Abstract
Thyroid hormone responsive protein Spot 14 has been consistently associated with de novo fatty acid synthesis activity in multiple tissues, including the lactating mammary gland, which synthesizes large quantities of medium chain fatty acids (MCFAs) exclusively via FASN. However, the molecular function of Spot14 remains undefined during lactation. Spot14-null mice produce milk deficient in total triglyceride and de novo MCFA that does not sustain optimal neonatal growth. The lactation defect was rescued by provision of a high fat diet to the lactating dam. Transgenic mice overexpressing Spot14 in mammary epithelium produced total milk fat equivalent to controls, but with significantly greater MCFA. Spot14-null dams have no diminution of metabolic gene expression, enzyme protein levels, or intermediate metabolites that accounts for impaired de novo MCFA. When [13C] fatty acid products were quantified in vitro using crude cytosolic lysates, native FASN activity was 1.6-fold greater in control relative to Spot14-null lysates, and add back of Spot14 partially restored activity. Recombinant FASN catalysis increased 1.4-fold and C = 14:0 yield was enhanced 4-fold in vitro following addition of Spot14. These findings implicate Spot14 as a direct protein enhancer of FASN catalysis in the mammary gland during lactation when maximal MCFA production is needed.
Keywords: fatty acid synthase catalysis, [13C] fatty acid quantification, medium chain fatty acids, lactation
Mammalian cells manufacture fatty acids de novo using a distinct pathway to synthesize fatty acids from acetyl and malonyl esters of CoA catalyzed by the dimer of FASN (EC 2.3.1.85) (1). FASN is absolutely essential for production of de novo fatty acids in mammals (2). The biological requirement for FASN is highlighted by the fact that FASN is detected in most normal human tissues and that homozygous deletion of FASN in the mouse results in embryonic lethality (3, 4). The importance of the de novo pathway is underscored by the complex biological functions of its fatty acid products, including biosynthesis of phospholipids, energy storage via esterification into triglycerides (TAGs), use as energy substrates for β-oxidation, providing both endocrine and nuclear hormone signaling ligands, and for critical posttranslational modifications of proteins (5–12). Perhaps the most demanding setting for FASN catalysis is during lactation, when extremely high quantities of de novo fatty acids are transmitted to the offspring as substrate for the growth of the young (13–15).
Regulation of the principle enzymes of de novo fatty acid synthesis, ATP citrate lyase (ACLY) (EC 2.3.3.8), acetyl-CoA carboxylase (ACC) (EC 6.4.1.2), and FASN, has been shown to occur at the level of enzyme gene expression (16), translation/degradation (17–19), phosphorylation (20–22), assembly into multimeric complexes (23–25), and allosteric activation/inhibition (18, 23, 26, 27). Recently, evidence for the control of de novo fatty acid synthesis by small effector proteins has emerged. Thyroid hormone responsive protein “Spot 14” (THRSP) (Spot14, S14) and only family member Spot14-related protein (Spot14-R) (MIG12, Mid1IP1) are postulated to function as effector proteins for the de novo fatty acid synthesis pathway (28, 29). Spot14 was discovered in 1981 and was established to be a small acidic protein of 17 kDa with an isoelectric point of 4.85 (30). It has no known enzymatic function, the primary sequence predicts no established functional domains, and the crystal structure has identified three antiparallel α-helices in an asymmetrical dimer (28, 31). Spot14 mRNA is sharply induced in the liver of streptozotocin-induced diabetic rats by insulin treatment, as well as in livers of fasted rats refed carbohydrates and acutely treated with thyroid hormone (32, 33). Its gene expression in the liver was decreased in rats fed a diet containing long chain PUFAs, and in the lactating mammary gland following administration of trans-10 cis-12 conjugated linoleic acid (34–36). In 2003, we observed that Spot14 was induced in unison with 75 metabolic genes in the mouse mammary gland at secretory activation (37). More recently, we showed regulation of Spot14 mRNA by sterol response element binding protein 1 in mammary epithelial cells (MECs) depleted of the adipose compartment, and that Spot14 mRNA in MECs was not suppressed by provision of a high fat diet (HFD) to the dam (13). Others have shown evidence, independent of rodent models, for Spot14 involvement during milk fat production in dairy cows (36, 38). Subsequent analysis of the Spot14-null mouse firmly established Spot14 as necessary for the de novo synthesis of medium chain fatty acids (MCFAs) characteristic of milk TAG (39). Hence, in lactation, Spot14 appears to enhance de novo synthesis of the MCFAs that contribute to the total milk energy content needed for neonatal growth.
Both activation and inhibition of de novo fatty acid synthesis by Spot14 have been reported, depending upon the lipogenic tissue type and the cellular context (28, 29, 31, 39–42). For example, the rate of de novo fatty acid synthesis in the Spot14-null mammary gland was decreased over 2-fold, implicating Spot14 as an activator of de novo fatty acid synthesis in this tissue (39). Conversely, Spot14-null hepatic de novo fatty acid synthesis was increased 2-fold under lipogenic stimuli, suggesting Spot14 is an inhibitor of this process in the liver (40). Coexpression of Spot14 with Spot14-R was determined to be inhibitory, as Spot14 heterodimerizes with Spot14-R to prevent Spot14-R association with ACC and decrease activation of the hepatic de novo fatty acid synthesis pathway (43). In adult neural stem cells, Knobloch et al. (29) demonstrated cytosolic localization and coimmunoprecipitation of Spot14 and Spot14-R, and that Spot14 overexpression decreased de novo synthesis of fatty acids in low proliferating cells. Colbert et al. (28) used liver-specific antisense Spot14 knock down in chow-fed mice resulting in approximately 30% decreased 3H2O incorporation into de novo lipid. This work was supported by efforts from Aipoalani et al. (41), who knocked down both Spot14 and Spot14-R in primary hepatocyte culture and observed a 65% reduction of [14C]-acetate incorporation into de novo fatty acids. Thus, overlapping and conditional roles for Spot14 regulation of fatty acid synthesis have been demonstrated (29, 41). In mammary cancer cells, antisense Spot14 knock down decreased 14C-acetate incorporation into lipid nearly 60%, supporting a role for Spot14 in de novo fatty acid synthesis activation (44). Cumulatively, these published findings indicate that Spot14 may both positively or negatively regulate the de novo fatty acid synthesis pathway depending on the tissue-specific or cellular context, as well as the presence of Spot14-R.
The lactating mammary gland lacks expression of Spot14-R, meaning that the MEC-specific function of Spot14 is independent from Spot14-R. In the lactating mammary gland, Spot14 is considered an activator of de novo fatty acid synthesis because the milk produced by Spot14-null dams contains 60% less MCFAs (39), which are unique products of mammary alveolar de novo fatty acid synthesis during lactation (15). Zhu et al. (39) determined that the activities of enzymes upstream of FASN in the biosynthetic pathway were present and functioning in the Spot14-null mammary gland during lactation, including malic enzyme, 6-phosphogluconate dehydrogenase, glucose-6-phosphate dehydrogenase, ACC, and FASN. Importantly, in vivo tracer studies using 3H2O incorporation into milk MCFA revealed that the rate of fatty acid synthesis in Spot14-null mice was decreased approximately 2.5-fold (39). Taken together, these observations indicate that the catalytic rate of FASN in the lactating mammary gland is diminished in the Spot14-null mouse.
Here we report that tissue-specific overexpression of Spot14 in MECs increased the amount of MCFA known to originate from de novo synthesis, without altering the percent milk fat by volume or influencing offspring growth. Spot14-null milk production was not diminished, but milk from Spot14-null dams had significantly reduced fat percentage leading to restricted pup growth. We found that this loss in total milk fat and subsequent inability to support litters was rescued by provision of a diet high in fat to the Spot14-null dams. Further, we tested the hypothesis that Spot14 increases the in vitro catalytic rate of both recombinant FASN and of native FASN in MEC cytosolic lysates using a recently developed method that specifically quantifies synthesis of [13C] fatty acid products (45). Together, these results suggest the molecular function of Spot14 in MECs is to enhance FASN catalysis during lactation when de novo fatty acid synthesis is needed to achieve optimal MCFA content in the milk.
MATERIALS AND METHODS
Animals
Mice were maintained in the Center for Comparative Medicine. The Institutional Animal Care and Use Committee of the University of Colorado Denver approved all animal procedures. C57Bl/6 and FVB control mice were purchased from Jackson Laboratories (Bar Harbor, Maine). Spot14-null mice maintained on the C57B6/J background were a kind gift of Dr. Cary Mariash, and details regarding generation of these mice are described elsewhere (40). The Transgenic and Knockout Core Facility at the University of Colorado Denver generated the transgenic mice overexpressing Spot14 driven by the mouse mammary tumor virus long terminal repeat (MMTV-Spot14). The pCMV-SPORT6 vector containing mouse Spot14 was purchased from Open Biosystems (Thermo Fisher Scientific, Waltham, MA), and PCR was used to add restriction enzyme sites and the C-terminal hemaglutinin A tag (forward, 5′-CAGT-GGA-TCC-GCC-ACC-ATG-CAA-GTG-CTA-ACG-AAA-CGC-TAT-CC; reverse, 5′-CAG-TGC-GAA-TTC-TCA-AGC-GTA-GTC-TGG-GAC-GTC-GTA-TGG-GTA-CAG-GAC-CTG-CCC-TGT-CAT-TTC-C). The PCR product was ligated into pcDNA3.1 with BamHI and EcoRI, excised with HindIII and EcoRI, and ligated into MMTV-SV40-Bssk, a gift from Dr. William Muller (McGill University). MMTV-Spot14 mice were genotyped using tail DNA with the following primers: forward, 5′-CAG-CGA-GGC-TGA-GAA-CGA-C-3′ reverse, 5′-TAG-TCT-GGG-ACG-TCG-TAT-GGG-TA-3′ to generate a product of 200 base pairs.
Mice were maintained on a standard day/night cycle. FVB, C57BL/6, Spot14-null, and MMTV-Spot14 females were mated with corresponding males of the same genetic background. Pregnancy day 1 was designated as the day vaginal plug was observed, and lactation day 1 was the day that pups were born. Litters were standardized to eight pups for all studies. For pup growth studies, litters from both backgrounds were cross-fostered to avoid genetic background differences. Litters were weighed daily between 9:00 AM and noon. FVB, C57B6 control, and Spot14-null mice were maintained on standard laboratory chow (5020, Lab Diets) that contained 9% (21.5% kcal) as fat. Specialized diets were purchased from Research Diets and contained 3.9% kcal fat, 12% kcal fat, and 46% kcal fat (Research Diets, Inc., New Brunswick, NJ), and dams were provided specialized diets 1 day prior to parturition and subsequently fed ad libitum.
