Abstract
A balanced t(1;11) translocation that transects the Disrupted in schizophrenia 1 (DISC1) gene shows genome-wide significant linkage for schizophrenia and recurrent major depressive disorder (rMDD) in a single large Scottish family, but genome-wide and exome sequencing-based association studies have not supported a role for DISC1 in psychiatric illness. To explore DISC1 in more detail, we sequenced 528 kb of the DISC1 locus in 653 cases and 889 controls. We report 2718 validated single-nucleotide polymorphisms (SNPs) of which 2010 have a minor allele frequency of <1%. Only 38% of these variants are reported in the 1000 Genomes Project European subset. This suggests that many DISC1 SNPs remain undiscovered and are essentially private. Rare coding variants identified exclusively in patients were found in likely functional protein domains. Significant region-wide association was observed between rs16856199 and rMDD (P=0.026, unadjusted P=6.3 × 10−5, OR=3.48). This was not replicated in additional recurrent major depression samples (replication P=0.11). Combined analysis of both the original and replication set supported the original association (P=0.0058, OR=1.46). Evidence for segregation of this variant with disease in families was limited to those of rMDD individuals referred from primary care. Burden analysis for coding and non-coding variants gave nominal associations with diagnosis and measures of mood and cognition. Together, these observations are likely to generalise to other candidate genes for major mental illness and may thus provide guidelines for the design of future studies.
Keywords: DISC1, recurrent major depressive disorder, sequencing
Introduction
Schizophrenia (SZ), bipolar disorder (BD) and recurrent major depressive disorder (rMDD) are common forms of serious mental illness, each with a strong and overlapping genetic component.1, 2, 3 Genome-wide linkage, association, cytogenetic, copy number variant and, more recently, sequencing studies establish that the genetic architecture of psychiatric illness is complex and that there is extensive genetic heterogeneity, which is incompletely defined or understood (reviewed in Sullivan et al.4). We previously reported a t(1;11) translocation in a single large Scottish family that showed genome-wide significant linkage for SZ, rMDD and jointly with BD.5 The t(1;11) translocation is balanced and structurally simple, but the outcome is genetically complex, disrupting the protein coding gene Disrupted in schizophrenia 1 (DISC1), the antisense non-coding gene Disrupted in schizophrenia 2 (DISC2) and the non-coding gene DISC1FP1, creating a DISC1/DISC1FP1 fusion transcript.6, 7, 8, 9, 10, 11
Several small independent studies have reported evidence for association of single DISC1 single-nucleotide polymorphisms (SNPs) (coding and non-coding) or haplotypes with SZ, BD, rMDD and other neuropsychiatric traits, including autism spectrum disorder, cognition, normative cognitive ageing, anxiety and structural and functional brain imaging phenotypes.9, 12, 13 Rare amino-acid substitution variants in DISC1 have been reported in cases of SZ,10, 11 BD,14 rMDD,13 autism spectrum disorder15 and agenesis of the corpus callosum,16 as has an increased burden of rare missense variants in exon 11 of DISC1 for schizoaffective disorder,17 and for DISC1 pathway genes in SZ.10 In contrast, a meta-analysis of all known common variants within the DISC locus, from a total of 11 626 cases and 15 237 controls that involved the testing of 1241 SNPs, found no evidence that common variants at the DISC locus are significantly associated with SZ.18 Moreover, the DISC1 locus has not reached genome-wide significance in large-scale meta-analyses of linkage studies of SZ,19 nor have its common variants in large-scale genome-wide association studies of SZ, BD or rMDD.20, 21, 22 A recent exon-based study that sequenced 2.7 kb of DISC1 in 727 cases of SZ and 733 controls found 32 rare alleles (minor allele frequency (MAF)<0.01) in SZ cases and 40 in European controls with no evidence for a significantly increased burden of likely pathogenic variants.23 DISC1, however, continues to feature strongly in attempts to assess genome-wide association results in terms of networks24 or in combination with known biological function.25, 26, 27
The biological functions of DISC1 fit well with current aetiological concepts in SZ-related major mental illness and cognition.28 DISC1 is a scaffold protein that interacts with, and modulates the activity of, multiple proteins with key roles in neurodevelopment, neurogenesis, neuronal migration, integration and signalling29, 30, 31 including the antidepressant and antipsychotic targets GSK3β32 and PDE4.33 Several common and rare amino-acid polymorphisms of DISC1 have predicted deleterious effects on protein function and demonstrable biological effects in experimental settings.31 The 704C allele is associated with reduced activity of ERK1 and Akt kinases, altered binding affinities of DISC1 for NDE1 and NDEL1 and variation in DISC1 oligomeric status.34, 35, 36, 37 The 607F allele results in (a) reduced binding and centrosomal localisation of PCM1, (b) reduced noradrenaline neurotransmitter release in SH-SY5Y cells,38 (c) altered mitochondrial trafficking39 and (d) a partial shift from neuronal to glial expression in the brain.40 Furthermore, Singh et al.41 reported that 607F impacts negatively on neural progenitor proliferation in E16 mouse brain, correlates with aberrant wnt signalling in human lymphoblasts, and is associated with a neurodevelopmental phenotype in morpholino mutant zebrafish. R37W lies within an arginine-rich nuclear localisation motif and a partially overlapping interaction domain for PDE442 and GSK3β.41, 43 The 37W allele shows reduced nuclear DISC1 expression and altered DISC1 regulation of ATF4, a critical modulator of cAMP signalling and mediator of the stress response.44
