Abstract
Autophagy is a well-conserved catabolic process, involving the degradation of a cell's own components through the lysosomal/vacuolar machinery. Autophagy is typically induced by nutrient starvation and has a role in nutrient recycling, cellular differentiation, degradation and programmed cell death. Another common response in eukaryotes is the extension of lifespan through dietary restriction (DR). We studied a link between DR and autophagy in the filamentous fungus Podospora anserina, a multicellular model organism for ageing studies and mitochondrial deterioration. While both carbon and nitrogen restriction extends lifespan in P. anserina, the size of the effect varied with the amount and type of restricted nutrient. Natural genetic variation for the DR response exists. Whereas a switch to carbon restriction up to halfway through the lifetime resulted in extreme lifespan extension for wild-type P. anserina, all autophagy-deficient strains had a shorter time window in which ageing could be delayed by DR. Under nitrogen limitation, only PaAtg1 and PaAtg8 mediate the effect of lifespan extension; the other autophagy-deficient mutants PaPspA and PaUth1 had a similar response as wild-type. Our results thus show that the ageing process impinges on the DR response and that this at least in part involves the genetic regulation of autophagy.
Keywords: dietary restriction, carbon and nitrogen restriction, ageing, autophagy, lifespan extension, Podospora anserina
1. Introduction
Calorie or dietary restriction (DR) refers to a dietary regimen in which an organism is subjected to a reduced food intake—either in calorific value or in specific food compounds—without causing malnutrition. DR has been shown to increase lifespan in a wide variety of organisms [1,2], even if applied temporarily [3,4]. The DR response is adaptive because it allows surviving lean periods by shifting the energy investment from germ line towards soma. As a result, reproduction is postponed until more favourable conditions arrive [5,6]. Despite this clear adaptive value of DR, little is known about the genetic and molecular bases that lead to the DR response. The lifespan extension effect can be viewed as postponement of ageing, but the age-independency of this response in, for instance, Drosophila refutes such a direct link to ageing mechanisms [7].
Both ageing and the presumed postponement of ageing by DR have been related to mitochondrial function. Harman's free radical theory focuses on the production of reactive oxygen species by the mitochondria, and their damaging effect on macromolecules as the major cause for ageing, and that as a result of DR the production of reactive oxygen species is reduced and hence lifespan is extended [8]. Alternative views focus on other DR mechanisms such as lowering the levels of glycation-mediated protein damage [9] or emphasize the multi-causal nature of ageing and suggest a genome-wide regulated response to DR [10].
One way to deal with age-related damage and possibly to reallocate resources in time of scarcity is autophagy. Vacuolar/lysosomal autophagic degradation is the major pathway for continuous turnover of damaged and obsolete macromolecules and organelles, and the involved genes have a high level of conservation throughout all eukaryotes [11,12]. Autophagy supposedly has a dual role, one linked to nutrient deprivation and recycling and to increase cellular lifespan, and the other linked to programmed cell death [12,13]. It has been shown that upon nitrogen restriction, genes involved in the autophagic pathway as well as genes involved in cellular proteolytic activity are induced [14–17].
Podospora anserina has been a model system for ageing since the 1950s [18]. Ageing in Podospora and some other filamentous fungi probably evolved due to the ephemerality of its habitat [19]. That is, P. anserina grows on herbivore dung and all natural isolates have a lifespan of only two to three weeks [20,21]. The senescence phenotype is characterized by a loss in reproductive potential, a reduction in growth rate and aerial hyphae production, abnormal branching and swelling of hyphal tips, and the accumulation of a dark lipofuscin ageing pigment [22–24]. Ageing in P. anserina is systemic, highly suppressive and infectious, and is driven by mitochondrial function and metabolism (for a review, see [25]). Major rearrangements of the mitochondrial genome—often extra-chromosomal amplifications—are associated with ageing in P. anserina; many of these are caused by regions inside the mitochondrial genome, known as the senDNAs [26–28]. In addition, extra-genomic elements such as the invertron-like mitochondrial pAL2-1 plasmid and homologues can interfere with lifespan, seemingly independent of other intrinsic and external lifespan-influencing conditions [21,29].
DR in the form of glucose restriction or other carbon sources such as fructose or acetate leads to extension of the vegetative lifespan in P. anserina, both in semi-synthetic and synthetic media [21,30]. This prolongation in lifespan of P. anserina can vary from two weeks up to 3 years [21,30–32], and while no reproduction takes place during this period, sexual and asexual reproduction are restored when normal nutritional conditions are available [31,32]. During carbon and nitrogen starvation the same genes in P. anserina are expressed as under heterokaryon/vegetative incompatibility reactions when fusion between incompatible genotypes leads to a programmed cell death response [15]. Fungal incompatibility reactions are characterized by a strong increase in the total cellular enzymatic proteolytic activity [14], but also in genes involved in the autophagic pathway [15,17]. Although a direct link to autophagy has not been established, given the similarities in the starvation and incompatible reactions it seems probable that autophagy could play a role.
The objectives of this study were to determine the effect of autophagy and age on the DR response in P. anserina. We used a well-defined synthetic medium with carbon and nitrogen restrictions to realize the DR regimes. We used a set of wild-type strains as well as a selection of laboratory strains and four knock-out mutants hampered in different genes involved in different stages of the autophagy pathway (ΔPaAtg1, ΔPaAtg8, ΔPaPspA and ΔPaUth1). We report here that in P. anserina, limitation of either carbon or nitrogen source results in a lifespan-extending DR effect in juvenile strains. However, in (pre)senescent (halfway through life and older) wild-type strains, this lifespan extension proved lost, in contrast to, e.g. effects observed in Drosophila melanogaster where DR reduces the short-term risks of death at least until the age of four weeks [3]. Although juvenile autophagy-deficient strains generally show a normal lifespan extension upon DR, the time period in which DR can extend lifespan proved to be severely reduced in the different autophagy-deficient mutants.
2. Material and methods
(a). Strains, mutants and genes
For testing the effect of reduction in either carbon or nitrogen sources on lifespan, we used the standard laboratory strains s and Cs [18] and several wild-type isolates (Wa32, Wa50, Wa70, Wa76 and Wa77; [20,21]).
For our studies on autophagy and mitophagy, we used P. anserina strains derived from the original strain s/Cs, mutated in genes that based on homologies and previous studies are involved in the autophagic degradation of mitochondria. The examined genes act at four different stages of the autophagy route.