Milk fat and fatty acid composition analysis
Milk was collected at lactation days 4 and 10 using vacuum suction, and milk fat analysis was conducted as previously described (46). Weigh-suckle-weigh experiments were conducted according to Dempsey et al. (47). Milk protein concentrations were determined using the Pierce 660 protein assay (Thermo Fisher Scientific). Milk lactose concentrations were quantified using the lactose assay kit according to the manufacturer’s instructions (Abcam, ab83384). HPLC grade reagents were purchased from Sigma-Aldrich, St. Louis, MO. Neutral lipid extraction was adapted from Hutchins et al. (48, 49), and pentafluorobenzyl derivatization was according to Zarini et al. (50). Mouse milk was thawed at 37°C for 10 min, vortexed, and 5 μl of whole milk was transferred to 2 ml Eppendorf tubes containing 0.5 ml of 2:1 v/v methanol/water; samples were acidified with 40 μl of 1M HCL, vortexed into solution, cleared by centrifugation at 10,000 g, and supernatant was transferred into borosilicate glass tubes. Milk fat was extracted twice using 1 ml of 2:1 v/v isooctane/ethyl acetate, and samples were vortexed vigorously, allowed to settle for 5 min at room temperature, and then centrifuged at 500 g for 1 min at room temperature. Samples were taken to dryness using a Savant SpeedVac concentrator and lipid pellets were resuspended in 0.5 ml of 100% methanol.
Blended internal standards (1 μg) containing [D]3-decanoic acid, [D]3-lauric acid, [D]3-myristic acid, 1,2,3,4-[13C]4-palmitic acid, [D]3-stearic acid, [D]2-oleic acid, [D]8-arachidonic acid, and [D]5-docosahexanoic acid (purchased from Sigma-Aldrich; Cambridge Isotopes, Andover, MA; or Cayman Chemical Co., Ann Arbor, MI with 99 atom % 13C and 99 atom % D, respectively) were added to each sample. Samples were saponified by addition of 0.5 ml of 1M NaOH at 37°C for 1 h mixing at 20 min intervals; and samples were acidified with 0.6 ml of 1M HCl to a pH between 2.0 and 4.0 and mixed by vortex. Saponified fatty acids were extracted twice with 1.0 ml of isooctane; 300 μl of the saponified fatty acid fraction was transferred to new glass tubes, and samples were taken to dryness under N2. Saponified fatty acid pellets were resuspended in 30 μl of 1% pentafluorobenzyl bromide in acetonitrile and 30 μl of 1% N,N-diisopropylethylamine in acetonitrile to initiate derivatization and incubated at room temperature for 30 min (50). Samples were dried under N2, resulting pellets were resuspended in 1 ml of isooctane, vortexed, and 50 μl was transferred into vials for GC-MS. Sample analysis was performed by negative ion chemical ionization GC-MS using the Finnigan DSQ GC-MS system (Thermo Finnigan, Thousand Oaks, CA), with a ZB-l column (15 m, 0.25 mm inner diameter, 0.10 mm film thickness; Phenomenex) using methane as reagent gas. The GC temperature gradient was 125 to 230°C at 20°C/min, 230 to 245°C at 5°C/min, and finally 245 to 300°C at 30°C/min, before being held at 310°C for 1 min. Data were acquired in full scan mode to identify fatty acids of acyl chain length from 10 to 22 carbons. Peak areas of the analyte or of the standard were measured, and the ratio of the area from the analyte-derived ion to that from the internal standard was calculated.
NMR metabolomic analysis
Lipid and aqueous metabolomic analysis of both whole gland and of MEC preparations were carried out according to Rudolph et al. (51). The MEC and tissue extracts were analyzed using a Bruker 500 MHz DRX NMR spectrometer. All metabolite concentrations were assessed as micromoles per gram of tissue.
Adipose depleted MECs
MECs were prepared as previously described (13) with the following modifications: mammary digestion buffer included 5 ml of Ham’s F12 medium (Life Technologies, Grand Island, NY), 5 mg trypsin (Gibco #840-7250), 10 mg collagenase (Boehringer #1088793), 50 mM NaF, and 5.0 mM sodium orthovanadate (heat activated). Tissues were minced and digested in 5 ml of buffer as previously described. After incubation for 25 min in a 37°C shaker at 200 rpm, 1 ml of FCS was added to the digested cell suspension, brought up to 15 ml with ice-cold PBS, and spun at <300 g for 10 min. Washing was repeated two additional times with PBS. Approximately 200 μl of the resulting 500 μl of packed organoids was lysed in 1 ml of TRIzol (Life Technologies) for RNA analysis and the other portion was lysed in 1 ml of lysis buffer for protein, consisting of 50 mM Tris (pH 7.4), 150 mM NaCl, 2.0 mM EDTA, 50 mM NaF, 5.0 mM sodium orthovanadate, 1% Triton X-100, 1% deoxycholate, and 0.1% SDS, to which 0.57 mM PMSF, 20 μl/ml EDTA-free Inhibitor cocktail (Roche), and 1.0 mM DTT was added. Samples were homogenized using a Brinkman Polytron, and lysate was centrifuged at 13,000 g for 10 min at 4°C. Protein concentrations were determined using Pierce 660 protein assay (Thermo Fisher Scientific).
Quantitative real-time PCR analysis
Supplementary Table II lists the primers, probes, and amplicon formula weights for all genes used in this study. The primers and probes were purchased for use with TaqMan reaction chemistry from Integrated DNA Technologies (Coralville, IA) or from Applied Biosystems (Life Technologies) with 5′ FAM fluorophore and either 3′ Black Hole or minor groove binding quenchers, respectively. The VERSO cDNA synthesis kit (Thermo Fisher Scientific) was used, and each reaction consisted of 0.5 μg total RNA in 11 μl of nuclease-free water, to which 1.0 μl of blended 3:1 v/v random hexamers to oligo dT was added and samples were incubated at 70°C for 5 min. Samples were collected with a quick spin, and 9 μl of a mix containing 4 μl 5× buffer, 2 μl of dNTP blend, 1 μl of RT enhancer, and 1 μl of VERSO enzyme was added per reaction. The cDNA was diluted to 50 μl, and 2.5 μl was used as template into the quantitative (q)PCR reaction (representing 25 ng total RNA per reaction). A qPCR master mix was made containing 12.5 μl of Absolute Fast qPCR Mix-Lox Rox (Thermo Fisher Scientific), 1.25 μl of 20× primer/probe mix (10 μM forward, 10 μM reverse, and 5 μM probe), and 6.25 μl of nuclease-free water per reaction well, to which 2.5 μl of cDNA per reaction was added. qPCR data were collected on the Applied Biosystems 7500 Fast or the 7900 HT thermocycler (Life Technologies). PCR products representing the unique product (the amplicon) for each gene were amplified from cDNA as above, and amplicons were purified using the Qiagen MinElute PCR purification kit (Qiagen). Concentrations of purified double stranded amplicons for each gene were measured using the NanoDrop (Thermo Fisher Scientific). The molarity of each amplicon was calculated using the equation:
where 308.95 g/mol is the average molecular weight of a nucleotide base and N represents the nucleotide length of the amplicon sequence (multiplied by 2 for double stranded). The resulting molar amount was converted to molecules/l by multiplying by 6.022 × 1023, and subsequently to molecules/μl by dividing by 1 × 106. Serial dilutions were performed to generate a standard regression curve from each of the amplicons that ranged from 1.204 × 107 amplicon molecules down to 7.7 × 102. All standard curves had near optimal slopes of −3.3, y-intercepts near 40, R correlations near 1, PCR efficiencies near 100%, and each target amplified within its standard curve. mRNA copy numbers were calculated using each amplicon standard regression curve from the known quantity of the amplicon.
Preparation of recombinant FASN
The DH10Bac bacterial stock (DH10-FASN-His) containing the poly-His-tagged FASN baculovirus construct was generously provided by Drs. Stuart Smith and Andrezj Witkowski (Children’s Hospital Oakland Research Institute). BD BaculoGold Bright DNA (Life Technologies) and 2 μg of purified FASN-His BacMid DNA were cotransfected into Sf9 cells to generate FASN-His baculovirus. Sf9 cells (500 × 106) were transduced with FASN-His baculovirus at an MOI of 2–3. Sf9 pellet was resuspended in 5 ml of buffer A containing 250 mM potassium phosphate (pH 7.0), 0.2 mM DTT, 10% glycerol, and 2× EDTA-free protease inhibitors (Roche Applied Science); passed through a glass dounce homogenizer using 25 strokes; and the cellular debris was cleared by ultracentrifugation at 100,000 g for 30 min. Nickel-NTA beads (5 ml) (Qiagen) were packed into a column, and the beads were equilibrated in 40 ml of buffer A. The cleared Sf9 lysate was added to the column by gravity drip, and the column was washed with 20 ml buffer A + 50 mM imidazole (repeated twice). FASN-His was eluted from nickel-NTA using 10 ml buffer A + 200 mM imidazole, and the eluate was transferred to dialysis tubing for equilibration in 1 l of buffer A + 2 mM DTT overnight at 4°C. Equilibrated FASN-His was concentrated in successive rounds using an Amicon Ultra-4 centrifugal filter unit (Millipore, Billerica, MA) at 3,000 g for 20 min at 4°C. FASN-His enzyme concentration was measured using a 280 nm extinction coefficient of 482,200 M−1cm−1 (dimer, without disulfides).
Preparation of recombinant Spot14
The pCMV-SPORT6 vector containing mouse Spot14 was purchased from Open Biosciences (Thermo Fisher Scientific), and the plasmid was purified using the Qiagen HiSpeed plasmid maxi kit (Qiagen) according to the manufacturer’s protocol. The purified plasmid containing mouse Spot14 was modified by PCR to introduce engineered restriction digestion cut sites into the sequence (forward: GGA-ATT-CCA-TAT-GAT-GCA-AGT-GCT-AAC-GAA-ACG-CTA-TCC; reverse: CGC-GGA-TCC-GCG-TCA-CTA-CAG-GAC-CTG-CCC-TGT-CAT-TTC). The PCR product was cleaned up using the Qiagen MinElute PCR purification kit according to manufacturer’s protocol (Qiagen) and was run on a 1% agarose gel to isolate the desired product. Gel-extracted PCR product (2 μg) and 2 ug of pET15b plasmid DNA were cut using NdeI and BamHI restriction enzymes in React buffer 3 in a total volume of 100 μl at 37°C overnight according to manufacturer’s instructions (Life Technologies). Digested DNA was cleaned up with the Qiagen DNA Mini purification kit (Qiagen), and DNA was ligated in a 15 μl volume using 8 μl of Spot14 insert, 1 μl vector, 3 μl 5× ligation buffer, 1 μl 10 mM ATP, and 1.5 μl T3 ligase at 10°C for 3 h. DH5α competent bacteria (25 μl) were transformed with ligated DNA (2 μl) by heat shock, single colonies were selected and screened for inclusion of the transgene, and a high expressing bacterial clone containing the pET15b-Spot14 plasmid was chosen. The plasmid was submitted for sequencing, and indicated that Spot14 had 100% identity to the original sequence. BL21 competent bacteria were transformed and plated, and single colonies were selected as above. Small preparations of single colonies were induced with IPTG, and production of the Spot14-His protein was evaluated by immunoblot using the anti-Spot14 antibody. The colony with the greatest amount of Spot14-His production was maintained for additional studies. BL21-Spot14-His bacteria were grown in 1 l of LB media under standard conditions containing 50 μg/ml ampicillin, were induced at an optical density of 0.6 with 0.5 mM IPTG, and the growth was maintained at 27°C overnight. Following protein induction, bacteria were collected by centrifugation at 6,000 g for 15 min and LB was decanted. Bacteria were resuspended in 40 ml of buffer containing 50 mM potassium phosphate (pH 7.0), 50 mM NaCl, 10 mM imidazole, and 1 mM DTT; and cells were lysed by sonication using 40 pulses at 90% power. Lysates were cleared by centrifugation at 15,000 g for 20 min, and the supernatant was transferred to a 50 ml conical tube. Purification of Spot14-His was performed essentially as the FASN-His protein, and the molecular weight was 19,388 g/mol and the extinction coefficient used was 12,090 M−1cm−1.
