Abstract
The heterodimeric nuclear factor 90/nuclear factor 45 complex (NF90/NF45) binds nucleic acids and is a multifunctional regulator of gene expression. Here we report that depletion of NF90/NF45 restores the expression of the p53 and p21 proteins in cervical carcinoma cells infected with high-risk human papillomaviruses (HPV). Knockdown of either NF90 or NF45 by RNA interference led to greatly elevated levels of p53 and p21 proteins in HPV-derived HeLa and SiHa cells, but not in other cancerous or normal cell lines. In HeLa cells, p21 mRNA increased concomitantly but the level of p53 mRNA was unaffected. RNA interference directed against p53 prevented the induction of both proteins. These results indicated that the up-regulation of p21 is due to p53-dependent transcription, whereas p53 is regulated post-transcriptionally. Proteasome-mediated turnover of p53 is accelerated by the HPV E6 and cellular E6AP proteins. We therefore examined the hypothesis that this pathway is regulated by NF90/NF45. Indeed, depletion of NF90 attenuated the expression of E6 RNA and inhibited transcription from the HPV early promoter, revealing a new role for NF90/NF45 in HPV gene expression. The transcription inhibition was largely independent of the reduction of P-TEFb levels caused by NF90 depletion. Consistent with p53 derepression, NF90/NF45-depleted HeLa cells displayed elevated PARP cleavage and susceptibility to camptothecin-induced apoptosis. We conclude that high-risk strains of HPV utilize the cellular NF90/NF45 complex for viral E6 expression in infected cervical carcinoma cell lines. Interference with NF90/NF45 function could assist in controlling cervical carcinoma.
Keywords: Apoptosis, human papillomavirus, transcription, p53, p21, NF90/NF45
Introduction
The cellular proteins p53 and p21 are crucial regulators of cell proliferation and genome stability (1-4). p21 is a cyclin-dependent kinase inhibitor that plays a pivotal role in cell cycle control. Largely regulated at the transcriptional level, it is induced by p53 as well as by p53-independent mechanisms (5). p53 is a potent tumor suppressor that activates p21 transcription in response to stress signals (6, 7) via two highly conserved p53-responsive promoter elements that bind p53 directly (5). p21 transcription is also induced by transcription factors such as SP1 and SP3 in response to external and internal stimuli. Increased levels of p21 bring about cell cycle arrest and senescence.
The p53 gene, TP53, is regulated at multiple levels, including by factors that stabilize the protein and prevent its degradation (3, 6, 8). Normally p53 is a short-lived protein, sometimes undetectable in cells except under stress conditions when it accumulates in the nucleus and functions as a transcription factor. Approximately 50% of primary tumors contain TP53 mutations, most of which lead to inactivation of the protein's DNA binding function and allow continued cell proliferation. p53 can bind to MDM2, which interacts with its activation domain and prevents it from activating downstream genes. MDM2 also functions as a ubiquitin-protein ligase targeting p53 for degradation by the proteasome (3, 6, 7, 9, 10).
Viral infection is a stressful event that induces p53 production (11, 12). Many DNA viruses that have the potential to cause cell transformation encode proteins that bind and inactivate p53 (6). The SV40 T-antigen and adenovirus E1B-55K proteins sequester p53 in non-functional complexes, whereas the human papilloma virus (HPV) E6 protein directs p53 to the proteasome for degradation (6, 13). HPV is a small non-enveloped dsDNA virus that establishes productive infections in stratified epithelium of skin and mucous membranes. High-risk HPVs, notably HPV-16 and -18, are implicated in the development of invasive cervical carcinomas. In most primary cervical carcinomas and cell lines established from them, such as HeLa and SiHa, HPV DNA is integrated into the cell genome and expresses HPV E6 and E7 proteins. These viral proteins are responsible for the malignant transformation of fibroblasts and keratinocytes (14-17).
Nuclear factor 90 (NF90) is a nucleic acid binding protein that forms a heterodimeric complex with nuclear factor 45 (NF45) (18). These proteins are products of the interleukin enhancer-binding factor-3 and -2 genes, ILF3 and ILF2, both of which are widespread in vertebrates. The NF90/NF45 core complex forms larger complexes together with NF110, another ILF3 isoform, and several additional proteins, and regulates cellular gene expression at several levels. It has been implicated in DNA metabolism (19, 20), transcription (21, 22), translation (23, 24), RNA export (25, 26) and microRNA biogenesis (27), and in the replication and gene expression of a number of viruses (28-34). NF90 contains two dsRNA-binding domains and an RGG motif with nucleic acid binding capability; a bipartite nuclear localization signal and a nuclear export signal which promote nucleo-cytoplasmic shuttling; and a DZF domain encompassing a NF45 homology domain necessary for NF45 binding (18, 35, 36). NF90 is largely nuclear during interphase (37) and its phosphorylation correlates with nucleo-cytoplasmic translocation (38-40). The stability of NF90 and NF110 is regulated by NF45, a shorter-lived protein that also participates in multiple cellular functions (18, 19, 22, 41).
In mice, knockout of the ILF3 gene results in prenatal (42) or perinatal (43) lethality due to muscle degeneration, apoptosis and respiratory failure (43). ILF3-/- mice exhibited an ∼50% decrease in p21 RNA and a corresponding reduction in p21 protein in neonatal skeletal muscle (43). Interactions between NF90 and the p21 3′UTR suggested regulation via mRNA stability (43), consistent with other reports of NF90-mRNA 3′UTR interactions (23, 24, 44-48). In HeLa cells, depletion of either NF90 or NF45 by RNA interference slows cell growth and leads to a multinucleated phenotype (18, 19). These observations led us to examine the effect of NF90/NF45 knockdown on p21 expression in HeLa cells.
In contrast to findings with ILF3-/- mice, we observed that depletion of NF90 or NF45 increased the expression of p21. Up-regulation occurred at both the protein and mRNA levels. Concomitantly the level of p53 protein increased dramatically although p53 mRNA levels were unchanged. We found that the increase in p21 expression is dependent on p53, and that the elevation of p53 expression in response to NF90/NF45 depletion is restricted to HPV-infected cervical carcinoma cells. These findings suggested that NF90/NF45 may be necessary for p53 destruction mediated by HPV E6. Accordingly, NF90/NF45 depletion reduced the level of E6 RNA and transcription driven by the HPV early promoter. Increased p53 can drive malignant cells to apoptosis especially if they are subjected to genotoxic stress (49). Correspondingly, NF90/NF45-depleted HeLa cells displayed an increase in apoptotic markers and in apoptosis resulting from treatment with the topoisomerase I inhibitor and anticancer drug camptothecin. Our data show that NF90/NF45 is required for effective transcription of the HPV E6 gene. The mechanism is partly, but not chiefly, dependent on the positive transcription elongation factor P-TEFb. Stabilizing p53 by inhibiting E6 production can sensitize even highly resistant malignant cells to apoptotic cell death caused by genotoxic drugs.
Results
Induction of p21 by NF90/NF45 depletion
Long-term depletion of the NF90/NF45 complex in HeLa cells leads to a multinucleated cell phenotype (18). We considered the possibility that p21 might be involved in this process because reduction in the level of p21 is associated with formation of multinucleated cells (50) and p21 expression is reduced in ILF3-/- mice (43). To examine the role of NF90 and NF45 in p21 regulation, we transfected HeLa cells with siRNA specific for NF90 (D3) or NF45 (D5) or with control siRNA (C). RT-PCR and immunoblotting analyses showed that the siRNAs specifically depleted their target RNAs (Fig. 1a) and proteins (Fig. 1b). In addition, NF45 was substantially depleted in D3-treated cells and NF90 was partially depleted in D5-treated cells (Fig. 1b). This co-regulation results from the mutual stabilization of the proteins in a heterodimeric NF90/NF45 complex (18). Remarkably, NF90/NF45 depletion by either D3 or D5 increased the level of p21 RNA and protein by up to 7 fold (Figs. 1a,b and 2a,b). Knockdown of the NF110 isoform did not alter p21 expression, however (Supplementary Fig. S1). Permanent cell lines depleted for NF90 and NF45 also exhibited p21 over-expression (data not shown). These observations imply that NF90/NF45 exerts a negative effect on p21 gene expression in HeLa cells, contrary to results obtained with ILF3-/- mice lacking both NF90 and NF110.
Fig. 1. Knockdown of NF90 or NF45 causes p21 up-regulation.
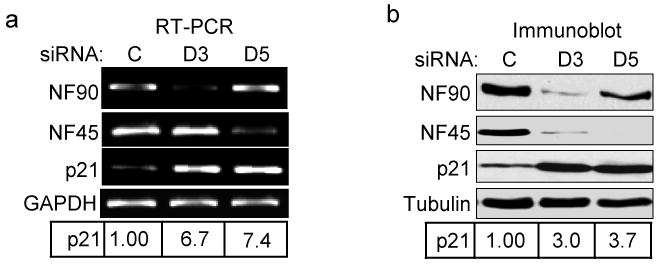
HeLa cells were transfected with siRNA directed against NF90 (D3) or NF45 (D5) or control sequence (C). (a) RT-PCR amplified products indicating levels of NF90, NF45, p21 and GAPDH RNA. (b) Immunoblots showing induction of p21 protein in NF90 and NF45 knockdown cells compared to tubulin. Relative expression of p21 RNA and protein in the experiments shown was normalized to GAPDH and tubulin, respectively.
Fig. 2. Induction of p53 and p21 by NF90/NF45 depletion in HeLa cells.