(i) PaUth1 putatively codes for a mitochondrial outer membrane protein that is part of the SUN superfamily. The homologous protein in Saccharomyces cerevisiae is UTH (YKR042W) which is (one of) the label(s) to mark mitochondria for mitophagy and is involved in the oxidative stress response [33,34]. A knock-out P. anserina strain blocked in the production of PaUTH1 (CAP67803) was made based on this sequence as described for other genes by El-Khoury et al. [35] in a strain lacking heterologous recombination thanks to the Ku mutation.
(ii) PaAtg1 codes for a serine/threonine protein kinase that is involved in the cytoplasm-to-vacuole transport and for the formation of autophagosomes [17]. Its product ATG1 is the homologue of yeast ATG1 (YGL180W).
(iii) PaAtg8 also has a similar-named homologue in yeast (YBL078C) and codes for a lipid-conjugated ubiquitin-like protein that connects to phosphatidylethanolamine, leading to the tethering and hemifusion of membranes of autophagosome and vacuole [16,36].
(iv) PaPspA codes for a subtilisin-like serine protease and is an orthologue of the vacuolar protease B PRB1 of S. cerevisiae (YEL060C), necessary for degradation within the vacuole [16].
ΔPaAtg1, ΔPaAtg8 and ΔPaPspA strains have kindly been provided by Dr Pinan-Lucarré and Dr Clavé from the Centre de Génétique Moléculaire of the Université de Bordeaux 2, in France. The four genes together are thus expected to cover several key processes in (mitochondrial) autophagy.
(b). Culture conditions and lifespan analysis
Podospora anserina synthetic medium (PASM) [21] was used in these experiments with as standard carbon and nitrogen sources, 2% (w/v) d-glucose and 0.1% (w/v) urea. d-glucose concentrations were varied from 0.02, 0.1, 0.2, 1, 2 to 4% (w/v), respectively, with a urea concentration of 0.1%. For the tests with different nitrogen amounts, 2% d-glucose was used in combination with 0.1, 0.01, 0.001 or 0.0001% (w/v) urea, respectively. The pH of all these media was set to 6.4. For germination of spores, PASM was supplemented with 0.06 M ammonium acetate; to induce reproduction and spore formation, PASM was supplemented with 0.5% (w/v) dried and ground horse dung. All incubations were done at 27°C in the dark, except for fertility tests that were done with a 12 hour L/12 D circadian rhythm.
Lifespans and growth rates were measured in days of growth with a continuous linear growth rate using 30 cm glass race tubes with 20 ml PASM medium. Tubes were inoculated using explants derived from 2- to 3-day-old cultures grown from spores on PASM with acetate. Growth was marked in 1- to 3-day intervals, and lifespans and linear growth rates were determined using at least five replicas per strain per condition. When race tubes were grown full, explants were cut from the mycelium at the distal ends of the tubes and transferred to new ones. For practical reasons, lifespan measurements were truncated after 90 days. Differences in lifespan and growth rates were analysed using a generalized linear model (GLM) in R. Subsequent pairwise interactions of the GLM models were done with the ghlt-function from the multcomp package. P-values reported from these pairwise interactions were adjusted for multiple comparison using the Tukey method.
(c). The effect of age on the response to carbon restriction
The different (wild-type and autophagy-deficient) strains were grown on Petri-dishes with standard PASM with 2% (w/v) d-glucose and 0.1% (w/v) urea. At different stages during life, explants were transferred from normal media to severe carbon-restricted conditions (PASM with 0.02% (w/v) d-glucose). Lifespan was measured in fivefold as described before. The experiments were truncated after 90 days which was set for the analyses as the maximum lifespan-extending effect.
(d). Microscopic analysis of the ΔPaUth1 mitophagy mutant
To study the effect of the ΔPaUth1 mutation on mitochondrial dynamics, we crossed the ΔPaUth1 mutant with a transformant strain, in which green fluorescent protein (GFP) is targeted to the mitochondria thanks to the mitochondrial-targeting sequence of Neurospora crassa ATP9 [37], and selected recombinant single-mating type offspring. The mitochondrial morphology was then visualized using confocal laser scanning microscopy (LSM 510, Zeiss, Germany).
3. Results
(a). Lifespan-extending effects of nitrogen restriction
Previously it has been shown that glucose restriction leads to a significant increase in lifespan [21,30–32]. For a constant starting concentration of 0.1% urea (w/v), our results indicate that in fact the relationship between lifespan and glucose concentration on a log scale is an inverse logarithmic function (figure 1a). To establish the effects of reduced concentrations of nitrogen on the lifespan of P. anserina, strain s was grown at concentrations of urea ranging from the standard 0.1 to 0.0001%. Unlike the inverse logarithmic relation found in glucose, a parabolic relationship between lifespan and the logarithm of the urea concentration was found (figure 1b). We observed that lifespan significantly increased for concentrations lower than 0.1% urea (figure 1b; χ2 = 14.154, d.f. = 3, p = 0.003). Lifespan of P. anserina peaked at both 0.01 and 0.001% urea, with a doubling in lifespan, whereas at 0.0001% lifespan only increased by 50% compared with the original 0.1% concentration (not significant after post hoc testing). Nitrogen restriction in P. anserina resulted also in several other phenotypic changes. Firstly, a significant 15% decrease in the growth rate was observed for the concentrations 0.01 and 0.001% urea compared with 0.1% urea (data not shown; χ2 = 20.757, d.f. = 3, p = 0.001). Secondly, mycelium density is decreased, while at the same time a strong increase in dark pigmentation is observed (figure 2). Thirdly, reduced fertility or even sterility was observed at the lower concentrations.
Figure 1.
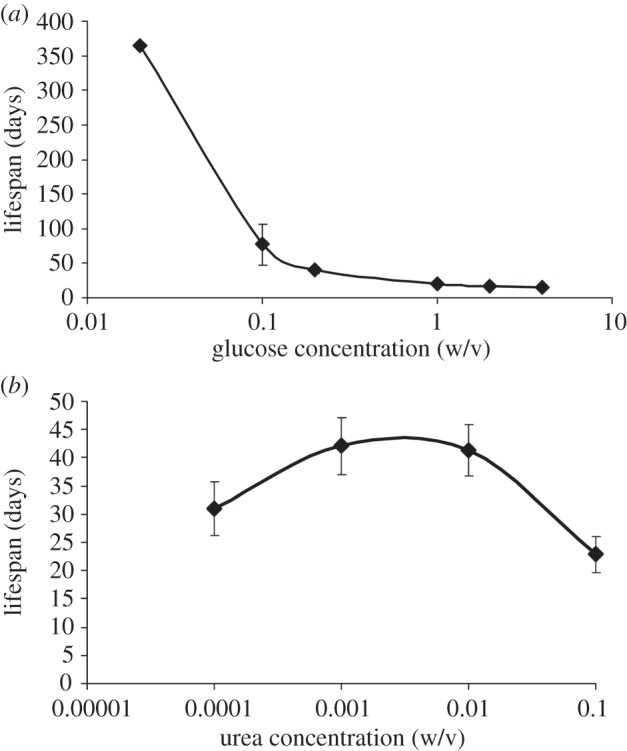
Effects of (a) d-glucose and (b) urea concentrations on the lifespan of P. anserina strain s. For the different d-glucose concentrations, the urea concentration was kept at 0.1% w/v; for the different urea concentrations the d-glucose level was kept at 2% w/v. The measurements of the five replicate lines growing on 0.02% glucose w/v and 0.1% urea w/v were truncated after 1 year of continuous growth, with all lines still alive.
