Abstract
Various molecular and cellular pathways are active in eukaryotes to control the quality and integrity of mitochondria. These pathways are involved in keeping a ‘healthy’ population of this essential organelle during the lifetime of the organism. Quality control (QC) systems counteract processes that lead to organellar dysfunction manifesting as degenerative diseases and ageing. We discuss disease- and ageing-related pathways involved in mitochondrial QC: mtDNA repair and reorganization, regeneration of oxidized amino acids, refolding and degradation of severely damaged proteins, degradation of whole mitochondria by mitophagy and finally programmed cell death. The control of the integrity of mtDNA and regulation of its expression is essential to remodel single proteins as well as mitochondrial complexes that determine mitochondrial functions. The redundancy of components, such as proteases, and the hierarchies of the QC raise questions about crosstalk between systems and their precise regulation. The understanding of the underlying mechanisms on the genomic, proteomic, organellar and cellular levels holds the key for the development of interventions for mitochondrial dysfunctions, degenerative processes, ageing and age-related diseases resulting from impairments of mitochondria.
Keywords: mitochondria, quality control, disease, ageing
1. Introduction
Maintaining the integrity of the individual functional cellular units of any biological system is the key for its proper function. Various pathways evolved to exert quality control (QC). The individual pathways are effective at different stages of QC from single amino acids to a protein, to organelles and whole cells. They constitute a complex network in which they appear to interact in a hierarchical order. Impairments in this network lead to adverse effects, including disease and ageing.
Mitochondria are eukaryotic organelles involved in various essential functions including energy transduction, iron/sulfur cluster synthesis, lipid metabolism and copper homeostasis. Mitochondria need an efficient QC system because they generate a dangerous superoxide anion radical as a by-product of respiration. The superoxide anion radical can be converted to other types of reactive oxygen species (ROS) [1,2]. ROS are essential at low levels for molecular signalling but increased levels are toxic since they can damage virtually all biomolecules including lipids, proteins and nucleic acids. In fact, the toxic features of ROS and their generation in mitochondria led to the suggestion that mitochondria are the ‘clock’ involved in determining the lifespan of biological systems [3]. The notion that mitochondrial ROS are the root cause of ageing is currently questioned because of many contradictory data that accumulated over the years [4,5]. However, a contributing role of ROS to ageing of biological systems and in onset of diseases is generally accepted [5–7].
In this review, we focus on the components of the mitochondrial QC system, their function and interactions. We describe the impact of genetic mutations on degenerative processes including biological ageing, neurodegenerative and age-related disorders and describe mutations that impair QC function.
2. Mutations in mtDNA in relation to ageing and disease
Mitochondria are semi-autonomous organelles. In mammals, an estimated 1200–1500 proteins of this organelle are encoded by both the nuclear DNA and the mitochondrial DNA (mtDNA) [8,9]. In humans, only 13 proteins involved in oxidative phosphorylation (OXPHOS) are encoded by the mtDNA along with a set of 22 tRNAs and the two rRNAs necessary for the mitochondrial protein synthesis. Preserving the integrity of the genetic information for mitochondrial proteins in both the nucleus as well as in mitochondria is essential and mtDNA mutations have been associated with complex neurodegenerative diseases [10,11]. mtDNA instability was found to be responsible for degenerative processes, first in lower organisms and later also in mammals [12–16]. Both point mutations and large-scale mtDNA events (table 1) typically accumulate during ageing. Keeping the mtDNA unchanged is a basic requirement for maintaining the mitochondrial ‘health’ over time.
Table 1.
Types of mitochondrial DNA rearrangements in neurodegenerative diseases and ageing [17]. Overview of the different types of mtDNA point mutations and mtDNA rearrangements reported in neurodegenerative diseases, mitochondrial diseases with neurological symptoms and ageing. The type of mutation, commonly affected tissue type, affected genes, heteroplasmy or homoplasmy status and the occurrence of the mutation type relative to other mutation types is given. M, mitotic; PM, post mitotic; NC, non-coding region; PC, protein coding region; tRNA, tRNA gene.
| mutation | involvement in neurodegenerative disease |
involvement in ageing |
reference | ||||||
|---|---|---|---|---|---|---|---|---|---|
| tissue | genes affected | hetero/homo | occurrence | tissue | genes affected | hetero/homo | occurrence | ||
| point mutations: transitions | |||||||||
| A>G | M/PM | NC/PC/tRNAa | hetero/homoa | common | M | no bias | hetero/homob | less common | [18] |
| G>A | M/PM | NC/PC/tRNAa | hetero/homoa | common | M | no bias | hetero/homob | most common | [18] |
| C>T | M/PM | NC/PC/tRNAa | hetero/homoa | less common | M | no bias | hetero/homob | less common | [18] |
| T>C | M/PM | NC/PC/tRNAa | hetero/homoa | common | M | no bias | hetero/homob | common | [18] |
| point mutations: transversions | |||||||||
| A>T | — | — | — | — | M | no bias | hetero/homob | very rare | [17] |
| A>C | — | — | — | — | M | no bias | hetero/homob | rare | [18] |
| C>A | M/PM | NC/PC/tRNAa | hetero/homoa | rare | M | no bias | hetero/homob | very rare | [18] |
| C>G | M/PM | NC/PC/tRNAa | hetero/homoa | rare | M | no bias | hetero/homob | very rare | [17] |
| G>C | — | — | — | — | M | no bias | hetero/homob | rare | [18] |
| G>T | M/PM | NC/PC/tRNAa | hetero/homoa | rare | M | no bias | hetero/homob | very rare | [17] |
| T>A | — | — | — | — | M | no bias | hetero/homob | rare | [17] |
| T>G | — | — | — | — | M | no bias | hetero/homob | rare | [18] |
| mtDNA rearrangements | |||||||||
| insertions | M/PM | NC/PC/tRNAa | hetero/homo | rare | M | no bias | hetero/homob | rare | [18] |
| small deletions | M/PM | NC/PC/tRNAa | hetero/homo | rare | M | no bias | hetero/homob | rare | [18] |
| single large deletions | M/PM | NC/PC/tRNAa | hetero/homo | common | PM | NC/PC/tRNA | hetero | common | [19,20] |
| multiple deletions | M/PM | NC/PC/tRNAa | hetero/homo | rare | PM | NC/PC/tRNA | hetero | rare | [21,22] |
| duplication | M/PM | NC/PC/tRNAa | hetero/homo | rare | PM | n.a. | n.a. | rare | [20] |
aThe affected gene and heteroplasmy level or homoplasmy is mutation specific.
bHomoplasmy might be caused by full clonal expansion of a heteroplasmic mutant.
(a). mtDNA reorganization in eukaryotic cell systems
The first molecular evidence for age-related reorganization of mtDNA is derived from the fungal ageing model Podospora anserina [23]. This fungus is characterized by a limited lifespan and a senescence syndrome [24]. Senescence turned out to be correlated with a massive reorganization of the mtDNA [12,25]. The mtDNA reorganization is very efficiently driven by an unusual mobile genetic element. In juvenile cultures, the genetic element is an integral part of the mtDNA [26,27]. In senescent cultures, it accumulates as a covalently closed circular DNA. It was termed plasmid-like (pl) DNA or αSen DNA [28,29]. Subsequently, the autonomous element reintegrates into the mtDNA leading to large reorganizations [30]. In senescent cultures, mtDNA reorganizations may result in deletion of large parts of the mtDNA [26,31]. These reorganizations are found in all wild-type strains of P. anserina and seem to be the cause of senescence. A number of modulators affect the efficiency and the rate of reorganization. Modulators are encoded by nuclear genes and by mitochondrial traits. One such mitochondrial maternally inherited trait was first described in P. anserina in a longevity mutant. The modulator, linear plasmid pAL2-1, encodes DNA and RNA polymerase and contains long terminal inverted repeats [32–37]. Remarkably, the presence of pAL2-1 delays the wild-type-specific, age-related mtDNA reorganization leading to a 12-fold lifespan extension.
Also in other filamentous fungi, in particular different Neurospora species, mtDNA reorganizations were linked to senescence and organismal death [38,39]. These mtDNA instabilities are linked to the activity of either circular mitochondrial plasmids, such as Mauriceville-1c and Varkud-1c, or linear plasmids, such as kalilo and maranhar [13,40,41]. The latter resemble in their structure the pAL2-1 invertron of P. anserina. Essentially, all elements act as efficient mutator elements resulting in mtDNA reorganization and accumulation of defective mtDNA leading to senescence and death.
(b). mtDNA reorganization in mammalian systems
First evidence for age-related mtDNA reorganizations in mammals was obtained by heteroduplex analysis of mtDNA isolated from rodents of different ages that revealed an age-dependent increase in mtDNA deletions and/or duplications [42]. Subsequently, age-related mtDNA deletions were reported in other organisms including humans [43–45]. The mtDNA deletions found in mice [46] and mitochondrial myopathy patients [47] affected only a subset of the mtDNA copies in the cell (heteroplasmy). Similar results were observed in a patient carrying the m.11778G > A mutation, causing Leber's hereditary optic neuropathy (MIM:535000) [48]. It is now recognized that only those mtDNA deletions in muscle that exceed a certain heteroplasmy threshold (proportion of mutated mtDNA molecules) cause a biochemical defect in OXPHOS enzyme activity [49]. For different mtDNA mutations in muscle, various heteroplasmy threshold effects have been observed, ranging from 67 ± 16% [50] to more than 90% [51]. These varying threshold effects are paralleled in other tissues [52–55] and transmitochondrial cybrid cell lines [56–58]. At present, the majority (73%) of reported pathogenic mtDNA mutations are heteroplasmic mutations [17].
The heteroplasmy threshold is determined by a combination of different levels of transcription, translation, OXPHOS enzyme activity and the biochemical effect on individual OXPHOS complexes. For example, between 50% and 80% inhibition of individual complexes is required before the respiratory rate of mitochondria is affected [59]. The threshold effect is tissue-specific owing to a different expression of mtDNA mutations and the tissue-specific dependency on mitochondrial respiration, as shown for mt-tRNA mutations in muscle, blood and urothelium [55]. These factors contribute to various clinical manifestations of mtDNA mutations in human disease [60,61] and ageing [62]. Similar effects of mtDNA mutations on mitochondrial integrity have been observed during ageing and in mitochondrial disease [18,63,64].
(c). Clonal expansion of heteroplasmic mtDNA mutations during ageing
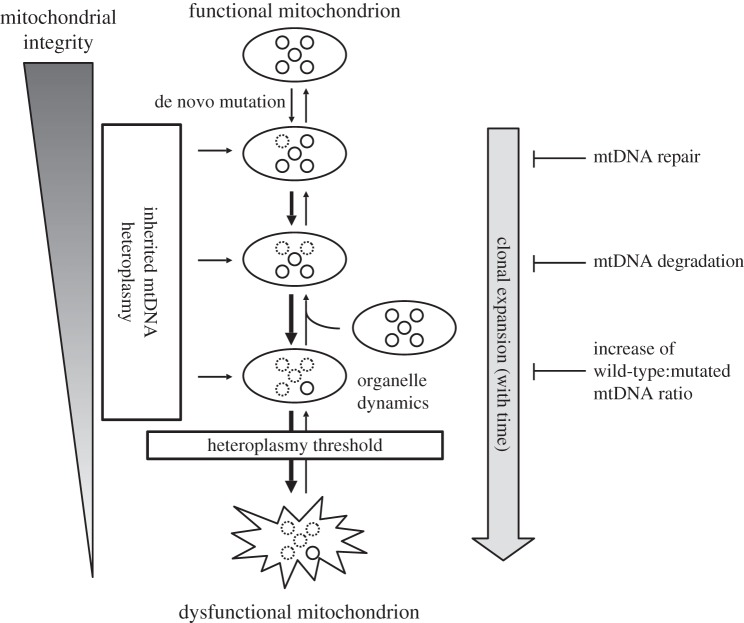
The occurrence of heteroplasmic mtDNA mutations in mitochondrial disease and accumulation of these mtDNA mutations with age [65,66] suggests that they are derived as clones from a single de novo mtDNA mutation. The process of increasing heteroplasmy with time is called clonal expansion and might occur owing to a selective advantage of damaged mtDNA molecules over healthy mtDNA genotypes (figure 1). Selective replication has been attributed to mtDNA size differences in single mtDNA deletion patients [48], intracellular heteroplasmy differences [67] or a reduced turnover rate of damaged mitochondria [68]. The latter two hypotheses view mitochondria as discrete organelles without the capability to exchange matrix content (figure 1) [69], but over longer periods of time mitochondrial fusion and fission may complement inter-mitochondrial differences. Clonal expansion of point mutations has also been observed in ageing mitotic tissues such as colon [62]. Recent modelling approaches support a size-based selective advantage, but only in long-lived species [70]. Another view is that clonal expansion is independent of a selection advantage of damaged mtDNA molecules, and that relaxed replication of mtDNA and constant turnover of mtDNA molecules causes random genetic drift of mtDNA mutations [71,72]. Random genetic drift requires a long lifespan for a single mtDNA mutation to occur and drift towards high-level heteroplasmy. For humans, these mutations would need to be inherited, subjected to a genetic bottleneck and purifying selection [73,74], or be fixed in early life [75]. In mammalian species, such as mice and rats, random genetic drift is not sufficient as a general mechanism to explain clonal expansion [76]. The mode of mtDNA selection is essential to understand the effect of clonally expanded mtDNA mutations on mitochondrial integrity during ageing and de novo mitochondrial disease.
Figure 1.
The influence of clonal expansion on mitochondrial integrity with time. Mutations can be introduced de novo or inherited in a number of mtDNA copies (heteroplasmic). Circles inside the mitochondrion are a representation of the mtDNA population for that particular mitochondrion. Normal mtDNA molecules are indicated with solid lines and mtDNA molecules with clonally expanded mutations are depicted with dashed lines. The clonal expansion of mutated mtDNA molecules dominates the expansion of normal mtDNA, indicated by thickness of the downward arrows. The mechanisms of mtDNA repair, selective/non-selective degradation of mtDNA and upregulated mtDNA replication to block clonal expansion at each level of mitochondrial integrity are indicated on the right.
