YMJ1 by replacing the genes with KanMX4 as described previously (56) and termed YMJ2 to YMJ10, respectively. The dna2-1, -4, -5, -7, -8, -9, -12, -13, -15, and -16 mutant alleles were isolated previously (35).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| YPH499 | MATa ade2–101 ura3–52 lys2–801 trp1–63 his3–200 leu2-Δ1 GAL+ | Ref. 103 |
| YJA2 | MATa ade2–101 ura3–52 lys2–801 trp1–63 his3–200 leu2-Δ1 GAL + dna2Δ405N | Ref. 31 |
| YJA1B | MATa ade2–101 ura3–52 lys2–801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL + dna2::HIS3 [pRS316-DNA2] | Ref. 58 |
| YJA1BJK65 | MATa ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 [pRS316-DNA2] [pRS314-dna2K1080E] | Ref. 58 |
| YMJ1 | MATa ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 [pRS316-DNA2] [pRS314-dna2K1080E] [pRS325-ADH1-RAD52] | This study |
| YMJ2 | MATa ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 rad50::KanMX4 [pRS316-DNA2] [pRS314-dna2K1080E] [pRS325-ADH1-RAD52] | This study |
| YMJ3 | MATa ade2–101 ura3–52 lys2–801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 rad51::KanMX4 [pRS316-DNA2] [pRS314-dna2K1080E] [pRS325-ADH1-RAD52] | This study |
| YMJ4 | MATa ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 rad54::KanMX4 [pRS316-DNA2] [pRS314-dna2K1080E] [pRS325-ADH1-RAD52] | This study |
| YMJ5 | MATa ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 rad55::KanMX4 [pRS316-DNA2] [pRS314-dna2K1080E] [pRS325-ADH1-RAD52] | This study |
| YMJ6 | MATa ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 rad57::KanMX4 [pRS316-DNA2] [pRS314-dna2K1081E] [pRS325-ADH1-RAD52] | This study |
| YMJ7 | MATa ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 rad59::KanMX4 [pRS316-DNA2] [pRS314-dna2K1082E] [pRS325-ADH1-RAD52] | This study |
| YMJ8 | MATa ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 elg1::KanMX4 [pRS316-DNA2] [pRS314-dna2K1082E] [pRS325-ADH1-RAD52] | This study |
| YMJ9 | MATa ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 rsc2::KanMX4 [pRS316-DNA2] [pRS314-dna2K1083E] [pRS325-ADH1-RAD52] | This study |
| YMJ10 | MATa ade2-101 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 GAL+ dna2::HIS3 rsc7::KanMX4 [pRS316-DNA2] [pRS314-dna2K1083E] [pRS325-ADH1-RAD52] | This study |
Chemicals, Nucleotides, and Enzymes
Methyl methanesulfonate was obtained from Sigma-Aldrich. Restriction endonucleases and DNA polymerases for PCR were purchased from either Enzynomics (Daejeon, Korea) or New England Biolabs (Beverly, MA). The pRS plasmids were purchased from New England Biolabs. The pET28 vectors used for protein expression in Escherichia coli were from Novagen (Darmstadt, Germany). Isopropyl β-d-1-thiogalactopyranoside and X-Gal were from ElpisBiotech (Daejeon, Korea). Imidazole was from Acros Organics (Geel, Belgium). A uracil analog, 5-fluorooritic acid, was obtained from Duchefa Biochemie (Haarlem, Netherlands). Antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX), Sigma-Aldrich, and Abcam (Cambridge, UK) or kindly provided (anti-H3 polyclonal antibodies) by Dr. Daeyoup Lee (Korea Advanced Institute of Science and Technology). The antibodies used are shown in Figs. 1, 3, and 4. Rad27, the yeast Fen1, was purified as described previously (17). Oligonucleotides and α-factor used in this study were synthesized commercially by Genotech (Daejeon, Korea) and Peptron (Deajeon, Korea), respectively.
FIGURE 1.
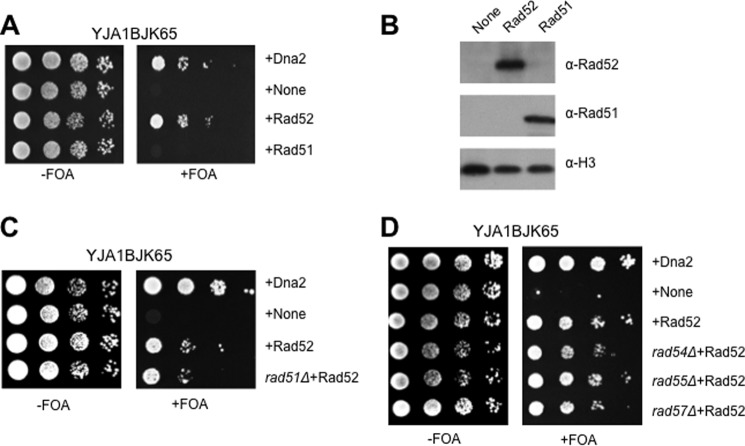
Rad52 suppresses the lethality of the helicase-deficient dna2-K1080E mutation in a Rad51-independent manner. A, YJA18JK65 (dna2::HIS3, pRS316-DNA2, and pRS314-dna2-K1080E) was transformed with pRS325-ADH1 plasmids expressing Dna2 (positive control), none (negative control), Rad52, or Rad51. Transformants were inoculated in SD-HWL liquid medium (2 ml) and grown for 2–3 days until saturated. Each saturated culture was spotted in 10-fold serial dilutions on SD-HWL plates without (−FOA) or with (+FOA) 5-fluorooritic acid (0.1%) followed by incubation for 5–6 days at 25 °C. B, expressions of Rad51 and Rad52 in each strain used in A were examined by Western blot using anti-Rad51 (α-Rad51; sc-133089, Santa Cruz Biotechnology) or anti-Rad52 antibodies (α-Rad52; sc-8939, Santa Cruz Biotechnology). Whole cell extracts were prepared by TCA precipitation as described under “Materials and Methods.” Histone H3 was used as the loading control. α-H3, antibodies against histone H3. C and D, the Rad52-dependent suppression of dna2-K1080E was examined in mutant cells lacking a gene belonging to the RAD52-epistatic group; Rad52 overexpression in rad51-null cells (C, indicated as rad51Δ+Rad52); Rad52 overexpression in rad54-, rad55-, and rad57-null cells (D, indicated as rad54Δ+Rad52, rad55Δ+Rad52, and rad57Δ+Rad52, respectively). Cells were grown and spotted as described for A.
FIGURE 3.
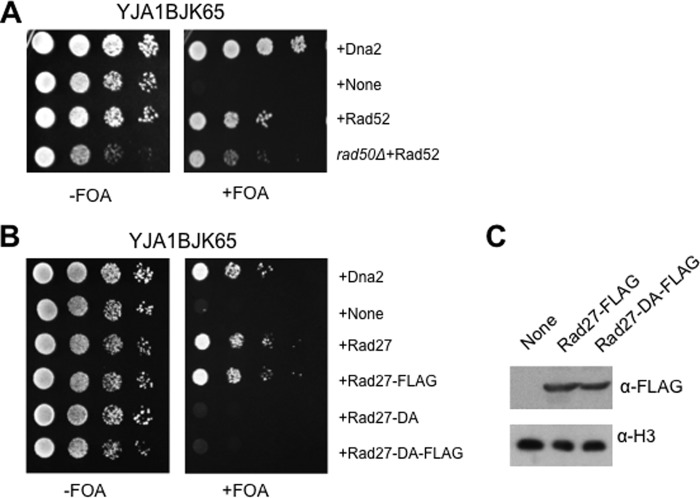
Defects of dna2-K1080E can be corrected by active Rad27. A, the Rad52-dependent suppression of dna2-K1080E was examined in rad50-null cells. Overexpression of Rad52 in rad50-null cells is indicated as rad50Δ+Rad52. Cells were grown and spotted as described in the legend for Fig. 1A. FOA, 5-fluorooritic acid. B, the YJA1BJK65 cells were transformed with pRS325-ADH1 plasmids expressing Rad27, Rad27-FLAG (Rad27 with a FLAG tag fused to its C terminus), Rad27-DA (Rad27-D179A; catalytically defective), and Rad27-DA-FLAG. Transformants were grown and spotted as described above in the legend for Fig. 1A. C, expression levels of Rad27-FLAG and Rad27-DA-FLAG were examined by Western blot using anti-FLAG antibody (α-FLAG; F-3165, Sigma-Aldrich). Histone H3 was used as the loading control. α-H3, antibodies against histone H3. The experiment was done as described in the legend for Fig. 1B.
FIGURE 4.
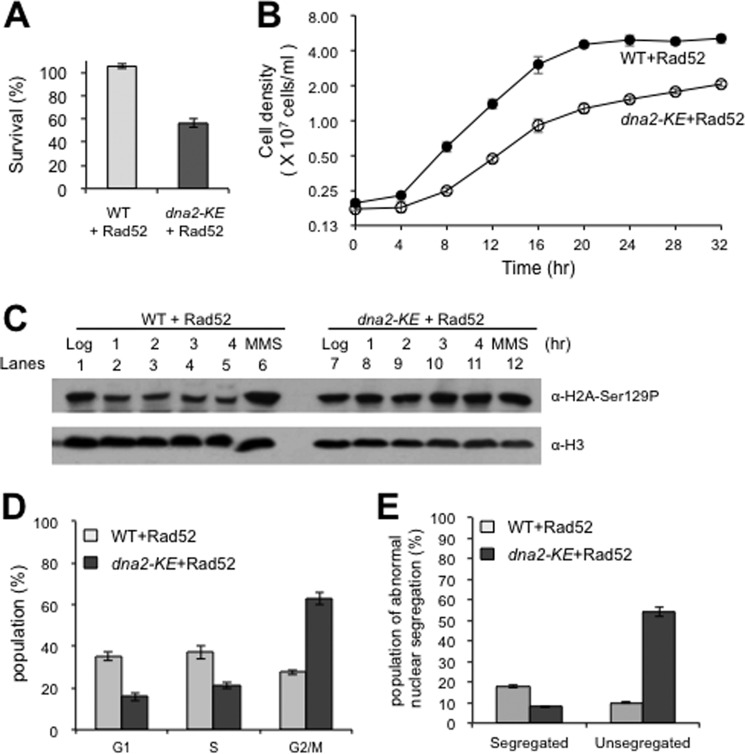
Overexpression of Rad52 rescues the lethality of dna2-K1080E, but it still displays growth defects. A and B, the survival (A) and growth (B) rates of wild type (WT+Rad52) and dna2-K1080E (dna2-KE+Rad52) cells overexpressing Rad52 were measured as described under “Materials and Methods.” The experiment was repeated three times, and the results are presented with error bars. C, the levels of H2A-Ser129 phosphorylation were examined. The logarithmically growing cells were treated with α-factor (3 μg/ml) followed by incubation at 25 °C for 4 h. The cells were withdrawn at the indicated time point, and the total cellular proteins were prepared as described under “Materials and Methods” followed by immunoblotting with α-H2A-Ser129P antibodies raised against phosphorylated serine 129 residue of H2A (ab17353, Abcam). Cells grown for 1 h in the presence of 0.033% methyl methanesulfonate were used as a positive control for the detection of phosphorylated H2A-Ser129. Histone H3 was used as loading control. α-H3, antibodies against histone H3. D, the relative distribution of the G1-, S-, and G2/M-phase cells in logarithmically growing cells was determined using microscopic analyses. Cells were categorized into three groups by their budding states: G1, unbudded cells; S, budding cells; G2/M, dumbbell-shaped cells. The experiment was repeated three times, and the results are presented with error bars. E, the morphology of nuclei was examined microscopically using cells stained with 4′,6-diamidino-2-phenylindole. The population of G2/M-phase cells with normal (two nuclei, segregated) and defective nuclear segregation (one nucleus, unsegregated) was determined. The experiment was repeated three times, and the results are presented with error bars.