In summary, whereas the primary genetic evidence from the original Scottish t(1;11) family was significant at the genome-wide significance level for both SZ and rMDD and the experimental evidence and biological plausibility of DISC1 remains strong, the evidence from subsequent linkage and candidate gene association studies is however inconsistent and not supported by genome-wide association studies or meta-analysis.12 To explore these contrasting findings, we aimed here to establish the nature and frequency of DISC1 genomic sequence variants, identify rare variants in putative functional domains, and test for effects of these on cognitive traits and the risk of psychiatric illness. We comprehensively sequenced 528 kb covering the entire DISC1 locus, including TRAX (also known as TSNAX) for which there is evidence for intergenic splicing with DISC145 and the intergenic region, which contains regulatory elements immediately 5' of DISC1.13, 46, 47, 48
Materials and methods
A full summary of the methods can be found in Supplementary Information. Briefly, all study participants gave signed consent for their data and samples to be used in studies that have been approved by the appropriate Research Ethics Committee or the GS access Committee. Genomic DNA from each individual was whole genome amplified in triplicate, the products pooled and amplified with primer pairs tiled across 528 kb of TRAX/DISC1 (hg18 chr1:229723339-230251606; hg19 chr1:231656716-232184983). For each sample, the pooled products were sheared, converted into paired-end Illumina libraries and sequenced on an Illumina GAII or HiSeq 2000 sequencer to >80% coverage and >30-fold depth. Sequences were aligned to the UCSC hg18 reference sequence, variants called using MAQ software49 and the variants in repeats removed. Ten percent of all remaining variants were validated using Sanger sequencing chemistry on an ABI3730 sequencer, and the derived information used to optimise the quality control filters. After quality control screening, all exonic and low frequency (MAF<1%) variants were also validated by Sanger chemistry sequencing as above. The variants were functionally annotated using SNPnexus50 (http://www.snp-nexus.org). Non-coding variants were annotated using the UCSC table browser for the following tracks: ‘RepeatMasker',51 ‘CpG island', ‘TFBS conserved', ‘7x Reg Potential' (which substantially overlaps with DNAse hypersensitivity sites) and/or ‘28-Way Most conserved—PlacMammal' (http://genome.ucsc.edu/). Sequence variants classified as coding were mapped to the DISC1 L isoform and potential pathogenicity ascribed using Pmut,52 Panther53 and PolyPhen-2.54 The coding sequence variants were mapped onto a list of known curated DISC1-interactor binding sites31 and with other functional elements (for example, phosphorylation sites55). Case–control association was tested on the combined case samples as well as individually for SZ, BP and rMDD using Fisher's exact test. Permutation was used to derive region-wide P-values and significance thresholds. Quantitative trait association analyses using LBC1936 samples were performed by linear regression of the trait residuals (adjusted for age and sex) on the number of minor alleles at each SNP, with empirical P-values estimated by permutation to avoid issues with the test statistic distribution caused by the combination of rare variants and slight deviations from normality in the phenotypes. All association analyses were performed using PLINK.56 Mark-recapture analysis followed the Lincoln-Petersen and Modified Petersen methods57, 58 with 95% confidence intervals calculated following Chapman.59 Burden analysis was performed in PLINK/SEQ to implement BURDEN and VTTEST with empirical P-values estimated using permutation. Genotyping of the replication and familial samples was performed by the Edinburgh Wellcome Trust Clinical Research Facility Genetics Core using TaqMan SNP genotyping assay C__33950433_10 with concurrent genotyping of known heterozygotes.
Data access
The accession numbers for sequence data are NCBI ss472328925—ss472331023.
Results
Sequence analysis
We sequenced 1542 Caucasians from Scotland comprising 240 cases of SZ, 221 cases of BD, 192 cases of rMDD and, as controls, 889 members of the Lothian Birth Cohort 1936 (LBC1936), which have been extensively phenotyped.60 Each sample was sequenced to >80% coverage at a minimum of 30-fold read depth by long-range PCR and sequencing on either Illumina GAII or HiSeq 2000 sequencers. To ensure a robust data set, all variants within repetitive regions were removed. Final quality score thresholds for the data were derived from capillary sequence validation of 10% of the remaining variants. All variants with an MAF <1% were validated by ABI3730 sequencing. After quality control, there was no evidence for sequencing bias between cases and controls (Supplementary Figure S1). Allele frequencies from our sample showed strong concordance to those from the European subset of the 1000 Genomes Project61 (Supplementary Figure S2). We report 2718 SNPs in the 1542 samples analysed, 708 at ⩾1% and 2010 at <1% MAF (Supplementary Table S1). Only 1027 of the 2718 SNPs (38%) were previously reported in the European subset of the 1000 Genomes Project.61
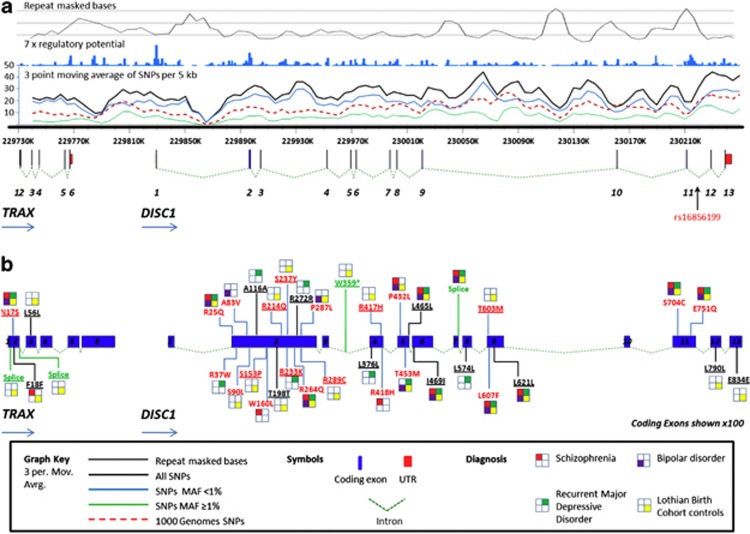
As defined and annotated by the UCSC genome browser (http://genome.ucsc.edu), 489 SNPs mapped to regions of regulatory potential, 177 to non-coding exons (including DISC2) and 36 to coding regions of exons. Of these 36 variants, 12 were synonymous changes, 23 were non-synonymous changes, with one producing a stop codon consistent with the DISC1 Es isoform (Figure 1; Supplementary Figure S3; Supplementary Tables S1 and S2). Supplementary Table S3 summarises the overlap between variants identified in this study and other DISC1 sequencing studies and relevant association studies.10, 11, 13, 14, 16, 17, 23, 46
Figure 1.
