Abstract
HIV-1 viral protein R (Vpr) can induce cell cycle arrest and cell death, and may be beneficial in cancer therapy to suppress malignantly proliferative cell types, such as adult T-cell leukemia (ATL) cells. In this study we examined the feasibility of employing the HIV-vpr gene, via targeted gene transfer, as a potential new therapy to kill ATL cells. We infected C8166 cells with a recombinant adenovirus carrying both vpr and GFP genes (rAd-vpr), as well as the vector control virus (rAd-vector). G2/M phase cell cycle arrest was observed in the rAd-vpr infected cells. Typical characteristics of apoptosis were detected in rAd-vpr infected cells, including sub-diploid peak exhibition in DNA content assay, the Hoechst 33342 accumulation, apoptotic body formation, mitochondrial membrane potential and plasma membrane integrity loss. The proteomic assay revealed apoptosis related protein changes, exhibiting the regulation of caspase-3 activity indicator proteins (vimentin and Rho GDP-dissociation inhibitor 2), mitochondrial protein (prohibitin) and other regulatory proteins. In addition, the up-regulation of anti-inflammatory redox protein, thioredoxin, was identified in the rAd-vpr infected group. Further supporting these findings, the increase of caspase 3&7 activity in the rAd-vpr infected group was observed. In conclusion, endogenous Vpr is able to kill HTLV-1 transformed C8166 cells, and may avoid the risks of inducing severe inflammatory responses through apoptosis-inducing and anti-inflammatory activities.
Keywords: HIV-1 Vpr, C8166 cells, apoptosis, proteomic, inflammation
Introduction
Adult T-cell leukemia (ATL) is a progressively debilitating disease, considered to be etiologically related to human T-cell leukemia virus-1 (HTLV-1) [1–3]. Although there are a number of ATL therapies available, such as chemotherapy, bone marrow transplantation, nucleoside analogs, interferon and monoclonal antibodies, their lack of effectiveness is still a major concern [2]. Patients treated with bone marrow transplantation frequently develop graft-versus-host disease (GVHD), and the survival rate has been reported less than 50% [4]. New therapeutic approaches are being developed with great effort, including gene therapy. Specifically, the strategy of targeted-transfer of suicidal genes into ATL cells has shown significant promise [2, 5].
As the only accessory protein incorporated in the HIV-1 virion, viral protein R (Vpr) is a cytotoxicity mediator, capable of inducing cell death in multiple human non-tumor cell types, including CD4+ T-cells [6], endothelial cells [7] and neurons [8], etc. This cytotoxicity involves both intracellular and extracellular pathways [9–11] and is pathogenically associated with AIDS symptoms, such as T-cell depletion, blood brain barrier integrity loss [12] and HIV-1 associated dementia (HAD) [13].
Vpr has been considered and tested as a tumor suppression drug to utilize both its cytotoxicity-inducing activity [14, 15] and ability to mediate cell cycle arrest [16]. It is known that HIV-1 infected CD4+ T-cells can be activated upon expression of viral proinflammatory proteins, such as trans-activator of transcription (Tat) [17] and gp120 [18], etc. However, prior to activation induced proliferation, infected T-cells can be arrested at the G2/M phase by Vpr [19–21]. This benefits HIV-1 remarkably by providing access to the energy, nucleus materials, transcription factors, ions and other mitogenesis related factors of the G2/M phase [22].
It is known that unrestrained proliferation is the most invasive phenotype of ATL cells and is correlated with the acute ATL progression [23, 24]. We hypothesized that, independent from virus infection, endogenous Vpr functionality will be preserved to induce cell cycle arrest and apoptosis of the HTLV-1 transformed cells. In this study, we successfully introduced the HIV-1 vpr gene into C8166 cells, an ATL cell line harboring defective HTLV-1, by employing an adenoviral vector delivery system. We demonstrate functionally endogenous Vpr expression, which was shown to induce cell cycle arrest, as well as apoptosis-associated cellular and proteomic alteration in C8166 cells. Our findings implicate significant potential for applying the HIV-1 vpr gene to future drug design and ATL gene therapy.
Materials and methods
Cells
The C8166 cell line, a kind gift from Dr. Yongtang Zheng, Kunming Institute of Zoology, Chinese Academy of Sciences, was maintained in suspension at 37°C with 5% CO2 in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2mM L-glutamine, 100 µg/ml streptomycin and 100 U/ml penicillin (Invitrogen, Beijing, China). The cell concentration was maintained at 1–1.5×106 cells/ml. The HEK293 cell line was acquired from the China Center of Type Culture Collection and cultured in 25- or 75-cm2 tissue culture flasks to reach 50–70% confluence before transfection.
Recombinant adenoviruses of rAd-vpr and rAd-vector
Adenoviral vectors expressing HIV-1 Vpr and green fluorescent protein (GFP) were constructed using the AdEasy Adenoviral Vector System (Stratagene, La Jolla, CA, USA) as described [25]. Briefly, the plasmid carrying vpr cDNA (GI: 28872817) was cloned into the shuttle vector pAdTrack-CMV which contained a GFP reporter gene. Then, the pAdTrack-CMV- vpr was linearized by digesting with restriction endonuclease Pme I, and transformed into E. coli BJ5183 cells containing adenoviral backbone plasmid pAdEasy-1 by electroporation. This generated recombinant plasmids, which were digested by Pac I and transfected into HEK293 cells with Lipofectamine 2000 reagent (Invitrogen) to package the rAd-vpr recombinant adenovirus. The rAd-vector recombinant adenovirus, which is a vector control of rAd-vpr, was obtained by the same method, except no vpr cloning was performed in the first step.
Infection
The viral stock concentrations of rAd-vector and rAd-vpr were adjusted to 1×109 plaque-forming units (pfu)/ml with fresh RPMI medium supplemented with 10% fetal bovine serum (FBS), 2mM L-glutamine, 100 µg/ml streptomycin and 100 U/ml penicillin (Invitrogen). Then, C8166 cells were infected with rAd-vector or rAd-vpr at 5 pfu/cell for 4 h. Cells were then washed twice with PBS (300×g, 10 min) to remove residual virus. The uninfected negative control (mock-infected group) was performed by mock-infecting cells with fresh RPMI. The infection rates of the 3 groups were determined by the GFP expression assay with a FACSCalibur flow cytometer (Becton Dickinson, Beijing, China) and analyzed by FCS-Express version 3.0 (De Novo Software, Los Angeles, CA, USA). To acquire the visualization confirmation, GFP expression was also observed by Nikon inverse phase contrast microscope (Nikon, Shanghai, China).