Figure 2.

Phenotype of P. anserina strain s on carbon (d-glucose; left) and nitrogen (urea; right) restriction, respectively. Growth front in the 0.001% (w/v) urea conditions marked with triangles. (Online version in colour.)
(b). Nutrient restriction in wild-types
Most previous work on DR in P. anserina has focused on a single strain, which has been maintained for many years in the laboratory. To establish whether these observations are not the result of adaptation to laboratory conditions, several wild-type strains were tested for their response to reduced concentrations of both glucose and urea (figure 3a,b, respectively). The lifespan of the wild-type strains increased in response to reduced glucose (figure 3a; χ2 = 5269.7, d.f. = 2, p < 0.001) and reduced concentrations of urea (figure 3b; χ2 = 108.4, d.f. = 2, p < 0.001) suggesting that the response to nutrient restriction universally applies to P. anserina strains. However, interestingly, the extent to which lifespan increased differed significantly between strains for glucose (figure 3a; χ2 = 272.0, d.f. = 4, p < 0.001) as well as urea restriction (figure 3b; χ2 = 73.17, d.f. = 3, p < 0.001). This is indicative of genetic variation for the DR responses. Indeed, we found a strong interaction between the lifespan of the strains and the glucose (figure 3a; χ2 = 532.7, d.f. = 8, p < 0.001) and urea (figure 3b; χ2 = 33.56, d.f. = 5, p < 0.001) environment.
Figure 3.
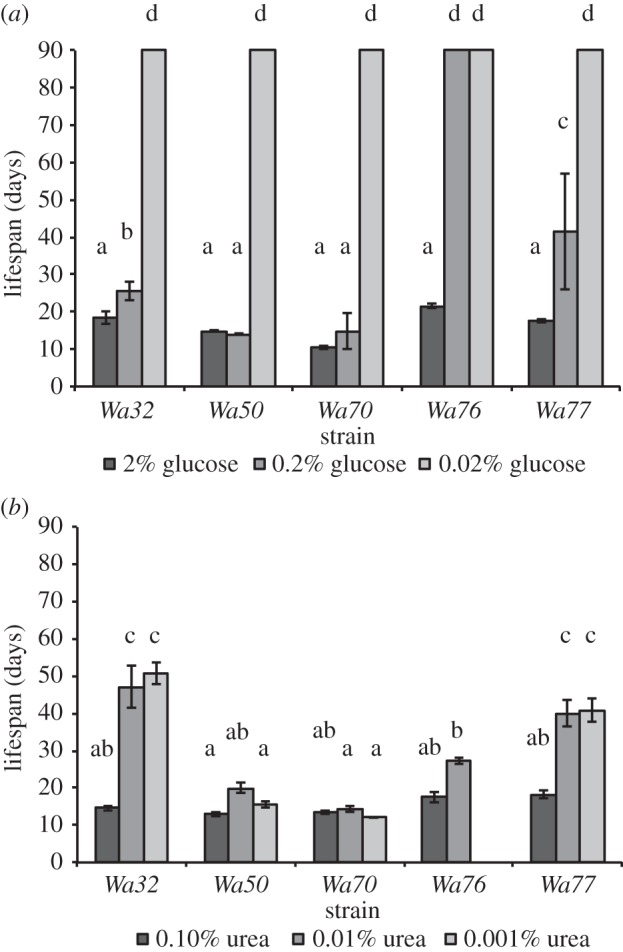
The lifespan of wild-type strains under glucose (a) and urea (b) restriction, respectively. The often small standard deviations are given for all columns, but when all strains lived longer than the cut-off point of 90 days, these are not visible in the graphs. The same letter above columns (a,b,c,d) indicates that these groups do not statistically differ in phenotype, while different letters indicate that these groups do.
(c). The role of autophagy in lifespan extension upon carbon and nitrogen restriction
To test whether the process of autophagy is important for the lifespan extension by carbon and/or nitrogen restriction, we tested several existing autophagy mutants (table 1). In addition, we created a knock-out mutant for the homologue of Uth1 from yeast involved in mitophagy and the oxidative stress response [34]. Phenotypically, the ΔPaUth1 is comparable to the wild-type when grown in a Petri dish, and even the ageing pigment lipofuscin is formed as normal prior to cell death; only the growth rate is lower (figure 4a). The putative mitophagy-deficient strain is fertile both as female and as male, though development of the fruiting bodies and spore formation are retarded in comparison with the parental strain. By contrast, the autophagy-deficient strains ΔPaAtg1, ΔPaAtg8 and ΔPaPspA as reported show a reduction in pigmentation and in hyphal density. These strains have lost their female fertility, but do produce spermatia enabling the backcrossing to the parental strain and selection of monokaryotic mutant lines for experiments [16,17].
Table 1.
List of the strains used in this study and their relevant characteristics. Symbol ‘+’ is normal compared with the wild-type.
| strain/mutation | parental strain | lifespan versus parent | growth rate versus parent | fertility ♀ | fertility ♂ | pigmentation | hyphal density | hyphal growth | reference |
|---|---|---|---|---|---|---|---|---|---|
| Wa32 | wild-type isolate | + | + | + | + | + | [20] | ||
| Wa50 | wild-type isolate | + | + | + | + | + | [20] | ||
| Wa70 | wild-type isolate | + | + | + | + | + | [20] | ||
| Wa76 | wild-type isolate | + | + | + | + | + | [20] | ||
| Wa77 | wild-type isolate | + | + | + | + | + | [20] | ||
| Cs | laboratory strain | + | + | + | + | + | [16] | ||
| ΔPaAtg1 | Cs | 15% reduced | infertile | + | reduced | + | + | [17] | |
| ΔPaAtg8 (idi-7) | Cs | 15% reduced | infertile | + | reduced | + | + | [16] | |
| ΔPaPspA (idi-6) | Cs | infertile | + | reduced | + | + | [16] | ||
| s | laboratory strain | + | + | + | + | + | [18] | ||
| ΔPaUth1 | s | 50% reduced | 50% reduced | ± delayed fruiting body/spore formation | + | + | + | abnormal | this paper |
Figure 4.