Alkaline Protein Extraction from S. cerevisiae
Expression levels of proteins in yeast were determined by alkaline protein extraction (57). Briefly, cells (5 × 107, total) were harvested and resuspended in lysis buffer (150 μl) (1.85 m NaOH and 7.5% β-mercaptoethanol) followed by incubation on ice for 10 min. Trichloroacetic acid (50% v/v, 150 μl) was added, and the mixture was incubated on ice for an additional 10 min. Samples were then centrifuged at 4 °C for 10 min at 14,000 rpm. Supernatants were discarded, and 1× SDS sample buffer (50 mm Tris-HCl, pH 7.5, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, and 0.01% bromphenol blue) was added to the precipitated materials. The samples were then neutralized with 1–2 μl of 2 m Tris-HCl, pH 8.0, boiled for 10 min, and subjected to electrophoresis in a 10% (Tris-glycine) SDS-PAGE in Tris-glycine buffer (25 mm Tris, 250 mm glycine, and 0.1% SDS, pH 8.5). Histone H3 used as a loading control was analyzed by a 10% (Tris-Tricine) SDS-PAGE using Tris-Tricine buffer (cathode buffer: 100 mm Tris, 100 mm Tricine, 0.1% SDS, pH 8.3; anode buffer: 200 mm Tris, pH 8.8). The levels of protein expression were examined by Western blot using antibodies as described for each experiment.
Purification of Recombinant Rad52 and Rad52-QDDD/AAAA
The RAD52 ORF was inserted into the pET28 vector, which expresses the inserted ORF as an N-terminal His6-tagged protein. The resulting pET28-RAD52 plasmid was transformed into E. coli BL21 (CodonPlus-RIL), and cells (0.5 liter) were grown at 25 °C until the A600 value reached 0.5. At this point, cells were induced with 1 mm isopropyl β-d-1-thiogalactopyranoside (final concentration) for 4 h. Cells (1 liter) were harvested by centrifugation, and the cell pellet was resuspended in 30 ml of buffer T200 (50 mm Tris-HCl, pH 8.0, 200 mm NaCl, 10% glycerol, 0.1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 0.1 mm benzamidine, 1.25 μg/ml leupeptin, and 0.625 μg/ml pepstatin A). The subscript number in T200 indicates the concentration of NaCl in mm. Then, cells were sonicated and centrifuged at 45,000 rpm for 30 min. The supernatant was loaded onto a 4-ml SP Sepharose column (GE Healthcare) that was pre-equilibrated with T200. The proteins were then eluted by a linear gradient (100 ml) of T200 to T800. Fractions that possessed protein peaks of Rad52 were collected and adjusted to 10 mm imidazole (final concentration) and 500 mm NaCl (final concentration) followed by loading onto a 1-ml Ni2+-nitrilotriacetic acid-agarose column (Qiagen, Valencia, CA) pre-equilibrated with T500 plus 10 mm imidazole. The column was then washed successively with 10-column volumes of T500 plus 10 mm imidazole and T500 plus 50 mm imidazole. The protein was eluted with T500 plus 500 mm imidazole. The eluate was subjected to glycerol gradient sedimentation (5 ml, 15–35% glycerol in buffer T500) at 45,000 rpm for 24 h in a SW55 Ti rotor. Fractions (250 μl each) were collected from the bottom of the tube. Peak fractions were stored at −80 °C. The vector to express Rad52-QDDD/AAAA was constructed similarly, and the proteins were purified using the same procedure as described above.
Preparation of DNA Substrates
The preparation of DNA substrates and their labeling at the 5′-end were as described (16). The structure and position of radioisotopic labels (indicated by asterisks in D and E) in substrates are shown in Fig. 5. To construct 5′-flap-structured substrates, the two oligonucleotides (referred to as upstream and downstream) were annealed to the same template oligonucleotide. The sequences of oligonucleotides used were as follows: the template oligonucleotide, 5′-GAA AAC ATT ATT AAT GGC GTC GAG CTA GGC ACA AGG CGA ACT GCT AAC GG-3′ (50 mer); the upstream oligonucleotide for Dna2 substrate, 5′-CCG TTA GCA GTT CGC CTT GTG CCT A-3′ (25 mer); the upstream oligonucleotide for Rad27 substrate, 5′-CCG TTA GCA GTT CGC CTT GTG CCT AG-3′ (26 mer); the downstream oligonucleotide for Dna2 substrate, 5′-TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT CGG ACG CTC GAC GCC ATT AAT AAT GTT TTC-3′ (60 mer); and the downstream oligonucleotide for Rad27 substrate, 5′-CGA ACA ATT CAG CGG CTT TAA CCG GAC GCT CGA CGC CAT TAA TAA TGT TTT C-3′ (52 mer).
FIGURE 5.
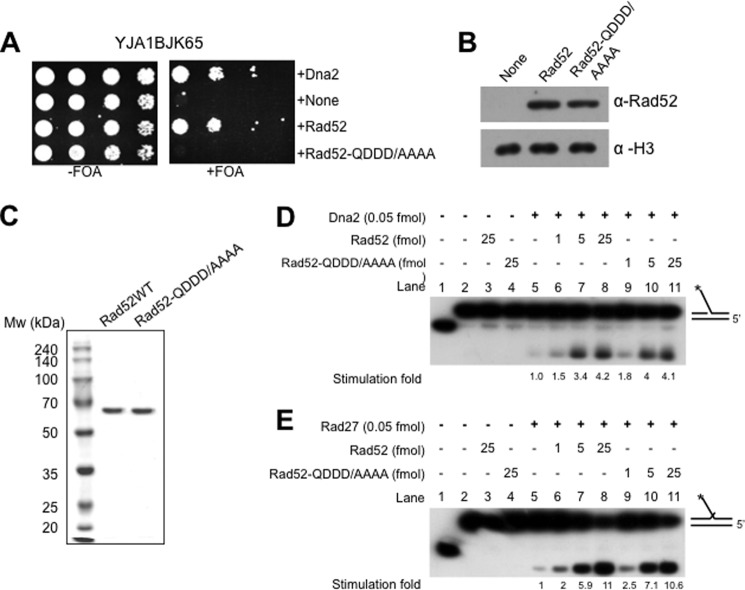
Rad52 stimulates endonuclease activities of Dna2 and Rad27. A, the pRS325-ADH1 plasmids expressing Dna2, Rad52, and Rad52-QDDD/AAAA were introduced into YJA1BJK65. None, empty vector (negative control). Transformants were grown and spotted as described in the legend for Fig. 1A. B, expression levels of wild type (Rad52) and mutant (Rad52-QDDD/AAAA) proteins were examined as described in the legend for Fig. 1B. C, SDS-PAGE analysis of purified recombinant Rad52 and Rad52-QDDD/AAAA proteins. Both proteins were purified as described under “Materials and Methods.” D and E, purified Rad52 proteins stimulate endonuclease activities of Dna2 and Rad27 in vitro. The substrates used in this experiment are indicated at the right of each panel. The asterisk represents the position of 32P radiolabel. The reaction mixtures were assembled with components as indicated above the gel and incubated at 37 °C for 30 min. The additions and omissions are indicated + and − signs, respectively. Cleavage products were analyzed on a 10% denaturing polyacrylamide gel and quantified as described under “Materials and Methods.” The stimulation fold obtained (with respect to Dna2 or Rad27 alone) by the addition of Rad52 proteins is indicated below the gel.
Nuclease Assays
Standard nuclease assays were performed in reaction mixtures (20 μl) containing 50 mm Tris-HCl, pH 7.8, 2 mm MgCl2, 2 mm dithiothreitol, 0.25 mg/ml bovine serum albumin, and DNA substrate (15 fmol). Reactions were incubated at 37 °C for 15 min followed by the addition of 4 μl of 6× stop solution (60 mm EDTA, pH 8.0, 40% sucrose, 0.6% SDS, 0.25% bromphenol blue, and 0.25% xylene cyanol). The products were subjected to electrophoresis for 30 min at 150 V in 0.5× TBE (45 mm Tris, 45 mm boric acid, and 1 mm EDTA). The gels were dried on a DEAE-cellulose paper and autoradiographed. Labeled DNA products were quantified with the use of a phosphorimaging device (Bas-1500, Fujifilm).
Measurement of Viability, Growth, and Doubling Time
Cells were grown to saturation at 25 °C for 2 days, and the cell density was measured by the use of a hemocytometer. The percentage of viable cells in the culture was determined as follows. Cells were diluted appropriately so that 400 cells were spread onto YPD plates followed by incubation at 25 °C for 3–4 days. The number of colonies formed on YPD plates was determined. Although the Rad52-mediated suppression of dna2-K1080E allowed cells to survive, the rescued cells were not as healthy as the wild type and were significantly larger. Therefore, the spectrophotometric determination of doubling time could not be accurate. Thus, we decided to determine the doubling time of live cells as follows. The culture of saturated cells obtained above was diluted (2 × 106 cells/ml) to a fresh medium followed by incubation at 25 °C. Samples were taken at 4-h intervals, properly diluted, and spread onto YPD plates followed by incubation at 25 °C for 3–4 days. Cell densities were determined based on the number of colonies formed. Doubling time was calculated using the software provided by Doubling Time. Each experiment was repeated three to four times, and the results are presented with error bars (Fig. 4B).
