Abstract
Glutathione transferases (GST) were purified from locally isolated bacteria, Acinetobacter calcoaceticus Y1, by glutathione-affinity chromatography and anion exchange, and their substrate specificities were investigated. SDS-polyacrylamide gel electrophoresis revealed that the purified GST resolved into a single band with a molecular weight (MW) of 23 kDa. 2-dimensional (2-D) gel electrophoresis showed the presence of two isoforms, GST1 (pI 4.5) and GST2 (pI 6.2) with identical MW. GST1 was reactive towards ethacrynic acid, hydrogen peroxide, 1-chloro-2,4-dinitrobenzene, and trans,trans-hepta-2,4-dienal while GST2 was active towards all substrates except hydrogen peroxide. This demonstrated that GST1 possessed peroxidase activity which was absent in GST2. This study also showed that only GST2 was able to conjugate GSH to isoproturon, a herbicide. GST1 and GST2 were suggested to be similar to F0KLY9 (putative glutathione S-transferase) and F0KKB0 (glutathione S-transferase III) of Acinetobacter calcoaceticus strain PHEA-2, respectively.
1. Introduction
Glutathione transferases (GSTs; EC2.5.1.18, formerly known as glutathione S-transferases) are a family of enzymes which catalyse the addition of the nucleophilic sulphur atom of glutathione (GSH) to the electrophilic centre of a large variety of hydrophobic molecules and thus make the conjugates more soluble and easily excreted from the cells. Apart from their catalytic role, GSTs are also capable of binding a number of endogenous and exogenous compounds noncatalytically. These include haem, bilirubin, hormones, flavonoids, fatty acids, and xenobiotics. GSTs have also been found to minimise the effects of oxygen toxicity and to mediate signal transduction during oxidative stress [1]. Therefore, GSTs are seen to play an important role in the detoxification of xenobiotics and endogenously toxic compounds.
GSTs are found in both eukaryotes and prokaryotes, and they can be divided into four families: cytosolic GST, mitochondrial GST, microsomal GST, and bacteria fosfomycin-resistant proteins [2]. In bacteria, four classes of cytosolic GSTs have been identified, namely, beta, chi, theta, and zeta. All beta class GSTs are characterized by the presence of cysteine residue in the active site that is responsible for the catalytic activity. The first GST of this class was purified from Proteus mirabilis [3]. The chi class is characterised by cyanobacterial GSTs of Thermosynechococcus elongatus BP-1 (TeGST) and Synechococcus elongatus PCC6301 (SeGST) [4]. The theta class of GSTs is represented by dichloromethane dehalogenase produced by methylotrophic bacteria [5], while the zeta class is represented by tetrachlorohydroquinone (TCHQ) dehalogenase [6].
GSTs are found intracellularly and they have two general functions: to detoxify toxic compounds in the cells and to maintain cellular sulfhydryl groups in their reduced forms [7]. Reports have shown that bacterial GSTs are involved in many distinct metabolic processes such as biotransformation, degradation, and reductive dechlorination, which are linked to biodegradation of xenobiotics, defence against oxidative stress, protection against chemicals, and resistance towards antimicrobial drugs [8]. Bacterial GSTs are also involved in the degradation of several monocyclic aromatic compounds such as toluene, xylenes, phenols, and atrazine [9].
The detoxification ability of GSTs confers much importance to this group of enzymes in agriculture; for example, GSTs in arthropods and plant pests may be the cause of their resistance to insecticides and herbicides. Due to this advantage GSTs have been considered as potential candidate for the development of herbicide and stress tolerant transgenic plants [10]. GSTs are also seen as potential candidates for biotechnological applications in bioremediation and toxicology. Compared to the use of chemical or physical methods, bioremediation (the use of relevant microbes and/or enzymes) is often a more effective and safer alternative to clean up contaminated environments [8]. Biosensor development is another application whereby GSTs can be used as a tool to check for xenobiotic-contaminated environments and samples. Optical biosensors based on immobilised GSTs have been developed to detect captan [11], acrylamide [12], malathion [13], and atrazine [14]. These are all based on the ability of particular isoforms which are reactive towards specific xenobiotics. In view of the important properties and applications of GSTs, a continuous screening of bacteria for GSTs with new catalytic abilities to detoxify harmful or persistent chemicals is therefore important. This study describes the screening of soil bacteria for the production of GSTs, the purification of the enzymes by affinity chromatography, and some characterization to evaluate their catalytic abilities.
