Abstract
Liver-specific overexpression of the insulin-like growth factor 2 (IGF2) mRNA binding protein p62/IGF2BP2-2 induces a fatty liver, which highly expresses IGF2. Because IGF2 expression is elevated in patients with steatohepatitis, the aim of our study was to elucidate the role and interconnection of p62 and IGF2 in lipid metabolism. Expression of p62 and IGF2 highly correlated in human liver disease. p62 induced an elevated ratio of C18:C16 and increased fatty acid elongase 6 (ELOVL6) protein, the enzyme catalyzing the elongation of C16 to C18 fatty acids and promoting nonalcoholic steatohepatitis in mice and humans. The p62 overexpression induced the activation of the ELOVL6 transcriptional activator sterol regulatory element binding transcription factor 1 (SREBF1). Recombinant IGF2 induced the nuclear translocation of SREBF1 and a neutralizing IGF2 antibody reduced ELOVL6 and mature SREBF1 protein levels. Concordantly, p62 and IGF2 correlated with ELOVL6 in human livers. Decreased palmitoyl-CoA levels, as found in p62 transgenic livers, can explain the lipogenic action of ELOVL6. Accordingly, p62 represents an inducer of hepatic C18 fatty acid production via a SREBF1-dependent induction of ELOVL6. These findings underline the detrimental role of p62 in liver disease.
Keywords: fatty acid elongase 6, sterol regulatory element binding transcription factor 1, insulin-like growth factor 2 signalling, hepatic steatosis
Nonalcoholic fatty liver disease (NAFLD) is considered to be the most common liver disorder in Western countries with a prevalence of 20–30% of the adult population (1, 2). There is a strong correlation between the characteristics of the metabolic syndrome, such as obesity and diabetes mellitus, and NAFLD/nonalcoholic steatohepatitis (NASH) (3).
The “two-hit” hypothesis represents a common model to describe the development and progression of fatty liver diseases. A simple steatosis can stand for the first step in early liver pathogenesis (4, 5). The progression from simple steatosis to NASH requires a “second hit” mediated by reactive oxygen species and release of inflammatory cytokines (6). This inflammatory environment can result in hepatic cirrhosis and finally in hepatocellular carcinoma (HCC) (7).
The development of hepatosteatosis can be induced by different mechanisms. The synthesis of lipids is regulated in a complex interplay induced by a set of lipogenic transcription factors, among which liver X receptor α (LXR-α/NR1H3), sterol regulatory element binding transcription factor 1 (SREBF1/SREBP1), and carbohydrate responsive element binding protein (ChREBP/MLXIPL) represent the most important ones (8). In this context, the fact that MLXIPL controls 50% of hepatic lipogenesis by regulating glycolytic and lipogenic gene expression (9) illustrates the importance of both insulin- and glucose-induced lipogenic pathways. One of the relevant inducers of lipid degradation in the liver is PPARA (10). Most importantly, there is a close interconnection between catabolic and anabolic pathways. In this context it is important to note that the mitochondrial β-oxidation pathway is negatively regulated by high malonyl-CoA levels (11, 12).
Besides the amount of lipids, which are relevant for pathophysiological actions, there is increasing evidence that the composition of lipids also has an impact on pathophysiology. In fact, human NAFLD is characterized by numerous changes in hepatic lipid composition and relative abundance of specific fatty acids (13, 14). Also hepatitis (B/C) has been described to strongly alter hepatic lipid content and composition (15, 16). Recently, the fatty acid elongase 6 (ELOVL6), which catalyzes the elongation of C16 to C18 fatty acids (17) and is a direct target of SREBF1 (18, 19), has been shown to promote NASH in mice and humans and to be overexpressed in a murine NASH model (20, 21). Interestingly, however, there is still a lack of understanding of the upstream signaling pathways that are responsible for SREBF1 activation and why elevated ELOVL6 increases total fatty acid production. Also, the role of ELOVL6 in HCC is as yet poorly understood and seems to depend on disease etiology (21–23).
We recently reported that a liver-specific overexpression of insulin-like growth factor 2 (IGF2) mRNA binding protein p62/IGF2BP2-2 induces steatosis in mice, coupled with high Igf2 expression and activation of the phosphoinositide 3-kinase/AKT-signaling pathway (24). Furthermore, p62 has been shown to promote NASH development (25). Most lipid species are elevated in p62-induced steatosis, with triglycerides showing the strongest increase (26). p62 was originally identified as an autoantigen overexpressed in about one-third to two-thirds of HCC patients and correlates with poor outcome (27–30). Interestingly, IGF2 is also overexpressed in NASH and HCC (31, 32), which is linked to p62 overexpression in HCC (27), and might be explained by the involvement of the IGF2 mRNA-binding proteins in RNA localization, stability, and translation (33). p62 is a splice variant of IGF2BP2 lacking exon 10, though exon 10 deletion does not affect the six characteristic RNA binding motifs (33). Recently, Li et al. (34) reported that IGF2BP2 binds and controls the translation of c-Myc, Sp1 transcription factor, and insulin-like growth factor 1 receptor. Binding affinities of IGF2BP2-2 to the respective mRNAs, however, are not described in the literature.
The aim of our study was to decipher the effects of p62 on lipid metabolism and to elucidate the influence of IGF2.
MATERIALS AND METHODS
Animals
All animal procedures were performed in accordance with the local animal welfare committee. Mice were kept under stable conditions regarding temperature, humidity, food delivery, and 12 h day/night rhythm. The p62 transgenic mice were established as described by Tybl et al. (24). The mice were euthanized at an age between 2.5 and 5 weeks.
Hyperinsulinemic euglycemic clamp study
Hepatic insulin sensitivity was determined by the hyperinsulinemic euglycemic clamp technique as described previously (35).
Human liver tissue
Thirty-five human liver tissues from patients undergoing surgical resection were analyzed in the framework of the project, which was authorized by the ethical committees of the Medical University of Graz (Reference number 1.0 24/11/2008) and the University of Heidelberg (Prof. Bannasch). Details on patient data are given in supplementary Table I.
Cell culture and transfection
HepG2 cells were cultured in RPMI-1640 (PAA, Cölbe, Germany) with supplementation of 10% (v/v) FCS (PAA), 1% (v/v) glutamine (PAA), and 1% (v/v) penicillin/streptomycin (PAA) at 37°C and 5% CO2. HepG2 overexpression and knockdown assays were performed according to Kessler et al. (27). For the detection of IGF2-mediated SREBF1 translocation, cells were treated for the indicated time with rhIGF-II (0.075 μg/ml, 292-G2; R&D Systems, Minneapolis, MN) or with IGF2 antibody (ab9574; Abcam, Cambridge, UK) as previously described (27).
Fatty acid measurement by GC-MS
Murine liver samples or HepG2 cells were lyophilized, hydrolyzed by the fatty acid methyl ester (FAME) method, and analyzed according to Bode et al. (36). Methyl-nonadecanoate (74208; Sigma Aldrich, Taufkirchen, Germany) was used as an internal standard. The method detects both free and bound free fatty acids.
Histochemistry
Hematoxylin/eosin staining and immunohistochemical detection of F4/80 of paraffin embedded sections were performed as previously reported (22, 24).
Real-time RT-PCR
Isolation of total RNA and reverse transcription was performed as described previously (37). RNA from human liver samples was isolated as previously described (27). Real-time RT-PCR was performed in an iQ5 cycler (Bio-Rad, Munich, Germany) or in a CFX96 cycler (Bio-Rad) with 5× HOT FIREPol® EvaGreen® qPCR Mix Plus (Solis BioDyne, Tartu, Estonia). All samples were estimated in triplicate. Primers and conditions are listed in supplementary Table II. Efficiency was determined for each experiment using a cDNA dilution series with a starting concentration equivalent to 0.5 μg RNA or with a standard dilution series as described previously (37). The relative gene expression was normalized to ACTB or 18s mRNA values.