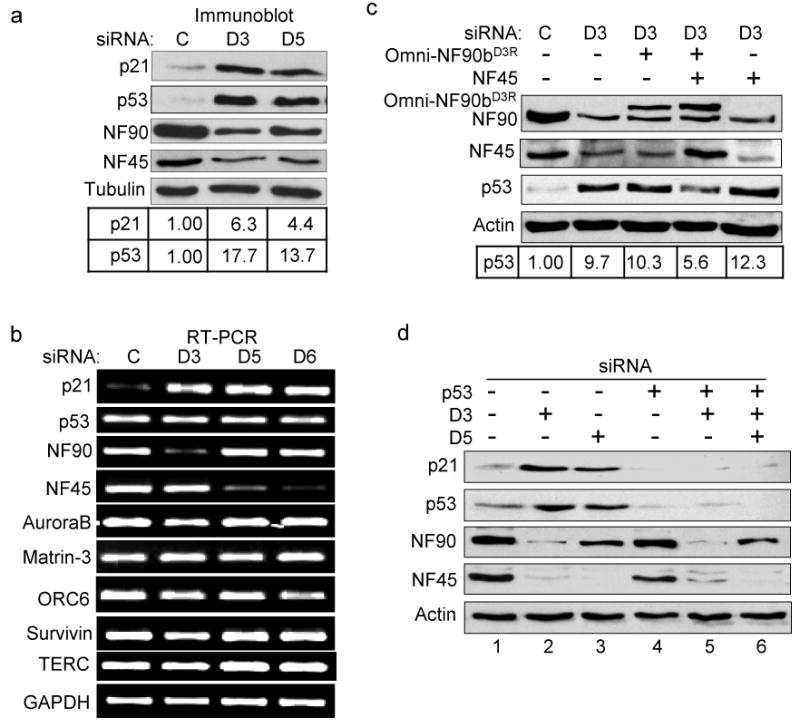
(a) Immunoblots showing the induction of p21 and p53 proteins in NF90 and NF45 knockdown cells. Relative expression of p21 and p53 proteins were normalized to tubulin. (b) RT-PCR analysis showing RNA levels of cellular genes in siRNA-transfected cells in NF90 and NF45 knockdown cells (D6 is a NF45-specific siRNA). (c) Expression of p53 depends on NF90/NF45. Immunoblots from cells transfected with siRNA to deplete the NF90/NF45 complex, in the absence or presence of plasmids expressing siRNA-resistant-NF90b (Omni-NF90bD3R) and over-expressing NF45. Relative expression of p53 protein was normalized to actin. (d) p53 regulates the expression of p21 in NF90/NF45-depleted cells. Immunoblots show that p53-specific siRNA prevents the induction of both p53 and p21 in cells transfected with D3 or D5.
NF90/NF45 regulates p53 in HeLa cells
p21 is under transcriptional regulation by p53, which is present at a low level in HeLa cells as a result of accelerated protein turnover (6, 13). We therefore examined the expression of p53 in NF90/NF45-depleted cells. Concomitant with the induction of p21, treatment with D3 or D5 dramatically increased the level of p53 by 13-18 fold (Fig. 2a). This action was not accompanied by an increase in p53 RNA, however (Fig. 2b). We also monitored the levels of several other RNAs encoding proteins involved in mitosis, particularly in abscission, cell architecture and the generation of multinucleated cells (50-53). No change was seen in the levels of RNAs encoding aurora B, matrin-3, ORC-6 or survivin, or the RNA component of telomerase, TERC (Fig. 2b). Similar results were obtained with D6, another siRNA that targets NF45 (Fig. 2b). As a further control, we conducted a “rescue” experiment. Expression of siRNA-resistant-NF90b (Omni-NF90bD3R), together with overexpression of NF45, suppressed the induction of p53 by D3 by about 50% (Fig. 2c). Neither alone was sufficient, indicating that the proteins are functioning as a complex. Taken together, these results show that depletion of the NF90/NF45 complex induces the expression of p21 RNA and protein, and of p53 protein.
Transcription of the p21 gene is modulated by p53 and by other factors. To determine whether the observed induction of p21 is due to the elevated level of p53, we simultaneously depleted cells for p53 and NF90/NF45. Immunoblotting showed that siRNA targeting p53 effectively abrogated p53 accumulation and eliminated the induction of p21 by either D3 or D5 (Fig. 2d; compare lanes 5 and 6 with lanes 2 and 3, respectively). Similar results were observed at the RNA level (data not shown). We conclude that NF90/NF45 exerts negative posttranscriptional control over p53 in HeLa cells, with a consequent secondary effect on p21 transcription.
Cell-type specificity
To assess the generality of this regulation, we evaluated the effect of NF90/NF45 on p21 and p53 levels in a panel of cell types. Cells were derived from cervical carcinoma (SiHa and C33A), osteosarcoma (U2OS), normal lung (WI38), and colon carcinoma (HCT 116). Immunoblotting showed that D3 depleted NF90 and NF45 in all cases, albeit to varying degrees, but only in SiHa cells were p21 and p53 induced (Fig. 3). Like HeLa cells, SiHa cells are derived from human cervix via transformation by high-risk human papillomaviruses (HPV-18 in HeLa and HPV-16 in SiHa). Notably, neither p53 nor p21 was induced in C33A cervical carcinoma cells, which are negative for HPV infection. These results implicate HPV in the NF90/NF45-mediated regulation of p21 and p53. Consistent with the p53 requirement for p21 induction observed above (Fig. 2d), NF90 knockdown by D3 failed to increase the level of p21 in p53-/- HCT 116 cells.
Fig. 3. Regulation of p21 and p53 expression by NF90/NF45 in HPV-infected cells.
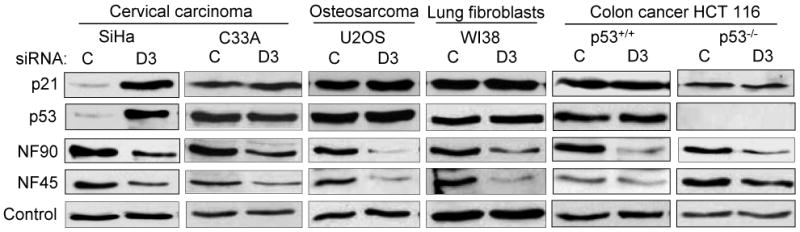
Immunoblots showing the induction of p21 and p53 in cervical carcinoma cells derived from HPV16 infection (SiHa) – but not in non-HPV cervical carcinoma cells (C33A), non-cervical carcinoma cells (U2OS, HCT116) or normal lung fibroblasts (WI38) – depleted for NF90/NF45 complex by D3 transfection.
NF90/NF45 regulates the expression of HPV E6
It is well established that HPV infection induces the rapid turnover of cellular p53 through interactions with the viral E6 oncogene and the cellular ubiquitin-protein ligase E6AP (54, 55). The E6 protein forms a complex with E6AP, also known as UBE3A, altering its substrate specificity and targeting p53 for degradation in the proteasome (Fig. 4a). We therefore examined the effect of NF90/NF45 depletion on E6 and E6AP. RT-PCR analysis showed that D3 siRNA reduced the expression of viral E6 RNA by ∼90% but had no effect on E6AP RNA (Fig. 4b, left panel). We were unable to visualize E6 protein by immunoblotting, but found E6AP protein to be unaffected by NF90 knockdown (Fig. 4b, right panel).
Fig. 4. NF90 regulates HPV E6 oncogene expression.
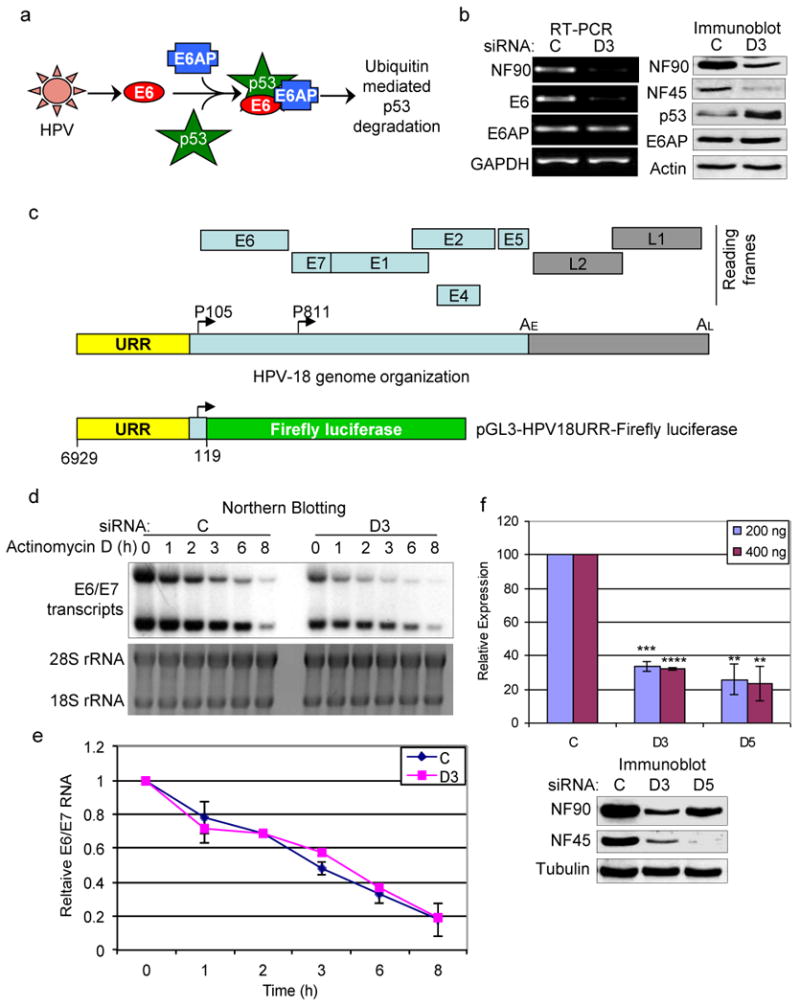
(a) Schematic showing HPV-mediated p53 degradation via E6 and E6AP. (b) Knockdown of NF90 suppresses the expression of HPV18 E6 RNA in HeLa cells. Left panel: RT-PCR indicating the levels of viral E6 and cellular E6AP RNA in siRNA-transfected cells. Right panel: immunoblots showing the expression of proteins. (c) Diagrams of HPV18 genome organization (linear form) and open reading frames (top) (105) and of pGL3-HPV18URR-Firefly luciferase reporter construct (bottom). URR, upstream regulatory region; P105, early promoter; P811, late promoter; AE, early polyadenylation site; AL, late polyadenylation site; E1, E2, E4, E5, E6 and E7: early genes; L1 and L2, late genes. (d) NF90/NF45 depletion does not destabilize E6 RNA. Top panel: northern blot showing E6/E7 RNA in siRNA-transfected HeLa cells treated with actinomycin D (0.5 μg/ml) compared to untreated cells (0 h time point). Lower panel: 18S and 28S ribosomal RNA stained with ethidium bromide. (e) RNA half-life estimation. Quantitation of E6/E7 transcripts from two independent northern blotting experiments as in panel (d). (f) Depletion of NF90/NF45 inhibits the HPV18 early promoter. Expression of Firefly luciferase from pGL3-HPV18URR-Firefly luciferase plasmid normalized to Renilla luciferase expression in HeLa cells transfected with C, D3 and D5 siRNA (n=4 independent experiments). Standard deviations and P values are relative to respective controls: **, < 0.01; ***, < 0.001; ****, < 0.0001. Similar results were obtained with 600 ng reporter plasmid. Immunoblot analysis for a representative experiment is shown.