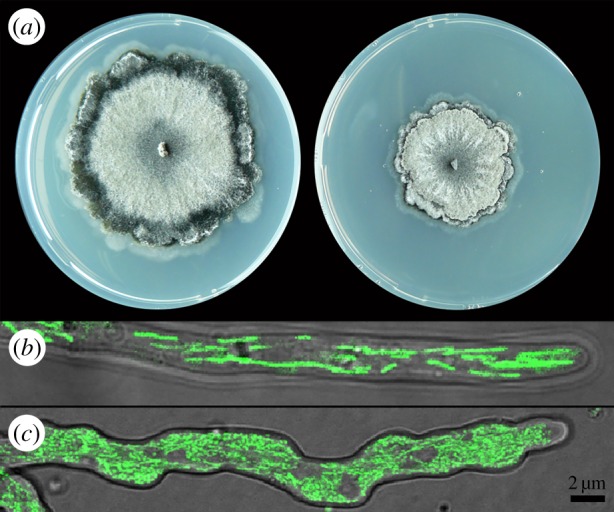
(a) Phenotype of a 5-day-old culture of wild-type collection strain s (left) and of the putative mitophagy-deficient ΔPaUth1 mutant (right) on PASM with 2% glucose and 0.1% urea. (b) Confocal laser microscopy picture of a hyphal tip of strain s with filamentous mitochondria expressing GFP on PASM with 2% glucose and 0.1% urea. (c) Part of an irregular formed hyphal tip of ΔPaUth1 mutant with diffuse mitochondria expressing GFP. (Online version in colour.)
Normal wild-type hyphae have a smooth shape in which the mitochondria form networks that undergo continuous fusion and fission (figure 4b). However, the ΔPaUth1 mutant shows irregular hyphal morphology (figure 4c) with often bulbous deformations at the hyphal tips and branching at sub-apical tip locations initiating further growth of the mycelial front. Occasionally, bursting hyphal tips were observed, even of young cultures (data not shown). This phenomenon could be an indication of early stage death, but this will require additional experimentation. The mitochondria of a ΔPaUth1 lineage crossed with strains expressing GFP in the mitochondria give a diffuse GFP signal throughout the cell without clear mitochondrial networks, but many punctuate units, potentially due to an imbalance between mitochondrial fission and fusion events, and more signal seems to be present within the cell, potentially due to the accumulation of undigested damaged mitochondria (figure 4c).
Explants of the different knock-out mutants derived from 2- to 3-day-old cultures (juvenile samples) were grown on media with normal and either reduced glucose or urea concentrations (figure 5a,b, respectively). The lifespan of mutants grown on glucose-restricted media significantly increased with decreasing glucose concentration (figure 5a; χ2 = 2401.48, d.f. = 2, p < 0.001) showing a lifespan that was similar to their respective wild-type. The exception was ΔPaUth1, which is the only strain in which severe carbon restriction (0.02% (w/v) glucose) does not result in an absolute DR lifespan of well over 90 days, but only of an average of 50 days (post hoc contrast p < 0.001).
Figure 5.
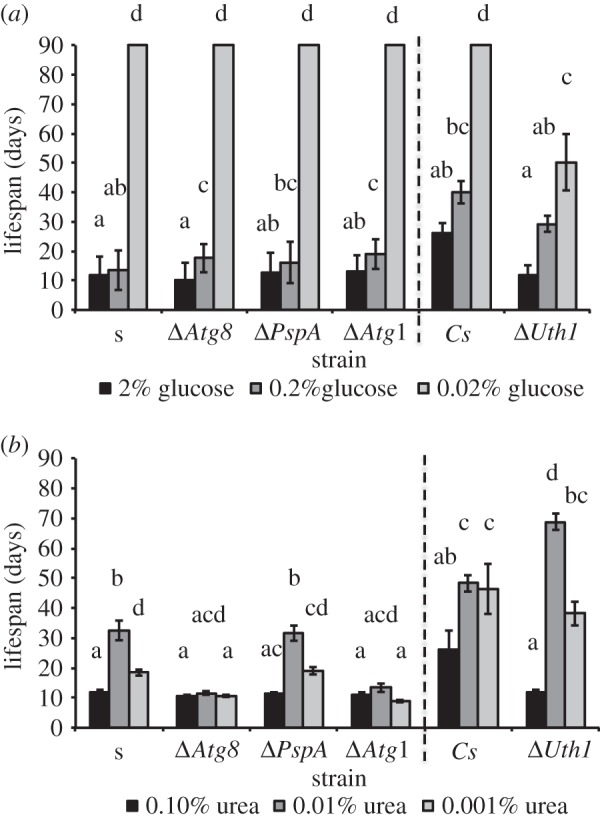
The lifespan of laboratory strains and derived autophagy-deficient mutants ΔPaAtg1, ΔPaAtg8, ΔPaPspA and ΔPaUth1 under glucose (a) and urea (b) restriction, respectively. All autophagy mutants are given to the right of the laboratory strain they originated from. The error bars indicate the standard deviation per experimental condition. Under experimental conditions in which all samples lived beyond 90 days there are no error bars, because the experiment was terminated at this point. The same letter above columns indicates that these groups do not statistically differ in phenotype.
The effect of nitrogen restriction strongly depended on the gene that was mutated (figure 5b; χ2 = 327.42, d.f. = 4, p < 0.001) and on the concentration of urea (figure 5b; χ2 = 201.52, d.f. = 2, p < 0.001). Interestingly, mutants ΔPaAtg1 and ΔPaAtg8 do not show the same lifespan increase as their ancestor. By contrast, the mutants ΔPaUth1 and ΔPaPspA behave similar to their respective ancestors with the longest lifespan at intermediate nitrogen restriction (figure 5b). However, the effect of nitrogen restriction in ΔPaUth1 was much more pronounced than in wild-type with a significantly longer lifespan at intermediate nitrogen restriction (0.01% urea) and a significantly shorter one at the standard growth conditions (figure 5b).