Preparation of nuclear extracts
Nuclear extracts from HepG2 cells were prepared as described previously (38).
Protein isolation and analysis by Western blot
Protein isolation from murine liver tissue was done according to Tybl et al. (24), whereas protein isolation from cells was according to Basirico et al. (39).
Protein separation and detection were performed as previously described (24). Information on the antibodies used and conditions can be found in supplementary Table III. The primary antibodies, anti-ELOVL6 and anti-α−tubulin, were purchased from Sigma Aldrich (PRS4571, T9026), anti-SREBF1 and anti-PPARA from Abcam (ab3259, ab8934), anti-FASN anti-lamin A/C antibody from Cell Signaling Technology (#3180, #2032; Danvers, MA), and anti-p62 antibody was kindly provided by Dr. Tan (TSRI, La Jolla, CA) (30, 40).
Palmitoyl-CoA extraction and analysis with ultra HPLC-MS/MS
Fresh snap-frozen liver tissue was immediately freeze-dried, homogenized, and stored at −80°C. Freeze-dried tissue (40 mg) was extracted for 30 min in 1 ml methanol containing n-heptadecanoyl-CoA as internal standard (with a final concentration of 500 nM in 150 μl final volume). The extract was centrifuged at 22,000 g for 10 min at 4°C. The supernatant was transferred to a new vial, dried under gaseous nitrogen, and reconstituted in 150 μl methanol/water [1:1 (v/v)]. The standard dilution series was made in methanol/water [1:1 (v/v)]. All steps were performed on ice.
The analyses were performed using a TSQ Access Max mass spectrometer equipped with an ESI source and a triple quadrupole mass detector (Thermo Finnigan, San Jose, CA). The MS detection was carried out in heated ESI mode, at a spray voltage of 4.5 kV, a probe temperature of 400°C, a nitrogen sheath gas pressure of 3.0 × 105 Pa, an auxiliary gas pressure of 1.0 × 105 Pa, a capillary temperature of 350°C, and a tube lens voltage of 114 V in negative ionization mode. Palmitoyl-CoA lithium salt (P9716-5MG) and n-heptadecanoyl-CoA lithium salt (H1385-5MG) were purchased from Sigma Aldrich.
Xcalibur software was used for data acquisition and plotting.
The chromatographic separation was carried out on an Accela UPLC, consisting of a quaternary pump, degasser, and autosampler (Thermo Finnigan) using an Accucore RP-MS column (150 × 2.1, 2.6 μm), with an injection volume of 25 μl.
The solvent system consisted of 10 mM ammonium acetate (A) and methanol (B).
HPLC method.
Gradient run of initial 65% of B in A and a flow of 600 µl/min. In 0.6 min the solvent mixture was changed to 100% of B, with a flow of 700 µl/min and kept for 3.4 min.
The amounts of palmitoyl-CoA and the internal standard n-heptadecanoyl-CoA were each determined in single reaction monitoring mode using the following transitions: i) palmitoyl-CoA: precursor ion m/z 1,005.992, product ion m/z 672.075, scan time 0.5 s, scan width m/z 3.000, collision voltage 44 V; and ii) heptadecanoyl-CoA (internal standard): precursor ion m/z 992.228, product ion m/z 926.442, scan time 0.5 s, scan width m/z 3.000, collision voltage 42 V.
Palmitoyl-CoA, with a retention time of 2.27 min, was quantified using the chromatographic peak area relative to the internal standard (retention time 2.17 min). The lower limit of quantitation was 2.1 nM. The measurement was performed with WT livers (n = 9) and livers of p62 transgenic animals (n = 10). Supplementary Fig. I shows the measurement of a representative palmitoyl-CoA dilution series. Supplementary Fig. II shows a representative measurement of palmitoyl-CoA extracted from mouse liver.
Statistical analysis
Results are expressed as mean ± SEM. The statistical significance was determined by independent two-sample t-test. Pearson’s correlation was used to test the relationship between p62, IGF2, ELOVL6, and FASN mRNA in human liver samples. The results were considered as statistically significant with P < 0.05.
RESULTS
Characterization of steatosis
Liver-specific overexpression of p62 has previously been shown to induce histologically detectable steatotic features in about 60% of the animals (24) (Fig. 1A).
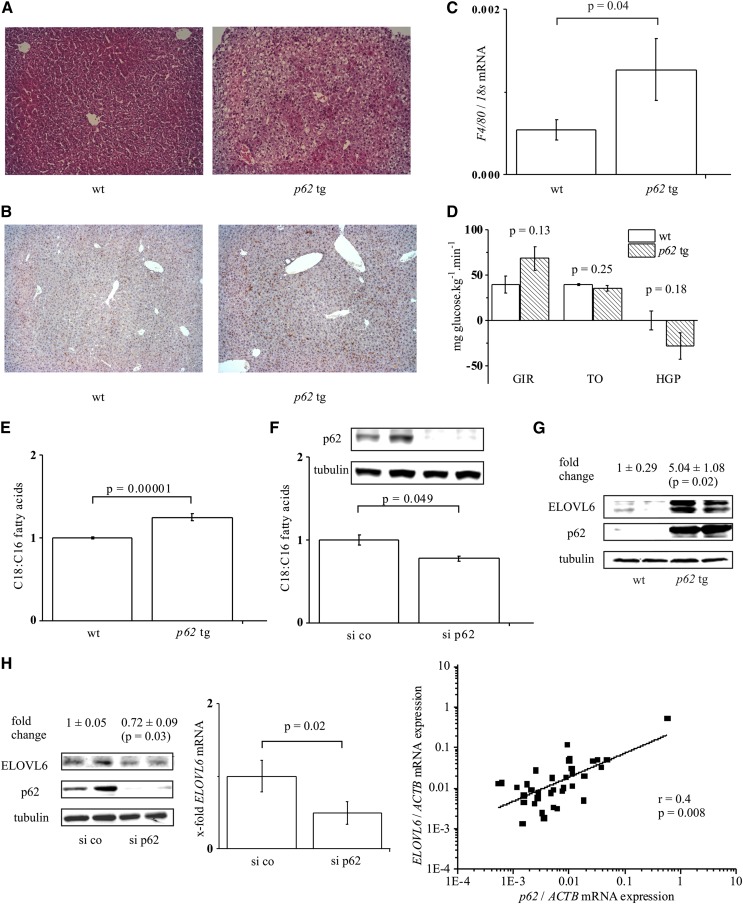
Fig. 1.
p62-Induced steatosis, C18:C16 ratio, and ELOVL6 expression. A: Hematoxylin/eosin staining of wild-type (wt) and p62 transgenic liver tissue (p62 tg) (original magnification 100×). B: F4/80 staining of wt and p62 tg liver tissue (original magnification 100×). C: F4/80 real-time RT-PCR of wild-type (wt) and p62 transgenic (p62 tg) livers (n = 13, each). D: Hyperinsulinemic euglycemic clamp study in wt (n = 3) and p62 tg (n = 4) mice: glucose infusion rate (GIR), glucose turnover (TO), and hepatic glucose production (HGP). Data show mean ± SEM. E: Hepatic C18:C16 fatty acid ratio: wt, n = 10; p62 tg, n = 19. F: C18:C16 fatty acid ratio and Western blot of the transfection control (72 h) in HepG2 cells transfected with random siRNA (si co) or p62 siRNA (si p62) (n = 3, duplicate). G: Representative ELOVL6 Western blot of wt and p62 tg mice (n = 5, each). H: ELOVL6 Western blot (n = 3, duplicate, left) and real-time RT-PCR (n = 2, triplicate, middle) of HepG2 cells transfected with si co or si p62 for ELOVL6. Right: Real-time RT-PCR for ELOVL6 and p62 of 35 liver tissues normalized on ACTB mRNA levels.