(d). Preserving mitochondrial integrity by mtDNA repair
DNA is subjected to damage during ageing (figure 1): spontaneous hydrolysis forming either apurinic/apyrimidinic sites or deaminated uracil sites, oxidative lesions, non-enzymatic alkylation of DNA, mismatched bases or single- and double-stranded breaks [77,78]. Formation of de novo mtDNA mutations is believed to be the result of increased mitochondrial oxidative stress and/or replication errors [79–81]. In order to prevent deleterious consequences of these mutations, cells may either repair arising mtDNA mutations or shift the heteroplasmy of existing mutations below the phenotypic threshold effect.
Not all DNA repair pathways known for nuclear DNA exist in mitochondria, although advances in the field of mtDNA repair have changed the perspective that mtDNA was hardly repaired at all. (For an extensive overview of mtDNA repair pathways, see [77,82,83]). Here, we focus on the role of mtDNA repair in ageing and disease.
Mitochondrial base excision repair (mtBER) is a pathway for repair of mtDNA oxidative lesions. Next to short-patch mtBER (repair of single bases), long-patch mtBER (removal of a string of several bases) is now also well documented in mitochondria. Mechanisms of nuclear BER (nBER) and mtBER are similar and many of the proteins active in the nucleus have also been identified in the mitochondria [84–86]. Many of the BER enzymes are essential for cellular viability, and conditional BER knockout models show severe mtDNA depletion [87–89].
The nucleus uses an array of DNA binding proteins with specialized tasks in DNA maintenance, but mitochondria use only their subset. For instance, POLG is the only polymerase responsible for DNA polymerase, 5′–3′ exonuclease, dRP-lyase and reverse transcriptase activities in mitochondria. It is therefore not surprising that POLG malfunction causes many clinical cases of mtDNA diseases [90,91]. Premature ageing and reduced lifespan in proofreading-deficient PolgA mutator mice were attributed to an increased somatic mtDNA mutation load promoting apoptosis [92,93]. However, these non-physiological levels have been criticized [94], as recent evidence shows that low-level transmitted heteroplasmic mtDNA variants (5–13%) also contribute to ageing in the mutator mouse.
MtDNA is clustered in protein-rich structures called nucleoids near the inner mitochondrial membrane, where it interacts with mitochondrial transcription factor A (TFAM) [95–97]. TFAM binding to damaged mtDNA could act as a recognition element for mtDNA repair enzymes. In an accelerated ageing rat model carrying the mtDNA deletion, TFAM overexpression in the inner ear led to a reduction of 8-oxoguanine DNA glycosylase OGG1, reduction of polymerase POLG activity and an increase in heteroplasmy [98]. BER enzymes appear to be regulated at the protein level. Whole tissue extracts of rat lens of two- and 14 months old animals contained similar mRNA levels of OGG1, APE1 and POLG, but a reduced protein expression under normoxic conditions [99]. Finally, OGG1 mRNA and protein downregulation were also observed in an accelerated senescence mouse model [100].
(e). Preserving mitochondrial integrity by mtDNA copy number regulation
MtDNA dynamics plays a significant role in preventing biochemical defects of mitochondrial mutations. Targeted degradation of damaged mtDNA molecules or increased mtDNA replication of wild-type molecules can shift the mtDNA mutation heteroplasmy below the phenotypic threshold level. Degradation of damaged mtDNA molecules was first suggested as the mechanism to remove deleterious mutations from the mtDNA population [101,102]. Further data supporting this claim only became available recently with two studies demonstrating that increased oxidative stress by hydrogen peroxide treatment leads to increased mtDNA degradation [103,104]. Surprisingly, alkylation of mtDNA did not have the same effect and abasic sites and single-stranded breaks were more predominant in mtDNA than oxidative lesions [104]. When these types of mtDNA damage are persistent, degradation of mtDNA is upregulated within several hours upon damage initiation [105]. This suggests that the mtDNA damage repair capacity is limited and the type and amount of mutagenic agents determines a cells' capacity to deal with mtDNA damage.
The amount of mtDNA decreases with age in various human tissues, including pancreatic islets [106], skeletal muscle [107], cerebral cortex and heart muscle albeit small [108]. These findings hold also for regions of the rat central nervous system [109]. Interestingly, various tissues in ageing mice do not show a reduction in mtDNA copy number [110], and the accumulation of mtDNA mutations in two epithelial lineages differed between mice and men [111]. These studies suggest that the impact of mitochondrial integrity on ageing may differ between species.
Segments of human muscle fibres demonstrating normal mitochondrial function contained a constant amount of wild-type mtDNA [51]. This finding supports the ‘maintenance of the wild-type’ hypothesis, which stipulates that cells require a minimal amount of wild-type mtDNA to maintain normal cellular function [112]. However, in human myocardium with an age-related loss in myocardial contractile force (MCF) and age-related increase of the mtDNA common deletion, mtDNA copy number remained constant [113]. The authors found a weak correlation between the level of mtDNA deletion and MCF loss. Clonal expansion of the mtDNA deletion is therefore not causative for age-related loss of tissue function owing to a compensatory replication of wild-type mtDNA. The importance of mtDNA copy number to support tissue function was also demonstrated in a large cohort study of blood mtDNA content in women undergoing ovarian hyperstimulation. MtDNA copy numbers were within normal range in non-responders to treatment, but severely decreased in patients showing premature ovarian failure [114]. Three independent studies that quantified mtDNA copy number in dominant optic atrophy (MIM:165500), a dominant inherited optic neuropathy caused by mutations in the OPA1 gene which leads to mitochondrial fragmentation, found contradicting results in mtDNA copy number [115–117]. Finally, leucocytes of patients with classical mitochondrial syndromes MELAS and MERRF also demonstrated age-related decline in mtDNA content, suggesting that beyond the phenotypic threshold level, cells can no longer compensate for deleterious mutations by increased mtDNA replication [118].
TFAM is the primary protein involved in mtDNA copy number regulation [119]. Loss of TFAM is embryonically lethal, characterized by delayed neural and cardiac tissue development [120]. There is growing evidence that mitochondrial proteases play an important role in TFAM-mediated mtDNA maintenance. Proteolytic control of TFAM by the mitochondrial LON protease regulates a stable TFAM : mtDNA ratio [121]. LON-depleted cells contained less oxidative lesions when exposed to H2O2 [122], presumably owing to increased levels of TFAM exerting a protective effect against oxidative stress [123]. In addition, phosphorylation of TFAM by protein kinase A impairs the DNA binding capacity of TFAM, leaving it vulnerable for degradation by the LON protease [124]. The mitochondrial matrix chaperone CLPX enhances DNA binding of TFAM independently of its protease activity in regulation of nucleoid size and mtDNA segregation [125]. This process may be co-facilitated by mtDNA D-loop binding of another AAA+ protein, ATAD3 [126], thereby linking mtDNA QC and mtDNA metabolism [127] to proteostatic mitochondrial QC.
3. Proteostasis in mitochondrial integrity and degenerative processes
Proteostasis of organellar proteins involves their biogenesis, trafficking across the membranes, their sorting, folding, assembly and degradation. Mitochondria contain different molecular pathways that participate in organellar proteostasis. Impairments of these pathways impact the integrity of mitochondria and affect the mitochondrial activity leading to degenerative processes. Error-free protein folding as well as prompt turnover and degradation of misfolded proteins is vital for organellar functions. Proteostasis counteracts toxic protein accumulation impacting the mitochondrial and cellular functions. Chaperones, including heat-shock proteins, are active in refolding of misfolded proteins. Different mitochondrial proteases degrade damaged proteins or proteins that are present in non-balanced quantities. The individual components of the QC system are effective in counteracting impairments caused by molecular damage via repair, refolding and reactivation of polypeptides to the native form. Degradation can also be used to protect mitochondria from damaged proteins. Proteostasis pathways are localized in all mitochondrial subcompartments (membranes, inter-membrane space and the mitochondrial matrix) and are essential for proper cellular functions (table 2).
Table 2.
Human mitochondrial proteostasis proteins and their association with disease. Proteins that have no known disease-causing mutations, but implicated in disease or ageing (e.g. up- or downregulation in mitochondria-deficient cells) are also indicated. ER, endoplasmic reticulum; IM, inner membrane; OM, outer membrane; IMS, intermembrane space.
| human gene | description | function | localization | disease | references |
|---|---|---|---|---|---|
| MSRA | mitochondrial peptide methionine sulfoxide reductase | reduction of methionine sulfoxide to methionine | matrix, cytosol | — | [128–130] |
| MSRB2 | methionine-R-sulfoxide reductase | reduction of methionine sulfoxide to methionine | matrix | — | [131] |
| MSRB3 | methionine-R-sulfoxide reductase | reduction of methionine sulfoxide to methionine | matrix, ER | deafness, autosomal recessive 74 (MIM:613718) | [132] |
| YME1L | ATP-dependent zinc metalloprotease | i-AAA subunit, proteolytic regulation of respiratory chain and OPA1 | IM (protruding IMS) | — | [133] |
| SPG7 | ATP-dependent zinc metalloprotease | m-AAA subunit, maturation and degradation of mitochondrial proteins | IM (protruding matrix) | spastic paraplegia 7 (MIM:607259) | [134] |
| AFG3L2 | ATP-dependent zinc metalloprotease | m-AAA subunit, maturation and degradation of mitochondrial proteins | IM (protruding matrix) | spinocerebellar ataxia 28 (MIM:610246); spastic ataxia autosomal recessive 5 (MIM:614487) | [135,136] |
| PHB, PHB2 | prohibitins | protein turnover, mitochondrial biogenesis and function | IM | — | [137,138]. |
| CLPP | component of a mitochondrial ATP-dependent proteolytic complex | unfolded-protein response stress signalling pathway | matrix | Perrault syndrome (MIM:614129) | [139] |
| CLPX | component of a mitochondrial ATP-dependent proteolytic complex | unfolded-protein response stress signalling pathway | matrix | forms a complex with CLPP | [140] |
| LONP1 | mitochondrial ATP-dependent protease Lon; serine protease 15 | degradation of misfolded, missorted, non-assembled and oxidized proteins; mtDNA regulation | matrix | protein levels affected in disease and disease models | [141] |
| MFN1 | transmembrane GTPase | mitochondrial fusion and distribution, forms heterocomplexes with MFN2 | OM | — | [142] |
| MFN2 | transmembrane GTPase | mitochondrial fusion and distribution, forms heterocomplexes with MFN2 | OM | Charcot–Marie–Tooth disease (MIM: 609260) | [143–145] |
| OPA1 | dynamin-related mitochondrial GTPase | regulation of mitochondrial network, OXPHOS and apoptosis | IM | optic atrophies (MIM:165500 and MIM:125250) | [146,147] |
| HSPA9 | HSP70 | protein import, chaperonin | matrix | — | [148] |
| HSPD1 | HSP60 | chaperonin | matrix | spastic paraplegia (SPG13; MIM:605280); MitCHAP60 disease (MIM:612233) | [149,150] |
(a). Protein repair by reduction of oxidized proteins
Mitochondria generate ROS as a consequence of oxidative metabolism, although the ROS production can increase greatly in pathological conditions [1]. The role of ROS is highlighted by the oxidized proteins that accumulate in aged cells and in age-related disorders [151]. Degradation of oxidized proteins becomes impaired with age [152]. Mitochondria can also reverse and repair certain types of protein oxidative damage with designated enzymatic systems that catalyse the regeneration of protein-bound oxidized cysteine and methionine [153].
Methionine residues can be oxidized to their sulfoxide forms as a result of oxidative damage [152]. This creates two forms of methionine sulfoxide, S and R. The two diastereomers can be reduced by the peptide methionine sulfoxide reductases A (MsrA) and B (Msr B). Both sulfoxide reductases are necessary for complete reduction [154,155]. In mammals, the system is active in mitochondria (MSRA and MSRB2/3) as well as outside the organelle [128,129]. This underscores the important role of these enzymes for the whole cell in both the maintenance of proteins under oxidative stress and the overall redox homeostasis [153]. The deletion of MSRA is known to negatively impact lifespan in mammals and enhance sensitivity to oxidative stress, resulting in accumulation of oxidized proteins and development of neurological disorders [130]. Consistent with the role of methionine sulfoxide reductases, the MSRB2 gene was shown to have beneficial effects on a human leukaemia cell line that missed the MSRA gene. MSRB2 overexpression protects human cells from H2O2-induced oxidative damage, generation of ROS, loss of mitochondrial membrane potential, protein carbonyl accumulation and apoptotic cell death, thus ensuring the mitochondrial integrity and cell survival by scavenging ROS [131]. The autosomal recessive null mutations in the human MSRB3 have been implicated in mitochondrial dysfunction leading to deafness (DFNB74 MIM:613718 [132]).
(b). Protein reconstitution by refolding
Mitochondria effectively prevent their deterioration by engaging molecular mechanisms of protein repair [156]. The majority of mitochondrial proteins are delivered and translocated to the matrix in their unfolded state [157] making them vulnerable to misfolding and aggregation owing to exposed hydrophobic regions. Mitochondrial chaperones mtHSP70, HSP60 and HSP10 facilitate correct protein folding [148,158–160]. The mammalian HSP60 (HSPD1) gene has been shown to be essential for cell survival [161]. Different mutations in the human mitochondrial chaperonin HSP60/HSPD1 are associated with two different disorders [162]: the dominantly inherited form of spastic paraplegia (SPG13; MIM:605280) [149] and an autosomal recessively inherited white matter disorder termed MitCHAP60 disease (MIM:612233) [150]. Interestingly, both disorders exclusively affect the central nervous system, and no other systems or organs. A mouse model with HSPD1 mutation has been developed to recapitulate clinical characteristic of SPG13 patients: swollen mitochondria in the corticospinal tracts, impaired ATP synthesis in the neocortex and spinal cord, and a pronounced defect in complex III assembly and activity [162]. At the molecular level, it appears that disease aetiology stems from haploinsufficiency of the HSPD1 chaperonin.