2. Materials and Methods
2.1. Chemicals
Unless otherwise stipulated, chemicals employed were of the highest grade obtainable. Dithiothreitol and phenylthiourea were from GE Healthcare. Centrifugal concentrators were obtained from Vivascience (Gottingen, Germany). Propoxur, isoproturon, fenoxaprop-ethyl, and clodinafop-propargyl were obtained from Sigma-Aldrich. Acrylamide, bisacrylamide, Biolyte ampholyte solution, and concentrated Coomassie Brilliant Blue protein reagent were obtained from BioRad Laboratories, Hercules, CA, USA. For isoelectric focusing, IPG strips were obtained from GE Healthcare. Buffer components were purchased from Sigma Chemical Co., St. Louis, USA.
2.2. Isolation, Screening, and Identification of GST-Expressing Bacteria
A quantity (1 g) of chemically contaminated soil collected from an illegal dump site located in University of Malaya, Kuala Lumpur, was placed in a sterile vial containing 10 mL of sterilized distilled water. The vial was shaken vigorously and left to stand overnight (18 hours) at 37°C in a water bath. The soil cultures were streaked on Nutrient Agar (Merck) plates and incubated for 18 hours at 37°C. Well separated colonies were purified on the same medium and maintained for screening for GST-expressing bacteria. Monochlorobimane (MCB) [15] was dissolved in acetonitrile at a concentration of 1 mg/mL (40 mM). Solutions were stored in a −20°C freezer and protected from light to avoid photolytic decomposition of MCB. To screen for GST-expressing bacteria, the MCB reagent was sprayed over each agar plate containing the bacterial isolates. Fluorescing colonies were visualized under long-wavelength (365 nm) UV light suggesting the presence of expressed GST. Isolates with intense fluorescence were grown, sedimented, and lysed (as mentioned in Section 2.4). The crude homogenate was tested for GST activity as described in Section 2.3. A positive isolate (with GST conjugating activity) was identified based on its 16S rRNA gene sequence.
2.3. Substrate Specificities
Enzymatic assays with 1-Chloro-2,4-dinitrobenzene (CDNB), ethacrynic acid (EA), sulfobromophthalein (BSP), p-nitrobenzyl chloride (NBC), trans-4-phenyl-3-buten-2-one (PBO) [16], and 3,4-Dichloronitrobenzene (DCNB) [17] were determined according to protocols described in the respective articles. Determination of glutathione peroxidase activity was performed by modifying the method described by Wendel [18]. Dichloromethane dehalogenase activity was determined using the method described by Sherratt et al. [19]. The ability to conjugate trans,trans-2,4-heptadienal was determined according to the method described by Brophy et al. [20].
2.4. Purification of Glutathione Transferases
A single colony of Acinetobacter calcoaceticus Y1 was picked and grown in nutrient broth (Oxoid) at 37°C for 18 hours after which the cells were pelleted by centrifugation (Beckman JA 7.5) at 3000 ×g for 20 minutes at 4°C. The cells were resuspended in 5 mL of 25 mM sodium phosphate buffer (pH 7.4), 1.0 mM EDTA, 0.1 mM DDT, 0.1 mM Phenylthiourea (PTU), and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). The suspended cells were disrupted by sonication (Powersonic 603) for 20 minutes at 4°C and centrifuged (Beckman 80 Ti) at 100,000 ×g for 30 minutes at 4°C. The supernatant was applied onto the glutathione (GSH) agarose matrix (Sigma-Aldrich) packed in a Tricorn 5/20 column (GE Healthcare) which was connected to the AKTA purifier equipped with a fraction collector. The flow rate was set at 0.3 mL/min. The enzymes were eluted with buffer A containing 10 mM of GSH. These purified enzyme solutions were concentrated and either used immediately for kinetic and substrate specificity determination or were stored at −20°C until further analysis. The GST isoforms were separated using GSH-affinity chromatography and ion exchange chromatography columns arranged in tandem. GSH-agarose column with bound GSTs was connected to DEAE Sepharose fast flow column (1 mL) (GE Healthcare). The two connected columns were washed with five bed volumes of buffer B (25 mM sodium phosphate buffer, pH 6.0), and the enzymes were subsequently eluted with buffer B containing 10 mM of GSH and the flow-through was collected. Any GST that remained bound to the ion exchange column was eluted using buffer B containing 50 M NaCl.
2.5. Protein Determination
Protein concentration was determined using Coomassie Brilliant Blue R-250, and bovine serum albumin was used as the standard [21]. Protein standards were prepared in triplicate. Aliquots of bovine serum albumin (BSA) stock (1 mg/mL) and sample were pipetted into test tubes and the total volume was made 100 μL. by addition of distilled water. To every sample and standard, 5 mL of Coomassie blue reagent was added, followed by vortexing. After 5 minutes the absorbance was read at 595 nm. The amounts of BSA in the standards were plotted against their average absorbance. The protein content of the samples was estimated from the standard curve.