Previous data suggested no distinct inflammation in p62 transgenic animals because we observed no elevated liver damage (24). Still, immunohistochemical staining and real-time RT-PCR revealed elevated levels of the macrophage marker F4/80 in p62 transgenic livers (Fig. 1B, C). Distinct leukocyte infiltrates (2–3 infiltrates per microscopic field, magnification 100×) were exhibited by 18.2% of p62 transgenic animals, but were not observed in WT animals.
Our previous data indicates slightly improved glucose tolerance of p62 transgenic animals (24). We now performed hyperinsulinemic euglycemic clamp analysis; both the higher glucose infusion rate and lower hepatic glucose production with unchanged glucose turnover indicated elevated hepatic insulin sensitivity. However, these results were not statistically significant (Fig. 1D).
Quantification of fatty acid composition revealed that transgenic animals, which exhibited a normal histology, also showed an increased fatty acid content (Table 1). Still, the fatty acid content was even higher in animals with a histologically proven steatosis (Table 1). Taking all p62 transgenic animals together, we observed a 1.66 ± 0.12-fold (P = 0.0007) increase of the total fatty acid content compared with WT animals. Having a closer look at the composition of the fatty acids, we observed that the chain length of fatty acids was different in p62 transgenic livers compared with WT tissue; steatotic livers exhibited an increased ratio of C18 to C16 fatty acids (Fig. 1E).
TABLE 1.
The p62-induced changes in murine hepatic fatty acids
| Fatty Acid | WT | Normal Histology, p62 Transgenic | Microvesicular Steatosis, p62 Transgenic | ||
| Micrograms per Milligram Dry Tissue | Micrograms per Milligram Dry Tissue | P versus WT | Micrograms per Milligram Dry Tissue | P versus WT | |
| 12:0 | 0.05 ± 0.05 | 0.26 ± 0.13 | 0.205 | 0.81 ± 0.18 | 0.0016 |
| 14:0 | 0.32 ± 0.11 | 0.65 ± 0.29 | 0.344 | 2.08 ± 0.38 | 0.0009 |
| 16:0 | 16.7 ± 0.77 | 21.82 ± 1.99 | 0.050 | 28.72 ± 1.75 | 0.00002 |
| 16:1 | 0.04 ± 0.03 | 0.10 ± 0.06 | 0.441 | 0.69 ± 0.12 | 0.0003 |
| 18:0 | 11.7 ± 0.20 | 14.04 ± 0.71 | 0.018 | 14.24 ± 0.54 | 0.0008 |
| 18:1 | 5.54 ± 0.47 | 9.98 ± 1.84 | 0.061 | 19.34 ± 1.76 | 0.000007 |
| 18:2 | 9.75 ± 0.62 | 16.69 ± 2.49 | 0.035 | 30.12 ± 2.76 | 0.00002 |
| 20:1 | n.d. | 0.06 ± 0.05 | 0.351 | 0.20 ± 0.09 | 0.0455 |
| 20:2 | 0.34 ± 0.07 | 0.43 ± 0.14 | 0.611 | 1.11 ± 0.17 | 0.0010 |
| 20:3 | 0.29 ± 0.07 | 0.61 ± 0.17 | 0.135 | 1.47 ± 0.19 | 0.00006 |
| 20:4 | 10.9 ± 0.26 | 10.17 ± 0.86 | 0.461 | 13.47 ± 0.65 | 0.0032 |
| 20:5 | n.d. | n.d. | — | 0.08 ± 0.08 | 0.3409 |
| 22:4 | 0.11 ± 0.07 | 0.16 ± 0.11 | 0.698 | 0.88 ± 0.21 | 0.0038 |
| 22:5 | 2.20 ± 0.92 | 0.98 ± 0.64 | 0.340 | 3.31 ± 0.66 | 0.3781 |
| 22:6 | 1.55 ± 0.52 | 1.00 ± 0.30 | 0.413 | 0.83 ± 0.22 | 0.2663 |
| Sum FA | 59.5 ± 2.57 | 76.94 ± 8.03 | 0.072 | 117.3 ± 9.76 | 0.000028 |
Livers of WT (n = 7) and p62 transgenic (n = 19, n = 8 normal histology, n = 11 microvesicular steatotic histology) mice were analyzed by GC-MS. P values indicate differences compared with values in livers of WT animals. n.d., not detectable.
In order to study the mechanisms responsible for hepatic lipid alterations, we employed HepG2 cells, which are able to develop cellular steatosis and are frequently used for respective mechanistic studies (15, 41–44). The fatty acid chain length, in fact, depended on the presence of p62; when p62 was knocked down in HepG2 cells, increased levels of C16 fatty acids were detectable (Table 2), so that the ratio of C18 to C16 fatty acids decreased (Fig. 1F). We therefore investigated the effect of p62 on ELOVL6, which catalyzes the elongation of C16 to C18 fatty acids (17).
TABLE 2.
The p62-induced changes in HepG2 fatty acids
| Fatty Acid | Random siRNA (μg/mg lyophilized cells) | p62 siRNA (μg/mg lyophilized cells) | P |
| 16:0 | 20.06 ± 0.91 | 29.21 ± 1.93 | 0.03 |
| 16:1 | 5.40 ± 1.32 | 8.01 ± 1.53 | 0.12 |
| 18:0 | 8.98 ± 1.3 | 10.04 ± 1.88 | 0.71 |
| 18:1 | 36.08 ± 5.17 | 40.58 ± 2.52 | 0.55 |
| 18:2 | 1.63 ± 0.21 | 1.31 ± 0.25 | 0.57 |
HepG2 cells treated with random siRNA and p62 siRNA were analyzed by GC-MS. P values indicate differences compared with random siRNA-treated HepG2 cells (n = 3, duplicate).
We observed that ELOVL6 is induced in a p62-dependent fashion; p62 transgenic animals displayed significantly increased levels of ELOVL6 protein (Fig. 1G). Vice versa, knockdown of p62 in HepG2 cells exhibited significantly reduced ELOVL6 mRNA and protein levels (Fig. 1H). The dependency of ELOVL6 expression on p62 in human livers was supported by a strong correlation of p62/ELOVL6 expression (Fig. 1H).
Expression of lipogenic regulators
Surprisingly, MLXIPL mRNA expression was significantly upregulated in p62 siRNA-treated HepG2 cells (Fig. 2A) and downregulated in cells overexpressing p62 (Fig. 2A). We could validate this effect in p62 transgenic animals in which MLXIPL was significantly downregulated (15 ± 8%, P = 0.0497, n = 5 in each group). Looking at NR1H3 mRNA expression as another lipogenic transcription factor in either p62 overexpressing or p62 siRNA cells, we did not observe any significant difference (Fig. 2A). The same was true for SREBF1c mRNA (Fig. 2A).
