SUMMARY
While virulence properties of Candida albicans, the most commonly isolated human fungal pathogen, are controlled by transcriptional and post-translational mechanisms, considerably little is known about the role of post-transcriptional, and particularly translational, mechanisms. We demonstrate that UME6, a key filament-specific transcriptional regulator whose expression level is sufficient to determine C. albicans morphology and promote virulence, has one of the longest 5′ untranslated regions (UTRs) identified in fungi to date, which is predicted to form a complex and extremely stable secondary structure. The 5′ UTR inhibits the ability of UME6, when expressed at constitutive high levels, to drive complete hyphal growth, but does not cause a reduction in UME6 transcript. Deletion of the 5′ UTR increases C. albicans filamentation under a variety of conditions but does not affect UME6 transcript level or induction kinetics. We show that the 5′ UTR functions to inhibit Ume6 protein expression under several filament-inducing conditions and specifically reduces association of the UME6 transcript with polysomes. Overall, our findings suggest that translational efficiency mechanisms, known to regulate diverse biological processes in bacterial and viral pathogens as well as higher eukaryotes, have evolved to inhibit and fine-tune morphogenesis, a key virulence trait of many human fungal pathogens.
INTRODUCTION
Candida albicans is the major cause of human fungal disease worldwide. While normally a commensal in the mammalian host, this organism is responsible for a wide range of mucosal and systemic infections (Odds, 1988, Calderone & Clancy, 2012). Immunocompromised individuals, including cancer patients on chemotherapy, organ transplant recipients, and recipients of artificial joints and prosthetic devices, are particularly susceptible (Dupont, 1995, Weig et al., 1998). Candida species are now the fourth-leading cause of hospital-acquired bloodstream infections in the U.S. with a ~40% mortality rate (Edmond et al., 1999, Wisplinghoff et al., 2004) and approximately $1 billion per year is spent in this country on antifungal therapies to treat systemic candidiasis (Miller et al., 2001).
C. albicans is known to possess a number of properties which contribute to virulence, including the ability to undergo a reversible morphological transition from single oval-shaped yeast cells to pseudohyphal and hyphal filaments (elongated cells attached end-to-end) in response to specific environmental cues in the host environment (eg: serum and body temperature, 37°C) (Odds, 1988, Calderone & Clancy, 2012). Hyphal filaments are known to play an important role in tissue invasion, lysis of macrophages as well as evasion of the host immune system (Kumamoto & Vinces, 2005). Several key experiments have also indicated that the C. albicans yeast-filament transition is required for virulence in a mouse model of systemic candidiasis (Lo et al., 1997, Saville et al., 2003, Braun & Johnson, 1997). Additional virulence-related processes include adhesion to host epithelial and endothelial cells, secretion of degradative enzymes, phenotypic switching and the ability to form biofilms on host surfaces as well as implanted medical devices. Both phenotypic switching and biofilm formation can lead to the development of antifungal drug resistance (Hoyer et al., 2008, Sundstrom, 1999, Calderone & Clancy, 2012, Douglas, 2002, Schaller et al., 2005).
Given the importance of the virulence properties described above for C. albicans pathogenicity, intense research efforts have focused on elucidating molecular mechanisms by which they are controlled, particularly in response to host environmental cues. Thus far, considerable progress has been made towards the identification and characterization of post-translational mechanisms. Both mitogen-activated protein (MAP) kinase and Ras cAMP-protein kinase A (PKA) signaling pathways have been shown to mediate C. albicans filamentation in response to a variety of environmental cues including starvation, serum and glucose (Biswas et al., 2007). Phosphorylation of septins and other targets by the Hgc1-Cdc28 cyclin/Cdk complex under filament-inducing conditions is important for the physical process of C. albicans hyphal development (Wang, 2009). Histone acetylation and/or deacetylation are also important for C. albicans stress adaptation, survival in macrophages, morphogenesis and phenotypic switching (Lopes da Rosa & Kaufman, 2012, Hnisz et al., 2010, Lopes da Rosa et al., 2010). In addition, ubiquitination and sumoylation have both been shown to control C. albicans morphogenesis, cell cycle progression and stress response (Leach et al., 2011b, Leach et al., 2011a, Leach & Brown, 2012).
Significant progress has also been made towards the identification of transcriptional mechanisms that control C. albicans virulence properties. A variety of transcriptional regulators of filamentous growth (eg: Efg1, Cph1, Nrg1) have been shown to function as downstream targets of the MAP kinase and/or Ras-cAMP-PKA signaling pathways described above (Biswas et al., 2007, Calderone & Clancy, 2012, Lu et al., 2011). Many of these regulators control the expression of adhesins and secreted degradative enzymes (Felk et al., 2002, Sohn et al., 2003, Kadosh & Johnson, 2005). In addition, transcriptional regulatory mechanisms are known to control phenotypic switching, biofilm formation, stress response, iron acquisition and the development of antifungal drug resistance (Sellam et al., 2012, Chen et al., 2011, Zordan et al., 2007, Nobile et al., 2012, Sanglard et al., 2009).
In contrast to post-translational and transcriptional mechanisms, considerably little is known about post-transcriptional mechanisms that control C. albicans virulence properties. A She3-dependent RNA transport system is important for both hyphal development and the transport of certain filament-specific transcripts to the hyphal tip (Elson et al., 2009). A nuclear localization mechanism has recently been shown to control Sef1, a transcriptional regulator important for virulence and iron uptake in the host (Chen & Noble, 2012). An mRNA stability mechanism is also known to regulate filamentation and virulence (Cleary et al., 2012) and the Ccr4-Pop2 mRNA deadenylase complex controls cell wall integrity, filamentation and antifungal drug resistance (Dagley et al., 2011, Tucker et al., 2001, Chen et al., 2001). Information regarding translational control of C. albicans virulence properties is particularly lacking. Interestingly, however, a recent whole-genome RNA-Seq analysis has revealed that several key transcripts encoding proteins involved in filamentation, biofilm formation, white-opaque phenotypic switching (important for mating), adhesion, degradation of host membrane proteins and other processes important for C. albicans pathogenesis have unusually long (> 500 bp) 5′ untranslated regions (UTRs) (Bruno et al., 2010); similar results were found in an independent experiment using high-resolution C. albicans tiling arrays (Sellam et al., 2010). Previous studies in other organisms have revealed that 5′ UTRs can play an important role reducing overall translational efficiency by a variety of mechanisms, including the formation of highly stable secondary structures which inhibit ribosome scanning and/or accessibility (Mignone et al., 2002).
In this study, we identify an exceptionally long 5′ UTR and examine the role that this element plays in controlling the expression of UME6, which encodes a critical filament-specific transcriptional regulator of the C. albicans morphological transition. UME6 is important for controlling the level and duration of filament-specific gene expression in response to filament-inducing conditions. Strains deleted for UME6 are defective for hyphal extension and attenuated for virulence in a mouse model of systemic candidiasis (Banerjee et al., 2008, Zeidler et al., 2009) and a recent study has specifically demonstrated that Ume6 protein levels play a critical role in these processes (Lu et al., 2013). We have also previously shown that expression levels of UME6 are sufficient to sequentially specify C. albicans yeast, pseudohyphal and hyphal morphologies in a dosage-dependent manner (Carlisle et al., 2009). Here, we demonstrate that the UME6 5′ UTR plays an important role in specifically inhibiting Ume6 protein expression, thus affecting the ability of this regulator to determine C. albicans morphology. We also specifically examine the role of this 5′ UTR in controlling UME6 at the level of translational efficiency and provide new information about translational regulation of morphological transitions in pathogenic fungi.
