Abstract
Separase, a protease encoded by the ESPL1 gene, cleaves the chromosomal cohesin during mitosis. Separase protein and transcripts are overexpressed in a wide range of human cancers (Meyer et al., Clin Cancer Res 2009; 15: 2703-2710). To investigate the physiological consequence of Separase overexpression in animals, we have generated a transgenic MMTVEspl1 mouse model that overexpresses Separase protein in the mammary glands. MMTV-Espl1 mice in a C57BL/6 genetic background develop aggressive, highly aneuploid, and estrogen receptor alpha positive (ERα+) mammary adenocarcinomas with an 80% penetrance. The mammary tumors caused by overexpression of Separase, alone or combined with p53 heterozygosity, in mammary epithelium mimic several aspects of the most aggressive forms of human breast cancer, including high levels of genetic instability, cell cycle defects, poor differentiation, distant metastasis, and metaplasia. Histopathologically, MMTV-Espl1 tumors are highly heterogeneous showing features of both luminal as well as basal subtypes of breast cancers, with aggressive disease phenotype. In addition to aneuploidy, Separase overexpression results in chromosomal instability (CIN) including premature chromatid separation (PCS), lagging chromosomes, anaphase bridges, micronuclei, centrosome amplification, multi nucleated cells, gradual accumulation of DNA damage, and progressive loss of tumor suppressors p53 and cadherin gene loci. These results suggest that Separase overexpressing mammary cells are not only susceptible to chromosomal missegregation-induced aneuploidy but also other genetic instabilities including DNA damage and loss of key tumor suppressor gene loci, which in combination can initiate tumorigenesis and disease progression.
Keywords: Separase, Espl1, Aneuploidy, p53, Cohesin, Chromosomal segregation, Mammary Cancer, Animal model
Introduction
Cohesin, an evolutionarily conserved protein complex, plays a pivotal role in chromosomal segregation. Cohesin complex holds two newly replicated sister chromatids together from S-phase until the end of metaphase to allow accurate segregation of chromatids into two daughter cells. At the onset of anaphase, Separase (encoded by the ESPL1 gene), an endopeptidase, is activated and cleaves the cohesin subunit Rad21 (also known as Scc1 or Mcd1 in budding yeast), which releases sister chromatid cohesion to allow chromosome disjunction (for a review see 1). Overexpression of Separase is a feature of many human tumors, including breast cancer, and has been reported to cause chromosomal missegregation and aneuploidy in in vitro tissue culture models 2,3. Compared to the matched normal breast tissue, more than 60% of human breast tumors overexpress Separase protein 2,3. Mining of the Oncomine database indicates a strong positive correlation between Separase mRNA expression and tumor grade as well as a strong negative correlation with disease-free and overall survival 2. How Separase overexpression-driven aneuploidy overcomes the threshold of tumor resisting forces within the cell and results in the initiation of tumor formation, and how other co-operating lesions further this process have not been investigated, and targeted Separase overexpression in the mammary gland provides an ideal model to probe the role of aneuploidy in mammary tumorigenesis.
Aneuploidy is a hallmark of human cancers and a leading cause of mental retardation and spontaneous miscarriages (http://cgap.nci.nih.gov/Chromosomes/Mitelman, 4,5). In contrast to aneuploidy (‘the state’ of having abnormal chromosomal number), chromosomal instability (CIN) — a high rate of gain or loss of whole or part of chromosomes during cell division is also observed in large proportion of human cancers. CIN is thought to drive continually evolving karyotypes and tumor heterogeneity 6-12. While CIN leads to aneuploidy, aneuploidy can occur without CIN. The link between CIN and aneuploidy is not fully understood, and the molecular mechanism by which CIN is driven is not fully known. In this context, the unsolved question that remains is how and whether aneuploidy and CIN predispose to tumorigenesis. In vitro studies suggest that aneuploidy could interfere with cell proliferation 13,14 and thereby would be selected against by the cellular surveillance mechanisms 15. However, further mutations or chromosomal alterations due to aneuploidy pressure 16 could allow cells to overcome this restriction and unleash their tumorigenic potential. It has been suggested that CIN would allow cells to overcome the negative effect of aneuploidy and promote tumorigenesis below a certain threshold 17. It is, however, not known when and how aneuploid cells acquire CIN phenotype, and whether aneuploidy drives CIN that ultimately overcomes the aneuploidy–induced growth disadvantages. Studies in budding yeast find that while aneuploidy alone, without any genetic mutations, can confer improved cellular growth under certain stress conditions; aneuploid cells in general divide less rapidly than normal diploid cells under conventional laboratory conditions 13,14,16,18, which is known as the “aneuploidy paradox” 15.
One of the major paths to aneuploidy is chromosomal missegregation. To probe the physiological consequences of Separase-overexpression induced chromosomal missegregation and resultant aneuploidy we have generated a MMTV-Espl1 transgenic mouse model with targeted overexpression of Separase in the mouse mammary epithelium. MMTV-Espl1 mice in C57/Bl6 genetic background develop aggressive, highly aneuploid, ERα+ mammary adenocarcinomas with an 80% penetrance and a median latency of 12 months. Separase overexpression results in aneuploidy and develops mammary tumors by acquiring additional cooperative lesions including DNA damage and progressive loss of p53. The mammary tumors caused by overexpression of Separase alone as well as combined with p53 mutant background mimic several aspects of the most aggressive forms of human breast cancer, and also provide a new model to study human breast cancers.
Results
Espl1 overexpression in mammary epithelium results in mammary tumorigenesis
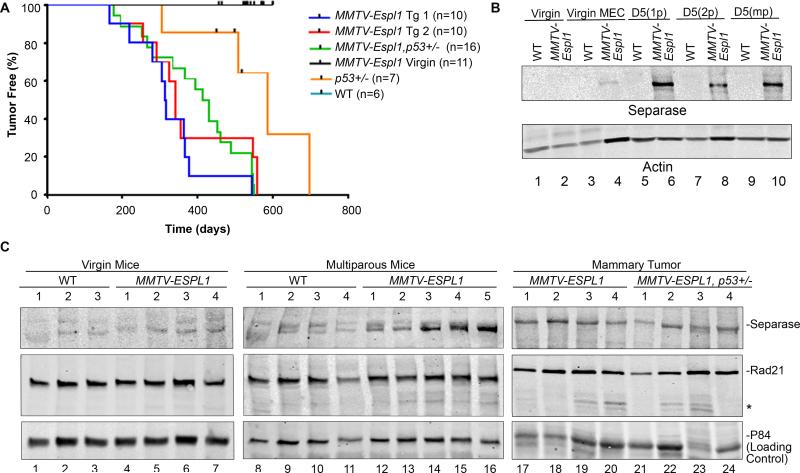
Expression of the mouse Espl1 (mEspl1) gene in C57BL/6 mice was placed under the regulation of the pregnancy hormone responsive mouse mammary tumor virus long term repeat (MMTV-LTR) promoter 19 to induce mammary-specific expression of the transgene encoding Separase protein (Fig. S1A). C57BL/6 genetic background was chosen for its rare incidence of spontaneous mammary tumor development 20, so that the specific effects of Separase-overexpression in the mammary epithelium over a long time could be monitored. Integration of mEspl1 transgene (Fig. S1B) and significant overexpression of Separase protein was confirmed in mammary glands of two independently established lines (Tg1 and Tg2) of transgenic mice compared to wild type (WT) litter mates following pregnancy and lactation (Fig. S1C). Mammary cells from Tg1 and Tg2 show 1 and 2 extra copies of the Separase gene (red signal), respectively, over the wild type littermate with two copies. Multiparous MMTV-Espl1 mice from both the lines (Tg1 and Tg2) developed mammary tumors with 80% penetrance and a median latency of 314 and 340 days, respectively (Fig. 1A, blue and red line, log-rank test p< 0.0001 compared to all other curves). Since there was no significant differences in the tumor profile between both the lines, Tg1 line was used for subsequent analysis. An additive effect of p53 heterozygosity with Separase overexpression in mammary tumor latency was not observed in this model, since multiparous MMTV-Espl1, p53 +/− mice developed mammary tumors with 100% penetrance and a median latency of 439 days (Fig. 1A, green line, log-rank test p=0.3503). No tumor was observed in the MMTV-Espl1 nulliparous mice with up to two years of age. In contrast to the nulliparous, the primiparous and multiparous MMTV-Espl1 mice and the resultant mammary tumors showed significantly increased expression of the Separase protein (Fig. 1B-C). There was no detectable difference in Separase expression in the nulliparous (virgin) transgenic mice tissue as compared to their virgin wild type littermates of same age (Fig. 1B-C), which is consistent with the previous report that MMTV expression is weak in the mammary gland in the absence of pregnancy-induced hormones 21,22. With increased Separase expression, cleavage of cohesin-Rad21 was noted in multiparous mammary glands and ~50% mammary tumors (Fig. 1C), suggesting differential enzymatic activity of the overexpressed Separase protein in MMTVEspl1 associated tumors. No significant overexpression of Separase was noted in tissues other than mammary gland of the MMTV-Espl1 multiparous mice (data not shown).