The reduced level of E6 RNA could be due to decreased transcription of the HPV genome (Fig. 4c), increased RNA degradation, or both. NF90 interacts with several mRNAs, including those encoding the HPV early proteins E6 and E7 (Supplementary Fig. S2), and it plays a role in RNA decay (23, 38, 40, 44, 46, 47, 56). To monitor RNA decay, RNA was isolated from control and D3-treated HeLa cells at varying times after inhibition of transcription by actinomycin D. Analysis by northern blotting resolved two bands containing E6 RNA, which had roughly equivalent intensity at time zero (Fig. 4d). Comparable to RT-PCR results (Fig. 4b), D3 reduced the overall level of E6 RNA by ∼66% in untreated cells (Fig. 4d; compare C and D3 at t=0 h). The upper band decayed somewhat faster than the lower band but the turnover rate was the same in control and D3-treated cells (t1/2 ∼3 h; Fig. 4d,e). These data indicate that NF90 does not significantly affect the stability of viral RNA.
To monitor transcription from the HPV promoter, we measured the effect of NF90/NF45 depletion on the expression of a firefly luciferase reporter driven by the HPV-18 upstream regulatory region (URR) responsible for E6 RNA synthesis (Fig. 4c). Renilla luciferase directed by the Rous sarcoma virus (RSV) promoter, which is not influenced by NF90 (21, 57), served as transfection control. At all concentrations tested, firefly luciferase expression was compromised in HeLa cells transfected with D3 or D5 siRNA by 60-70% when compared to control cells (Fig. 4f). No significant inhibition was observed in HPV-negative C33A cells (data not shown). The attenuated expression in NF90/NF45-depleted HeLa cells corresponds to the reduced level of E6/E7 RNA in these cells, indicating that HPV transcription is impaired. We therefore conclude that NF90/NF45 up-regulates the activity of the HPV early promoter.
Involvement of P-TEFb
The cellular transcription factor P-TEFb, composed of cyclin T1 and CDK9, is important for HPV E6/E7 expression (58). Furthermore, recent data from our lab showed that its cyclin T1 subunit is regulated by NF90 (24). These findings suggested that P-TEFb may be involved in the regulation of E6 expression by NF90/NF45. To evaluate this possibility, we first examined the effects of siRNA-mediated cyclin T1 depletion. Immunoblotting showed that cyclin T1 siRNA reduced the level of cyclin T1 and consequently of CDK9 (24), but not of NF90, by 72 h post-transfection (Fig. 5a). Notably, both p53 and p21 proteins were induced, compatible with a role for P-TEFb in HPV transcription (58). As expected (24), D3 also down-regulated cyclin T1 and CDK9 although to a slightly lesser degree. These observations are consistent with the hypothesis that P-TEFb mediates the action of NF90/NF45 on the HPV promoter, but do not prove it.
Fig. 5. Regulation of E6 expression by P-TEFb-dependent and -independent mechanisms.
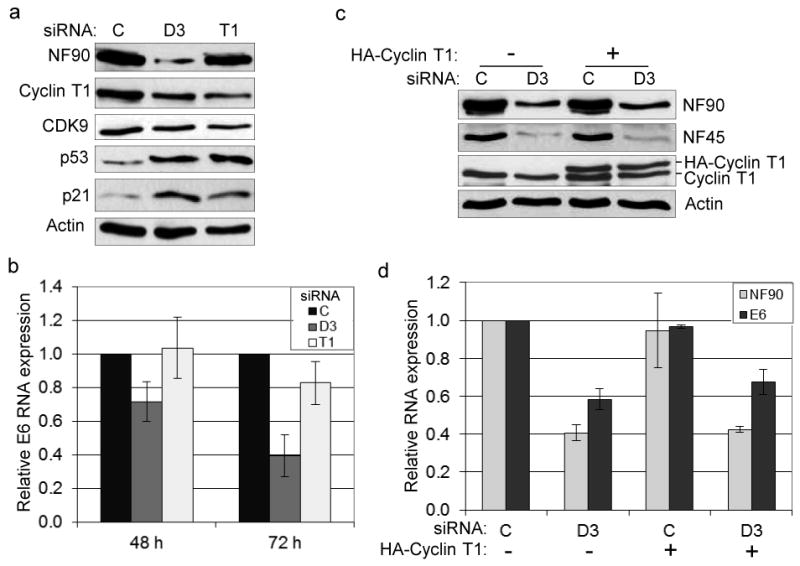
(a) Immunoblots showing P-TEFb protein levels (cyclin T1 and CDK9) in HeLa cells treated with C, D3 and T1 (cyclin T1) siRNAs at 72 h post transfection. (b) qRT-PCR analysis of E6 RNA in cells treated as in panel (a) (n=3 independent experiments). (c) Expression of HA-cyclin T1 in NF90 knockdown cells. Immunoblots show the expression of NF90, NF45 and cyclin T1 in HeLa cells transfected with siRNA and pcDNA3.0 HA-cyclin T1. (d) qRT-PCR analysis of E6 RNA in cells transfected with siRNA and pcDNA3.0 HA-cyclin T1 as in panel (c) (n=2 independent experiments).
We next conducted qRT-PCR analysis to compare the effects of NF90 and cyclin T1 depletion on E6 RNA at different time points. The results confirmed that D3 inhibited E6 RNA expression (by ∼30% at 48 h and ∼60% at 72 h), but cyclin T1 siRNA had no measurable effect on E6 RNA at 48 h and reduced it only slightly (by ∼18%) at 72 h (Fig. 5b). These kinetic data imply that NF90 exerts a separate action on the HPV promoter, in addition to a relatively modest action mediated via cyclin T1.
A prediction of this scenario is that cyclin T1 over-expression should only partially rescue the inhibition of E6 expression brought about by D3. Transfection of cells with a vector expressing hemagglutinin (HA)-tagged cyclin T1 that is resistant to NF90 depletion (24) increased the total levels of cyclin T1 by ∼2 fold, exceeding its level in untreated cells (Fig. 5c). The levels of NF90 and NF45 were unchanged (Fig. 5c), and cyclin T1 over-expression did not increase the level of NF90 RNA (Fig. 5d). The level of E6 RNA was also unaffected by cyclin T1 over-expression in control siRNA-treated cells (Fig. 5d). In D3-treated cells, however, E6 RNA increased to a small but significant extent (by 15%; P = 0.028). We therefore conclude that NF90/NF45 regulates HPV transcription by both P-TEFb-dependent and P-TEFb-independent mechanisms, and that the latter mechanism predominates under these conditions.
NF90/NF45 depletion enhances drug-induced apoptosis
p53 is responsible for critical cell functions including apoptosis, which is suppressed in HPV-transformed cell lines. Since TP53 is wild-type in these cells (59) reactivation of p53 expression sensitizes them to apoptosis induced by anticancer drugs such as cisplatin and camptothecin (60, 61). Because p53 expression is markedly increased in NF90/NF45-depleted HeLa cells, we examined their susceptibility to apoptosis. Inspection under the microscope disclosed changes in the overall morphology of cells in D3-treated cultures compared to controls (Fig. 6a). Many cells were rounded and refractory, exhibiting features such as nuclear condensation and nuclear fragmentation that are characteristic of apoptosis, while few such cells were seen in cultures transfected with control siRNA. Treatment with 10 μM camptothecin, a topoisomerase I inhibitor that induces apoptosis (60), markedly increased the number of rounded cells in control cultures and especially in D3-treated cultures (Fig. 6a).
Fig. 6. Depletion of NF90/NF45 sensitizes cells to camptothecin-induced apoptosis.