RESULTS
Analysis of the C. albicans UME6 upstream region identifies an unusually long 5′ UTR
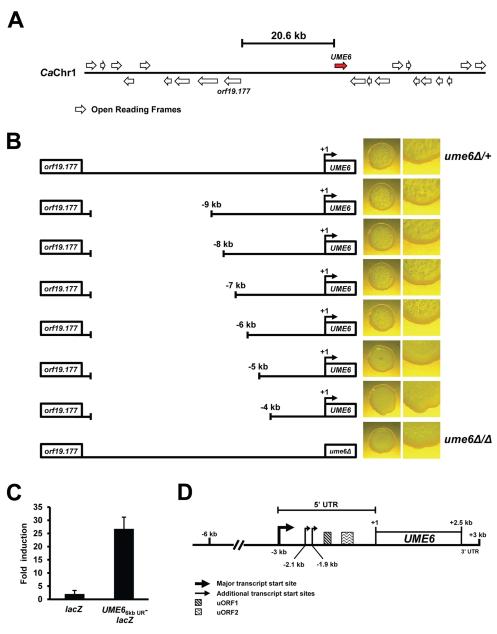
Given the importance of UME6 for C. albicans morphology determination and virulence, we initially sought to identify and characterize upstream regulatory elements that control expression of this transcription factor. We immediately noticed the presence of an unusually long intergenic region (20.6 kb) between UME6 and the nearest upstream gene, orf19.177, on C. albicans chromosome 1 (Figure 1A). In order to identify upstream elements in the intergenic region important for UME6 function, we first generated a ume6Δ/+ strain in which one allele of the UME6 coding sequence, along with 19.3 kb of upstream sequence, was deleted and replaced with a HIS1 marker (this strain was phenotypically equivalent to a strain deleted for one copy of only the UME6 open reading frame). Next, a variety of deletions of varying lengths were generated in the upstream intergenic region of the second UME6 allele. Morphology phenotypes of all deletion strains, along with ume6Δ/+ and ume6Δ/Δ control strains, were compared on solid medium in the presence of serum at 37°C, a strong filament-inducing condition (Figure 1B). Consistent with previous observations (Banerjee et al., 2008), the ume6Δ/Δ mutant was significantly defective for filamentation relative to the ume6Δ/+ strain. We observed that deletion strains containing only 4 kb and 5 kb of the upstream UME6 intergenic region showed a filamentation defect equivalent to that of the ume6Δ/Δ mutant. In contrast, strains containing 6 kb or greater of the UME6 upstream region showed a level of filamentation similar to that of the ume6Δ/+ parent strain. These results indicate that at least 6 kb of the upstream region is required for UME6 function with respect to filamentation and suggest that critical promoter elements are located in this region. In order to test this hypothesis, a C. albicans strain was generated in which the 6 kb UME6 upstream region was placed upstream of a heterologous Streptomyces thermophilus lacZ reporter gene. As shown in Figure 1C, the 6 kb UME6 upstream region was sufficient to drive ~25-fold transcriptional induction of the lacZ reporter in the presence of serum at 37°C vs. 30°C only. In contrast, the lacZ reporter alone did not show significant induction. These results indicate that the 6 kb UME6 upstream region contains important promoter elements and suggest that deletion strains containing only 5 kb and 4 kb of upstream sequence are defective for filamentation because they lack these elements. In order to map the transcriptional initiation site within this region, we performed a 5′ RACE (rapid amplification of cDNA ends) analysis using cDNA prepared from wild-type cells grown in the presence of serum at 37°C. This analysis revealed the presence of three transcript start sites located at positions −3041 bp, −2126 bp and −1923 bp relative to the translational initiation codon (+1) (Figures 1D and S1). These findings are consistent with a previous report which, using RNA-Seq analysis, indicated that the UME6 5′ UTR is greater than 1500 bp in length (Bruno et al., 2010). Based on the total size of the UME6 transcript, as determined by Northern analysis, as well as known sizes of UME6 open reading frame and 3′ UTR (Braun et al., 2005, Bruno et al., 2010), the major transcription start site is located at 3041 bp upstream of the UME6 start codon. UME6 transcripts whose sizes are consistent with 2.1 kb and 1.9 kb transcription start sites were not detected by Northern analysis, suggesting that these are minor start sites. The 3041 bp 5′ UTR of UME6 is much longer than the average 5′ UTR (~150 bp, based on our calculations) in C. albicans (Bruno et al., 2010) and, based on a search of the UTRdb database (Grillo et al., 2010), is one of the longest 5′ UTRs identified in fungi to date.
Figure 1.
Characterization of the UME6 upstream region identifies an exceptionally long 5′ UTR. (A) Schematic representation of the UME6 locus, and surrounding region, based on Candida Genome Database (http://www.candidagenome.org/) annotation (Braun et al., 2005). (B) 3 × 103 cells of the indicated strains were spotted on solid YEPD + 10% serum medium, grown at 37°C for 3 days and visualized by light microscopy. Images on the right are enlarged to show spot edges. Please note that only a single UME6 allele is shown in the schematic representations; the second allele, along with 19.3 kb of the UME6 upstream intergenic region, has been deleted in all strains except ume6Δ/Δ. (C) C. albicans strains bearing the indicated reporter constructs were grown overnight in YEPD at 30°C and diluted into pre-warmed YEPD + 10% serum at 37°C (filament-inducing conditions) or YEPD only at 30°C (non-filament-inducing conditions). At 30 minutes post-induction, cells were harvested for total RNA preparation and cDNA synthesis. lacZ expression levels were determined by qRT-PCR and normalized to levels of an ACT1 internal control. Fold induction was determined by dividing normalized lacZ values in induced cells by those observed in cells grown under non-filament-inducing conditions. Data shown represents the average of three biological replicates run in technical duplicate (mean ± SEM). (D) Schematic representation of the immediate UME6 upstream intergenic region (note: not drawn exactly to scale). Transcript start sites were determined by 5′ RACE analysis using cDNA prepared from wild-type (SC5314) cells harvested following induction in YEPD + 10% serum at 37°C.
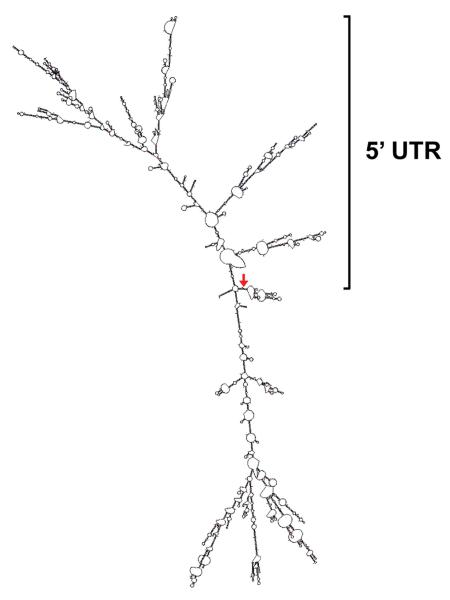
An in silico analysis of the UME6 5′ UTR sequence revealed the presence of two upstream open reading frames (uORFs) which are 39 bp and 102 bp in length and located at −1519 bp and −685 bp, respectively, upstream from the UME6 start codon (Figure 1D). In order to perform a predicted structure analysis of the UME6 transcript we used the RNA folding program mFold (http://mfold.rna.albany.edu/?q=mfold/) (Zuker, 2003). This program is widely used and was selected because it performs calculations based on Minimum Folding Energy (MFE) algorithms, which are typically more accurate in determining major substructures of RNAs with low folding energies. As indicated in Figure 2, an mFold analysis of the full-length UME6 transcript sequence predicted that the 5′ UTR forms a complex secondary structure. The 5′ UTR alone is predicted to be extremely stable with a very low folding free energy (ΔG) of −468.1 kcal/mol; the full-length UME6 transcript has a predicted ΔG of −1017.7 kcal/mol. An independent analysis using RNAFold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) (Zuker & Stiegler, 1981), a different MFE-based RNA structural prediction program, predicted similar structures and folding energies. The average folding free energy for 5′ UTRs of transcripts encoding transcription factors, growth factors, receptors and other regulatory proteins across species is −50 kcal/mol (Davuluri et al., 2000). Both uORFs and stable secondary structures are characteristic of 5′ UTRs which are known to play important roles in translational regulation (Mignone et al., 2002) and their apparent presence in this 5′ UTR suggested that UME6 may be controlled by such a mechanism.
Figure 2.
The UME6 5′ UTR is predicted to form a complex and highly stable secondary structure. A predicted structure analysis of the full-length UME6 transcript sequence, including the 5′ UTR, was performed using Mfold (http://mfold.rna.albany.edu/?q=mfold/), an RNA folding software program that predicts single-stranded RNA minimum folding free energies (Zuker, 2003). The sequence was folded using default parameters (37°C and 1M NaCl) and the predicted secondary structure is shown. Red arrow indicates the translation start site.
Deletion of the UME6 5′ UTR increases C. albicans filamentous growth but does not affect UME6 transcript level or induction kinetics
In order to determine whether the long 5′ UTR was important for the ability of UME6 to control C. albicans filamentous growth, we generated a strain in which both copies of the UME6 5′ UTR were deleted. The 5′ UTR deletion spanned from positions −3011 bp to −47 bp upstream of the UME6 start codon and this sequence was replaced with a 34 bp FRT site as well as XhoI and NotI restriction sites (see Supporting Information for details as well as a complete sequence of the fusion joint). Importantly, the complete UME6 open reading frame (ORF), as well as 46 bp immediately upstream of the UME6 AUG start codon, were left intact. As shown in Figure 3A, the UME6 5′ utrΔ/Δ strain showed a mild increase in filamentation, relative to that of the wild-type control strain, when cells were grown on solid YEPD medium in the presence of serum at 37°C. The increased filamentous growth phenotype of the UME6 5′ utrΔ/Δ mutant was more pronounced on solid Spider (nitrogen and carbon starvation) and Lee’s pH 6.8 media, but was not observed when cells were grown on solid YEPD medium at 30°C for 3 days (non-filament-inducing conditions). The UME6 5′ utrΔ/Δ mutant also showed greater filamentation than that of a wild-type control strain in liquid Spider medium at 37°C (data not shown). These results indicate that the UME6 5′ UTR functions to inhibit C. albicans filamentation in response to a variety of filament-inducing conditions.
Figure 3.
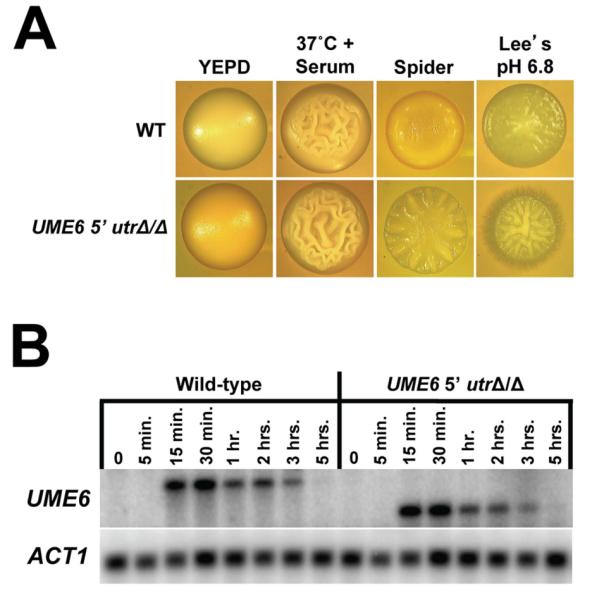
Deletion of the UME6 5′ UTR increases C. albicans filamentation but does not affect UME6 transcript level or induction kinetics. (A) Colony morphologies of the indicated strains grown on solid non-filament-inducing medium (YEPD) or on the indicated solid filament-inducing media. Colonies were grown for 2 days at 30°C on YEPD and Spider media, 2 days at 37°C on YEPD + 10% serum medium, 3 days at 30°C on Lee’s pH 6.8 medium and visualized by light microscopy. DK318 was used as the wild-type control strain. (B) The indicated strains were grown in YEPD medium at 30°C and diluted into prewarmed filament-inducing (YEPD + 10% serum at 37°C) medium. Cells were harvested at the indicated time points for total RNA preparation. Northern analysis was performed using 2.5 μg of RNA from each sample and the indicated probes. ACT1 is included as a loading control. Please note that the UME6 transcript size is reduced due to deletion of the 5′ UTR.