Western blotting
Cells were lysed by RIPA buffer (Sigma-Aldrich, St. Louis, MO, USA) containing a 1× protease inhibitor cocktail (Biovision, Mountain View, CA, USA). Cell lysates containing equal amounts of protein were fractionated by 12% SDS-PAGE gels and transferred onto PVDF membranes (Millipore, Bedford, USA). Membranes were blocked by 5% non-fat milk in PBS, and incubated overnight at 4°C with goat anti-Vpr polyclonal Ab (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Horseradish peroxidase (HRP)-conjugated donkey anti-goat IgG Ab (1:1000; Jackson Immunoresearch, West Grove, PA, USA) was used to bind the primary Ab. HRP activity was visualized by an enhanced chemiluminescence detection procedure (Pierce, Rockford, IL, USA).
DNA content determination
Cells were harvested and permeabilized by cold 70% ethanol (−20°C) containing 10% FBS for 30 min. After 2 cold PBS washes, cells were labeled by propidium iodide (PI; Sigma-Aldrich) staining solution (50 µg/ml PI and 1% Triton-100 in PBS) for 15 min at room temperature. Cells were subsequently analyzed by flow cytometry where intensities at the FL2-A channel, reflecting DNA content, were used to analyze cell cycle phases and differentiate dead cells at a sub-diploid peak. Cell cycle and cell death populations were analyzed by the ModFit LT version 3.1 (Verity Software House, Topsham, ME, USA).
Mitochondrial membrane potential
Cells were stained with mitochondrial membrane potential sensitive probe, JC-1 (5,5',6,6'-tetrachloro-1,1',3,3'- tetra-ethylbenzimidazolylcarbocyanine iodide) (Invitrogen), at a final concentration of 2 µM in PBS for 15 min at 37°C. After two PBS washes (300×g, 5 min), labeled cells were observed by LSM 510 META laser scan confocal microscope (Carl Zeiss, Guangzhou, Guangdong, China) and analyzed by a FACSCalibur flow cytometer (BD). The aggregate red form of JC-1 has absorption/emission maxima of 585/590 nm, while the green monomeric form has 510/527 nm. Images acquired from confocal microscopy were measured and calculated for mean fluorescence intensities (MFI) using ImageJ software version 1.41 (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2008.). For flow cytometry, JC-1 aggregate was measured at the FL-2 channel, and green fluorescent (both JC-1 monomer and/or GFP) at the FL-1 channel. FACS data were analyzed by FCS-Express version 3.0 (De Novo).
Apoptosis and plasma membrane integrity assay
Apoptosis was analyzed by Hoechst 33342 (Ho)/PI confocal microscopy and flow cytometry. Cells were labeled with Ho (Sigma-Aldrich) at a final concentration of 10 µg/ml in PBS for 10 min at 37°C. After being washed once with PBS (300×g, 5 min), cells were then stained for 5 min in PI solution (Sigma-Aldrich; 1 µg/ml in PBS) at room temperature to indicate plasma membrane integrity. This method differs from the DNA content assay in that the cells were not fixed, and Triton-100 is absent from the PI solution. Cells were observed by LSM 510 META laser scan confocal microscope (Carl Zeiss) and the MFI of Ho was measured by ImageJ 1.41 software (Rasband, W.S.). Labeled cells were also analyzed with a FACSAria flow cytometer by exciting Ho at 405 nm and PI at 488 nm lasers. FACS data were analyzed by FCS-Express version 3.0 (De Novo).
Two-dimensional electrophoresis
Cells were lysed in IPG Rehydration Buffer (Bio-Rad, Beijing, China) for 30 min and the supernatant was clarified by ultracentrifugation. Protein concentration was determined with the 2D Quant Kit (GE Healthcare, Piscataway, NJ, USA). In the first dimension (IEF), 1 mg protein in 350 µl IPG Rehydration Buffer (Bio-Rad) was loaded on 17-cm IPG Strips (pH 4–7, Bio-Rad) using passive rehydration at 20°C, and then isoelectrically focused in Protean IEF cell (Bio-Rad) for a total of 6×104 Volt-hour. Gel strips were reduced with equilibration buffer I (375 mM Tris-HCl pH 8.8, 6 M urea, 2% SDS, 20% glycerol and 2% DTT) for 15 min, and alkylated with equilibration buffer II (375 mM Tris-HCl pH 8.8, 6 M urea, 2% SDS, 20% glycerol and 4% iodoacetamide) for another 15 min. Gel strips were transferred onto 10% SDS polyacrylamide gels to perform a second dimensional separation with a current setting of 15 mA per gel for the initial 1 h, and 25 mA/gel thereafter. Gels were then stained by Colloidal Coomassie and scanned with a GS-800 calibrated densitometer (Bio-Rad). Gel images were analyzed by PDQuest software version 7.4.0 (Bio-Rad).
In-gel digestion
Protein spots of interest were dissected into gel plugs, which were washed with ultra-pure water and destained by 50% acetonitrile (ACN) and 25 mM NH4HCO3. After being dehydrated with ACN and dried by cold trapped vacuum centrifugation, gel plugs were rehydrated with 10 ul trypsin (Promega, Beijing, China) solution (20 ng/µl trypsin dissolved in 10% ACN/25 mM NH4HCO3) and incubated at 37°C overnight. Peptides were then extracted with 0.1% trifluroacetic acid (TFA) and 60% ACN [26, 27].
Mass spectrometry
The tryptic digested samples were first loaded onto a liquid chromatography (LC) C18 pre-column (Agilent, Santa Clara, CA, USA) and washed for 5 min with loading solvent (3% ACN/97% H2O/0.1% formic acid/0.01% TFA). The samples were then injected onto a 15 cm Vydac 218ms column (Grace, Deerfield, IL, USA) at a flow rate of 180 nl/min. A linear gradient was performed from Solution A (5% ACN/0.1% formic acid) to Solution B (60% ACN/0.1% formic acid) over a period of 30 min, and then ramped to 90% Solution B in 0.1 min and re-equilibrated for 15 min. The LC eluent was sprayed at 2000 V directly into a Q-STAR XL mass spectrometer (MS; Applied Biosystems, Foster City, CA, USA). MS/MS data were recorded and subjected to Mascot analysis (Matrix Science, London, UK; http://www.matrixsciences.com) allowing database searching for protein identification [27, 28].