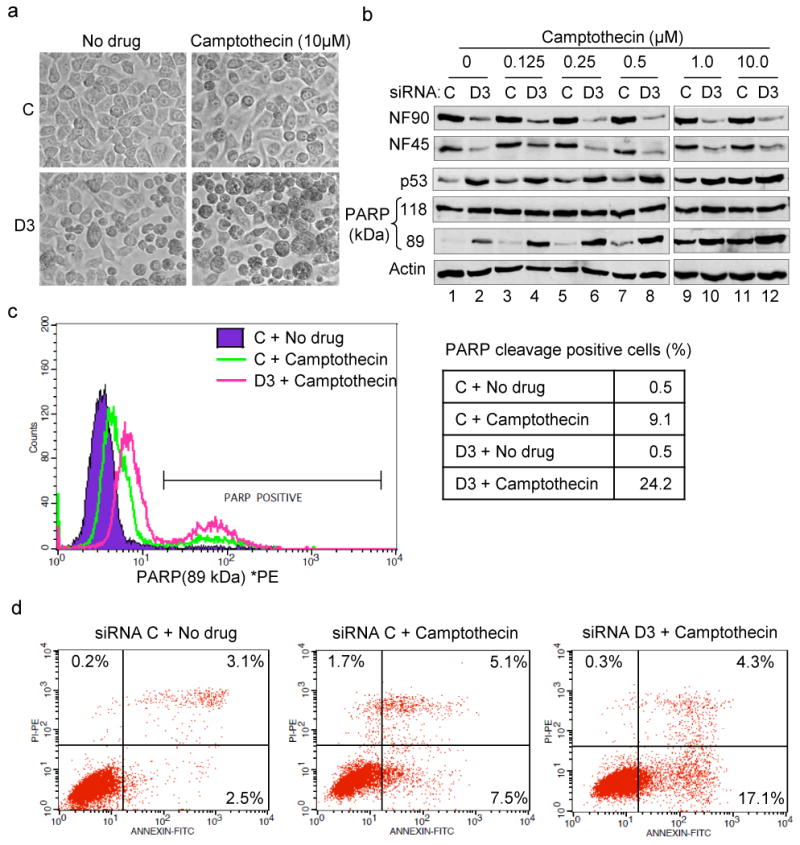
(a) Morphology of HeLa cells treated with siRNA and camptothecin. Phase-contrast images were observed with 20X objective. (b) Knockdown of NF90/NF45 enhances camptothecin-induced PARP cleavage and p53 induction. Immunoblot analysis of proteins in siRNA-transfected-HeLa cells treated with increasing concentration of camptothecin for 14 h. (c) In vivo PARP cleavage in cells treated with siRNA and camptothecin as in panel (a). Cells were stained with phycoerythrin (PE)-labeled PARP (89 kDa) antibody and analyzed by flow cytometry. Percentage of cells in each culture that were PARP-positive was the mean of triplicate analyses. Essentially superimposable profiles were obtained with control and D3-treated cultures in the absence of camptothecin. (d) NF90 depletion enhances camptothecin-induced early apoptosis. Cells treated with siRNA and camptothecin as in panel (a) were stained with fluorescein isothiocyanate (FITC)-labeled annexin V and propidium iodide. Dot plots represent the cells as analyzed by flow cytometry. Control and D3-treated cultures gave indistinguishable plots in the absence of camptothecin. Percentages of cells in each quadrant (top left, necrosis; top right, late apoptosis; bottom right, early apoptosis) are mean values of triplicate analyses.
Cleavage of poly ADP-ribose polymerase (PARP) by activated caspase-3 and caspase-7 occurs as a late event in the apoptotic pathway, generating 24-kDa and 89-kDa PARP fragments. D3 induced PARP cleavage, detected by immunoblotting for the 89-kDa fragment (Fig. 6b, compare lanes 1 and 2), and similar results were obtained with D5 (data not shown). Cleavage increased in D3-treated cells in the presence of low concentrations of camptothecin (lanes 3-6), and became detectable in control cells as well as D3-treated cells at higher drug concentrations (>0.5 μM; lanes 7-12). D3 cells contained more 89-kDa PARP than control cells irrespective of drug concentration, and the level of cleaved PARP invariably correlated with the induction of p53 (Fig. 6b). These data indicate a synergistic activation of the cleavage pathway by the drug and NF90/NF45 depletion.
The induction of PARP cleavage was also examined by flow cytometry (Fig. 6c). Cells were incubated in the presence or absence of camptothecin, then immunostained with phycoerythrin (PE)-labeled antibody specific for the 89-kDa PARP fragment. D3 increased the staining for cleaved PARP only slightly on its own, presumably because a small increase in cleaved PARP in a large number of cells is undetectable above the fluorescence background. In camptothecin-treated cells D3 increased the intensity of staining as well as the proportion of strongly staining cells (from ∼9% to ∼24%; Fig. 6c). Similar results were obtained with Annexin V, which detects phosphotidylserine on the outer cell membrane, an early marker for apoptosis (62). In the absence of camptothecin ∼2.5% of the cells were Annexin V-positive; in the presence of camptothecin, ∼7.5% of control cells and ∼17% of D3-treated cells were positive for this marker (Fig. 6d). These results show that D3 sensitized cells to camptothecin-induced apoptosis by ∼3 fold, indicating that NF90/NF45 depletion induces both p53 and apoptosis in cervical cancer cell lines.
Discussion
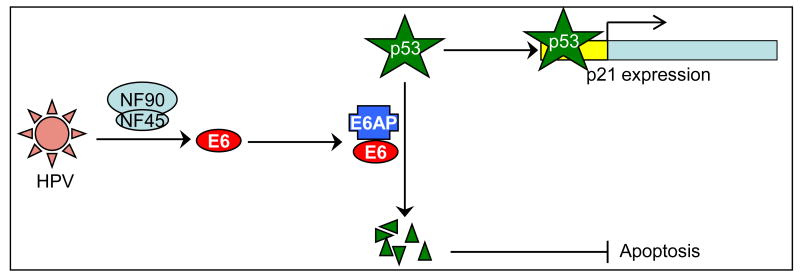
Results presented here show that the NF90/NF45 complex plays an important role in HPV early gene expression and hence in viral control over cell fate (apoptosis) via the key cellular regulators p53 and p21. High-risk HPVs exploit the NF90/NF45 complex to transcribe their E6 oncogene; the E6 protein, together with E6AP, induces degradation of p53; and functions mediated by p53 are suppressed, including p21 expression and apoptosis (Fig. 7). NF90/NF45 is a multifunctional adaptor complex that interacts with DNA, RNA and protein ligands and affects several cellular pathways. We therefore speculate that NF90/NF45 integrates signals from multiple sources and facilitates HPV-induced oncogenesis.
Fig. 7. Model for NF90/NF45-dependent p53 regulation in HPV-infected cells.
The NF90/NF45 complex mediates expression of the E6 oncogene from the viral genome. With E6AP, the viral E6 protein induces degradation of p53 by the ubiquitin-proteasome degradation pathway. Degradation of p53 results in the suppression of apoptosis and reduced p21 expression.
NF90/NF45 functions in transcription
Our analysis indicates that NF90/NF45 regulates HPV E6 transcription predominantly through a P-TEFb-independent mechanism, and to a lesser extent via P-TEFb. NF90 and NF45 participate in gene expression pathways at many steps, including transcription. Initially purified as DNA binding proteins, they interact with the ARRE element of the interleukin-2 promoter and stimulate its transcription (63). Subsequently, the NF90/NF45 complex has been shown to regulate the activity of several cellular and viral promoters (21) through direct interactions with specific promoter sequences (22, 47, 64, 65). NF90/NF45 can also act indirectly, via other proteins that modulate chromatin structure and transcription (20, 24, 57, 66-69). Efficient transcription by RNA polymerase II depends on its phosphorylation by P-TEFb (70), and NF90 regulates P-TEFb by enhancing expression of the cyclin T1 subunit of this elongation factor (24). First shown to be required for processive transcription of the HIV-1 genome (71), P-TEFb has recently been implicated in the regulation of HPV gene expression. It is recruited to the HPV genome through interactions with the Brd4 protein that are modulated by the viral E2 product (58).
While the precise action of NF90/NF45 at the HPV promoter has yet to be identified, one possible mechanism involves the multifunctional factor Yin Yang-1 (YY1) for which there are binding sites in the HPV URR (72-74). YY1 acts as a repressor or activator depending on the promoter context (67, 72, 73, 75, 76) and serves to recruit the histone deacetylase complex and protein arginine methyltransferase 1 (PRMT1). ILF3-derived proteins interact with PRMT1, enhance its methylase activity (77, 78), and favor YY1-PRMT1 interactions that stimulate transcription (67), although the data for NF90 itself are not unequivocal. The HPV URR also contains sites for several other factors, however, and NF90 engages in numerous interactions with transcriptional activators and co-activators, so further studies are needed to characterize the contributions of these sites and factors to the mechanism of NF90 action at the HPV promoter.
Regulation of p53, p21 and apoptosis
Depletion of NF90 or NF45 induces the expression of p21 RNA and protein in HPV-transformed cells, whereas depletion of NF110 was ineffective (Fig. S1). NF90 and NF110 differ substantially at their C termini and in their interactions with several ligands (18, 20, 79-81). Accordingly, different consequences ensue from specific knockdown of the two ILF3 isoforms, either singly or in combination (18). For example, NF90 depletion leads to reduced DNA synthesis and a slow-growth phenotype whereas NF110 knockdown does not (18). Such differential effects could well account for the discrepancy between our observations and those obtained with homozygous knockout mice which lack both NF90 and NF110 (43). p21 is expressed during terminal differentiation (82, 83) and its expression was impaired in ILF3 knockout pups with profound abnormalities in muscle development (43). Although p21 can be regulated in a variety of ways, our observations indicate that NF90/NF45 affects p21 transcription mediated by p53 in HeLa cells.
p53 is a pivotal regulator of cellular activities including proliferation (1, 84). Functional loss of p53, due to protein degradation or to TP53 mutations, is a frequent event in cancer development (6) and a number of viruses escape cell cycle regulation by targeting p53 (59, 85-87). In cells infected with HPV, a leading cause of cervical cancer, the viral oncoprotein E6 binds to (wild-type) p53 and induces its degradation via E6AP-mediated polyubiquitination (13, 54, 55, 88). Our results demonstrate the role of NF90/NF45 in suppressing p53 expression in cell lines originating from HPV-infected tumors. Inhibition of E6 expression by NF90/NF45 knockdown represses the degradation of p53, and restores cell functions including apoptosis and the p53-dependent transcription of genes such as p21 (58, 89). p53 is required for efficient activation of apoptosis following irradiation and treatment with chemotherapeutic agents (90). Activation of p53-dependent apoptosis induces Bax expression, resulting in cytochrome c release from mitochondria and caspase-3 activation (91-93). Activated caspase-3 executes apoptosis by inducing cleavage of cellular substrates including PARP (93) and NF90/NF45 depletion in HeLa cells resulted in the accumulation of p53 and the 89-kDa cleavage product of PARP. Expression of p53 sensitizes cells to drug-induced apoptosis (49); correspondingly, camptothecin amplified the effect of NF90/NF45 depletion on PARP cleavage, indicating activation of p53-dependent apoptosis.