Many UTRs (primarily 3′ UTRs) are known to control the level of their respective transcripts (Mignone et al., 2002). In order to determine whether the UME6 5′ UTR plays a role in controlling UME6 transcript level and/or induction kinetics, a serum and temperature-induction time course experiment was carried out using both UME6 5′ utrΔ/Δ and wild-type control strains. Cells from each strain were harvested at the zero time point, as well as various post-induction time points, and total RNA was prepared for Northern analysis. As shown in Figure 3B, UME6 induction kinetics, as well as overall transcript level, in the UME6 5′ utrΔ/Δ strain appeared nearly identical to those observed in the wild-type control strain. We have also observed that both of these strains express UME6 at an equivalent level when cells are grown in Spider medium at 37°C (Figure S2).
The UME6 5′ UTR inhibits the ability of UME6, when expressed at constitutive high levels, to drive complete hyphal growth but does not cause a reduction in UME6 transcript
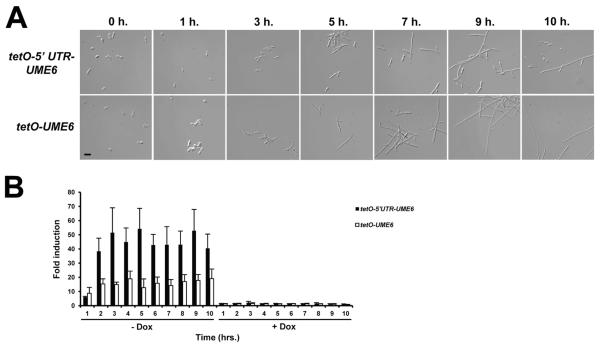
We have previously demonstrated that constitutive high-level expression of UME6 is sufficient to drive nearly complete hyphal growth of C. albicans in the absence of filament-inducing conditions (Carlisle et al., 2009). This result was obtained using a strain in which the E. coli tet operator (tetO) was placed upstream of the start codon for one allele of UME6 (the 5′ UTR was not present). In the absence of doxycycline (Dox, a tetracycline derivative), a transactivator binds tetO and UME6 is expressed at high constitutive levels; in the presence of Dox, the UME6 allele is shut off. When the tetO-UME6 strain is initially grown in the presence of Dox and then transferred to medium lacking Dox, C. albicans cells sequentially transition from yeast to pseudohyphae to a nearly complete hyphal population over a time course in the absence of filament-inducing conditions. In order to determine the effect of the UME6 5′ UTR on the ability of UME6 levels to specify C. albicans morphology, a similar strain was generated in which the tet operator was placed immediately upstream of the 3 kb 5′ UTR. Both tetO-5′ UTR-UME6 and tetO-UME6 strains were grown under non-filament-inducing conditions in the presence of Dox, and then cultures were diluted into medium lacking Dox, as described above. As a control, cultures from both strains were also diluted into medium containing Dox. Cell morphology of both strains was examined at various time points following Dox depletion. By the 3 hour time point, the tetO-5′ UTR-UME6 strain grew as pseudohyphae (Figure 4A). We also observed that this strain showed a higher proportion of yeast cells (32%) compared to that of the tetO-UME6 strain (14%). By the 10 hour time point, the tetO-UME6 strain had transitioned to a nearly complete hyphal population (note: in our previous study (Carlisle et al., 2009) this transition was completed within 9 hours, most likely due to differences in culture volume) whereas the tetO-5′ UTR-UME6 strain grew as a mixture of yeast, pseudohyphae and hyphae. Even following growth overnight in the absence of Dox, the tetO-5′ UTR-UME6 strain showed a mixed population of cell morphologies and was unable to transition completely to hyphae (Figure S3). Also, consistent with previous observations (Carlisle et al., 2009, Carlisle & Kadosh, 2010), control cells of both strains grown in the presence of Dox remained in the yeast morphology (data not shown). Overall, these results indicate that the 5′ UTR is important for the ability of UME6 expression levels to specify C. albicans morphology.
Figure 4.
The 5′ UTR inhibits UME6-driven hyphal growth in the absence of filament-inducing conditions but does not cause a reduction in UME6 transcript levels. (A) The indicated strains were grown overnight in YEPD medium at 30°C in the presence of 1 μg mL−1 Dox, washed twice with ddH2O and inoculated into fresh prewarmed YEPD medium at 30°C in the absence of Dox. Cell aliquots were harvested at the indicated time points, fixed with 4.5% formaldehyde, washed twice with 1X PBS and visualized by DIC microscopy. Bar = 10 μm. (B) Total RNA was isolated from cells grown as described in (A) as well as cells inoculated into fresh prewarmed YEPD medium at 30°C in the presence of 1 μg mL−1 Dox as a control. UME6 transcript levels were determined by qRT-PCR analysis and normalized to those of an ACT1 internal control. Fold induction was determined by dividing normalized UME6 expression values for each time point by the normalized UME6 expression value for the zero time point. Data shown represents the average of three biological replicates run in technical duplicate (mean ± SEM).
In order to examine whether the 5′ UTR affects C. albicans morphology determination by causing a reduction in UME6 transcript levels, total RNA was prepared from cells of both tetO-5′UTR-UME6 and tetO-UME6 strains at each time point of the time course experiment described above and used for quantitative RT-PCR analysis. As shown in Figure 4B, in the tetO-UME6 strain the UME6 transcript was induced 8.7-fold at the 1-hour time point following Dox depletion and remained induced at a high level (12- to 19-fold) from the 2-hour time point through the remainder of the time course. UME6 was expressed at roughly equivalent levels in both the tetO-5′ UTR-UME6 and tetO-UME6 strains at the 1 hour time point in the absence of Dox. Interestingly, over the remaining time points in the absence of Dox, UME6 levels in the tetO-5′ UTR-UME6 strain were consistently higher (38- to 54-fold induced) than those of the tetO-UME6 strain. As expected, UME6 was not expressed to a significant degree in either strain when cells were grown in the presence of Dox. Overall, these results clearly indicate that the inability of the tetO-5′ UTR-UME6 strain to drive complete hyphal growth in the absence of Dox cannot be attributed to a reduction in UME6 transcript levels.
The UME6 5′ UTR functions to inhibit translational efficiency of UME6
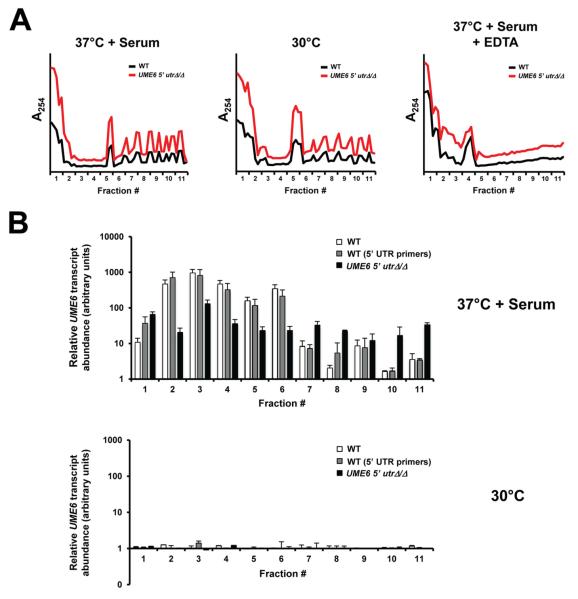
Given the failure of the 5′ UTR to cause a reduction in UME6 transcript level or control UME6 induction kinetics, we hypothesized that this region may control the translational efficiency of UME6. This hypothesis was supported by previous reports documenting the role of 5′ UTR regions in translational regulation, as well as our observation that the UME6 5′ UTR is predicted to possess a highly stable secondary structure and contains two uORFs, all of which have been associated with translational control in prior studies (Pickering & Willis, 2005, Mignone et al., 2002). In order to test this hypothesis, we performed a polysome profiling assay. Wild-type and UME6 5′ utrΔ/Δ strains showed similar polysome profiles when cells were grown under filament-inducing (serum at 37°C) and non-filament-inducing (30°C) conditions (Figure 5A). In addition, treatment with EDTA, a known inhibitor of polysome formation, disrupted the polysome profile in both strains, as expected. As indicated in Figure 5B, the UME6 transcript generally showed significantly greater association with the polysome fractions of the UME6 5′ utrΔ/Δ vs. wild-type control strain when cells were induced by serum at 37°C. The greatest differences in association (~ 10-fold) were observed in the final fractions (10 and 11), which are richest in polysomes. The UME6 transcript of the wild-type strain generally showed a similar abundance, regardless of whether qRT-PCR primers for the open reading frame or 5′ UTR were used for detection, indicating that the 5′ UTR was still present. In addition, an ACT1 control transcript generally did not show large differences in abundance between the wild-type and UME6 5′ utrΔ/Δ strains (Figure S4). As expected, we also observed an overall shift in transcript abundance from polysome-bound to unbound fractions upon treatment with EDTA (data not shown). Also consistent with previous observations that UME6 is transcriptionally induced in a filament-specific manner (Banerjee et al., 2008, Zeidler et al., 2009), very low levels of UME6 transcript were observed in all fractions when cells of both wild-type and UME6 5′ utrΔ/Δ strains were grown under non-filament-inducing conditions (30°C) (Figure 5B). These results strongly suggest that the 5′ UTR functions to inhibit UME6 translational efficiency by reducing association of the UME6 transcript with polysomes under filament-inducing conditions.