(d). The effect of age on the magnitude of the dietary restriction response
Carbon restriction can increase the lifespan of both wild-type and autophagy/mitophagy-deficient mutants in juvenile samples. But how effective is this carbon restriction response, when encountering lean periods later in life? To answer this question both culture collection strain s and the four autophagy-deficient mutants ΔPaAtg1, ΔPaAtg8, ΔPaPspA and ΔPaUth1 were grown to different relative ages on 2% (w/v) glucose and subsequently transferred to severe carbon-restricted conditions (0.02% (w/v) glucose).
The relative response of wild-type strain s to a switch to severe carbon-restricted conditions was largest: up to a relative age of 30% of the lifespan all lines could be rescued by carbon restriction and transplants lived at least 90 days. Up to halfway through its lifespan, 50% of the transplants showed the typical DR response and after that the effect of DR diminished (fitted survival curves are given in figure 6; see the electronic supplementary material, figure S1 for the individual replicate data points). However, all three autophagy-deficient mutants, ΔPaAtg1, ΔPaAtg8 and ΔPaPspA, lost their ability to extend lifespan in response to DR at a significantly earlier relative age compared with the wild-type. Mutant ΔPaUth1 shows this effect even stronger as already young strains (relative age 0) have a reduced lifespan extension on severe carbon-restricted medium (see above, figure 5a).
Figure 6.
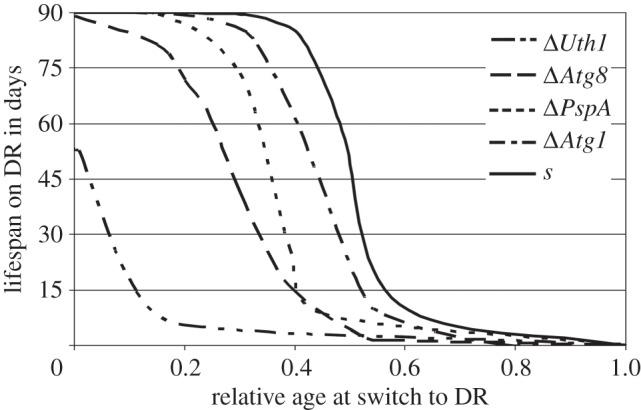
Fitted curves of the DR response at relative ages at onset of DR (PASM with 0.02% w/v glucose) for the autophagy-deficient mutants (ΔPaUth1, ΔPaAtg8, ΔPaPspA andΔPaAtg1), and the wild-type parental strain s (in order of appearance from left to right; growth experiments were stopped after 90 days, p-values fitted curves of less than 0.001). Refer to the electronic supplementary material, figure S1 for the individual replicate data points.
4. Discussion
Extension of lifespan by DR appears to be evolutionarily conserved as it is found in a large range of organisms, from yeasts to potentially primates and humans [1–4,6]. However, the mechanisms through which this occurs in the various organisms are largely unknown. Starvation and other stress conditions induce autophagy, a large-scale lysosomal degradation of (damaged) cytoplasmic content including organelles such as mitochondria. Therefore, a more efficient recycling of damaged mitochondria could be a mechanism to extend lifespan during DR. Here, we show that in the filamentous fungus P. anserina, for which mitochondria have been unambiguously implicated in ageing, DR (both C- and N-restriction) early in life has a profound effect on lifespan, for which autophagy is not required. By contrast, however, for later-onset DR autophagy appears to be essential.
(a). Dietary restriction
Restriction in the amount of available carbon or nitrogen can both lead to lifespan extension—or reduced death rates—in the filamentous fungus P. anserina. There is a maximum increase in lifespan at an optimum amount of urea in the medium that ranges between 0.001 and 0.01% (w/v); further reduction of nitrogen source decreases lifespan by starvation. However, reducing the carbon source results in maximum lifespan at the lowest measured concentration; strains growing on 0.02% (w/v) glucose have been shown to grow continuously for over 3.5 years, more than 50 times the life expectancy of two to three weeks under standard nutrient-rich conditions [31,32]. The typical mitochondrial genome rearrangements correlated with senescence in P. anserina on normal media are also observed in carbon- and nitrogen-restricted conditions. However, for the severe carbon-restricted conditions where in the long-lived strains the mitochondrial stability is enhanced, the prominent rearrangements due to senDNAα are absent and only occasional rearrangements due to other known senDNA regions are observed (for Wa32, [31]; for mutants, AD van Diepeningen 2009, personal observation).
DR extends healthy lifespan in diverse organisms and reduces fecundity under those conditions. In P. anserina, carbon-restricted conditions may reduce reproduction up until effective sterility, but once conditions are favourable again the fertility is often restored and thus reproduction happens well beyond the period in which Podospora reproduces under normal nutritional conditions [31,32]. Grandison et al. [38] showed in D. melanogaster that specific amino acids may restore fertility even under DR conditions, indicating that a somatic lifespan extension through DR may well cause malnutrition with respect to fertility, and that lifespan and reproduction thus do not rely fully on the same specific resources. DR in P. anserina does not only result in reduction in fertility, but also in less dense mycelium with increased (carbon restriction) or decreased (nitrogen restriction) growth rates. Pigmentation under nitrogen restriction is normal or even increased, whereas under carbon restriction it is absent, indicating different responses within the cell to reduction in different food components. These differences are likely related to the changes in mycelial growth and growth rates (‘thinning’ and ‘condensing’ effects).
(b). Autophagy
Autophagy is one of the mechanisms proposed to be involved in the degradation and recycling of (damaged) cell components and thus may have a role in the DR responses. Autophagy was found to be involved in, but neither necessary nor sufficient to cause lifespan extension in C. elegans [13]. Under normal culture conditions, several P. anserina autophagy mutants have no or little adverse effects on lifespan or growth rate although they are infertile [16,17]. However, the ΔPaUth1 mutant shows severe effects on cell and mitochondrion morphology, growth rate and lifespan, while this mutant is still able to reproduce (though sexual spore formation is delayed). In yeast, it was found that UTH1 acts on various cellular pathways beside mitophagy such as the response to oxidative stress [39], mitochondrial biogenesis [40] and apoptosis [41]. The phenotypes of the mutant described in our paper may be due to pleiotropic effects of these or other processes. Moreover, it is not the only gene involved in mitophagy [42].