Figure 1. Overexpression of Separase induces mammary tumor in MMTV-Espl1 mice.
(A) Kaplan–Meier analysis for tumor onset and survival for wild type (WT), MMTV-Espl1 (Tg1 and Tg2), MMTV-Espl1 virgin, MMTV-Espl1, p53+/−, and p53+/− genotypes. The number of mice used (n) for each genotype is shown in the bracket. Log-rank test p=0.3503. (B) Expression of Separase protein in MMTV-Espl1 virgin [lanes 1, 2: protein from whole cell lysate; lanes 3, 4: protein from primary mammary epithelial cells (MEC) at passage zero], primiparous at day 5 lactating (D5 1p), and multi-parous at day 5 lactating (D5 2p and D5 mp) mice. (C) Expression of Separase and its cohesin substrate Rad21 in virgin, multiparous WT and MMTV-Espl1 mammary gland of 7.5 months old mice, and in the mammary tumors from MMTV-Espl1 and MMTV-Espl1, p53+/− multiparous animals (n=3-5/group). p84 nuclear matrix protein is shown to compare loading. *anticipated Rad21 cleavage band.
Histopathological characterization of MMTV-Espl1 tumors in relation to human breast cancers
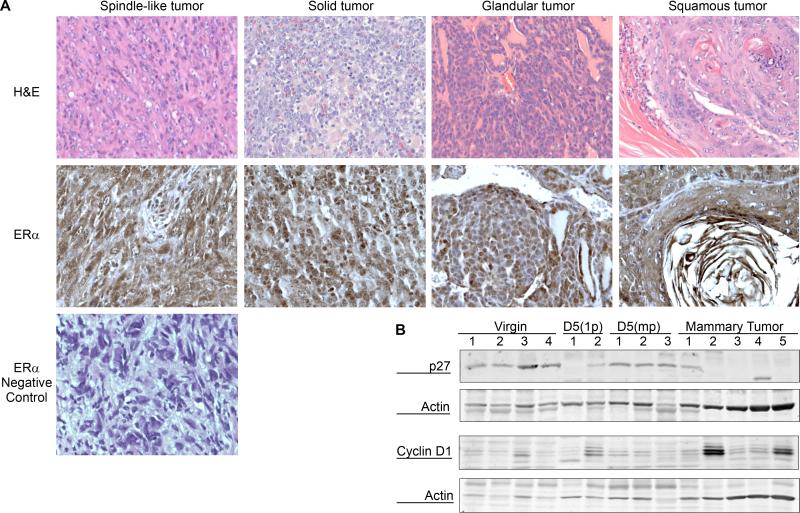
Four distinct histological subtypes of the mammary tumors were observed by immunohistochemical staining (spindle-like, solid, glandular and squamous, Fig. 2A). All mammary carcinomas irrespective of their histological appearance expressed varying levels of ERα nuclear and cytoplasmic staining (Fig. 2A). Over 40% of these tumors also showed higher expression of Cyclin D1 in their well differentiated regions, a common prognostic marker for human ERα positive breast tumors 23-25 and loss of cyclin dependent kinase (CDK) inhibitor p27 (CDKN1B) (Fig. 2B). Loss of p27 has been associated with Tamoxifen resistance in ERα positive human breast cancers 26-28 and is also an independent marker for more invasive breast carcinomas in humans 29.
Figure 2. Histopathological characterization of MMTV-Espl1 tumors.
(A) Four histological subtypes (spindle-like, solid, glandular, and squamous) of MMTV-Espl1 multiparous mouse mammary tumors were observed. Representative Hematoxylin and eosin (H&E) stained images (top panel) and ERα positive nuclear and cytoplasmic staining (middle panel) is shown. A spindle-like tumor section is shown for the negative staining using Rabbit IgG and secondary antibody treatment (bottom panel) indicating the absence of positive, brown nuclear and cytoplasmic staining specific for ERα (B) Western blot showing loss of cyclin dependent kinase (CDK) inhibitor p27 expression in 4 out of 5, and higher expression of Cyclin D1 in 2 out of 5 of mammary tumors compared to multiparous mammary glands. β-Actin is shown to compare loading.
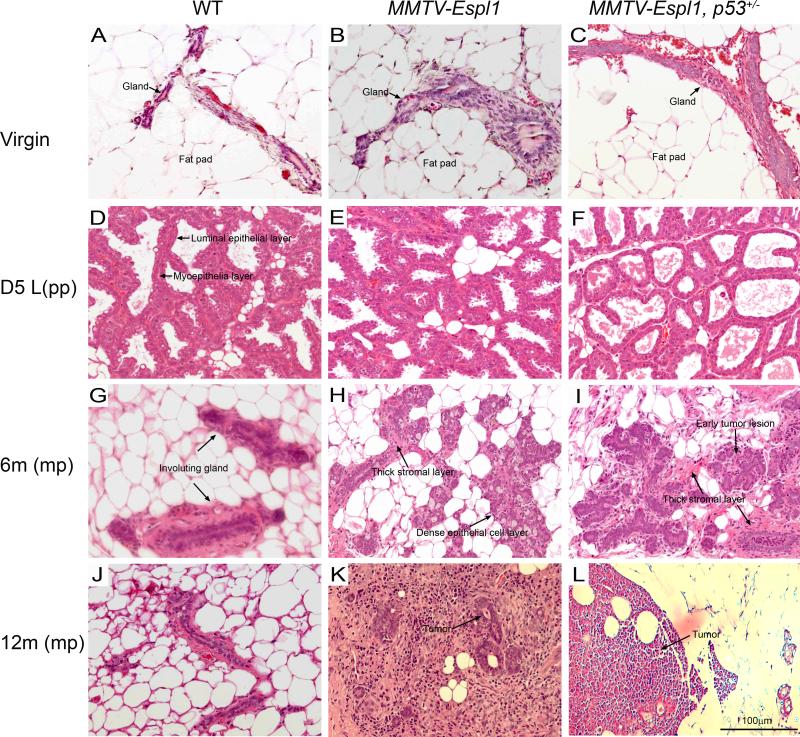
Early in our studies, we noted that the tumors were formed in the transgenic mice that underwent cycles of pregnancy and lactation. In contrast, no tumors were observed in virgin transgenic mice. To examine the histopathological effects of Separase overexpression we performed mammary gland biopsies (n=3 mice) over a time period (virgin, 5 days post first lactation, 6 and 12 months post first lactation in mice force weaned at day one). Glands from MMTV-Espl1 and MMTV-Espl1, p53 +/− mice appeared histologically normal and comparable to the control WT littermate cohorts at the virgin and day 5 post first lactation states (Fig. 3A-F). At 6 months of age, post lactating mammary glands of these mice force weaned on day one appeared severely hyperplastic and had significantly delayed involution compared to their wild type cohorts (Fig. 3G-I). They were also hyper-proliferative at this stage with visible mitotic figures and pleomorphic nuclei. At approximate 12 months of age, both the MMTV-Espl1 and MMTV-Espl1, p53 +/− mice developed small but significant masses of mammary hyperplasia and large amounts of immune reaction as well as the presence of hyper proliferative stroma (Fig. 3J-L).
Figure 3. Histological characterization and progression of mammary tumorigenesis in MMTV-Espl1 transgenic mammary glands.
Mammary morphologies in Hematoxylin and eosin (H&E)-stained sections of Virgin (A, B, C) , Day five (D5) lactation (D, E, F), D5 post force weaned at D1, 6 month old mice (G, H, I) and D5 post force weaned at D1, 12month old mice (J, K, L) of wild type (WT), MMTV-Espl1 and MMTV-Espl1, p53+/− genotype are shown. Mammary glands from virgin WT, MMTV-Espl1 and MMTV-Espl1, p53+/− mice display a normally sparse and isolated mammary gland epithelium within large areas of mammary fat pad. MMTV-Espl1 and MMTV-Espl1, p53+/− mice also display normal looking mammary gland architecture at day 5 lactation (E, F) with regular luminal epithelial ductal structure and a thin myoepithelial layer surrounding the luminal cells comparable to WT littermate mice (D, arrows). The epithelial cells in multi-pregnant 6 month old MMTV-Espl1 mice at day five post day one force weaned mice display dense epithelial cell clusters (H, I, arrows) and thick stromal layers (H, I, arrows) compared to WT litter mate controls that display sparse mammary epithelial cells and involuting mammary glands at this stage (G). Mammary duct hyperplasia and early tumor lesions also develop in 6 month old multiparous MMTV-Espl1, p53+/− mice (I, arrows). At 12 months post day one lactation 80% MMTV-Espl1 mice and 100% MMTV-Espl1, p53+/− mice show mammary tumor development with palpable mammary tumor formation (K, L, arrows). (n = 3 for each genotype. Representative images are shown for all animals.)