HeLa cells are highly resistant to apoptosis and expression of p53 is not sufficient to kill them (94). Correspondingly, p53 induction by NF90/NF45 knockdown did not elicit a detectable increase in apoptosis as measured by flow cytometric analysis of PARP cleavage or Annexin V staining. Although this could reflect a characteristic of single-cell assays, which require a threshold level for detection, it is also consistent with the need for a second signal such as that provided by camptothecin. Similarly, p53 induction by E6 knockdown elicited little cell death unless complemented by the drug celecoxib (94). It is also notable that the slow-growth phenotype of HeLa cells depleted for NF90/NF45 (18) is not rescued by p53 knockdown (Fig. S3), consistent with the pleiotropic functions of NF90 and NF45 in gene expression and cell metabolism.
NF90/NF45 and virus infection
Our data appear to be the first to establish a connection between NF90/NF45 and HPV gene expression. Human NF90/NF45 participates in numerous biological processes that impinge on virus infection, including the cell cycle (18, 95) and antiviral defense mechanisms (39, 96-98), and it plays a role in the life cycle of several viruses (28, 29, 34, 99). In transient expression assays, NF90 in association with NF45 inhibited transcription from the adenovirus-2 major late promoter, but activated transcription from the cytomegalovirus immediate early promoter (21) as with the HPV early promoter (Fig. 4f). Interactions of NF90 and NF45 with viral proteins that affect viral replication have been observed in influenza virus H5N1, Ebola virus and infectious bursal disease virus (32-34). NF90 is present in protein complexes binding to the non-translated regions (NTRs) of bovine viral diarrhea virus (BVDV), hepatitis C virus (HCV), rhinovirus, and dengue virus, where it functions in viral replication and gene expression (28, 29, 100, 101). Interactions with BVDV and HCV NTRs induce the formation of circular genome structures essential for translation and replication. NF90/NF45 binds to the dengue virus 3′ NTR, contributing to the production of infectious virus particles, and it also interacts with hepatitis B virus transcripts (102) and with adenovirus VA RNA (103). By analogy, the interaction of NF90 with E6/E7 RNA (Fig. S2) could imply its involvement in post-transcriptional processes, such as the regulation of HPV gene expression, that await definition.
In conclusion, NF90/NF45 is a multi-domain complex that participates in numerous cellular processes and is harnessed by many viruses to promote their replication. Our findings have revealed a novel role for this complex in HPV-induced oncogenesis and p53-mediated apoptosis. Specifically, the NF90/NF45 complex acts as a positive regulator of HPV E6 expression, and NF90/NF45 depletion results in the accumulation of active p53. Targeting the function of NF90/NF45 could therefore be exploited as a means to combat viral infection and cancer.
Materials and Methods
Cell culture and transient expression assays
Cell lines C33A, HeLa, SiHa, U2OS and WI38 lines were obtained from ATCC and HCT116 cells from Dr. Betsy Barnes (UMDNJ-NJMS). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Sigma) and 1% penicillin/streptomycin (Invitrogen). Transfection was performed with 1.5-2.5 × 105 cells using 30 or 50 nM of small interfering RNA (siRNA) and INTERFERin (Polyplus-transfection). At 48 h or 72 h posttransfection, cells were washed with phosphate buffered saline (PBS; Cellgro) and harvested by trypsinization. siRNA sequences used for knockdown experiments were control (C): On-TARGETplus Non-Targeting siRNA (Ambion); D3 (NF90) and D5 (NF45) (18); D6 (NF45), 5′-ACTTTGATATCCAAATGGAGT-3′; p53, 5′-GACTCCAGTGGTAATCTACT-3′. For over-expression studies, at 8 h post siRNA transfection, cells were transfected with 1 μg of pcDNA3.1-siRNA-resistant-NF90b (Omni-NF90bD3R), pcDNA3.1-NF45 or pcDNA3.0-HA-cyclin T1 (24) using jetPEI (Polyplus-transfection).
Western blotting
Cell extract was prepared (18) and analyzed by immunoblotting (19) using antibodies directed against NF90 (DRBP76; BD Biosciences), NF45 (Everest), p21 (Ab-1; Calbiochem, and C-19; Santa Cruz), p53 (Ab-6; Calbiochem), E6AP (E4; Santa Cruz), cyclin T1 (T18; Santa Cruz), CDK9 (C-20; Santa Cruz), 89-kDa N terminus PARP (BD Biosciences), full-length PARP-1/2 (H-250; Santa Cruz), tubulin (Sigma), and actin (Santa Cruz).
Reverse transcription polymerase chain reaction (RT-PCR)
Total cellular RNA was isolated using RNeasy RNA isolation kit (Qiagen). Complementary DNA (cDNA) synthesis and PCR were performed in a single reaction tube with 50 ng of RNA using the Qiagen OneStep RT-PCR kit (Qiagen) with annealing at 62°C for 1 min. Amplified DNA was resolved by electrophoresis in a 1.5% agarose gel. Primer sequences were as follows. NF90: forward primer (FP) 5′GATGGCAATTCATTCGAGGC3′, reverse primer (RP) 5′GCAAGAGCAGCGTAGGCCTTC3′; NF45: FP 5′GAATCAGGACCTGGCTCCCA3′, RP 5′GGTCAAGGATCCAGGGTGTG3′; p21: FP 5′GTCAGTTCCTTGTGGAGCCG3′, RP 5′CTCCAGTGGTGTCTCGGTG3′; p53: FP 5′GTTTCCGTCTGGGCTTCTTGC3′, RP 5′ACGCAAATTTCCTTCCACTCGG3′; E6AP: FP 5′CGCATGTACAGTGAACGAAGA3′, RP 5′CTTTGGAAACACCTCCCTCA3′; HPVE6: FP 5′CCAGAAACCGTTGAATCCAG3′, RP 5′GTTGGAGTCGTTCCTGTCGT3′; Cyclin T1: FP 5′ATGGAGGGAGAGAGGAAGAAC3′, RP 5′CTACCTTGATGACATGTTCCA3′; AuroraB: FP 5′TCCGGAAAGAGCCTGTCACC3′, RP 5′CACAGACCAGCCGAAGTCAG3′; Matrin-3: FP 5′GATGCAATGGCAATGGTTGACC3′, RP 5′TCGTCTCCCACTGAACTGCC3′; ORC6: FP 5′GGACAGCAGGTCGACAGAG3′, RP 5′AGCCCACTGCCCAGACTTAG3′; Survivin: FP 5′GAACTGGCCCTTCTTGGAGG3′, RP 5′GTGGCACCAGGGAATAAACCC3′; TERC: FP 5′CGCGCTGTTTTTCTCGCTGAC3′, RP 5′GACTCGCTCCGTTCCTCTTC3′; GAPDH: FP 5′CTGGTAAAGTGGATATTGTTG3′, RP 5′GAGGCTGTTGTCATACTTCTC3′.
Quantitative real-time PCR (qRT-PCR)
cDNA was synthesized from 2 μg of RNA hybridized with 0.5 μg of oligo dT (15-mer; Sigma). Reverse transcription was performed at 37°C for 1 h in the presence or absence of Moloney murine leukemia virus reverse transcriptase (Promega). qPCR was performed in duplicate 25 μl reactions on 1/8th of the cDNA using 0.3 μM primers and Maxima SYBR Green/ROX (Fermentas) PCR master mix in StepOnePlus Real-Time PCR system (Applied Biosystems).
RNA turnover assay
At 72 h after siRNA transfection, cells were treated with 0.5 μg/ml of actinomycin D and total cellular RNA was isolated at intervals. Denatured RNA (10 μg) was subjected to formaldehyde-agarose gel electrophoresis, transferred to GeneScreenPlus hybridization membrane and crosslinked to the membrane by UV irradiation (UVXL-1000, Fischer Scientific). The immobilized RNA was hybridized with random-prime labeled E6 probe at 42°C and radioactive signals were detected by imaging (Typhoon 8600 scanner, Molecular Dynamics) and autoradiography.
Luciferase assays
HeLa cells (2 × 105) transfected with C, D3 and D5 siRNA were transfected 48 h later with 200 or 400 ng of pGL3-HPV18URR-Firefly luciferase (104) and 200 ng of RSV-Renilla luciferase (21) using jetPEI transfection reagent. Luciferase expression was analyzed 24 h after transfection, using the Dual-Luciferase Reporter assay system (Promega).
Flow cytometry
At 72 h post siRNA transfection, 2 × 105 cells were treated with 10 μM camptothecin (MP Biomedicals) or left untreated for 14 h. The cells were harvested by trypsinization and washed twice with ice-cold PBS at (1000x g, 5 min, 4°C). For the analysis of PARP cleavage, cells were incubated with phycoerythrin (PE)-labeled mouse anti-cleaved PARP (89-kDa) antibody (BD Biosciences). For analysis of apoptosis-specific phosphatidylserine, cells were incubated with fluorescein isothiocyanate (FITC) conjugated Annexin V (BD Biosciences) and propidium iodide (PI; BD Biosciences). Stained cells were analyzed by FACSCalibur (BD Biosciences).