(c). Protein degradation
Mitochondrial proteases comprise another part of the QC system which copes with protein damage. Mitochondrial proteome turnover appears to be prevalent in mammalian cells and proteolysis occurs continuously in every mitochondrion under physiological and pathological conditions [163] by means of autophagy, through mitochondrial proteases [164] and the ubiquitin–proteasome system [165]. Subunits of the respiratory chain are encoded in both mitochondrial and nuclear genomes, therefore they are vulnerable to an imbalance in gene expression that could lead to dysfunctional respiratory chain complexes. Protein degradation is a mitochondrial mechanism that can potentially not only eliminate damaged proteins, but can also restore the balance between mtDNA- and nuclear-encoded subunits of the respiratory chain. Turnover rates vary greatly for different proteins and even for subunits of the same respiratory complex [166]. We used the data of Price et al. [167] to calculate protein half-lives in mammalian brain. In general, mitochondrial proteins in mammalian brain have exceptionally long half-lives (median 16 days, avg. = 20.5, s.d. = 20.2), two times longer compared to proteins that localize to the nucleus (median 7 days, avg. = 11.8, s.d. = 23.8) and all other proteins surveyed (8 days, avg. = 12.4, s.d. = 21.4). The half-lives and lower mitochondrial turnover rate are evolutionarily conserved [166]. The rate depends most likely on biological properties of the proteins and the role they play in mitochondria [163].
(i). The inner membrane proteases: i-AAA and m-AAA protease
The mitochondrial inner membrane embeds the oxidative phosphorylation chain essential for proper respiratory function. Surveillance of protein quality as well as regulation of biogenesis of the inner membrane subunits, vital for mitochondrial function, is carried out by m-AAA and i-AAA proteases. Misfolded or damaged proteins are degraded to peptides, which are then either exported from the organelle or degraded further to amino acids by various oligopeptidases [168]. Two ATP-dependent protease enzymes are embedded in the inner mitochondrial membrane. The ATPases associated with diverse cellular activities (AAA) domain-containing peptidases form oligomers and expose their catalytic centre to the opposite sides of the inner membrane. i-AAA proteases protrude into the inter-membrane space and m-AAA protease is directed to the matrix. Both proteases regulate biogenesis of mitochondrial proteins [133], exert chaperone-like properties, monitor the folding state of solvent-exposed domains and specifically degrade non-native membrane proteins [169,170]. Despite different localization of the active sites i-AAA and m-AAA proteases can partially complement each others function in fungi. This implies overlapping specificities of both enzymes. Additionally, human i-AAA and m-AAA are able to rescue fungal deletion strains [134,164].
Mitochondrial i-AAA protease is an oligomeric enzyme composed of YME1L subunits. Human YME1L proteolytically regulates respiratory chain biogenesis as evidenced by excessive accumulation of non-assembled respiratory chain subunits (Ndufb6, ND1 and Cox4) upon YME1L depletion [133]. The loss of YME1L leads to reduced cell proliferation, apoptotic resistance, altered mitochondrial ultrastructure, diminished rotenone-sensitive respiration and increased sensitivity to oxidative damage [133].
A recent study with P. anserina revealed an important impact of PaIAP, the homologue of YME1L, on ageing [171]. Deletion of PaIap led to a temperature-dependent phenotype. Unexpectedly, at 27°C growth temperature, lifespan was strongly increased. At 37°C growth temperature, a reduction in lifespan and impairments in fruiting body formation and spore germination were observed. Significantly, in wild-type strains, PaIAP abundance is strongly increased. In addition, two other QC proteins, PaCLPP and PaHSP60, were also found to be abundant in wild-type strains at 37°C. Moreover, while the total abundance of OXPHOS complexes was essentially unchanged in the mutant, a shift towards the formation of stable respiratory supercomplexes and a destabilization of complex V dimers was found in the mutant. Overall, the data identified a function of PaIAP to cope with temperature stress in the fungus that, in contrast to humans, is not able to control temperature in its vegetation body.
In contrast to i-AAA protease, m-AAA proteases expose their active sites towards the matrix space. In humans, m-AAA proteases are present either as a homo-oligomer with AFG3L2 alone or a hetero-oligomer together with its homolog paraplegin (encoded by SPG7 gene). The composition of m-AAA protease is variable in different tissues [134]. m-AAA proteases are responsible for removal of damaged or misfolded proteins and proteolytic activation of essential mitochondrial proteins as well as autocatalytic processing [172]. In mammals, m-AAA protease has been implicated in maturation of MRPL32 essential for ribosomal assembly [173], maturation of cytochrome c peroxidase [134], OPA1 processing [174,175], as well as surveillance and degradation of unfolded and damaged proteins [168].
Heterozygous missense mutations in the gene coding for m-AAA subunit AFG3L2 cause dominant hereditary ataxia SCA28 (MIM:610246), mostly due to altered proteolytic activity manifesting itself as Purkinje neuron degeneration [135]. Another genetic variant, a homozygous missense AFG3L2 mutation, results in hypomorphic allele and impaired ability of AFG3L2 to assemble homo- and hetero-oligomeric m-AAA complexes (with and without paraplegin), resulting in low levels of functionally active protease complexes and a functional paraplegin defect [136]. This molecular defect manifests itself as early onset spastic ataxia-neuropathy (SPG5, MIM:614487) with features characteristic for mitochondrial disorders [136]. Yet another molecular mechanism that underlies neurodegeneration caused by AFG3L2 mutations is the fragmentation of the mitochondrial network [176] and abnormal distribution of mitochondria in neurons [177]. Defective mitochondrial protein synthesis has been proposed as the molecular mechanism of fragmentation in Purkinje neurons [177]. The fragmentation leaves many mitochondria without ER connections limiting the Ca2+ distribution along the mitochondrial network [176].
Mutations in the paraplegin gene cause axonal degeneration in hereditary spastic paraplegia (SPG7, MIM:607259) [178], with phenotypic consequences likely partially alleviated by the presence of AFG3L2 homo-oligomer and the presence of m-AAA protease activity [134]. Both the age of onset and severity of the symptoms are highly variable even among related individuals [178,179] but are generally characterized by progressive weakness and spasticity of the lower limbs owing to degeneration of corticospinal axons with optic, cortical and cerebellar atrophy [178]. Neurodegeneration is preceded by mitochondrial morphological abnormalities in axons [180,181] with impaired axonal transport as a possible cause of axonal degeneration [181]. Next to the axonal phenotype, decreased complex I activity has been observed in fibroblasts of some patients [182]. In such cases, the negative impact of the paraplegin mutation may be twofold: on the one hand, paraplegin dysfunction results in impaired complex I activity resulting in increased ROS production, on the other hand, proper degradation of ROS-damaged mitochondrial proteins is impaired in the paraplegin mutant [164]. Respiratory complex instability is fully realized in a double paraplegin-Afg3l2 mouse mutant that shows, next to neuronal degeneration, also loss of mtDNA and respiratory complexes instability [168].
(ii). Prohibitins
Human mitochondrial prohibitins PHB1 and PHB2 assemble into a ring-like macromolecular structure in the inner mitochondrial membrane and act as protein and lipid scaffolds [183]. Prohibitins are implicated in diverse cellular processes, from mitochondrial biogenesis to a role in cell death and replicative senescence [184]. Prohibitins regulate the turnover of membrane proteins by the m-AAA protease [185], act as chaperone proteins in the mitochondria, and stabilize and protect unassembled membrane proteins until the assembly of respiratory complexes is complete [186,187]. Their expression increases in situations of imbalance between nuclear- and mitochondrial-encoded OXPHOS proteins in mammals [137,186,188]. Prohibitins function in the stabilization of the mtDNA in mitochondrial nucleoids [189,190]. Mammalian cell senescence is accompanied by reduced expression of PHB proteins, correlated with a heterogeneous decline in mitochondrial membrane potential during ageing [137]. Abnormal prohibitin levels have been reported in Parkinson's disease [191]. Prohibitins influence mitochondrial inner membrane fusion and cristae morphogenesis by stabilization of OPA1 [192]. Loss of PHB2 in mouse forebrain leads to extensive neurodegeneration associated with aberrant mitochondria and hyperphosphorylation of the microtubule-associated protein tau [138]. In aged PHB2-deficient neurons, mitochondrial genome destabilization and respiratory deficiencies are observed [138].
(iii). Matrix peptidases CLPP and CLPX
The mitochondrial matrix peptidase CLPP and ATPase/chaperone CLPX form together a proteasome-like hetero-oligomeric cylinder that cleaves unfolded substrates. The specificity of this proteolytic chamber is provided by CLPX that translocates substrates [140]. In mammals, induction of CLPP is observed upon accumulation of unfolded proteins supporting the protein's role in cell stress and protein QC [193]. This observation is consistent with experiments in Caenorhabditis elegans, where the loss of CLPP modulates mitochondrial unfolded protein response (UPRmt) [194,195] and the filamentous fungus P. anserina that shows a surprising healthy lifespan increase upon a deletion of the CLPP gene when grown under standard growth conditions [196]. Remarkably, the lifespan extension phenotype can be reverted by the expression of the human CLPP gene indicating a strong functional conservation of the fungal and the human protease. In humans, recessive CLPP mutations were observed in the Perrault variant of ovarian failure and sensorineural hearing loss [139]. In an independent study, the disease has been faithfully recapitulated in a mouse model that enabled detailed molecular and phenotypic studies [197]. CLPP-deficient mice accumulated mitochondrial chaperones as well as cytosolic proteolytic machinery as a probable compensatory effort to prevent further adverse effect of CLPP-deficiency. Owing to this compensatory mechanism rather subtle bioenergetic deficits were observed, despite many-fold elevated mtDNA levels. The disease mechanism likely involves deficient turnover of mitochondrial components, sustained inflammation with induction of T-lymphocytes in the spleen and resulting severe growth retardation with other age-associated phenotypes [197].
(iv). LON protease
LON protease is an ATP-dependent protease that plays a crucial role in protein QC in the mitochondrial matrix. This protease is the homo-oligomeric enzyme involved in the degradation of misfolded, missorted, non-assembled and oxidized proteins [198]. Each subunit of the protein homo-oligomer contains a substrate binding domain, an AAA motif and the proteolytic domain. Degradation of folded proteins requires ATP for the unfolding of substrates [198]. In contrast to the matrix peptidase CLPP, a number of LON substrates have been identified. In yeast, the LON homologue PIM1 was reported to degrade a subunit of matrix processing peptidase MPPβ, a subunit of the F1F0ATPase, and a number of other proteins, the majority of which are metabolic enzymes or subunits of the respiratory chain. An age-related change of LON abundance and activity was reported as being tissue-specific. While a decrease of LON activity was found in liver of old rats, LON activity remained the same in the hearts [199,200]. In patients with hereditary spastic paraplegia (SPG13), a decline in LON and CLPP levels was reported [201]. Additionally, mitochondrial LON has been implicated in six other diseases with its expression levels significantly up- and downregulated [141]: myoclonic epilepsy and ragged-red (MERRF) fibres syndrome (MIM:545000), myopathy, encephalopathy, lactic acidosis, stroke-like episodes (MELAS) syndrome (MIM:540000) and Friedreich ataxia (MIM:229300).
A clear effect of the experimental modulation of LON levels on organismal ageing was demonstrated in P. anserina. A deletion of the gene, PaLon1, coding for the mitochondrial LON was found to reduce the lifespan of the fungus [202]. Accordingly, constitutive overexpression resulted in an increased health span. PaLon over-expressors were improved in mitochondrial function (i.e. respiration), lower levels of glycoxidized proteins, reduced carbonylation of mitochondrial aconitase and a reduced generation of hydrogen peroxide [203].
4. Mitochondrial dynamics
Mitochondria are dynamic cellular units that constantly change their morphology by fission and fusion. These two processes are genetically controlled and are effective in keeping a ‘healthy’ population of mitochondria. Mitochondrial dynamics is thought to be indispensable once the molecular QC systems comprised by repair enzymes, proteases and chaperones are overwhelmed in their capacity and damage passes certain thresholds [204–206]. In particular, separation of parts of a mitochondrion with a local accumulation of damage can lead to mitochondria with higher mtDNA quality. The mitochondrion can subsequently ‘grow’ by biogenesis and divide again to generate a fully functional population of mitochondria [204]. The damaged mitochondria that remain are subsequently removed by mitophagy. A recent study revealed that sensing of damaged mitochondria requires interaction between autophagy and fission machineries. In yeast, ATG32, which binds to the mitochondrial outer membrane and recruits ATG11, interacts with components of the fission machinery DNM1 and FIS1. Subsequent fission of the mitochondrion and delivery of the part marked by the ATG32/11 to the vacuole leads to the degradation of the organelle [207]. In addition, translocation of DRP1, the homologue of DNM1, to mitochondria [208] and subsequent mitochondrial fragmentation are associated with apoptosis [209,210].
First data about such a role of mitochondrial dynamics were obtained for the two fungal ageing models P. anserina and S. cerevisiae [211]. In this study, it was reported that mitochondria of juvenile cultures are filamentous while those from senescent cultures are punctate. In P. anserina, this age-related change in mitochondrial structure is linked to an increase of transcripts of Dnm1 coding for a GTPase that is essential for mitochondrial fission. Deletion of the Dnm1 gene in yeast and P. anserina leads to an increase in health span. However, it appears that the lifespan increase occurs only under certain conditions (e.g. laboratory conditions) as demonstrated for the i-AAA protease and CLPP in P. anserina. Under variable natural conditions, the fission of mitochondria may be beneficial or indispensable in certain developmental stages. This can be concluded from studies in mice in which the deletion of the Dnm1 homologue (Drp1) was found to be lethal [212]. It appears that Drp1 is dispensable for proliferation and viability of cells, but is essential at some time during development and organogenesis. This conclusion was validated in a study in which deletion of Drp1 was restricted to the neural system. Such strains died soon after birth owing to brain hypoplasia and increased apoptosis in brain. In mammals, mitochondrial fission appears to be an example of antagonistic pleiotropy theory [213]. The theory states that genes coding for components that are beneficial or essential early in life will be selected for—even if they are disadvantageous later in life.