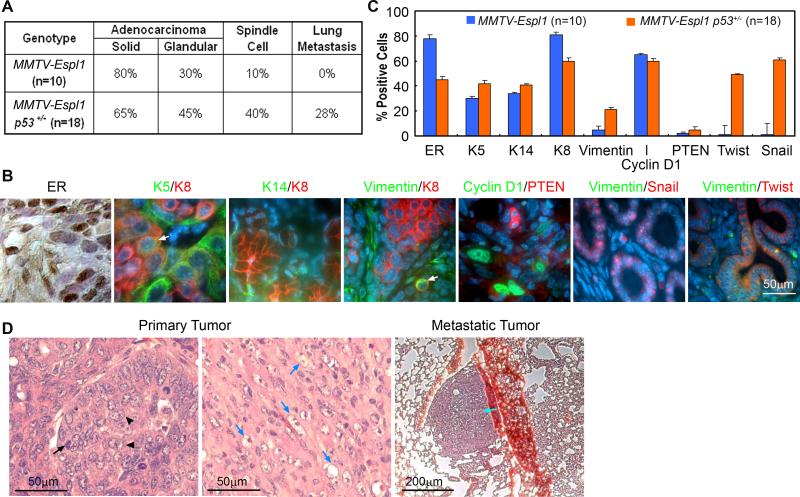
To identify key features similar to human breast cancers, we performed histological examination and marker analysis on mammary tumors obtained from the MMTV-Espl1 and MMTV-Espl1, p53+/− mice. Both genotypes developed high grade mammary adenocarcinomas with both inter- and intra-tumor heterogeneity indicative of highly perturbed cellular differentiation. Regardless of genotype, the tumors exhibited mixed morphologies summarized in Fig. 4A. All mammary carcinomas, irrespective of their histological appearance, expressed varying levels of ERα and luminal epithelial markers, including Keratin-8 and Cyclin D1 (Fig. 4B), with abundant expression in well differentiated tumor regions but greatly diminished or absent in poorly differentiated areas. Nests of carcinoma cells variably expressed basal/myoepithelial lineage markers (Keratins-5, -14, Vimentin, Fig. 4B, quantified in Fig. 4C). Occasional co-expression of basal and luminal markers (Keratin-5 and Keratin-8) were observed in small regions of some of these tumors, suggesting the presence of undifferentiated cell populations (Fig. 4B, arrow).
Figure 4. MMTV-Espl1 transgenic mice have highly heterogeneous ERα positive mammary tumors with variable markers for luminal and basal epithelial cells.
(A) Occurrence of each tumor morphology/feature for all tumors within a genotypic class (n = number of tumors; p=0.0633, Fisher's exact test). (B) Representative immunohistochemical and immunofluorescence staining for ERα brown), Keratin-5 (K5, green), Keratin-8 (K8, red), Keratin-14 (K14, green), Vimentin (green), Cyclin D1 (green), PTEN (red), Snail and Twist (red). Keratin-8 (green) and Keratin-5 (red) label luminal and myoepithelial cells, respectively, with regions of co-expressing cells (arrow). Metaplastic tumor cells show dual staining of Keratin-8 and the mesenchymal marker, Vimentin (arrow). (C) The quantification (% positive cells) for all markers, (p<0.0001) from 10 MMTV-Espl1 and 18 MMTV-Espl1 p53+/− tumors. Approximate 100 cells from each tumor were analyzed. (D) Solid morphology of the primary tumors, with high levels of mitosis (arrows) and pleomorphic nuclei (arrowheads). Homogeneous, fusiform, spindloid cells of a primary carcinosarcoma type mammary tumor entrap carcinomatous cells are shown (blue arrows). Pulmonary metastases (cyan arrow) were observed in MMTV-Espl1, p53+/− tumors. Because the tumors showed mixed morphological phenotypes, regardless of genotype, the occurrence of each morphology/feature for all tumors within a genotypic class was tabulated (A, C).
Both MMTV-Espl1 and MMTV-Espl1, p53+/− mice (1/10 vs. 8/18 cases, p=0.0633, Fisher's exact test) developed regions of carcinosarcomas, which are also called “EMT tumors” (Epithelial to Mesenchymal Transition) 30. These tumors have characteristic faintly-staining, fusiform, spindloid cells (Fig. 4D, blue arrows), and are reminiscent of mammary tumors in p53 mutant mice. In some of these carcinosarcomas we observed dual expression of mesenchymal (Vimentin) and epithelial (Keratin-8) markers, which are mutually exclusive lineage markers in the normal gland (Fig. 4B, arrow). Also about 30% of the MMTV-Espl1, p53+/− tumors (5/18) has significant expression of Twist and Snail, two prominent transcription factors that have been implicated to regulate epithelial to mesenchymal transition (Fig. 4B). Unlike MMTV-Espl1 tumors, distant metastases were observed only in the lungs of MMTV-Espl1, p53+/− (5 of 18 cases, not all of them overlapping with the expression of Twist and Snail as stated above) (Fig. 4D, cyan arrow), which is notable since relatively few transgenic mammary tumor models metastasize 31.
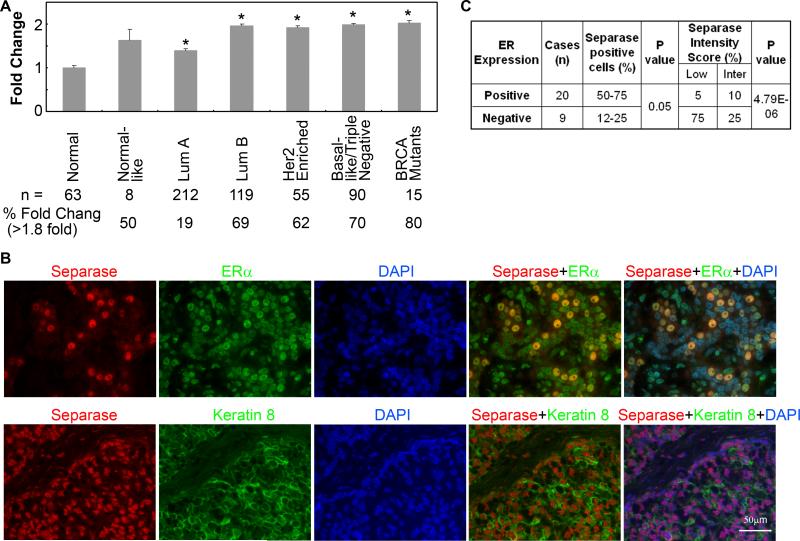
Analysis of the TCGA data 32 indicates that over 53% of breast tumors overexpress ESPL1 transcript. With the exception of Luminal-A tumors all other major subtypes (Luminal-B, Basal and Triple negative breast cancer) have an average of two fold increase in the abundance of ESPL1 transcript (Fig. 5A). As shown in Figure 5B, in comparison to ERα-negative breast tumors, Separase protein is significantly overexpressed in human ERα positive breast tumors expressing luminal epithelial markers like Keratin-8/18. Due to the small number of tumors in this array, we didn’t classify these tumors further into Luminal-A or -B or TNBC subtypes.
Figure 5. Expression of Separase transcript and protein in human breast cancer.
(A) Analysis of ESPL1 transcript that encodes Separase protein using the TCGA breast cancer data sets 32 showing overexpression of Separase mRNA in various tumor subtypes. Bottom panel shows the total number of tumors used in the analysis and % of tumors that express Separase transcripts >1.8 times compared to the normal breast tissues. (A) Representative tissue microarrays (TMA) of human breast cancers that reveal strong Separase expression and nuclear co-localization with ERα (top panel) in pre-dominantly cytokeratin-8/18 positive tumors (bottom panel). (B) Percentage of Separase positive cells and the corresponding intensity scores of Separase expression in ERα positive and negative tumors in the human TMA.
In summary, MMTV-Espl1 tumors represent a great deal of inter- and intra-tumor heterogeneity, with histological features comprising of both luminal (Keratin-8/18 and Cyclin D1) and basal (Keratins-5, -14, Vimentin) subtypes. However, irrespective of their histological appearance, these tumors are of high grade, solid morphology and express varying levels of ERα. The presence of multiple and sometimes overlapping cell lineage molecular markers like Keratin 8/18 and Vimentins indicated a highly perturbed differentiation status among the MMTV-Espl1 tumor cells, also a characteristic feature of some human breast cancers (e.g. Claudin low tumors) 33,34 as well as mammary tumors arising in some mouse models 35.