Standard and quantitative immunoblotting
MEC lysates were prepared as above and proteins were resolved using standard SDS PAGE techniques, transferred to normal or low fluorescence PVDF membrane (Millipore), and incubated overnight at 4°C with primary antibodies. Antibodies were purchased from Cell Signaling Technology: ACC (#3676), pACC ser79 (#3661), ACLY (#4332), pACLY ser454 (#4331), AMPK (#2603), pAMPK thr172 (#2535), and CypA (#2175); from Santa Cruz Biotechnology: FASN (SC-20140), goat anti-rabbit-HRP (SC-2004), KRT18 (SC-28264), and SLC25A1 (sc-86392); from Fitzgerald Industries International: PLIN1 (70R-PR004), PLIN2 (20R-AP002); from Abcam: Spot14 (ab56193); CSN2 was a kind gift of Margaret Neville (University of Colorado); and thioesterase 2 (TE2) was a kind gift of Stuart Smith (CHORI, San Francisco, CA). Recombinant FASN and Spot14 proteins were used to generate a standard curve to quantify the amount of FASN or Spot14 present in MEC lysates. Triplicate dilutions ranging from 0.15 to 1.0 μg of FASN or Spot14 protein were run using standard SDS PAGE techniques, transferred to low fluorescence PVDF membrane, and incubated overnight at 4°C with primary antibodies as above. Quantitative immunoblots were incubated in 1:15,000 goat anti-rabbit IRDye 800CW secondary (LI-COR Biosciences) for 2 h at room temperature. Infrared immunoblots were scanned using the LI-COR Odyssey system and data were analyzed using LI-COR Image Studio 2.0 software (LI-COR Biosciences). Amounts of endogenous FASN were calculated according to the following equation:
FASN activity assay tracing [13C] substrates
Reactions were performed according to Rudolph et al. (45) with the following modifications. Reactions were performed in a total volume of 900 μl in assay buffer consisting of 0.1M potassium phosphate (pH 6.8), 1.0 mM DTT, and 1.0 mM EDTA. Recombinant FASN (5 μg/900 μl mixture) was incubated with or without Spot14 in reaction buffer for 10 min at room temperature. FASN from control and Spot14-null MEC crude tissue lysates was quantified relative to the recombinant FASN regression curve, and volumes of cytosolic extract equaling 5 μg total FASN were incubated with 900 μl assay buffer. Individual reactions were performed one by one. NADPH (200 mM) (Sigma-Aldrich) was added to individual reactions and incubated for 2 min at room temperature, and subsequent catalysis was initiated in individual reactions by then adding a blend of 1,2-[13C]2-acetyl-CoA and 1,2,3-[13C]3-malonyl-CoA (99 atomic % 13C; Sigma-Aldrich). Following initiation, 200 μl aliquots were collected at 20, 40, 60, and 80 s, and each time point was quenched by adding reactants to 400 ul of 100% methanol and vortexing vigorously. [13C] fatty acid products were then extracted and prepared for GC-MS as described above.
RESULTS
HFD restores and high carbohydrate diet impairs growth of pups reared by Spot14-null dams
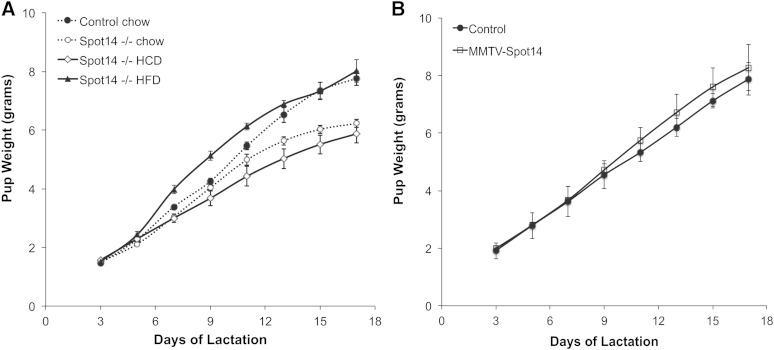
The TAG macronutrient content in milk provides the primary source of calories needed for the growth of the offspring, particularly in mice that produce about 30% milk fat by volume. Maternal HFD has been shown to suppress de novo fatty acid synthesis in mammary alveolar cells during lactation, while a high carbohydrate/low fat diet stimulates this pathway (13, 52). Zhu et al. (39) showed that the lactation phenotype in Spot14-null dams results in reduced growth of the offspring, and we have confirmed these published data by observing reduced growth of offspring reared by Spot14-null dams (Fig. 1A). To eliminate any effect due to the genetic background of the pups, both control and Spot14-null dams were cross-fostered with wild-type pups, and litters were standardized to seven or eight pups. Extending those findings, Spot14-null mothers were also provided a high carbohydrate/low fat diet to stimulate de novo fatty acid synthesis 1 day prior to parturition, and the lactation defect worsened (Fig. 1A; high carbohydrate diet (HCD), diamonds; 3.9% kcal fat). The Spot14-null lactation phenotype was rescued by maternal provision of a diet rich in fat 1 day prior to parturition (HFD, triangles; 46% kcal fat) (Fig. 1A). These results indicate that Spot14-null dams consuming normal chow or a HCD cannot generate enough milk energy density to sustain the optimal growth of their offspring, but that incorporation of maternal dietary fat into the milk TAG restores this energy deficit.
Fig. 1.
Pup weights of offspring from control, Spot14-null, and MMTV-Spot14 mothers. A: Litters were standardized to seven or eight wild-type pups per dam, and n = 3–5 dams per genotype and diet. Dams were provided standard chow, HCD (3.9% kcal fat), or HFD (46% kcal fat) 1 day prior to parturition. Growth of pups reared by Spot14-null dams on the chow diet (open circles) diverged from that of controls (solid circles) at lactation day 11 (P ≤ 0.05). Maternal HFD (solid triangles) provided 1 day prior to parturition restored the pup growth deficit observed in offspring suckling Spot14-null dams provided chow (open circles). Pups reared by dams provided HCD grew at the lowest rate. B: Pup growth of offspring reared on control or dams overexpressing Spot14 driven by a mammary epithelium specific promoter (MMTV-Spot14); the difference is not significant.
Mammary epithelial Spot14 overexpression does not impair mammary gland development or lactation
To evaluate the effect of transgenic Spot14 on mammary gland development and lactation, mice were generated that overexpress Spot14 driven by the mammary epithelial specific MMTV LTR. Spot14 transgene expression was confirmed by immunohistochemistry directed against the HA tag engineered into the transgene (supplementary Fig. IA). Transgene expression was also quantified by qPCR, and revealed that the highest MMTV-Spot14 expression was detected in the mammary gland, followed by occasional detection of the transgene in the lung of some mice; however, no expression of the transgene was detected in the ovary, uterus, heart, hypothalamus/pituitary, or liver (supplementary Fig. IB). Qualitative evaluation of whole mounts from virgin day 5 and nulliparous mammary glands indicated subtle differences in branching may exist in MMTV-Spot14 females, but terminal end buds were hollow and appeared similar to controls (supplementary Fig. IC, D). Examination of whole-mounted mammary glands from mid-pregnant and mid-lactating control and MMTV-Spot14 females, as well as analysis of histological sections at pregnant day 14 and lactation day 4, revealed no obvious differences in gland morphology and organization (supplementary Fig. ID, E). Consistent with competent development and function of the mammary gland was the normal growth of offspring reared by MMTV-Spot14 transgenic dams (Fig. 1B). Although the weights diverged slightly upward for pups reared by the MMTV-Spot14 dams, no significant difference in pup growth was observed, suggesting that MMTV-Spot14 mothers do not supply excessive nutrients to the offspring.
Spot14-null dams, but not MMTV-Spot14 dams, produce lipid-poor milk
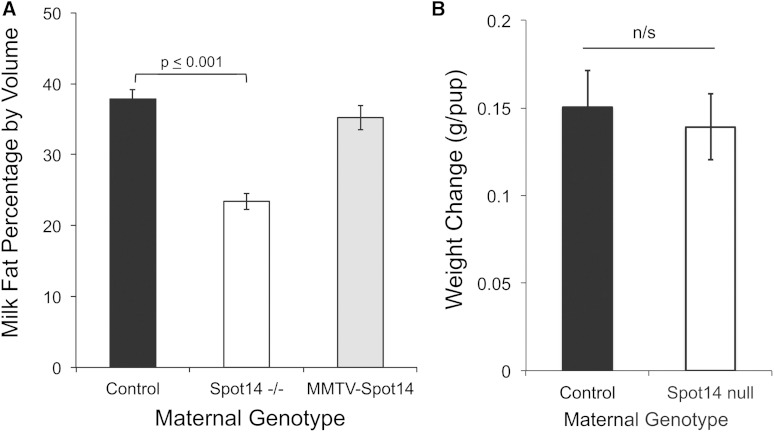
To determine the influence of Spot14 on milk fat content, control, Spot14-null, and MMTV-Spot14 transgenic milk was collected at lactation day 4 and the fat content was analyzed as previously described (46). Control dams synthesized milk containing approximately 35% milk fat by wet volume, however the fat content in milk from Spot14-null mice was significantly decreased, approximately 1.6-fold (P = 0.001) (Fig. 2A). In contrast to the Spot14-null dams, milk fat percentage from the MMTV-Spot14 dams was equivalent to controls, indicating that MEC overexpression of Spot14 did not influence total TAG amount. The protein and lactose contents of the Spot14-null milk and the MMTV-Spot14 milk were not altered relative to control milk (data not shown). A weigh-suckle-weigh approach was used to determine the amount of milk produced in a 2 h period (47). Milk production in the Spot14-null dams was equivalent to controls, suggesting milk synthesis and secretion is not obstructed (Fig. 2B).
Fig. 2.