2.6. 2-Dimensional Gel Electrophoresis and Isoelectrofocusing
Isoelectric focusing was conducted by using 2-D electrophoresis. The first dimension electrophoresis was carried out on a Multiphor II (GE Healthcare), and the second dimension electrophoresis was carried out in a Biorad Mini Protean II electrophoresis tank. Samples (the purified GST) for separation in the first dimension were mixed with 8 M urea, 2% CHAPS, 0.15% DTT, 30 mM thiourea, 2% Biolyte pH 3–10, and traces of Bromophenol Blue and were applied to a 7 cm, pH 3–10 IPG strip (Amersham Bioscience) during rehydration of the strip. The first dimension was set at a constant voltage of 200 V (1st stage, 1 min), followed by a gradient increase to 3500 V (2nd stage, 1 : 30 hr) and followed by constant 3500 V (3rd stage, 1 : 30 hr). Subsequently, proteins in the gel were reduced with DTT and alkylated with iodoacetamide. Electrophoresis in the second dimension was then run in 12% polyacrylamide gel in the presence of 0.1% SDS at 150 V. Sodium dodecyl sulfate polyacrylamide electrophoresis (SDS-PAGE) was performed according to the method of Laemmli [22].
To determine the pI of the sample protein, isoelectrofocusing was performed. The system used was Novex IEF Gels using the XCell SureLock Mini-Cell. The running buffers used were IEF Cathode Buffer (Invitrogen) and IEF Anode Buffer (Invitrogen). Protocols followed the manufacturer's recommendation. The running condition of the system was set at a constant voltage of 100 V for the first 1 hour, then 200 V for the next 1 hour and followed by 500 V for the last 30 minutes. Once the system stopped running, the plate was removed from the electrophoresis unit. The IEF gel was first fixed in 12% TCA for 30 minutes before staining with Coomassie Blue G-250 [23] and silver [24]. The stained gel was scanned using Image Scanner III (GE Healthcare) and visualized and analysed with Image Master Software.
2.7. In-Gel Digestion and Peptide Fingerprint Analysis
Protein spots (1 mm3) were excised and transferred into precleaned 0.5 mL capped tubes. The gels were destained two times by incubating in 200 μL of 200 mM ammonium bicarbonate in 50% (v/v) acetonitrile for 45 minutes at 37°C [25]. The gel pieces were then dried using a CentriVap concentrator at room temperature for 10 minutes.
The dehydrated gel slices were rehydrated with 10 μL of the trypsin solution (0.02 μg/μL) for one hour at room temperature. An additional 50 μL of 40 mM ammonium bicarbonate in 10% (v/v) acetonitrile was added and incubated for 16 to 18 hours at 37°C. The extract was transferred into a new tube. The generated peptides were extracted from the gel pieces by treatment with 50 μL 0.1% (v/v) trifluoroacetic acid twice for 45 minutes each at 37°C. The extracts were combined with the primary supernatant. The volume of the extract was reduced in vacuo to not more than 50% of total volume followed by fingerprinting analysis using MALDI-TOF (ABI 4800 PLUS). The protein was identified using Matrix Science MASCOT search engine.
2.8. Thin Layer Chromatography (TLC)
The conjugation of GSH to Propoxur, Isoproturon, Fenoxaprop-ethyl, and Clodinafop-propargyl was performed using the purified GST isoforms. All the reactions were performed in a volume of 3 mL at 25°C for 20 minutes. An aliquot of reaction mixtures were loaded on a 0.2 mm thick TLC silica gel plate (Merck) and developed using butanol/acetic acid/water (12 : 3 : 5; v/v/v) for 2 hours [26]. The air-dried plate was subsequently stained with ninhydrin (0.25%, w/v, in acetone). After 15 min, coloured spots were observed and marked.
3. Results and Discussion
The screening of GST-expressing bacteria from soil is possible by using MCB [5] as shown in this study. The 16S rRNA gene is highly conserved in bacteria; therefore its sequence is commonly used to identify bacterial isolates [27]. By analysing its 16S rRNA gene sequence, the GST-expressing bacteria isolated in this study were identified and named as Acinetobacter calcoaceticus Y1. Crude activity on EA conjugation was detected from the lysed culture of this isolate, confirming the presence of GSTs in the cells. A single band protein was visualized on SDS-PAGE after purification using GSH-affinity chromatography. The molecular weight (MW) was estimated to be 23 kDa (Figure 1(a)) which was within the range of GSTs molecular weights. A further 2-D gel analysis revealed that the band consisted of two isoforms of GSTs (Figure 1(b)) estimated by vertical isoelectric focusing to have pI values of 4.5 and 6.2, respectively (Figure 1(c)). The presence of multiple isoforms of GSTs is rarely documented in bacteria compared to insects or human. The functions of both isoforms are of great interest; therefore we attempted to purify and characterize each isoform.
Figure 1.