TRAX/DISC1 (Disrupted in schizophrenia 1) genomic and exon structure: alignment of coding and regulatory variants. (a) Three-period moving average of all single-nucleotide polymorphisms (SNPs) identified per 5 kb across the region in this study with TRAX/DISC1 intron/exons structure given to scale. Total SNP number (black), those with a minor allele frequency (MAF) of <1% SNPs (blue), those ⩾1% MAF (green), rs16856199 (arrow). For comparison, the number of SNPs identified in the 1000 genomes (red, dashed) and the number of bases repeat masked (top black) and 7x regulatory potential (top blue) are also shown. Exon and intron structure of TRAX and DISC1 are drawn to scale. (b) The position and diagnoses of exonic or regulatory SNPs. SNPs not seen previously (underlined), synonymous SNPs (black) and non-synonymous SNPs (red), stop or putative splice SNPs (green). Novel SNPs not previously reported in the European samples of the 1000 Genomes Project (v3.20101123) or the NHLBI GO Exome Sequencing Project (ESP6500) or relevant sequencing and association studies10, 16, 17, 64, 65 are underlined.
Association and segregation of common variants with psychiatric illness and related quantitative traits
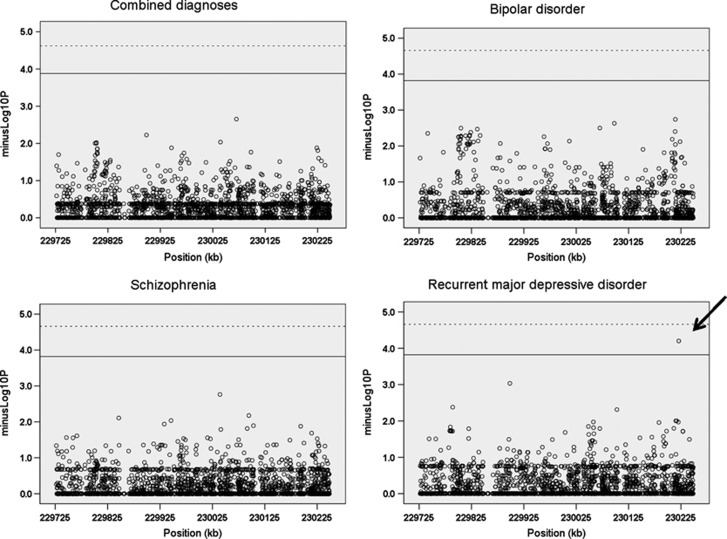
Genome-wide association studies of SZ, BP and rMDD are most consistent with a polygenic liability for common variants, but they also imply that there is real ‘missing' genetic variation, which is most likely due to risk variants having low frequency in the population. To test for evidence of DISC1 association, we applied the Fisher's exact test across all variants and all diagnoses (Figure 2). There was no evidence for SNP association at genome-wide levels of significance for any diagnosis when considered separately or combined, nor was there evidence for locus-wide association of variants with SZ or BP. We did detect a novel, locus-wide empirical association P=0.026 (OR=3.48, 95% CI=1.95–6.23, unadjusted P=6.3 × 10−5) between intronic variant rs16856199 and rMDD. We speculated that individual risk alleles might be predicted to segregate with disease in families. Twelve additional family members were available for genotyping for four rs16856199 carriers. The rs16856199 risk allele segregated with rMDD in all four families (Supplementary Figure S4).
Figure 2.
Region-wide association analysis for schizophrenia, bipolar and recurrent major depressive disorder. Nominal P-values for Fisher's exact tests are plotted against genomic location (hg18) across the TRAX/DISC1 (Disrupted in schizophrenia 1) locus. Reference lines represent 1% (dashed) and 5% (solid) region-wide empirical thresholds. Only the association of rs16856199 and recurrent major depressive disorder remains significant at the 5% threshold (arrow).
We next tested for association of rs1685199 with depression in three additional sample sets: a group of individuals referred from primary care to a hospital outpatient clinic (n=467 rMDD patients), and two population-based samples drawn from primary care as part of the Generation Scotland: Scottish Family Health Study consisting of 645 cases with rMDD and 690 cases with single episode MDD. All three groups were compared with 4017 controls drawn exclusively from Generation Scotland: Scottish Family Health Study (Supplementary additional text and Table S4). No significant association was seen with any individual replication set or all three combined (best P=0.088). Analysis of all three rMDD sets, both the original set and the two rMDD replication sets, was supportive of association (1112 rMDD, 4017 controls; P=0.0058, OR=1.46, 95%CI=1.12–1.91). Combined analysis of both sets of individuals referred from primary care showed stronger nominal association (P=0.00065, OR=1.76, 95%CI=1.27–2.44). No association was seen in the combined analysis of both the rMDD and MDD population-based replication sets (P=0.41). The risk allele for rs16856199 did not segregate with rMDD in 10 families of carriers identified from the Generation Scotland replication sample (Supplementary Figure S5). This suggests that there is increased evidence for association of rs16856199 in the more severely affected individuals.
SNP rs16856199 is on the Affymetrix 6.0 array, but the best tagging SNP on the Illumina 660W-Quad, Human Hap, Human1M-Duo arrays is rs16856189. SNP rs16856189 has an r2 of 0.27 with rs16856199, which may explain in part why this association has not been reported previously in genome-wide association studies.22, 62, 63 SNP rs6678723, which lies 2.1 kb distal to this SNP within intron 11, showed the most significant association of DISC1 in the recent mega-analysis of depression (P=0.0092).20
The LBC1936 has quantitative measures of symptoms of anxiety, depression and the personality trait of neuroticism, plus psychometrically tested measures of cognitive ability (fluid (age sensitive) and crystallised (non-age sensitive)) and cognitive ageing,60, 64 which have been shown to be highly heritable and polygenic.65, 66 Association of these traits with DISC1 was tested by linear regression analyses, co-varied for age at testing and sex (Supplementary Figures S6–S8). There were no region-wide significant findings for any of these quantitative traits.