Percentage of milk fat by volume at lactation day 4. A: Milk was collected and analyzed by creamocrit as described. Spot14-null (Spot14−/−) milk fat was reduced approximately 35% relative to controls, while milk fat percentage from dams overexpressing Spot14 in the mammary epithelium (MMTV-Spot14) was not different from controls. All animals were maintained on standard rodent chow. B: Weigh-suckle-weigh analysis of total milk production was not different in control and Spot14-null mice.
The NMR metabolomic lipid analysis of the Spot14-null MECs, depleted of adipocytes and luminal milk content (53), was consistent with reduced milk lipid by volume, indicated by nearly 30% decreased intracellular TAG content (Table 1; P = 0.01). Similar results were obtained with analysis of the whole mammary gland (Table 1). The PUFA components in the Spot14-null MECs and whole gland were also significantly decreased, but the MUFA:PUFA ratio was unchanged (Table 1). No differences were observed for the phospholipids quantified. These results indicate that the Spot14-null lactation phenotype is due to production of lipid-poor milk, rather than defective lactation that could result from impaired gland development, impaired secretory activation, or obstructed secretion as is seen in other models. This observation is further supported by the ability of the HFD to compensate for the lipid-poor milk produced by Spot14-null mothers provided the control diet (Fig. 1A; open circles vs. triangles).
TABLE 1.
Metabolomic profiling of Spot14-null and control adipose-depleted MECs and whole mammary gland
| Adipose Depleted MECs | Whole Mammary Gland | |||||||||
| Control | SEM | S14−/− | SEM | t Test | Control | SEM | S14−/− | SEM | t Test | |
| Lipids | ||||||||||
| TAGs | 50.8 | 3.8 | 36.1 | 3.4 | 0.009 | 131.5 | 8.0 | 93.8 | 9.0 | 0.01 |
| MUFAs | 30.1 | 5.9 | 22.7 | 3.4 | 0.14 | 90.7 | 5.9 | 82.6 | 7.8 | 0.22 |
| PUFAs | 60.8 | 5.8 | 48.8 | 3.8 | 0.05 | 154.2 | 6.6 | 112.1 | 9.4 | 0.003 |
| Total fatty acids (α CH2) | 152.9 | 1.2 | 111.0 | 0.9 | 0.01 | 424.6 | 2.7 | 289.0 | 2.5 | 0.01 |
| Total lipids (CH2n) | 490.2 | 3.8 | 354.7 | 2.8 | 0.009 | 1330.6 | 7.7 | 912.8 | 7.6 | 0.002 |
| Total lipids (omega CH3) | 173.0 | 1.3 | 135.5 | 0.8 | 0.02 | 472.8 | 2.4 | 321.7 | 2.6 | 0.002 |
| Cholesterol | 3.7 | 0.2 | 3.9 | 0.2 | 0.20 | 8.5 | 0.8 | 7.5 | 0.2 | 0.14 |
| MUFA:PUFA | 0.50 | 0.07 | 0.46 | 0.06 | 0.33 | 0.59 | 0.11 | 0.74 | 0.06 | 0.35 |
| Phospholipids | ||||||||||
| Glycerophospholipids | 14.3 | 1.0 | 16.2 | 0.9 | 0.10 | 26.4 | 1.2 | 25.6 | 1.6 | 0.35 |
| Phosphatidylethanolamine | 2.1 | 0.2 | 2.7 | 0.3 | 0.08 | 3.7 | 0.3 | 4.0 | 0.3 | 1.06 |
| Phosphatidylcholine | 4.6 | 0.4 | 5.4 | 0.4 | 0.09 | 8.5 | 0.5 | 8.4 | 0.5 | 0.99 |
| Total cholines | 6.9 | 0.5 | 7.8 | 0.4 | 0.09 | 12.4 | 0.6 | 11.9 | 0.7 | 0.31 |
| Lipid-31P scans | ||||||||||
| Phosphatidylinositol | 12.3 | 0.9 | 11.9 | 2.0 | 0.35 | 7.9 | 0.5 | 9.1 | 0.4 | 0.35 |
| Phosphatidylethanolamine | 3.1 | 0.5 | 3.4 | 1.2 | 0.29 | 1.5 | 0.2 | 1.5 | 0.1 | 0.32 |
| Phosphatidylcholine | 21.3 | 2.1 | 19.9 | 3.0 | 0.20 | 12.0 | 0.9 | 13.9 | 0.6 | 0.45 |
Lipid metabolites were quantified using NMR from Spot14-null (S14−/−) and control adipose-depleted MECs and whole mammary glands at lactation day 10. Significant differences were observed in concentrations of TAG, PUFA, and lipids containing α-methylene, acyl-methylene, and omega methyl groups in both MECs and whole mammary gland. Units are in micromoles per gram. Boldface denotes significantly different metabolites as determined by Welch’s t-test.
Overexpression of Spot14 increases MCFA composition of milk TAG
In contrast to the adipose tissue and the liver, de novo synthesis of fatty acids in the mammary gland generates great amounts of MCFA that are defined by the saturated acyl chain length ranging between 10 and 14 carbons (C = 10 to 14) (54). Conversely, the long chain fatty acids in milk TAG, those with an acyl chain length of C ≥ 16 carbons, have been shown to originate from either dietary sources or from mobilization of TAG reserves of the adipose tissue (15). Accordingly, “preformed” fatty acids originate from sources other than the de novo fatty acid pathway of the MEC and possess an acyl chain length of C ≥ 16 carbons. Fatty acids with C = 16 can be synthesized de novo in the MEC, or they might be synthesized by the liver and adipose tissue, or they could be present in the diet. Therefore, C = 16 represents fatty acids derived from both de novo and preformed sources.
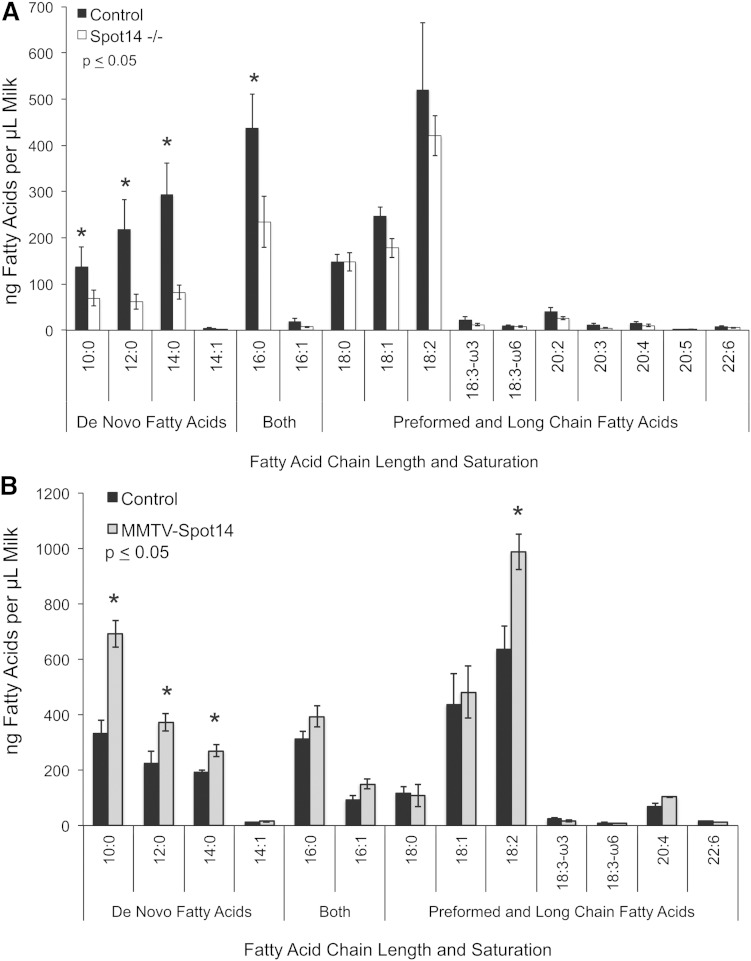
Milk was collected from control, Spot14-null, and MMVT-Spot14 overexpressing dams as previously described (46)and the neutral lipids were extracted, saponified with NaOH to release individual fatty acids, and the fatty acid composition was quantified by GC-MS according to previous methods (45, 48, 50). Loss of Spot14 resulted in significantly decreased MCFA from the milk TAG at lactation day 10 (Fig. 3A), and although the PUFA appeared to be decreased, the difference did not reach significance. These data were consistent with the NMR lipid analysis of extracts prepared from whole mammary glands or MECs. Conversely, overexpression of Spot14 in MECs from transgenic mice resulted in significantly increased MCFA in milk TAG at lactation day 4 (Fig. 3B). This finding indicates that overexpression of Spot14 in vivo during lactation is sufficient to elevate the de novo synthesized MCFA products incorporated into milk TAG, especially in early lactation when MCFAs are not yet largely present (13). Interestingly, the levels of linoleic acid (18:2) and arachidonic acid (20:4) were also elevated in milk from MMTV-Spot14 transgenic mice. Although this result is consistent with loss of PUFA in Spot14-null tissues and milk, we do not understand the functional relevance (if any) of this observation at present. Collectively, when Spot14 was lost during lactation, the de novo MCFA yield was diminished, and when Spot14 was artificially overexpressed in vivo, the de novo MCFA yield was increased.
Fig. 3.
Milk fatty acid composition. A: Spot14-null (Spot14−/−) and control dams were milked at lactation day 10 when contribution of de novo synthesized fatty acid contribution in milk is maximal. Total milk lipid was extracted and saponified into individual fatty acids that were quantified by GC-MS. MCFAs (C = 10:0–14:0) and C = 16:0, known to originate from de novo synthesis, were all significantly reduced in Spot14−/− milk. B: MMTV-Spot14 and control dams were milked at lactation day 4 when de novo synthesized fatty acid contribution in milk is moderate. Fatty acid composition was quantified as in (A). Milk from MMTV-Spot14 overexpressing dams contained significantly greater de novo fatty acids (C = 10:0–16:0) than controls. Linoleic and arachidonic acids (18:2 and 20:4) were also significantly increased in MMTV-Spot14 milk at lactation day 4. (*) denotes significantly different fatty acids by Welch’s t test.