Three large GTPase proteins, MFN1, MFN2 (localized in the outer membrane) and OPA1 (inner membrane), regulate mitochondrial dynamics by mitochondrial fusion [214]. MFN1 and MFN2 are anchored to the mitochondrial outer membrane and mediate mitochondrial fusion by tethering outer membranes of opposing mitochondria [142]. Mutations in MFN2 cause Charcot–Marie–Tooth disease type 2A (CMT2A) [143] likely owing to loss of protein function [144] resulting in a defect of transport of mitochondria and their distribution [145]. OPA1 is another large GTPase that regulates mitochondrial dynamics. OPA1 functions in the fusion of the inner membrane and cristae remodelling. Mutations in OPA1 cause autosomal dominant optic atrophy, a degenerative disease of the optic nerve [146,147].
In general, mitochondrial dynamics is accepted to be an effective way to control mitochondrial quality. Fusion of mitochondria is thought to improve overall quality of the mitochondrial population by content mixing of fully functional mitochondria with defective organelles enabling protein complementation, mtDNA repair and equal distribution of metabolites [215]. Fission of mitochondria is important during growth and development to increase the number of mitochondria. On the other hand, fission is an important mechanism to separate damaged parts of the mitochondrial network from parts that are less affected [216]. Mitochondrial dynamics, however, may only be beneficial to a system under conditions of moderate damage. A recent mathematical model in which mitochondrial damage by ROS, mitochondrial dynamics, biogenesis and mitophagy were integrated suggested a reduction of fission/fusion as beneficial in cells with damaged mitochondria after passing a critical threshold of impairment [204].
5. Autophagy
Autophagy is a cellular recycling mechanism that sequesters cellular components into vesicles, termed autophagosomes, which subsequently deliver their cargo to lysosomes in animal systems and to vacuoles in fungi and plants. The role of autophagy as a QC system is to break down damaged molecules or whole organelles. Its function in ageing is currently intensely investigated [217–222]. Autophagy is induced in several C. elegans longevity mutants in which different molecular pathways are affected, [223] and during ageing of P. anserina [222]. In mice, it was found that the overexpression of Atg5, a gene coding for an essential component of autophagosome formation, leads to an extension of lifespan [224]. In addition, in P. anserina, evidence arises that impairments in specific pathways (e.g. knockout of genes coding for scavenging enzymes) appear to be compensated by increased autophagy [219]. Such responses and cross talks complicate the analysis of defined experiments because they can lead to counterintuitive or controversial results (e.g. no effect of the deletion of a gene coding for a QC pathway). One type of selective autophagy, termed mitophagy, specifically delivers mitochondria to lysosomes or vacuoles. In mammalian cells, several components including NIX [225], PARKIN [226] and PINK1 [227] have been associated with the degradation of mitochondria. PINK1 is a serine/threonine kinase, which, in functional mitochondria, is localized in the inner mitochondrial membrane. In impaired mitochondria, a dissipation of the mitochondrial membrane potential leads to the translocation of PINK to the outer mitochondrial membrane. There, PINK1 together with PARKIN promote segregation of damaged mitochondria from the mitochondrial network [228]. The PINK1 kinase phosphorylates the E3-ubiquitin ligase which subsequently ubiquitinates several proteins including the voltage-dependent anion channel 1 [229] and the two mitofusins MFN1 and MFN2 [230–232]. Ubiquitinated outer membrane proteins subsequently are degraded by the cytosolic proteasome. Impairments in this system are associated with the development of Parkinson's disease. According to the model, mitophagy interacts with mitochondrial dynamics and delivers mitochondria with impaired function that became separated from the mitochondrial network and are unable to fuse with other mitochondria due to reduced membrane potential [216,233]. At this time, information on the impact of autophagy and mitophagy on biological ageing is sparse and remains to be unravelled in more detail.
6. Apoptosis
In multicellular organisms, apoptosis is a type of programmed cell death (PCD) that is essential for proper development and organogenesis. In addition, it is part of the cellular QC network that eliminates severely damaged cells [234]. Interestingly, PCD is also found in unicellular organisms such as yeast, or in multicellular fungi such as P. anserina [235,236]. In the fungal systems, the pathways controlling PCD are less complex than in mammals. While in fungi there is no extrinsic pathway known to control apoptosis, there is an intrinsic pathway, in which mitochondria play a key role. In this pathway, the release of apoptogens cytochrome c and apoptosis inducing factors as well as the induction of a mitochondrial permeability transition pore (mPTP) initiates apoptosis. The intrinsic pathway is active in lower systems such as fungi as well as in mammals. Other components of apoptosis are specific to higher organisms, for example, the mitochondrial outer membrane pore has not been identified in fungi so far [235,237–239].
One aspect that is not sufficiently investigated in mammals is the role of PCD on organismal ageing. In fungi, the induction of PCD appears to be the final executor of organismal ageing. Recent studies with P. anserina and mice revealed a role of the mPTP in ageing [237,240,241]. In the fungal system, mitochondria from senescent individuals contain three times more cyclophilin D (CypD), a mitochondrial peptidyl–prolyl–cis,trans-isomerase that is a regulator of a protein complex in the inner mitochondrial membrane pore and a part of the mPTP [240]. Binding to still undetermined proteins, probably ATPase complexes, leads to membrane opening and the induction of PCD leading to death of the cell [242]. The time of induction can be accelerated by the overexpression of CypD and decelerated again by the application of cyclosporine A. During ageing of P. anserina, the ultrastructure of the inner mitochondrial membrane becomes strongly remodelled [237]. In juvenile cultures, tubular cristae are found, while during ageing the inner membrane retracts and a reticulate network of membranes is formed. Major structure-building components are ATPase dimers at the site of the strongest cristae curvature. According to a model, these ATPase dimers are speculated to bind CypD during ageing, thereby giving rise to the severe membrane changes and finally to the disruption of mitochondria and the release of apoptogens [242]. In how far such processes are also occurring during ageing in other organisms, in particular in mammals, is currently unsolved. At least, the structure-forming function of ATPase dimers appears to be conserved from yeast to mammals [243]. Moreover, in mice an increase of CypD was reported in gastrocnemius muscle in aged individuals. By contrast, in the mitochondrial ‘mutator mouse’ with accelerated ageing a significant depletion of CypD in quadriceps and gastrocnemius muscle has been reported [244]. CypD is deacetylated in the vicinity of a functional site by NAD+-dependent deacetylase SIRT3. SIRT3-deficient mice show signs of accelerated ageing and mitochondrial swelling due to increased mPTP opening [245]. These mice are characterized by cardiac hypothropy, fibrosis and were hypersensitive to heat stress. Overall, it appears that apoptosis linked to mitochondrial pathways is relevant for skeletal and heart muscle function and age- and cardiac failure-related deaths in humans.
7. Conclusions and perspectives
The importance of mitochondrial QC is demonstrated by the expression of a variety of human diseases, often manifested later in life, and effects of impairments of QC systems on ageing processes. Degenerative disease and ageing have been associated with the increased generation of ROS and perturbations in cellular redox status [141]. Oxidative phosphorylation, a major source of ROS in the cell, exposes mitochondria to the risk of oxidative damage that may result in organellar imbalance at the level of mtDNA, residues, proteins and the whole organelle. But mitochondria, although being the main source of ROS, also contain important lines of defence against the oxidative damage and are equipped with protective pathways. Understanding the effects of ROS accumulation, the regulation and crosstalk of different mitochondrial QC pathways holds the key to understanding the mechanisms leading to degenerative diseases and ageing.
Apart from the molecular machineries described above, there are other systems that participate in mitochondrial QC, but which have not yet been directly linked to disease and ageing processes. One mechanism that recently was reported in yeast to play a role in mitochondrial QC is mitochondria-associated degradation (MAD). This pathway is active on proteins located and translocated to the outer mitochondrial membrane. The underlying pathway is similar to ER-associated degradation involved in the degradation of proteins by the ubiquitin proteasome system [246]. The system is dependent on the ubiquitination of the proteins to be degraded by E3 ligates at the outer mitochondrial membrane. A number of different proteins with this activity have been identified. Ubiquitinated proteins are subsequently extracted out of the membrane via cdc48/p97 protein complexes and present the ubiquitinated protein for degradation to the proteasome. A number of mitochondrial proteins including DRP1/DNM1, MFN1 and MFN2 as part of the mitochondrial fission/fusion machinery have been demonstrated to be ubiquitinated [246,247]. However, the relevance of MAD in respect to ageing is not established so far.
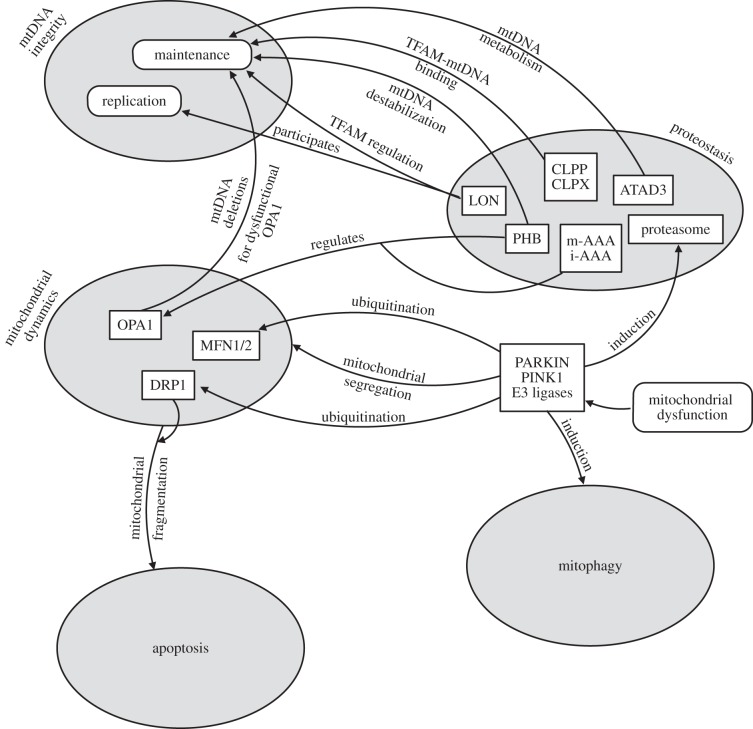
Overall, different pathways that are active at the level of single molecules (DNA, proteins), of organelles (mitophagy), as well as whole cells (programmed cell death), raise questions about redundancy of their components, their hierarchy and regulation (figure 2). Currently, it is believed that pathways of ‘higher-order’ (e.g. mitophagy) become activated when ‘lower-order’ pathways (e.g. reduction of ROS-damaged residues) are overwhelmed in their capacity. This concept [205,206,248], although intuitive, is not rigorously proven. However, it does offer explanation for counterintuitive results obtained in specific experiments. For instance, the deletion of the gene coding for mitochondrial superoxide dismutase does not lead to a reduced lifespan in P. anserina. The induction of mitophagy to get rid of damaged mitochondria as they accumulate faster would be a possible way to ‘heal’ the impaired ROS scavenging capacity in this mutant.
Figure 2.
Interactions between quality control levels in mitochondria. Five QC levels are represented by grey ovals corresponding to mtDNA integrity, proteostasis, dynamics, mitophagy and apoptosis. Proteins (rectangles) and processes (rounded corners) that interact between levels of QC hierarchy are shown. Arrows indicate their influence on other components of QC as described in the text.
It appears that the ‘lower-’ and ‘higher-order’ QC pathways do not work as separate entities, but rather resemble a network with hierarchies intrinsically connected and highly intervening with each other (figure 2). A striking example of interactions between different levels of QC is the mitochondrial matrix LON protease. On the one hand, the protease functions in proteolytic and protein-regulatory functions, on the other hand, it plays an important role in mtDNA metabolism, regulating the mtDNA copy number [121]. In mammals, LON protease additionally interacts with mtDNA regions in nucleoids and is involved in mtDNA transcription and replication [122,141].
Other types of cross-level QC interactions are i-AAA and m-AAA peptidases and prohibitins that primarily degrade and regulate OXPHOS proteins. Additionally, they process and regulate OPA1 thus influencing mitochondrial fission and mitochondrial dynamics [144]. Conversely, OPA1 ‘plus’ patients harbour mtDNA deletions in their muscle cells, suggesting a role for mitochondrial fusion in maintaining mtDNA integrity [249,250]. These proteins clearly cross the hierarchy boundaries of mitochondrial QC pathways.
Acknowledgements
The authors would like to thank Florence van Tienen for comments on the manuscript.
Funding statement
This work was supported by Horizon grant (050-71-053) from the Netherlands Organization for Scientific Research (NWO; R.S.) and the CSBR (Centres for Systems Biology Research) initiative from NWO (no: CSBR09/013V; R.S., M.N.) and by the German Research Foundation (DFG; Os75/13-1, Os75/15-1, H.D.O).