Figure 5.
The UME6 5′ UTR functions to inhibit translational efficiency. The indicated strains were grown overnight in YEPD medium at 30°C and diluted into prewarmed YEPD at 30°C (non-filament-inducing conditions) or YEPD + 10% serum at 37°C (filament-inducing conditions). At 30 minutes following serum and temperature induction, cells were treated with 0.1 mg mL−1 cycloheximide, lysed (in the presence or absence of 25 mM EDTA for cells grown in YEPD + 10% serum at 37°C), and subjected to sucrose gradient centrifugation for polysome isolation. (A) Polysome profiles across sucrose gradients for the indicated strains grown in YEPD at 30°C or YEPD + 10% serum at 37°C (+/− EDTA treatment) are shown; please note that profiles for each strain are offset. (B) RNA was extracted from each fraction of the indicated sucrose gradients to determine the abundance of UME6 transcript by qRT-PCR analysis. Data shown represents normalized mean UME6 transcript levels based on two independent experiments (± SEM). Please note that in the presence of serum at 37°C there is a statistically significant increase in UME6 transcript abundance in UME6 5′ utrΔ/Δ vs. wild-type (WT) strains for the polysome fractions 7,10 and 11 (p ≤ 0.01 as determined by Student’s t test).
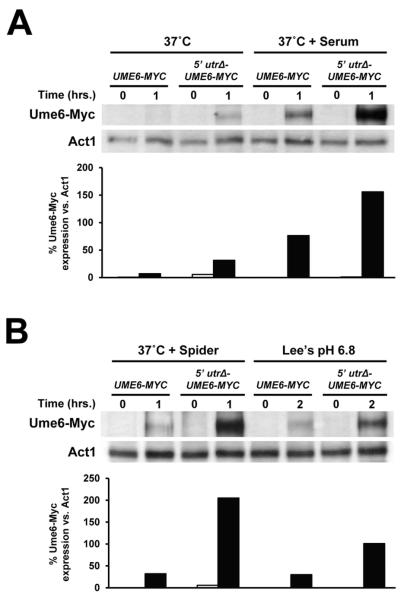
In order to confirm that the UME6 5′ UTR functions to inhibit Ume6 protein expression, the 5′ UTR was deleted in a strain expressing Myc-tagged Ume6 and cells were induced to form filaments by growth at 37°C in the presence of serum. As shown in Figure 6A, Ume6-Myc showed a significantly greater induction in the 5′ utrΔ-UME6-MYC strain compared to that observed in the UME6-MYC control strain. We also observed increased expression of Ume6-Myc in the 5′ utrΔ-UME6-MYC vs. UME6 MYC strain in the presence other filament-inducing conditions, including 37°C only, Spider at 37°C and Lee’s pH 6.8 media (Figures 6A and 6B). Interestingly, the extent to which Ume6 levels rose upon deletion of the 5′ UTR appeared to vary between filament-inducing conditions (eg: compare 37°C + Spider and Lee’s pH 6.8). These differences were reproducible based on multiple replicates (two replicates for 37°C and 37°C + Serum and three replicates for 37°C + Spider and Lee’s pH 6.8), suggesting that translational inhibition by the 5′ UTR may, to some degree, be modulated by environmental signals which control C. albicans filamentation. A Northern analysis indicated that for all filament-inducing conditions there was not a significant difference in the UME6 transcript level in UME6 MYC vs. 5′ utrΔ-UME6-MYC strains (Figure S5). As expected, UME6 levels were generally higher in the strongest filament-inducing condition, serum at 37°C, compared to those observed in Spider at 37°C, Lee’s pH 6.8 and 37°C only, which are weaker inducing conditions. Overall, these findings suggest that reduced translational efficiency directed by the UME6 5′ UTR results in a significant decrease in Ume6 protein expression in the presence of a variety of filament-inducing conditions.
Figure 6.
The UME6 5′ UTR inhibits Ume6 protein expression under a variety of filament-inducing conditions. The indicated strains were grown overnight in YEPD at 30°C and diluted 1:100 into pre-warmed YEPD at 37°C (37°C) and YEPD + 10% serum at 37°C (37°C + Serum) (A) or Spider at 37°C (37°C + Spider) and Lee’s pH 6.8 medium at 30°C (Lee’s pH 6.8) (B). Cells were harvested for protein isolation at the indicated time points and protein levels were determined by Western analysis. Act1 is shown as a loading control. Densitometry quantitation of Ume6-Myc expression vs. Act1 is shown below each respective Western (sample order is the same as that listed for each blot). White bars = 0 hour time point. Black bars = 1 hour time point (or 2 hour time point for the Lee’s pH 6.8 condition).
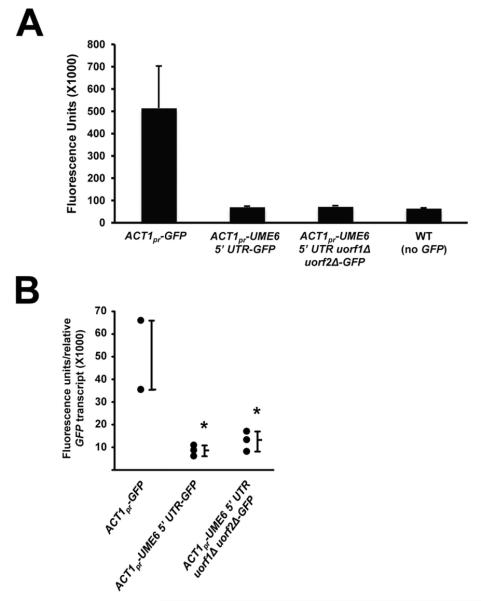
To determine whether the UME6 5′ UTR was sufficient to inhibit translation, we placed the 5′ UTR immediately upstream of a heterologous GFPγ reporter gene driven by a constitutive ACT1 promoter. ACT1pr-UME6 5′ UTR-GFP and ACT1pr-GFP strains were grown under non-filament inducing conditions and GFP protein expression was quantified by fluorometry. As shown in Figures 7A and S6, the 5′ UTR caused a significant reduction in GFP protein levels. Indeed, the ACT1pr-UME6 5′ UTR- GFP strain showed fluorescence values equivalent to those of a wild-type control strain which does not express GFP. In addition, removal of both uORFs did not affect the ability of the UME6 5′ UTR to inhibit GFP protein expression. Importantly, the ratio of GFP protein to transcript (as determined by qRT-PCR) was significantly higher in the ACT1pr- GFP strain when compared to that of the ACT1pr-UME6 5′ UTR-GFP and ACT1pr-UME6 5′ UTR uorf1Δ uorf2Δ-GFP strains (Figure 7B). A Northern analysis also confirmed that the GFP transcript was expressed at equivalent levels in all three of these strains and not expressed in a wild-type control strain (Figure S7). These results strongly suggest that the UME6 5′ UTR is sufficient to inhibit translation, but not transcription, via a uORF-independent mechanism when placed in the context of a heterologous promoter.
Figure 7.
The UME6 5′ UTR is sufficient to inhibit translation of a heterologous GFP reporter. (A) Cells of the indicated strains were grown in YNB minimal medium at 30°C. Fluorescence units were determined by dividing the fluorescence value (485 nm excitation, 535 nm emission) by the optical density (600 nm) of each sample. Data shown represents the average of three biological replicates (mean ± SEM). (B) Cells from part (A) were harvested for RNA isolation and GFP transcript levels were measured by qRT-PCR. Fluorescence units determined in (A) were normalized to the relative GFP expression of each biological replicate. Data shown is for three biological replicates of each strain. Statistical significance was determined by one-way ANOVA analysis. * = p< 0.01 compared to ACT1pr-GFP.
DISCUSSION
In C. albicans, the most commonly isolated human fungal pathogen, a variety of post-translational and/or transcriptional mechanisms are known to be involved in the regulation of morphogenesis, adhesion, secretion of degradative enzymes, biofilm formation, phenotypic switching and other virulence properties. However, significantly less is known about post-transcriptional, and especially translational, mechanisms that control these processes (although several genes are known to be translationally regulated during filamentation in the non-pathogenic model yeast Saccharomyces cerevisiae (Park et al., 2006)). Here, we describe a 5′ UTR-mediated translational efficiency mechanism that plays an important role in inhibiting C. albicans morphogenesis by controlling the expression of Ume6, a key filament-specific transcription factor. We provide several lines of evidence to support this mechanism: 1) deletion of the UME6 5′ UTR causes increased filamentation and hyphal growth but does not affect the induction kinetics or level of the UME6 transcript, 2) the 5′ UTR inhibits the ability of UME6, when expressed at constitutive high levels, to drive complete hyphal formation but does not cause a reduction in UME6 transcript, 3) the 5′ UTR specifically inhibits the ability of the UME6 transcript to associate with polysomes and also inhibits Ume6 protein expression in the presence of a variety of filament-inducing conditions, 4) the UME6 5′ UTR is sufficient to inhibit translation, but not transcription, of a heterologous reporter gene. In addition, the UME6 5′ UTR is exceptionally long and contains several features which have previously been associated with translational control (Mignone et al., 2002, Pickering & Willis, 2005).