Estimating the net pool of DISC1 variants
To estimate the effective pool of common and rare sequence variants in the European population, we applied a ‘mark-recapture' approach (see Supplementary methods) to our data and that of the 1000 Genomes Project (v3.20101123)67 after appropriate checks on read depth and Sanger sequence validation (Supplementary Table S5; Supplementary Figure 9). The total number of DISC1 SNPs ⩾1% MAF was estimated at 905 (95%CI=905±5), of which 901 (99.5%) are known (Supplementary Table S6). The number of rare variants (<1% MAF) is less confidently predicted, but is likely to be substantially higher (95%CI=3777±252) (Supplementary Table S6). Thus, despite the ∼2500 European genomes in which the DISC1 locus has been completely sequenced and the 2305 rare DISC1 variants now known, ∼40% or more remain to be discovered, and will be essentially ‘private'.
Rare amino-acid substitutions
Of the 17 rare coding SNPs previously reported for DISC1,10, 16, 17, 68, 69 we identified 12 (70.6%) plus an additional 8 non-synonymous variants, of which 5 are also absent from the European samples of both the 1000 Genomes Project (v3.20101123)61 and the Exome Variant Server (NHLBI GO Exome Sequencing Project (ESP), Seattle, WA; http://evs.gs.washington.edu/EVS/; September, 2012), and previous DISC1 sequencing studies.10, 11, 13, 14, 16, 17, 23, 46, 70 (Figure 1; Supplementary Figure S3; Supplementary Tables S2 and S3).
Five variants, R37W, T453M, T603M, L607F and S704C, are predicted to be deleterious by all three applied prediction algorithms (PolyPhen-2, Pmut and Panther; Supplementary Table S2). From this set, the functional effects of the common variants L607F (rs6675281) and S704C (rs821616) have been well documented in the literature, as mentioned earlier. R37W lies within a defined nuclear localisation signal71 and PDE4B binding site72 and is seen in a single case of rMDD (discussed at the end of this section). T453M is present at low frequency in cases and control individuals, both in this study and others (Supplementary Table S3). T603M was only identified in a single control, but Song et al.11 reported a T603I variant in a schizophrenic individual that was absent in their set of control individuals (Supplementary Table S3).
Five variants were only observed in affected individuals from our study (R37W, A83V, W160L, R233K and R418H), but not in 889 control individuals. Four of these non-synonymous case-only singletons are located in the largest coding exon, DISC1 exon 2, and the remaining variant is in DISC1 exon 4 (Figure 1). A83V was seen in a single individual with BD; this variant is predicted to be deleterious by PolyPhen-2 and Pmut and has been shown to affect wnt signalling.41 It was however observed at low frequency in controls and in individuals with partial agenesis of the corpus callosum in previous studies (Supplementary Table S3).11, 14, 16 Apart from R37W and A83V, none of the three remaining non-synonymous variants, W160L, R233K and R418H, are consistently predicted by the three prediction algorithms to have functional effects (Supplementary Table S2). 160L and 418H were detected in single SCZ individuals and have also previously been reported in individuals with SCZ; but 160L has also been detected in control individuals.10 The variant 233K has not been previously reported, and was identified in an individual with rMDD. No non-synonymous variants in TRAX were found in cases only. A single stop mutation was identified in a control individual and produces an alternative stop site for the DISC1 Es isoform. These variants can now be tested for potential impact on DISC1 biophysical properties, protein interaction and biological function.29, 30, 31
Evidence for familial segregation was sought for five rare exonic variants where additional family members were available, but none segregated perfectly or unequivocally with diagnosis (Supplementary Figure 9). Of note however was the identification of the non-synonymous amino-acid variant R37W (rs137948488), first reported68 in a subject with SZ, and seen here in a single case of rMDD. R37 is strictly conserved among orthologues and recent publications, including our own, have demonstrated biological effects of 37W on DISC1 interactions,32, 73 and shown a dominant-negative effect on the sub-cellular distribution of DISC1.44 Five additional family members of the 37W carrier were available for genotyping diagnosed with rMMD, generalised anxiety disorder, bipolar II or no psychiatric diagnosis at the time of assessment. The R37W mutation was present in relatives with rMDD and generalised anxiety disorder, but not in a relative with bipolar II, or any unaffected individual (Figure 3).
Figure 3.
Segregation of the R37W polymorphism with psychiatric diagnoses in a small Scottish family. The proband of the family is indicated (arrow). The codon containing the T allele encodes for the amino acid tryptophan (W) and the codon containing the A allele encodes arginine (R). rMDD, recurrent major depressive disorder; BP2, bipolar II.
Burden analysis for putative functional variants
To explore the burden of SNPs of potential functional significance, all variants with MAF <1% were first validated by ABI3730 sequencing. There was no significant overall difference in the number of singleton variants (Supplementary Figure S10) or in the overall number of minor alleles by diagnosis (see Supplementary Methods). SNPs were classified on the basis of bioinformatic annotation into seven functional classes: those in exons including untranslated exons, coding sequence, non-synonymous coding SNPs, conserved regions, regions with regulatory potential, conserved transcription factor binding sites and CpG islands (see Materials and methods and Supplementary Table S7). The empirical P-values for the burden analysis were obtained by permutation correcting for the multiple thresholds tested, but not for the multiple functional subgroups or diagnostic classes (SZ, BP and rMDD and all cases combined), therefore all results are reported as nominal significance values (Supplementary Table S8; Supplementary Methods). Details of nominally significant results are given in Table 1. For rMDD only, there was a nominally significant (P=0.044) excess of minor alleles for SNPs with regulatory potential across all frequencies, and for rare SNPs in conserved regions with MAF ⩽0.18% (P=0.022). Nominally significant association was found in the LBC1936 data between the burden of minor alleles across all frequencies for SNPs in conserved transcription factor binding sites and increased symptoms of depression, a measure of depressed mood at the time of testing (Table 1). In addition, a nominally significant increase in burden of minor alleles for SNPs in CpG islands or coding SNPs was observed with Moray House Test scores, measures of cognitive ability (Supplementary Table S9). Summaries of the nominally significant results are given in Table 1.