Decreased MFCA in Spot14-null milk does not result from reduced lipogenic mRNA, proteins, and enzyme phosphorylation status
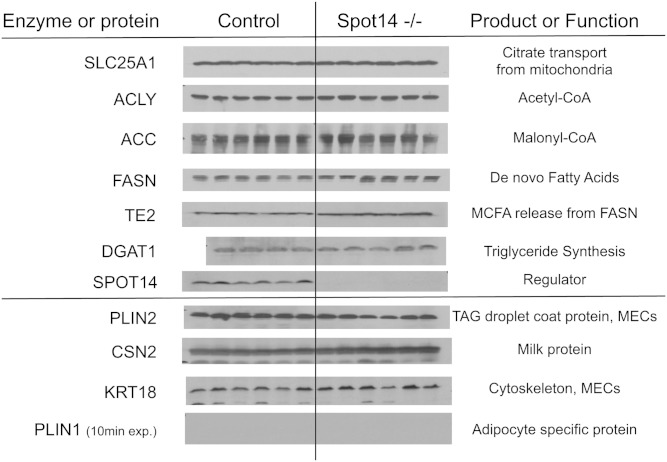
The de novo fatty acid pathway is regulated at multiple levels including gene expression, enzyme abundance, and posttranslational control, such as phosphorylation. Control and Spot14-null MECs depleted of mammary adipose were prepared in order to avoid confounding analysis due to adipocyte contamination (53). Gene expression was quantified for multiple members of the glycolytic and fatty acid synthesis pathways at lactation days 4 and 10 in these MECs. mRNA levels were equivalent for the 18 metabolic pathway genes evaluated, and some mRNAs were significantly increased (supplementary Fig. II). This finding indicated that loss of Spot14 in MECs did not influence expression of these genes. Overexpression of Spot14 in the MMTV-Spot14 MECs also did not alter mRNA levels of ACLY, ACC, or FASN (data not shown). Analysis of total protein levels in adipose-depleted MECs from control and Spot14-null mice showed no noteworthy differences for the mitochondrial citrate transporter (SLC25A1), ACLY, ACC, FASN, or diacylglycerol O-acyltransferase (DGAT)1 (Fig. 4). Loss of any of these proteins could account for a reduction in intermediate metabolites that supply substrate for FASN catalysis; and, if FASN levels were decreased in Spot14-null MECs, then reduced MCFA might result. Importantly, levels of TE2 (EC = 3.1.2.14), the MEC-specific enzyme responsible for releasing MCFA products from FASN during lactation (55), was maintained at the mRNA level (supplementary Fig. I, Lac 10) and appeared slightly greater at the protein level (Fig. 4). While a reduction of TE2 could account for the diminished MCFA observed in the Spot14-null milk, no difference in TE2 mRNA or protein was detected.
Fig. 4.
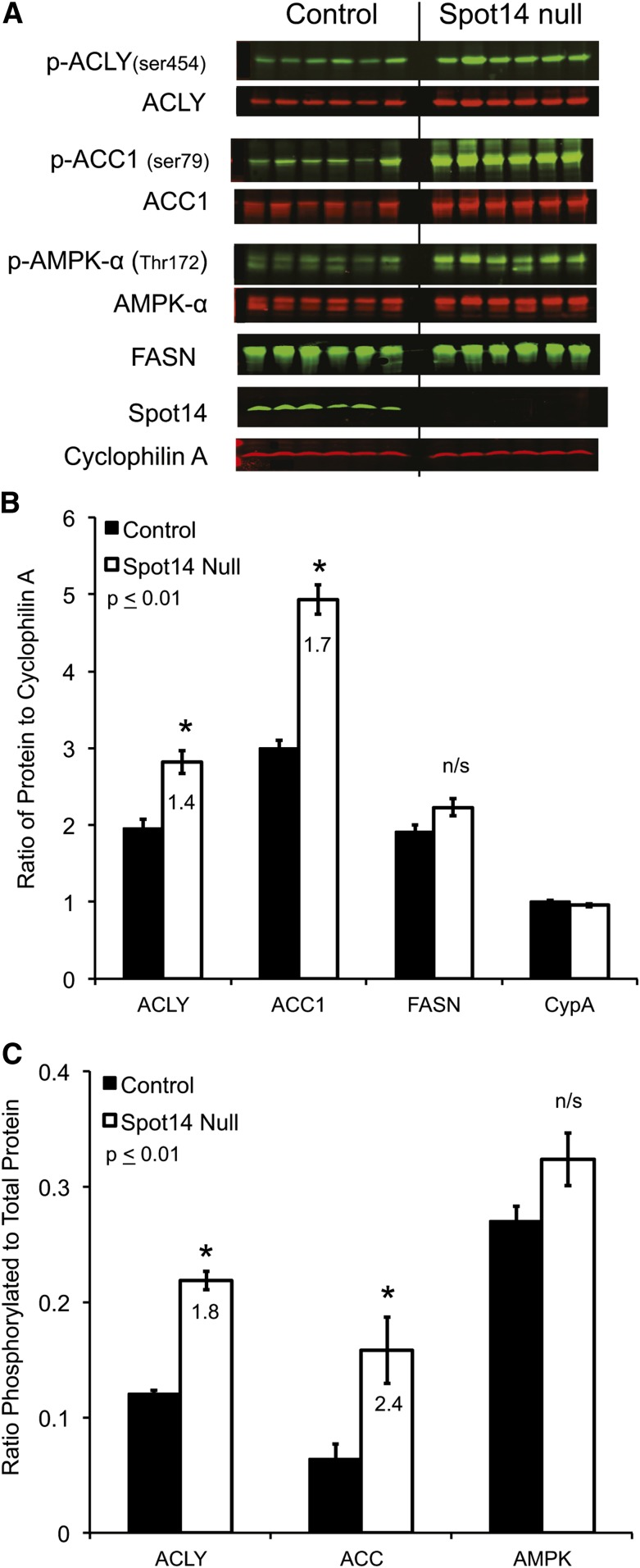
Immunoblot for de novo fatty acid synthesis pathway proteins. MECs depleted of the adipose compartment (MECs) were isolated from Spot14-null (Spot14−/−) and control dams at lactation day 10. The mitochondrial citrate transporter (SLC25A1), ACLY, ACC, FASN, and DGAT1 were equivalent in Spot14−/− relative to controls. TE2 was slightly increased in Spot14−/− MECs. The proteins of the de novo fatty acid synthesis pathway are intact, which does not account for the decreased MCFA composition in Spot14-null milk. Perilipin 2 (PLIN2, adipophilin), β casein (CSN2), and cytokeratin 18 (KRT18) are MEC-specific proteins; and, perilipin 1 (PLIN1) is an adipose-specific protein whose absence indicates sufficient depletion of adipocyte compartment during the MEC preparation.
Quantitative immunoblots were used to evaluate the total abundance of ACLY and ACC; both were significantly increased in Spot14-null MECs (1.4- and 1.7-fold, respectively), and total FASN trended upward but did not reach significance (Fig. 5A, B). In addition to total enzyme levels, phosphorylation of the de novo fatty acid synthesis enzymes potently regulates their activity. Phosphorylation of ACLY at Ser454 has been shown to activate the enzyme (20); while, phosphorylation of ACC at Ser79 is associated with catalytic inhibition in vitro (21). The phosphorylation status of ACLY and ACC was evaluated using quantitative immunoblotting (Fig. 5C). Interestingly, phosphorylated ACLY (activated) and phosphorylated ACC (inhibited) were both significantly increased in Spot14-null MECs (1.8- and 2.4-fold, respectively). Regulation of FASN activity by phosphorylation is beginning to emerge. It has been reported that mTORC mediates threonine phosphorylation at positions 1029 and 1033, which negatively regulates FASN in the mouse liver (12). Currently, no commercial antibodies directed against the phosphorylated epitopes of FASN exist; its analysis requires immunoprecipitation of FASN and use of phosphorylated threonine-specific antibodies (12). No difference was observed in the phosphorylated threonine form of immunoprecipitated FASN using an anti-phospho-threonine antibody that is not site specific (supplementary Fig. IVB).
Fig. 5.
Quantitative immunoblots from day 10 control and Spot14-null MECs. A: Scanned fluorescent images of total and phosphorylated forms of ACLY, ACC, and AMPK, with cyclophilin A as a loading control. The presence of Spot14 denotes genotype. B: Background adjusted intensities plotted as the ratio of total ACLY, ACC, and FASN to cyclophilin A loading control. Total ACLY and ACC are significantly increased in the Spot14-null MECs. C: The ratio of phosphorylated to total ACLY, ACC, and AMPK was calculated from the background adjusted intensities. Phosphorylated ACLY and ACC are significantly increased and phosphorylated AMPK trends upward in the Spot14-null MECs. *P ≤ 0.01.
Intermediate metabolites are not decreased in Spot14-null MECs
The phosphorylation status of ACLY and ACC suggested that the MECs from the Spot14-null dams should have increased acetyl-CoA (the product of ACLY) and decreased malonyl-CoA (the product of ACC). NMR metabolomics was used to quantify 35 aqueous metabolites from both the MECs and whole mammary gland lysates (WTL; contains luminal milk and adipose contents) of control and Spot14-null dams at lactation day 10, and revealed that acetyl-CoA was significantly increased (supplementary Table I). Malonyl-CoA levels were not significantly different in Spot14-null glands, despite the increased inhibitory phosphorylation of ACC (Fig. 5C). Hence, adequate levels of both acetyl-CoA and malonyl-CoA substrates required for FASN catalysis were present in Spot14-null glands, indicating that loss of intermediate metabolites do not account for the low MCFA present in Spot14-null milk. These observations, in combination with published data that demonstrated a 2.5-fold reduced rate of 3H2O incorporation into de novo milk lipids (39), led to the hypothesis that FASN activity must be reduced in the Spot14-null lactating mammary gland.
Spot14 enhances recombinant FASN catalysis in vitro
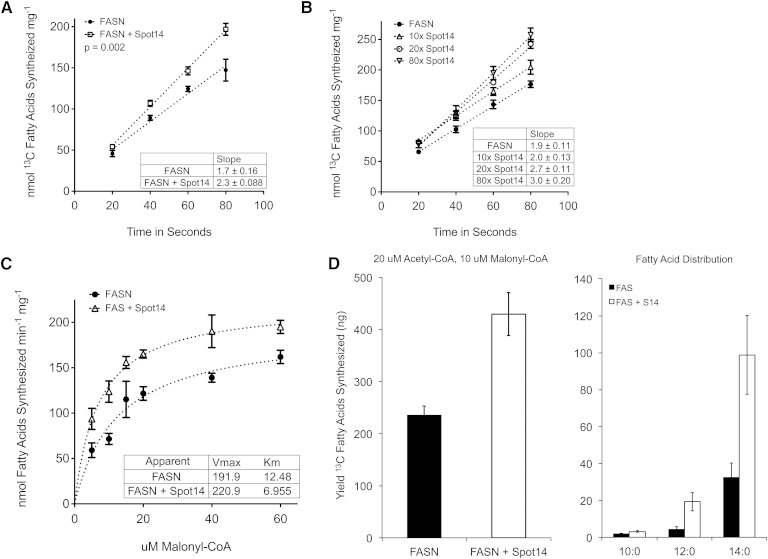
In order to test the hypothesis that Spot14 may directly increase the activity of FASN, recombinant Spot14 and FASN were prepared and FASN kinetic assays were conducted, as previously described with minor modifications (45). Using [13C]2-acetyl-CoA and [13C]3-malonyl-CoA as substrates, uniformly [13C] incorporated fatty acid products of FASN catalysis were quantified using GC-MS (45). Equimolar substrate concentrations of 20 μM were selected in order to preserve steady state kinetics at 1,000-fold excess of FASN (20 nM), and so that the blended substrates would initiate fatty acid synthesis with an equal probability for FASN to bind either a priming substrate (acetyl-CoA) or an elongation substrate (malonyl-CoA) (56, 57). The addition of Spot14, in 40× molar excess to the reaction mixture, increased FASN activity from 1.697 nmol [13C] fatty acids synthesized sec−1 mg−1 to 2.335 nmol [13C] fatty acids synthesized sec−1 mg−1, using the sum of C = 10–18 acyl chain length [13C] fatty acid products (Fig. 6A). In an independent series of experiments, FASN activity was enhanced with increasing concentrations of Spot14 present in the reaction mixture using 20 μM [13C]2-acetyl-CoA and 40 μM [13C]3-malonyl-CoA (Fig. 6B). Relative to FASN alone, addition of 10×, 20×, and 80× molar excess Spot14 increased FASN activity 7, 30, and 38%, respectively.