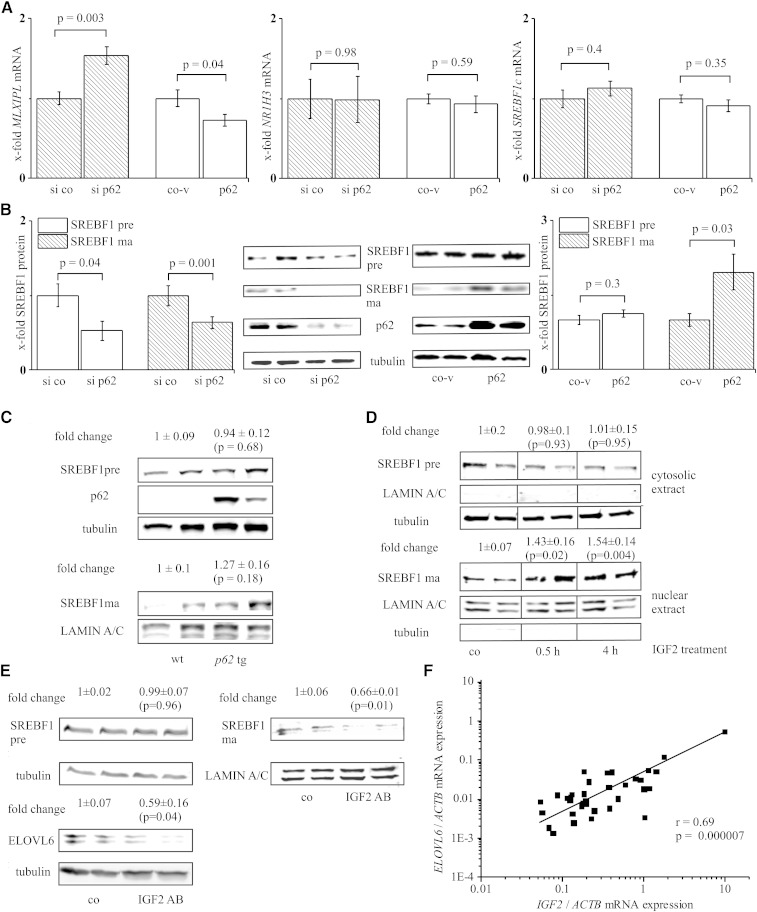
Fig. 2.
Implications of p62 on lipogenic pathways. Real-time RT-PCR (n = 3–6, triplicate) (A) or Western blot (n = 3, duplicate) (B) of HepG2 cells transfected with either p62 siRNA (si p62) or p62 overexpression vector (p62) and their respective controls (si co, co-v). A: MLXIPL, NR1H3, and SREBF1c mRNA. B: Precursor SREBF1 (SREBF1 pre) and mature SREBF1 (SREBF1 ma). C: SREBF1 pre and SREBF1 ma Western blot of wild-type (wt) and p62 transgenic (p62 tg) livers (n = 6). D: SREBF1 pre and SREBF1 ma Western blot on cytosolic and nuclear proteins of HepG2 cells treated with IGF2 (n = 9, duplicate). E: SREBF1 pre and SREBF1 ma and ELOVL6 Western blot on cytosolic and nuclear proteins of HepG2 cells (co) incubated with neutralizing IGF2 antibody (IGF2 AB, 48 h) (n = 4, duplicate). F: Real-time RT-PCR for ELOVL6 and p62 of 35 liver tissues normalized on ACTB mRNA levels.
In addition to transcriptional regulation (45, 46), SREBF1 can be activated by cleavage from its precursor to its mature form upon insulin treatment (38). Because p62 has been demonstrated to upregulate IGF2 in both mouse (24) and human livers (27), we hypothesized that p62 regulates SREBF1 on a protein level. We observed significantly increased levels of the mature isoform of SREBF1 after overexpression of p62 in HepG2 cells (Fig. 2B). The SREBF1 mature form in p62 transgenic mice behaved similarly, but quantified values were not statistically significant (Fig. 2C). The precursor did not show an induction in p62 overexpressing HepG2 cells (Fig. 2B) or in p62 transgenic mice (Fig. 2C). Knockdown of p62 by siRNA resulted in reduced levels of both the SREBF1 precursor and the mature form of SREBF1 (Fig. 2B).
Suggesting IGF2 to be responsible for SREBF1 activation, we treated HepG2 cells with IGF2, and indeed observed higher amounts of mature SREBF1 in their nuclei (Fig. 2D). Antagonization of IGF2 activity with an IGF2-specific antibody in HepG2 cells reduced the levels of ELOVL6 and the nuclear form of SREBF1 compared with their respective controls (Fig. 2E). In human liver tissue, in which p62 and ELOVL6 correlated, we also observed a distinct correlation of IGF2 and ELOVL6 expression levels (Fig. 2F). In the same tissues (n = 35), we could confirm a correlation between p62 and IGF2 (r = 0.41, P = 0.02) as recently reported for a cohort of HCC patients (27).
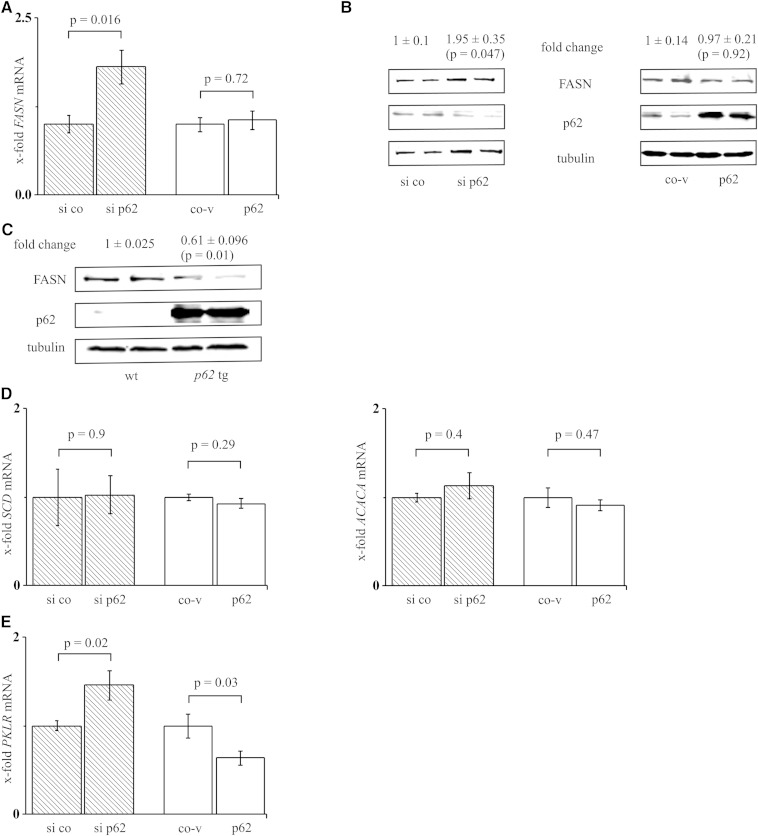
FASN, as an important lipogenic enzyme, also displays a target of SREBF1 activation, which is why we hypothesized that p62-induced lipid accumulation might be mediated via FASN induction. Interestingly, however, FASN mRNA and protein levels were not changed upon p62 overexpression and increased upon p62 knockdown (Fig. 3A, B). Concordantly, the p62 transgenics revealed reduced levels of FASN protein (Fig. 3C). The analysis of human liver samples showed no correlation between p62 and FASN mRNA (r = −0.1, P = 0.58). To reassess the unforeseen behavior of the direct SREBF1 target FASN, we had a closer look at stearoyl-CoA desaturase (delta-9-desaturase) (SCD1, SCD) and acetyl-CoA carboxylase α (ACACA, ACC) mRNA, which are SREBF1 targets and important enzymes in lipogenesis. Interestingly, neither p62 knockdown nor p62 overexpression revealed changes in SCD or ACACA mRNA levels (Fig. 3D). Because most lipogenic genes are coordinately regulated by SREBF1 and MLXIPL (47), we speculated that inversely regulated MLXIPL action might reverse the action of activated SREBF1. Therefore, we had a closer look at pyruvate kinase, liver and red blood cell (PKLR, L-PK) expression, which is exclusively regulated by MLXIPL (48). Indeed, p62 regulation of PKLR mRNA exhibited the same expression pattern compared with MLXIPL mRNA; PKLR mRNA was increased after p62 knockdown and decreased after p62 overexpression (Fig. 3E).
Fig. 3.