Bioinformatics
Bioinformatics analyses of the proteomic data were assisted by Swiss-Prot Protein Knowledgebase (http://www.expasy.ch/sprot/) [26, 27]. The protein functionality was referred to the Swiss-Prot/TrEMBL searching entry based on the access number acquired by mass spectrometry. Molecular weight (Mw) and isoelectric point (pI) were calculated with the Compute pI/Mw tool. The physical protein gel positions, detected by 2DE and mass spectrometry, were compared with and validated by the gel images of similar studies from other groups via searching SWISS-2DPAGE two-dimensional polyacrylamide gel electrophoresis database (http://www.expasy.org/ch2d/).
Reverse Transcription PCR
RNA was extracted from cells by using the TRIzol reagent (Invitrogen). Reverse Transcription PCR (RT PCR) was performed using the OneStep RT-PCR Kit (QIAGEN, Shanghai, China) according to the manufacturer’s protocol. Briefly, the reaction mixture (25 µl final volume) was made by combining 5 µl RT-PCR buffer (5×), 10 mM of dNTPs, 10 µM of Primers, 1µl Enzyme Mix, 1µg RNA and DEPC water. The PCR program for each amplification cycle was 50°C/30 min, 95°C/15 min, 94°C/45 s, 58°C /45 s and 72°C/45 s. Thirty cycles were performed for each PCR assay, followed by a final extension at 72°C for 10 min and holding at 4°C. Primers used were: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), sense 5'-GTCTTCACCACCATGGAGAAGGCTG-3' and anti-sense primer 5'-TGAGGTCCACCACCCTGTTGCTGTA-3'; 14-3-3, sense 5'- ATGGCTGCCAAAGTGTTTGAGTCC-3' and anti-sense primer 5'-TCACTGGGGCAGCTGGAGGA-3'; Glutathione S-transferase P1(GSTP1), sense 5'-ATGCCGCCCTACACCGTGGTCTAT-3' and anti-sense primer 5'-TCACTGTTTCCCGTTGCCATTGA-3'. The amplified products were separated electrophoretically in 1% agarose gels, stained with ethidium bromide and visualized by UV light. Optical density (OD) of each DNA band was quantified by ImageJ 1.41 software (Rasband, W.S.) and normalized to ODGAPDH.
Caspases 3&7 activity assay
The red fluorescent-labeled inhibitors of caspases (FLICA) kit (Immunochemistry Technologies, LLC, Bloomington, MN, USA) was employed to evaluate intracellular activity of caspases 3&7. FLICA is specific for active caspases - it will not react with pro-caspases nor inactive forms of the enzyme (manufacturer’s manual). The labeling method was previously described [27]. Briefly, 1×106 cells from each group were washed twice with PBS (300×g, 5 min) and labeled with 1× FLICA working solution in a final volume of 300 µl for 1 h at 37 °C. After 2 washes with 1× washing solution, cells were observed by laser confocal microscopy for active caspase 3&7 (red) and GFP expression (green).
Statistics
To determine the statistical differences between groups, one-way ANOVA analysis was applied and Bonferroni post hoc test was employed. Values were given as mean ± standard error of the mean (SEM). Statistical analysis was done with GraphPad Prism version 5.0 (GraphPad, San Diego, CA, USA). Statistical significance was accepted at P<0.05.
Results
Functional Vpr expressed in C8166 cells
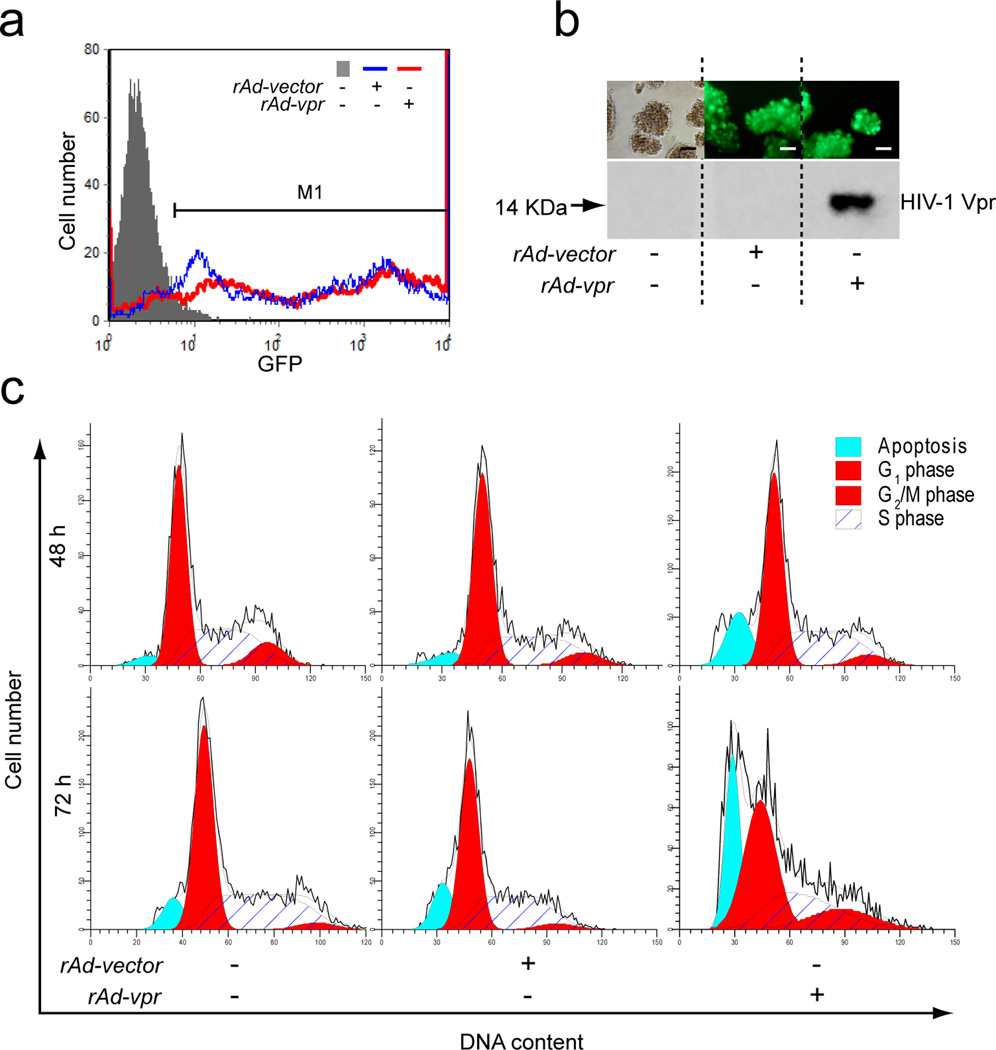
Recombinant adenoviral vector delivery is one of the most efficient and widely used approaches to introduce exogenous genes into target cells [29, 30]. By employing the AdEasy-1 technique, we successfully constructed the recombinant adenoviruses of rAd-vpr, which expresses Vpr and GFP independently, as well as the vector control, rAd-vector, which expresses GFP only. Based on the GFP reporter protein expression, we observed high infectious capacities of both rAd-vpr and rAd-vector in the leukemia cell line, C8166 cells (Fig. 1a). At 24 h post-infection, GFP positive cells in both rAd-vpr and rAd-vector groups represented >80% of all cell populations, with fluorescence intensity normalization to the mock-infected group (Fig. 1a).