The mutants deficient at different positions in the mainstream autophagy pathway all show a lifespan-extending effect of carbon restriction, but two—ΔPaAtg1 and ΔPaAtg8—fail to do so under nitrogen-restricted conditions, and thus seem to mediate the effect of nitrogen restriction on lifespan extension. The ΔPaUth1 mutant gave a smaller response to carbon-restricted conditions than its parental strain, but a larger response on nitrogen restriction suggesting that under these conditions UTH1 may actually be involved in processes with an adverse effect on lifespan. Our results indicate that the tested set of different genes from the complex autophagy pathway can be involved in different ways in the response to carbon and/or to nitrogen restriction in P. anserina.
The P. anserina mutant ΔPaUth1 has other characteristics than the ‘youthfull’ Uth1 mitophagy mutant of S. cerevisiae: Uth1 has been identified in yeast based on its enhanced stress resistance and its longer lifespan [33,34], but notably our ΔPaUth1 strain has a shorter lifespan. Microscopically, Uth1-deficient yeasts show no (clear) abnormalities in either cell or mitochondrial morphology [34]. However, the ΔPaUth1 P. anserina strain has a very irregular growth and shows many malformed hyphal tips, while its mitochondrial networks are severely reduced, and many punctuate mitochondria are present. These punctuate mitochondria may be the result of a lack of autophagic degradation as they may be the units normally tagged for the autophagy pathway and are reminiscent of aged cells [43].
Clearly, autophagy is not the only mechanism involved in DR responses, nor is the involvement the same when different resources are limited. Proteolytic enzymes might well provide complementary and partly redundant mechanisms. In P. anserina, autophagy has been linked not only to DR, but also to reactions that resemble programmed cell death when two different strains undergo hyphal fusions (a.k.a. heterokaryon or vegetative incompatibility reactions; [15–17]). Also, proteolytic activity has been shown to increase during such incompatibility reactions [14]. In turn, programmed cell death in yeast has been linked to regulating mitochondrial fission proteins [44], which lead us back to the observed punctuated mitochondria in our ΔPaUth1 strain. Thus, ΔPaUth1 may interfere with the mitochondrial fusion and fission processes.
(c). Timing of dietary restriction
DR leads to lifespan extension through a still to be unravelled mechanism and generally has the consequence of severe but often reversible reductions in fecundity. But when should one start with DR to have an optimal lifespan? In D. melanogaster, DR has been shown to be effective already after 48 h and reduced the short-time risk of death [3]. Note though, that in this study the effect was measured until the age of 28 days, which is well before the mortality rate starts to increase under the standard culture conditions for Drosophila. Here, for P. anserina however, we investigated the DR response over the whole lifespan, and show that only when a wild-type strain is switched to DR in the first half of its life when it is also still fully fertile, will it acquire the near ‘immortal’ lifespan typical for growth under DR. Later in life, a switch to DR results only in a slight increase in life expectancy of a mere few days.
Our data thus show that the DR response is age-dependent in P. anserina. It is not clear why Podospora is only showing such a DR response early in life. Maybe its growth on herbivore dung, a short-lived substrate, has dictated strong selection for early reproduction and an early potential DR response. Under such a scenario, selection for an effective late-life DR response would have been weak as at that point in life the fungus has normally already reproduced. Such weak selection would allow accumulation of mutations and/or damage to the cellular machinery essential for the response with age, both within the lifespan of the fungus as well as over evolutionary time. Combining this reasoning with the age-dependent responses of the wild-type and mutant strains results in the model depicted in figure 7.
Figure 7.
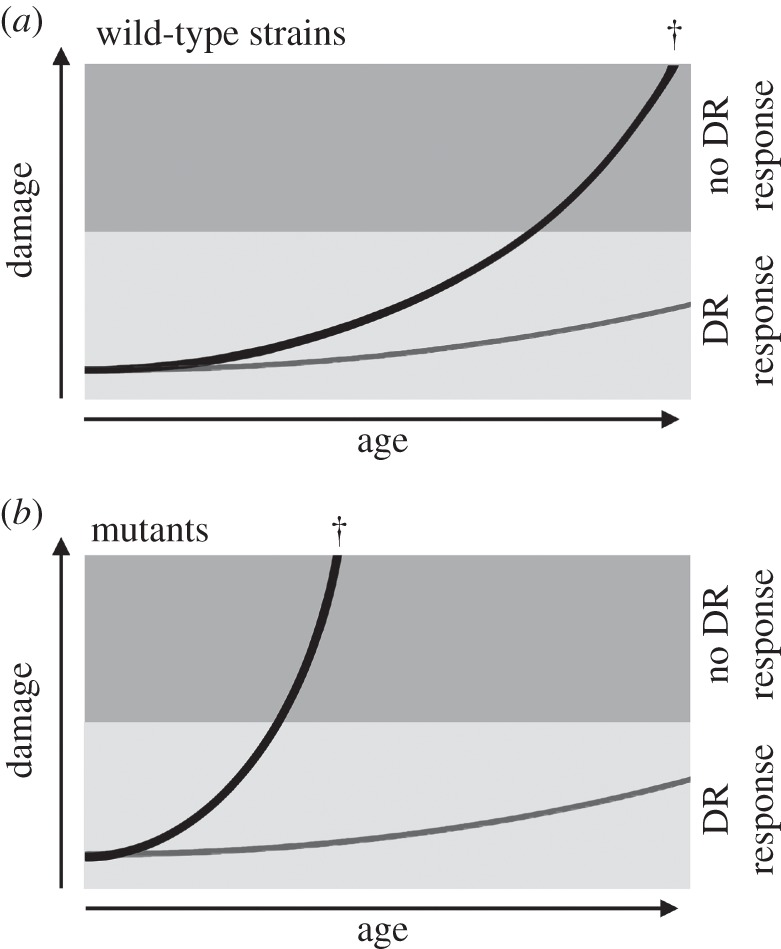
Models of damage accumulation over time for wild-type (a) and autophagy mutant (b) strains growing under nutrient-rich (black line) and calorie-restricted (grey line) conditions. The shaded areas depict the damage levels in which a lifespan extending DR effect is (light shade) or is no longer (darker shade) possible. Normal accumulation of damage under nutrient-rich conditions will lead to a threshold level of damage after which cell death (cross symbol) becomes inevitable. Below such damage levels DR may postpone senescence phenomena and cell death for a long time. Mutants impaired in their autophagy pathway will show faster ageing processes under nutrient-rich conditions, resulting in a lack of response to DR conditions earlier in the life time of the mutants.
In this model, in a wild-type strain grown under dietary-rich conditions damage will be accumulating with age, finally resulting in cell death (figure 7a, black line). Switching to dietary-restricted conditions will result in lifespan extension up to a certain point in life, a threshold, after which there will be too much accumulated damage and switching will only result in a very limited lifespan-extending response. The same strain grown under DR conditions will not accumulate as much damage and will reach the critical accumulated damage for DR response and cell death at a much later time point, if at all (figure 7a, grey line).