Genomic instability accumulates in the mammary epithelial cells of MMTV-Espl1 mice prior to tumorigenesis
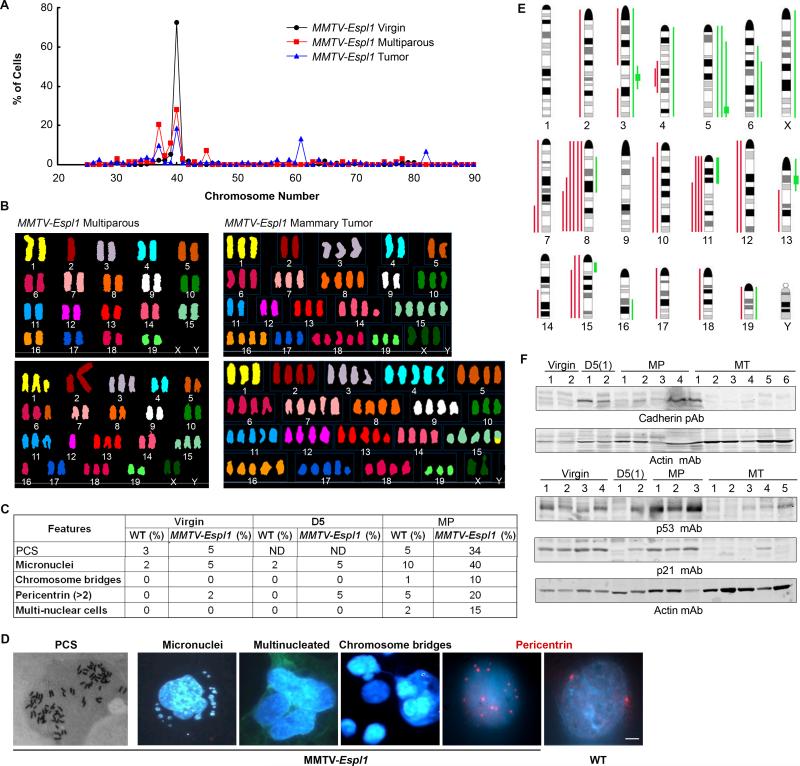
Since aberrant Separase expression can affect the proper removal of chromosomal cohesion, resulting in missegregation leading to aneuploidy, we examined the status of chromosome number in the mammary epithelia of the MMTV-Espl1 mice at all stages of mammary gland development, including the virgin and post lactation (multiparous) glands and the mammary tumors. This was done by Giemsa staining of metaphase chromosomes obtained from primary mammary epithelial cell cultures at passage one to avoid any culture related artifacts. We found that both the Tg1 and Tg2 lines of the MMTV-Espl1 mice progressively accumulated similar level and distribution of aneuploidy in the parous mammary epithelia compared to wild type litter mate mice at similar age and reproductive stages (Fig. 6A for Tg1; Fig S2 for Tg2). The mammary tumors that developed in the parous mice are invariably aneuploid. In the parous mammary epithelia 25-30% of the metaphases were found to be hypodiploid (over 20% cells with 37 chromosomes) indicating both chromosome gain and loss after chronic Separase induction in vivo. The chromosomes in the Separase overexpressed cells are found to be thinner suggesting premature sister chromatid separation (PCS). No detectable changes in chromosome number were observed in the control animals. Metaphase spread analysis of the MMTV-Espl1 tumors indicated 100% aneuploidy with chromosome number ranging 26-87, with three prominent aneuploid peaks with 37, 61 and 82 chromosomes (Fig 6A-B). To further evaluate specific chromosomal aberrations and the aneuploidy in the multiparous mammary glands and mammary tumors in these mice, we performed spectral karyotyping (SKY) on the same metaphase chromosome preparation (three independent mice, 5-10 cells/mammary gland) described above that were used for karyotyping. SKY analysis showed significant clonal chromosomal gains and losses as well as translocations in the multiparous mammary epithelia and mammary tumors (Fig. 6B). Spectral karyotypic analysis of the MMTV-Espl1 multiparous mammary gland revealed clonal loss of one copy of chromosome 2 and both the copies of chromosome X {37,−X,−X,−2[3]/38,−10.−16[1]/61,XXX,+1,+7,−17,−17[1]/80,XXXX[2]} (Fig. 6B top-left panel). Clonal Robertsonian translocation between two chromosome 2 (Fig. 6B bottom-left panel), and trisomies for chromosome 1, 3, 6, 11, 13, 15 and 18, and monosomy for chromosome 12, and 16 were noted in genomic compliment –40-44,XX,+2,ROB(2;2),+3,+6,−11,−12,−12,+13,+15,−16,+18[cp4]/65,XXX,+1,ROB(2;2),+3,+5,−6,−6,+8,+13,+13,+13,+14,+14,+15,−17,−17,−19[1]. The spectral analysis identified a karyotype 61-74,XXX,−2,−4,−5,+8,−9,DEL(11A2),−12,+DEL(14B),+15,+15,+16,+18,+18,+18,+19[cp6]; 82,XX,−X,−X,−1,+4,+7,−10,−11,DEL(13B),+DER(15)T(1?;15E),+16,+DEL(17B),−19[1] in genomic compliment of mammary tumors from the Separase overexpressed mice (Fig.6B right panel).
Figure 6. MMTV-Espl1 mammary epithelium display high levels of genetic instability.
(A) Distribution of chromosome number measured using Giemsa stained metaphase chromosomes (n=100) from passage one primary mouse mammary epithelial cells from 7-9 months old MMTV-Espl1 virgin, multiparous (MP, with > 2 litters) mice and mammary tumors. (B) Representative spectral karyotyping (SKY) images for multiparous MMTV-Espl1 mouse mammary epithelial cells and mammary tumors cells grown at passage one (MT) that was used in (A). (C) Summary of additional features of chromosomal instability in various reproductive stages of MMTV-Espl1 mice mammary epithelial cells used in (A) with representative images of these features [premature chromatid separation (PCS), micronuclei, multinucleated cells, chromosome bridges, multiple centrosomes] shown in (D). (E) Gains and losses in chromosomes in multiparous mammary glands and mammary tumors assayed by comparative genomic hybridization (CGH) array. Green and red lines adjacent to the ideograms indicate relative gain or loss, respectively. (F) Western blot showing progressive loss of Pan Cadherin proteins (Chr 8) and p53 protein (Chr 11) in virgin (nulliparous), day five lactation, primiparous [D5(1)] and multi-parous (MP) mammary glands, and mammary tumors (MT) in MMTV-Espl1 mice. Levels of p21 (cyclin-dependent kinase inhibitor) protein corresponding to p53 protein levels in mammary tumors (MT) are shown. mAb: monoclonal antibody, pAb, rabbit polyclonal antibody.
To assess the effect of Separase overexpression on sister chromatid cohesion and the nature of defects causing aneuploidy in Separase overexpressing cells, we examined the level of PCS prior to exit from mitosis, and formation of anaphase bridge, a form of genomic instability. PCS consists of separate and splayed chromatids with discernible centromeres and involves all or most chromosomes of a metaphase. Using the above criteria PCS was examined in metaphase chromosomes from mammary epithelial cells. Compared to the parous WT littermate controls, mammary epithelial cells from the parous MMTV-Espl1 mice showed significant rise (34%) in PCS, indicating increased chromosomal missegregation induced by the overexpressed Separase (Fig. 6C-D).
We also examined formation of anaphase bridges, aberrant number of centrosomes, micronuclei, multi-nucleated cells, forms of genomic instability, in 500 DAPI stained nuclei of multiparous MMTV-Espl1 and WT mammary epithelial cells from the littermate control mice. Primary mammary epithelial cell cultures at passage zero from MMTV-Espl1 multiparous mice display significant increase in the appearance of micro-nuclei, multi-nucleated cells, chromosome bridges, and aberrant numbers of centrosomes that were only seen in insignificant amounts in the WT and virgin MMTV-Espl1 age matched litter mate mice (Fig. 6C-D) suggesting that overexpressed Separase results in errors in cytokinesis and chromosomal instability leading to aneuploidy.