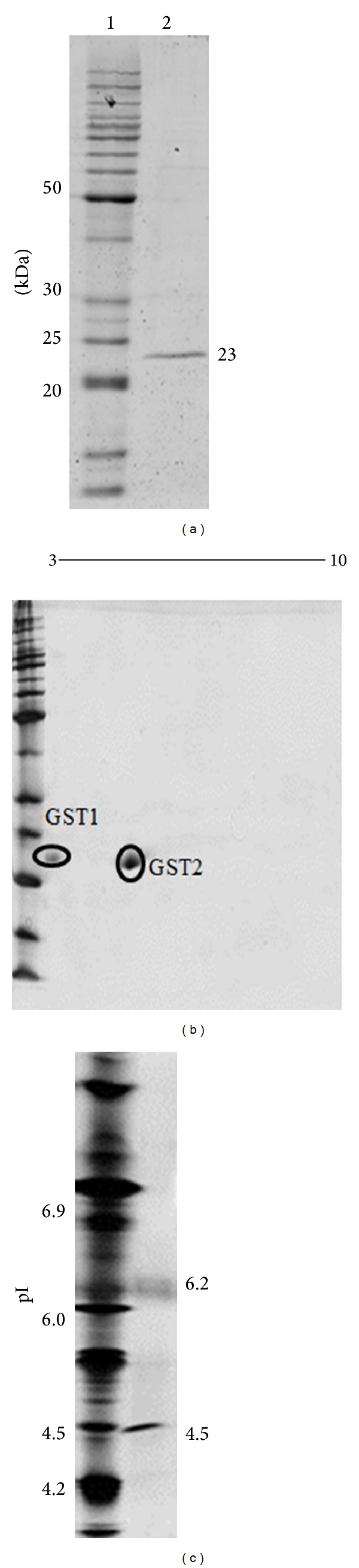
(a) SDS-PAGE of purified GSTs from Acinetobacter calcoaceticus Y1. Lane 1 shows the benchmark standard marker (Invitrogen). Lane 2 shows the presence of single band GST with MW estimated at 23 kDa (10 μg). (b) 2-D gel electrophoresis (IPG 3–10) of purified GSTs resolved into two isoforms (30 μg). (c) Isoelectric-focusing of the purified GST (10 μg). The calculated pIs are 4.5 and 6.2 for GST1 and GST2, respectively. SERVA IEF marker (Invitrogen) was used for pI estimation. Gels were stained with colloidal coomassie blue (a and b) and silver (c).
Both isoforms were separated by using CM and DEAE Sepharose fast flow columns (GE Healthcare). Only “flow-through” was collected from both columns and the eluate was discarded. When the CM Sepharose column was used, isoform pI 4.5 would come out in the “flow-through” while isoform pI 6.2 would appear in the second eluate generated by 10 mM of sodium chloride (NaCl). On the other hand, when the DEAE Sepharose column was used, isoform pI 6.2 would come out in the “flow-through” while isoform pI 4.5 would appear in the second eluate generated by 10 mM of sodium chloride (NaCl). However, neither isoform pI 6.2 nor isoform pI 4.5 from the second eluate fractions showed any GSH-conjugation activity. There was absence of band when both isoforms were run on SDS-PAGE (data not shown). We suggest that the isoforms which were eluted from both columns using NaCl solution might have experienced denaturation as a result of distortion of the ionic interaction within the protein. According to Date and Dominy [28], salts such as NaCl might influence protein stability through electrostatic mechanisms as well as through nonpolar Hofmeister effect. For convenience of discussion isoform pI 4.5 and isoform pI 6.2 were designated as GST1 and GST2, respectively.
GST1 showed a single band on SDS-PAGE which was consistent with the result shown in Figure 1(a) where a single band also occurred at 23 kDa. On the other hand, GST2 displayed different physical characteristic compared to GST1. GST2 tended to form aggregation of which a single band at estimated 110–115 kDa was observed instead of at 23 kDa. Extracellular aggregation of GSTs had not been reported so far but intracellular aggregation of GSTs had been reported for human Pi (π) class GST (hGSTP) [29] and Onchocerca volvulus GST (OvGST) [30]. Intracellular protein aggregation is mainly caused by posttranslational modification of protein through the process of glycosylation and phosphorylation.