The fact that autophagy-deficient strains of P. anserina show a reduced lifespan-extending effect of a switch to DR suggests that during early onset DR in the fungus P. anserina autophagy forestalls ageing by preventing the critical accumulation of highly suppressive damaged mitochondria leading to a senescent state that is culminating in cell death (figure 7b, grey line). A switch to DR late in life apparently cannot revert this ageing process by autophagic removal of accumulated damage in P. anserina. In the model, an autophagy-deficient mutant will accumulate damaged cell components faster and thus also get to the threshold point earlier after which DR will no longer be leading to a strong lifespan-extending response (figure 7b, black line). Critically, grown under DR conditions from the start there will be little difference between mutant and wild-type strains (figure 7a,b).
Our results on the involvement of autophagy genes in the reproduction and lifespan response to DR thus indicate that there is both a direct, for nitrogen restriction, and indirect, for the late-onset response, connection. We hypothesize that the DR response depends on an intact (mitochondrial) cellular machinery as the DR effects are limited to the first half of the lifespan and are truncated in all the autophagy mutants. As a consequence, our results are the first to indicate that the normal ageing process and the response to DR conditions are linked, at least in P. anserina.
Supplementary Material
Acknowledgements
We thank Dr Bérangère Pinan-Lucarre and Dr Corinne Clavé from the Centre de Génétique Moléculaire of the Université de Bordeaux 2, in France for kindly providing the three P. anserina autophagy-deficient strains.
Funding statement
This research was part of the MiMage project on the role of mitochondria in conserved mechanisms of ageing and supported by funding from the Community's Sixth Framework Programme (LSHM-CT-2004-512020). This work was further supported by the European Union's FP7 Programme (IDEAL FP7/2007-2011/259679, http://www.ideal-ageing.eu/, to B.J.Z.).
References
- 1.Lane MA, Black A, Handy A, Tilmont EM, Ingram DK, Roth GS. 2001. Caloric restriction in primates. Ann. NY Acad. Sci. 928, 287–295. ( 10.1111/j.1749-6632.2001.tb05658.x) [DOI] [PubMed] [Google Scholar]
- 2.Weindruch RH, Walford RL. 1988. The retardation of aging and disease by dietary restriction. Springfield, IL: CC Thomas. [Google Scholar]
- 3.Mair W, Goymer P, Pletcher SD, Partridge L. 2003. Demography of dietary restriction and death in Drosophila. Science 301, 1731–1733. ( 10.1126/science.1086016) [DOI] [PubMed] [Google Scholar]
- 4.Partridge L, Gems D. 2006. Beyond the evolutionary theory of ageing, from functional genomics to evo-gero. Trends Ecol. Evol. 21, 334–340. ( 10.1016/j.tree.2006.02.008) [DOI] [PubMed] [Google Scholar]
- 5.Guarente L, Kenyon C. 2000. Genetic pathways that regulate ageing in model organisms. Nature 408, 255–262. ( 10.1038/35041700) [DOI] [PubMed] [Google Scholar]
- 6.Shanley DP, Kirkwood TB. 2000. Calorie restriction and aging: a life-history analysis. Evolution 54, 740–750. [DOI] [PubMed] [Google Scholar]
- 7.Mair W, Piper MDW, Partridge L. 2005. Calories do not explain extension of lifespan by dietary restriction in Drosophila. PLoS Biol. 3, e223 ( 10.1371/journal.pbio.0030223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harman D. 1981. The aging process. Proc. Natl Acad. Sci. USA 78, 7124–7128. (doi:10.1073/pnas.78.11.7124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thornalley PJ, Langborg A, Minhas HS. 1999. Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose . Biochem. J. 344, 109–116. ( 10.1042/0264-6021:3440109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koubova J, Guarente L. 2003. How does calorie restriction work? Genes Dev. 17, 313–321. ( 10.1101/gad.1052903) [DOI] [PubMed] [Google Scholar]
- 11.Levine B, Klionsky DJ. 2004. Development by self-digestion, molecular mechanisms and biological functions of autophagy. Dev. Cell. 6, 463–477. ( 10.1016/S1534-5807(04)00099-1) [DOI] [PubMed] [Google Scholar]
- 12.Shintani T, Klionsky DJ. 2004. Autophagy in health and disease, a double-edged sword. Science 306, 990–995. ( 10.1126/science.1099993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. 2008. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 4, e24 ( 10.1371/journal.pgen.0040024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bégueret J. 1972. Protoplasmic incompatibility: possible involvement of proteolytic enzymes. Nature 235, 56–58. ( 10.1038/newbio235056a0) [DOI] [PubMed] [Google Scholar]
- 15.Démenthon K, Paoletti M, Pinan-Lucarré B, Loubradou-Bourges N, Sabourin M, Saupe SJ, Clavé C. 2003. Rapamycin mimics the incompatibility reaction in the fungus Podospora anserina. Eukaryotic Cell 2, 238–246. ( 10.1128/EC.2.2.238-246.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinan-Lucarré B, Paoletti M, Dementhon K, Coulary-Salin B. 2003. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol. Microbiol. 47, 321–333. ( 10.1046/j.1365-2958.2003.03208.x) [DOI] [PubMed] [Google Scholar]
- 17.Pinan-Lucarré B, Balguerie A, Clavé C. 2005. Accelerated cell death in Podospora autophagy mutants. Eukaryotic Cell 4, 1765–1774. ( 10.1128/EC.4.11.1765-1774.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizet G. 1953. Sur la longévité des souches de P. anserine. CR. Acad. Sci. 237, 1106–1109. [PubMed] [Google Scholar]
- 19.Geydan TD, Debets AJM, Verkley GJ, van Diepeningen AD. 2012. Correlated evolution of senescence and ephemeral substrate use in the Sordariomycetes. Mol. Ecol. 21, 2816–2828. ( 10.1111/j.1365-294X.2012.05569.x) [DOI] [PubMed] [Google Scholar]