RESULTS
Rad52 Acts as a Multicopy Suppressor of dna2-K1080E
As a means of discovering novel factors involved in Okazaki fragment processing, we have continued to identify genetic suppressors that can rescue the growth defects of dna2 mutations. We isolated a DNA fragment containing the open reading frame of RAD52 that suppressed the lethal phenotype of dna2-K1080E, a helicase-defective allele of DNA2 (32, 55) using the same screening method as described previously (58–60). Subsequent subcloning analyses using the multicopy pRS325-ADH1 vector (containing the 2μ origin and constitutive ADH1 promoter) confirmed that overexpression of Rad52 alone was sufficient and necessary to suppress dna2-K1080E mutation (Fig. 1A). Interestingly, overexpression of Rad51, which is loaded onto ssDNA by Rad52 to form a nucleofilament for canonical HR, failed to suppress dna2-K1080E (Fig. 1A). We examined the expression levels of Rad51 and Rad52 by Western blotting using monoclonal and polyclonal antibodies, respectively, against native forms of each protein, and we detected both Rad51 and Rad52 proteins when they were expressed exogenously (Fig. 1B).
A marked difference in the abilities of Rad51 and Rad52 to suppress dna2-K1080E prompted us to further verify that the suppression is independent of RAD51. To this end, we overexpressed Rad52 in the rad51-null strain (termed YMJ3; see Table 1 for genotype) and found that the suppression is independent of RAD51 (Fig. 1C). This result suggests that the suppression observed is primarily due to a function of Rad52. We also investigated whether RAD54, RAD55, and RAD57, which work cooperatively with Rad51 in Rad51-dependent HR, are also involved in the suppression. Rad54 interacts with Rad51 nucleoprotein filaments and promotes the unwinding of duplex DNA to facilitate pairing between the recipient and the incoming ssDNA (3, 47). Rad55 forms a heterodimer complex with Rad57, and this complex stabilizes Rad51 nucleoprotein filaments (3, 47). Considering that they all are related to a Rad51 function, it is predicted that overexpression of Rad52 is able to suppress dna2-K1080E in the absence of Rad54, Rad55, or Rad57. To address this issue, we examined the influence of Rad52 overexpression in the rad54-, rad55-, and rad57-null strains (YMJ4, YMJ5, and YMJ6, respectively). Consistent with this prediction, we found that the Rad52-mediated suppression was not affected by the deletion of RAD54, RAD55, and RAD57 (Fig. 1D). These observations indicate that the suppression is independent of the formation of Rad51 nucleoprotein filament and its invasion to homologous sequence.
Rad52 Fails to Suppress the N-terminally Deleted dna2 but Is Able to Suppress Mutant dna2 Alleles in Catalytic Domains
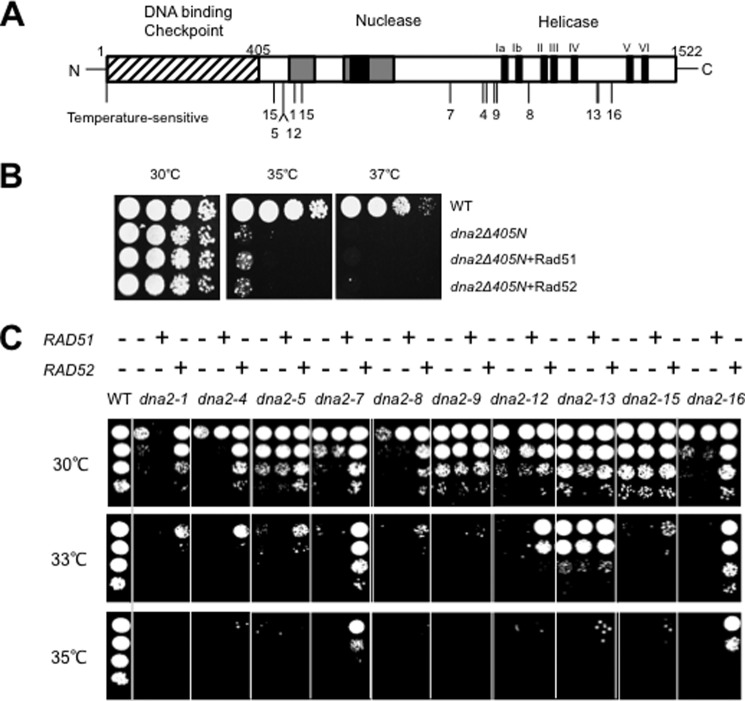
To confirm that the suppression is not restricted to the helicase-negative mutant dna2 allele, we examined whether all ts mutant alleles of dna2 isolated previously (35) could be suppressed by overexpression of Rad52 but not Rad51. These ts dna2 mutants contained one or two aa substitutions, largely in the nuclease or helicase domain, and some of them (dna2-1, dna2-8, and dna2-16) displayed methyl methanesulfonate sensitivity (Fig. 2A). In this test, we also included the dna2Δ405N allele, which is devoid of the N-terminal 405-aa domain that confers a ts phenotype. Neither Rad51 nor Rad52 suppressed the ts growth defect of dna2Δ405N upon overexpression (Fig. 2B). It was shown that the N-terminal 405-aa deletion is not a loss-of-function mutation in enzymatic activity; the Dna2Δ405N mutant enzyme is catalytically as active or more active than wild type Dna2 under some circumstances (28, 31). This observation suggests that the types of DNA damages caused by dna2Δ405N are not critically dependent on the elevated levels of Rad51 or Rad52 for repair. In contrast, most ts dna2 mutations mapped in the catalytic domains were suppressed specifically by overexpression of Rad52 but not by Rad51, although the levels of suppression varied (Fig. 2C). Among them, the dna2-7 and dna2-16 mutants were most efficiently rescued by Rad52 overexpression (Fig. 2C). These findings indicate that the Rad52-dependent, but Rad51-independent, pathway contributes to the repair of DNA damages that could occur in catalytically defective dna2 cells.
FIGURE 2.
Overexpression of Rad52 suppresses temperature sensitivity of dna2 mutant alleles. A, a schematic representation of Dna2. Locations of three domains and various ts dna2 alleles isolated previously by others (35) are as indicated. The mutant alleles include dna2-1 (P504S), -4 (D1015N P1031L), -5 (H471Y), -7 (G913D), -8 (H1129Y), -9 (A1036V P1031S), -12 (H471Y), -13 (P1311L, T1312I), -15 (G446A, R521K), and -16 (G1350E). Amino acid substitutions are indicated in parentheses. The N-terminal 405-aa domain is indicated by a hatched box. The conserved parts of the Dna2 nuclease domain are indicated by gray boxes. The black box in the nuclease domain denotes the RecB homology region. The seven motifs (denoted Ia to VI) common to helicase superfamily I are indicated by black strips. B, overexpression of Rad51 and Rad52 was examined in dna2Δ405N cells. The pRS325-ADH1, pRS325-ADH1-RAD51, and pRS325-ADH1-RAD52 plasmids were introduced in dna2Δ405N cells (indicated as dna2Δ405N dna2Δ405N+RAD51, and dna2Δ405N+RAD52, respectively). Transformants were grown in liquid SD-L medium and spotted in 10-fold serial dilutions in YPD plates followed by incubation at 30, 35, and 37 °C for 3 days. C, pRS325-ADH1-RAD51 or pRS325-ADH1-RAD52 plasmid was introduced into the ts dna2 mutants shown in A. Transformants were grown in liquid SD-L medium and spotted in 10-fold serial dilutions in YPD plates and incubated at 30, 33, and 35 °C for 3 days.
Overexpression of Rad52 Suppresses the Defects Associated Specifically with Processing of Okazaki Fragments
Dna2 plays multiple roles during DNA metabolisms by participating in the processing of Okazaki fragments, long-range resection of DSB repair, and S-phase checkpoint activation. Among these, Okazaki fragment processing is an essential process, depending on the coordinated interplay between the N-terminal DNA-binding domain and the endonuclease and helicase activities of Dna2. Because the helicase activity of Dna2 was shown to be dispensable for DSB resection or checkpoint activation as described above, it is assumed that Dna2 helicase activity participates exclusively in DNA replication by resolving the structured flaps present in a subset of Okazaki fragments. Nevertheless, it is still possible that the helicase function of Dna2 could contribute to DSB resection under specific circumstances, for example, when DSB occurs in structure-forming sequences. However, we wanted to confirm that the lethality of dna2-K1080E is not caused by the failure of DSB resection in the absence of the helicase activity of Dna2. To this end, we constructed a dna2-K1080E rad50Δ strain (YMJ2). In the absence of a functional copy of RAD50, Dna2 could not be recruited to the sites of DSB for the subsequent long-range resection, because the Mre11-Rad50-Xrs2 (MRX) complex bound to DSB provides a structural platform to recruit the Sgs1-Dna2 resection machinery (61). As shown in Fig. 3A, the overexpression of Rad52 allowed the dna2-K1080E rad50Δ double mutant cells to grow, suggesting that the suppression of dna2-K1080E by Rad52 is independent of the MRX complex required for the Dna2-mediated long-range resection. It should be noted that the control rad50Δ (rad50Δ dna2Δ + pRS316-DNA2 + pRS314-dna2-K1080E) cells displayed a slight growth defect compared with RAD50 control cells. Hence, we concluded that the Rad52-dependent suppression of dna2-K1080E occurs in the absence of Dna2-mediated DSB resection.
Then we examined whether RAD27, encoding the alternative nuclease yeast FEN1 for 5′-flap cleavage during Okazaki fragment processing, could rescue the lethality of dna2-K1080E upon overexpression. If the lethality of dna2-K1080E was attributed to the reduced efficiency of Okazaki fragment processing, overexpression of Rad27 could suppress the lethal phenotype of dna2-K1080E. In support of this assumption, overexpression of Rad27 (and Rad27-FLAG, a C-terminally FLAG epitope-tagged Rad27) efficiently suppressed the lethality of dna2-K1080E (Fig. 3B). In contrast, overexpression of Rad27-D179A (Rad27DA, a catalytically dead enzyme with an Asp to Ala substitution at amino acid 179) (62–64) could not suppress dna2-K1080E (Fig. 3B), although the expression level of this mutant protein was comparable with that of wild type Rad27 (Fig. 3C). Our finding that the catalytically active Rad27 only was able to suppress dna2-K1080E underscores the importance of endonucleolytic cleavage of the 5′-flap. Therefore, we concluded that the helicase function of Dna2 contributes critically to Okazaki fragment processing and that the lethality of dna2-K1080E is most likely caused by defective Okazaki fragment processing.