We then performed Comparative Genomic Hybridization (CGH) analysis of mammary tumors as well as mammary glands from multiparous MMTV-Espl1 mice (Fig. 6E). CGH analysis indicated losses in chromosomes 8 (common region of loss 8C1-E2 encompassing multiple cadherin genes in eight out of nine tumors and in one out of six multiparous mammary glands examined), chromosome 11 (common region 11B1-E2 encompassing the p53 gene locus in three out of nine tumors and one out of six multiparous mammary glands examined), and gain in chromosome 15 (common region 15B1-F3 in one out nine tumors examined) as well as some high level amplifications in regions 3F2, 5G2, 13A3 (Fig. 6E, Table S1). The common regions lost on chromosome 8 encompassed many potential tumor suppressor genes and cell cycle regulators including several genes from the Cadherin super family (Cdh1, Cdh3, Cdh5, Cdh8 and Cdh11), Rbl2, Aktip, Ctcf and Gadd45gip1. The progressive loss of expression of p53 and Cadherin proteins in a set of the mammary tumors was confirmed using Western blot analysis in the mammary glands and tumors from these mice (Fig. 6F). Further pathway analysis showed down-regulation of p21, a p53 transcriptional target in four out of five of these tumors (Fig. 6F). Overall these results re-affirm the heterogeneity and mixed lineage of these tumors with loss of key tumor suppressor genes. Further analysis of specific clones within these tumors would be necessary for a deeper understanding of the role of loss of each of these genes in tumor progression.
Increased early cellular proliferation coupled with accumulation of DNA damage causes rapid tumor progression in multiparous transgenic mammary epithelium
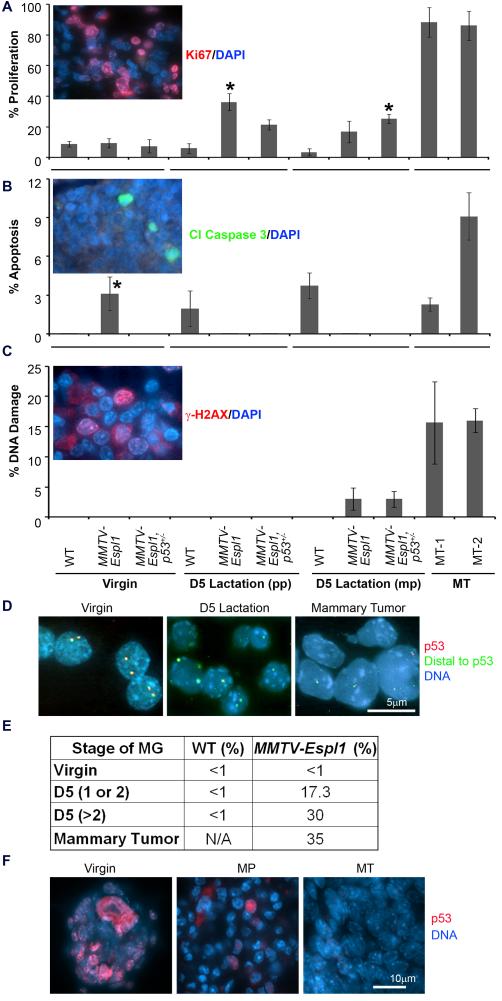
Since progressive accumulation of high levels of aneuploidy in multiparous but not the virgin MMTV-Espl1 mice developed mammary tumor, we hypothesized that co-operating lesions were necessary for tumor initiation. As a logical first step, we examined the levels of cellular proliferation, apoptosis and DNA damage in the mammary epithelium of the transgenic animals. Separase overexpressing cells clearly showed a higher level of proliferation in terms of Ki67 staining than the WT mammary glands when compared at day 5 post single lactation as well as in multiparous mice at six months of age (Fig. 7A, p<0.0005, star). However, no significant change in proliferation was observed in the virgin mice. On the other hand, Separase overexpression elicited p53-dependent cell death in the virgin transgenic mice (Fig. 7B, p<0.0005, star). Apoptotic cell death was low or absent in the MMTV-Espl1, p53 +/− mice at all stages of mammary gland development. Since a recent report 36 indicates that chromosomal missegregation can directly cause DNA damage, we assayed aneuploid multiparous mammary glands and tumors from the transgenic mice for γ-H2AX foci, a surrogate marker for the detection of DNA damage. Mammary epithelia from the multiparous MMTV-Espl1 mice as well as mammary tumors from these mice showed significant rise in DNA damage compared to their virgin and WT littermate cohorts (Fig. 7C, p<0.0005). These results suggest that multiple cycles of proliferation is one of the co-operating events in Separase overexpression induced mammary tumorigenesis, resulting in the gradual accumulation of cells with missegregated chromosomes and damaged DNA.
Figure 7. Effect of Separase overexpression on cellular proliferation, apoptosis, DNA damage, and progressive loss of p53 locus.
(A) The mean percentage of Ki67 positive proliferative cells (red fluorescence, inset; blue fluorescence shows the nucleus) in day five (D5) lactating mammary glands from primiparous (pp) or multiparious (mp) animals, and in mammary tumors (MT). (*significantly different from corresponding wild type littermate, p<0.0005). (B) Assay of apoptotic cell death using cleaved Caspase 3 staining (green fluorescence, inset). (C) Assay of DNA damage using γH2AX staining (red fluorescence, inset). (n=3 animals used for each genotype, and ~200 cells per animals were counted). (D) Representative fluorescent in situ hybridization (FISH) images using p53 gene (red fluorescence) and DNA distal to p53 gene (green fluorescence) as probes on touch preparations of MMTV-Espl1 mouse mammary glands [virgin (n=200), D5 lactation(n=300) and mammary tumor (n=500)]. (E) Progressive heterozygous loss of p53 in all samples except in virgin mice, quantified from (D). Number of parity (e.g. 1, 2, >2) are shown in bracket. (F) Representative immunofluorescence staining of p53 protein (red fluorescence) showing progressive loss of p53 in mammary glands from virgin and multi-parous (MP) animal and a near complete loss in mammary tumors (MT).
Loss of p53 locus further contributes to genetic instability and tumor progression
Since CGH analysis of multiparous MMTV-Espl1 mammary glands as well as mammary tumors from these mice indicated frequent loss of chromosome 11 flanking the p53 gene, we assayed the status of p53 in various stages of mammary gland development in these mice. We performed Fluorescent In situ Hybridization (FISH) analysis on MMTV-Espl1 mouse mammary glands from virgin, primiparous and multiparous mice and on the mammary tumors using Trp53 gene containing BAC clone (RP23-114M1). DNA segments distal to the p53 gene on chromosome 11 (RP23-422L16) was used as a control to assay the loss of p53 gene. Our FISH analysis detected p53 loss in all stages of mammary gland development except in the virgin mice. The percentage of cells losing a single copy of the p53 gene increased from 15% in the primiparous mice to near 35% in the multiparous mice and the mouse mammary tumors (Fig. 7D-E, n= 5 animals for each subset of mice, at least 100 cells from primary passage one mammary epithelial cultures counted for each mouse). Immunofluorescence analysis indicated the progressive loss of p53 protein in the mammary epithelia of these mice (Fig. 7F). Collectively these results suggest that progressive loss of the tumor suppressor function of p53 may be a co-operating lesion in Separase overexpression-induced mammary tumorigenesis.
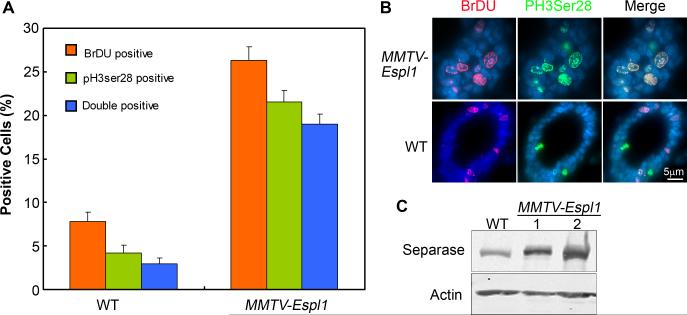
To further investigate if cells in Separase overexpressing mice cycle normally soon after the MMTV promoter expression peaked in mid-pregnancy, mice (virgin transgenic) were injected with Estrogen (E) + Progesterone (P) for 2 days to simulate the day 12 pregnancy stage. One hour after BrdU injection, mammary glands were harvested. Tissue sections were stained for BrdU and phosphorylated serine 28 of histone H3 (PH3), a well-established mitotic marker. The percentage of BrdU labeled cells was significantly higher (5 fold increase) in Separase overexpressing mice compared to wild type controls (Fig. 8A-B). PH3 staining was observed in about 20% of the mammary cells, which was significantly higher than that observed in WT littermate mice (~5%), (Fig. 8A, p<0.0005). To our surprise, a significant percentage of PH3 positive cells (>10% vs. 2% in WT) were also positive for BrdU within the same nucleus (Fig. 8A-B). Normally these stains are restricted to two independent and distinct stages of the cell cycle (S phase and late G2/M phase, respectively). No significant apoptosis was observed at this stage of mammary gland development. Combined, these results suggested that Separase overexpressing mice displayed inappropriate DNA replication indicated by a significantly increased percentage of BrdU labeled cells as well as premature chromatin condensation even before the completion of S phase (indicated by the double labeled cells), which could enhance genetic instability and cooperate in tumorigenesis.