How exactly does the 5′ UTR function to inhibit translational efficiency of UME6? Based on previous studies (Mignone et al., 2002, Pickering & Willis, 2005), four possible mechanisms may explain our observations. First, as indicated by our in silico analysis, the UME6 5′ UTR contains two putative uORFs. Because, in eukaryotes, the small ribosomal subunit typically initiates translation at the first scanned AUG codon, the large majority of translation in the 5′ UTR may be initiating at the uORFs, rather than at the UME6 start codon, depending on the context of ribosome capture. In this case, translation of UME6 would occur as a result of either leaky ribosome scanning or re-initiation. However, our observation that removal of both uORFs does not significantly affect the ability of the UME6 5′ UTR to inhibit translation appears to exclude this mechanism.
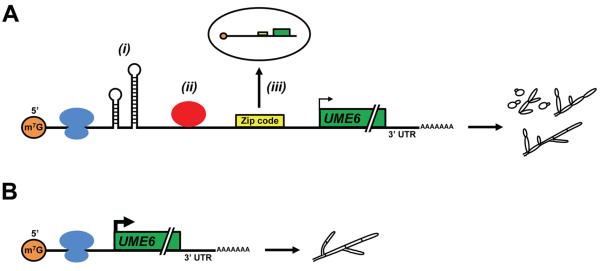
A second mechanism by which the 5′ UTR may control UME6 translational efficiency is by forming an extremely stable secondary structure (Figure 8). Stable 5′ UTR secondary structures have previously been shown to inhibit the ability of ribosomes to access and/or efficiently scan mRNA transcripts and reach the start codon (Mignone et al., 2002, Pickering & Willis, 2005). This mechanism is supported by our in silico analysis indicating the UME6 5′ UTR is predicted to form a complex and highly stable secondary structure with very low folding free energy. The predicted free energy of the 5′ UTR is about 9 times the free energy required for hairpin structures to block ribosome scanning (−50 kcal/mol) (Pelletier & Sonenberg, 1985, Kozak, 1989). Unfortunately, due to the exceptionally large size and predicted complexity of the UME6 5′ UTR, standard approaches (eg: compensatory base change and chemical probing experiments) to determine directly whether its secondary structure is important for controlling translational efficiency are not feasible. A deletion series analysis of the 5′ UTR suggested the possible involvement of certain regions in translational control, but was generally difficult to interpret since many of the partial deletion mutants also showed alterations in transcript levels when compared to those observed in UME6-MYC and UME6-5′ utrΔ-MYC strains (data not shown). In addition, partial deletions in the 5′ UTR caused significant changes in predicted overall secondary structure, which further complicated the analysis and made it difficult to determine the role of specific native 5′ UTR structures in translational control.
Figure 8.
Model for inhibition of UME6-driven hyphal formation in C. albicans by the long 5′ UTR. (A) In the presence of the 5′ UTR, scanning and/or access by ribosomes (shown in blue) can be inhibited by the formation of highly stable secondary structures causing a reduction in translational efficiency (i). Translation can also be inhibited by RNA-binding proteins (shown in red) which bind to the 5′ UTR and block access by ribosomes and/or induce formation of secondary structures (ii). Finally, a zip code sequence in the 5′ UTR may direct localization of the UME6 mRNA to a P-body or other distal cellular compartment (iii). As a consequence of one or more of these mechanisms (they are not mutually exclusive), UME6 translation will be significantly reduced (small arrow) and cells will form a mixed population of yeast, pseudohyphae and hyphae upon constitutive high-level expression of the UME6 transcript. (B) In the absence of the long 5′ UTR, translation of UME6 (large arrow) is not inhibited by any of the mechanisms described above and constitutive high-level UME6 expression causes cells to form a nearly complete hyphal population. Please note: we cannot exclude the possibility that the 5′ UTR may function to inhibit translation of UME6 by alternative mechanisms which are not described in part (A).
A third mechanism involves trans-acting RNA-binding proteins, which may bind to specific structural or sequence elements located within the UME6 5′ UTR. These factors can compete with ribosomes for access to the transcript or induce secondary structures which inhibit ribosome scanning (Figure 8) (Adeli, 2011). Another related possibility is that the UME6 5′UTR functions to prevent the formation of an internal ribosomal entry site (IRES) which allows ribosomes to directly enter the transcript at the start codon, instead of scanning from the 5′ end. However, IRES elements have typically been identified in viral and mammalian, rather than yeast, mRNAs (Kozak, 2001, Mignone et al., 2002, Pickering & Willis, 2005).
A fourth mechanism for UME6 5′ UTR-mediated translational inhibition may involve alternative mRNA localization (Figure 8). 5′ UTRs have been shown to affect the subcellular localization of their respective transcripts and C. albicans is known to transport certain mRNAs in a She3-dependent manner to the apical tip of hyphal filaments (Mignone et al., 2002, Elson et al., 2009). P-bodies, known to be important for mRNA degradation and storage, have recently been shown to accumulate during C. albicans hyphal development (Jung & Kim, 2011). Localization of the UME6 transcript to a P-body or other cellular compartment could therefore possibly lead to restricted translation at specific subcellular locations or storage for translation at a later time. The RNA predicted structural analysis, in combination with our demonstration that the 5′ UTR inhibits association of UME6 mRNA with polysomes, strongly suggests that the secondary structure and/or RNA-binding protein mechanisms play an important role. Importantly, the mechanisms described above are not mutually exclusive and could act either alone, in combination, or in conjunction with alternative mechanisms, to control the translational efficiency of UME6.
In many eukaryotic systems, post-transcriptional control mechanisms play an important role in rapidly fine-tuning the expression of genes involved in important developmental processes and the mechanism we have described here appears to be no exception. UME6 encodes a key filament-specific transcriptional regulator which controls C. albicans morphology and virulence, as well as the level and duration of filament-specific gene expression. UME6 also serves as an important downstream target for multiple filamentous growth signaling pathways and we have previously shown that the morphology of C. albicans cells is exquisitely sensitive to UME6 transcript levels (Banerjee et al., 2008, Carlisle et al., 2009, Zeidler et al., 2009). Our observations that the UME6 5′ utrΔ/Δ strain shows a generally mild increase in filamentation only in the presence of filament-inducing conditions and that constitutive high-level expression of UME6 in the presence of the 5′ UTR generates a mixed population of cell morphologies (rather than all yeast) suggest that the 5′ UTR-mediated translational efficiency mechanism serves to rapidly control and fine-tune Ume6 expression levels. Consistent with this observation, we have found that the level of translational inhibition directed by the UME6 5′ UTR can vary in the presence of different filament-inducing conditions, suggesting that translation of Ume6 can be modulated by environmental signals. Because UME6 transcript levels are very low in non-filament-inducing conditions, it was not possible to determine whether the 5′ UTR affects UME6 translation under these conditions. However, our GFP reporter experiment does suggest that the 5′ UTR can inhibit translation under non-filament-inducing conditions as well. A recent report has indicated that stabilization of Ume6 protein by multiple filamentous growth signaling pathways is critical for C. albicans hyphal development and maintenance (Lu et al., 2013). In this respect, the 5′ UTR-mediated translational inhibition mechanism may serve to rapidly reduce Ume6 protein levels, thus preventing unnecessary hyphal growth until appropriate host environmental cues are present.
Our results also suggest that the 5′ UTR may possess elements that can increase UME6 mRNA levels, at least when UME6 is expressed from a heterologous tet operator under non-filament-inducing conditions. Consistent with this observation, a recent report indicates that Ume6 can bind to its own upstream region in the vicinity of the 5′ UTR to increase transcription in a positive feedback loop (Lu et al., 2013). In addition, a transcription factor important for temperature-induced C. albicans morphogenesis, Hms1, has recently been shown to bind the UME6 5′ UTR and induce UME6 expression (Shapiro et al., 2012). Our finding that natural UME6 transcript levels are not altered upon deletion of the 5′ UTR is most likely explained by the observation that a significant number of additional transcriptional regulators appear to play an important role in the activation of UME6 (likely via the promoter) under filament-inducing conditions (Zeidler et al., 2009). Alternatively, changes in chromatin structure resulting from introduction of the 5′ UTR in the context of the tetO cassette could possibly account for increased UME6 transcript levels in the tetO-5′ UTR-UME6 vs. tetO-UME6 strain. Given the importance of Ume6 for determining morphology and controlling filament- and virulence-specific gene expression, it is not surprising that C. albicans has evolved multiple mechanisms (transcriptional, translational and post-translational) to carefully adjust the levels of this regulator under a variety of different environmental conditions.