Table 1 indicates that both GST1 and GST2 had highest affinity towards ethacrynic acid compared to the other substrates. This was an early indication that both isoforms behave similar to a Pi (π) class GSTs [31]. Although 1-Chloro-2,4-dinitrobenzene is a common substrate for GSTs from other organisms, both isoforms from the A. calcoaceticus Y1 isolated in this study recorded a considerably low activity towards the substrate. This characteristic is in line with bacterial GSTs. Our study showed that GST1 reacted with hydrogen peroxide, indicating that it is a selenium-dependent glutathione peroxidase [32]. In contrast, GST2 was not shown to be catalytically active towards hydrogen peroxide and cumene hydroperoxides. The conjugation of both isoforms with a lipid peroxidation product such as trans,trans-hepta-2,4-dienal also suggested that both the isoforms may play a role in combating oxidative stress due to lipid peroxidation. No dichloromethane dehalogenase activity was seen for both isoforms, implying that they were not of the theta class GST. Inference had been made on the ability of GSTs to degrade morpholine [33]. In this study, the ability of both isoforms to conjugate pesticides was also investigated. Our findings showed that both were unable to conjugate propoxur, fenoxaprop-ethyl, and clodinafop-propargyl. However, GST2 was able to conjugate isoproturon (Figure 2), a feature not seen with GST1. Allocati et al. [8] had reported that bacterial GSTs were able to detoxify several classes of herbicides. Early studies have linked conjugating activity with DCNB with insecticide resistance, particularly in insects. However, later studies showed that this was not necessarily correlated; for example, an epsilon class of GST in fruit flies, CG16936, [34] and in Anopheles gambiae, GSTE1-1z, [35] had conjugating activity towards DCNB but did not confer insecticide resistance. Our findings showed that GST2 had no activity with DCNB but was able to selectively conjugate isoproturon, suggesting its specific functional role in the cells. The substrate specificities of both isoforms were different and thus it is believed that they were of two existing homodimers.
Table 1.
Specific activities of GST1 and GST2 towards selected substrates.
| Substrate | Specific activity (μmol/min/mg) | |
|---|---|---|
| GST1 | GST2 | |
| Ethacrynic acid | 24.91 ± 1.24 | 15.71 ± 2.53 |
| Hydrogen peroxide | 3.93 ± 0.38 | nd |
| 1-Chloro-2,4-dinitrobenzene | 0.61 ± 0.122 | 0.542 ± 0.127 |
| trans,trans-Hepta-2,4-dienal | 0.11 ± 0.02 | 0.084 ± 0.01 |
| 1,2-Dichloro-4-nitrobenzene | Nd | nd |
| p-nitrophenyl chloride | Nd | nd |
| Sulfobromophthalein | Nd | nd |
| trans-4-Phenyl-3-butene-2-one | Nd | nd |
| Cumene hydroperoxide | Nd | nd |
| Dichloromethane | Nd | nd |
nd: not detected.
Figure 2.
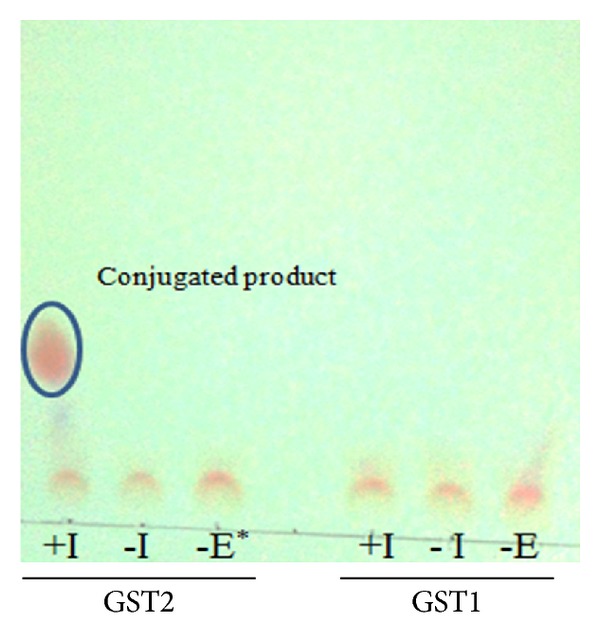
Thin layer chromatography indicating conjugation of isoproturon by GST2 (isoform pI 6.2) (circled spot). No conjugated product was seen when GST1 (isoform pI 4.5) was tested. +I = with isoproturon, −I = without isoproturon, −E = without enzyme.
Due to lack of bacterial genome databases, we were unable to significantly identify each isoform as glutathione transferase through MALDI-TOF mass spectrometry analysis. The putative glutathione transferases of Acinetobacter calcoaceticus strain PHEA-2 [36] which were, however, retrieved from http://www.uniprot.org/uniprot/ are tabulated in Table 2. It shows that the strain has 10 putative GSTs of which two of the expressed proteins have close characteristics to GST1 and GST2. As shown in Table 2, The UniProt identifiers of F0KLY9 (putative glutathione S-transferase) and F0KKB0 (Glutathione S-transferase III) are similar in MW and pI values to GST1 and GST2, respectively. Nevertheless, more confirmative evaluation is warranted for the identification of the isoforms, such as N-terminal sequencing.
Table 2.