References
- 1.Adam-Vizi V, Chinopoulos C. 2006. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol. Sci. 27, 639–645. ( 10.1016/j.tips.2006.10.005) [DOI] [PubMed] [Google Scholar]
- 2.Koopman WJ, Nijtmans LG, Dieteren CE, Roestenberg P, Valsecchi F, Smeitink JA, Willems PHGM. 2010. Mammalian mitochondrial complex I: biogenesis, regulation, and reactive oxygen species generation. Antioxid. Redox Signal. 12, 1431–1470. ( 10.1089/ars.2009.2743) [DOI] [PubMed] [Google Scholar]
- 3.Harman D. 1972. The biologic clock: the mitochondria? J. Am. Geriatr. Soc. 20, 145–147. [DOI] [PubMed] [Google Scholar]
- 4.Lapointe J, Stepanyan Z, Bigras E, Hekimi S. 2009. Reversal of the mitochondrial phenotype and slow development of oxidative biomarkers of aging in long-lived Mclk1+/− mice. J. Biol. Chem. 284, 20 364–20 374. ( 10.1074/jbc.M109.006569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blagosklonny MV. 2008. Aging: ROS or TOR. Cell Cycle 7, 3344–3354. ( 10.4161/cc.7.21.6965) [DOI] [PubMed] [Google Scholar]
- 6.Harman D. 1992. Role of free radicals in aging and disease. Ann. N.Y. Acad. Sci. 673, 126–141. ( 10.1111/j.1749-6632.1992.tb27444.x) [DOI] [PubMed] [Google Scholar]
- 7.Harman D. 2006. Alzheimer's disease pathogenesis: role of aging. Ann. N.Y. Acad. Sci. 1067, 454–460. ( 10.1196/annals.1354.065) [DOI] [PubMed] [Google Scholar]
- 8.Meisinger C, Sickmann A, Pfanner N. 2008. The mitochondrial proteome: from inventory to function. Cell 134, 22–24. ( 10.1016/j.cell.2008.06.043) [DOI] [PubMed] [Google Scholar]
- 9.Pagliarini DJ, et al. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123. ( 10.1016/j.cell.2008.06.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koopman WJ, Distelmaier F, Smeitink JA, Willems PH. 2013. OXPHOS mutations and neurodegeneration. EMBO J. 32, 9–29. ( 10.1038/emboj.2012.300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koopman WJ, Willems PH, Smeitink JA. 2012. Monogenic mitochondrial disorders. N. Engl. J. Med. 366, 1132–1141. ( 10.1056/NEJMra1012478) [DOI] [PubMed] [Google Scholar]
- 12.Esser K, Kück U, Stahl U, Tudzynski P. 1981. Mitochondrial DNA and senescence in Podospora anserina. Curr. Genet. 4, 83 ( 10.1007/BF00376791) [DOI] [PubMed] [Google Scholar]
- 13.Bertrand H, Griffiths AJ, Court DA, Cheng CK. 1986. An extrachromosomal plasmid is the etiological precursor of kalDNA insertion sequences in the mitochondrial chromosome of senescent Neurospora. Cell 47, 829–837. ( 10.1016/0092-8674(86)90525-8) [DOI] [PubMed] [Google Scholar]
- 14.Linnane AW, Baumer A, Maxwell RJ, Preston H, Zhang CF, Marzuki S. 1990. Mitochondrial gene mutation: the ageing process and degenerative diseases. Biochem. Int. 22, 1067–1076. [PubMed] [Google Scholar]
- 15.Cortopassi GA, Shibata D, Soong NW, Arnheim N. 1992. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc. Natl Acad. Sci. USA 89, 7370–7374. ( 10.1073/pnas.89.16.7370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piko L, Hougham AJ, Bulpitt KJ. 1988. Studies of sequence heterogeneity of mitochondrial DNA from rat and mouse tissues: evidence for an increased frequency of deletions/additions with aging. Mech. Ageing Dev. 43, 279–293. ( 10.1016/0047-6374(88)90037-1) [DOI] [PubMed] [Google Scholar]
- 17.MITOMAP. 2013. A human mitochondrial genome database. See http://www.mitomap.org.
- 18.Greaves LC, Elson JL, Nooteboom M, Grady JP, Taylor GA, Taylor RW, Mathers JC, Kirkwood TBL, Turnbull DM. 2012. Comparison of mitochondrial mutation spectra in ageing human colonic epithelium and disease: absence of evidence for purifying selection in somatic mitochondrial DNA point mutations. PLoS Genet. 8, e1003082 ( 10.1371/journal.pgen.1003082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell GR, Reeve A, Ziabreva I, Polvikoski TM, Taylor RW, Reynolds R, Turnbull DM, Mahad DJ. 2013. Mitochondrial DNA deletions and depletion within paraspinal muscles. Neuropathol. Appl. Neurobiol. 39, 377–389. ( 10.1111/j.1365-2990.2012.01290.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damas J, Samuels DC, Carneiro J, Amorim A, Pereira F. 2014. Mitochondrial DNA rearrangements in health and disease: a comprehensive study. Hum. Mutat. 35, 1–14. ( 10.1002/humu.22452) [DOI] [PubMed] [Google Scholar]
- 21.Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM. 2006. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am. J. Hum. Genet. 79, 469–480. ( 10.1086/507132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Costa CK, Kiyomoto BH, Schmidt B, Oliveira AS, Gabbai AA, Tengan CH. 2007. Age-related mitochondrial DNA point mutations in patients with mitochondrial myopathy. J. Neurol. Sci. 263, 139–144. ( 10.1016/j.jns.2007.07.006) [DOI] [PubMed] [Google Scholar]
- 23.Osiewacz HD, Hamann A, Zintel S. 2013. Assessing organismal aging in the filamentous fungus Podospora anserina. Methods Mol. Biol. 965, 439–462. ( 10.1007/978-1-62703-239-1_29) [DOI] [PubMed] [Google Scholar]
- 24.Rizet G. 1953. [Impossibility of obtaining uninterrupted and unlimited multiplication of the ascomycete Podospora anserina]. C.R. Hebd. Seances Acad. Sci. 237, 838–840. [In French.] [PubMed] [Google Scholar]
- 25.Belcour L. 1981. Mitochondrial DNA and senescence in Podospora anserina. Curr. Genet. 4, 81–82. ( 10.1007/BF00376790) [DOI] [PubMed] [Google Scholar]
- 26.Kück U, Stahl U, Esser K. 1981. Plasmid-like DNA is part of mitochondrial DNA in Podospora anserina. Curr. Genet. 3, 151–156. ( 10.1007/BF00365719) [DOI] [PubMed] [Google Scholar]
- 27.Osiewacz HD, Esser K. 1984. The mitochondrial plasmid of Podospora anserina: a mobile intron of a mitochondrial gene. Curr. Genet. 8, 299–305. ( 10.1007/BF00419728) [DOI] [PubMed] [Google Scholar]
- 28.Stahl U, Lemke PA, Tudzynski P, Kück U, Esser K. 1978. Evidence for plasmid like DNA in a filamentous fungus, the ascomycete Podospora anserina. Mol. Gen. Genet. 162, 341–343. ( 10.1007/BF00268860) [DOI] [PubMed] [Google Scholar]
- 29.Cummings DJ, Belcour L, Grandchamp C. 1979. Mitochondrial DNA from Podospora anserina. II. Properties of mutant DNA and multimeric circular DNA from senescent cultures. Mol. Gen. Genet. 171, 239–250. ( 10.1007/BF00267578) [DOI] [PubMed] [Google Scholar]
- 30.Sellem CH, Lecellier G, Belcour L. 1993. Transposition of a group II intron. Nature 366, 176–178. ( 10.1038/366176a0) [DOI] [PubMed] [Google Scholar]
- 31.Kück U, Esser K. 1982. Genetic map of mitochondrial DNA in Podospora anserina. Curr. Genet. 5, 143–147. ( 10.1007/BF00365705) [DOI] [PubMed] [Google Scholar]
- 32.Osiewacz HD, Hermanns J, Marcou D, Triffi M, Esser K. 1989. Mitochondrial DNA rearrangements are correlated with a delayed amplification of the mobile intron (plDNA) in a long-lived mutant of Podospora anserina. Mutat. Res. 219, 9–15. ( 10.1016/0921-8734(89)90036-2) [DOI] [PubMed] [Google Scholar]
- 33.Hermanns J, Osiewacz HD. 1992. The linear mitochondrial plasmid pAL2–1 of a long-lived Podospora anserina mutant is an invertron encoding a DNA and RNA polymerase. Curr. Genet. 22, 491–500. ( 10.1007/BF00326415) [DOI] [PubMed] [Google Scholar]
- 34.Hermanns J, Asseburg A, Osiewacz HD. 1995. Evidence for giant linear plasmids in the ascomycete Podospora anserina. Curr. Genet. 27, 379–386. ( 10.1007/BF00352108) [DOI] [PubMed] [Google Scholar]
- 35.Hermanns J, Debets F, Hoekstra R, Osiewacz HD. 1995. A novel family of linear plasmids with homology to plasmid pAL2–1 of Podospora anserina. Mol. Gen. Genet. 246, 638–647. ( 10.1007/BF00298971) [DOI] [PubMed] [Google Scholar]
- 36.Hermanns J, Osiewacz HD. 1996. Induction of longevity by cytoplasmic transfer of a linear plasmid in Podospora anserina. Curr. Genet. 29, 250–256. ( 10.1007/BF02221555) [DOI] [PubMed] [Google Scholar]
- 37.Hermanns J, Asseburg A, Osiewacz HD. 1994. Evidence for a life span-prolonging effect of a linear plasmid in a longevity mutant of Podospora anserina. Mol. Gen. Genet. 243, 297–307. ( 10.1007/BF00301065) [DOI] [PubMed] [Google Scholar]
- 38.Griffiths AJ. 1992. Fungal senescence. Annu. Rev. Genet. 26, 351–372. ( 10.1146/annurev.ge.26.120192.002031) [DOI] [PubMed] [Google Scholar]
- 39.Osiewacz HD. 1990. Molecular analysis of aging processes in fungi. Mutat. Res. 237, 1–8. ( 10.1016/0921-8734(90)90026-N) [DOI] [PubMed] [Google Scholar]
- 40.Griffiths AJ, Bertrand H. 1984. Unstable cytoplasms in Hawaiian strains of Neurospora intermedia. Curr. Genet. 8, 387–398. ( 10.1007/BF00419828) [DOI] [PubMed] [Google Scholar]
- 41.Myers CJ, Griffiths AJ, Bertrand H. 1989. Linear kalilo DNA is a Neurospora mitochondrial plasmid that integrates into the mitochondrial DNA. Mol. Gen. Genet. 220, 113–120. ( 10.1007/BF00260864) [DOI] [PubMed] [Google Scholar]
- 42.Piko L, Bulpitt KJ, Meyer R. 1984. Structural and replicative forms of mitochondrial DNA in tissues from adult and senescent BALB/c mice and Fischer 344 rats. Mech. Ageing Dev. 26, 113–131. ( 10.1016/0047-6374(84)90170-2) [DOI] [PubMed] [Google Scholar]
- 43.Linnane AW, Marzuki S, Ozawa T, Tanaka M. 1989. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet 1, 642–645. ( 10.1016/S0140-6736(89)92145-4) [DOI] [PubMed] [Google Scholar]
- 44.Melov S, Hertz GZ, Stormo GD, Johnson TE. 1994. Detection of deletions in the mitochondrial genome of Caenorhabditis elegans. Nucleic Acids Res. 22, 1075–1078. ( 10.1093/nar/22.6.1075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kadenbach B, Müller-Höcker J. 1990. Mutations of mitochondrial DNA and human death. Naturwissenschaften 77, 221–225. ( 10.1007/BF01138485) [DOI] [PubMed] [Google Scholar]
- 46.Boursot P, Yonekawa H, Bonhomme F. 1987. Heteroplasmy in mice with deletion of a large coding region of mitochondrial DNA. Mol. Biol. Evol. 4, 46–55. [DOI] [PubMed] [Google Scholar]
- 47.Holt IJ, Harding AE, Morgan-Hughes JA. 1988. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331, 717–719. ( 10.1038/331717a0) [DOI] [PubMed] [Google Scholar]
- 48.Wallace DC. 1989. Mitochondrial DNA mutations and neuromuscular disease. Trends Genet. 5, 9–13. ( 10.1016/0168-9525(89)90005-X) [DOI] [PubMed] [Google Scholar]
- 49.Sciacco M, Bonilla E, Schon EA, DiMauro S, Moraes CT. 1994. Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Hum. Mol. Genet. 3, 13–19. ( 10.1093/hmg/3.1.13) [DOI] [PubMed] [Google Scholar]
- 50.Mancuso M, et al. 2013. Phenotypic heterogeneity of the 8344A>G mtDNA ‘MERRF’ mutation. Neurology 80, 2049–2054. ( 10.1212/WNL.0b013e318294b44c) [DOI] [PubMed] [Google Scholar]
- 51.Durham SE, Samuels DC, Cree LM, Chinnery PF. 2007. Normal levels of wild-type mitochondrial DNA maintain cytochrome c oxidase activity for two pathogenic mitochondrial DNA mutations but not for m.3243A→G. Am. J. Hum. Genet. 81, 189–195. ( 10.1086/518901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Al-Dosary M, Whittaker RG, Haughton J, McFarland R, Goodship J, Turnbull DM, Taylor RW. 2009. Neuromuscular disease presentation with three genetic defects involving two genomes. Neuromuscul. Disord. 19, 841–844. ( 10.1016/j.nmd.2009.10.001) [DOI] [PubMed] [Google Scholar]
- 53.Yu-Wai-Man P, Lai-Cheong J, Borthwick GM, He L, Taylor GA, Greaves LC, Taylor RW, Griffiths PG, Turnbull DM. 2010. Somatic mitochondrial DNA deletions accumulate to high levels in aging human extraocular muscles. Invest. Ophthalmol. Vis. Sci. 51, 3347–3353. ( 10.1167/iovs.09-4660) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blakely EL, Trip SA, Swalwell H, He L, Wren DR, Rich P, Turnbull DM, Omer SE, Taylor RW. 2009. A new mitochondrial transfer RNAPro gene mutation associated with myoclonic epilepsy with ragged-red fibers and other neurological features. Arch. Neurol. 66, 399–402. ( 10.1001/archneurol.2008.576) [DOI] [PubMed] [Google Scholar]
- 55.Blackwood JK, Whittaker RG, Blakely EL, Alston CL, Turnbull DM, Taylor RW. 2010. The investigation and diagnosis of pathogenic mitochondrial DNA mutations in human urothelial cells. Biochem. Biophys. Res. Commun. 393, 740–745. ( 10.1016/j.bbrc.2010.02.072) [DOI] [PubMed] [Google Scholar]
- 56.Bai Y, Shakeley RM, Attardi G. 2000. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol. Cell Biol. 20, 805–815. ( 10.1128/MCB.20.3.805-815.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayashi J, Ohta S, Kikuchi A, Takemitsu M, Goto Y, Nonaka I. 1991. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc. Natl Acad. Sci. USA 88, 10 614–10 618. ( 10.1073/pnas.88.23.10614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pye D, et al. 2006. Production of transmitochondrial cybrids containing naturally occurring pathogenic mtDNA variants. Nucleic Acids Res. 34, e95 ( 10.1093/nar/gkl516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rossignol R, Malgat M, Mazat JP, Letellier T. 1999. Threshold effect and tissue specificity. Implication for mitochondrial cytopathies. J. Biol. Chem. 274, 33 426–33 432. ( 10.1074/jbc.274.47.33426) [DOI] [PubMed] [Google Scholar]
- 60.Taylor RW, Turnbull DM. 2005. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6, 389–402. ( 10.1038/nrg1606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McFarland R, Turnbull DM. 2009. Batteries not included: diagnosis and management of mitochondrial disease. J. Intern. Med. 265, 210–228. ( 10.1111/j.1365-2796.2008.02066.x) [DOI] [PubMed] [Google Scholar]
- 62.Krishnan KJ, Greaves LC, Reeve AK, Turnbull DM. 2007. Mitochondrial DNA mutations and aging. Ann. N.Y. Acad. Sci. 1100, 227–240. ( 10.1196/annals.1395.024) [DOI] [PubMed] [Google Scholar]