Effect of p62 on lipogenic genes. A: FASN real-time RT-PCR of HepG2 cells transfected with either p62 siRNA (si p62, n = 3, triplicate) or p62 overexpression vector (p62, n = 5, triplicate) and their respective controls (si co, co-v). B: FASN Western blot of HepG2 cells transfected with si p62 or p62 (n = 3, duplicate). C: Representative FASN Western blot of wild-type (wt) and p62 transgenic (p62 tg) livers (n = 5, each). D: SCD and ACACA real-time RT-PCR of HepG2 cells transfected with either p62 siRNA (n = 3, triplicate) or p62 overexpression vector (n = 5, triplicate) and their respective controls. E: PKLR real-time RT-PCR of HepG2 cells transfected with either p62 siRNA (n = 4, triplicate) or p62 overexpression vector (n = 5, triplicate) and their respective controls.
Regulation of lipolytic pathways
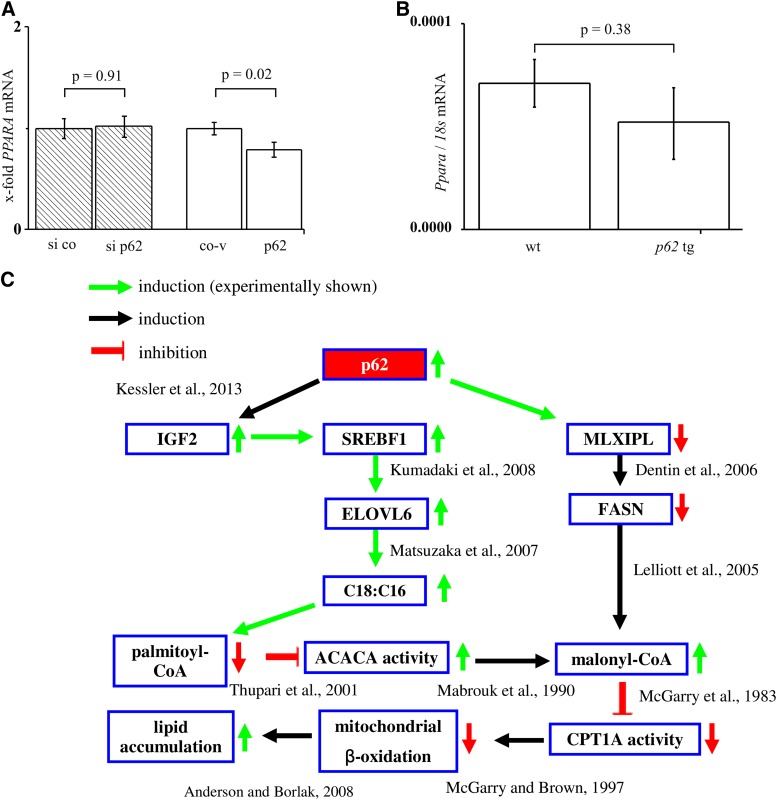
Our data suggested a distinct action of p62 on fatty acid composition (i.e., chain length), but no effect on lipogenic enzymes. We therefore suggested that the elevated levels of lipids in p62 livers might instead be facilitated by decreased β-oxidation. We therefore tested PPARA expression upon both p62 knockdown and overexpression in human hepatoma cells and in p62 transgenic mice. The data revealed a downregulation of PPARA mRNA after p62 overexpression in cells (Fig. 4A) and a lack of effect in the p62 transgenic mouse model (Fig. 4B). Therefore, the investigation of PPARA expression does not provide conclusive data on a potentially decreased β-oxidation due to p62.
Fig. 4.
Implications of p62 on lipolytic pathways and summary. A: PPARA real-time RT-PCR of HepG2 cells transfected with either p62 siRNA (si p62, n = 3, triplicate) or p62 overexpression vector (p62, n = 5, triplicate) and their respective controls (si co, co-v). B: PPARA real-time RT-PCR of wild-type (wt) and p62 transgenic (p62 tg) livers (n = 5, each). C: The p62 overexpression decreases FASN expression by MLXIPL depletion and induces IGF2 expression. Reduced FASN levels lead to malonyl-CoA accumulation. IGF2 promotes the maturation of SREBF1, which increases ELOVL6 expression, leading to an increased C18 to C16 ratio. The reduced inhibition of palmitoyl-CoA on ACACA activity elevates malonyl-CoA levels, which inhibit the CPT1A-activity and therefore mitochondrial β-oxidation.
Mitochondrial β-oxidation is regulated by carnitine palmitoyltransferase 1A (CPT1A) activity (Fig. 4C), which is controlled by malonyl-CoA. Malonyl-CoA levels depend on the palmitoyl-CoA-mediated inhibition of ACACA. Malonyl-CoA levels were below the detection limit of ultra HPLC-MS/MS analysis. Still, reduced palmitoyl-CoA levels (37 ± 12%), as found in p62 transgenic animals, suggested that attenuated mitochondrial β-oxidation is responsible for ELOVL6-induced steatosis.
DISCUSSION
The p62-induced steatosis is characterized by an increase in almost all lipid classes, with the most distinct effect on triglycerides (26). Accordingly, we here observed an elevated abundance of almost all fatty acids (Table 1). Due to unchanged serum cholesterol and triglyceride levels in p62 transgenics compared with WT animals (24), it is unlikely that hepatic lipid accumulation is due to reduced lipid export from the liver or increased lipid uptake.
Excess hepatic lipid incorporation is often associated with inflammatory events, which link a simple steatosis to NASH (6). We here show that p62 transgenic mice also display inflammatory signs, as validated by increased F4/80 and leukocyte infiltrates. The inflammation is rather mild, though, and does not result in elevated transaminase levels (24), which might also be linked to cytoprotective actions of p62 (27). Despite lipid accumulation and an inflammatory environment, development of insulin resistance is absent in p62 transgenic mice, confirming our previous data (24), which suggest slightly elevated glucose tolerance.
In addition to the general increase in lipids in p62 transgenic livers, we could observe an increased ratio of C18 to C16 fatty acids. Accordingly, ELOVL6, which catalyzes the elongation of C16 to C18 fatty acids (17), is increased in p62 transgenic mice. Our results from HepG2 cells and human liver tissues are in line with these findings. Recently, Matsuzaka et al. (20) reported that overexpression of ELOVL6 promotes NASH in mice and humans. What is more, ELOVL6 expression specifically characterizes steatotic events being linked to inflammation (21, 22). This is in line with the finding that p62 transgenic mice exhibit increased levels of ELOVL6 and also show signs of liver inflammation.
The pathways that are responsible for the upregulation of ELOVL6 as a pathophysiological regulator of liver disease are as yet unknown. SREBF1 is an important transcription factor that regulates lipid metabolism and contributes to the pathophysiology of the metabolic syndrome (49, 50). Our data demonstrate a p62-dependent cleavage of SREBF1 into its active form. SREBF1 gene expression, as well as its cleavage-induced activation, is enhanced by insulin, leading to its binding to sterol-response elements, which are located in the promoters of its target genes (51). HepG2 cells, in which the transcriptional regulator of SREBF1 NR1H3 was activated, displayed increased SREBF1c mRNA levels (52–54), whereas SREBF1a expression, the dominant isoform in cultured hepatocytes (55), was not regulated by NR1H3 activation (54, 56, 57). Because neither p62 knockdown nor p62 overexpression showed any effect on NR1H3 expression, it is hardly surprising that SREBF1c mRNA levels were not affected by p62.
IGF1 and insulin treatment have been shown to induce SREBF1 in sebocytes via activation of the insulin and IGF1 receptors (58). IGF2 binds to insulin receptor, IGF1-receptor, and IGF2-receptor with moderate to high affinity (45). Human hepatoma cell lines overexpressing p62 and p62 transgenic animals have been shown to express high levels of IGF2 mRNA (24, 27). We here report a p62-mediated induction of SREBF1 and ELOVL6, which depends on IGF2 expression. This causal link is supported by the correlation of p62, IGF2, and ELOVL6 expression in human liver tissue.