Fig. 1.
Functional Vpr expressed endogenously in C8166 cells. a GFP expression in mock-, rAd-vector and rAd-vpr infected groups. Cells with either mock or viral infections were cultured for 24 h. GFP was detected at FL1 channel in flowcytometry and data shown are representative of 3 independent experiments. M1 region shows GFP positive cells. b Representative images of GFP expression in different groups (scale bar = 10 µm), followed by western blotting confirmation of HIV-1 Vpr in C8166 cells. c Cell cycle and cell death of C8166 cells. Cells were stained with propidium iodide and analyzed for DNA content to determine cell populations at 48 and 72 h after mock, rAd-vector and rAd-vpr infection. In the order of increasing DNA content, dead cells are shown in the sub-diploid peaks, G1 phase populations in diploid peaks, S phase cells in the plateau area and G2/M cells in the polyploidy peaks.
GFP positivity in both viral infection groups was confirmed by fluorescence observation, compared with non-fluorescent visualization of the mock infection group (Fig. 1b). Immunoblotting validation was subsequently performed and the specific Vpr protein band was detected by western blotting only in the rAd-vpr group (Fig. 1b), ruling out the possibility of selective GFP expression.
To confirm and characterize the functionality of endogenous Vpr expressed by adenoviral vector delivery in leukemia cells, we examined cell cycle and cell death of C8166 cells at 48 and 72 h post-infection (Fig. 1c). Cells in the rAd-vpr group showed G2/M phase (16.9%) cell cycle arrest at 72 h post-infection, 4.3-fold higher than those in the rAd-vector group (3.9%), and 4.7-fold higher than the mock-infected group (3.6%) (Fig. 1c). Cell death rates in the rAd-vpr group showed a longitudinal increase from 13.7% at 48 h to 24.1% at 72 h, higher at both time points than those in the rAd-vector group, which were 6.4% and 14.1%, respectively (Fig. 1c). Cell death rates for the mock-infected group showed a modest increase from 3.6% to 8.4% (Fig. 1c).
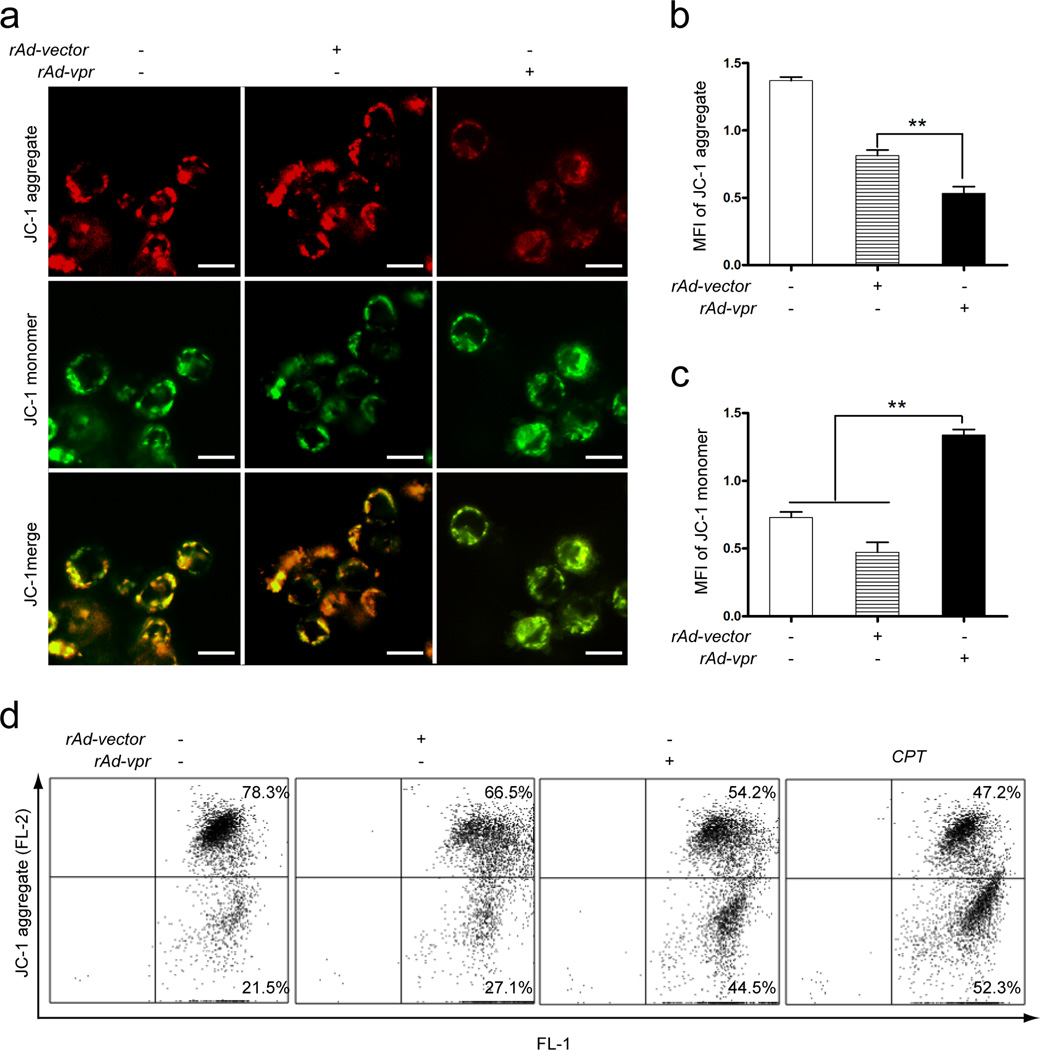
Mitochondrial membrane potential loss in C8166 cells
Confocal microscopy observations indicated the ratio changes of red and green fluorescence in mock-, rAd-vpr and rAd-vector infected groups (Fig. 2a). An equal proportion of red and green fluorescence results in yellow images; however, to either increase the green or decrease the red fluorescence will output greener images. In the context of JC-1 staining, either an increase of JC-1 monomer (green) or decrease of JC-1 aggregate (red) implies the loss of mitochondrial membrane potential (MMP) [31, 32]. Fig. 2a shows that the merged image of the rAd-vpr group is greener than rAd-vector and the mock-infected groups. The MFI of JC-1 aggregate in the rAd-vpr group (50.2 ± 7.0) was significantly lower than that in the rAd-vector (101.0 ± 12.1) (P < 0.05, n=3) and the mock-infected group (113.9 ± 13.0) (P < 0.05, n=3) (Fig. 2b). Furthermore, the MFI of JC-1 monomer in the rAd-vpr group (87.3 ± 12.1) was significantly higher than that in the rAd-vector group (46.7 ± 6.3) (P < 0.05, n=3) (Fig. 2c).