Figure 8. MMTV-Espl1 mice display early cell cycle defects and co-localization of BrdU positive (S phase) and PH3 (Ser28, mitotic phase) cells.
(A-B) Mid-pregnant mammary gland sections were stained for BrdU (red fluorescence) and Phospho Histone H3 (Serine 28, green fluorescence). Representative images are shown for MMTV-Espl1 and wild type (WT) mice (B) and quantified in (A) as a percentage of total cells positive for DAPI (p<0.0005). (C) Western blot shows increased Separase expression in MMTV-Espl1 mice mid-pregnant glands compared to WT. (n=3 animals used for each genotype, and ~200 cells per animal were analyzed).
Discussion
Using a MMTV-Espl1 transgenic mouse model, we provide here the direct evidence that overexpression of Separase protein alone in the mouse mammary gland is sufficient to generate aneuploidy and facilitate tumorigenesis. Our studies also demonstrate that chromosomal instability, manifested as increased chromosomal number (aneuploidy), monosomy, trisomy, translocation and other chromosomal defects is associated with tumor development. Overexpression of Separase also induces a number of CIN phenotypes including appearance of micro-nuclei, multi-nucleated cells, chromosome bridges, aberrant numbers of centrosomes, and PCS. Up to 30% of PCS, and formation of anaphase bridges from lagging chromosomes were observed in approximately 10% of Separase overexpressed cells while <1% bridges were observed in the control cells. These findings suggest that hyperactive Separase results in prematurely segregated chromosomes and lagging chromosomes at anaphase which is indicative of chromosomal instability and development of aneuploidy.
In this study, we have also established early aneuploidy as a contributor to tumorigenesis. Most importantly our studies indicate some key and potential events that co-operate with aneuploidy to overcome cell intrinsic defense mechanisms and allow tumor initiation and progression within the mammary gland environment. MMTV-Espl1 mice in a C57BL/6 genetic background develop aggressive, highly aneuploid, mammary adenocarcinomas with an 80% penetrance, and with median latency of 12 months. Previously we reported that acute overexpression of Separase in vitro from a Tet-promoter in the diploid FSK3 cells, an immortalized mouse mammary epithelial line with a p53 mutant background, can induce aneuploidy within five days of Separase induction 3. Using orthotopic mouse mammary transplant model, we also demonstrated that turning on the expression of Separase in these p53 mutant FSK3 cells for 3-4 weeks resulted in the formation of aneuploid tumors in the mammary gland. Thus the question is why does it take one year to develop mammary tumors in the MMTV-Espl1 transgenic mice? The answer to this question is likely that constitutive Separase overexpression-induced aneuploidy in mammary epithelium alone may not be sufficient to give rise to tumorigenesis, and may require additional lesions. Our studies suggest that some of these lesions include but may not be limited to: (1) increased proliferation of genetically unstable cells through multiple cycles of pregnancy and lactation, (2) gradual accumulation of DNA damage and CIN in Separase overexpressing cells, and (3) progressive loss of p53 and cadherin gene loci resulting in diminished tumor suppressor functions. However, as a paradox, compared to MMTV-Espl1 mice MMTV-Espl1, p53 +/− double mutant mice had no synergistic effect on tumor latency but achieved a greater extent of tumor progression resulting in distant metastasis to the lungs in ~30% of mice. This is not an unlikely phenotype since the apoptotic function of p53 has been previously reported to be the primary function that is selectively lost, enabling carcinoma progression in various tissue types 37. The increased levels of p53-dependent apoptosis in the MMTV-Espl1 virgin mice protecting them from tumor development further support this hypothesis. It is conceivable therefore, that all other tumor suppressor functions of wild type p53 are preserved in the MMTV-Espl1 mice until there is selective pressure to lose single copies of this gene at a later stage (in the multiparous mice) allowing tumor initiation, but not progression to metastasis. In contrast, in the MMTV-Espl1, p53 +/− mice, the germline loss of one copy of p53 significantly reduces its pleiotropic tumor suppressor functions (for example cell cycle arrest, senescence, angiogenesis etc), resulting in an early weakened genetic make up thereby enabling progression to distant metastasis. Alternatively, these results may also suggest a role of cellular defense mechanisms independent of p53 to prevent tumor formation in aneuploid cells of the MMTV-Espl1 mouse mammary epithelium that are potentially perturbed or lost in the MMTV-Espl1, p53 +/− gland resulting in tumor metastasis. We have previously reported that the hormonal (progesterone and estrogen) stimulation of p53-null mice mammary glands not only induces aneuploidy, but also results in the overexpression of Separase, a key chromosomal separation protein, and Mad2, a mitotic checkpoint protein 38. Since overexpression of Separase either constitutionally or conditionally can cause aneuploidy independent of hormonal stimulation, Separase induction is therefore likely to be a key mechanism in both the hormone-sensitive and hormone-resistant mammary epithelium.
The role of aneuploidy in carcinogenesis has been an area of much controversy and still remains an unresolved issue in tumor cell biology 39,40. Studies in budding yeast find that while aneuploidy alone, without any genetic mutations, can confer improved cellular growth under certain stress conditions, aneuploid cells in general divide less rapidly than normal diploid cells under conventional laboratory conditions 16,18. In contrast with these studies, we however do not observe any significant change in proliferation levels in the virgin MMTV-Espl1 mice. In fact, we observe higher levels of apoptosis in this scenario, possibly suggesting that any Separase overexpressing cells undetected by our Western blot analysis that could have lead to tumor formation were being eliminated by key checkpoints within the cell, perhaps by an active p53. This result may explain the “aneuploidy paradox” whereby greater levels of aneuploidy have been associated with “reduced fitness” in yeast 13,15. It is interesting to note that there are two predominant populations of aneuploid cells in the multiparous mammary epithelium with 37 and 45 chromosomes, respectively. The cells with 37 chromosomes have loss of one copy of chromosome 2 and both copies of X, while the cells with 45 chromosomes have a Robertsonian translocation of chromosome 2, and trisomy for chromosomes 1, 3, 6, 11, 13, 15, and 18. Considering that the cohesin subunit SA2 is localized in chromosome X and has been shown to play critical role in aneuploidy and tumorigenesis 41 prompts us to speculate that loss of chromosomes, particularly chromosome-X and −2 may play important role early in the aneuploidy and transformation, and to overcome the aneuploidy pressure and to sustain the transformed cells, cells may force to acquire additional chromosomes. Our studies suggest that in a mammalian system cellular checkpoints possibly counterbalance the presence of aneuploid and less proliferative cells by partially eliminating them via an active apoptotic route. This dynamic balance maintained within the cells allows for the slow accumulation of aneuploidy but does not allow for tumor formation on its own. Recent studies have also indicated that aneuploidy alone can promote tumorigenesis in humans in specific cellular contexts 41.
Our studies show that overexpression of Separase alone is sufficient to cause hyperproliferation in the hormone stimulated (estrogen and progesterone) mid-pregnant mammary epithelium, potentially a novel function of Separase. Separase overexpression results inappropriate DNA replication as well as premature chromatin condensation even before the completion of S phase, indicating a defect in mitotic entry and exit. Previous studies using Separase mutant zebrafish model have reported accumulation of double labeled (BrdU and PH3, markers of S and M phases respectively) cells 42, suggesting that Separase plays an important role in the mitotic processes, potentially in mitotic entry and exit. Studies in yeast have also confirmed that Separase is part of the mitotic exit complex 43. It is therefore possible that, Separase overexpression in the presence of mitogenic hormones shorten the cell cycle by accelerating mitotic phases resulting in hyperproliferation and genomic instability leading to tumorigenesis.