Our data reveal an upregulation of MLXIPL by p62 knockdown, while SREBF1 is inactivated. Vice versa, a MLXIPL downregulation occurs after p62 overexpression, while SREBF1 is activated. In fact, the literature reports that SREBF1 overexpression reduces MLXIPL expression (41). Because MLXIPL knockout animals show improved plasma glucose control (48, 59), the improved glucose tolerance exhibited in p62 transgenic animals (24) can be explained by MLXIPL downregulation and increased Igf2 levels.
Although SREBF1 can induce FASN (41), we observed that p62 decreased FASN levels, while SREBF1 was activated. The effects of p62-induced SREBF1 activation seem to be abrogated by the parallel decreased MLXIPL expression, which is in line with the finding that 50% of lipogenic gene expression is associated to MLXIPL (9). Interestingly, the p62 model is characterized by a higher sensitivity of ELOVL6 toward SREBF1 activation than to MLXIPL depletion. FASN, on the other hand, is more strongly affected by attenuated MLXIPL levels. In this context, Yu, Maguire, and Alwine (60) recently reported that human fibroblasts, in which MLXIPL was knocked down by a lentiviral short hairpin RNA plasmid, showed strongly decreased levels of FASN mRNA expression, but no effect on ELOVL6 expression, suggesting that MLXIPL shows stronger transcriptional activation toward FASN than to ELOVL6. PKLR, which is uniquely induced by MLXIPL (61), is reversely regulated by p62 and is therefore convergent to the MLXIPL expression.
Despite the effect of p62 on FASN in HepG2 cells and in the murine mouse model, we could not detect any correlation between p62 and FASN expression in human liver samples. Also, published data show no elevation of FASN in human NAFLD (62). Concordantly, Donnelly et al. (63) reported that de novo lipogenesis contributes to elevated hepatic lipids only to a minor extent, as found in human NAFLD.
In line with these findings, the literature describes that liver-specific knockout of FASN does not rescue the animals from the development of a fatty liver (64) and Jones et al. (65) recently reported the development of a steatosis in TSC22D4 overexpressing mice despite decreased FASN mRNA levels. Most interestingly, animals overexpressing ELOVL6 also show increased liver triglycerides and at the same time reduced FASN expression (20). Therefore, fatty acid synthesis does not appear to be the pivotal step in p62-mediated steatosis development.
One of the important inducers of peroxisomal and mitochondrial β-oxidation pathways in the liver is PPARA (10). PPARA agonists, like fenofibrate, reduce steatosis in mice with a hereditary fatty liver (66). PPARA is downregulated in p62 overexpressing HepG2 cells, but not in the p62 transgenics, suggesting that p62 does not facilitate lipolysis via PPARA. We suggest that p62 induces high fatty acid levels due to elevated malonyl-CoA levels. Indeed, the mitochondrial β-oxidation pathway, as the central lipolytic pathway, is negatively regulated by high malonyl-CoA levels (67). ELOVL6 can elevate malonyl-CoA because it reduces the levels of its substrate, palmitoyl-CoA, in p62 transgenic mice (Fig. 4C). Because fatty acids in p62 transgenic mice are mostly bound in triglycerides (26) and only free fatty acids are converted to acyl-CoAs (68), triglyceride-bound palmitic acid cannot be converted into palmitoyl-CoA. Whereas high levels of palmitoyl-CoA inhibit the activity of the enzyme responsible for malonyl-CoA synthesis (69), i.e., ACACA (67), low levels promote the generation of malonyl-CoA via ACACA. Consequently, the mitochondrial β-oxidation is reduced due to malonyl-CoA-mediated attenuation in CPT1A activity (70) (Fig. 4C). Pharmacological inhibition of CPT1A is associated with the development of steatosis and steatohepatitis (71, 72). Furthermore, a tamoxifen-induced steatosis in rats, which strongly inhibited FASN expression, was characterized by an accumulation of malonyl-CoA and therefore decreased CPT1A activity (73) (Fig. 4C).
In summary, our data provide evidence of p62 as an inducer of ELOVL6, a pathophysiological promoter of NASH. ELOVL6 overexpression results in a subsequent production of a deleterious fatty acid profile, which finally induces hepatic steatosis (Fig. 4C). This study underlines the detrimental role of p62 in liver disease.
Supplementary Material
Footnotes
Abbreviations:
- ACACA/ACC
- acetyl-CoA carboxylase α
- ChREBP/MLXIPL
- carbohydrate responsive element binding protein
- CPT1A
- carnitine palmitoyltransferase 1A
- ELOVL6
- fatty acid elongase 6
- HCC
- hepatocellular carcinoma
- IGF2
- insulin-like growth factor 2
- LXR-α/NR1H3
- liver X receptor α
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- PKLR
- pyruvate kinase, liver and red blood cells
- SREBF1/SREBP1
- sterol regulatory element binding transcription factor 1
This work was funded, in part, by the Graduiertenförderung of Saarland University (S.L.), an European Association for the Study of the Liver Dame Sheila Sherlock Fellowship, the Boehringer Ingelheim Fonds, and by the research committee of Saarland University (61-cl/Anschub2012) (S.M.K.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures and three tables.
REFERENCES
- 1.de Alwis N. M., Day C. P. 2008. Non-alcoholic fatty liver disease: the mist gradually clears. J. Hepatol. 48(Suppl 1): S104–S112. [DOI] [PubMed] [Google Scholar]
- 2.Erickson S. K. 2009. Nonalcoholic fatty liver disease. J. Lipid Res. 50(Suppl): S412–S416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams L. A., Waters O. R., Knuiman M. W., Elliott R. R., Olynyk J. K. 2009. NAFLD as a risk factor for the development of diabetes and the metabolic syndrome: an eleven-year follow-up study. Am. J. Gastroenterol. 104: 861–867. [DOI] [PubMed] [Google Scholar]
- 4.Adams L. A., Angulo P., Lindor K. D. 2005. Nonalcoholic fatty liver disease. CMAJ. 172: 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angulo P. 2002. Nonalcoholic fatty liver disease. N. Engl. J. Med. 346: 1221–1231. [DOI] [PubMed] [Google Scholar]
- 6.Day C. P. 2006. From fat to inflammation. Gastroenterology. 130: 207–210. [DOI] [PubMed] [Google Scholar]
- 7.Malhi H., Gores G. J. 2008. Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Semin. Liver Dis. 28: 360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musso G., Gambino R., Cassader M. 2009. Recent insights into hepatic lipid metabolism in non-alcoholic fatty liver disease (NAFLD). Prog. Lipid Res. 48: 1–26. [DOI] [PubMed] [Google Scholar]
- 9.Iizuka K., Horikawa Y. 2008. ChREBP: a glucose-activated transcription factor involved in the development of metabolic syndrome. Endocr. J. 55: 617–624. [DOI] [PubMed] [Google Scholar]
- 10.Lefebvre P., Chinetti G., Fruchart J. C., Staels B. 2006. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J. Clin. Invest. 116: 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakil S. J., Abu-Elheiga L. A. 2009. Fatty acid metabolism: target for metabolic syndrome. J. Lipid Res. 50(Suppl): S138–S143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGarry J. D., Mills S. E., Long C. S., Foster D. W. 1983. Observations on the affinity for carnitine, and malonyl-CoA sensitivity, of carnitine palmitoyltransferase I in animal and human tissues. Demonstration of the presence of malonyl-CoA in non-hepatic tissues of the rat. Biochem. J. 214: 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puri P., Wiest M. M., Cheung O., Mirshahi F., Sargeant C., Min H. K., Contos M. J., Sterling R. K., Fuchs M., Zhou H., et al. 2009. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 50: 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puri P., Baillie R. A., Wiest M. M., Mirshahi F., Choudhury J., Cheung O., Sargeant C., Contos M. J., Sanyal A. J. 2007. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 46: 1081–1090. [DOI] [PubMed] [Google Scholar]