- 20.van der Gaag M, Debets AJM, Osiewacz HD, Hoekstra RF. 1998. The dynamics of pAL2–1 homologous linear plasmids in Podospora anserina. Mol. Gen. Genet. 258, 521–529. ( 10.1007/s004380050763) [DOI] [PubMed] [Google Scholar]
- 21.van Diepeningen AD, Debets AJM, Slakhorst SM, Hoekstra RF. 2008. Mitochondrial pAL2–1 plasmid homologues are senescence factors in Podospora anserina independent of intrinsic senescence. Biotechnol. J. 3, 791–802. ( 10.1002/biot.200800005) [DOI] [PubMed] [Google Scholar]
- 22.Griffiths AJF. 1992. Fungal senescence. Annu. Rev. Genet. 26, 351–372. ( 10.1146/annurev.ge.26.120192.002031) [DOI] [PubMed] [Google Scholar]
- 23.Silar P, Lalucque H, Vierny C. 2001. Cell degeneration in the model system Podospora anserina. Biogerontology 2, 1–17. ( 10.1023/A:1010000816277) [DOI] [PubMed] [Google Scholar]
- 24.Tudzynski P, Stahl U, Kück U, Esser K. 1980. Senescence in Podospora anserina. Gerontologist 20, 215. [Google Scholar]
- 25.Lorin S, Dufour E, Sainsard-Chanet A. 2006. Mitochondrial metabolism and aging in the filamentous fungus Podospora anserina. Biochim. Biophys. Acta 1757, 604–610. ( 10.1016/j.bbabio.2006.03.005) [DOI] [PubMed] [Google Scholar]
- 26.Belcour L, Sainsard-Chanet A, Sellem CH. 1994. Mobile group II introns, DNA circles, reverse transcriptase and senescence. Genetica 93, 225–228. ( 10.1007/BF01435254) [DOI] [PubMed] [Google Scholar]
- 27.Jamet-Vierny C, Begel O, Belcour L. 1980. Senescence in Podospora anserina—amplification of a mitochondrial-DNA sequence. Cell 21, 189–194. ( 10.1016/0092-8674(80)90126-9) [DOI] [PubMed] [Google Scholar]
- 28.Stahl U, Lemke PA, Tudzynski P, Kuck U, Esser K. 1978. Evidence for plasmid like DNA in a filamentous fungus, ascomycete Podospora anserina. Mol. Gen. Genet. 162, 341–343. ( 10.1007/BF00268860) [DOI] [PubMed] [Google Scholar]
- 29.Hermanns J, Osiewacz HD. 1996. Induction of longevity by cytoplasmic transfer of a linear plasmid in Podospora anserina. Curr Genet. 29, 250–256. ( 10.1007/BF02221555) [DOI] [PubMed] [Google Scholar]
- 30.Maas MFPM, de Boer HJ, Debets AJM, Hoekstra RF. 2004. The mitochondrial plasmid pAL2–1 reduces calorie restriction mediated lifespan extension in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41, 865–871. ( 10.1016/j.fgb.2004.04.007) [DOI] [PubMed] [Google Scholar]
- 31.van Diepeningen AD, et al. 2010. Calorie restriction causes healthy lifespan extension in the filamentous fungus Podospora anserina. Mech. Ageing Dev. 131, 60–68. ( 10.1016/j.mad.2009.12.002) [DOI] [PubMed] [Google Scholar]
- 32.van Diepeningen AD, Slakhorst SM, Koopmanschap AB, Ikink GJ, Debets AJM, Hoekstra RF. 2010. Calorie restriction in the filamentous fungus Podospora anserina. Exp. Gerontol. 45, 516–525. ( 10.1016/j.exger.2010.01.002) [DOI] [PubMed] [Google Scholar]
- 33.Camougrand N, Kissová I, Velours G, Manon S. 2004. Uth1p: a yeast mitochondrial protein at the crossroads of stress, degradation and cell death. FEMS Yeast Res. 5, 133–140. ( 10.1016/j.femsyr.2004.05.001) [DOI] [PubMed] [Google Scholar]
- 34.Kissová I, Deffieu M, Manon S, Camougrand N. 2004. Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 279, 39 068–39 074. ( 10.1074/jbc.M406960200) [DOI] [PubMed] [Google Scholar]
- 35.El-Khoury R, Sellem CH, Coppin E, Boivin A, Maas MFPM, Debuchy R, Sainsard-Chanet A. 2008. Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr. Genet. 53, 249–258. ( 10.1007/s00294-008-0180-3) [DOI] [PubMed] [Google Scholar]
- 36.Nakatogawa H, Ichimura Y, Ohsumi Y. 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178. ( 10.1016/j.cell.2007.05.021) [DOI] [PubMed] [Google Scholar]
- 37.Sellem CH, Marsy S, Boivin A, Lemaire C, Sainsard-Chanet A. 2007. A mutation in the gene encoding cytochrome c1 leads to a decreased ROS content and to a long-lived phenotype in the filamentous fungus Podospora anserina. Fung. Genet. Biol. 44, 648–658. ( 10.1016/j.fgb.2006.09.005) [DOI] [PubMed] [Google Scholar]
- 38.Grandison RC, Piper MDW, Partridge L. 2009. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila . Nature 462, 1061–1064. ( 10.1038/nature08619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandara PDS, Flattery-O'Brien JA, Grant CM, Dawes IW. 1998. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Curr. Genet. 34, 259–268. ( 10.1007/s002940050395) [DOI] [PubMed] [Google Scholar]
- 40.Camougrand NM, Mouassite M, Velours GM, Guérin MG. 2000. The ‘SUN’ family: UTH1, an ageing gene, is also involved in the regulation of mitochondria biogenesis in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 375, 154–160. ( 10.1006/abbi.1999.1655) [DOI] [PubMed] [Google Scholar]
- 41.Camougrand N, Grelaud-Coq A, Marza E, Priault M, Bessoule JJ, Manon S. 2003. The product of the UTH1 gene, required for Bax-induced cell death in yeast, is involved in the response to rapamycin. Mol. Microbiol. 47, 495–506. ( 10.1046/j.1365-2958.2003.03311.x) [DOI] [PubMed] [Google Scholar]
- 42.Kissová I, Salin B, Schaeffer J, Bhatia S, Manon S, Camougrand N. 2007. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy 3, 329–336. [DOI] [PubMed] [Google Scholar]
- 43.Bossy-Wetzel E, Barsoum MJ, Godzik A, Schwarzenbacher R, Lipton SA. 2003. Mitochondrial fission in apoptosis, neurodegeneration and aging. Curr. Opin. Cell Biol. 15, 706–716. ( 10.1016/j.ceb.2003.10.015) [DOI] [PubMed] [Google Scholar]
- 44.Fannjiang Y, Chen WC, Lee SJ, Qi B, Pevsner J, Caffery JM, Hill RB, Basanez G, Hardwick JM. 2004. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 18, 2785–2797. ( 10.1101/gad.1247904) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