Table 1. Summary of nominally significant burden results.
| Case–control | Trait | SNP subset | Test | Average excess of variants in cases | Optimised MAF threshold (N. SNP) | P-value |
|---|---|---|---|---|---|---|
| rMDD | RegPot | BURDEN | 1.39 | — | 0.044 | |
| rMDD | PhastCon | VTTEST | 1.20 | 0.0018 (31) | 0.022 | |
|
Quantitative traits |
Trait |
SNP subset |
Test |
Effect size (beta) |
Optimised MAF threshold (N. SNP) |
P-value |
| Symptoms of depressiona | TFBS | BURDEN | 0.062 | — | 0.032 | |
| Moray House Test at age 70b | PhastCon | VTTEST | −0.074 | 0.0013 (21) | 0.030 | |
| Moray House Test at age 70 adjusted for the Moray House Test score at age 11b | CpG | VTTEST | −0.064 | 0.00060 (2) | 0.047 | |
| Moray House Test at age 11b | Coding | VTTEST | −0.101 | 0.00070 (12) | 0.028 | |
| Moray House Test at age 11b | CpG | VTTEST | −0.097 | 0.31(3) | 0.040 |
Abbreviations: MAF, minor allele frequency; rMDD, recurrent major depressive disorder; SNPs, single-nucleotide polymorphisms.
See main text for trait descriptions and Supplementary Table S8 for results on all diagnoses and traits. Quantitative trait analysis was one-tailed under the hypothesis that an increased burden of minor alleles would reduce scores for cognitive traits and increase scores for anxiety, depression and neuroticism.
References for all quantitative traits are available in the open-access Lothian Birth Cohort protocol paper (Deary et al.60). Coding: SNPs in protein-coding regions of exons including both synonymous and non-synonymous SNPs. PCon, SNPs within regions of conservation in placental mammals (PhastCon, UCSC). RegPot: SNPs with putative regulatory potential (UCSC, 7 x regulatory potential). CpG: SNPs in CpG islands commonly associated with promoter regions.
Tested using the Hospital Anxiety Depression Scales.
The Moray House Test is the general cognitive test—mostly a verbal reasoning, IQ-type test—that was used in the Scottish Mental Survey 1947.
Discussion
Diagnoses of SZ, BP and rMDD were all present in the original Scottish family carrying the translocation that disrupts DISC1. All three DSMIV diagnoses have a strong and overlapping genetic component, but robust statistical analysis of gene-level contributions to risk are complicated by extensive genetic heterogeneity within and between diagnoses.4 We have provided the most comprehensive landscape of genetic variation at the DISC1 locus to date in patients with this spectrum of psychiatric illness and in healthy population controls with quantitative measures of mood and cognition. Comparison between our sequencing study and that of the 1000 Genomes Project confirms that current genome-wide association studies effectively captures the majority of common (but not rare) variants in the European population. Our sample size is large by current sequencing study standards, but we lack power to detect genome-wide significant P-values for either common or rare variants (see Supplementary Information and Supplementary Figure S11 for further details and also Kiezun et al.).74 Indeed, the predicted abundance of independent rare variants at this (and any other given) locus makes it highly improbable that any one will contribute to illness in the population at a frequency that will be statistically significant, given the numbers of patients we can afford to analyse by direct sequencing.74
We observed no evidence for association at the whole-genome level of statistical significance between individual rare or common variants and either psychiatric illness or cognition. This is consistent with recent findings,74 which suggest that much larger samples would be required to detect such associations. Burden analysis of multiple rare and/or deleterious putative functional variants also failed to show association with these traits. We do report both functional and putative regulatory variants that are both individually, and by functional classification, nominally associated with rMDD and/or cognitive ability at the locus-wide level of significance.
Our study identified a novel association between intronic SNP rs16856199 and rMDD in hospital-referral subjects. Segregation with diagnosis in the relatives of these probands corroborated the association, but further studies are required to understand the lack of replication in population-based cohorts with depression. This may be due to inherent differences between patients recruited from hospital-referral compared with those from population-based cohorts. Cohorts from primary care are more likely to have a family history of depression,75 and may have more physical and psychiatric comorbidity in general. Conversely, the population-based sample may have shorter, less severe episodes76 than the hospital-based cohorts.77 However, given the modestly significant P-value for rMDD in the discovery cohort, the number of psychiatric traits examined and the lack of replication, it is possible that the observed association is due to chance.
The nominal associations of the burden of common (threshold MAF=30.7%) and rare potentially regulatory variants (threshold MAF=0.060%) to measures of cognitive ability merit further study. A yet-to-be-defined subset of these is likely to have critical roles is spatial and temporal regulation of transcription and splicing.46, 78 This highlights the need for annotation tools with improved predictive value for non-coding variants.79
More importantly, our study demonstrates that substantial coding and non-coding genetic variation at the DISC1 locus remains undiscovered. Despite sequencing over 1500 subjects, we have probably captured only ∼40% of the extant DISC1 variants in just the European population. Crowley et al.23 sequenced 2.7 kb of DISC1 exons and 5' and 3' regulatory sequence in 1460 samples of European or African origin. We observed 13/38 (34%) of the variants genotyped in the replication phase, supporting the argument for an abundance of rare variants.