Interestingly, a recent whole-genome RNA-Seq analysis has indicated that, similar to UME6, a significant number of C. albicans genes involved in a wide variety of processes important for pathogenicity possess unusually long (> 500 bp) 5′ UTRs (Bruno et al., 2010). Many of these genes encode transcriptional regulators that control filamentous growth, biofilm formation, white-opaque switching and/or antifungal drug resistance (eg: EFG1, CPH1, RFG1, CZF1, FKH2, SFL1, CRZ1, UPC2, GCN4, SIR2) (Stoldt et al., 1997, Ramage et al., 2002, Sonneborn et al., 1999, Liu et al., 1994, Kadosh & Johnson, 2001, Brown et al., 1999, Vinces & Kumamoto, 2007, Zordan et al., 2007, Bensen et al., 2002, Li et al., 2007, Onyewu et al., 2004, Santos & de Larrinoa, 2005, Silver et al., 2004, Tripathi et al., 2002, Perez-Martin et al., 1999). Several genes encoding adhesins of the α-agglutinin-like (ALS) family (ALS4, ALS5, ALS9), which are important for interaction of C. albicans with host cells (Hoyer, 2001), secreted aspartyl proteases (SAP1, SAP2) and lipases (LIP4, LIP8) , important for the ability of C. albicans to degrade host cell membranes (Schaller et al., 2003, Hube et al., 2000), a superoxide dismutase (SOD4) involved in responding to oxidative stress in the host (Martchenko et al., 2004, Frohner et al., 2009), and a key regulator of iron-uptake genes and gastrointestinal commensalism (SFU1) (Chen et al., 2011) also have long 5′ UTRs. Finally, several genes that play critical roles in the mechanics of C. albicans hyphal development (HGC1, CDC24, RGA2) and cell cycle control (CLN3, CLB4) fall in this category as well (Zheng et al., 2004, Zheng et al., 2007, Bassilana et al., 2003, Chapa y Lazo et al., 2005, Bensen et al., 2005, Bachewich & Whiteway, 2005, Wang, 2009). While not all of these genes may necessarily be controlled by a translational efficiency mechanism, the presence of a long 5′ UTR upstream of so many genes involved in virulence-related processes suggests, based on our findings, that this mechanism may play a significant role in controlling and fine-tuning C. albicans pathogenicity in the host.
There is also evidence to suggest that 5′ UTR-mediated translational efficiency mechanisms may play an evolutionarily conserved role in the regulation of morphogenesis and pathogenicity in non-albicans Candida species. Many of these species possess UME6 orthologs and the synteny of the long upstream intergenic region is conserved in Candida dubliniensis, Candida tropicalis, Candida parapsilosis, Candida guilliermondii and Candida lusitaniae (Candida Gene Order Browser, http://cgob.ucd.ie/), but has diverged in the non-pathogenic model yeast S. cereviseae. In addition, a recent whole-genome RNA-Seq experiment has identified over 250 C. parapsilosis genes with 5′ UTRs greater than 500 bp in length, many of which appear to be involved in filamentous growth and pathogenicity (Guida et al., 2011) and several of which encode orthologs of the C. albicans genes with long 5′ UTRs discussed above (including UME6).
In bacterial and viral pathogens, 5′ UTR-mediated translational efficiency mechanisms have been shown to control a variety of biological processes, several of which are important for virulence. In Listeria monocytogenes, 5′ UTRs are important for controlling the translation of key virulence factors involved in the production of listeriolysin O (Johansson et al., 2002, Wong et al., 2004, Shen & Higgins, 2005). The Haemophilus influenzae sxy gene, which encodes an important regulator of DNA uptake, is also known to be translationally regulated by a 5′ UTR which forms an inhibitory secondary structure (Cameron et al., 2008). In several viral pathogens, including poliovirus and Hepatitis C virus, 5′ UTRs contain IRES sequences important for controlling translational efficiency and viral replication (Balvay et al., 2009). In higher eukaryotes, UTRs are known to mediate translational control of a wide variety of genes that function in diverse cellular processes, including cell cycle, stress response, oncogenesis, fertilization and development (Pickering & Willis, 2005, Chatterjee & Pal, 2009). 5′ UTRs have also been shown to affect the expression of genes associated with a number of human diseases including breast cancer, Alzheimer’s disease, and bipolar affective disorder (BPAD) (Chatterjee & Pal, 2009). Our findings are significant because they suggest that in the major human fungal pathogen C. albicans, a 5′ UTR-mediated translational efficiency mechanism has evolved to inhibit and fine-tune morphogenesis, a key developmental process important for pathogenicity in the host environment.
Given that 5′ UTRs are likely to control the expression of a variety of important regulators of C. albicans morphology and/or virulence, what mechanisms are responsible for mediating translational control? How exactly do translational mechanisms modulate and/or fine-tune the expression of key virulence factors in response to host environmental conditions? Do certain components of the translation machinery respond to specific environmental cues? Is there crosstalk between translational mechanisms and known transcriptional and post-translational mechanisms which have previously been shown to control virulence properties? How and why did translational mechanisms apparently evolve to control so many genes associated with fungal pathogenicity? It is hoped that future research in this area will help to address these questions and shed more light on the important, but poorly understood, role that translational mechanisms may play in controlling a variety of processes important for fungal pathogenesis.
MATERIALS AND METHODS
Strains and DNA Constructions
A complete listing of strains used in this study is shown in Table S1. A detailed description of the plasmids and methods used to generate additional strains is provided in the Supplemental Materials and Methods section. All primers used for plasmid and strain constructions are described in Table S2.
Media and Growth Conditions
Standard non-filament-inducing growth conditions were YEPD (yeast extract-peptonedextrose) medium at 30°C. Induction of filamentous growth by 10% serum at 37°C was performed as described previously (Banerjee et al., 2008). Spider and Lee’s media were prepared as previously described (Lee et al., 1975, Liu et al., 1994). Induction of filamentous growth in Spider medium at 37°C was performed by first growing strains overnight in YEPD medium at 30°C. Cells were then washed and resuspended in 10 mL YEPD medium or Spider medium at a concentration of 1 × 106 cells mL−1. 10 μL of cells from each suspension were inoculated into 50 mL pre-warmed YEPD medium at 30°C or Spider medium at 37°C, respectively. Cultures were grown for 36 hours and cells were harvested for RNA extraction and microscopic analysis. DK318 was used as the wild-type control for the filament induction and polysome profiling experiments. The C. albicans morphological transition time course experiment was performed by initially growing tetO-UME6 and tetO-5′UTR-UME6 strains overnight at 30°C in YEPD medium + 1.0 μg mL−1 Dox to OD600 ~ 0.5. 50 mL aliquots of cells from each strain were next washed once in prewarmed YEPD medium at 30°C and used to inoculate 1.5 L of YEPD medium in the presence or absence of 1.0 μg/mL Dox. Cultures were grown at 30°C and cells were harvested for RNA extraction at each hour for 10 hours. Cells for the zero hour time point were collected from the tetO-UME6 overnight culture just prior to washing. For the 5′ RACE analysis, cells were induced with serum at 37°C as described previously (Banerjee et al., 2008) and harvested at 30 min. (for identification of the −2126 transcript start site) or 1 hour (for identification of the −1923 and −3041 transcript start sites) for total RNA and cDNA preparation. For the GFP expression experiment (Figures 7, S6 and S7) strains were grown overnight in YNB minimal medium at 30°C, diluted into fresh YNB minimal medium the next day, and grown for 3 hours at 30°C, as described by Wolyniak and Sundstrom (Wolyniak & Sundstrom, 2007); CAF2-1 was used as the wild-type control strain. For the Ume6-Myc expression experiment (Figure 6) strains were grown overnight in YEPD at 30°C and diluted 1:10 into the indicated pre-warmed media. Cells for the 0 hr. time point sample of this experiment were harvested immediately prior to dilution.
RNA Preparation and Analysis
Total RNA preparation and Northern analysis were performed as described previously (Banerjee et al., 2008). Primers used for Northern probes are described in Table S2. RNA from the polysome profiling experiments was extracted with 1:1 phenol:chloroform, precipitated overnight at −20°C with 2 volumes of 70% ethanol in 2.5M LiCl, washed with 70% ethanol and resuspended in nuclease-free water prior to qRT-PCR analysis (del Prete et al., 2007). RNA for qRT-PCR analysis and for 5′ RACE analysis was prepared using the SV Total RNA Isolation Kit (Promega) according to the manufacturer’s directions with the following modification: cells were resuspended in 225 μL buffer RLT and placed in a bead beater for 2.5 minutes (yeast) or 5 minutes (hyphae); cells were rested on ice for 1 minute per every 2.5 minutes in the bead beater.
5′ RACE Analysis
5′ RACE analysis was performed using an Ambion FirstChoice® RLM-RACE kit (Applied Biosystems). Briefly, total RNA was first treated with calf intestinal phosphatase (CIP) to remove 5′ phosphates from rRNA, tRNA, degraded mRNA and genomic DNA. Next, the RNA was treated with tobacco acid pyrophosphatase (TAP) to remove cap structures from the full-length mRNAs. A 45 bp 5′ RACE adaptor was then ligated to the decapped mRNA. Following a random-primed reverse transcription reaction, the 5′ end of the UME6 transcript was then amplified by nested PCR. All 5′ RACE PCR products were run on 0.8% agarose gels and directly sequenced to determine size and identity.
Real-time Quantitative RT-PCR Analysis
cDNA for qRT-PCR analysis was prepared from 2 μg of total RNA treated with DNAse I (Invitrogen) using an Applied Biosystems – High Capacity cDNA Reverse Transcription Kit, according to the manufacturer’s instructions. Real-time PCR was performed in duplicate in 96-well plates using the Chromo4 Four-Color Real-Time PCR Detection System (Bio-Rad). PCR reactions were carried out in 25 μL volumes containing 5 μL 1:25 diluted cDNA (original cDNA volume was 20 μL), 12.5 μL GoTaq qPCR Master Mix (Promega) and 4.3 μL dH2O. Primers used for qRT-PCR analysis are described in Table S2. Real-time PCR was performed using the following cycling conditions: Step 1: 95 °C for 2 minutes, Step 2: 95 °C for 30 seconds, Step 3: annealing temperature (determined for each primer pair) for 1 minute, Step 4: read plate, Step 5: repeat steps 2-4 for 39 times, Step 6: 72 °C for 5 minutes, Step 7: Melting Curve 50 °C – 95 °C every 0.4 °C, hold 1 second and read plate. Standard curves were generated using 7 serial dilutions of a pool of cDNA from each experiment to determine primer efficiency. Expression levels of each gene were normalized to levels of an internal ACT1 control using the Pfaffl method (Pfaffl, 2001). For the polysome profiling experiment, spike-in mRNA (Solaris) was added to each sample at a final concentration of 1X prior to RNA isolation and expression levels of UME6 were normalized to spike-in mRNA using the Pfaffl method.