Supplementary Material
Fig. S1. Expression of p21 is induced by knockdown of NF90 but not of NF110. Immunoblot analysis of p21, NF90, NF45 and NF110 in HeLa cells transfected with siRNA directed against NF90 (D3) or NF110 (D4). Note that D4 causes a small decrease in NF90 secondary to the knockdown of NF110 and destabilization of a fraction of NF45. The drop in NF90 level is limited by the differential amounts of the proteins (NF110 < NF90) and relative stability of NF90 (half-life ∼2.4 days, compared to ∼1 hr for NF45; ref. 18).
Fig. S2. Endogenous E6 and E7 RNA preferentially interact with NF90. Whole cell extract from HeLa cells was immunoprecipitated with 2 μg of the antibodies shown. RNA was isolated and RT-PCR was performed with E6 and E7 specific primers.
Fig. S3. NF90 regulates cell proliferation in HeLa cells. (a) Proliferation rates of cells depleted for NF90 (D3), p53, or both. Cultures were monitored for cell proliferation for 6 days after siRNA transfection (DAT). (b) Immunoblot (IB) analysis of p21, p53, NF90 and tubulin in siRNA-transfected HeLa cells.
Acknowledgments
We thank Dr. Edward Goodwin for pGL3-HPV18URR-Firefly luciferase plasmid and Dr. Betsy Barnes for HCT 116 cell lines. We are indebted to Dr. Raymond Birge and Dr. Edouard Azzam for discussions and helpful suggestions. The early stages of this work were supported by NIH grant R01 AI034552 to MBM.
References
- 1.Molchadsky A, Rivlin N, Brosh R, Rotter V, Sarig R. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis. 2010;31(9):1501–8. doi: 10.1093/carcin/bgq101. Epub 2010/05/28. [DOI] [PubMed] [Google Scholar]
- 2.Zheltukhin AO, Chumakov PM. Constitutive and induced functions of the p53 gene. Biochemistry (Mosc) 2010;75(13):1692–721. doi: 10.1134/s0006297910130110. Epub 2011/03/23. [DOI] [PubMed] [Google Scholar]
- 3.Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2(12):a000893. doi: 10.1101/cshperspect.a000893. Epub 2010/05/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gartel AL. p21(WAF1/CIP1) and cancer: a shifting paradigm? Biofactors. 2009;35(2):161–4. doi: 10.1002/biof.26. Epub 2009/05/19. [DOI] [PubMed] [Google Scholar]
- 5.Gartel AL, Tyner AL. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp Cell Res. 1999;246(2):280–9. doi: 10.1006/excr.1998.4319. Epub 1999/02/02. [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–10. doi: 10.1038/35042675. Epub 2000/12/01. [DOI] [PubMed] [Google Scholar]
- 7.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol. 2009;1(5):a001883. doi: 10.1101/cshperspect.a001883. Epub 2010/01/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dey A, Verma CS, Lane DP. Updates on p53: modulation of p53 degradation as a therapeutic approach. Br J Cancer. 2008;98(1):4–8. doi: 10.1038/sj.bjc.6604098. Epub 2008/01/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki K, Matsubara H. Recent advances in p53 research and cancer treatment. J Biomed Biotechnol. 2011;2011:978312. doi: 10.1155/2011/978312. Epub 2011/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010;24(15):1580–9. doi: 10.1101/gad.1941710. Epub 2010/08/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Austin D, Baer A, Lundberg L, Shafagati N, Schoonmaker A, Narayanan A, et al. p53 Activation following Rift Valley Fever Virus Infection Contributes to Cell Death and Viral Production. PLoS One. 2012;7(5):e36327. doi: 10.1371/journal.pone.0036327. Epub 2012/05/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munoz-Fontela C, Pazos M, Delgado I, Murk W, Mungamuri SK, Lee SW, et al. p53 serves as a host antiviral factor that enhances innate and adaptive immune responses to influenza A virus. J Immunol. 2011;187(12):6428–36. doi: 10.4049/jimmunol.1101459. Epub 2011/11/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tommasino M, Accardi R, Caldeira S, Dong W, Malanchi I, Smet A, et al. The role of TP53 in Cervical carcinogenesis. Hum Mutat. 2003;21(3):307–12. doi: 10.1002/humu.10178. Epub 2003/03/06. [DOI] [PubMed] [Google Scholar]
- 14.Chow LT, Broker TR, Steinberg BM. The natural history of human papillomavirus infections of the mucosal epithelia. APMIS. 2010;118(6-7):422–49. doi: 10.1111/j.1600-0463.2010.02625.x. Epub 2010/06/18. [DOI] [PubMed] [Google Scholar]
- 15.Narisawa-Saito M, Kiyono T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: roles of E6 and E7 proteins. Cancer Sci. 2007;98(10):1505–11. doi: 10.1111/j.1349-7006.2007.00546.x. Epub 2007/07/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas M, Narayan N, Pim D, Tomaic V, Massimi P, Nagasaka K, et al. Human papillomaviruses, cervical cancer and cell polarity. Oncogene. 2008;27(55):7018–30. doi: 10.1038/onc.2008.351. Epub 2008/11/26. [DOI] [PubMed] [Google Scholar]
- 17.Howie HL, Katzenellenbogen RA, Galloway DA. Papillomavirus E6 proteins. Virology. 2009;384(2):324–34. doi: 10.1016/j.virol.2008.11.017. Epub 2008/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan D, Altan-Bonnet N, Parrott AM, Arrigo CJ, Li Q, Khaleduzzaman M, et al. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol Cell Biol. 2008;28(14):4629–41. doi: 10.1128/MCB.00120-08. Epub 2008/05/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shamanna RA, Hoque M, Lewis-Antes A, Azzam EI, Lagunoff D, Pe'ery T, et al. The NF90/NF45 complex participates in DNA break repair via nonhomologous end joining. Mol Cell Biol. 2011;31(23):4832–43. doi: 10.1128/MCB.05849-11. Epub 2011/10/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karmakar S, Mahajan MC, Schulz V, Boyapaty G, Weissman SM. A multiprotein complex necessary for both transcription and DNA replication at the beta-globin locus. EMBO J. 2010;29(19):3260–71. doi: 10.1038/emboj.2010.204. Epub 2010/09/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reichman TW, Muniz LC, Mathews MB. The RNA binding protein nuclear factor 90 functions as both a positive and negative regulator of gene expression in mammalian cells. Mol Cell Biol. 2002;22:343–56. doi: 10.1128/MCB.22.1.343-356.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiesler P, Haynes PA, Shi L, Kao PN, Wysocki VH, Vercelli D. NF45 and NF90 regulate HS4-dependent interleukin-13 transcription in T cells. J Biol Chem. 2010;285(11):8256–67. doi: 10.1074/jbc.M109.041004. Epub 2010/01/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuwano Y, Pullmann R, Jr, Marasa BS, Abdelmohsen K, Lee EK, Yang X, et al. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 2010;38(1):225–38. doi: 10.1093/nar/gkp861. Epub 2009/10/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoque M, Shamanna RA, Guan D, Pe'ery T, Mathews MB. HIV-1 replication and latency are regulated by translational control of cyclin T1. J Mol Biol. 2011;410(5):917–32. doi: 10.1016/j.jmb.2011.03.060. Epub 2011/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gwizdek C, Ossareh-Nazari B, Brownawell AM, Evers S, Macara IG, Dargemont C. Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J Biol Chem. 2004;279(2):884–91. doi: 10.1074/jbc.M306808200. [DOI] [PubMed] [Google Scholar]
- 26.Urcuqui-Inchima S, Castano ME, Hernandez-Verdun D, St-Laurent G, 3rd, Kumar A. Nuclear Factor 90, a cellular dsRNA binding protein inhibits the HIV Rev-export function. Retrovirology. 2006;3:83. doi: 10.1186/1742-4690-3-83. Epub 2006/11/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, et al. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol. 2009;29(13):3754–69. doi: 10.1128/MCB.01836-08. Epub 2009/04/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomila RC, Martin GW, Gehrke L. NF90 binds the dengue virus RNA 3′ terminus and is a positive regulator of dengue virus replication. PLoS One. 2011;6(2):e16687. doi: 10.1371/journal.pone.0016687. Epub 2011/03/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isken O, Grassmann CW, Sarisky RT, Kann M, Zhang S, Grosse F, et al. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 2003;22(21):5655–65. doi: 10.1093/emboj/cdg562. Epub 2003/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isken O, Grassmann CW, Yu H, Behrens SE. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA. 2004;10(10):1637–52. doi: 10.1261/rna.7290904. Epub 2004/09/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrill MK, Gromeier M. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J Virol. 2006;80(14):6936–42. doi: 10.1128/JVI.00243-06. Epub 2006/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stricker R, Behrens SE, Mundt E. Nuclear factor NF45 interacts with viral proteins of infectious bursal disease virus and inhibits viral replication. J Virol. 2010 doi: 10.1128/JVI.02506-09. Epub 2010/08/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P, Song W, Mok BW, Zhao P, Qin K, Lai A, et al. Nuclear factor 90 negatively regulates influenza virus replication by interacting with viral nucleoprotein. J Virol. 2009;83(16):7850–61. doi: 10.1128/JVI.00735-09. Epub 2009/06/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shabman RS, Leung DW, Johnson J, Glennon N, Gulcicek EE, Stone KL, et al. DRBP76 associates with Ebola virus VP35 and suppresses viral polymerase function. J Infect Dis. 2011;204(Suppl 3):S911–8. doi: 10.1093/infdis/jir343. Epub 2011/10/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reichman TW, Mathews MB. The NF90 Family of Double-stranded RNA Binding Proteins: Regulators of Viral and Cellular Function. In: Bradshaw R, Dennis E, editors. Cell Signaling Handbook. Academic Press; 2003. pp. 335–42. [Google Scholar]