The level of sequence variation identified in our study is unlikely to be exceptional, and indeed is consistent with evidence emerging from other genome sequencing studies.74 Consequently, it will be challenging to demonstrate robust (replicated) association by statistical evidence alone in case–control studies, exceptionally so with the numbers of patients that are currently affordable for sequencing. The original t(1;11) family illustrates the added issue of variable penetrance and cross-boundary diagnosis for a given mutation: ∼70% of carriers had SZ, BP or rMDD, but ∼30% had no formal psychiatric diagnosis, yet t(1;11) carriers, including both affected and unaffected, had ERP P300 measures in the range typical of individuals with SZ.5 The original identification of 37W in a case of SZ and here in a case of rMDD (and two offspring, one with rMDD, the other generalised anxiety disorder) may suggest variable penetrance of this biologically functional variant.44 Of note, the 37W variant was not observed in 10 000 control individuals,11 the 1000 Genomes project,61 the NHLBI GO Exome Sequencing Project (ESP), nor any of our 889 control samples. These findings on R37W reinforce the probable importance of this domain for DISC1 subcellular distribution and binding of interacting proteins31, 44 and add to the weight of evidence for other functional studies of DISC1 amino-acid substitutions.41 Each observed amino-acid substitution provides a similar opportunity to tease out the relationships between genotype and phenotype and between structure and function.29, 31 Overall, these results demonstrate a high level of sequence variation in DISC1, a subset of which may contribute to psychiatric disorder in some individuals who will be typically rare in the population precluding classical statistical analysis and requiring biological validation. This predicts a population-specific contribution of rare casual variants to risk.80 Our results indicate the potential value of sequencing non-coding regions of the genome, which may harbour disease-associated regulatory variants. Our findings of both functional and putative regulatory variants nominally associated with depression and cognitive ability merit replication in independent samples and biological exploration.
Acknowledgments
We acknowledge the contribution of Prof Walter M Muir (deceased) for the collection of the clinical samples used in this study. We thank Gail Davies, Dave Liewald, Lee Murphy, Alan Maclean, Pamela Linksted, Archie Campbell, Shona Kerr, Andrew McQuillan and Stuart Macgregor for help in preparing samples, data and statistical advice and Generation Scotland for access to GS:SFHS samples and data. We thank the LBC1936 cohort members. We thank the LBC1936 study secretary Paula Davies and Alan Gow, Janie Corley and Caroline Brett, for cognitive data collection and data entry. We thank the nurses and staff at the WTCRF where the medical phenotypic data and DNA were collected. We thank the staff at the Lothian Health Board, and the staff at the SCRE Centre, University of Glasgow. The LBC1936 research was supported by a programme grant from Research Into Ageing and continues with programme grants from Help the Aged/Research Into Ageing (Disconnected Mind). The research was done within the University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, supported by the BBSRC, EPSRC, ESRC and MRC, as part of the cross-council Lifelong Health and Wellbeing Initiative (G0700704/84698). Generation Scotland receives core support from the Chief Scientist Office of the Scottish Government Health Directorates CZD/16/6/2 and the Scottish Funding Council. We are grateful to the GS Executive Committee Professors Andrew D Morris, Blair H Smith, Anna F Dominiczak, David J Porteous and Drs Lynne J Hocking and Sandosh Padmanabhan and all the families who took part, the general practitioners and the Scottish School of Primary Care for their help in recruiting them, and the whole Generation Scotland team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, health-care assistants and nurses. We thank Sharon Mitchell and Carolann Gundersen for help in the preparation of this manuscript. This work was supported by a gift from T and V Stanley to CSHL, the UK Medical Research Council (G0902166), the Wellcome Trust (WT 008179), the Chief Scientists Office of the Scottish Government Department of Health (CZD/16/6/), a Scottish Funding Council Strategic Research Development Grant award (HR03006) and a Lifelong Health and Wellbeing (G0700704/84698). AMcR and PMV acknowledge support from the Australian National Health and Medical Research Council.
Author contributions
DJP, PT and WRM designed the study, interpreted the data and wrote the paper. WJM, DJM, AMM and DHB were responsible for the psychiatric evaluation, selection and provision of case samples. DJP, PT, IJD, DLM, AMM and DHB were responsible for the provision of GS:SFHS samples and associated phenotypic measures. SEH, JMS and IJD were responsible for the provision of LBC1936 samples and associated phenotypic measures. JSP developed the sample processing methodology and edited the paper. MK developed the sequence analysis pipeline and conducted the quality control of the sequence data and edited the paper. PT, AFM and PMV conducted the statistical analysis. KR, DCS, SM, SC and PT conducted the bioinformatics analysis. All authors contributed to revise the paper.