List of putative glutathione transferases of Acinetobacter calcoaceticus (strain PHEA-2) retrieved from http://www.uniprot.org/uniprot/. pI and MW values were computed at Compute pI/Mw (http://www.expasy.org/tools/). Bold indicates that F0KLY9 (putative glutathione S-transferase) and F0KKB0 (glutathione S-transferase III) were similar in MW and pI values to GST1 and GST2, respectively.
| UniProt identifiers | Protein name | Gene name | a.a.* length | pI/MW |
|---|---|---|---|---|
| F0KI99 | Putative glutathione S-transferase | gstB BDGL_000451 | 203 | 4.91/22987.64 |
| F0KLY9 | Putative glutathione S-transferase | erd13 BDGL_003315 | 201 | 4.85/22385.40 |
| F0KKB0 | Glutathione S-transferase III | gst3 BDGL_000699 | 214 | 5.97/24579.78 |
| F0KI95 | Putative glutathione S-transferase | yliJ BDGL_000447 | 208 | 5.19/24435.28 |
| F0KGP1 | Glutathione transferase FosA | fosA BDGL_002726 | 135 | 5.34/15740.79 |
| F0KH55 | Putative glutathione S-transferase | BDGL_001572 | 228 | 6.01/26244.78 |
| F0KL12 | Glutathione S-transferase-like protein | yghU BDGL_000797 | 282 | 5.62/31834.09 |
| F0KLP2 | Glutathione S-transferase | BDGL_000872 | 246 | 5.52/28865.77 |
| F0KND6 | Putative glutathione S-transferase | yfcG BDGL_003500 | 206 | 5.13/23912.28 |
| F0KJF2 | Glutathione S-transferase | gst3 BDGL_003061 | 222 | 6.79/26004.25 |
*a.a.: amino acid.
An amino acid sequence similarity check indicated that both isoforms (F0KLY9 and F0KKB0) have no similarity to any class of GSTs in the database. In most GST classes the N-terminal tyrosine residue is the active residue that interacts with GSH to stabilise the thiolate anion. In theta and zeta classes the role is carried out by serine residue, while in omega and beta classes cysteine is the active residue [2], at most of the time in positions 5 and 6.
In Table 3, the retrieved amino acid sequence of F0KLY9 (putative glutathione S-transferase) indicates the presence of tyrosine residue at position 6. However, none of the abovementioned amino acids known to be active residues was found in the N-terminal sequence of F0KKB0 (Glutathione S-transferase III). Besides the N-terminal amino acids, C-terminal histidine-106 and lysine-107 had been reported to contribute to the interaction with GSH [37]. The same behaviour was seen in Drosophila melanogaster GSTD3 where distal amino acids that are catalytically essential for conjugation (unpublished data from our laboratory). Therefore, to identify the catalytically active residues in F0KKB0 (Glutathione S-transferase III) is of a great interest. Both putative GSTs were however not identified to any class of GSTs when submitted to BLAST (http://blast.ncbi.nlm.nih.gov/).
Table 3.
Protein (FASTA) sequences for F0KLY9 (putative glutathione S-transferase) and F0KKB0 (glutathione S-transferase III) were retrieved from http://www.uniprot.org/uniprot/. Bold and italic is the identified active residue (Y, S, or C at positions 5 or 6) which is present in F0KLY9 (putative glutathione S-transferase) amino acid sequence.
| UniProt Identifiers | Amino acid sequences |
|---|---|
| F0KLY9 |
MSLKLYTNKESRGVVIDWLLVELGVECERIEVAYKTEMKSPEYLKLN PFGKVPVLVDGDVVIYELGAICAYLADKFSDKGLAPALDDPKRGLYYR WLFLMAGPWEAAGVDKALGIEVSPEQKMFVGYGDYNDAYQALVQGL SEANPYVCGEQFTAADVSVGAMLLWQLKMNAIESHPAITRYVETIKQR EGLKQSTMGQLL |
| F0KKB0 | MSIILHHLNASRSFRILWLLEEINQPYELKSYFRDKTTNLAPQELKNIHP LGKSPVIELNGKVIAESGAIVEILIEKFAPQLMPAKDSDSYLDYLQWVH FSESSAMVPYLLNIFNSIELKNGTQLKFLDQYAHTELDKVFSYLDQQLV GKKFLVGNSLTGADFMIGFVVYGLINSLNIRSKYLNIEQYVKSLENLES WQKAMSIEQNLHHQTNA |
4. Conclusion
A locally isolated bacterium, identified as Acinetobacter calcoaceticus Y1, was found to produce a GST with a molecular weight of 23 kDa (based on SDS-PAGE). Through the use of two-dimensional chromatography, the band was found to compose of two isoforms. Isoelectric focusing determined that the isoforms, named GST1 and GST2, have pI values of 4.5 and 6.2, respectively. Both isoforms were able to conjugate with substrates involved in lipid peroxidation, and GST1 had shown peroxidase activity. These suggest that GST1 and GST2 may have roles in combating oxidative stress. GST2 was shown to specifically conjugate isoproturon. The behaviour could protect the bacteria against harmful toxic herbicide. Both GST1 and GST2 closely resemble putative GSTs of Acinetobacter calcoaceticus (strain PHEA-2), F0KLY9 (Putative glutathione S-transferase) and F0KKB0 (Glutathione S-transferase III), respectively. The findings of this study concur that GST-expressing bacteria are found in the soil and that they might involve in the detoxification and degradation of xenobiotics including pesticides. Further studies on bacterial GSTs are warranted because of the potential of soil bacteria as natural and ecofriendly agents in environmental bioremediation. GSTs alone would however not be sufficiently effective for a broad-based xenobiotic biotransformation, perhaps engineered fusion proteins with several distinct catalytic capabilities should be one of the promising approaches. All in all, GSTs and their genes are promising tools to develop applications in biological treatment of environmental and industrial pollutants.