- 63.Greaves LC, Barron MJ, Plusa S, Kirkwood TB, Mathers JC, Taylor RW, Turnbull DM. 2010. Defects in multiple complexes of the respiratory chain are present in ageing human colonic crypts. Exp. Gerontol. 45, 573–579. ( 10.1016/j.exger.2010.01.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taylor RW, et al. 2003. Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 112, 1351–1360. ( 10.1172/JCI19435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fayet G, Jansson M, Sternberg D, Moslemi AR, Blondy P, Lombes A, Fardeau M, Oldfors A. 2002. Ageing muscle: clonal expansions of mitochondrial DNA point mutations and deletions cause focal impairment of mitochondrial function. Neuromuscul. Disord. 12, 484–493. ( 10.1016/S0960-8966(01)00332-7) [DOI] [PubMed] [Google Scholar]
- 66.Nekhaeva E, Bodyak ND, Kraytsberg Y, McGrath SB, Van Orsouw NJ, Pluzhnikov A, Wei JY, Vijg J, Khrapko K. 2002. Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues. Proc. Natl Acad. Sci. USA 99, 5521–5526. ( 10.1073/pnas.072670199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoneda M, Chomyn A, Martinuzzi A, Hurko O, Attardi G. 1992. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyopathy. Proc. Natl Acad. Sci. USA 89, 11 164–11 168. ( 10.1073/pnas.89.23.11164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Grey AD. 1997. A proposed refinement of the mitochondrial free radical theory of aging. Bioessays 19, 161–166. ( 10.1002/bies.950190211) [DOI] [PubMed] [Google Scholar]
- 69.Chan DC. 2006. Mitochondria: dynamic organelles in disease, aging, and development. Cell 125, 1241–1252. ( 10.1016/j.cell.2006.06.010) [DOI] [PubMed] [Google Scholar]
- 70.Kowald A, Dawson M, Kirkwood TB. 2014. Mitochondrial mutations and ageing: can mitochondrial deletion mutants accumulate via a size based replication advantage? J. Theor. Biol. 340C, 111–118. ( 10.1016/j.jtbi.2013.09.009) [DOI] [PubMed] [Google Scholar]
- 71.Elson JL, Samuels DC, Turnbull DM, Chinnery PF. 2001. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am. J. Hum. Genet. 68, 802–806. ( 10.1086/318801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coller HA, Bodyak ND, Khrapko K. 2002. Frequent intracellular clonal expansions of somatic mtDNA mutations: significance and mechanisms. Ann. N.Y. Acad. Sci. 959, 434–447. ( 10.1111/j.1749-6632.2002.tb02113.x) [DOI] [PubMed] [Google Scholar]
- 73.Cree LM, Samuels DC, de Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HM, Chinnery PF. 2008. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat. Genet. 40, 249–254. ( 10.1038/ng.2007.63) [DOI] [PubMed] [Google Scholar]
- 74.Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, Larsson N-G. 2008. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 6, e10 ( 10.1371/journal.pbio.0060010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sudo H, et al. 2008. Fetal-juvenile origins of point mutations in the adult human tracheal-bronchial epithelium: absence of detectable effects of age, gender or smoking status. Mutat. Res. 646, 25–40. ( 10.1016/j.mrfmmm.2008.08.016) [DOI] [PubMed] [Google Scholar]
- 76.Kowald A, Kirkwood TB. 2013. Mitochondrial mutations and aging: random drift is insufficient to explain the accumulation of mitochondrial deletion mutants in short-lived animals. Aging Cell 12, 728–731. ( 10.1111/acel.12098) [DOI] [PubMed] [Google Scholar]
- 77.Alexeyev M, Shokolenko I, Wilson G, LeDoux S. 2013. The maintenance of mitochondrial DNA integrity: critical analysis and update. Cold Spring Harb. Perspect. Biol. 5, a012641 ( 10.1101/cshperspect.a012641) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilson DM, III, Bohr VA. 2007. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst.) 6, 544–559. ( 10.1016/j.dnarep.2006.10.017) [DOI] [PubMed] [Google Scholar]
- 79.de Souza-Pinto NC, et al. 2001. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res. 61, 5378–5381. [PubMed] [Google Scholar]
- 80.Evans MD, Dizdaroglu M, Cooke MS. 2004. Oxidative DNA damage and disease: induction, repair and significance. Mutat. Res. 567, 1–61. ( 10.1016/j.mrrev.2003.11.001) [DOI] [PubMed] [Google Scholar]
- 81.Zheng W, Khrapko K, Coller HA, Thilly WG, Copeland WC. 2006. Origins of human mitochondrial point mutations as DNA polymerase gamma-mediated errors. Mutat. Res. 599, 11–20. ( 10.1016/j.mrfmmm.2005.12.012) [DOI] [PubMed] [Google Scholar]
- 82.Liu P, Demple B. 2010. DNA repair in mammalian mitochondria: much more than we thought? Environ. Mol. Mutagen. 51, 417–426. [DOI] [PubMed] [Google Scholar]
- 83.Boesch P, Weber-Lotfi F, Ibrahim N, Tarasenko V, Cosset A, Paulus F, Lightowlers RN, Dietrich A. 2011. DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim. Biophys. Acta 1813, 186–200. ( 10.1016/j.bbamcr.2010.10.002) [DOI] [PubMed] [Google Scholar]
- 84.Chattopadhyay R, et al. 2006. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 34, 2067–2076. ( 10.1093/nar/gkl177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lakshmipathy U, Campbell C. 1999. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell Biol. 19, 3869–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takao M, Aburatani H, Kobayashi K, Yasui A. 1998. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 26, 2917–2922. ( 10.1093/nar/26.12.2917) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fung H, Demple B. 2005. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell 17, 463–470. ( 10.1016/j.molcel.2004.12.029) [DOI] [PubMed] [Google Scholar]
- 88.Hance N, Ekstrand MI, Trifunovic A. 2005. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum. Mol. Genet. 14, 1775–1783. ( 10.1093/hmg/ddi184) [DOI] [PubMed] [Google Scholar]
- 89.Shokolenko IN, Fayzulin RZ, Katyal S, McKinnon PJ, Wilson GL, Alexeyev MF. 2013. Mitochondrial DNA ligase is dispensable for the viability of cultured cells but essential for mtDNA maintenance. J. Biol. Chem. 288, 26 594–26 605. ( 10.1074/jbc.M113.472977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stumpf JD, Copeland WC. 2011. Mitochondrial DNA replication and disease: insights from DNA polymerase gamma mutations. Cell Mol. Life Sci. 68, 219–233. ( 10.1007/s00018-010-0530-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tang S, Wang J, Lee NC, Milone M, Halberg MC, Schmitt ES, Craigen WJ, Zhang W, Wong L-JC. 2011. Mitochondrial DNA polymerase gamma mutations: an ever expanding molecular and clinical spectrum. J. Med. Genet. 48, 669–681. ( 10.1136/jmedgenet-2011-100222) [DOI] [PubMed] [Google Scholar]
- 92.Kujoth GC, et al. 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309, 481–484. ( 10.1126/science.1112125) [DOI] [PubMed] [Google Scholar]
- 93.Trifunovic A, et al. 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423. ( 10.1038/nature02517) [DOI] [PubMed] [Google Scholar]
- 94.Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. 2007. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat. Genet. 39, 540–543. ( 10.1038/ng1988) [DOI] [PubMed] [Google Scholar]
- 95.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. 2003. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell 14, 1583–1596. ( 10.1091/mbc.E02-07-0399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spelbrink JN. 2009. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life 62, 19–32. ( 10.1002/iub.282) [DOI] [PubMed] [Google Scholar]
- 97.Hensen F, Cansiz S, Gerhold JM, Spelbrink JN. 2013. To be or not to be a nucleoid protein: a comparison of mass-spectrometry based approaches in the identification of potential mtDNA-nucleoid associated proteins. Biochimie 100, 219–226. ( 10.1016/j.biochi.2013.09.017) [DOI] [PubMed] [Google Scholar]
- 98.Zhong Y, et al. 2011. Mitochondrial transcription factor A overexpression and base excision repair deficiency in the inner ear of rats with d-galactose-induced aging. FEBS J. 278, 2500–2510. ( 10.1111/j.1742-4658.2011.08176.x) [DOI] [PubMed] [Google Scholar]
- 99.Zhang Y, Ouyang S, Zhang L, Tang X, Song Z, Liu P. 2010. Oxygen-induced changes in mitochondrial DNA and DNA repair enzymes in aging rat lens. Mech. Ageing Dev. 131, 666–673. ( 10.1016/j.mad.2010.09.003) [DOI] [PubMed] [Google Scholar]
- 100.Tian F, Tong TJ, Zhang ZY, McNutt MA, Liu XW. 2009. Age-dependent down-regulation of mitochondrial 8-oxoguanine DNA glycosylase in SAM-P/8 mouse brain and its effect on brain aging. Rejuvenation Res. 12, 209–215. ( 10.1089/rej.2009.0849) [DOI] [PubMed] [Google Scholar]
- 101.Clayton DA, Doda JN, Friedberg EC. 1974. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl Acad. Sci. USA 71, 2777–2781. ( 10.1073/pnas.71.7.2777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mita S, Monnat RJ, Jr, Loeb LA. 1988. Resistance of HeLa cell mitochondrial DNA to mutagenesis by chemical carcinogens. Cancer Res. 48, 4578–4583. [PubMed] [Google Scholar]
- 103.Furda AM, Marrangoni AM, Lokshin A, Van Houten B. 2012. Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction. DNA Repair (Amst.) 11, 684–692. ( 10.1016/j.dnarep.2012.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. 2009. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 37, 2539–2548. ( 10.1093/nar/gkp100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shokolenko IN, Wilson GL, Alexeyev MF. 2013. Persistent damage induces mitochondrial DNA degradation. DNA Repair (Amst.) 12, 488–499. ( 10.1016/j.dnarep.2013.04.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cree LM, Patel SK, Pyle A, Lynn S, Turnbull DM, Chinnery PF, Walker M. 2008. Age-related decline in mitochondrial DNA copy number in isolated human pancreatic islets. Diabetologia 51, 1440–1443. ( 10.1007/s00125-008-1054-4) [DOI] [PubMed] [Google Scholar]
- 107.Short KR, Bigelow ML, Kahl J, Singh R, Coenen-Schimke J, Raghavakaimal S, Nair KS. 2005. Decline in skeletal muscle mitochondrial function with aging in humans. Proc. Natl Acad. Sci. USA 102, 5618–5623. ( 10.1073/pnas.0501559102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Frahm T, Mohamed SA, Bruse P, Gemund C, Oehmichen M, Meissner C. 2005. Lack of age-related increase of mitochondrial DNA amount in brain, skeletal muscle and human heart. Mech. Ageing Dev. 126, 1192–1200. ( 10.1016/j.mad.2005.06.008) [DOI] [PubMed] [Google Scholar]
- 109.McInerny SC, Brown AL, Smith DW. 2009. Region-specific changes in mitochondrial D-loop in aged rat CNS. Mech. Ageing Dev. 130, 343–349. ( 10.1016/j.mad.2009.01.008) [DOI] [PubMed] [Google Scholar]
- 110.Masuyama M, Iida R, Takatsuka H, Yasuda T, Matsuki T. 2005. Quantitative change in mitochondrial DNA content in various mouse tissues during aging. Biochim. Biophys. Acta 1723, 302–308. ( 10.1016/j.bbagen.2005.03.001) [DOI] [PubMed] [Google Scholar]
- 111.Greaves LC, Barron MJ, Campbell-Shiel G, Kirkwood TB, Turnbull DM. 2011. Differences in the accumulation of mitochondrial defects with age in mice and humans. Mech. Ageing Dev. 132, 588–591. ( 10.1016/j.mad.2011.10.004) [DOI] [PubMed] [Google Scholar]
- 112.Chinnery PF, Samuels DC. 1999. Relaxed replication of mtDNA: a model with implications for the expression of disease. Am. J. Hum. Genet. 64, 1158–1165. ( 10.1086/302311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miller F, Nagley P, Mariani JA, Ou R, Liu VW, Zhang C, Linnane AW, Pepe S, Rosenfeldt F. 2009. Age-related decline in stress responses of human myocardium may not be explained by changes in mtDNA. Mech. Ageing Dev. 130, 742–747. ( 10.1016/j.mad.2009.09.003) [DOI] [PubMed] [Google Scholar]
- 114.Bonomi M, et al. 2012. Blood cell mitochondrial DNA content and premature ovarian aging. PLoS ONE 7, e42423 ( 10.1371/journal.pone.0042423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim JY, Hwang JM, Ko HS, Seong MW, Park BJ, Park SS. 2005. Mitochondrial DNA content is decreased in autosomal dominant optic atrophy. Neurology 64, 966–972. ( 10.1212/01.WNL.0000157282.76715.B1) [DOI] [PubMed] [Google Scholar]
- 116.Sitarz KS, et al. 2012. OPA1 mutations induce mtDNA proliferation in leukocytes of patients with dominant optic atrophy. Neurology 79, 1515–1517. ( 10.1212/WNL.0b013e31826d5f60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Iommarini L, Maresca A, Caporali L, Valentino ML, Liguori R, Giordano C, Carelli V. 2012. Revisiting the issue of mitochondrial DNA content in optic mitochondriopathies. Neurology 79, 1517–1519. ( 10.1212/WNL.0b013e31826d5f72) [DOI] [PubMed] [Google Scholar]
- 118.Liu CS, et al. 2006. Alteration in the copy number of mitochondrial DNA in leukocytes of patients with mitochondrial encephalomyopathies. Acta Neurol. Scand. 113, 334–341. [DOI] [PubMed] [Google Scholar]
- 119.Ekstrand MI, et al. 2004. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 13, 935–944. ( 10.1093/hmg/ddh109) [DOI] [PubMed] [Google Scholar]
- 120.Silva JP, Larsson NG. 2002. Manipulation of mitochondrial DNA gene expression in the mouse. Biochim. Biophys. Acta 1555, 106–110. ( 10.1016/S0005-2728(02)00263-3) [DOI] [PubMed] [Google Scholar]
- 121.Matsushima Y, Goto Y, Kaguni LS. 2010. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM). Proc. Natl Acad. Sci. USA 107, 18 410–18 415. ( 10.1073/pnas.1008924107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lu B, et al. 2007. Roles for the human ATP-dependent Lon protease in mitochondrial DNA maintenance. J. Biol. Chem. 282, 17 363–17 374. ( 10.1074/jbc.M611540200) [DOI] [PubMed] [Google Scholar]
- 123.Xu S, et al. 2009. Overexpression of Tfam protects mitochondria against beta-amyloid-induced oxidative damage in SH-SY5Y cells. FEBS J. 276, 3800–3809. ( 10.1111/j.1742-4658.2009.07094.x) [DOI] [PubMed] [Google Scholar]