WRM has participated in Illumina sponsored meetings over the past four years and received travel reimbursement and an honorarium for presenting at these events.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Gottesman II, Laursen TM, Bertelsen A, Mortensen PB. Severe mental disorders in offspring with 2 psychiatrically ill parents. Arch Gen Psychiatry. 2010;67:252–257. doi: 10.1001/archgenpsychiatry.2010.1. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eykelenboom JE, Briggs GJ, Bradshaw NJ, Soares DC, Ogawa F, Christie S, et al. A t(1;11) translocation linked to schizophrenia and affective disorders gives rise to aberrant chimeric DISC1 transcripts that encode structurally altered, deleterious mitochondrial proteins. Hum Mol Genet. 2012;21:3374–3386. doi: 10.1093/hmg/dds169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Zhou X, Chen Q, Schaukowitch K, Kelsoe JR, Geyer MA. Insoluble DISC1-Boymaw fusion proteins generated by DISC1 translocation. Mol Psychiatry. 2010;15:669–672. doi: 10.1038/mp.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone M, Thomson PA, Hall J, McIntosh AM, Lawrie SM, Porteous DJ. DISC1 in schizophrenia: genetic mouse models and human genomic imaging. Schizophrenia Bull. 2011;37:14–20. doi: 10.1093/schbul/sbq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens LN, De Rijk P, Reumers J, Van den Bossche MJ, Glassee W, De Zutter S, et al. Sequencing of DISC1 pathway genes reveals increased burden of rare missense variants in schizophrenia patients from a northern Swedish population. PLoS ONE. 2011;6:e23450. doi: 10.1371/journal.pone.0023450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Li W, Feng J, Heston LL, Scaringe WA, Sommer SS. Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem Biophys Res Commun. 2008;367:700–706. doi: 10.1016/j.bbrc.2007.12.117. [DOI] [PubMed] [Google Scholar]
- Bradshaw NJ, Porteous DJ. DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology. 2012;62:1230–1241. doi: 10.1016/j.neuropharm.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carless MA, Glahn DC, Johnson MP, Curran JE, Bozaoglu K, Dyer TD, et al. Impact of DISC1 variation on neuroanatomical and neurocognitive phenotypes Mol Psychiatry 2011161096–1104.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Li W, Noltner K, Yan J, Green E, Grozeva D, et al. Identification of high risk DISC1 protein structural variants in patients with bipolar spectrum disorder. Neurosci Lett. 2010;486:136–140. doi: 10.1016/j.neulet.2010.09.027. [DOI] [PubMed] [Google Scholar]
- Williams JM, Beck TF, Pearson DM, Proud MB, Cheung SW, Scott DAA. 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. Am J Med Genet A. 2009;149A:1758–1762. doi: 10.1002/ajmg.a.32941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbun N, Li J, O'Driscoll MC, Strominger Z, Wakahiro M, Rider E, et al. Genetic and functional analyses identify DISC1 as a novel callosal agenesis candidate gene. Am J Med Genet A. 2011;155A:1865–1876. doi: 10.1002/ajmg.a.34081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EK, Grozeva D, Sims R, Raybould R, Forty L, Gordon-Smith K, et al. DISC1 exon 11 rare variants found more commonly in schizoaffective spectrum cases than controls. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:490–492. doi: 10.1002/ajmg.b.31187. [DOI] [PubMed] [Google Scholar]
- Mathieson I, Munafo MR, Flint J. Meta-analysis indicates that common variants at the DISC1 locus are not associated with schizophrenia. Mol Psychiatry. 2012;17:634–641. doi: 10.1038/mp.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MY, Levinson DF, Faraone SV, Suarez BK, DeLisi LE, Arinami T, et al. Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Mol Psychiatry. 2009;14:774–785. doi: 10.1038/mp.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium PG A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2012;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley JJ, Hilliard CE, Kim Y, Morgan MB, Lewis LR, Muzny DM, et al. Deep resequencing and association analysis of schizophrenia candidate genes. Mol Psychiatry. 2012;18:138–140. doi: 10.1038/mp.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Wang L, Fanous AH, Pato CN, Edwards TL, Zhao Z. Network-assisted investigation of combined causal signals from genome-wide association studies in schizophrenia. PLoS Comput Biol. 2012;8:e1002587. doi: 10.1371/journal.pcbi.1002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- Tiwary BK. The severity of mental disorders is linked to interaction among candidate genes. Integr Biol (Camb) 2012;4:1096–1101. doi: 10.1039/c2ib20066j. [DOI] [PubMed] [Google Scholar]
- Insel TR. Rethinking schizophrenia. Nature. 2010;468:187–193. doi: 10.1038/nature09552. [DOI] [PubMed] [Google Scholar]
- Porteous DJ, Millar JK, Brandon NJ, Sawa A. DISC1 at 10: connecting psychiatric genetics and neuroscience. Trends Mol Med. 2011;17:699–706. doi: 10.1016/j.molmed.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci. 2011;12:707–722. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares DC, Carlyle BC, Bradshaw NJ, Porteous DJ. DISC1: Structure, Function, and Therapeutic Potential for Major Mental Illness. ACS Chem Neurosci. 2011;2:609–632. doi: 10.1021/cn200062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Numakawa T, Ohnishi T, Kumamaru E, Yagasaki Y, Ishimoto T, et al. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum Mol Genet. 2006;15:3024–3033. doi: 10.1093/hmg/ddl244. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Tomoda T, Chang J, Takaki M, Zhan C, Morita M, et al. DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Hum Mol Genet. 2006;15:3313–3323. doi: 10.1093/hmg/ddl407. [DOI] [PubMed] [Google Scholar]
- Burdick KE, Kamiya A, Hodgkinson CA, Lencz T, DeRosse P, Ishizuka K, et al. Elucidating the relationship between DISC1, NDEL1, and NDE1 and the risk for schizophrenia: Evidence of epistasis and competitive binding. Hum Mol Genet. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliveld SR, Hendriks P, Michel M, Sajnani G, Bader V, Trossbach S, et al. Oligomer assembly of the C-terminal DISC1 domain (640–854) is controlled by self-association motifs and disease-associated polymorphism S704C. Biochemistry. 2009;48:7746–7755. doi: 10.1021/bi900901e. [DOI] [PubMed] [Google Scholar]
- Eastwood SL, Hodgkinson CA, Harrison PJ. DISC-1 Leu607Phe alleles differentially affect centrosomal PCM1 localization and neurotransmitter release. Mol Psychiatry. 2009;14:556–557. doi: 10.1038/mp.2009.13. [DOI] [PubMed] [Google Scholar]