Finally our model has recapitulated some key features of human ER positive breast cancers, a category of tumors that have very few existing mouse models. The mammary tumors that arise in this MMTV-Espl1 mouse are aggressive, display a high degree of intratumoral heterogeneity of which the most differentiated subset retains ER expression, and also has a high degree of genomic instability and DNA repair deficits. However, additional analyses of the TCGA data represents a discrepancy between the level of ESPL1 transcript in the TCGA data set, which suggests that this is highly expressed in basal tumors, and the level of Separase protein expression in our small tumor array, which suggest that it is lowly expressed in ER negative tumors. There can be several explanations: 1) post-transcriptional silencing of the ESPL1 transcript resulting in the down-regulation of Separase expression in ER negative human tumors. In this context it can be noted that either Separase overexpression or its knock down has similar effect resulting in chromosomal missegregation and tumorigenesis 3,44. 2) It is also possible that a more complex genetic interplay between key tumor suppressors like p53 and Brca1 regulate Separase expression in specific subtypes of human cancers. And finally, according to a recent finding ~20% of TBNC tumors have low level expression of ER transcripts 45, and it is possible that the basal tumors that express ER are the ones that over express Separase. It is noteworthy that majority of the MMTV-Espl1, p53 +/− tumors in our mouse model also show significant features of human breast cancers undergoing EMT and/or basal like breast cancers, including metaplastic histology and co-expression of Cytokeratins and Vimentin. As reported before by others, these EMT like tumors continue to show strong cytoplasmic expression of ERα46 . In view of the complex nature of aneuploidy, the highly heterogeneous phenotypes of Separase tumors are not surprising. Future studies are warranted to examine the role of Separase-induced aneuploidy either as a cause or consequence of tumor heterogeneity by analyzing specific genetic changes occurring in the sub-clones of MMTV-Espl1 tumor populations.
One ERα+ breast cancer subtype that does show DNA repair defects is BRCA2 mutation associated breast cancer 47. It is also interesting to note that, >80% of human BRCA mutant tumors overexpress Separase transcripts. Therefore, it would be interesting not only to compare the BRCA mutant tumors with the MMTV-Espl1 tumors, but also explore a role of Separase overexpression, if any, in the loss of BRCA tumor suppressor function in human tumors. Future experiments are also required to ascertain the ER dependence of the MMTV-Espl1 tumors. We hope our model will provide preclinical platforms to test the backlog of therapeutic compounds in the developmental pipeline in vivo, since the successful treatment of malignant breast cancers requires overcoming the formidable challenges presented by tumor heterogeneity and the influence of complex tumor microenvironment milieu. Furthermore, MMTV-Espl1 transgenic mice provide a new experimental model to study the molecular mechanisms of aneuploidy and mammary carcinogenesis.
Materials and Methods
Derivation of MMTV-Espl1 transgenic mice
The full length mouse Espl1 cDNA was cloned into EcoR1 site of MMTV-KbPAII plasmid, and the resultant MMTV-Espl1 plasmid (pDP1090) was used to generate the transgenic mice. Transgenic founders were generated on a C57/BL6 mouse background by pronuclear injection at the Baylor College of Medicine mouse transgenic core. MMTV-Espl1 transgenic mice were genotyped by PCR amplification of a 500-bp transgene fragment using the oligo pair: 5′-GTGTTATACAGGTCCCTCTCCTTTCGTGAA-3′ and 5′-CCTCCATATAACATGAATTTTACAATAGCGAA-3′. Cycling profile was 94°C for 3min. 30 sec.; 35 cycles of 94°C for 1 min., 58°C for 1min. 50 sec., and 72°C for 50 sec.; and final incubation of 72°C for 5 min. A total of four independent transgenic mouse lines were generated. DNA analysis revealed integration of the transgene into the mouse genome. Two lines (Tg1, Tg2) were selected for follow-up studies, with very similar results.
Transgenic breeding strategies
To study the effect of p53 inactivation on mammary tumorigenesis, MMTV-Espl1 mice were mated to p53 mutant mice in C57/BL6 genetic background (p53+/−, Harlan Laboratories, Houston, TX). p53 genotypes were determined by PCR. Cycling variables were as for the MMTV-Espl1 reaction. We produced female mice with the genotypes MMTV-Espl1, p53+/− and MMTV-Espl1, p53−/−. The non-transgenic female littermates (p53+/+, p53+/− or p53−/−) served as controls. All mice underwent at least one cycle of lactation to induce Espl1 expression. Parturition of the first litter was designated as Day 1 for all aging studies. Due to early development of lymphoma, p53−/− mice were not used in our studies.
Histopathology and apoptosis assays
A portion of each mammary sample was fixed overnight in 10% phosphate-buffered formalin, transferred to 70% ethanol, and then embedded in paraffin. Samples were sectioned for 10 successive layers at 5-μm intervals and stained with hematoxylin and eosin for histopathologic examination using routine and established methods. Apoptosis levels were assessed by immunofluorescence staining of cleaved-Caspase 3 (Asp175) (Cell Signaling Technology, catalogue # 9661, Danvers, MA). Differences in apoptosis levels between mice with different genotypes were evaluated by Student's t test (p < 0.05 was deemed statistically significant).
Immunohistochemistry and immunofluorescence
Immunohistochemical analysis was done on formalin-fixed paraffin sections. Antigen retrieval for all antibodies was done by boiling the slides in citrate buffer (pH 6.0; Zymed, San Francisco, CA) for 15 min. Endogenous peroxidase activity was quenched with 10-minute incubation in 3% H2O2 in methanol. The panel of antibodies and the dilutions used for immunostaining were α-cytokeratins 8/18 (Ker8/18, 1:450, Progen, Heidelberg, Germany), α-cytokeratin 5 (K5, 1:8000, Covance, Princeton, NJ), anti–phosphorylated histone H3 (1:100, Upstate, Waltham, MA), Vimentin (1:500, Fitzgerald, Acton, MA), Pan-Cadherin (1:500, Cell Signaling Technology, Danvers, MA), Keratin 14 (1:500), ERα (H-184: sc-7207, Santa Cruz Biotechnology, INC.) and Pericentrin (1:500, Abcam, Cambridge, UK). Detection for all antibodies was done using the Vector ABC Elite kit and a Vector 3,3′-Diaminobenzidine kit for substrate detection (Vector Laboratories, Burlingame, CA). All immunofluorescence reactions were done using AlexaFlour-conjugated secondary antibodies (AlexaFluor 488 and 594, Life Technologies, Grand Island, NY). Slides were counterstained with 4',6-diamidino-2-phenylindole (DAPI) and mounted using Hardset Mounting Media (Vector Laboratories).
Western blot analysis
Western blot analysis was done as described before 38. Antibodies used were Separase (1:1000, Abnova, Taipei, Taiwan), Tubulin (1:5000, Santa Cruz), p27 (1:500, Santa Cruz), Cyclin D1 (1:500, Thermo Scientific, Fremont, CA), Actin (1:5000, Sigma, St. Louis, MO), p53 (1:500, Cell Signaling Technology, Danvers, MA), p21 (1:500, Abcam, Cambridge, UK), and nuclear matrix protein p84 (1:1000, GeneTex, Irvine, CA).
Comparative Genomic Hybridization (CGH) analysis
High-molecular weight DNA was extracted from transgenic and littermate control mouse mammary tissue and mammary tumors by standard methods and subjected to CGH according to the previously published method 48. The metaphase preparations were captured and processed using the Quantitative Image Processing System (QUIPS) (Applied Imaging, Santa Clara, CA). Copy number changes were detected based on the variance of the red:green ratio profile from the standard of 1. Ratio values of 1.2 and 2.0 were defined as thresholds for gains and amplifications, respectively and losses were defined as ratio of 0.8 or lower.
Primary mammary epithelial cell culture, chromosome spread and karyotype analysis
Primary mammary epithelial cells were cultured as described before 49. Enrichment of epithelial cells was obtained by multiple short high speed spins and discarding of supernatant containing fibroblasts and adipose cells. Cells were grown at passage zero to 60% confluence, before adding colcemid for three hours resulting in accumulation of mitotic cells. The cells were then fixed in 3:1 methanol – acetic acid solution for karyotyping. Slides with cells were stained with Giemsa and at least 100 metaphases per animal were counted for karyotype analysis.
Spectral karyotyping (SKY) analysis
The cocktail of mouse chromosome paints was obtained from Applied Spectral Imaging (ASI, Vista, CA). Hybridization and detection were carried out according to the manufacturer's protocol. Chromosomes were counterstained with DAPI. Images were acquired with a SD300H Spectra cube (ASI) mounted on a Zeiss Axioplan II microscope using a custom designed optical filter (SKY-1) (Chroma Technology, Brattleboro, VT), and analyzed using SKY View 2.1.1 software (ASI, Vista, CA). At least ten cells were analyzed per sample for SKY. At least two mice per time point were used for each time point analyzed (multi-pregnant and mammary tumor).
Fluorescence in situ hybridization (FISH)
FISH analysis was performed using spectrum labeled BAC clone (RP23-37N7) containing mEspl1 gene, BAC clones RP23-114M1 (Chromosome 11 for the p53 gene) and RP23-422L16 for a control region distal to p53 gene. The probes were labeled by nick translation using Spectrum Red and/or Spectrum Green (Abbott Laboratories, Des Plaines, IL). Hybridization and detection were performed according to the manufacturer's protocols. The slides were counterstained with DAPI and the images were captured using Nikon E800 microscope equipped with a cooled-charge coupled devices (CCD) camera. The cells were analyzed using Quips Pathvision (Applied Imaging, Santa Clara, CA).