Acknowledgments
This work was funded by the University of Malaya Postgraduate Research Fund (PPP: PV034/2012A) and the Ministry of Higher Education under the Fundamental Research Grant (FRGS) (FP023/2009).
Conflict of Interests
The authors declare that there is no conflict of interests.
References
- 1.Adler V, Yin Z, Fuchs SY, et al. Regulation of JNK signaling by GSTp. EMBO Journal. 1999;18(5):1321–1334. doi: 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochemical Journal. 2001;360(1):1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Ilio C, Aceto A, Piccolomini R, et al. Purification and characterization of three forms of glutathione transferase from Proteus mirabilis. Biochemical Journal. 1988;255(3):971–975. doi: 10.1042/bj2550971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiktelius E, Stenberg G. Novel class of glutathione transferases from cyanobacteria exhibit high catalytic activities towards naturally occurring isothiocyanates. Biochemical Journal. 2007;406(1):115–123. doi: 10.1042/BJ20070328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bader R, Leisinger T. Isolation and characterization of the Methylophilus sp. strain DM11 gene encoding dichloromethane dehalogenase/glutathione S-transferase. Journal of Bacteriology. 1994;176(12):3466–3473. doi: 10.1128/jb.176.12.3466-3473.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anandarajah K, Kiefer PM, Jr., Donohoe BS, Copley SD. Recruitment of a double bond isomerase to serve as a reductive dehalogenase during biodegradation of pentachlorophenol. Biochemistry. 2000;39(18):5303–5311. doi: 10.1021/bi9923813. [DOI] [PubMed] [Google Scholar]
- 7.Cacciatore I, Cornacchia C, Pinnen F, Mollica A, Di Stefano A. Prodrug approach for increasing cellular glutathione levels. Molecules. 2010;15(3):1242–1264. doi: 10.3390/molecules15031242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allocati N, Federici L, Masulli M, Di Ilio C. Glutathione transferases in bacteria. FEBS Journal. 2009;276(1):58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- 9.Vuilleumier S. Bacterial glutathione S-transferases: what are they good for? Journal of Bacteriology. 1997;179(5):1431–1441. doi: 10.1128/jb.179.5.1431-1441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chronopoulou EG, Labrou NE. Glutathione transferases: emerging multidisciplinary tools in red and green biotechnology. Recent Patents on Biotechnology. 2009;3(3):211–223. doi: 10.2174/187220809789389135. [DOI] [PubMed] [Google Scholar]
- 11.Choi JW, Kim YK, Song SY, Lee IH, Lee WH. Optical biosensor consisting of glutathione-S-transferase for detection of captan. Biosensors and Bioelectronics. 2003;18(12):1461–1466. doi: 10.1016/s0956-5663(03)00124-6. [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa K, Miwa S, Tajima T, Tsutsumiuchi K, Taniguchi H, Miwa J. A rapid and inexpensive method to screen for common foods that reduce the action of acrylamide, a harmful substance in food. Toxicology Letters. 2007;175(1–3):82–88. doi: 10.1016/j.toxlet.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Kapoli P, Axarli IA, Platis D, et al. Engineering sensitive glutathione transferase for the detection of xenobiotics. Biosensors and Bioelectronics. 2008;24(3):498–503. doi: 10.1016/j.bios.2008.06.037. [DOI] [PubMed] [Google Scholar]
- 14.Andreou VG, Clonis YD. Novel fiber-optic biosensor based on immobilized glutathione S-transferase and sol-gel entrapped bromcresol green for the determination of atrazine. Analytica Chimica Acta. 2002;460(2):151–161. [Google Scholar]
- 15.Eklund BI, Edalat M, Stenberg G, Mannervik B. Screening for recombinant glutathione transferases active with monochlorobimane. Analytical Biochemistry. 2002;309(1):102–108. doi: 10.1016/s0003-2697(02)00258-0. [DOI] [PubMed] [Google Scholar]
- 16.Habig WH, Pabst MJ, Jakoby WB. Glutathione S transferases. The first enzymatic step in mercapturic acid formation. Journal of Biological Chemistry. 1974;249(22):7130–7139. [PubMed] [Google Scholar]
- 17.Motoyama N, Dauterman WC. Purification and properties of housefly glutathione S-transferase. Insect Biochemistry. 1977;7(4):361–369. [Google Scholar]