- 15.Miyoshi H., Moriya K., Tsutsumi T., Shinzawa S., Fujie H., Shintani Y., Fujinaga H., Goto K., Todoroki T., Suzuki T., et al. 2011. Pathogenesis of lipid metabolism disorder in hepatitis C: polyunsaturated fatty acids counteract lipid alterations induced by the core protein. J. Hepatol. 54: 432–438. [DOI] [PubMed] [Google Scholar]
- 16.Kim K. H., Shin H. J., Kim K., Choi H. M., Rhee S. H., Moon H. B., Kim H. H., Yang U. S., Yu D. Y., Cheong J. 2007. Hepatitis B virus X protein induces hepatic steatosis via transcriptional activation of SREBP1 and PPARgamma. Gastroenterology. 132: 1955–1967. [DOI] [PubMed] [Google Scholar]
- 17.Matsuzaka T., Shimano H., Yahagi N., Kato T., Atsumi A., Yamamoto T., Inoue N., Ishikawa M., Okada S., Ishigaki N., et al. 2007. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat. Med. 13: 1193–1202. [DOI] [PubMed] [Google Scholar]
- 18.Kumadaki S., Matsuzaka T., Kato T., Yahagi N., Yamamoto T., Okada S., Kobayashi K., Takahashi A., Yatoh S., Suzuki H., et al. 2008. Mouse Elovl-6 promoter is an SREBP target. Biochem. Biophys. Res. Commun. 368: 261–266. [DOI] [PubMed] [Google Scholar]
- 19.Wang C. Y., Stapleton D. S., Schueler K. L., Rabaglia M. E., Oler A. T., Keller M. P., Kendziorski C. M., Broman K. W., Yandell B. S., Schadt E. E., et al. 2012. Tsc2, a positional candidate gene underlying a quantitative trait locus for hepatic steatosis. J. Lipid Res. 53: 1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuzaka T., Atsumi A., Matsumori R., Nie T., Shinozaki H., Suzuki-Kemuriyama N., Kuba M., Nakagawa Y., Ishii K., Shimada M., et al. 2012. Elovl6 promotes nonalcoholic steatohepatitis. Hepatology. 56: 2199–2208. [DOI] [PubMed] [Google Scholar]
- 21.Muir K., Hazim A., He Y., Peyressatre M., Kim D. Y., Song X., Beretta L. 2013. Proteomic and lipidomic signatures of lipid metabolism in NASH-associated hepatocellular carcinoma. Cancer Res. 73: 4722–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kessler S. M., Simon Y., Gemperlein K., Gianmoena K., Cadenas C., Zimmer V., Pokorny J., Barghash A., Helms V., van Rooijen N., et al. 2014. Fatty acid elongation in non-alcoholic steatohepatitis and hepatocellular carcinoma. Int. J. Mol. Sci. 15: 5762–5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessler S. M., Laggai S., Barghash A., Helms V., Kiemer A. K. Lipid metabolism signatures in NASH-associated HCC - letter. Cancer Res. Epub ahead of print. April 28, 2014; 10.1158/0008-5472.CAN-13-2852. [DOI] [PubMed] [Google Scholar]
- 24.Tybl E., Shi F. D., Kessler S. M., Tierling S., Walter J., Bohle R. M., Wieland S., Zhang J., Tan E. M., Kiemer A. K. 2011. Overexpression of the IGF2-mRNA binding protein p62 in transgenic mice induces a steatotic phenotype. J. Hepatol. 54: 994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon Y., Kessler S. M., Bohle R. M., Haybaeck J., Kiemer A. K. 2014. The insulin-like growth factor 2 (IGF2) mRNA-binding protein p62/IGF2BP2-2 as a promoter of NAFLD and HCC? Gut. 63: 861–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laggai S., Simon Y., Ranssweiler T., Kiemer A. K., Kessler S. M. 2013. Rapid chromatographic method to decipher distinct alterations in lipid classes in NAFLD/NASH. World J. Hepatol. 5: 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessler S. M., Pokorny J., Zimmer V., Laggai S., Lammert F., Bohle R. M., Kiemer A. K. 2013. IGF2 mRNA binding protein p62/IMP2-2 in hepatocellular carcinoma: antiapoptotic action is independent of IGF2/PI3K signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 304: G328–G336. [DOI] [PubMed] [Google Scholar]
- 28.Qian H. L., Peng X. X., Chen S. H., Ye H. M., Qiu J. H. 2005. p62 Expression in primary carcinomas of the digestive system. World J. Gastroenterol. 11: 1788–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Chan E. K. 2002. Autoantibodies to IGF-II mRNA binding protein p62 and overexpression of p62 in human hepatocellular carcinoma. Autoimmun. Rev. 1: 146–153. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J. Y., Chan E. K., Peng X. X., Tan E. M. 1999. A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. J. Exp. Med. 189: 1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tovar V., Alsinet C., Villanueva A., Hoshida Y., Chiang D. Y., Sole M., Thung S., Moyano S., Toffanin S., Minguez B., et al. 2010. IGF activation in a molecular subclass of hepatocellular carcinoma and pre-clinical efficacy of IGF-1R blockage. J. Hepatol. 52: 550–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiappini F., Barrier A., Saffroy R., Domart M. C., Dagues N., Azoulay D., Sebagh M., Franc B., Chevalier S., Debuire B., et al. 2006. Exploration of global gene expression in human liver steatosis by high-density oligonucleotide microarray. Lab. Invest. 86: 154–165. [DOI] [PubMed] [Google Scholar]
- 33.Christiansen J., Kolte A. M., Hansen T., Nielsen F. C. 2009. IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. J. Mol. Endocrinol. 43: 187–195. [DOI] [PubMed] [Google Scholar]
- 34.Li Z., Gilbert J. A., Zhang Y., Zhang M., Qiu Q., Ramanujan K., Shavlakadze T., Eash J. K., Scaramozza A., Goddeeris M. M., et al. 2012. An HMGA2-IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev. Cell. 23: 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanthier N., Molendi-Coste O., Horsmans Y., van Rooijen N., Cani P. D., Leclercq I. A. 2010. Kupffer cell activation is a causal factor for hepatic insulin resistance. Am. J. Physiol. Gastrointest. Liver Physiol. 298: G107–G116. [DOI] [PubMed] [Google Scholar]
- 36.Bode H. B., Ring M. W., Schwar G., Kroppenstedt R. M., Kaiser D., Muller R. 2006. 3-Hydroxy-3-methylglutaryl-coenzyme A (CoA) synthase is involved in biosynthesis of isovaleryl-CoA in the myxobacterium Myxococcus xanthus during fruiting body formation. J. Bacteriol. 188: 6524–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiemer A. K., Senaratne R. H., Hoppstadter J., Diesel B., Riley L. W., Tabeta K., Bauer S., Beutler B., Zuraw B. L. 2009. Attenuated activation of macrophage TLR9 by DNA from virulent mycobacteria. J. Innate Immun. 1: 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azzout-Marniche D., Becard D., Guichard C., Foretz M., Ferre P., Foufelle F. 2000. Insulin effects on sterol regulatory-element-binding protein-1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem. J. 350: 389–393. [PMC free article] [PubMed] [Google Scholar]
- 39.Basirico L., Morera P., Lacetera N., Ronchi B., Nardone A., Bernabucci U. 2011. Down-regulation of hepatic ApoB100 expression during hot season in transition dairy cows. Livest. Sci. 137: 49–57. [Google Scholar]