Statistical Analysis
Expression of Separase protein in human breast tumor specimen was compared to the matched normal tissues using a set of paired tests including paired t-test, ranksum and signed rank test, wherever applicable. Ranksum and signed rank tests are more robust to departures from normality, particularly for small sample size. Unpaired one sample t-test was also performed to test if Separase expression is statistically significant in tumor specimens where no matched normal tissue is available. For the unpaired analysis, mean expression of Separase from the available 30 normal specimens was used to compare the tumors. Expression of Separase transcripts in breast cancer was analyzed by examining molecular profiles of TCGA primary breast tumors and normal breast tissue 32. In total, the dataset includes expression profiles of 507 breast tumor samples and 63 normal breast tissue samples. Agilent custom 244K whole genome microarrays were used for expression profiling, and we downloaded and analyzed normalized data (level 3) from TCGA. TCGA used a 50-gene PAM50 model 50 to classify tumors into five subtypes, and expression profiles were available for 90 Basal-like, 55 HER2-enriched, 212 Luminal A, 119 Luminal B and 8 Normal-like tumors. In addition, 15 profiled tumors had evidence for deleterious mutations in BRCA1 or BRCA2, including 2 tumors with multiple alterations. Separase expression was normalized in tumors relative to normal, and when expression was elevated, significance was estimated using a heteroscedastic two-sample t-test against expression profiles in normal samples.
Supplementary Material
Acknowledgements
This study was supported by the grants from the National Cancer Institute (1RO1 CA109330 and 1RO1 CA109478) to D. Pati.
References
- 1.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 2.Meyer R, Fofanov V, Panigrahi AK, Merchant F, Zhang N, Pati D. Overexpression and mislocalizaion of the chromosomal segregation protein Separase in multiple human cancers. Clin Cancer Res. 2009;15:2703–2710. doi: 10.1158/1078-0432.CCR-08-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang N, Ge G, Meyer R, Sethi S, Basu D, Pradhan S, et al. Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci U S A. 2008;105:13033–13038. doi: 10.1073/pnas.0801610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13:501–514. doi: 10.1038/embor.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–538. doi: 10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao C, Furge K, Koeman J, Dykema K, Su Y, Cutler ML, et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci U S A. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heilig CE, Loffler H, Mahlknecht U, Janssen JW, Ho AD, Jauch A, et al. Chromosomal instability correlates with poor outcome in patients with myelodysplastic syndromes irrespectively of the cytogenetic risk group. J Cell Mol Med. 2010;14:895–902. doi: 10.1111/j.1582-4934.2009.00905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuukasjarvi T, Karhu R, Tanner M, Kahkonen M, Schaffer A, Nupponen N, et al. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 1997;57:1597–1604. [PubMed] [Google Scholar]
- 11.McClelland SE, Burrell RA, Swanton C. Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle. 2009;8:3262–3266. doi: 10.4161/cc.8.20.9690. [DOI] [PubMed] [Google Scholar]
- 12.Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci U S A. 2009;106:8671–8676. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 14.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pariente N. A balancing act: focus on aneuploidy. EMBO Rep. 2012;13:472. doi: 10.1038/embor.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berman J. Evolutionary genomics: When abnormality is beneficial. Nature. 2010;468:183–184. doi: 10.1038/468183a. [DOI] [PubMed] [Google Scholar]
- 19.Ornitz DM, Moreadith RW, Leder P. Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc Natl Acad Sci U S A. 1991;88:698–702. doi: 10.1073/pnas.88.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackburn AC, McLary SC, Naeem R, Luszcz J, Stockton DW, Donehower LA, et al. Loss of heterozygosity occurs via mitotic recombination in Trp53+/- mice and associates with mammary tumor susceptibility of the BALB/c strain. Cancer Res. 2004;64:5140–5147. doi: 10.1158/0008-5472.CAN-03-3435. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto KR. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- 22.Sonnenberg A, van BP, Hilgers J, Schuuring E, Nusse R. Oncogene expression during progression of mouse mammary tumor cells; activity of a proviral enhancer and the resulting expression of int-2 is influenced by the state of differentiation. EMBO J. 1987;6:121–125. doi: 10.1002/j.1460-2075.1987.tb04728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnold A, Papanikolaou A. Cyclin D1 in breast cancer pathogenesis. J Clin Oncol. 2005;23:4215–4224. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 24.Lacroix M, Leclercq G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol. 2004;219:1–7. doi: 10.1016/j.mce.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Lacroix M, Toillon RA, Leclercq G. Stable ‘portrait’ of breast tumors during progression: data from biology, pathology and genetics. Endocr Relat Cancer. 2004;11:497–522. doi: 10.1677/erc.1.00758. [DOI] [PubMed] [Google Scholar]
- 26.Cariou S, Donovan JC, Flanagan WM, Milic A, Bhattacharya N, Slingerland JM. Down-regulation of p21WAF1/CIP1 or p27Kip1 abrogates antiestrogen-mediated cell cycle arrest in human breast cancer cells. Proc Natl Acad Sci U S A. 2000;97:9042–9046. doi: 10.1073/pnas.160016897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 28.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han S, Park K, Kim HY, Lee MS, Kim HJ, Kim YD. Reduced expression of p27Kip1 protein is associated with poor clinical outcome of breast cancer patients treated with systemic chemotherapy and is linked to cell proliferation and differentiation. Breast Cancer Res Treat. 1999;55:161–167. doi: 10.1023/a:1006258222233. [DOI] [PubMed] [Google Scholar]
- 30.Cardiff RD. The pathology of EMT in mouse mammary tumorigenesis. J Mammary Gland Biol Neoplasia. 2010;15:225–233. doi: 10.1007/s10911-010-9184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fantozzi A, Christofori G. Mouse models of breast cancer metastasis. Breast Cancer Res. 2006;8:212. doi: 10.1186/bcr1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janssen A, van der BM, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 37.Lu X, Yang C, Yin C, Van DT, Simin K. Apoptosis is the essential target of selective pressure against p53, whereas loss of additional p53 functions facilitates carcinoma progression. Mol Cancer Res. 2011;9:430–439. doi: 10.1158/1541-7786.MCR-10-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pati D, Haddad BR, Haegele A, Thompson H, Kittrell FS, Shepard A, et al. Hormone-induced chromosomal instability in p53-null mammary epithelium. Cancer Res. 2004;64:5608–5616. doi: 10.1158/0008-5472.CAN-03-0629. [DOI] [PubMed] [Google Scholar]
- 39.Ganmore I, Smooha G, Izraeli S. Constitutional aneuploidy and cancer predisposition. Hum Mol Genet. 2009;18:R84–R93. doi: 10.1093/hmg/ddp084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panigrahi AK, Pati D. Road to the crossroads of life and death: Linking sister chromatid cohesion and separation to aneuploidy, apoptosis and cancer. Crit Rev Oncol Hematol. 2009;72:181–193. doi: 10.1016/j.critrevonc.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solomon DA, Kim T, az-Martinez LA, Fair J, Elkahloun AG, Harris BT, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shepard JL, Amatruda JF, Finkelstein D, Ziai J, Finley KR, Stern HM, et al. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes Dev. 2007;21:55–59. doi: 10.1101/gad.1470407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- 44.Mukherjee M, Ge G, Zhang N, Huang E, Nakamura LV, Minor M, et al. Separase loss of function cooperates with the loss of p53 in the initiation and progression of T- and B-cell lymphoma, leukemia and aneuploidy in mice. PLoS ONE. 2011;6:e22167. doi: 10.1371/journal.pone.0022167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18:123–133. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radaelli E, Damonte P, Cardiff RD. Epithelial-mesenchymal transition in mouse mammary tumorigenesis. Future Oncol. 2009;5:1113–1127. doi: 10.2217/fon.09.93. [DOI] [PubMed] [Google Scholar]
- 47.Stefansson OA, Jonasson JG, Johannsson OT, Olafsdottir K, Steinarsdottir M, Valgeirsdottir S, et al. Genomic profiling of breast tumours in relation to BRCA abnormalities and phenotypes. Breast Cancer Res. 2009;11:R47. doi: 10.1186/bcr2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 49.Medina D, Kittrell FS. In: “Establishment of Mouse Mammary Cell Lines,” in Methods in mammary Gland Biology and Breast Cancer Research. Ip MM, Asch BB, editors. Kluwer Academic Press; New York: 2000. pp. 137–145. [Google Scholar]
- 50.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.