Fig. 6.
Recombinant Spot14 increases the activity of recombinant FASN in vitro. A: FASN activity increased 1.4-fold with addition of Spot14 in 40× molar excess in the presence of 20 μM [13C]2-acetyl-CoA and [13C]3-malonyl-CoA. Uniformly incorporated [13C] fatty acid products of enzyme catalysis were quantified using GC-MS. B: FASN activity was increased in a Spot14 concentration-dependent manner. C: Spot14 increased the apparent Vmax and apparent Km for the overall reaction using constant 20 μM [13C]2-acetyl-CoA and 200 mM NADPH, while varying the amount of [13C]3-malonyl-CoA. D: At the 20:10 μM ratio of [13C]2-acetyl-CoA to [13C]3-malonyl-CoA, the total [13C] fatty acid yield was increased 1.8-fold in the presence of Spot14, and the proportion of C = 10–14 [13C] fatty acids was increased nearly 4-fold over FASN alone.
The reaction velocity was determined using varied amounts of [13C]3-malonyl-CoA with constant 20 μM [13C]2-acetyl-CoA and 200 mM NADPH (Fig. 6C). In the presence of Spot14, the reaction velocity was greater than FASN alone, reaching an apparent Vmax of 220 nmol [13C] fatty acids synthesized min−1 mg−1 compared with 191 nmol [13C] fatty acids synthesized min−1 mg−1. Interestingly, the apparent Km was reduced nearly 45% from 12.5 μM to 6.9 μM [13C]3-malonyl-CoA in the presence of Spot14. Although kinetics under steady-state catalysis have been shown to follow a typical overall Michaelis-Menten rate law for each of the three substrates, acetyl-CoA, malonyl-CoA, and NADPH (57), the apparent Km data must be interpreted for the overall reaction kinetics and not the individual reactions leading to palmitate, such as substrate loading/unloading or acyl hydrolysis. The reduction in apparent Km would be consistent with release of fatty acid products shorter than C = 16:0 (C ≤ 14:0) from the multicomponent enzyme. Accordingly, the distribution of [13C] MCFA (C = 10–14) was increased nearly 4-fold relative to FASN alone (Fig. 6D), and the total [13C] fatty acid yield increased 1.8-fold over the entire reaction. Overall, addition of Spot14 to recombinant FASN in vitro enhanced the rate of catalysis by increasing the product yield of C = 10–14 [13C] fatty acids.
Native FASN activity is reduced in crude lysates from Spot14-null MECs
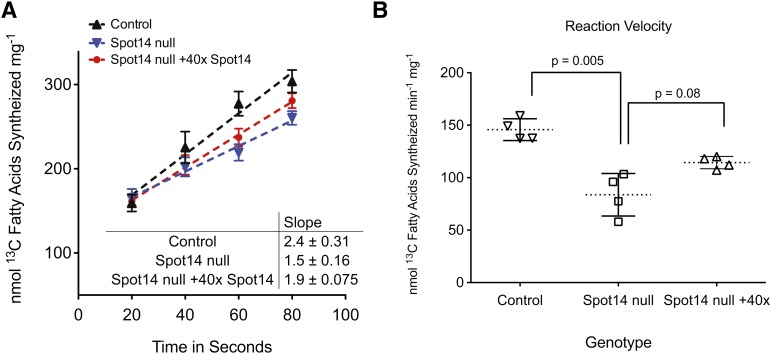
The MECs from lactation day 10 control and Spot14-null dams were prepared, and crude cytosolic extracts were generated for FASN activity assays as previously described (45). MECs were prepared from the whole mammary gland to avoid any FASN and Spot14 that could be contributed by mammary adipocytes present in the whole tissue (53). For the purpose of the kinetic calculations, the amount of cytosolic FASN per microgram of total cytosolic protein from control and Spot14-null MECs was quantified relative to the regression curve using quantitative immunoblotting for recombinant FASN (supplementary Fig. III). Based on the calculated amount of FASN per microgram of MEC total cytosolic protein, equivalent amounts of FASN, as a function of cytosolic lysate volume, were loaded into each activity assay. FASN activity assays were performed as above using 20 μM [13C]2-acetyl-CoA and 20 μM [13C]3-malonyl-CoA substrate concentrations. The activity of FASN was significantly less in cytosolic lysates from Spot14-null MECs relative to control lysates containing Spot14 (1.5 ± 0.16 nmol vs. 2.4 ± 0.31 nmol [13C] fatty acids synthesized sec−1 mg−1; Fig. 7A); and, the reaction velocity in control lysates containing Spot14 was significantly increased 1.6-fold relative to Spot14-null lysates (Fig. 7B). When recombinant Spot14 was added to the Spot14-null lysates in 40 molar excess, a 20% increase in the catalytic rate was observed (1.9 ± 0.08 nmol vs. 1.5 ± 0.16 nmol [13C] fatty acids synthesized sec−1 mg−1; Fig. 7A); however, the reaction velocity increase did not reach significance (P = 0.08; Fig. 7B). These observations suggest that recombinant Spot14 added back to Spot14-null MEC lysates could enhance the rate of native FASN catalysis relative to Spot14-null lysates; however, alternative forms of regulation may influence FASN catalysis because Spot14 add back could not fully restore activity.
Fig. 7.
Native FASN activity is decreased in Spot14-null MEC cytosolic lysates. A: [13C] fatty acid products of enzyme catalysis were quantified using GC-MS. FASN activity increased 1.6-fold in control lysates that contained Spot14 relative to Spot14-null lysates, and add back of Spot14 in 40 molar excess to null lysates increased catalysis approximately 20% under reaction conditions containing 20 μM [13C]2-acetyl-CoA and [13C]3-malonyl-CoA. B: Cytosolic lysates from control MECs had a significantly increased reaction velocity for overall enzyme kinetics (P = 0.005). Add back of recombinant Spot14 in 40× molar excess to null lysates trended higher but did not reach significance (P = 0.08).
DISCUSSION
Analysis of Spot14-null and transgenic mice overexpressing Spot14 during lactation was conducted to investigate the molecular function of Spot14 in a tissue type that is devoid of confounding influences of Spot14-R. In contrast to other mouse models with severe lactation failure, such as the DGAT 1-null, prolactin receptor +/−, STAT5a-null, ELF5 +/−, α-lactalbumin-null, and galanin-null mice (58–63), the lactation phenotype in Spot14-null dams is mild and no lactation defect was observed for MMTV-Spot14 overexpressing dams (Fig. 1A, B). Consistent with this mild phenotype is the observation that Spot14-null dams produce milk and lactate sufficiently (Fig. 2B), but that the total lipid content of the milk is reduced (Fig. 2A). Pup growth deficit was rescued simply by providing a HFD to the lactating dams (Fig. 1A, HFD). While reduced milk TAG would be consistent with lower levels of DGAT1, no differences in DGAT1 mRNA (supplementary Fig. II) or protein levels (Fig. 4) were observed. The MCFA (C = 10:0 to 14:0) unique to mammary epithelium de novo fatty acid synthesis were significantly reduced by 60% in Spot14-null milk (Fig. 3A). Importantly, none of the mRNA and total protein levels for de novo fatty acid synthesis pathway enzymes were lower in Spot14-null MECs, that could account for the reduction of MCFA, revealing that Spot14 does not regulate mRNA or protein abundance in lactating MECs.
We have found that Spot14 stimulates the in vitro activity of recombinant FASN (Fig. 6A) and native FASN present in MEC cytosolic lysates (Fig. 7A). The in vitro results are consistent with the in vivo observations from the Spot14-null and the MMTV-Spot14 overexpressing mice. Loss of Spot14 function resulted in significantly decreased MCFA (Fig. 3A), while MMTV-Spot14 overexpressing dams produced milk with significantly increased MCFA (Fig. 3B). MCFAs were increased in the MMTV-Spot14 dams at lactation day 4, a time when the de novo pathway is not yet maximal (13), indicating that Spot14 overexpression stimulated de novo fatty acid synthesis without influencing levels of preformed fatty acids. Moreover, overexpression of Spot14 increased MCFA and altered the composition of fatty acids present in the milk TAG (Fig. 3B) without affecting the total amount of TAG in the milk (Fig. 2A). Together, these data are in agreement with work that showed the in vivo rate of 3H2O incorporation into de novo synthesized lipids was decreased 2.5-fold in mammary glands of Spot14-null dams (39). Collectively, these experiments suggest the lactating mammary epithelium has evolved a tissue-specific molecular function for Spot14 as an enhancer of FASN catalysis.
Kim et al. (43) demonstrated that Spot14-R facilitated multimerization of ACC in vitro; however, they observed no Spot14 effect on the assembly or activity of ACC. We could not rule out the possibility that Spot14 may function to facilitate dimer formation or stabilization of FASN to stimulate catalysis in vivo. Nondenaturing native gels indicated that FASN existed as a dimer in both control and Spot14-null samples (supplementary Fig. IV). With the exception of TE2, the independent enzyme that converts product specificity of FASN from C = 16:0 to MCFA (15, 64), little is understood regarding proteins that might alter catalytic rate and fatty acid product distribution. Consistent with increased product release, the presence of Spot14 in vitro resulted in an approximately 4-fold increase in C = 14:0 (Fig. 6D) and a nearly 2-fold decrease of apparent Km (Fig. 6C). Spot14 itself does not have hydrolase activity using the model TE2 substrate myristoyl-CoA (supplementary Fig. IIID), and Spot14 did not bind acetyl- or malonyl-CoA substrates (not shown). These results suggest that Spot14 might augment release of fatty acids via the resident thioesterase domain of FASN. Naggert et al. (65) showed that the resident thioesterase domain from FASN relies entirely on its covalent linkage to FASN for function, as the recombinant “free” thioesterase domain had no activity toward S-acyl-FASN. We speculate that Spot14 could foster a more productive interaction of the resident thioesterase domain with the acyl carrier domain of FASN to stimulate release of its fatty acid products.