Polysome Profiling Analysis
Cells were treated with 0.1 mg mL−1 cycloheximide and incubated on ice for 5 min. Next, cells were washed twice in lysis buffer (20 mM Tris-Cl, pH 8.0, 140 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 1% Triton X-100, 0.1 mg mL−1 cycloheximide, 1 mg mL−1 heparin), lysed by vortexing with 2/3 volume beads for 4 × 20 seconds and centrifuged 5 minutes at 4500 rpm at 4°C. Fifty OD260 units of supernatant were loaded on the top of a 10-50% continuous sucrose gradient and centrifuged at 35,000 rpm for 160 minutes at 4°C in a SW41 rotor (Beckman Coulter). Fractions were collected manually in 1 mL aliquots for RNA isolation or 200 μL aliquots to monitor OD254 absorbance. RNA was isolated from each fraction for qRT-PCR analysis. For the EDTA release assay, following treatment with 0.1 mg mL−1 cycloheximide, 25 mM EDTA was added to the lysis buffer and sucrose gradient.
Western Analysis
Protein isolation was performed as described previously (Cao et al., 2006). 20 μg of total protein extract was separated by 8% (for Ume6-Myc) or 12% (for Act1) SDS-PAGE and transferred to a PVDF membrane (Invitrogen). Membranes were blocked with 5% milk in 1X PBS with 0.01% Tween-20, incubated with primary antibody for Myc (Cell Signaling Technology #2272) or actin (Sigma #A5060) overnight at 4°C, washed three times in 1 × PBS with 0.01% Tween-20, then incubated with HRP-conjugated goat anti-rabbit secondary antibody (Zymed). The ECL system (GE Healthcare) was used for detection. Densitometry quantitation of Western blots was performed using GelQuant.NET software provided by biochemlabsolutions.com.
Fluorometry
Cellular fluorescence levels were quantified as described previously (Wolyniak & Sundstrom, 2007) using COSTAR 96-well plates and a Biotek Synergy 2 microplate reader. Plate wells contained 1.25 × 107 cells of each strain. Fluorometry assays were performed in biological triplicate and technical duplicate. Fluorescence values for each sample were normalized to cell density as determine by OD600.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Brian Wickes and other members of the San Antonio Center for Medical Mycology for fruitful discussions and advice during the course of the experiments and for useful comments and suggestions on this manuscript. We especially thank Terri Kinzy (Rutgers Robert Wood Johnson Medical School) as well as Chris Browne and Andrew Link (Vanderbilt University Medical Center) for useful comments and suggestions regarding the polysome profiling analysis. We are grateful to Alistair Brown (University of Aberdeen, United Kingdom), Hironobu Nakayama and Mikio Arisawa (Nippon Roche Research Center, Kamakura, Japan), James Konopka (Stony Brook University) and Haoping Liu (University of California, Irvine) for plasmids and/or strains. We also thank Ian Morris for assistance with statistical analysis and microscopy and Haoping Liu and Yang Lu for assistance and advice with Western analysis. D.S.C. was supported by a COSTAR training grant (National Institute of Dental and Craniofacial Research Grant T32DE14318) as well as a Ruth L. Kirschstein National Research Service Award for Individual Predoctoral Fellows (National Institute of Dental and Craniofacial Research Grant F31DE021930). D.K. was supported by National Institute of Allergy and Infectious Diseases Grant 5RO1AI083344 in addition to a Voelcker Young Investigator Award from the Max and Minnie Tomerlin Voelcker Fund. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institute of Dental and Craniofacial Research or the National Institute of Health.
REFERENCES
- Adeli K. Translational control mechanisms in metabolic regulation: critical role of RNA binding proteins, microRNAs, and cytoplasmic RNA granules. Am J Physiol Endocrinol Metab. 2011;301:E1051–1064. doi: 10.1152/ajpendo.00399.2011. [DOI] [PubMed] [Google Scholar]
- Bachewich C, Whiteway M. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot Cell. 2005;4:95–102. doi: 10.1128/EC.4.1.95-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. Structural and functional diversity of viral IRESes. Biochimica et biophysica acta. 2009;1789:542–557. doi: 10.1016/j.bbagrm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, Lopez-Ribot JL, Kadosh D. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Molecular biology of the cell. 2008;19:1354–1365. doi: 10.1091/mbc.E07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M, Blyth J, Arkowitz RA. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot Cell. 2003;2:9–18. doi: 10.1128/EC.2.1.9-18.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Clemente-Blanco A, Finley KR, Correa-Bordes J, Berman J. The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Molecular biology of the cell. 2005;16:3387–3400. doi: 10.1091/mbc.E04-12-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Filler SG, Berman J. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot Cell. 2002;1:787–798. doi: 10.1128/EC.1.5.787-798.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor. TUP1 Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- Braun BR, van Het Hoog M, d’Enfert C, Martchenko M, Dungan J, Kuo A, Inglis DO, Uhl MA, Hogues H, Berriman M, Lorenz M, Levitin A, Oberholzer U, Bachewich C, Harcus D, Marcil A, Dignard D, Iouk T, Zito R, Frangeul L, Tekaia F, Rutherford K, Wang E, Munro CA, Bates S, Gow NA, Hoyer LL, Kohler G, Morschhauser J, Newport G, Znaidi S, Raymond M, Turcotte B, Sherlock G, Costanzo M, Ihmels J, Berman J, Sanglard D, Agabian N, Mitchell AP, Johnson AD, Whiteway M, Nantel A. A human-curated annotation of the Candida albicans genome. PLoS genetics. 2005;1:36–57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DH, Jr., Giusani AD, Chen X, Kumamoto CA. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Molecular microbiology. 1999;34:651–662. doi: 10.1046/j.1365-2958.1999.01619.x. [DOI] [PubMed] [Google Scholar]
- Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 2010;20:1451–1458. doi: 10.1101/gr.109553.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone RA, Clancy CJ. Candida and Candidiasis. ASM Press; Washington, D.C.: 2012. [Google Scholar]
- Cameron AD, Volar M, Bannister LA, Redfield RJ. RNA secondary structure regulates the translation of sxy and competence development in Haemophilus influenzae. Nucleic acids research. 2008;36:10–20. doi: 10.1093/nar/gkm915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Lane S, Raniga PP, Lu Y, Zhou Z, Ramon K, Chen J, Liu H. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Molecular biology of the cell. 2006;17:295–307. doi: 10.1091/mbc.E05-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, Lopez-Ribot JL, Kadosh D. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:599–604. doi: 10.1073/pnas.0804061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle PL, Kadosh D. Candida albicans Ume6, a filament-specific transcriptional regulator, directs hyphal growth via a pathway involving Hgc1 cyclin-related protein. Eukaryot Cell. 2010;9:1320–1328. doi: 10.1128/EC.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapa y Lazo B, Bates S, Sudbery P. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot Cell. 2005;4:90–94. doi: 10.1128/EC.4.1.90-94.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Pal JK. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- Chen C, Noble SM. Post-transcriptional regulation of the Sef1 transcription factor controls the virulence of Candida albicans in its mammalian host. PLoS pathogens. 2012;8:e1002956. doi: 10.1371/journal.ppat.1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Pande K, French SD, Tuch BB, Noble SM. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell host & microbe. 2011;10:118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. Journal of molecular biology. 2001;314:683–694. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- Cleary IA, Lazzell AL, Monteagudo C, Thomas DP, Saville SP. BRG1 and NRG1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence. Molecular microbiology. 2012;85:557–573. doi: 10.1111/j.1365-2958.2012.08127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagley MJ, Gentle IE, Beilharz TH, Pettolino FA, Djordjevic JT, Lo TL, Uwamahoro N, Rupasinghe T, Tull DL, McConville M, Beaurepaire C, Nantel A, Lithgow T, Mitchell AP, Traven A. Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Molecular microbiology. 2011;79:968–989. doi: 10.1111/j.1365-2958.2010.07503.x. [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Zhang MQ. CART classification of human 5′ UTR sequences. Genome Res. 2000;10:1807–1816. doi: 10.1101/gr.gr-1460r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Prete MJ, Vernal R, Dolznig H, Mullner EW, Garcia-Sanz JA. Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. Rna. 2007;13:414–421. doi: 10.1261/rna.79407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas LJ. Medical importance of biofilms in Candida infections. Rev Iberoam Micol. 2002;19:139–143. [PubMed] [Google Scholar]
- Dupont PF. Candida albicans, the opportunist. A cellular and molecular perspective. J Am Podiatr Med Assoc. 1995;85:104–115. doi: 10.7547/87507315-85-2-104. [DOI] [PubMed] [Google Scholar]
- Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- Elson SL, Noble SM, Solis NV, Filler SG, Johnson AD. An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth. PLoS genetics. 2009;5:e1000664. doi: 10.1371/journal.pgen.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felk A, Kretschmar M, Albrecht A, Schaller M, Beinhauer S, Nichterlein T, Sanglard D, Korting HC, Schafer W, Hube B. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infection and immunity. 2002;70:3689–3700. doi: 10.1128/IAI.70.7.3689-3700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohner IE, Bourgeois C, Yatsyk K, Majer O, Kuchler K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Molecular microbiology. 2009;71:240–252. doi: 10.1111/j.1365-2958.2008.06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, Pesole G. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic acids research. 2010;38:D75–80. doi: 10.1093/nar/gkp902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guida A, Lindstadt C, Maguire SL, Ding C, Higgins DG, Corton NJ, Berriman M, Butler G. Using RNA-seq to determine the transcriptional landscape and the hypoxic response of the pathogenic yeast Candida parapsilosis. BMC genomics. 2011;12:628. doi: 10.1186/1471-2164-12-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Majer O, Frohner IE, Komnenovic V, Kuchler K. The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS pathogens. 2010;6:e1000889. doi: 10.1371/journal.ppat.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. 2001;9:176–180. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Green CB, Oh SH, Zhao X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family--a sticky pursuit. Medical mycology. 2008;46:1–15. doi: 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube B, Stehr F, Bossenz M, Mazur A, Kretschmar M, Schafer W. Secreted lipases of Candida albicans: cloning, characterisation and expression analysis of a new gene family with at least ten members. Arch Microbiol. 2000;174:362–374. doi: 10.1007/s002030000218. [DOI] [PubMed] [Google Scholar]
- Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Jung JH, Kim J. Accumulation of P-bodies in Candida albicans under different stress and filamentous growth conditions. Fungal genetics and biology. 2011;48:1116–1123. doi: 10.1016/j.fgb.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Molecular and cellular biology. 2001;21:2496–2505. doi: 10.1128/MCB.21.7.2496-2505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Molecular biology of the cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Molecular and cellular biology. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. New ways of initiating translation in eukaryotes? Molecular and cellular biology. 2001;21:1899–1907. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- Leach MD, Brown AJ. Posttranslational modifications of proteins in the pathobiology of medically relevant fungi. Eukaryot Cell. 2012;11:98–108. doi: 10.1128/EC.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach MD, Stead DA, Argo E, Brown AJ. Identification of sumoylation targets, combined with inactivation of SMT3, reveals the impact of sumoylation upon growth, morphology, and stress resistance in the pathogen Candida albicans. Molecular biology of the cell. 2011a;22:687–702. doi: 10.1091/mbc.E10-07-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach MD, Stead DA, Argo E, MacCallum DM, Brown AJ. Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Molecular microbiology. 2011b;79:1574–1593. doi: 10.1111/j.1365-2958.2011.07542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KL, Buckley HR, Campbell CC. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- Li Y, Su C, Mao X, Cao F, Chen J. Roles of Candida albicans Sfl1 in hyphal development. Eukaryot Cell. 2007;6:2112–2121. doi: 10.1128/EC.00199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lopes da Rosa J, Boyartchuk VL, Zhu LJ, Kaufman PD. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1594–1599. doi: 10.1073/pnas.0912427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes da Rosa J, Kaufman PD. Chromatin-mediated Candida albicans virulence. Biochimica et biophysica acta. 2012;1819:349–355. doi: 10.1016/j.bbagrm.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Su C, Solis NV, Filler SG, Liu H. Synergistic Regulation of Hyphal Elongation by Hypoxia, CO2, and Nutrient Conditions Controls the Virulence of Candida albicans. Cell host & microbe. 2013;14:499–509. doi: 10.1016/j.chom.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Su C, Wang A, Liu H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS biology. 2011;9:e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martchenko M, Alarco AM, Harcus D, Whiteway M. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Molecular biology of the cell. 2004;15:456–467. doi: 10.1091/mbc.E03-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome biology. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LG, Hajjeh RA, Edwards JE., Jr. Estimating the cost of nosocomial candidemia in the United States. Clin Infect Dis. 2001;32:1110. doi: 10.1086/319613. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Candida and Candidosis. Baillière Tindall; London: 1988. p. 468. [Google Scholar]
- Onyewu C, Wormley FL, Jr., Perfect JR, Heitman J. The calcineurin target, Crz1, functions in azole tolerance but is not required for virulence of Candida albicans. Infection and immunity. 2004;72:7330–7333. doi: 10.1128/IAI.72.12.7330-7333.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YU, Hur H, Ka M, Kim J. Identification of translational regulation target genes during filamentous growth in Saccharomyces cerevisiae: regulatory role of Caf20 and Dhh1. Eukaryot Cell. 2006;5:2120–2127. doi: 10.1128/EC.00121-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Perez-Martin J, Uria JA, Johnson AD. Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J. 1999;18:2580–2592. doi: 10.1093/emboj/18.9.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Semin Cell Dev Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Ramage G, VandeWalle K, Lopez-Ribot J, Wickes B. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002;214:95. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- Sanglard D, Coste A, Ferrari S. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 2009;9:1029–1050. doi: 10.1111/j.1567-1364.2009.00578.x. [DOI] [PubMed] [Google Scholar]
- Santos M, de Larrinoa IF. Functional characterization of the Candida albicans CRZ1 gene encoding a calcineurin-regulated transcription factor. Curr Genet. 2005;48:88–100. doi: 10.1007/s00294-005-0003-8. [DOI] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M, Bein M, Korting HC, Baur S, Hamm G, Monod M, Beinhauer S, Hube B. The secreted aspartyl proteinases Sap1 and Sap2 cause tissue damage in an in vitro model of vaginal candidiasis based on reconstituted human vaginal epithelium. Infection and immunity. 2003;71:3227–3234. doi: 10.1128/IAI.71.6.3227-3234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M, Borelli C, Korting HC, Hube B. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses. 2005;48:365–377. doi: 10.1111/j.1439-0507.2005.01165.x. [DOI] [PubMed] [Google Scholar]
- Sellam A, Hogues H, Askew C, Tebbji F, van Het Hoog M, Lavoie H, Kumamoto CA, Whiteway M, Nantel A. Experimental annotation of the human pathogen Candida albicans coding and noncoding transcribed regions using high-resolution tiling arrays. Genome biology. 2010;11:R71. doi: 10.1186/gb-2010-11-7-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellam A, Tebbji F, Whiteway M, Nantel A. A novel role for the transcription factor Cwt1p as a negative regulator of nitrosative stress in Candida albicans. PloS one. 2012;7:e43956. doi: 10.1371/journal.pone.0043956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Sellam A, Tebbji F, Whiteway M, Nantel A, Cowen LE. Pho85, Pcl1, and Hms1 signaling governs Candida albicans morphogenesis induced by high temperature or Hsp90 compromise. Curr Biol. 2012;22:461–470. doi: 10.1016/j.cub.2012.01.062. [DOI] [PubMed] [Google Scholar]
- Shen A, Higgins DE. The 5′ untranslated region-mediated enhancement of intracellular listeriolysin O production is required for Listeria monocytogenes pathogenicity. Molecular microbiology. 2005;57:1460–1473. doi: 10.1111/j.1365-2958.2005.04780.x. [DOI] [PubMed] [Google Scholar]
- Silver PM, Oliver BG, White TC. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot Cell. 2004;3:1391–1397. doi: 10.1128/EC.3.6.1391-1397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K, Urban C, Brunner H, Rupp S. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Molecular microbiology. 2003;47:89–102. doi: 10.1046/j.1365-2958.2003.03300.x. [DOI] [PubMed] [Google Scholar]
- Sonneborn A, Tebarth B, Ernst JF. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infection and immunity. 1999;67:4655–4660. doi: 10.1128/iai.67.9.4655-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom P. Adhesins in Candida albicans. Current opinion in microbiology. 1999;2:353–357. doi: 10.1016/S1369-5274(99)80062-9. [DOI] [PubMed] [Google Scholar]
- Tripathi G, Wiltshire C, Macaskill S, Tournu H, Budge S, Brown AJ. Gcn4 co ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 2002;21:5448–5456. doi: 10.1093/emboj/cdf507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Vinces MD, Kumamoto CA. The morphogenetic regulator Czf1p is a DNA-binding protein that regulates white opaque switching in Candida albicans. Microbiology. 2007;153:2877–2884. doi: 10.1099/mic.0.2007/005983-0. [DOI] [PubMed] [Google Scholar]
- Wang Y. CDKs and the yeast-hyphal decision. Current opinion in microbiology. 2009;12:644–649. doi: 10.1016/j.mib.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Weig M, Gross U, Muhlschlegel F. Clinical aspects and pathogenesis of Candida infection. Trends Microbiol. 1998;6:468–470. doi: 10.1016/s0966-842x(98)01407-3. [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- Wolyniak MJ, Sundstrom P. Role of actin cytoskeletal dynamics in activation of the cyclic AMP pathway and HWP1 gene expression in Candida albicans. Eukaryot Cell. 2007;6:1824–1840. doi: 10.1128/EC.00188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Bouwer HG, Freitag NE. Evidence implicating the 5′ untranslated region of Listeria monocytogenes actA in the regulation of bacterial actin-based motility. Cell Microbiol. 2004;6:155–166. doi: 10.1046/j.1462-5822.2003.00348.x. [DOI] [PubMed] [Google Scholar]
- Zeidler U, Lettner T, Lassnig C, Muller M, Lajko R, Hintner H, Breitenbach M, Bito A. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 2009;9:126–142. doi: 10.1111/j.1567-1364.2008.00459.x. [DOI] [PubMed] [Google Scholar]
- Zheng X, Wang Y, Wang Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004;23:1845–1856. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XD, Lee RT, Wang YM, Lin QS, Wang Y. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 2007;26:3760–3769. doi: 10.1038/sj.emboj.7601814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS biology. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic acids research. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.