Overexpression of Rad52 Allows dna2-K1080E Cells to Grow but Does Not Fully Restore Their Defects
To investigate the influence of Rad52 overexpression on cell growth, we examined the viability of wild type and dna2-K1080E cells overexpressing Rad52 that grew mitotically (indicated as WT+Rad52 and dna2-KE+Rad52, respectively) as described under “Materials and Methods.” Overexpression of Rad52 allowed approximately half (∼57%) of the dna2-K1080E mutant cells to form visible colonies (Fig. 4A). Subsequently, we measured the doubling time of both WT+Rad52 and dna2-KE+Rad52 cells and found that the dna2-KE+Rad52 cells grew significantly more slowly compared with the WT+Rad52 cells. The doubling times of the wild type and mutant cells were ∼3.0 and 4.2 h, respectively, in the presence of Rad52 overexpression (Fig. 4B). Considering that the repair of HO-induced DSB takes 2–4 h to complete (65), this result raises the possibility that the structure of DNA damages or the repair pathway could differ from HO-induced DSB and thus does not rely on long-range or extensive DSB resection for repair.
The slow growth and reduced viability observed with dna2-KE+Rad52 cells indicate that they are still defective in overall DNA replication and repair process. Thus, it is highly likely that the genomic DNA in the mutant cells may not be intact and contains damages. To address this issue, we decided to investigate the extent of serine 129 phosphorylation of H2A (H2A-Ser129), a landmark event indicative of DSB occurrence in vivo (66). When we examined asynchronous logarithmically growing cells (0.4 < A600 < 0.6), no significant difference was observed on the levels of H2A-Ser129 phosphorylation between WT+Rad52 and dna2-KE+Rad52 (Fig. 4C, compare lanes 1 and 7). We then treated cells with α-factor, which arrests cells at the G1/S boundary, and found that the extent of H2A-Ser129 phosphorylation markedly (>70%) decreased in WT+Rad52, whereas no significant change was observed in dna2-KE+Rad52 (Fig. 4C, compare lanes 2–5 with 8–11). This result suggests that cell cycle progression is retarded in dna2-KE+Rad52 cells because of persistent DNA damages that can be converted into DSBs.
We also analyzed under a microscope the morphology of both wild type and dna2-K1080E cells overexpressing Rad52 as described previously (67, 68). Microscopic analyses revealed significant differences in size and morphology between WT+Rad52 and dna2-KE+Rad52 cells; dna2-KE+Rad52 cells were considerably larger than wild type (data not shown), and the population of dumbbell-shaped (G2/M phase) cells increased markedly (2.3 times) compared with WT+Rad52 (Fig. 4D). In addition, we found that the duplicated nuclei of dumbbell-shaped cells were not properly segregated to daughter cells, accumulating in the bud neck in dna2-KE+Rad52, although nuclei were normally segregated to daughter cells in WT+Rad52. > 50% of the cells in dna2-KE+Rad52 had an unsegregated single nucleus in the bud neck, whereas ∼10% of the WT+Rad52 cells displayed such a phenotype (Fig. 4E). This result is in keeping with our prediction that the dna2-K1080E cells still possess some problems in their DNA, despite the fact that overexpression of Rad52 renders dna2-K1080E cells viable.
Direct Stimulation of Endonuclease Activity of Dna2 and Rad27 by Rad52 Is Not the Underlying Mechanism for the Suppression of dna2-K1080E
The simplest explanation for the suppression mechanism could be the direct stimulation of nuclease activity of either Dna2 or Rad27 or both by Rad52, as Rad27 and nuclease-proficient Dna2K1080E/Dna2Δ405N enzymes, upon overexpression, were able to suppress the lethal phenotype of dna2-K1080E (21, 31, 33, 60). Alternatively, the Rad52-mediated suppression of dna2 mutations could be explained by efficient recombination-mediated repair of damaged DNA lesions generated by dna2 mutations. To distinguish between these two possibilities, we constructed another pRS325-ADH1 vector that expresses the Rad52-QDDD-308-311-AAAA (Rad52-QDDD/AAAA) mutant protein. It was shown that Rad52-QDDD/AAAA did not support HR because of its reduced recombination mediator activity and inability to interact with RPA, although it retained normal DNA binding and annealing activities in the absence of RPA (50). Interestingly, we found that overexpression of Rad52-QDDD/AAAA failed to suppress the lethality of dna2-K1080E (Fig. 5A). This was not due to the failure of protein expression in vivo, as the expression level of Rad52-QDDD/AAAA was comparable to that of wild type Rad52 (Fig. 5B).
These data suggest that the suppression observed requires a recombination function of the Rad52 protein. However, we cannot exclude the possibility that the failure suppression was due to the failure of the mutant Rad52 to stimulate the nuclease activities of Dna2 or Rad27. To rule out this possibility, we purified both Rad52 and Rad52-QDDD/AAAA from E. coli to near homogeneity (Fig. 5C) as described under “Materials and Methods.” Neither the wild type nor the mutant Rad52 proteins showed any detectable nuclease activity (Fig. 5D, lanes 3 and 4). Using these preparations, we first carried out nuclease assays with Dna2 and found that both Rad52 and Rad52-QDDD/AAAA significantly stimulated the Dna2-catalyzed cleavage of the 5′-flap substrate to a comparable extent (>4-fold with 25 fmol of Rad52 proteins) (Fig. 5D, lanes 6–8 and 9–11, respectively). Next, we examined the influence of Rad52 and Rad52-QDDD/AAAA on the endonuclease activity of Rad27. Rad27 was also stimulated markedly (∼11-fold) by Rad52 (Fig. 5E, lanes 5–8) almost to the same extent as by Rad52-QDDD/AAAA (Fig. 5E, compare lanes 6–8 with 9–11). These results suggest that stimulation in vitro of the endonuclease activity of either Dna2 or Rad27 is dispensable for the suppression in vivo of the lethal phenotype of dna2-K1080E. Another possible explanation for the failure of the mutant Rad52 to suppress dna2-K1080E in vivo is an impairment of a cryptic function of Rad52 in the mutant Rad52 used that, together with its Dna2/Rad27 stimulation activity, is needed for the suppression. We also investigated the possibility, although highly remote, that the Rad52-QDDD/AAAA mutant did not stimulate the endonuclease activity of the Dna2-K1080E enzyme, unlike wild-type Rad52. We found that both wild type and mutant Rad52-QDDD/AAAA protein stimulated the Dna2-K1080E-catalyzed cleavage reaction to a comparable extent (data not shown).
We were also interested in the effect of Rad51 on the endonuclease activity of Dna2 or Rad27 because of its proximity to Rad52 in the Rad51-dependent recombination pathway. We repeated the nuclease assays using purified Rad51 protein and found that the endonuclease activities of Dna2 and Rad27 were stimulated but less efficiently by Rad51 than by Rad52 (2.7- and 4.6-fold, respectively, with 100 fmol of Rad51; data not shown). This is in keeping with our finding above that the stimulation of the endonuclease activities of Dna2 or Rad27 is not responsible for the suppression of dna2-K1080E.
The Rad52-dependent Suppression of dna2-K1080E Requires the DNA Annealing Activity of Rad52
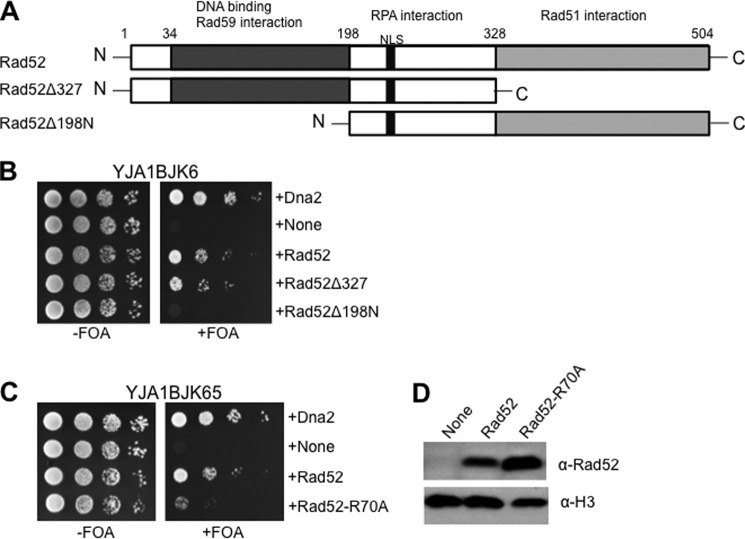
The suppression of dna2-K1080E in a manner dependent on Rad52, but not Rad51, suggests that the recombination mediator activity of Rad52, which functions to nucleate the Rad51 recombinase onto RPA-coated ssDNA, is dispensable for the suppression observed. If this is the case, it is predicted that the Rad52 mutant protein devoid of the recombination mediator activity could still suppress dna2-K1080E as efficiently as wild type Rad52. To prove this point, we constructed expression vectors that expressed Rad52Δ327 and Rad52Δ198N, the two truncated versions of Rad52, as illustrated in Fig. 6A. Rad52Δ327 lacks the C-terminal one-third region of Rad52 required for the interaction with Rad51 (40, 41), but it is able to support Rad51-independent recombination events such as break-induced replication (BIR) and SSA (69). In support of our prediction, we found that overexpression of Rad52Δ327 could suppress dna2-K1080E as efficiently as wild type (Fig. 6B), confirming again that suppression does not rely on the function of Rad51. In strong contrast, overexpression of Rad52Δ198N, which lacked the N-terminal 198-aa region, failed to suppress the lethality of dna2-K1080E (Fig. 6B). As the N-terminal part of Rad52 is required to facilitate the annealing of two complementary ssDNA (70), this result indicates that the DNA annealing activity of Rad52 is essential for the efficient repair of DNA damages caused by dna2-K1080E mutation. We were not able to show the expression of Rad52Δ327 because the available polyclonal antibodies were raised against a peptide mapped near the C terminus of yeast Rad52. We also attempted to detect the Rad52Δ198N using the same antibody but in vain; this may have been due to the low expression level or rapid degradation of this mutant protein. Nevertheless, the results obtained with overexpression of truncated Rad52 fragments support the idea that the C-terminal region (aa 328–504) of Rad52 is utterly dispensable for the suppression of dna2-K1080E.