Fig. 2.
Mitochondrial membrane potential loss was induced by endogenous Vpr expression in C8166 cells. Cells with mock- and viral infections as well as CPT (0.5 µmol/l) treatment were cultured for 24 h prior to JC-1 staining. a JC-1 aggregate and monomer observation. Images shown are representative observations from 3 independent experiments. Red images indicate the JC-1 aggregate fluorescence from healthy mitochondria, while green images exhibit cytosolic JC-1 monomers. Merged images indicated the co-localization of JC-1 aggregates and monomers. Scale bar = 10 µm. b MFI quantification of JC-1 aggregate in mock-, rAd-vector and rAd-vpr infected groups. c MFI quantification of JC-1 monomer in different groups. d MMP loss assay by FACS. Data are shown as mean ± SEM. One-way-ANOVA analysis, n = 3. ** P<0.01.
In the flow cytometry assay, the cells with decreased MMP were located in the lower right panel (“apoptotic region”) of each result window (Fig. 2d). Compared with the mock-infected group, there was a more than 30% increase of low MMP cells in the CPT group (Fig. 2d). In the rAd-vpr infected group, there were over 17% more cells with low MMP than that in the rAd-vector infected group (Fig. 2d). These results are consistent with those from the confocal microscopy analysis (Fig. 2a &b).
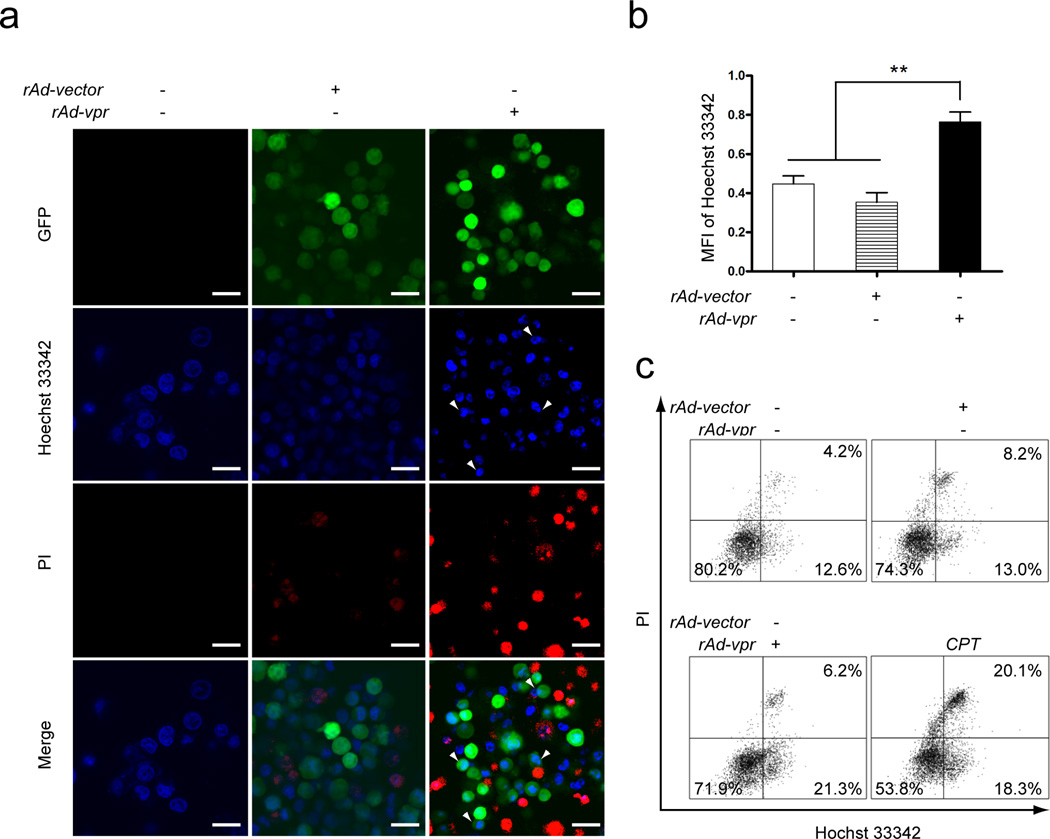
Apoptosis and plasma membrane integrity loss in C8166 cells
Apoptotic cells accumulate Ho more rapidly than do viable non-apoptotic cells, making them more brightly fluorescent and condensed compared to normal [33]. These apoptotic morphologies (Fig. 3a, Hoechst 33342), as well as apoptotic bodies (Fig. 3a, arrow heads), were prevalently observed in the rAd-vpr group. PI positivity, a characteristic phenomenon in late stage apoptosis or necrosis, was also observed in non-apoptotic body forming cells (Fig. 3a). The MFI of Ho in the rAd-vpr group (0.76±0.05) was significantly higher than that in the rAd-vector (0.35±0.05) and the mock-infected (0.45±0.04) groups (P < 0.01, n=3) (Fig. 3b).
Fig. 3.
Apoptosis and plasma membrane integrity changes induced by endogenously expressed Vpr in C8166 cells. Mock-, rAd-vector and rAd-vpr infected as well as CPT (0.5 µmol/l) treated cells were cultured for 24 h and subjected to Hoechst 33342 and PI double staining. a Observations of apoptotic bodies and plasma membrane integrity changes. Images shown are representative observations from 3 independent experiments. Green images indicate GFP positive cells, blue images show Hoechst 33342 fluorescence, and red images exhibit PI staining of nucleic acids. Arrow heads indicate condensed DNA and apoptotic bodies. Scale bar = 20 µm. b MFI quantification of Hoechst 33342 staining in mock-, rAd-vector and rAd-vpr infected groups. c FACS analysis of both Hoechst 33342 and PI staining. Data are shown as mean ± SEM. One-way-ANOVA analysis, n = 3. ** P<0.01.