- 124.Lu B, et al. 2013. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol. Cell 49, 121–132. ( 10.1016/j.molcel.2012.10.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kasashima K, Sumitani M, Endo H. 2012. Maintenance of mitochondrial genome distribution by mitochondrial AAA+ protein ClpX. Exp. Cell Res. 318, 2335–2343. ( 10.1016/j.yexcr.2012.07.012) [DOI] [PubMed] [Google Scholar]
- 126.He J, et al. 2007. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 176, 141–146. ( 10.1083/jcb.200609158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.He J, et al. 2012. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 40, 6109–6121. ( 10.1093/nar/gks266) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim HY, Gladyshev VN. 2004. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol. Biol. Cell 15, 1055–1064. ( 10.1091/mbc.E03-08-0629) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vougier S, Mary J, Friguet B. 2003. Subcellular localization of methionine sulphoxide reductase A (MsrA): evidence for mitochondrial and cytosolic isoforms in rat liver cells. Biochem. J. 373, 531–537. ( 10.1042/BJ20030443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. 2001. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl Acad. Sci. USA 98, 12 920–12 925. ( 10.1073/pnas.231472998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cabreiro F, Picot CR, Perichon M, Castel J, Friguet B, Petropoulos I. 2008. Overexpression of mitochondrial methionine sulfoxide reductase B2 protects leukemia cells from oxidative stress-induced cell death and protein damage. J. Biol. Chem. 283, 16 673–16 681. ( 10.1074/jbc.M708580200) [DOI] [PubMed] [Google Scholar]
- 132.Ahmed ZM, et al. 2011. Functional null mutations of MSRB3 encoding methionine sulfoxide reductase are associated with human deafness DFNB74. Am. J. Hum. Genet. 88, 19–29. ( 10.1016/j.ajhg.2010.11.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stiburek L, Cesnekova J, Kostkova O, Fornuskova D, Vinsova K, Wenchich L, Houstek J, Zeman J. 2012. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol. Biol. Cell 23, 1010–1023. ( 10.1091/mbc.E11-08-0674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Koppen M, Metodiev MD, Casari G, Rugarli EI, Langer T. 2007. Variable and tissue-specific subunit composition of mitochondrial m-AAA protease complexes linked to hereditary spastic paraplegia. Mol. Cell Biol. 27, 758–767. ( 10.1128/MCB.01470-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Di Bella D, et al. 2010. Mutations in the mitochondrial protease gene AFG3L2 cause dominant hereditary ataxia SCA28. Nat. Genet. 42, 313–321. ( 10.1038/ng.544) [DOI] [PubMed] [Google Scholar]
- 136.Pierson TM, et al. 2011. Whole-exome sequencing identifies homozygous AFG3L2 mutations in a spastic ataxia-neuropathy syndrome linked to mitochondrial m-AAA proteases. PLoS Genet. 7, e1002325 ( 10.1371/journal.pgen.1002325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Coates PJ, et al. 2001. Mammalian prohibitin proteins respond to mitochondrial stress and decrease during cellular senescence. Exp. Cell Res. 265, 262–273. ( 10.1006/excr.2001.5166) [DOI] [PubMed] [Google Scholar]
- 138.Merkwirth C, Martinelli P, Korwitz A, Morbin M, Bronneke HS, Jordan SD, Rugarli EI, Langer T. 2012. Loss of prohibitin membrane scaffolds impairs mitochondrial architecture and leads to tau hyperphosphorylation and neurodegeneration. PLoS Genet. 8, e1003021 ( 10.1371/journal.pgen.1003021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jenkinson EM, et al. 2013. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am. J. Hum. Genet. 92, 605–613. ( 10.1016/j.ajhg.2013.02.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Baker TA, Sauer RT. 2012. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta 1823, 15–28. ( 10.1016/j.bbamcr.2011.06.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Venkatesh S, Lee J, Singh K, Lee I, Suzuki CK. 2012. Multitasking in the mitochondrion by the ATP-dependent Lon protease. Biochim. Biophys. Acta 1823, 56–66. ( 10.1016/j.bbamcr.2011.11.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. 2004. Structural basis of mitochondrial tethering by mitofusin complexes. Science 305, 858–862. ( 10.1126/science.1099793) [DOI] [PubMed] [Google Scholar]
- 143.Zuchner S, et al. 2004. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot–Marie–Tooth neuropathy type 2A. Nat. Genet. 36, 449–451. ( 10.1038/ng1341) [DOI] [PubMed] [Google Scholar]
- 144.Otera H, Mihara K. 2011. Molecular mechanisms and physiologic functions of mitochondrial dynamics. J. Biochem. 149, 241–251. ( 10.1093/jb/mvr002) [DOI] [PubMed] [Google Scholar]
- 145.Cartoni R, Martinou JC. 2009. Role of mitofusin 2 mutations in the physiopathology of Charcot–Marie–Tooth disease type 2A. Exp. Neurol. 218, 268–273. ( 10.1016/j.expneurol.2009.05.003) [DOI] [PubMed] [Google Scholar]
- 146.Alexander C, et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26, 211–215. ( 10.1038/79944) [DOI] [PubMed] [Google Scholar]
- 147.Delettre C, et al. 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26, 207–210. ( 10.1038/79936) [DOI] [PubMed] [Google Scholar]
- 148.Kang PJ, Ostermann J, Shilling J, Neupert W, Craig EA, Pfanner N. 1990. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348, 137–143. ( 10.1038/348137a0) [DOI] [PubMed] [Google Scholar]
- 149.Hansen JJ, et al. 2002. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am. J. Hum. Genet. 70, 1328–1332. ( 10.1086/339935) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Magen D, et al. 2008. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am. J. Hum. Genet. 83, 30–42. ( 10.1016/j.ajhg.2008.05.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Scherz-Shouval R, Elazar Z. 2007. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 17, 422–427. ( 10.1016/j.tcb.2007.07.009) [DOI] [PubMed] [Google Scholar]
- 152.Friguet B, Bulteau AL, Petropoulos I. 2008. Mitochondrial protein quality control: implications in ageing. Biotechnol. J. 3, 757–764. ( 10.1002/biot.200800041) [DOI] [PubMed] [Google Scholar]
- 153.Petropoulos I, Friguet B. 2005. Protein maintenance in aging and replicative senescence: a role for the peptide methionine sulfoxide reductases. Biochim. Biophys. Acta 1703, 261–266. ( 10.1016/j.bbapap.2004.08.018) [DOI] [PubMed] [Google Scholar]
- 154.Sharov VS, Ferrington DA, Squier TC, Schoneich C. 1999. Diastereoselective reduction of protein-bound methionine sulfoxide by methionine sulfoxide reductase. FEBS Lett. 455, 247–250. ( 10.1016/S0014-5793(99)00888-1) [DOI] [PubMed] [Google Scholar]
- 155.Grimaud R, et al. 2001. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 276, 48 915–48 920. ( 10.1074/jbc.M105509200) [DOI] [PubMed] [Google Scholar]
- 156.Baker MJ, Tatsuta T, Langer T. 2011. Quality control of mitochondrial proteostasis. Cold Spring Harb. Perspect. Biol. 3, a007559 ( 10.1101/cshperspect.a007559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Marom M, Azem A, Mokranjac D. 2011. Understanding the molecular mechanism of protein translocation across the mitochondrial inner membrane: still a long way to go. Biochim. Biophys. Acta 1808, 990–1001. ( 10.1016/j.bbamem.2010.07.011) [DOI] [PubMed] [Google Scholar]
- 158.Koll H, Guiard B, Rassow J, Ostermann J, Horwich AL, Neupert W, Hartl F-U. 1992. Antifolding activity of hsp60 couples protein import into the mitochondrial matrix with export to the intermembrane space. Cell 68, 1163–1175. ( 10.1016/0092-8674(92)90086-R) [DOI] [PubMed] [Google Scholar]
- 159.Horwich AL, Weber-Ban EU, Finley D. 1999. Chaperone rings in protein folding and degradation. Proc. Natl Acad. Sci. USA 96, 11 033–11 040. ( 10.1073/pnas.96.20.11033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, Hayer-Hartl M. 2001. Dual function of protein confinement in chaperonin-assisted protein folding. Cell 107, 223–233. ( 10.1016/S0092-8674(01)00517-7) [DOI] [PubMed] [Google Scholar]
- 161.Christensen JH, Nielsen MN, Hansen J, Fuchtbauer A, Fuchtbauer EM, West M, Corydon TJ, Gregersen N, Bross P. 2010. Inactivation of the hereditary spastic paraplegia-associated Hspd1 gene encoding the Hsp60 chaperone results in early embryonic lethality in mice. Cell Stress Chaperones 15, 851–863. ( 10.1007/s12192-010-0194-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Magnoni R, Palmfeldt J, Christensen JH, Sand M, Maltecca F, Corydon TJ, West M, Casari G, Bross P. 2013. Late onset motoneuron disorder caused by mitochondrial Hsp60 chaperone deficiency in mice. Neurobiol. Dis. 54, 12–23. ( 10.1016/j.nbd.2013.02.012) [DOI] [PubMed] [Google Scholar]
- 163.Lau E, Wang D, Zhang J, Yu H, Lam MP, Liang X, Zong N, Kim T-Y, Ping P. 2012. Substrate- and isoform-specific proteome stability in normal and stressed cardiac mitochondria. Circ. Res. 110, 1174–1178. ( 10.1161/CIRCRESAHA.112.268359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Koppen M, Langer T. 2007. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit. Rev. Biochem. Mol. Biol. 2, 221–242. ( 10.1080/10409230701380452) [DOI] [PubMed] [Google Scholar]
- 165.Clarke KJ, Adams AE, Manzke LH, Pearson TW, Borchers CH, Porter RK. 2012. A role for ubiquitinylation and the cytosolic proteasome in turnover of mitochondrial uncoupling protein 1 (UCP1). Biochim. Biophys. Acta 1817, 1759–1767. ( 10.1016/j.bbabio.2012.03.035) [DOI] [PubMed] [Google Scholar]
- 166.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. 2013. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc. Natl Acad. Sci. USA 110, 6400–6405. ( 10.1073/pnas.1221132110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. 2010. Analysis of proteome dynamics in the mouse brain. Proc. Natl Acad. Sci. USA 107, 14 508–14 513. ( 10.1073/pnas.1006551107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Martinelli P, Rugarli EI. 2010. Emerging roles of mitochondrial proteases in neurodegeneration. Biochim. Biophys. Acta 1797, 1–10. ( 10.1016/j.bbabio.2009.07.013) [DOI] [PubMed] [Google Scholar]
- 169.Leonhard K, Stiegler A, Neupert W, Langer T. 1999. Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature 398, 348–351. ( 10.1038/18704) [DOI] [PubMed] [Google Scholar]
- 170.Arnold I, Langer T. 2002. Membrane protein degradation by AAA proteases in mitochondria. Biochim. Biophys. Acta 1592, 89–96. ( 10.1016/S0167-4889(02)00267-7) [DOI] [PubMed] [Google Scholar]
- 171.Weil A, Luce K, Dröse S, Wittig I, Brandt U, Osiewacz HD. 2011. Unmasking a temperature-dependent effect of the P. anserina i-AAA protease on aging and development. Cell Cycle 10, 4280–4290. ( 10.4161/cc.10.24.18560) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Koppen M, Bonn F, Ehses S, Langer T. 2009. Autocatalytic processing of m-AAA protease subunits in mitochondria. Mol. Biol. Cell 20, 4216–4224. ( 10.1091/mbc.E09-03-0218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T. 2005. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123, 277–289. ( 10.1016/j.cell.2005.08.003) [DOI] [PubMed] [Google Scholar]
- 174.Ishihara N, Fujita Y, Oka T, Mihara K. 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25, 2966–2977. ( 10.1038/sj.emboj.7601184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ehses S, et al. 2009. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187, 1023–1036. ( 10.1083/jcb.200906084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Maltecca F, De Stefani D, Cassina L, Consolato F, Wasilewski M, Scorrano L, Rizzuto R, Casari G. 2012. Respiratory dysfunction by AFG3L2 deficiency causes decreased mitochondrial calcium uptake via organellar network fragmentation. Hum. Mol. Genet. 21, 3858–3870. ( 10.1093/hmg/dds214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Almajan ER, et al. 2012. AFG3L2 supports mitochondrial protein synthesis and Purkinje cell survival. J. Clin. Invest. 122, 4048–4058. ( 10.1172/JCI64604) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Casari G, et al. 1998. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 93, 973–983. ( 10.1016/S0092-8674(00)81203-9) [DOI] [PubMed] [Google Scholar]
- 179.Warnecke T, Duning T, Schwan A, Lohmann H, Epplen JT, Young P. 2007. A novel form of autosomal recessive hereditary spastic paraplegia caused by a new SPG7 mutation. Neurology 69, 368–375. ( 10.1212/01.wnl.0000266667.91074.fe) [DOI] [PubMed] [Google Scholar]
- 180.Crosby AH, Proukakis C. 2002. Is the transportation highway the right road for hereditary spastic paraplegia? Am. J. Hum. Genet. 71, 1009–1016. ( 10.1086/344206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ferreirinha F, et al. 2004. Axonal degeneration in paraplegin-deficient mice is associated with abnormal mitochondria and impairment of axonal transport. J. Clin. Invest. 113, 231–242. ( 10.1172/JCI200420138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Arnoldi A, et al. 2008. A clinical, genetic, and biochemical characterization of SPG7 mutations in a large cohort of patients with hereditary spastic paraplegia. Hum. Mutat. 29, 522–531. ( 10.1002/humu.20682) [DOI] [PubMed] [Google Scholar]
- 183.Tatsuta T, Model K, Langer T. 2005. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 16, 248–259. ( 10.1091/mbc.E04-09-0807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Artal-Sanz M, Tavernarakis N. 2009. Prohibitin and mitochondrial biology. Trends Endocrinol. Metab. 20, 394–401. ( 10.1016/j.tem.2009.04.004) [DOI] [PubMed] [Google Scholar]