- 18.Wendel A. Glutathione peroxidase. Methods in Enzymology C. 1981;77:325–333. doi: 10.1016/s0076-6879(81)77046-0. [DOI] [PubMed] [Google Scholar]
- 19.Sherratt PJ, Pulford DJ, Harrison DJ, Green T, Hayes JD. Evidence that human class Theta glutathione S-transferase T1-1 can catalyse the activation of dichloromethane, a liver and lung carcinogen in the mouse: comparison of the tissue distribution of GST T1-1 with that of classes Alpha, Mu and Pi GST in human. Biochemical Journal. 1997;326(3):837–846. doi: 10.1042/bj3260837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brophy PM, Southan C, Barrett J. Glutathione transferases in the tapeworm Moniezia expansa . Biochemical Journal. 1989;262(3):939–946. doi: 10.1042/bj2620939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9(6):255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 24.Swain M, Ross NW. A silver stain protocol for proteins yielding high resolution and transparent background in sodium dodecyl sulfate-polyacrylamide gels. Electrophoresis. 1995;16(6):948–951. doi: 10.1002/elps.11501601159. [DOI] [PubMed] [Google Scholar]
- 25.Alias Z, Clark AG. Studies on the glutathione S-transferase proteome of adult Drosophila melanogaster. Responsiveness to chemical challenge. Proteomics. 2007;7(19):3618–3628. doi: 10.1002/pmic.200700070. [DOI] [PubMed] [Google Scholar]
- 26.Ben-Arie N, Khen M, Lancet D. Glutathione S-transferases in rat olfactory epithelium: purification, molecular properties and odorant biotransformation. Biochemical Journal. 1993;292(2):379–384. doi: 10.1042/bj2920379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. Journal of Clinical Microbiology. 2007;45(9):2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Date MS, Dominy BN. Modeling the influence of salt on the hydrophobic effect and protein fold stability. Communications in Computational Physics. 2011;13(1):90–106. [Google Scholar]
- 29.Ranganathan PN, Whalen R, Boyer TD. Characterization of the molecular forms of glutathione S-transferase P1 in human gastric cancer cells (Kato III) and in normal human erythrocytes. Biochemical Journal. 2005;386(3):525–533. doi: 10.1042/BJ20041419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liebau E, Wildenburg G, Walter RD, Henkle-Duhrsen K. A novel type of glutathione S-transferase in Onchocerca volvulus. Infection and Immunity. 1994;62(11):4762–4767. doi: 10.1128/iai.62.11.4762-4767.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang HL, Zeng QY, Nie LJ, Zhu SG, Zhou XW. Purification and characterization of a novel glutathione S-transferase from Atactodea striata. Biochemical and Biophysical Research Communications. 2003;307(3):626–631. doi: 10.1016/s0006-291x(03)01221-x. [DOI] [PubMed] [Google Scholar]
- 32.Thomson CD. Selenium-dependent and non-selenium-dependent glutathione peroxidase in human tissues of New Zealand residents. Biochemistry International. 1985;10(4):673–679. [PubMed] [Google Scholar]
- 33.Emtiazi G, Saleh T, Hassanshahian M. The effect of bacterial glutathione S-transferase on morpholine degradation. Biotechnology Journal. 2009;4(2):202–205. doi: 10.1002/biot.200800238. [DOI] [PubMed] [Google Scholar]
- 34.Alias Z, Clark AG. Adult Drosophila melanogaster glutathione S-transferases: effects of acute treatment with methyl parathion. Pesticide Biochemistry and Physiology. 2010;98(1):94–98. [Google Scholar]
- 35.Ortelli F, Rossiter LC, Vontas J, Ranson H, Hemingway J. Heterologous expression of four glutathione transferase genes genetically linked to a major insecticide-resistance locus from the malaria vector Anopheles gambiae . Biochemical Journal. 2003;373(3):957–963. doi: 10.1042/BJ20030169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhan Y, Yan Y, Zhang W, et al. Genome sequence of Acinetobacter calcoaceticus PHEA-2, isolated from industry wastewater. Journal of Bacteriology. 2011;193(10):2672–2673. doi: 10.1128/JB.00261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casalone E, Allocati N, Ceccarelli I, et al. Site-directed mutagenesis of the Proteus mirabilis glutathione transferase B1-1 G-site. FEBS Letters. 1998;423(2):122–124. doi: 10.1016/s0014-5793(98)00080-5. [DOI] [PubMed] [Google Scholar]