In each flow cytometry result panel of Fig. 3c, the viable cells are located in the lower left panel, apoptotic cells in the lower right panel, as well as necrotic and late stage apoptotic cells in the upper right panel. Compared with the mock-infected group, there was a more than 6% increase of apoptotic cells and a 15% increase in necrotic cells in the CPT group (Fig. 3c). In the rAd-vpr infected group, there were over 8% more apoptotic cells than that in the rAd-vector infected group (Fig. 3c). These results are consistent with those from confocal microscopy analysis (Fig. 3a &b).
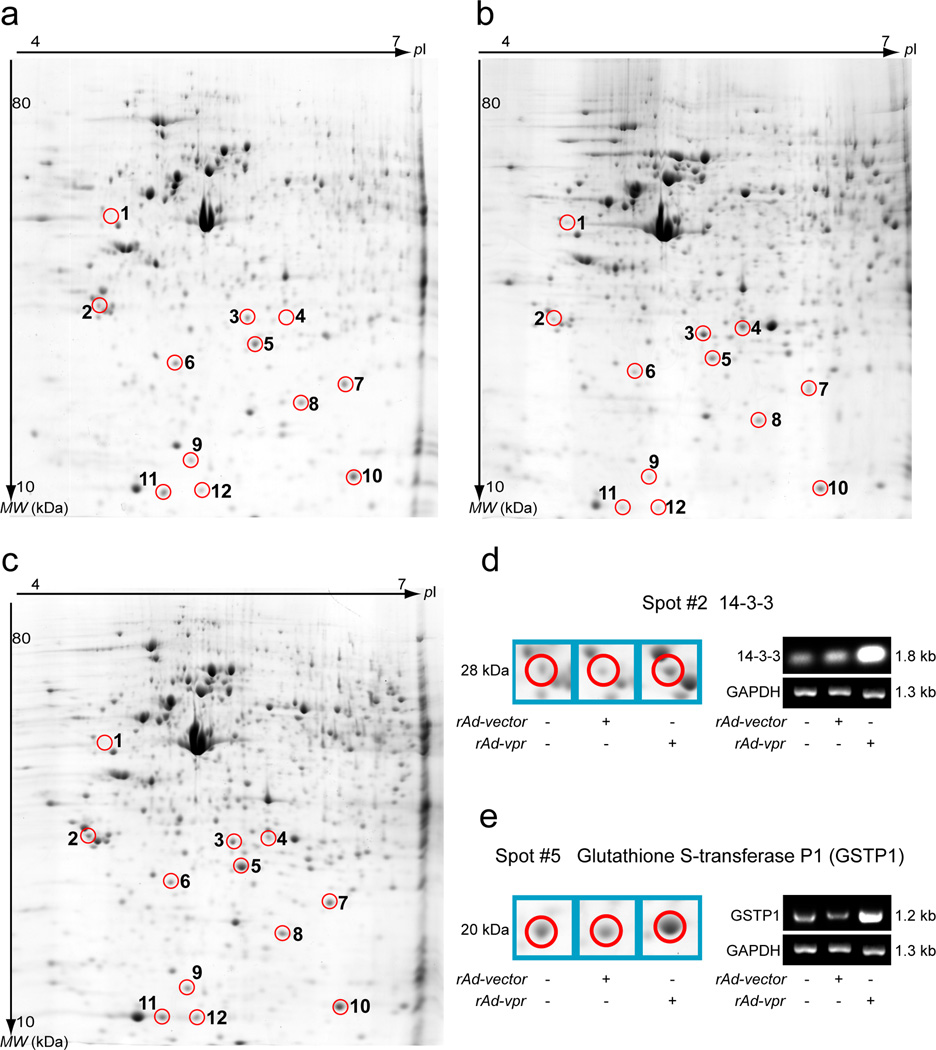
Proteome alterations in C8166 cells
Representative 2DE analysis images are shown as mock (Fig. 4a), rAd-vector (Fig. 4b) and rAd-vpr (Fig. 4c) groups. After comparing images of all 3 groups, the spots, which were subsequently identified by mass spectrometry (Table 1), are circled and numbered (Fig. 4a, 4b and 4c).
Fig. 4.
Two-dimensional electrophoresis analysis of endogenous Vpr induced C8166 cell death. Cells were cultured 48 h after mock or viral infection before lysing. Protein separation of cell lysates obtained by 2DE gels and images were acquired after post-staining with colloidal Coomassie Blue. After analysis and normalization by PDQuest software, images are shown for mock- (a), rAd-vector (b), and rAd-vpr (c) infected C8166 cells. Circled spots with identical spot numbers across gels indicate proteins at the same gel positions. The numbering is also consistent with Table 1, which provides protein identifications generated by mass spectrometry. RT-PCR was performed on the 14-3-3 (d) and GSTP1 (e) proteins, and sample loading was controlled by GAPDH mRNA quantification, respectively.
Table 1.
Proteome changes of endogenous HIV-1 Vpr induced C8166 cell death.