- 36.Barber GN. The NFAR's (nuclear factors associated with dsRNA): evolutionarily conserved members of the dsRNA binding protein family. RNA Biol. 2009;6(1):35–9. doi: 10.4161/rna.6.1.7565. Epub 2008/12/25. [DOI] [PubMed] [Google Scholar]
- 37.Parrott AM, Walsh MR, Reichman TW, Mathews MB. RNA binding and phosphorylation determine the intracellular distribution of nuclear factors 90 and 110. J Mol Biol. 2005;348(2):281–93. doi: 10.1016/j.jmb.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 38.Pei Y, Zhu P, Dang Y, Wu J, Yang X, Wan B, et al. Nuclear export of NF90 to stabilize IL-2 mRNA is mediated by AKT-dependent phosphorylation at Ser647 in response to CD28 costimulation. J Immunol. 2008;180(1):222–9. doi: 10.4049/jimmunol.180.1.222. Epub 2007/12/22. [DOI] [PubMed] [Google Scholar]
- 39.Harashima A, Guettouche T, Barber GN. Phosphorylation of the NFAR proteins by the dsRNA-dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. Genes Dev. 2010;24(23):2640–53. doi: 10.1101/gad.1965010. Epub 2010/12/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu P, Jiang W, Cao L, Yu W, Pei Y, Yang X, et al. IL-2 mRNA stabilization upon PMA stimulation is dependent on NF90-Ser647 phosphorylation by protein kinase CbetaI. J Immunol. 2010;185(9):5140–9. doi: 10.4049/jimmunol.1000849. Epub 2010/09/28. [DOI] [PubMed] [Google Scholar]
- 41.Graber TE, Baird SD, Kao PN, Mathews MB, Holcik M. NF45 functions as an IRES trans-acting factor that is required for translation of cIAP1 during the unfolded protein response. Cell Death Differ. 2010;17(4):719–29. doi: 10.1038/cdd.2009.164. Epub 2009/11/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeifer I, Elsby R, Fernandez M, Faria PA, Nussenzveig DR, Lossos IS, et al. NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc Natl Acad Sci U S A. 2008;105(11):4173–8. doi: 10.1073/pnas.0711222105. Epub 2008/03/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, et al. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J Biol Chem. 2005;280(19):18981–9. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- 44.Vumbaca F, Phoenix KN, Rodriguez-Pinto D, Han DK, Claffey KP. Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol Cell Biol. 2008;28(2):772–83. doi: 10.1128/MCB.02078-06. Epub 2007/11/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shim J, Lim H, RY J, Karin M. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol Cell. 2002;10:1331–44. doi: 10.1016/s1097-2765(02)00730-x. [DOI] [PubMed] [Google Scholar]
- 46.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, Yang X, et al. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol Cell Biol. 2008;28(14):4562–75. doi: 10.1128/MCB.00165-08. Epub 2008/05/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi L, Godfrey WR, Lin J, Zhao G, Kao PN. NF90 regulates inducible IL-2 gene expression in T cells. J Exp Med. 2007;204(5):971–7. doi: 10.1084/jem.20052078. Epub 2007/05/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corso C, Pisapia L, Citro A, Cicatiello V, Barba P, Cigliano L, et al. EBP1 and DRBP76/NF90 binding proteins are included in the major histocompatibility complex class II RNA operon. Nucleic Acids Res. 2011;39(16):7263–75. doi: 10.1093/nar/gkr278. Epub 2011/06/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowe SW, Bodis S, McClatchey A, Remington L, Ruley HE, Fisher DE, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266(5186):807–10. doi: 10.1126/science.7973635. Epub 1994/11/04. [DOI] [PubMed] [Google Scholar]
- 50.Li F, Ackermann EJ, Bennett CF, Rothermel AL, Plescia J, Tognin S, et al. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol. 1999;1(8):461–6. doi: 10.1038/70242. Epub 1999/12/10. [DOI] [PubMed] [Google Scholar]
- 51.Eldridge AG, Loktev AV, Hansen DV, Verschuren EW, Reimann JD, Jackson PK. The evi5 oncogene regulates cyclin accumulation by stabilizing the anaphase-promoting complex inhibitor emi1. Cell. 2006;124(2):367–80. doi: 10.1016/j.cell.2005.10.038. Epub 2006/01/28. [DOI] [PubMed] [Google Scholar]
- 52.Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci U S A. 2003;100(16):9150–5. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nair JS, Ho AL, Tse AN, Coward J, Cheema H, Ambrosini G, et al. Aurora B kinase regulates the postmitotic endoreduplication checkpoint via phosphorylation of the retinoblastoma protein at serine 780. Mol Biol Cell. 2009;20(8):2218–28. doi: 10.1091/mbc.E08-08-0885. Epub 2009/02/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–36. doi: 10.1016/0092-8674(90)90409-8. Epub 1990/12/21. [DOI] [PubMed] [Google Scholar]
- 55.Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10(13):4129–35. doi: 10.1002/j.1460-2075.1991.tb04990.x. Epub 1991/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol Cell. 2004;13(1):101–11. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 57.Nie Y, Ding L, Kao PN, Braun R, Yang JH. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol Cell Biol. 2005;25(16):6956–63. doi: 10.1128/MCB.25.16.6956-6963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan J, Li Q, Lievens S, Tavernier J, You J. Abrogation of the Brd4-positive transcription elongation factor B complex by papillomavirus E2 protein contributes to viral oncogene repression. J Virol. 2010;84(1):76–87. doi: 10.1128/JVI.01647-09. Epub 2009/10/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheffner M, Munger K, Byrne JC, Howley PM. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci U S A. 1991;88(13):5523–7. doi: 10.1073/pnas.88.13.5523. Epub 1991/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kreis NN, Sanhaji M, Kramer A, Sommer K, Rodel F, Strebhardt K, et al. Restoration of the tumor suppressor p53 by downregulating cyclin B1 in human papillomavirus 16/18-infected cancer cells. Oncogene. 2010;29(41):5591–603. doi: 10.1038/onc.2010.290. Epub 2010/07/28. [DOI] [PubMed] [Google Scholar]
- 61.Tan S, Hougardy BM, Meersma GJ, Schaap B, de Vries EG, van der Zee AG, et al. Human papilloma virus 16 e6 RNA interference enhances Cisplatin and death receptor-mediated apoptosis in human cervical carcinoma cells. Mol Pharmacol. 2012;81(5):701–9. doi: 10.1124/mol.111.076539. Epub 2012/02/14. [DOI] [PubMed] [Google Scholar]
- 62.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184(1):39–51. doi: 10.1016/0022-1759(95)00072-i. Epub 1995/07/17. [DOI] [PubMed] [Google Scholar]
- 63.Corthésy B, Kao PN. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J Biol Chem. 1994;269:20682–90. [PubMed] [Google Scholar]
- 64.Ranpura SA, Deshmukh US, Reddi PP. NF45 and NF90 in Murine Seminiferous Epithelium: Potential Role in SP-10 Gene Transcription. J Androl. 2008;29:186–97. doi: 10.2164/jandrol.107.003756. [DOI] [PubMed] [Google Scholar]
- 65.Sakamoto S, Morisawa K, Ota K, Nie J, Taniguchi T. A binding protein to the DNase I hypersensitive site II in HLA-DR alpha gene was identified as NF90. Biochemistry. 1999;38(11):3355–61. doi: 10.1021/bi982099g. Epub 1999/03/17. [DOI] [PubMed] [Google Scholar]
- 66.Shi L, Qiu D, Zhao G, Corthesy B, Lees-Miller S, Reeves WH, et al. Dynamic binding of Ku80, Ku70 and NF90 to the IL-2 promoter in vivo in activated T-cells. Nucleic Acids Res. 2007;35(7):2302–10. doi: 10.1093/nar/gkm117. Epub 2007/03/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, et al. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003;17(8):1019–29. doi: 10.1101/gad.1068003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bose SK, Sengupta TK, Bandyopadhyay S, Spicer EK. Identification of Ebp1 as a component of cytoplasmic bcl-2 mRNP (messenger ribonucleoprotein particle) complexes. Biochem J. 2006;396(1):99–107. doi: 10.1042/BJ20051548. Epub 2006/01/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–40. doi: 10.1038/nature03120. Epub 2004/11/09. [DOI] [PubMed] [Google Scholar]
- 70.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxl-terminal domain kinase. J Biol Chem. 1996;271:27176–83. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 71.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, et al. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11(20):2622–32. doi: 10.1101/gad.11.20.2622. Epub 1997/10/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lace MJ, Isacson C, Anson JR, Lorincz AT, Wilczynski SP, Haugen TH, et al. Upstream regulatory region alterations found in human papillomavirus type 16 (HPV-16) isolates from cervical carcinomas increase transcription, ori function, and HPV immortalization capacity in culture. J Virol. 2009;83(15):7457–66. doi: 10.1128/JVI.00285-09. Epub 2009/05/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bauknecht T, Jundt F, Herr I, Oehler T, Delius H, Shi Y, et al. A switch region determines the cell type-specific positive or negative action of YY1 on the activity of the human papillomavirus type 18 promoter. J Virol. 1995;69(1):1–12. doi: 10.1128/jvi.69.1.1-12.1995. Epub 1995/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bednarek PH, Lee BJ, Gandhi S, Lee E, Phillips B. Novel binding sites for regulatory factors in the human papillomavirus type 18 enhancer and promoter identified by in vivo footprinting. J Virol. 1998;72(1):708–16. doi: 10.1128/jvi.72.1.708-716.1998. Epub 1998/01/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pajunk HS, May C, Pfister H, Fuchs PG. Regulatory interactions of transcription factor YY1 with control sequences of the E6 promoter of human papillomavirus type 8. J Gen Virol. 1997;78(Pt 12):3287–95. doi: 10.1099/0022-1317-78-12-3287. Epub 1997/12/24. [DOI] [PubMed] [Google Scholar]