- 185.Steglich G, Neupert W, Langer T. 1999. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell Biol. 19, 3435–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nijtmans LG, Artal SM, Grivell LA, Coates PJ. 2002. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell Mol. Life Sci. 59, 143–155. ( 10.1007/s00018-002-8411-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mishra S, Murphy LC, Murphy LJ. 2006. The prohibitins: emerging roles in diverse functions. J. Cell Mol. Med. 10, 353–363. ( 10.1111/j.1582-4934.2006.tb00404.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Da Cruz S, Parone PA, Gonzalo P, Bienvenut WV, Tondera D, Jourdain A, Quadroni M, Martinou J-C. 2008. SLP-2 interacts with prohibitins in the mitochondrial inner membrane and contributes to their stability. Biochim. Biophys. Acta 1783, 904–911. ( 10.1016/j.bbamcr.2008.02.006) [DOI] [PubMed] [Google Scholar]
- 189.Bogenhagen DF, Rousseau D, Burke S. 2008. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 283, 3665–3675. ( 10.1074/jbc.M708444200) [DOI] [PubMed] [Google Scholar]
- 190.Kasashima K, Sumitani M, Satoh M, Endo H. 2008. Human prohibitin 1 maintains the organization and stability of the mitochondrial nucleoids. Exp. Cell Res. 314, 988–996. ( 10.1016/j.yexcr.2008.01.005) [DOI] [PubMed] [Google Scholar]
- 191.Ferrer I, Perez E, Dalfo E, Barrachina M. 2007. Abnormal levels of prohibitin and ATP synthase in the substantia nigra and frontal cortex in Parkinson's disease. Neurosci. Lett. 415, 205–209. ( 10.1016/j.neulet.2007.01.026) [DOI] [PubMed] [Google Scholar]
- 192.Merkwirth C, et al. 2008. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 22, 476–488. ( 10.1101/gad.460708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. 2002. A mitochondrial specific stress response in mammalian cells. EMBO J. 21, 4411–4419. ( 10.1093/emboj/cdf445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. 2007. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell 13, 467–480. ( 10.1016/j.devcel.2007.07.016) [DOI] [PubMed] [Google Scholar]
- 195.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. 2010. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol. Cell 37, 529–540. ( 10.1016/j.molcel.2010.01.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fischer F, Weil A, Hamann A, Osiewacz HD. 2013. Human CLPP reverts the longevity phenotype of a fungal ClpP deletion strain. Nat. Commun. 4, 1397 ( 10.1038/ncomms2397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gispert S, et al. 2013. Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum. Mol. Genet. 22, 4871–4887. ( 10.1093/hmg/ddt338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bota DA, Davies KJ. 2002. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 4, 674–680. ( 10.1038/ncb836) [DOI] [PubMed] [Google Scholar]
- 199.Bakala H, Delaval E, Hamelin M, Bismuth J, Borot-Laloi C, Corman B, Friguet B. 2003. Changes in rat liver mitochondria with aging. Lon protease-like activity and Nɛ-carboxymethyllysine accumulation in the matrix. Eur. J. Biochem. 270, 2295–2302. ( 10.1046/j.1432-1033.2003.03598.x) [DOI] [PubMed] [Google Scholar]
- 200.Delaval E, Perichon M, Friguet B. 2004. Age-related impairment of mitochondrial matrix aconitase and ATP-stimulated protease in rat liver and heart. Eur. J. Biochem. 271, 4559–4564. ( 10.1111/j.1432-1033.2004.04422.x) [DOI] [PubMed] [Google Scholar]
- 201.Hansen J, Corydon TJ, Palmfeldt J, Durr A, Fontaine B, Nielsen MN, Christensen JH, Gregersen N, Bross P. 2008. Decreased expression of the mitochondrial matrix proteases Lon and ClpP in cells from a patient with hereditary spastic paraplegia (SPG13). Neuroscience 153, 474–482. ( 10.1016/j.neuroscience.2008.01.070) [DOI] [PubMed] [Google Scholar]
- 202.Adam C, Picard M, Dequard-Chablat M, Sellem CH, Hermann-Le Denmat S, Contamine V. 2012. Biological roles of the Podospora anserina mitochondrial Lon protease and the importance of its N-domain. PLoS ONE 7, e38138 ( 10.1371/journal.pone.0038138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Luce K, Osiewacz HD. 2009. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat. Cell Biol. 11, 852–858. ( 10.1038/ncb1893) [DOI] [PubMed] [Google Scholar]
- 204.Figge MT, Reichert AS, Meyer-Hermann M, Osiewacz HD. 2012. Deceleration of fusion-fission cycles improves mitochondrial quality control during aging. PLoS Comput. Biol. 8, e1002576 ( 10.1371/journal.pcbi.1002576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Tatsuta T, Langer T. 2008. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 27, 306–314. ( 10.1038/sj.emboj.7601972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fischer F, Hamann A, Osiewacz HD. 2012. Mitochondrial quality control: an integrated network of pathways. Trends Biochem. Sci. 37, 284–292. ( 10.1016/j.tibs.2012.02.004) [DOI] [PubMed] [Google Scholar]
- 207.Mao K, Wang K, Liu X, Klionsky DJ. 2013. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev. Cell 26, 9–18. ( 10.1016/j.devcel.2013.05.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. 2001. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256. ( 10.1091/mbc.12.8.2245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. 2001. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 1, 515–525. ( 10.1016/S1534-5807(01)00055-7) [DOI] [PubMed] [Google Scholar]
- 210.Cereghetti GM, Costa V, Scorrano L. 2010. Inhibition of Drp1-dependent mitochondrial fragmentation and apoptosis by a polypeptide antagonist of calcineurin. Cell Death Differ. 17, 1785–1794. ( 10.1038/cdd.2010.61) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Scheckhuber CQ, Erjavec N, Tinazli A, Hamann A, Nystrom T, Osiewacz HD. 2007. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 9, 99–105. ( 10.1038/ncb1524) [DOI] [PubMed] [Google Scholar]
- 212.Ishihara N, et al. 2009. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11, 958–966. ( 10.1038/ncb1907) [DOI] [PubMed] [Google Scholar]
- 213.Williams GC. 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411. ( 10.2307/2406060) [DOI] [Google Scholar]
- 214.Santel A, Fuller MT. 2001. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114, 867–874. [DOI] [PubMed] [Google Scholar]
- 215.Chen H, Chan DC. 2009. Mitochondrial dynamics: fusion, fission, movement, and mitophagy in neurodegenerative diseases. Hum. Mol. Genet. 18, R169–R176. ( 10.1093/hmg/ddp326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Twig G, et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446. ( 10.1038/sj.emboj.7601963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. 2005. Autophagy and aging: the importance of maintaining ‘clean’ cells. Autophagy 1, 131–140. ( 10.4161/auto.1.3.2017) [DOI] [PubMed] [Google Scholar]
- 218.Kraft C, Deplazes A, Sohrmann M, Peter M. 2008. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10, 602–610. ( 10.1038/ncb1723) [DOI] [PubMed] [Google Scholar]
- 219.Philipp O, Hamann A, Servos J, Werner A, Koch I, Osiewacz HD. 2013. A genome-wide longitudinal transcriptome analysis of the aging model Podospora anserine. PLoS ONE 8, e83109 ( 10.1371/journal.pone.0083109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Schmid D, Munz C. 2007. Immune surveillance via self digestion. Autophagy 3, 133–135. [DOI] [PubMed] [Google Scholar]
- 221.Kikuma T, Ohneda M, Arioka M, Kitamoto K. 2006. Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae. Eukaryot. Cell 5, 1328–1336. ( 10.1128/EC.00024-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Knuppertz L, Hamann A, Pampaloni F, Stelzer E, Osiewacz HD. 2013. Identification of autophagy as a longevity assurance mechanism in the aging model Podospora anserina. Autophagy 10, 822–834. ( 10.4161/auto.28148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lapierre LR, et al. 2013. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4, 2267 ( 10.1038/ncomms3267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pyo JO. 2013. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun. 4, 2300 ( 10.1038/ncomms3300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. 2008. Essential role for Nix in autophagic maturation of erythroid cells. Nature 454, 232–235. ( 10.1038/nature07006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Narendra D, Tanaka A, Suen DF, Youle RJ. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803. ( 10.1083/jcb.200809125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298 ( 10.1371/journal.pbio.1000298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ashrafi G, Schwarz TL. 2013. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20, 31–42. ( 10.1038/cdd.2012.81) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. 2010. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131. ( 10.1038/ncb2012) [DOI] [PubMed] [Google Scholar]
- 230.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. 2010. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 19, 4861–4870. ( 10.1093/hmg/ddq419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Glauser L, Sonnay S, Stafa K, Moore DJ. 2011. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem. 118, 636–645. ( 10.1111/j.1471-4159.2011.07318.x) [DOI] [PubMed] [Google Scholar]
- 232.Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, McBride HM, Park DS, Fon EA. 2012. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 13, 378–385. ( 10.1038/embor.2012.14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Weber TA, Reichert AS. 2010. Impaired quality control of mitochondria: aging from a new perspective. Exp. Gerontol. 45, 503–511. ( 10.1016/j.exger.2010.03.018) [DOI] [PubMed] [Google Scholar]
- 234.Kelly GL, Strasser A. 2011. The essential role of evasion from cell death in cancer. Adv. Cancer Res. 111, 39–96. ( 10.1016/B978-0-12-385524-4.00002-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Madeo F, Herker E, Wissing S, Jungwirth H, Eisenberg T, Frohlich KU. 2004. Apoptosis in yeast. Curr. Opin. Microbiol. 7, 655–660. ( 10.1016/j.mib.2004.10.012) [DOI] [PubMed] [Google Scholar]
- 236.Hamann A, Brust D, Osiewacz HD. 2008. Apoptosis pathways in fungal growth, development and ageing. Trends Microbiol. 16, 276–283. ( 10.1016/j.tim.2008.03.003) [DOI] [PubMed] [Google Scholar]
- 237.Brust D, Daum B, Breunig C, Hamann A, Kühlbrandt W, Osiewacz HD. 2010. Cyclophilin D links programmed cell death and organismal aging in Podospora anserina. Aging Cell 9, 761–775. ( 10.1111/j.1474-9726.2010.00609.x) [DOI] [PubMed] [Google Scholar]
- 238.Kroemer G, Galluzzi L, Brenner C. 2007. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87, 99–163. ( 10.1152/physrev.00013.2006) [DOI] [PubMed] [Google Scholar]
- 239.Brust D, Hamann A, Osiewacz HD. 2010. Deletion of PaAif2 and PaAmid2, two genes encoding mitochondrial AIF-like oxidoreductases of Podospora anserina, leads to increased stress tolerance and lifespan extension. Curr. Genet. 56, 225–235. ( 10.1007/s00294-010-0295-1) [DOI] [PubMed] [Google Scholar]
- 240.Groebe K, et al. 2007. Differential proteomic profiling of mitochondria from Podospora anserina, rat and human reveals distinct patterns of age-related oxidative changes. Exp. Gerontol. 42, 887–898. ( 10.1016/j.exger.2007.07.001) [DOI] [PubMed] [Google Scholar]
- 241.Marzetti E, Wohlgemuth SE, Lees HA, Chung HY, Giovannini S, Leeuwenburgh C. 2008. Age-related activation of mitochondrial caspase-independent apoptotic signaling in rat gastrocnemius muscle. Mech. Ageing Dev. 129, 542–549. ( 10.1016/j.mad.2008.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Daum B, Walter A, Horst A, Osiewacz HD, Kühlbrandt W. 2013. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc. Natl Acad. Sci. USA 110, 15 301–15 306. ( 10.1073/pnas.1305462110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Davies KM, Strauss M, Daum B, Kief JH, Osiewacz HD, Rycovska A, Zickermann V, Kühlbrandt W. 2011. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl Acad. Sci. USA 108, 14 121–14 126. ( 10.1073/pnas.1103621108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hiona A, et al. 2010. Mitochondrial DNA mutations induce mitochondrial dysfunction, apoptosis and sarcopenia in skeletal muscle of mitochondrial DNA mutator mice. PLoS ONE 5, e11468 ( 10.1371/journal.pone.0011468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hafner AV, et al. 2010. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY) 2, 914–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Taylor EB, Rutter J. 2011. Mitochondrial quality control by the ubiquitin-proteasome system. Biochem. Soc. Trans. 39, 1509–1513. ( 10.1042/BST0391509) [DOI] [PubMed] [Google Scholar]
- 247.Jeon HB, Choi ES, Yoon JH, Hwang JH, Chang JW, Lee EK, Choi HW, Park Z-Y, Yoo YJ. 2007. A proteomics approach to identify the ubiquitinated proteins in mouse heart. Biochem. Biophys. Res. Commun. 357, 731–736. ( 10.1016/j.bbrc.2007.04.015) [DOI] [PubMed] [Google Scholar]
- 248.Korolchuk VI, Menzies FM, Rubinsztein DC. 2010. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 584, 1393–1398. ( 10.1016/j.febslet.2009.12.047) [DOI] [PubMed] [Google Scholar]
- 249.Amati-Bonneau P, et al. 2008. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain 131, 338–351. ( 10.1093/brain/awm298) [DOI] [PubMed] [Google Scholar]
- 250.Hudson G, et al. 2008. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain 131, 329–337. ( 10.1093/brain/awm272) [DOI] [PubMed] [Google Scholar]