| Spot No. |
MASCOT Scores a |
Access No.b |
Protein Name |
Mw (kDa) |
pI c | Sequences of Peptides Identified d | Fold Changes e |
|---|---|---|---|---|---|---|---|
| 1 | 224 | P48616 | Vimentin | 54 | 5.06 | K.NLQEAEEWYKSK.F R.ISLPLPNFSSLNLR.E K.FADLSEAANRNNDALR.Q R.QVQSLTCEVDALKGTNESLER.Q K.TVETRDGQVINETSQHHDDLE.- R.LLQDSVDFSLADAINTEFKNTR.T |
−21.9 |
| 2 | 408 | P27348 | 14-3-3 protein | 28 | 4.68 | R.NLLSVAYK.N K.DSTLIMQLLR.D R.YLAEVACGDDR.K K.YLIANATNPESK.V R.SICTTVLELLDK.Y K.QTIDNSQGAYQEAFDISK.K R.KQTIDNSQGAYQEAFDISK.K K.TAFDEAIAELDTLNEDSYK.D |
+7.0 |
| 3 | 561 | P35232 | Prohibitin | 30 | 5.57 | R.NVPVITGSK.D K.EFTEAVEAK.Q R.FDAGELITQR.E K.DLQNVNITLR.I R.VLPSITTEILK.S R.IFTSIGEDYDER.V K.LEAAEDIAYQLSR.S R.KLEAAEDIAYQLSR.S R.NITYLPAGQSVLLQLPQ.- R.AATFGLILDDVSLTHLTFGK.E K.FGLALAVAGGVVNSALYNVDAGHR.A R.GVQDIVVGEGTHFLIPWVQKPIIFDCR.S |
−1.6 |
| 4 | 82 | P02787 | Serotransferrin [Fragment] | 79 | 6.81 | K.YLGEEYVK.A K.SASDLTWDNLK.G |
−2.8 |
| 5 | 107 | PO9211 | Glutathione S-transferase P | 24 | 5.43 | K.ASCLYGQLPK.F M.PPYTVVYFPVRGR.C K.ALPGQLKPFETLLSQNQGGK.T |
+4.3 |
| 6 | 94 | P52566 | Rho GDP-dissociation inhibitor 2 | 23 | 5.10 | K.TLLGDGPVVTDPK.A K.TLLGDGPVVTDPKAPNVVVTR.L |
+5.1 |
| 7 | 288 | Q99497 | Protein DJ-1 | 20 | 6.33 | K.VTVAGLAGK.D K.DGLILTSR.G R.DVVICPDASLEDAKK.E K.GAEEMETVIPVDVMR.R K.GLIAAICAGPTALLAHEIGFGSK.V K.EGPYDVVVLPGGNLGAQNLSESAAVK.E |
+5.2 |
| 8 | 293 | P15531 | Nucleoside diphosphate kinase A | 17 | 5.83 | R.GLVGEIIK.R R.GDFCIQVGR.N K.DRPFFAGLVK.Y K.FMQASEDLLK.E R.TFIAIKPDGVQR.G R.NIIHGSDSVESAEK.E R.VMLGETNPADSKPGTIR.G K.YMHSGPVVAMVWEGLNVVK.T |
+2.2 |
| 9 | 90 | O75347 | Tubulin-specific chaperone A | 13 | 5.25 | R.LVLDSVKLEA.- R.LEAAYLDLQR.I R.RLEAAYLDLQR.I |
+12.0 |
| 10 | 136 | Q01469 | Fatty acid-binding protein | 15 | 5.34 | K.ELGVGIALR.K K.MGAMAKPDCIITCDGK.N K.TTQFSCTLGEKFEETTADGR.K |
+2.3 |
| 11 | 215 | PO9382 | Galectin-1 | 15 | 5.34 | K.SFVLNLGK.D K.LPDGYEFK.F K.DSNNLCLHFNPR.F R.LNLEAINYMAADGDFK.I R.EAVFPFQPGSVAEVCITFDQANLTVK.L |
+45.6 |
| 12 | 245 | Q5REA8 | Thioredoxin domain-containing protein 17 | 14 | 5.40 | K.TIFAYFTGSK.D K.NLKVTAVPTLLK.Y K.TIFAYFTGSKDAGGK.S K.VTAVPTLLKYGTPQK.L K.SWCPDCVQAEPVVR.E R.YEEVSVSGFEEFHR.A |
+10.3 |
Note:
Only proteins with significant Mascot database matches (P<0.05) and scores (>68) are listed.
Swissprot database access number (http://ca.expasy.org/sprot/).
Theoretical isoelectric point calculated by Swissprot database at http://ca.expasy.org/sprot/.
Peptides identified by LC/MS/MS for the specific proteins.
Positive numbers show flod changes of up-regulation comparing the ad-vector infection group with ad-vpr infected group; negative numbers show down-regulation.
A comparison of the rAd-vector and rAd-vpr groups’ protein identification and fold changes are shown in Table 1. Endogenous Vpr expression altered the C8166 cell proteome through the down-regulation of both structural (vimentin and prohibitin) and regulatory proteins (serotransferrin fragment), as well as up-regulation of regulatory proteins [14-3-3, GSTP1, galectin-1, Rho GDP-dissociation inhibitor 2 (Rho GDI2), tubulin-specific chaperone A and fatty acid-binding protein], redox proteins (protein DJ-1 and thioredoxin) and nucleoside synthesis related enzyme (nucleoside diphosphate kinase A).
To validate the proteome alterations detected by 2DE analysis and mass spectrometry, 14-3-3 and GSTP1 mRNA were analyzed by RT PCR (Fig.4d and 4e). Comparing the rAd-vector and rAd-vpr groups, 14-3-3 showed 2.3-fold up-regulation after normalizing to the GAPDH band (OD14-3-3/ODGAPDH) (Fig. 4d), indicating a consistently changing trend of 7.0-fold upregulation at protein level detected by 2DE and mass spectrometry (Fig. 4d and Table 1). Similarly, GSTP1 showed 3.6-fold up-regulation at the mRNA level, while exhibiting a 4.3-fold up-regulation at the protein level in 2DE and mass spectrometry (Fig. 4e and Table 1).
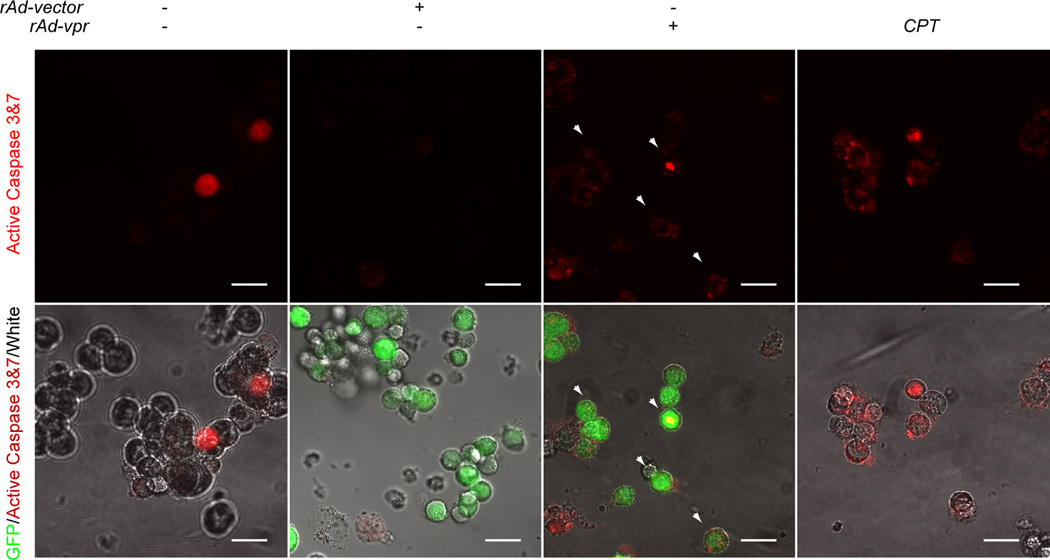
Caspase activity and co-localization with GFP
With the detection of apoptosis, especially caspase-3, related protein changes in the preoteomic assay, we further validated the caspases 3&7 activity regulations. We used exactly the same settings for the confocal observation of caspases 3&7 activity. The caspases 3&7 negative cells are dark, while the positive cells are red in the single-channel images (Fig. 5; upper row). Furthermore, the co-localization of active caspases 3&7 and cells were shown in the merged images of Fig. 5. In the CPT treated group, cells showed brighter active caspases 3&7 staining than that in the mock-infected group (Fig. 5). Compared with the rAd-vector group, more co-localization of GFP and active caspases 3&7 were observed in the rAd-vpr group (Fig. 5, arrow heads).