This observation could also indicate that expression of Spot14 in the lactating MEC may function to support the intervention of TE2 to liberate high quantities of MCFA, rather than acting predominantly with the resident TE domain of FASN. At the present time, we cannot distinguish between Spot14 interaction with endogenous resident TE domain of FASN or if Spot14 enhances TE2 interaction to release MFCA from FASN in vivo. FASN was immunoprecipitated from control and Spot14-null mammary gland lysates, but neither Spot14 nor TE2 were detected in the FASN pull-down, indicating that any potential FASN/Spot14/TE2 complexes associate transiently. That TE2 was not detected in FASN pull-down was consistent with previous work that failed to observe a stable FASN/TE2 complex from rat mammary gland lysates (66). In that study, Mikkelsen et al. (66), used a clever experimental approach to demonstrate that a stable complex formed only when FASN was acylated on the 4′-phosphopantetheine thiol group of the ACP domain that was actively engaged in chain elongation, after which, the complex rapidly dissociated following acyl hydrolysis. It is possible that active acyl chain elongation on FASN is also required for association of Spot14 with FASN, but this hypothesis was not addressed in the present studies.
When recombinant Spot14 was added back to null cytosolic lysates, the catalytic rate was not fully restored to that from control lysates containing native Spot14 (Fig. 7A). Given the increased phosphorylation of ACLY and ACC in Spot14-null MECs (Fig. 5C), we hypothesized that FASN could be more highly phosphorylated in Spot14-null MECs during lactation. Phosphorylation of FASN at Thr1029 and Thr1033 (mouse) by mTORC1 has been shown to inhibit FASN activity (12); however, no significant difference in total phosphorylated threonine was observed using an anti-phosphothreonine antibody (supplementary Fig. IVB). This observation indicates that the regulation of FASN by mTORC1 may not be a feature in lactating MECs as was observed in liver (12). Site-specific antibodies are needed to properly analyze the phosphorylation status of FASN as it pertains to enzyme activity. The fact that addition of recombinant Spot14 to null lysates did not restore FASN catalysis to levels observed in control lysates suggests that other proteins and/or posttranslational modifications may regulate FASN in vivo to preserve the dynamic balance between preformed fatty acids and those synthesized by FASN.
It is interesting to note that none of the aqueous intermediate metabolites of glycolysis and the TCA cycle were decreased in Spot14-null MECs (supplementary Table I); consistent with the previous observation that enzyme activity upstream of FASN using in vitro assays was intact in the Spot14-null mammary gland (39). Furthermore, a small number of these metabolites were significantly increased in the Spot14-null MECs, including acetyl-CoA, aromatic amino acids, glutamate, pyruvate, and succinate; while malonyl-CoA trended higher in Spot14-null mammary glands (supplementary Table I). This observation indicates that ample substrate is present in vivo for FASN catalysis, and supports the conclusion that Spot14 functions to enhance the rate of FASN catalysis during lactation in vivo. Lipogenesis in the lactating mammary gland represents a highly specialized function that maintains the dynamic balance of preformed fatty acids with activity of the de novo fatty acid synthesis pathway to preserve a relatively constant milk fat content unique to each mammalian species. Our findings implicate Spot14 as a direct protein enhancer of FASN, enhancing MCFA synthesis, a function that appears to be unique and critical to the mammary gland during lactation.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. S. Smith and A. Witkowski (Children’s Hospital Oakland Research Institute, Oakland, CA) for their generosity, for their many thoughtful discussions regarding this work, and for the FASN baculovirus needed to conduct this research. The authors also thank Dr. Robert Murphy and Christopher Johnson for use of their mass spectrometer and expertise, supported by Colorado Clinical and Translational Sciences Institute 5UL1RR025780.
Footnotes
Abbreviations:
- ACC
- acetyl-CoA carboxylase
- ACLY
- ATP citrate lyase
- DGAT
- diacylglycerol O-acyltransferase
- HCD
- high carbohydrate diet
- HFD
- high-fat diet
- MCFA
- medium chain fatty acid
- MEC
- mammary epithelial cell
- MMTV LTR
- Mouse Mammary Tumor Virus Long Terminal Repeat
- Spot14-R (MIG12)
- Spot14-related protein
- TE2
- thioesterase 2
- THRSP
- thyroid hormone responsive protein Spot 14
- TAG
- triglyceride (triacylglyceride)
The University of Colorado Cancer Center Protein Production/Tissue Culture/Monoclonal Antibody Shared Resource produced recombinant FASN from baculovirus expression vectors, supported by P30 CA046934. The UCCC/CTSA Imaging and Metabolomics Shared Resources used to collect NMR data, supported by UL1 TR001082. M.C.R. was supported by DoD Breast Cancer Research predoctoral trainee award BC810596 and National Institutes of Health NORC Grant P30 DK048520; S.M.A. was supported by National Institutes of Health Grant PO1-HD38129; N.J.S. was supported by National Institutes of Health Grants P30 CA046934 and UL1 RR025780 (CTSA); and E.A.W. was supported by DoD Breast Cancer Research Program BC098051.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures and two tables.
REFERENCES
- 1.Smith S. 1994. The animal fatty acid synthase: one gene, one polypeptide, seven enzymes. FASEB J. 8: 1248–1259. [PubMed] [Google Scholar]
- 2.Smith S., Witkowski A., Joshi A. K. 2003. Structural and functional organization of the animal fatty acid synthase. Prog. Lipid Res. 42: 289–317. [DOI] [PubMed] [Google Scholar]
- 3.Chirala S. S., Chang H., Matzuk M., Abu-Elheiga L., Mao J., Mahon K., Finegold M., Wakil S. J. 2003. Fatty acid synthesis is essential in embryonic development: fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc. Natl. Acad. Sci. USA. 100: 6358–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jayakumar A., Tai M. H., Huang W. Y., al-Feel W., Hsu M., Abu-Elheiga L., Chirala S. S., Wakil S. J. 1995. Human fatty acid synthase: properties and molecular cloning. Proc. Natl. Acad. Sci. USA. 92: 8695–8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudolph M. C., Monks J., Burns V., Phistry M., Marians R., Foote M. R., Bauman D. E., Anderson S. M., Neville M. C. (2010) Sterol regulatory element binding protein and dietary lipid regulation of fatty acid synthesis in the mammary epithelium. Am. J. Physiol. Endocrinol. Metab. 299: E918–E927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kliewer S. A., Sundseth S. S., Jones S. A., Brown P. J., Wisely G. B., Koble C. S., Devchand P., Wahli W., Willson T. M., Lenhard J. M., et al. 1997. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc. Natl. Acad. Sci. USA. 94: 4318–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross J., Najjar A. M., Sankaranarayanapillai M., Tong W. P., Kaluarachchi K., Ronen S. M. 2008. Fatty acid synthase inhibition results in a magnetic resonance-detectable drop in phosphocholine. Mol. Cancer Ther. 7: 2556–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rioux V., Legrand P. 2007. Saturated fatty acids: simple molecular structures with complex cellular functions. Curr. Opin. Clin. Nutr. Metab. Care. 10: 752–758. [DOI] [PubMed] [Google Scholar]
- 9.Cao H., Gerhold K., Mayers J. R., Wiest M. M., Watkins S. M., Hotamisligil G. S. 2008. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 134: 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhajda F. P. 2000. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 16: 202–208. [DOI] [PubMed] [Google Scholar]
- 11.Kuhajda F. P. 2006. Fatty acid synthase and cancer: new application of an old pathway. Cancer Res. 66: 5977–5980. [DOI] [PubMed] [Google Scholar]
- 12.Jensen-Urstad A. P., Song H., Lodhi I. J., Funai K., Yin L., Coleman T., Semenkovich C. F. 2013. Nutrient-dependent phosphorylation channels lipid synthesis to regulate PPARα. J. Lipid Res. 54: 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudolph M. C., Monks J., Burns V., Phistry M., Marians R., Foote M. R., Bauman D. E., Anderson S. M., Neville M. C. 2010. Sterol regulatory element binding protein and dietary lipid regulation of fatty acid synthesis in the mammary epithelium. Am. J. Physiol. Endocrinol. Metab. 299: E918–E927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith S., Watts R., Dils R. 1968. Quantitative gas-liquid chromatographic analysis of rodent milk triglycerides. J. Lipid Res. 9: 52–57. [PubMed] [Google Scholar]
- 15.Smith S. 1980. Mechanism of chain length determination in biosynthesis of milk fatty acids. J. Dairy Sci. 63: 337–352. [DOI] [PubMed] [Google Scholar]
- 16.Horton J. D., Goldstein J. L., Brown M. S. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson D. M., Lyons R. T., Scott D. F., Muto Y. 1972. Synthesis and degradation of the lipogenic enzymes of rat liver. Adv. Enzyme Regul. 10: 187–204. [DOI] [PubMed] [Google Scholar]
- 18.Majerus P. W., Kilburn E. 1969. Acetyl coenzyme A carboxylase. The roles of synthesis and degradation in regulation of enzyme levels in rat liver. J. Biol. Chem. 244: 6254–6262. [PubMed] [Google Scholar]
- 19.Flick P. K., Chen J., Vagelos P. R. 1977. Effect of dietary linoleate on synthesis and degradation of fatty acid synthetase from rat liver. J. Biol. Chem. 252: 4242–4249. [PubMed] [Google Scholar]
- 20.Potapova I. A., El-Maghrabi M. R., Doronin S. V., Benjamin W. B. 2000. Phosphorylation of recombinant human ATP:citrate lyase by cAMP-dependent protein kinase abolishes homotropic allosteric regulation of the enzyme by citrate and increases the enzyme activity. Allosteric activation of ATP:citrate lyase by phosphorylated sugars. Biochemistry. 39: 1169–1179. [DOI] [PubMed] [Google Scholar]
- 21.Ha J., Daniel S., Broyles S. S., Kim K. H. 1994. Critical phosphorylation sites for acetyl-CoA carboxylase activity. J. Biol. Chem. 269: 22162–22168. [PubMed] [Google Scholar]
- 22.Jin Q., Yuan L. X., Boulbes D., Baek J. M., Wang Y. N., Gomez-Cabello D., Hawke D. H., Yeung S. C., Lee M. H., Hortobagyi G. N., et al. 2010. Fatty acid synthase phosphorylation: a novel therapeutic target in HER2-overexpressing breast cancer cells. Breast Cancer Res. 12: R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guy P. S., Cohen P., Hardie D. G. 1981. Purification and physicochemical properties of ATP citrate (pro-3S) lyase from lactating rat mammary gland and studies of its reversible phosphorylation. Eur. J. Biochem. 114: 399–405. [DOI] [PubMed] [Google Scholar]