- Atkin TA, MacAskill AF, Brandon NJ, Kittler JT.Disrupted in Schizophrenia-1 regulates intracellular trafficking of mitochondria in neurons Mol Psychiatry 201116122–124.121. [DOI] [PubMed] [Google Scholar]
- Eastwood SL, Walker M, Hyde TM, Kleinman JE, Harrison PJ. The DISC1 Ser704Cys substitution affects centrosomal localization of its binding partner PCM1 in glia in human brain. Hum Mol Genet. 2010;19:2487–2496. doi: 10.1093/hmg/ddq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, De Rienzo G, Drane L, Mao Y, Flood Z, Madison J, et al. Common DISC1 polymorphisms disrupt Wnt/GSK3beta signaling and brain development. Neuron. 2011;72:545–558. doi: 10.1016/j.neuron.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling.[see comment] Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, et al. Disrupted in Schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK32/2-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavasi EL, Ogawa F, Porteous DJ, Millar JK. DISC1 variants 37W and 607F disrupt its nuclear targeting and regulatory role in ATF4-mediated transcription. Hum Mol Genet. 2012;21:2779–2792. doi: 10.1093/hmg/dds106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Christie S, Semple CA, Porteous DJ. Chromosomal location and genomic structure of the human translin-associated factor X gene (TRAX; TSNAX) revealed by intergenic splicing to DISC1, a gene disrupted by a translocation segregating with schizophrenia. Genomics. 2000;67:69–77. doi: 10.1006/geno.2000.6239. [DOI] [PubMed] [Google Scholar]
- Hennah W, Porteous D. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS ONE. 2009;4:e4906. doi: 10.1371/journal.pone.0004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson PA, Macintyre DJ, Hamilton G, Dominiczak A, Smith BH, Morris A, et al. Association of DISC1 variants with age of onset in a population-based sample of recurrent major depression. Mol Psychiatry. 2012;18:745–747. doi: 10.1038/mp.2012.117. [DOI] [PubMed] [Google Scholar]
- Walker RM, Hill AE, Newman AC, Hamilton G, Torrance HS, Anderson SM, et al. The DISC1 promoter: characterization and regulation by FOXP2. Hum Mol Genet. 2012;21:2862–2872. doi: 10.1093/hmg/dds111. [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelala C, Khan A, Lemoine NR. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25:655–661. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel S. Using and understanding RepeatMasker. Methods Mol Biol. 2012;859:29–51. doi: 10.1007/978-1-61779-603-6_2. [DOI] [PubMed] [Google Scholar]
- Ferrer-Costa C, Gelpi JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CGJ. The yearly immigration of young plaice into the Limfjord from the German Sea. Danish Biol Station. 1896;6:1–77. [Google Scholar]
- Lincoln FC.Calculating Waterfowl Abundances on the Basis of Banding Returns 1930, US Department of Agriculture Circular; 118, 1–4. [Google Scholar]
- Chapman DG. Some properties of the hypergeometric distribution with applications to zoological sample censuses. University of California Press: Berkeley; 1951. [Google Scholar]
- Deary IJ, Gow AJ, Taylor MD, Corley J, Brett C, Wilson V, et al. The Lothian Birth Cohort 1936: a study to examine influences on cognitive ageing from age 11 to age 70 and beyond. BMC Geriatr. 2007;7:28. doi: 10.1186/1471-2318-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TGP A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry. 2012;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Gow AJ, Harris SE, Hayward C, Allerhand M, Starr JM, et al. Cognitive ability at age 11 and 70 years, information processing speed, and APOE variation: the Lothian Birth Cohort 1936 study. Psychol Aging. 2009;24:129–138. doi: 10.1037/a0014780. [DOI] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ, Yang J, Davies G, Harris SE, Tenesa A, Liewald D, et al. Genetic contributions to stability and change in intelligence from childhood to old age. Nature. 2012;482:212–215. doi: 10.1038/nature10781. [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Li W, Feng J, Heston LL, Scaringe WA, Sommer SS. Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem Biophys Res Commun. 2008;367:700–706. doi: 10.1016/j.bbrc.2007.12.117. [DOI] [PubMed] [Google Scholar]
- Song W, Li W, Noltner K, Yan J, Green E, Grozeva D, et al. Identification of high risk DISC1 protein structural variants in patients with bipolar spectrum disorder. Neurosci Lett. 2010;486:136–140. doi: 10.1016/j.neulet.2010.09.027. [DOI] [PubMed] [Google Scholar]
- Hennah W, Thomson P, McQuillin A, Bass N, Loukola A, Anjorin A, et al. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14:865–873. doi: 10.1038/mp.2008.22. [DOI] [PubMed] [Google Scholar]
- Sawamura N, Ando T, Maruyama Y, Fujimuro M, Mochizuki H, Honjo K, et al. Nuclear DISC1 regulates CRE-mediated gene transcription and sleep homeostasis in the fruit fly Mol Psychiatry 2008131138–1148.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch H, Mackie S, Collins DM, Hill EV, Bolger GB, Klussmann E, et al. Isoform-selective susceptibility of DISC1/phosphodiesterase-4 complexes to dissociation by elevated intracellular cAMP levels. J Neurosci. 2007;27:9513–9524. doi: 10.1523/JNEUROSCI.1493-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, De Rienzo G, Drane L, Mao Y, Flood Z, Madison J, et al. Common DISC1 polymorphisms disrupt Wnt/GSK3beta signaling and brain development. Neuron. 72:545–558. doi: 10.1016/j.neuron.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, McLaren PJ, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Wells JE, Joyce PR, Bushnell JA, Mulder RT, Oakley-Browne MA. Family history of depression in clinic and community samples. J Affect Disord. 1996;40:159–168. doi: 10.1016/0165-0327(96)00056-0. [DOI] [PubMed] [Google Scholar]
- Eaton WW, Anthony JC, Gallo J, Cai G, Tien A, Romanoski A, et al. Natural history of Diagnostic Interview Schedule/DSM-IV major depression. The Baltimore Epidemiologic Catchment Area follow-up. Arch Gen Psychiatry. 1997;54:993–999. doi: 10.1001/archpsyc.1997.01830230023003. [DOI] [PubMed] [Google Scholar]
- Solomon DA, Keller MB, Leon AC, Mueller TI, Shea MT, Warshaw M, et al. Recovery from major depression. A 10-year prospective follow-up across multiple episodes. Arch Gen Psychiatry. 1997;54:1001–1006. doi: 10.1001/archpsyc.1997.01830230033005. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu AB, Le-Niculescu H. The P-value illusion: how to improve (psychiatric) genetic studies. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:847–849. doi: 10.1002/ajmg.b.31076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.