FIGURE 6.
The Rad52-dependent suppression of dna2-K1080E depends on the DNA annealing activity of Rad52. A, schematic representation of Rad52. Domains required for DNA binding and interactions with Rad59, RPA, and Rad51 are as shown. The two truncated fragments, Rad52Δ327 and Rad52Δ198N, are as indicated. NLS, nuclear localization signal. B, the YJA1BJK56 strain was transformed with each of the five pRS325-ADH1 plasmids expressing Dna2, empty vector (None), Rad52, Rad52Δ327, and Rad52Δ198N. The resulting transformants were grown and spotted as described in the legend for Fig. 1A. C, the pRS325-ADH1 plasmids expressing the proteins indicated at the right of the figure were transformed into YJA1BJK65. Transformants were grown and spotted as described in the legend for Fig. 1A. D, expression levels of Rad52 and Rad52-R70A proteins were examined as described in legend for Fig. 1B. Histone H3 was used as the loading control. α-H3, antibodies against histone H3.
As shown above, the DNA annealing activity, but not the recombination mediator activity, of Rad52 was essential for the suppression of dna2-K1080E mutation. The result obtained using the Rad52-QDDD/AAAA mutant protein needs further clarification, as Rad52-QDDD/AAAA is impaired in both its recombination mediator activity and its interaction with RPA (50). We were interested in the influence of Rad52 on RPA-governed events such as DNA annealing. It was shown that RPA interferes with spontaneous annealing between two complementary ssDNA, which can be overcome by the addition of wild type Rad52 (44, 51). To this end, we decided to construct an expression vector that expresses the Rad52-R70A mutant protein, which has a specific defect, i.e. impairment in annealing RPA-coated ssDNA, but is normal in recombination mediator and DNA binding activities (51). When the Rad52-R70A mutant protein was overexpressed in dna2-K1080E, it suppressed the lethality of dna2-K1080E poorly, as shown in Fig. 6C, unlike wild type Rad52. This was not due to the failure of Rad52-R70A expression, because its expression level was higher than the wild type (Fig. 6D). This result indicates that the repair of DNA damages formed in dna2-K1080E mutant cells is critically dependent on a step that requires the annealing of RPA-coated complementary ssDNA.
Rad52 Works in Conjunction with Rad59 to Rescue dna2-K1080E Lethality
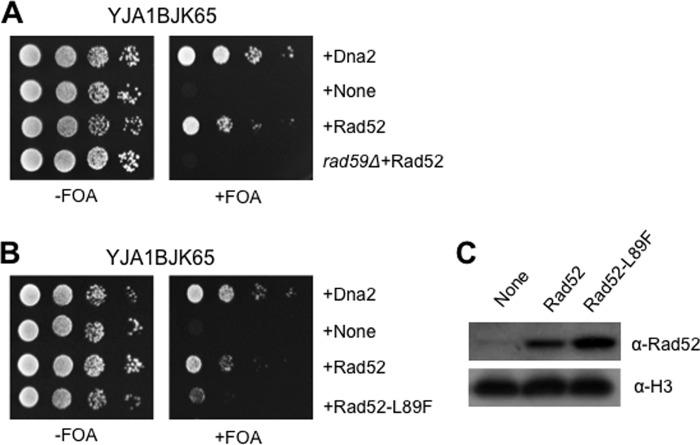
One additional important factor involved in Rad51-independent HR is Rad59. Rad59 shares significant homology with the N-terminal part of Rad52, and it possesses DNA annealing activity (71, 72). Rad59 was first isolated in an effort to identify genes that play roles in Rad51-independent spontaneous mitotic recombination between inverted repeats (71). Strong support for the importance of Rad59 in Rad51-independent recombination is provided by several genetic and biochemical observations. First, the rad51Δ rad59Δ double mutant displays severely reduced recombination rates that are comparable with those observed with rad52Δ (71, 73). Second, overexpression of Rad52 is able to restore the DNA repair or recombination defects caused by the rad59 mutations, suggesting that Rad59 has overlapping roles with Rad52 (71, 74). Third, it has been shown that Rad59 physically interacts with Rad52 (72). These results together strongly support the possibility that the suppression of dna2-K1080E by RAD52 requires a functional copy of RAD59. To verify this likelihood, we examined the influence of Rad52 overexpression in the rad59-null strain, YMJ7. As shown in Fig. 7A, the overexpression of Rad52 failed to suppress the lethal phenotype of dna2-K1080E in the absence of Rad59.
FIGURE 7.
Rad59 is essential for Rad52-mediated suppression of dna2-K1080E. A, the Rad52-dependent suppression of dna2-K1080E was examined in rad59-null cells. Overexpression of Rad52 in rad59-null cells is indicated as rad59Δ+Rad52. Transformants were grown and spotted as described above in legend for Fig. 1C. FOA, 5-fluorooritic acid. B, the pRS325-ADH1 plasmids expressing Dna2, Rad52, and Rad52-L89F were introduced into YJA1BJK65, and the resulting transformants were grown and spotted as described in the legend for Fig. 1A. C, expression levels of Rad52 and Rad52-L89F proteins were examined as described in legend for Fig. 1B. Histone H3 was used as loading control. α-H3, antibodies against histone H3.
We attempted to further confirm this observation using the rad52-L89F mutant, which displays phenotypes similar to rad59Δ, most likely due to its inability to bind Rad59 (73). The intrachromosomal recombination rate between two inverted repeats is similarly diminished in rad51Δ rad52-L89F and rad51Δ rad59Δ or rad52Δ (73). To test whether Rad59 is involved in the Rad52-dependent suppression of dna2-K1080E, we constructed a plasmid overexpressing the rad52-L89F allele. The result was that the overexpression of Rad52-L89F poorly suppressed dna2-K1080E lethality, as shown in Fig. 7B, although the expression level of Rad52-L89F was higher (∼2-fold) than that of wild type Rad52 (Fig. 7C). This result indicates that the physical interaction of Rad52 with Rad59 and/or their cooperative function may be important for the suppression of the dna2-K1080E defect.
Suppression of dna2-K1080E by Rad52 Depends on Several Factors Required for Sister Chromatid Cohesion
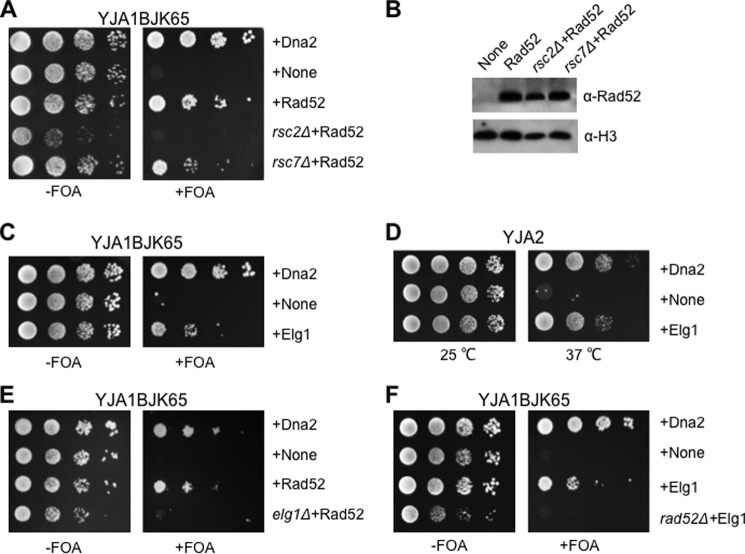
Previous studies on HR revealed several recombination pathways that do not require Rad51; they include SSA and BIR, although a majority of BIR is dependent on Rad51 (43, 46, 75–77). In addition, a subset of sister chromatid recombination (SCR) events, which is facilitated by the RSC chromatin-remodeling complex, also can occur independently of Rad51 (78, 79). This Rad51-independent SCR pathway has been shown to require Rsc2 and Rsc7, which are the two critical components of the RSC chromatin-remodeling complex (79). For this reason, we were interested in the involvement of the RSC complex in the suppression of dna2-K1080E. When Rad52 was overexpressed in rsc2Δ and rsc7Δ mutant cells (YMJ9 and YMJ10, respectively), the Rad52-dependent suppression of dna2-K1080E was totally abrogated in rsc2Δ cells but only partially in rsc7Δ cells (Fig. 8A). The levels of Rad52 expression in rsc2Δ and rsc7Δ cells were comparable with that of the wild type (Fig. 8B), indicating that the abrogation of suppression observed in rsc2Δ was not due to the lack of Rad52 protein expression (Fig. 8B). Therefore, we concluded that Rsc2 plays a crucial role in the Rad52-dependent suppression of dna2-K1080E.
FIGURE 8.
Rsc2 and Elg1 are essential for the Rad52-dependent suppression of dna2-K1080E. A, the Rad52-dependent suppression of dna2-K1080E was examined in rsc2- and rsc7-null cells. Overexpression of Rad52 in rsc2- or rsc7-null cells is indicated as rsc2Δ+Rad52 and rsc7Δ+Rad52, respectively. FOA, 5-fluorooritic acid. B, expression levels of Rad52 proteins were examined in the absence of Rsc2 or Rsc7 as described in the legend for Fig. 1B. C, overexpression of Elg1 suppresses the lethality of dna2-K1080E. The pRS325-ADH1 plasmids expressing Dna2 and Elg1 were introduced into YJA1BJK65 (dna2::HIS3, pRS314-dna2-K1080E, and pRS316-DNA2), and the resulting transformants were grown and spotted as described in the legend for Fig. 1A. D, the pRS325-ADH1 plasmids expressing Dna2 and Elg1 were introduced into YJA2 (dna2Δ405N). Cells were grown until saturated and spotted on an SD-L plate followed by incubation at 25 and 37 °C for 3 days. E, the Rad52-dependent suppression of dna2-K1080E was examined in elg1-null cells as described in the legend for Fig. 1C. Overexpression of in elg1-null cells is indicated as elg1Δ+Rad52. F, the Elg1-dependent suppression of dna2-K1080E was examined in rad52-null cells. Overexpression of Elg1 in rad52-null cells is indicated as rad52Δ+Elg1.