- 24.Landman A. D., Darkshinamurti K. 1975. Acetyl-Coenzyme A carboxylase. Role of the prosthetic group in enzyme polymerization. Biochem. J. 145: 545–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi A. K., Rangan V. S., Smith S. 1998. Differential affinity labeling of the two subunits of the homodimeric animal fatty acid synthase allows isolation of heterodimers consisting of subunits that have been independently modified. J. Biol. Chem. 273: 4937–4943. [DOI] [PubMed] [Google Scholar]
- 26.Vagelos P. R., Alberts A. W., Martin D. B. 1963. Studies on the mechnism of activation of acetyl coenzyme A carboxylase by citrate. J. Biol. Chem. 238: 533–540. [PubMed] [Google Scholar]
- 27.Numa S., Ringelmann E., Lynen F. 1965. On inhibition of acetyl-CoA-carboxylase by fatty acid-coenzyme A compounds [article in German]. Biochem. Z. 343: 243–257. [PubMed] [Google Scholar]
- 28.Colbert C. L., Kim C. W., Moon Y. A., Henry L., Palnitkar M., McKean W. B., Fitzgerald K., Deisenhofer J., Horton J. D., Kwon H. J. 2010. Crystal structure of Spot 14, a modulator of fatty acid synthesis. Proc. Natl. Acad. Sci. USA. 107: 18820–18825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knobloch M., Braun S. M., Zurkirchen L., von Schoultz C., Zamboni N., Arauzo-Bravo M. J., Kovacs W. J., Karalay O., Suter U., Machado R. A., et al. 2013. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature. 493: 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seelig S., Liaw C., Towle H. C., Oppenheimer J. H. 1981. Thyroid hormone attenuates and augments hepatic gene expression at a pretranslational level. Proc. Natl. Acad. Sci. USA. 78: 4733–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaFave L. T., Augustin L. B., Mariash C. N. 2006. S14: insights from knockout mice. Endocrinology. 147: 4044–4047. [DOI] [PubMed] [Google Scholar]
- 32.Jump D. B., Bell A., Lepar G., Hu D. 1990. Insulin rapidly induces rat liver S14 gene transcription. Mol. Endocrinol. 4: 1655–1660. [DOI] [PubMed] [Google Scholar]
- 33.Mariash C. N., Seelig S., Schwartz H. L., Oppenheimer J. H. 1986. Rapid synergistic interaction between thyroid hormone and carbohydrate on mRNAS14 induction. J. Biol. Chem. 261: 9583–9586. [PubMed] [Google Scholar]
- 34.Foretz M., Foufelle F., Ferre P. 1999. Polyunsaturated fatty acids inhibit fatty acid synthase and spot-14-protein gene expression in cultured rat hepatocytes by a peroxidative mechanism. Biochem. J. 341: 371–376. [PMC free article] [PubMed] [Google Scholar]
- 35.Kadegowda A. K., Connor E. E., Teter B. B., Sampugna J., Delmonte P., Piperova L. S., Erdman R. A. 2010. Dietary trans fatty acid isomers differ in their effects on mammary lipid metabolism as well as lipogenic gene expression in lactating mice. J. Nutr. 140: 919–924. [DOI] [PubMed] [Google Scholar]
- 36.Harvatine K. J., Bauman D. E. 2006. SREBP1 and thyroid hormone responsive spot 14 (S14) are involved in the regulation of bovine mammary lipid synthesis during diet-induced milk fat depression and treatment with CLA. J. Nutr. 136: 2468–2474. [DOI] [PubMed] [Google Scholar]
- 37.Rudolph M. C., McManaman J. L., Hunter L., Phang T., Neville M. C. 2003. Functional development of the mammary gland: use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J. Mammary Gland Biol. Neoplasia. 8: 287–307. [DOI] [PubMed] [Google Scholar]
- 38.Bauman D. E., Harvatine K. J., Lock A. L. 2011. Nutrigenomics, rumen-derived bioactive fatty acids, and the regulation of milk fat synthesis. Annu. Rev. Nutr. 31: 299–319. [DOI] [PubMed] [Google Scholar]
- 39.Zhu Q., Anderson G. W., Mucha G. T., Parks E. J., Metkowski J. K., Mariash C. N. 2005. The Spot 14 protein is required for de novo lipid synthesis in the lactating mammary gland. Endocrinology. 146: 3343–3350. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Q., Mariash A., Margosian M. R., Gopinath S., Fareed M. T., Anderson G. W., Mariash C. N. 2001. Spot 14 gene deletion increases hepatic de novo lipogenesis. Endocrinology. 142: 4363–4370. [DOI] [PubMed] [Google Scholar]
- 41.Aipoalani D. L., O’Callaghan B. L., Mashek D. G., Mariash C. N., Towle H. C. 2010. Overlapping roles of the glucose-responsive genes, S14 and S14R, in hepatic lipogenesis. Endocrinology. 151: 2071–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donnelly C., Olsen A. M., Lewis L. D., Eisenberg B. L., Eastman A., Kinlaw W. B. 2009. Conjugated linoleic acid (CLA) inhibits expression of the Spot 14 (THRSP) and fatty acid synthase genes and impairs the growth of human breast cancer and liposarcoma cells. Nutr. Cancer. 61: 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim C. W., Moon Y. A., Park S. W., Cheng D., Kwon H. J., Horton J. D. 2010. Induced polymerization of mammalian acetyl-CoA carboxylase by MIG12 provides a tertiary level of regulation of fatty acid synthesis. Proc. Natl. Acad. Sci. USA. 107: 9626–9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martel P. M., Bingham C. M., McGraw C. J., Baker C. L., Morganelli P. M., Meng M. L., Armstrong J. M., Moncur J. T., Kinlaw W. B. 2006. S14 protein in breast cancer cells: direct evidence of regulation by SREBP-1c, superinduction with progestin, and effects on cell growth. Exp. Cell Res. 312: 278–288. [DOI] [PubMed] [Google Scholar]
- 45.Rudolph M. C., Karl Maluf N., Wellberg E. A., Johnson C. A., Murphy R. C., Anderson S. M. 2012. Mammalian fatty acid synthase activity from crude tissue lysates tracing (13)C-labeled substrates using gas chromatography-mass spectrometry. Anal. Biochem. 428: 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwertfeger K. L., McManaman J. L., Palmer C. A., Neville M. C., Anderson S. M. 2003. Expression of constitutively activated Akt in the mammary gland leads to excess lipid synthesis during pregnancy and lactation. J. Lipid Res. 44: 1100–1112. [DOI] [PubMed] [Google Scholar]
- 47.Dempsey C., McCormick N. H., Croxford T. P., Seo Y. A., Grider A., Kelleher S. L. 2012. Marginal maternal zinc deficiency in lactating mice reduces secretory capacity and alters milk composition. J. Nutr. 142: 655–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hutchins P. M., Barkley R. M., Murphy R. C. 2008. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J. Lipid Res. 49: 804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutchins P. M., Moore E. E., Murphy R. C. 2011. Electrospray MS/MS reveals extensive and nonspecific oxidation of cholesterol esters in human peripheral vascular lesions. J. Lipid Res. 52: 2070–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zarini S., Gijon M. A., Folco G., Murphy R. C. 2006. Effect of arachidonic acid reacylation on leukotriene biosynthesis in human neutrophils stimulated with granulocyte-macrophage colony-stimulating factor and formyl-methionyl-leucyl-phenylalanine. J. Biol. Chem. 281: 10134–10142. [DOI] [PubMed] [Google Scholar]
- 51.Rudolph M. C., McManaman J. L., Phang T., Russell T., Kominsky D. J., Serkova N. J., Stein T., Anderson S. M., Neville M. C. 2007. Metabolic regulation in the lactating mammary gland: a lipid synthesizing machine. Physiol. Genomics. 28: 323–336. [DOI] [PubMed] [Google Scholar]
- 52.Wahlig J. L., Bales E. S., Jackman M. R., Johnson G. C., McManaman J. L., Maclean P. S. 2012. Impact of high-fat diet and obesity on energy balance and fuel utilization during the metabolic challenge of lactation. Obesity (Silver Spring). 20: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rudolph M. C., Wellberg E. A., Anderson S. M. 2009. Adipose-depleted mammary epithelial cells and organoids. J. Mammary Gland Biol. Neoplasia. 14: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith S., Dils R. 1966. Factors affecting the chain length of fatty acids synthesised by lactating-rabbit mammary glands. Biochim. Biophys. Acta. 116: 23–40. [DOI] [PubMed] [Google Scholar]
- 55.Libertini L. J., Smith S. 1978. Purification and properties of a thioesterase from lactating rat mammary gland which modifies the product specificity of fatty acid synthetase. J. Biol. Chem. 253: 1393–1401. [PubMed] [Google Scholar]
- 56.Soulié J. M., Sheplock G. J., Tian W. X., Hsu R. Y. 1984. Transient kinetic studies of fatty acid synthetase. A kinetic self-editing mechanism for the loading of acetyl and malonyl residues and the role of coenzyme A. J. Biol. Chem. 259: 134–140. [PubMed] [Google Scholar]
- 57.Cox B. G., Hammes G. G. 1983. Steady-state kinetic study of fatty acid synthase from chicken liver. Proc. Natl. Acad. Sci. USA. 80: 4233–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ormandy C. J., Binart N., Kelly P. A. 1997. Mammary gland development in prolactin receptor knockout mice. J. Mammary Gland Biol. Neoplasia. 2: 355–364. [DOI] [PubMed] [Google Scholar]
- 59.Zhou J., Chehab R., Tkalcevic J., Naylor M. J., Harris J., Wilson T. J., Tsao S., Tellis I., Zavarsek S., Xu D., et al. 2005. Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. EMBO J. 24: 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naylor M. J., Ginsburg E., Iismaa T. P., Vonderhaar B. K., Wynick D., Ormandy C. J. 2003. The neuropeptide galanin augments lobuloalveolar development. J. Biol. Chem. 278: 29145–29152. [DOI] [PubMed] [Google Scholar]
- 61.Liu X., Robinson G. W., Wagner K. U., Garrett L., Wynshaw-Boris A., Hennighausen L. 1997. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 11: 179–186. [DOI] [PubMed] [Google Scholar]
- 62.Stacey A., Schnieke A., Kerr M., Scott A., McKee C., Cottingham I., Binas B., Wilde C., Colman A. 1995. Lactation is disrupted by alpha-lactalbumin deficiency and can be restored by human alpha-lactalbumin gene replacement in mice. Proc. Natl. Acad. Sci. USA. 92: 2835–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cases S., Zhou P., Shillingford J. M., Wiseman B. S., Fish J. D., Angle C. S., Hennighausen L., Werb Z., Farese R. V., Jr 2004. Development of the mammary gland requires DGAT1 expression in stromal and epithelial tissues. Development. 131: 3047–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith S., Ryan P. 1979. Asynchronous appearance of two enzymes concerned with medium chain fatty acid synthesis in developing rat mammary gland. J. Biol. Chem. 254: 8932–8936. [PubMed] [Google Scholar]
- 65.Naggert J., Witkowski A., Wessa B., Smith S. 1991. Expression in Escherichia coli, purification and characterization of two mammalian thioesterases involved in fatty acid synthesis. Biochem. J. 273: 787–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mikkelsen J., Witkowski A., Smith S. 1987. Interaction of rat mammary gland thioesterase II with fatty acid synthetase is dependent on the presence of acyl chains on the synthetase. J. Biol. Chem. 262: 1570–1574. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.