- 40.Lu M., Nakamura R. M., Dent E. D., Zhang J. Y., Nielsen F. C., Christiansen J., Chan E. K., Tan E. M. 2001. Aberrant expression of fetal RNA-binding protein p62 in liver cancer and liver cirrhosis. Am. J. Pathol. 159: 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dubuquoy C., Robichon C., Lasnier F., Langlois C., Dugail I., Foufelle F., Girard J., Burnol A. F., Postic C., Moldes M. 2011. Distinct regulation of adiponutrin/PNPLA3 gene expression by the transcription factors ChREBP and SREBP1c in mouse and human hepatocytes. J. Hepatol. 55: 145–153. [DOI] [PubMed] [Google Scholar]
- 42.Lee H., Park M. T., Choi B. H., Oh E. T., Song M. J., Lee J., Kim C., Lim B. U., Park H. J. 2011. Endoplasmic reticulum stress-induced JNK activation is a critical event leading to mitochondria-mediated cell death caused by β-lapachone treatment. PLoS ONE. 6: e21533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinciguerra M., Carrozzino F., Peyrou M., Carlone S., Montesano R., Benelli R., Foti M. 2009. Unsaturated fatty acids promote hepatoma proliferation and progression through downregulation of the tumor suppressor PTEN. J. Hepatol. 50: 1132–1141. [DOI] [PubMed] [Google Scholar]
- 44.Tripathy S., Jump D. B. 2013. Elovl5 regulates the mTORC2-Akt-FOXO1 pathway by controlling hepatic cis-vaccenic acid synthesis in diet-induced obese mice. J. Lipid Res. 54: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chao W., D’Amore P. A. 2008. IGF2: epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev. 19: 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen G., Liang G., Ou J., Goldstein J. L., Brown M. S. 2004. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc. Natl. Acad. Sci. USA. 101: 11245–11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Postic C., Girard J. 2008. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J. Clin. Invest. 118: 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dentin R., Benhamed F., Hainault I., Fauveau V., Foufelle F., Dyck J. R., Girard J., Postic C. 2006. Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes. 55: 2159–2170. [DOI] [PubMed] [Google Scholar]
- 49.Ahmed M. H., Byrne C. D. 2007. Modulation of sterol regulatory element binding proteins (SREBPs) as potential treatments for non-alcoholic fatty liver disease (NAFLD). Drug Discov. Today. 12: 740–747. [DOI] [PubMed] [Google Scholar]
- 50.Shimomura I., Bashmakov Y., Horton J. D. 1999. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J. Biol. Chem. 274: 30028–30032. [DOI] [PubMed] [Google Scholar]
- 51.Ferré P., Foufelle F. 2007. SREBP-1c transcription factor and lipid homeostasis: clinical perspective. Horm. Res. 68: 72–82. [DOI] [PubMed] [Google Scholar]
- 52.Kim K. H., Lee G. Y., Kim J. I., Ham M., Won Lee J., Kim J. B. 2010. Inhibitory effect of LXR activation on cell proliferation and cell cycle progression through lipogenic activity. J. Lipid Res. 51: 3425–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang M., Sun S., Wu T., Zhang L., Song H., Hao W., Zheng P., Xing L., Ji G. 2013. Inhibition of LXRα/SREBP-1c-mediated hepatic steatosis by Jiang-Zhi granule. Evid. Based Complement. Alternat. Med. 2013: 584634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshikawa T., Shimano H., Amemiya-Kudo M., Yahagi N., Hasty A. H., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., et al. 2001. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell. Biol. 21: 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimomura I., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. 1997. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 99: 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeBose-Boyd R. A., Ou J., Goldstein J. L., Brown M. S. 2001. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc. Natl. Acad. Sci. USA. 98: 1477–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 14: 2819–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith T. M., Gilliland K., Clawson G. A., Thiboutot D. 2008. IGF-1 induces SREBP-1 expression and lipogenesis in SEB-1 sebocytes via activation of the phosphoinositide 3-kinase/Akt pathway. J. Invest. Dermatol. 128: 1286–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iizuka K., Miller B., Uyeda K. 2006. Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am. J. Physiol. Endocrinol. Metab. 291: E358–E364. [DOI] [PubMed] [Google Scholar]
- 60.Yu Y., Maguire T. G., Alwine J. C. 2014. ChREBP, a glucose-responsive transcriptional factor, enhances glucose metabolism to support biosynthesis in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA. 111: 1951–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Denechaud P. D., Bossard P., Lobaccaro J. M., Millatt L., Staels B., Girard J., Postic C. 2008. ChREBP, but not LXRs, is required for the induction of glucose-regulated genes in mouse liver. J. Clin. Invest. 118: 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caballero F., Fernandez A., De Lacy A. M., Fernandez-Checa J. C., Caballeria J., Garcia-Ruiz C. 2009. Enhanced free cholesterol, SREBP-2 and StAR expression in human NASH. J. Hepatol. 50: 789–796. [DOI] [PubMed] [Google Scholar]
- 63.Donnelly K. L., Smith C. I., Schwarzenberg S. J., Jessurun J., Boldt M. D., Parks E. J. 2005. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Invest. 115: 1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chakravarthy M. V., Pan Z., Zhu Y., Tordjman K., Schneider J. G., Coleman T., Turk J., Semenkovich C. F. 2005. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 1: 309–322. [DOI] [PubMed] [Google Scholar]
- 65.Jones A., Friedrich K., Rohm M., Schäfer M., Algire C., Kulozik P., Seibert O., Müller-Decker K., Sijmonsma T., Strzoda D., et al. 2013. TSC22D4 is a molecular output of hepatic wasting metabolism. EMBO Mol. Med 5: 294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harano Y., Yasui K., Toyama T., Nakajima T., Mitsuyoshi H., Mimani M., Hirasawa T., Itoh Y., Okanoue T. 2006. Fenofibrate, a peroxisome proliferator-activated receptor alpha agonist, reduces hepatic steatosis and lipid peroxidation in fatty liver Shionogi mice with hereditary fatty liver. Liver Int. 26: 613–620. [DOI] [PubMed] [Google Scholar]
- 67.Thupari J. N., Pinn M. L., Kuhajda F. P. 2001. Fatty acid synthase inhibition in human breast cancer cells leads to malonyl-CoA-induced inhibition of fatty acid oxidation and cytotoxicity. Biochem. Biophys. Res. Commun. 285: 217–223. [DOI] [PubMed] [Google Scholar]
- 68.Soupene E., Kuypers F. A. 2008. Mammalian long-chain acyl-CoA synthetases. Exp. Biol. Med. (Maywood). 233: 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mabrouk G. M., Helmy I. M., Thampy K. G., Wakil S. J. 1990. Acute hormonal control of acetyl-CoA carboxylase. The roles of insulin, glucagon, and epinephrine. J. Biol. Chem. 265: 6330–6338. [PubMed] [Google Scholar]
- 70.McGarry J. D., Brown N. F. 1997. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur. J. Biochem. 244: 1–14. [DOI] [PubMed] [Google Scholar]
- 71.Anderson N., Borlak J. 2008. Molecular mechanisms and therapeutic targets in steatosis and steatohepatitis. Pharmacol. Rev. 60: 311–357. [DOI] [PubMed] [Google Scholar]
- 72.Koteish A., Diehl A. M. 2001. Animal models of steatosis. Semin. Liver Dis. 21: 89–104. [DOI] [PubMed] [Google Scholar]
- 73.Lelliott C. J., Lopez M., Curtis R. K., Parker N., Laudes M., Yeo G., Jimenez-Linan M., Grosse J., Saha A. K., Wiggins D., et al. 2005. Transcript and metabolite analysis of the effects of tamoxifen in rat liver reveals inhibition of fatty acid synthesis in the presence of hepatic steatosis. FASEB J. 19: 1108–1119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.