Because Rsc2, a critical component for sister chromatid cohesion establishment (80, 81), is essential in the Rad52-dependent suppression of dna2-K1080E, it is possible that other cohesion establishment factors could also contribute to the suppression of dna2-K1080E. For example, Elg1 is a well known factor that is required for the establishment of sister chromatid cohesion (82, 83); deletion of ELG1 results in a higher frequency of precocious sister chromatid separation (82). In addition, Elg1 genetically interacts with Dna2, and a synthetic lethal interaction between elg1Δ and dna2-2 is reported (9). Consistent with these observations, we isolated Elg1 as a dna2-K1080E suppressor as shown in Fig. 8C. In addition, the overexpression of Elg1 resulted in the efficient suppression of the ts growth defect of dna2Δ405N (Fig. 8D). We also examined the influence of Rad52 overexpression on the growth of the elg1-null strain (YMJ8) and discovered that the Rad52-dependent suppression of dna2-K1080E requires a functional copy of ELG1 because Rad52 overexpression failed to suppress the lethality of dna2-K1080E in the absence of Elg1 (Fig. 8E). Interestingly, the suppression of dna2-K1080E by Elg1 overexpression was not observed in the absence of Rad52 (Fig. 8F), indicating the functional interaction between the two proteins in repair of damages during lagging strand DNA synthesis. We discuss how they work together below.
DISCUSSION
In this study, we found that Rad52 upon overexpression rescued the lethality of dna2-K1080E, a DNA helicase-negative dna2 allele, and showed that Rad52-mediated recombination could play an important role in rectifying faulty Okazaki fragment processing. The suppression observed was not attributable to the ability of Rad52 to stimulate the endonuclease activities of Dna2 or Rad27 but to the bona fide recombination function of Rad52. This finding suggests that stimulation of the nuclease activity of Rad27 (Mgs1 and Mus81), Dna2 (RPA), or both (Vts1, Mph1, and Rad27) might not be the only mechanism by which suppression occurred (17, 58–60, 64, 84). Previously, it was shown that the overexpression of nuclease-attenuated (34) or helicase-negative mutant Dna2 enzymes (60) or of wild type Rad27 (21) allows several dna2 mutant cells to grow, indicating that the enhanced nuclease activity of Dna2 or Fen1 is the most plausible means to suppress the dna2 mutations. One likely scenario accounting for our finding that robust stimulation in vitro of the Dna2 and Rad27 activities by Rad52 did not result in suppression in vivo is that the interactions between Rad52 and Dna2/Rad27 may be restricted in vivo. Alternatively, these levels of stimulation may not be enough to rescue dna2-K1080E. We, however, prefer the first possibility for the following reason: in many cases, the 5–10-fold stimulation of either Dna2 or Rad27 is sufficient to suppress growth defects of dna2 alleles (58, 60, 84). Therefore, we believe that the underlying mechanism for the Rad52-mediated suppression of dna2-K1080E is due to the increased levels of recombination events.
Because the overexpression of catalytically active Rad27 suppressed the dna2-K1080E lethal phenotype (Fig. 3B), the lethality of dna2-K1080E could be attributed to the failure of 5′-flap cleavage, most likely leading to the generation of long flaps, which have a greater potential to form structured ones. This is consistent with the results obtained from our previous in vitro studies that Dna2-K1080E is not able to cleave 5′-flaps containing hairpin structures (32). This is also in keeping with the observation that overexpression of RPA suppresses dna2-K1080E (32); RPA is known to have the ability to destabilize short duplexes such as hairpins in 5′-flaps. Previously, it has been shown that Rad27 disengages nonproductively bound Dna2 molecules, facilitating Dna2 recycling (85). Theoretically, the Rad27-mediated recycling of Dna2 could lead to a net increase in the effective concentrations of Dna2 in cells, which in turn could lead to the suppression of dna2-K1080E. Our finding that nuclease-dead Rad27-DA, however, was not able to suppress dna2-K1080E (Fig. 3B) rules out the possibility that the suppression observed by Rad27 is due to the ability of Rad27 to dissociate Dna2 from DNA. It was shown that Dna2Δ405N, mutant Dna2 lacking the N-terminal 405-aa domain, was not able to recognize and bind to secondary structured flaps and that the inability of Dna2Δ405N to target to a secondary structured flap rendered this mutant enzyme severely defective in resolving secondary structure (28). Although Dna2-K1080E and Dna2Δ405N are similar with respect to their inability to process secondary structured flaps, dna2-K1080E cells displayed much more severe growth defects than those of dna2Δ405N; the former are lethal, whereas the latter grow in a temperature-sensitive manner. One conceivable explanation for this is as follows. In cells producing Dna2Δ405N, the unprocessed secondary structured flaps could be processed with the aid of other factors, such as Mph1 helicase, which may constitute redundant pathways in parallel with Dna2. In contrast, Dna2-K1080E is supposed to remain firmly bound to the structured 5′-flap, forming a nonproductive DNA-protein complex that interferes with the access of other enzymes required to process the flap. Analogous to and in support of this possibility, it has been shown that rad27-p, a mutant allele defective in PCNA binding, ameliorates the growth defect of nuclease-dead rad27-n when both mutations are combined (63). It was interpreted that the Rad27-n,p allows Okazaki fragments to be processed by other alternative nucleases such as Dna2 or even by the 3′ to 5′ exonuclease activity of Pol δ (63). Recently, it was shown that Dna2 is involved in the activation of the Mec1 kinase, a sensor protein for the intra-S-phase checkpoint activation (27, 28). Therefore, it is possible that Dna2-K1080E in a complex with the 5′-structured flap could cause cell death due to the hyperactivation of the intra-S-phase checkpoint. This possibility remains to be tested.
Our results shown above support the idea that the helicase activity of Dna2 is important in the replication of DNA sequences or chromosomal regions that can be converted into secondary or higher ordered structures when replication forks pass. They may include ribosomal and telomeric repeats and other repeats containing palindromes or simple trinucleotide repeats. Results from several studies have revealed that Dna2 is implicated in the replication or the maintenance of telomeric repeat sequences or rDNA arrays (37, 86–90). For example, the dna2-2 mutant displayed elevated rates of DSB-associated recombination and increased levels of replication pause and fork convergence at the rDNA region (86, 91). Moreover, Dna2 is maximally associated with rDNA during S phase (86), supporting the possibility that Dna2 is importantly involved in rDNA replication. Therefore, the helicase activity of Dna2 becomes more important particularly when replication forks proceed through such structure-forming DNA regions. It would be interesting to examine which parts of chromosomes become unstable in the absence of the helicase activity of Dna2.
There have been a number of experimental data suggesting that Rad51-independent HR could play a crucial role in the repair of DNA damages associated with lagging strand DNA replication. It was shown that the pol12-100, cdc9-1, cdc2-1 mutants, which have defects in lagging strand DNA replication, accumulated significant amounts of recombination intermediates during S phase in rDNA, whereas the recombination intermediates were not observed in pol2-1 and dpb2-1 mutants, which are defective in leading strand DNA replication (92). These recombination intermediates were formed in a manner dependent on Rad52 but not on Rad51 and its other paralogs (92). In addition, it was reported that Rad52, but not Rad51, is associated with stalled replication forks in rDNA (93). The genetic data that rad59Δ or rad59-K166A is synthetic lethal or sick with rad27Δ or cdc9-1 (52, 94, 95) also support the importance of Rad59-dependent recombination in the repair of lagging strand associated damages. Our findings strongly suggest that the Rad52/Rad59-dependent pathway is a highly preferred choice, whereas the Rad51-dependent pathway is negatively regulated during DNA replication. The genetic observation that rad59-Y92A cells display increases in Rad51-dependent HR events (94) raises the possibility that the two pathways are tightly regulated. For example, Rad59 could play a role in the prevention of unscheduled Rad51-dependent recombination. In addition, it has been shown that during S phase, sumoylated PCNA recruits Srs2, which functions to dismantle Rad51 from nucleofilament, leading to suppression of unwanted HR (96, 97). Considering that the Rad51-dependnet HR requires many more accessory factors and a longer time to complete, the Rad51-independent HR-mediated repair of DNA damages occurring during DNA replication, particularly in lagging strand, could be a preferred choice.
Previous studies have revealed that SSA and BIR could occur in the absence of Rad51 function, although a majority of BIR occurs more efficiently in the presence of Rad51 (43, 46, 75–77). We examined whether SSA and BIR could contribute to the Rad52-dependent suppression of dna2-K1080E and found that the deletion of Rad10, Saw1, and Tid1, important factors for SSA or BIR (76, 98, 99), did not affect the suppression (data not shown). It has also been shown that a subset of SCR events, which is facilitated by the RSC complex, could occur independently of Rad51 (78, 79). We found that the suppression was completely abolished in rsc2Δ and elg1Δ cells, raising the possibility that the same mechanism for SCR is responsible, at least in part, for the repair of faulty flaps. We also investigated whether Ctf4 and Ctf18, two other sister chromatid cohesion establishment factors, had a role in the suppression of dna2-K1080E by Rad52 overexpression. It has been found that ctf4 shows synthetic lethality or synthetic growth defects when combined with dna2 ts mutant alleles (35), and CTF18 has been isolated as a multicopy suppressor of dna2Δ405N (data not shown). However, we were not able to determine the requirement of Ctf4 and Ctf18 for the Rad52-mediated suppression, because overexpression of Rad52 rendered ctf4Δ and ctf18Δ cells inviable (data not shown).