- 76.Lopez-Saavedra A, Gonzalez-Maya L, Ponce-de-Leon S, Garcia-Carranca A, Mohar A, Lizano M. Functional implication of sequence variation in the long control region and E2 gene among human papillomavirus type 18 variants. Arch Virol. 2009;154(5):747–54. doi: 10.1007/s00705-009-0362-4. Epub 2009/04/02. [DOI] [PubMed] [Google Scholar]
- 77.Tang J, Kao PN, Herschman HR. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J Biol Chem. 2000;275:19866–76. doi: 10.1074/jbc.M000023200. [DOI] [PubMed] [Google Scholar]
- 78.Cazanove O, Batut J, Scarlett G, Mumford K, Elgar S, Thresh S, et al. Methylation of Xilf3 by Xprmt1b alters its DNA, but not RNA, binding activity. Biochemistry. 2008;47(32):8350–7. doi: 10.1021/bi7008486. Epub 2008/07/19. [DOI] [PubMed] [Google Scholar]
- 79.Chen T, Brownawell AM, Macara IG. Nucleocytoplasmic shuttling of JAZ, a new cargo protein for exportin-5. Mol Cell Biol. 2004;24(15):6608–19. doi: 10.1128/MCB.24.15.6608-6619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp Cell Res. 2007;313(20):4196–207. doi: 10.1016/j.yexcr.2007.07.020. Epub 2007/09/04. [DOI] [PubMed] [Google Scholar]
- 81.Ulke-Lemee A, Trinkle-Mulcahy L, Chaulk S, Bernstein NK, Morrice N, Glover M, et al. The nuclear PP1 interacting protein ZAP3 (ZAP) is a putative nucleoside kinase that complexes with SAM68, CIA, NF110/45, and HNRNP-G. Biochim Biophys Acta. 2007;1774(10):1339–50. doi: 10.1016/j.bbapap.2007.07.015. Epub 2007/09/25. [DOI] [PubMed] [Google Scholar]
- 82.Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, et al. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267(5200):1018–21. doi: 10.1126/science.7863327. Epub 1995/02/17. [DOI] [PubMed] [Google Scholar]
- 83.Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, et al. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9(8):935–44. doi: 10.1101/gad.9.8.935. Epub 1995/04/15. [DOI] [PubMed] [Google Scholar]
- 84.Collavin L, Lunardi A, Del Sal G. p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ. 2010;17(6):901–11. doi: 10.1038/cdd.2010.35. Epub 2010/04/10. [DOI] [PubMed] [Google Scholar]
- 85.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278(5701):261–3. doi: 10.1038/278261a0. Epub 1979/03/15. [DOI] [PubMed] [Google Scholar]
- 86.Lan KH, Sheu ML, Hwang SJ, Yen SH, Chen SY, Wu JC, et al. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene. 2002;21(31):4801–11. doi: 10.1038/sj.onc.1205589. Epub 2002/07/09. [DOI] [PubMed] [Google Scholar]
- 87.Wang XW, Gibson MK, Vermeulen W, Yeh H, Forrester K, Sturzbecher HW, et al. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res. 1995;55(24):6012–6. Epub 1995/12/15. [PubMed] [Google Scholar]
- 88.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75(3):495–505. doi: 10.1016/0092-8674(93)90384-3. Epub 1993/11/05. [DOI] [PubMed] [Google Scholar]
- 89.Chang JT, Kuo TF, Chen YJ, Chiu CC, Lu YC, Li HF, et al. Highly potent and specific siRNAs against E6 or E7 genes of HPV16- or HPV18-infected cervical cancers. Cancer Gene Ther. 2010;17(12):827–36. doi: 10.1038/cgt.2010.38. Epub 2010/10/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362(6423):847–9. doi: 10.1038/362847a0. Epub 1993/04/29. [DOI] [PubMed] [Google Scholar]
- 91.McCurrach ME, Connor TM, Knudson CM, Korsmeyer SJ, Lowe SW. bax-deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc Natl Acad Sci U S A. 1997;94(6):2345–9. doi: 10.1073/pnas.94.6.2345. Epub 1997/03/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brantley-Finley C, Lyle CS, Du L, Goodwin ME, Hall T, Szwedo D, et al. The JNK, ERK and p53 pathways play distinct roles in apoptosis mediated by the antitumor agents vinblastine, doxorubicin, and etoposide. Biochem Pharmacol. 2003;66(3):459–69. doi: 10.1016/s0006-2952(03)00255-7. Epub 2003/08/09. [DOI] [PubMed] [Google Scholar]
- 93.Lee CH, Lim H, Moon S, Shin C, Kim S, Kim BJ, et al. Novel anticancer agent, benzyldihydroxyoctenone, isolated from Streptomyces sp. causes G1 cell cycle arrest and induces apoptosis of HeLa cells. Cancer Sci. 2007;98(6):795–802. doi: 10.1111/j.1349-7006.2007.00473.x. Epub 2007/04/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saha B, Adhikary A, Ray P, Saha S, Chakraborty S, Mohanty S, et al. Restoration of tumor suppressor p53 by differentially regulating pro- and anti-p53 networks in HPV-18-infected cervical cancer cells. Oncogene. 2012;31(2):173–86. doi: 10.1038/onc.2011.234. Epub 2011/07/19. [DOI] [PubMed] [Google Scholar]
- 95.Smith NL, Miskimins WK. Phosphorylation at serine 482 affects stability of NF90 and its functional role in mitosis. Cell Prolif. 2011;44(2):147–55. doi: 10.1111/j.1365-2184.2011.00742.x. Epub 2011/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krasnoselskaya-Riz I, Spruill A, Chen YW, Schuster D, Teslovich T, Baker C, et al. Nuclear factor 90 mediates activation of the cellular antiviral expression cascade. AIDS Res Hum Retroviruses. 2002;18(8):591–604. doi: 10.1089/088922202753747941. [DOI] [PubMed] [Google Scholar]
- 97.Patel RC, Vestal DJ, Xu Z, Bandyopadhyay S, Guo W, Erme SM, et al. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J Biol Chem. 1999;274(29):20432–7. doi: 10.1074/jbc.274.29.20432. Epub 1999/07/10. [DOI] [PubMed] [Google Scholar]
- 98.Parker LM, Fierro-Monti I, Mathews MB. Nuclear factor 90 is a substrate and regulator of the eukaryotic initiation factor 2 kinase double-stranded RNA-activated protein kinase. J Biol Chem. 2001;276(35):32522–30. doi: 10.1074/jbc.M104408200. Epub 2001/07/05. [DOI] [PubMed] [Google Scholar]
- 99.Agbottah ET, Traviss C, McArdle J, Karki S, St Laurent GC, 3rd, Kumar A. Nuclear Factor 90(NF90) targeted to TAR RNA inhibits transcriptional activation of HIV-1. Retrovirology. 2007;4:41. doi: 10.1186/1742-4690-4-41. Epub 2007/06/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Isken O, Baroth M, Grassmann CW, Weinlich S, Ostareck DH, Ostareck-Lederer A, et al. Nuclear factors are involved in hepatitis C virus RNA replication. RNA. 2007;13(10):1675–92. doi: 10.1261/rna.594207. Epub 2007/08/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Merrill MK, Dobrikova EY, Gromeier M. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J Virol. 2006;80(7):3147–56. doi: 10.1128/JVI.80.7.3147-3156.2006. Epub 2006/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shin HJ, Kim SS, Cho YH, Lee SG, Rho HM. Host cell proteins binding to the encapsidation signal epsilon in hepatitis B virus RNA. Arch Virol. 2002;147:471–91. doi: 10.1007/s007050200001. [DOI] [PubMed] [Google Scholar]
- 103.Liao HJ, Kobayashi R, Mathews MB. Novel functions of adenovirus-associated RNAs: Purification and characterization of RNA binding proteins. Proc Natl Acad Sci USA. 1998;95 doi: 10.1073/pnas.95.15.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goodwin EC, Naeger LK, Breiding DE, Androphy EJ, DiMaio D. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J Virol. 1998;72(5):3925–34. doi: 10.1128/jvi.72.5.3925-3934.1998. Epub 1998/04/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang X, Meyers C, Wang HK, Chow LT, Zheng ZM. Construction of a full transcription map of human papillomavirus type 18 during productive viral infection. J Virol. 2011;85(16):8080–92. doi: 10.1128/JVI.00670-11. Epub 2011/06/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression of p21 is induced by knockdown of NF90 but not of NF110. Immunoblot analysis of p21, NF90, NF45 and NF110 in HeLa cells transfected with siRNA directed against NF90 (D3) or NF110 (D4). Note that D4 causes a small decrease in NF90 secondary to the knockdown of NF110 and destabilization of a fraction of NF45. The drop in NF90 level is limited by the differential amounts of the proteins (NF110 < NF90) and relative stability of NF90 (half-life ∼2.4 days, compared to ∼1 hr for NF45; ref. 18).
Fig. S2. Endogenous E6 and E7 RNA preferentially interact with NF90. Whole cell extract from HeLa cells was immunoprecipitated with 2 μg of the antibodies shown. RNA was isolated and RT-PCR was performed with E6 and E7 specific primers.
Fig. S3. NF90 regulates cell proliferation in HeLa cells. (a) Proliferation rates of cells depleted for NF90 (D3), p53, or both. Cultures were monitored for cell proliferation for 6 days after siRNA transfection (DAT). (b) Immunoblot (IB) analysis of p21, p53, NF90 and tubulin in siRNA-transfected HeLa cells.