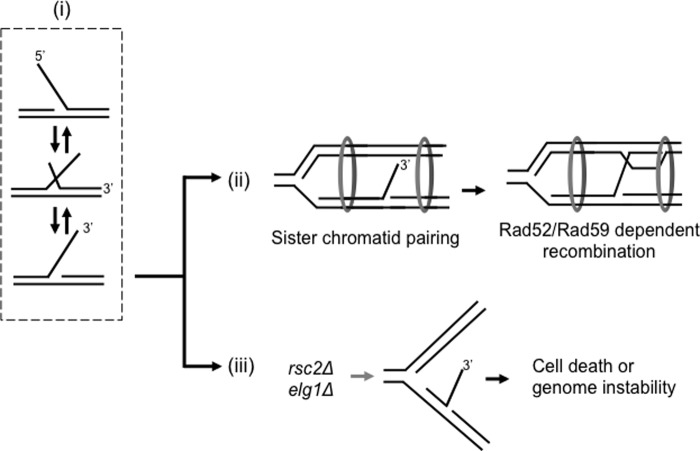
One question that arises is how the Rad52-dependent recombination is initiated to repair the problematic flaps that have not been appropriately processed due to a defective processing enzyme such as helicase-deficient Dna2. For the HR-mediated repair of general DSB or collapsed replication forks, the initial step is the generation of 3′-ssDNA overhangs by the combined action of the MRX complex and DSB resection machineries (23, 47, 100). Contrary to this, we found that the overexpression of Rad52 in a rad50-null (Fig. 3A) or rpd3-null (data not shown) strain defective in DSB resection (61, 101) resulted in efficient suppression of dna2-K1080E. These findings suggest that the 3′-ssDNA overhangs may not be generated by the canonical DSB resection. One likely alternative mechanism for the generation of 3′-ssDNA would be flap equilibration, the spontaneous process of competitive annealing between the two 5′- and 3′-strands. The 5′-flaps, presumably with hairpin or other aberrant structures and thus longer than the average ones, are expected to accumulate in dna2-K1080E, and they can be converted to 3′-flaps through energetically allowed flap equilibration (Fig. 9, step i). One role of Rad52 in this spontaneous process is to facilitate the interconversion between the 5′- and 3′-flaps by participating in the following process. The RPA-coated long 5′-flaps could recruit Rad52, because RPA-coated ssDNA recruits Rad52 through specific protein-protein interactions (50). Then, the 5′-flap-bound Rad52 proteins could rapidly generate 3′-flaps by facilitating the annealing of the 5′-flap to the template using the strand exchange activity of Rad52 (102). As a result of flap equilibration, the newly generated 3′-flap could form a complex with Rad52 molecules and invade a homologous region in the sister chromatid, readily available because of the close proximity by sister chromatid cohesion (Fig. 9, step ii). This proximity may allow the 3′-ssDNA to find its homologous sequence without the aid of Rad51. Subsequently, the strand exchange activity of Rad52 could be used to promote strand invasion between the invading 3′-ssDNA and donor sister chromatid to establish D-loop structure. This later step may not be efficient without the proper paring of the two sister chromatids (Fig. 9, step iii). An alternative fate of the 3′-flaps formed via flap equilibration is that they could be newly processed by a 3′-endonuclease such as Mus81 or its related complexes. This is in keeping with the observation that the overexpression of Mus81-Mms4 suppresses the lethality of dna2-K1080E (59).
FIGURE 9.
A model for Rad52/59-mediated repair of faulty Okazaki fragment processing. i, unprocessed 5′-flaps can be converted to 3′-flaps via a process called “flap equilibration” (see “Discussion” for details). ii, the 3′-flaps can invade the homologous DNA sequences located on a sister chromatid with the aid of Rad52 and Rad59 proteins. The sister chromatid pairing appears critical for this invasion step. iii, in the absence of cohesion establishment factors, sister chromatids are not able to pair properly, causing the failure of error-free recombination-mediated repair of erroneous flaps, which in turn leads to cell death or genome instability.
Acknowledgment
We are grateful to the members of our laboratory for critical reading of the manuscript.
This work was supported by National Research Foundation of Korea Grant 2012R1A2A2A01047260 from the Ministry of Education, Science, and Technology.
- Pol
- polymerase
- PCNA
- proliferating cell nuclear antigen
- DSB
- double strand break
- aa
- amino acid
- ts
- temperature-sensitive
- HR
- homologous recombination
- SSA
- single strand annealing
- MRX
- Mre11-Rad50-Xrs2
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- BIR
- break-induced replication
- SCR
- sister chromatid recombination
- RPA
- replication protein A.
REFERENCES
- 1. Haber J. E. (1999) Sir-Ku-itous routes to make ends meet. Cell 97, 829–832 [DOI] [PubMed] [Google Scholar]
- 2. Kowalczykowski S. C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25, 156–165 [DOI] [PubMed] [Google Scholar]
- 3. Krogh B. O., Symington L. S. (2004) Recombination proteins in yeast. Annu. Rev. Genet. 38, 233–271 [DOI] [PubMed] [Google Scholar]
- 4. Alvaro D., Lisby M., Rothstein R. (2007) Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 3, e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hanawalt P. C. (2007) Paradigms for the three Rs: DNA replication, recombination, and repair. Mol. Cell 28, 702–707 [DOI] [PubMed] [Google Scholar]
- 6. Li X., Heyer W. D. (2008) Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 18, 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maher R. L., Branagan A. M., Morrical S. W. (2011) Coordination of DNA replication and recombination activities in the maintenance of genome stability. J. Cell. Biochem. 112, 2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hübscher U., Seo Y. S. (2001) Replication of the lagging strand: a concert of at least 23 polypeptides. Mol. Cells 12, 149–157 [PubMed] [Google Scholar]
- 9. Budd M. E., Tong A., Peng X., Polaczek A., Boone A., Campbell J. L. (2005) A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 1, 634–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loeillet S., Palancade B., Cartron M., Thierry A., Richard G. F., Dujon B., Doye V., Nicolas A. (2005) Genetic network interactions among replication, repair, and nuclear pore deficiencies in yeast. DNA Repair 4, 459–468 [DOI] [PubMed] [Google Scholar]
- 11. Kang Y. H., Lee C. H., Seo Y. S. (2010) Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 45, 71–96 [DOI] [PubMed] [Google Scholar]
- 12. Garg P., Burgers P. M. (2005) DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 40, 115–128 [DOI] [PubMed] [Google Scholar]
- 13. Zheng L., Shen B. (2011) Okazaki fragment maturation: nucleases take centre stage. J. Mol. Cell. Biol. 3, 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Balakrishnan L., Bambara R. A. (2013) Flap endonuclease 1. Annu. Rev. Biochem. 82, 119–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burgers P. M. (2009) Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 284, 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bae S. H., Seo Y. S. (2000) Characterization of the enzymatic properties of the yeast Dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 275, 38022–38031 [DOI] [PubMed] [Google Scholar]
- 17. Bae S. H., Bae K. H., Kim J. A., Seo Y. S. (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412, 456–461 [DOI] [PubMed] [Google Scholar]
- 18. MacNeill S. A. (2001) DNA replication: partners in the Okazaki two-step. Curr. Biol. 11, R842–844 [DOI] [PubMed] [Google Scholar]
- 19. Ayyagari R., Gomes X. V., Gordenin D. A., Burgers P. M. (2003) Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 20. Jin Y. H., Ayyagari R., Resnick M. A., Gordenin D. A., Burgers P. M. (2003) Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′-5′ exonuclease activities of Pol δ in the creation of a ligatable nick, J. Biol. Chem. 278, 1626–1633 [DOI] [PubMed] [Google Scholar]
- 21. Budd M. E., Campbell J. L. (1997) A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17, 2136–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang H. Y., Choi E., Bae S. H., Lee K. H., Gim B. S., Kim H. D., Park C., MacNeill S. A., Seo Y. S. (2000) Genetic analyses of Schizosaccharomyces pombe dna2(+) reveal that Dna2 plays an essential role in Okazaki fragment metabolism. Genetics 155, 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G. (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double strand break ends. Cell 134, 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J. L., Kowalczykowski S. C. (2010) DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467, 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nimonkar A. V., Genschel J., Kinoshita E., Polaczek P., Campbell J. L., Wyman C., Modrich P., Kowalczykowski S. C. (2011) BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Symington L. S., Gautier J. (2011) Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 [DOI] [PubMed] [Google Scholar]
- 27. Kumar S., Burgers P. M. (2013) Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 27, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee C. H., Lee M., Kang H. J., Kim D. H., Kang Y. H., Bae S. H., Seo Y. S. (2013) The N-terminal 45-kDa domain of Dna2 endonuclease/helicase targets the enzyme to secondary structure DNA. J. Biol. Chem. 288, 9468–9481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Budd M. E., Campbell J. L. (1995) A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. U.S.A. 92, 7642–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bae S. H., Choi E., Lee K. H., Park J. S., Lee S. H., Seo Y. S. (1998) Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 273, 26880–26890 [DOI] [PubMed] [Google Scholar]
- 31. Bae S. H., Kim J. A., Choi E., Lee K. H., Kang H. Y., Kim H. D., Kim J. H., Bae K. H., Cho Y., Park C., Seo Y. S. (2001) Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease. Nucleic Acids Res. 29, 3069–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bae S. H., Kim D. W., Kim J., Kim J. H., Kim D. H., Kim H. D., Kang H. Y., Seo Y. S. (2002) Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem. 277, 26632–26641 [DOI] [PubMed] [Google Scholar]
- 33. Budd M. E., Choe Wc., Campbell J. L. (2000) The nuclease activity of the yeast Dna2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 275, 16518–16529 [DOI] [PubMed] [Google Scholar]
- 34. Lee K. H., Kim D. W., Bae S. H., Kim J. A., Ryu G. H., Kwon Y. N., Kim K. A., Koo H. S., Seo Y. S. (2000) The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 28, 2873–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Formosa T., Nittis T. (1999) Dna2 mutants reveal interactions with Dna polymerase α and Ctf4, a Pol α accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics 151, 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen X., Niu H., Chung W. H., Zhu Z., Papusha A., Shim E. Y., Lee S. E., Sung P., Ira G. (2011) Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat. Struct. Mol. Biol. 18, 1015–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Masuda-Sasa T., Polaczek P., Peng X. P., Chen L., Campbell J. L. (2008) Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition. J. Biol. Chem. 283, 24359–24373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sung P. (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272, 28194–28197 [DOI] [PubMed] [Google Scholar]
- 39. Song B., Sung P. (2000) Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J. Biol. Chem. 275, 15895–15904 [DOI] [PubMed] [Google Scholar]
- 40. Milne G. T., Weaver D. T. (1993) Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 7, 1755–1765 [DOI] [PubMed] [Google Scholar]
- 41. Krejci L., Song B., Bussen W., Rothstein R., Mortensen U. H., Sung P. (2002) Interaction with Rad51 is indispensable for recombination mediator function of Rad52. J. Biol. Chem. 277, 40132–40141 [DOI] [PubMed] [Google Scholar]
- 42. Mortensen U. H., Bendixen C., Sunjevaric I., Rothstein R. (1996) DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. U.S.A. 93, 10729–10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shinohara A., Shinohara M., Ohta T., Matsuda S., Ogawa T. (1998) Rad52 forms ring structures and cooperates with RPA in single-strand DNA annealing. Genes Cells 3, 145–156 [DOI] [PubMed] [Google Scholar]
- 44. Sugiyama T., New J. H., Kowalczykowski S. C. (1998) DNA annealing by Rad52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 95, 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nimonkar A. V., Sica R. A., Kowalczykowski S. C. (2009) Rad52 promotes second-end DNA capture in double-stranded break repair to form complement-stabilized joint molecules. Proc. Natl. Acad. Sci. U.S.A. 106, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pâques F., Haber J. E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Symington L. S. (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Van Dyck E., Stasiak A. Z., Stasiak A., West S. C. (2001) Visualization of recombination intermediates produced by RAD52-mediated single-strand annealing. EMBO Rep. 2, 905–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sugiyama T., Kowalczykowski S. C. (2002) Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 277, 31663–31672 [DOI] [PubMed] [Google Scholar]
- 50. Plate I., Hallwyl S. C., Shi I., Krejci L., Müller C., Albertsen L., Sung P., Mortensen U. H. (2008) Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J. Biol. Chem. 283, 29077–29085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shi I., Hallwyl S. C., Seong C., Mortensen U., Rothstein R., Sung P. (2009) Role of the Rad52 amino-terminal DNA binding activity in DNA strand capture in homologous recombination. J. Biol. Chem. 284, 33275–33284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Symington L. S. (1998) Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26, 5589–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Debrauwère H., Loeillet S., Lin W., Lopes J., Nicolas A. (2001) Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. U.S.A. 98, 8263–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fiorentino D. F., Crabtree G. R. (1997) Characterization of Saccharomyces cerevisiae dna2 mutants suggests a role for the helicase late in S phase. Mol. Biol. Cell 8, 2519–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Budd M. E., Choe Wc., Campbell J. L. (1995) DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem. 270, 26766–26769 [DOI] [PubMed] [Google Scholar]
- 56. Wach A., Brachat A., Pöhlmann R., Philippsen P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10, 1793–1808 [DOI] [PubMed] [Google Scholar]
- 57. Riezman H., Hase T., van Loon A. P., Grivell L. A., Suda K., Schatz G. (1983) Import of proteins into mitochondria: a 70 kilodalton outer membrane protein with a large carboxy-terminal deletion is still transported to the outer membrane. EMBO J. 2, 2161–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kang Y. H., Kang M. J., Kim J. H., Lee C. H., Cho I. T., Hurwitz J., Seo Y. S. (2009) The MPH1 gene of Saccharomyces cerevisiae functions in Okazaki fragment processing. J. Biol. Chem. 284, 10376–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kang M. J., Lee C. H., Kang Y. H., Cho I. T., Nguyen T. A., Seo Y. S. (2010) Genetic and functional interactions between Mus81-Mms4 and Rad27. Nucleic Acids Res. 38, 7611–7625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee C. H., Shin Y. K., Phung T. T., Bae J. S., Kang Y. H., Nguyen T. A., Kim J. H., Kim D. H., Kang M. J., Bae S. H., Seo Y. S. (2010) Involvement of Vts1, a structure-specific RNA-binding protein, in Okazaki fragment processing in yeast. Nucleic Acids Res. 38, 1583–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shim E. Y., Chung W. H., Nicolette M. L., Zhang Y., Davis M., Zhu Z., Paull T. T., Ira G., Lee S. E. (2010) Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29, 3370–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shen B., Nolan J. P., Sklar L. A., Park M. S. (1997) Functional analysis of point mutations in human flap endonuclease-1 active site. Nucleic Acids Res. 25, 3332–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gary R., Park M. S., Nolan J. P., Cornelius H. L., Kozyreva O. G., Tran H. T., Lobachev K. S., Resnick M. A., Gordenin D. A. (1999) A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell. Biol. 19, 5373–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Munashingha P. R., Lee C. H., Kang Y. H., Shin Y. K., Nguyen T. A., Seo Y. S. (2012) The trans-autostimulatory activity of Rad27 suppresses dna2 defects in Okazaki fragment processing. J. Biol. Chem. 287, 8675–8687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chung W. H., Zhu Z., Papusha A., Malkova A., Ira G. (2010) Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 6, e1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van Attikum H., Gasser S. M. (2005) The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell Biol. 6, 757–765 [DOI] [PubMed] [Google Scholar]
- 67. Hollenhorst P. C., Bose M. E., Mielke M. R., Müller U., Fox C. A. (2000) Forkhead genes in transcriptional silencing, cell morphology and the cell cycle: overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics 154, 1533–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. IJpma A. S., Greider C. W. (2003) Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tsukamoto M., Yamashita K., Miyazaki T., Shinohara M., Shinohara A. (2003) The N-terminal DNA-binding domain of Rad52 promotes RAD51-independent recombination in Saccharomyces cerevisiae. Genetics 165, 1703–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Singleton M. R., Wentzell L. M., Liu Y., West S. C., Wigley D. B. (2002) Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl. Acad. Sci. U.S.A. 99, 13492–13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bai Y., Symington L. S. (1996) A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10, 2025–2037 [DOI] [PubMed] [Google Scholar]
- 72. Davis A. P., Symington L. S. (2001) The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159, 515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cortés-Ledesma F., Malagón F., Aguilera A. (2004) A novel yeast mutation, rad52-L89F, causes a specific defect in Rad51-independent recombination that correlates with a reduced ability of Rad52-L89F to interact with Rad59. Genetics 168, 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Feng Q., Düring L., de Mayolo A. A., Lettier G., Lisby M., Erdeniz N., Mortensen U. H., Rothstein R. (2007) Rad52 and Rad59 exhibit both overlapping and distinct functions. DNA Repair 6, 27–37 [DOI] [PubMed] [Google Scholar]
- 75. Malkova A., Ivanov E. L., Haber J. E. (1996) Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. U.S.A. 93, 7131–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ira G., Haber J. E. (2002) Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22, 6384–6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pohl T. J., Nickoloff J. A. (2008) Rad51-independent interchromosomal double-strand break repair by gene conversion requires Rad52 but not Rad55, Rad57, or Dmc1. Mol. Cell. Biol. 28, 897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liefshitz B., Kupiec M. (2011) Roles of RSC, Rad59, and cohesin in double-strand break repair. Mol. Cell. Biol. 31, 3921–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Oum J. H., Seong C., Kwon Y., Ji J. H., Sid A., Ramakrishnan S., Ira G., Malkova A., Sung P., Lee S. E., Shim E. Y. (2011) RSC facilitates Rad59-dependent homologous recombination between sister chromatids by promoting cohesin loading at DNA double-strand breaks. Mol. Cell. Biol. 31, 3924–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Huang J., Hsu J. M., Laurent B. C. (2004) The RSC nucleosome-remodeling complex is required for Cohesin's association with chromosome arms. Mol. Cell 13, 739–750 [DOI] [PubMed] [Google Scholar]
- 81. Huang J., Laurent B. C. (2004) A Role for the RSC chromatin remodeler in regulating cohesion of sister chromatid arms. Cell Cycle 3, 973–975 [PubMed] [Google Scholar]
- 82. Maradeo M. E., Skibbens R. V. (2009) The Elg1-RFC clamp-loading complex performs a role in sister chromatid cohesion. PLoS One 4, e4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Parnas O., Zipin-Roitman A., Mazor Y., Liefshitz B., Ben-Aroya S., Kupiec M. (2009) The ELG1 clamp loader plays a role in sister chromatid cohesion. PLoS One 4, e5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kim J. H., Kang Y. H., Kang H. J., Kim D. H., Ryu G. H., Kang M. J., Seo Y. S. (2005) In vivo and in vitro studies of Mgs1 suggest a link between genome instability and Okazaki fragment processing. Nucleic Acids Res. 33, 6137–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Stewart J. A., Campbell J. L., Bambara R. A. (2009) Significance of the dissociation of Dna2 by flap endonuclease 1 to Okazaki fragment processing in Saccharomyces cerevisiae. J. Biol. Chem. 284, 8283–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hoopes L. L., Budd M., Choe W., Weitao T., Campbell J. L. (2002) Mutations in DNA replication genes reduce yeast life span. Mol. Cell. Biol. 22, 4136–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lin W., Sampathi S., Dai H., Liu C., Zhou M., Hu J., Huang Q., Campbell J., Shin-Ya K., Zheng L., Chai W., Shen B. (2013) Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 32, 1425–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chai W., Zheng L., Shen B. (2013) DNA2, a new player in telomere maintenance and tumor suppression. Cell Cycle 12, 1985–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tomita K., Kibe T., Kang H. Y., Seo Y. S., Uritani M., Ushimaru T., Ueno M. (2004) Fission yeast Dna2 is required for generation of the telomeric single-strand overhang. Mol. Cell. Biol. 24, 9557–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Choe W., Budd M., Imamura O., Hoopes L., Campbell J. L. (2002) Dynamic localization of an Okazaki fragment processing protein suggests a novel role in telomere replication. Mol. Cell. Biol. 22, 4202–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Weitao T., Budd M., Hoopes L. L., Campbell J. L. (2003) Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem. 278, 22513–22522 [DOI] [PubMed] [Google Scholar]
- 92. Zou H., Rothstein R. (1997) Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90, 87–96 [DOI] [PubMed] [Google Scholar]
- 93. Irmisch A., Ampatzidou E., Mizuno K., O'Connell M. J., Murray J. M. (2009) Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J. 28, 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liddell L. C., Manthey G. M., Owens S. N., Fu B. X., Bailis A. M. (2013) Alleles of the homologous recombination gene, RAD59, identify multiple responses to disrupted DNA replication in Saccharomyces cerevisiae. BMC Microbiol. 13, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Nguyen H. D., Becker J., Thu Y. M., Costanzo M., Koch E. N., Smith S., Myung K., Myers C. L., Boone C., Bielinsky A. K. (2013) Unligated Okazaki fragments induce PCNA ubiquitination and a requirement for Rad59-dependent replication fork progression. PLoS One. 8, e66379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., Klein H., Ellenberger T., Sung P. (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 [DOI] [PubMed] [Google Scholar]
- 97. Veaute X., Jeusset J., Soustelle C., Kowalczykowski S. C., Le Cam E., Fabre F. (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 [DOI] [PubMed] [Google Scholar]
- 98. Li F., Dong J., Eichmiller R., Holland C., Minca E., Prakash R., Sung P., Yong Shim E., Surtees J. A., Eun Lee S. (2013) Role of Saw1 in Rad1/Rad10 complex assembly at recombination intermediates in budding yeast. EMBO J. 32, 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Li F., Dong J., Pan X., Oum J. H., Boeke J. D., Lee S. E. (2008) Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol. Cell 30, 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mimitou E. P., Symington L. S. (2009) DNA end resection: many nucleases make light work. DNA Repair 8, 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Robert T., Vanoli F., Chiolo I., Shubassi G., Bernstein K. A., Rothstein R., Botrugno O. A., Parazzoli D., Oldani A., Minucci S., Foiani M. (2011) HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 471, 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bi B., Rybalchenko N., Golub E. I., Radding C. M. (2004) Human and yeast Rad52 proteins promote DNA strand exchange. Proc. Natl. Acad. Sci. U.S.A. 101, 9568–9572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]