Fig. 5.
Endogenously expressed Vpr induces up-regulation of caspase 3&7 activity in C8166 cells. Mock-, rAd-vector and rAd-vpr infected as well as CPT (0.5 µmol/l) treated cells were cultured for 24 h before being labeled by red FLICA. Images in the upper row show the active caspases 3&7 in the C8166 cells of different groups, while those in the lower row indicate the merged images of GFP, active caspases 3&7 and white field. Scale bar = 20µm.
Discussion
In this study, we examined the function of the HIV-1 vpr gene and demonstrated that endogenously expressed Vpr-mediated apoptosis in HTLV-1 transformed C8166 cells. This implicates a promising potential in developing new therapies for leukemia, while incorporating further progressions of targeted gene delivery. In this case, the viability of this approach requires safe and efficient vectors that can recognize selective cellular markers on ATL cells, carry and deliver functional genes and support subsequent endogenous protein expression [34]. Although clinical studies have shown severe host inflammatory responses induced by human adenoviral-vector administration [35], progress was made to improve the safety and gene transfer specificity of the vector by modifying capsid [5] or promoter genes [36]. Equally important are the development of new and effective vector systems over the past decade, including HIV-based vectors [2], monoclonal antibody-based vectors [37, 38] and murine leukemia virus (MLV)-based replication-competent retroviral vectors [39]. These advancements continue to facilitate the evaluation of a variety of candidate ATL cell suppression genes; specifically, herpes simplex virus-thymidine kinase (HSV-TK) [40], tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) [36] and Il2RG gene [41].
Our data suggests that Vpr is able to induce cell cycle G2/M phase arrest in C8166 cells, which is significantly beneficial in containing malignant proliferation. Various mechanisms have been proposed to explain the Vpr-mediated cell cycle arrest; however, given the differences in cell type [7, 42, 43], anatomical loci [44] and host cell species [45, 46] examined, a definitive theory is yet to be recognized. The proteomic data of this study indicates that cell cycle related protein up-regulation, especially that of 14-3-3, GSTP1 and galectin-1, may play important roles in Vpr-induced ATL cell cycle arrest. In human T-cells, 14-3-3 participates in Vpr-induced G2/M arrest through the binding of cell cycle related proteins, such as Cdk1, Cdc25C, and CyclinB1 [47]. In malignant cells, including breast cancer cells [48], HeLa cells [49] and human colon cancer cells [50], drug treatment was reported to cause 14-3-3 up-regulation. GSTP1 is a regulator of mitogen-activated protein kinases (MAPK), and is correlated with cell cycle G1 phase arrest in drug treated cancer cells [51]. Galectin-1 has been shown to suppress tumor cell growth by activating the cyclin-dependent kinase inhibitors (CKI) p21 and p27 [52]. In our previous study, we observed similar proteomic alteration of these 3 proteins and reported their possible roles in astrocyte-induced viral replication inhibition in HIV-1 infected microglia [27]. Taken together, these data imply that Vpr induced alteration of the 3 regulatory proteins examined could be a common phenomenon in both cancer cell cycle arrest and HIV-1 infection.
We demonstrated that apoptosis is the major mechanism in Vpr-mediated C8166 cell death, evidenced by sub-diploid peak detection in DNA content assay, increased accumulation of Ho in cell nuclei and apoptotic body formation, as well as the up-regulation of caspase-3 expression and caspases 3&7 activation. Apoptosis is generally recognized as a safe and controlled pathway to clear unwanted cells, such as T cells and neutrophils, with minimal inflammation [53]. By avoiding acute collapse of the plasma membrane, unlike necrosis, apoptotic cells undergo programmed disassociation via the formation of apoptotic bodies that are subsequently cleared by macrophages [54]. This mode of operation supported the use of vpr in ATL gene therapy, thereby decreasing the risk of host inflammatory responses.
We believe that mitochondrial dysfunction, as demonstrated in this study, may influence Vpr-mediated C8166 cell apoptosis. Mitochondrial membrane potential loss results in the release of cytochrome c (Cyt c), an activator of apoptosis-specific endonuclease, caspase-3 [55]. In addition, functional mitochondria are essential for stabilizing intracellular ion concentrations by maintaining plasma membrane ion channels, such as Ca2+ channel [56]. However, during the later stages of apoptosis or necrosis, devastating mitochondrial dysfunction is associated with the loss of plasma membrane integrity [57]. This same phenomenon was observed in the Vpr-mediated cell apoptosis demonstrated in this study. Furthermore, the changes in regulation of 2 endpoint proteins, vimentin and Rho GDI2, shown in our results further support our rationale. Vimentin is a substrate of caspase-3 [58]. As a processor protein, p26 can be cleaved by caspase-3 to form Rho GDI2 [59]. Thus, both down-regulation of vimentin and up-regulation of Rho GDI2 in the rAd-vpr infected group strongly implicates the caspase-3 pathway activation. This was confirmed by the caspases 3&7 activity and GFP co-expression assays in this study. Furthermore, Choi et al reported comparable phenomena of caspase-3 mediated Rho GDI2 processing during proteasome inhibitor-mediated HL-60 and K562 leukemia cell apoptosis [60]. Supportively, prohibitin, which is a mitochondrial protein [61], was also found down-regulated in this study. These data suggest a direct correlation of mitochondrial membrane potential loss with endogenous Vpr-induced HTLV-1 transformed C8166 cell apoptosis.
These results led us to conclude the consequences of other Vpr-induced regulatory protein changes in C8166 cells: they tend to increase the apoptotic signaling. For example, serotransferrin has shown protective functions in cancer cells [62]; it was found down-regulated in this study. Galectin-1 [63] and fatty acid-binding protein [64] are apoptosis inducers in various cell types; here they were shown up-regulated. Tubulin-specific chaperone A is the first component of the β-tubulin folding pathway [65]; the up-regulation exhibited in this study implies the blockage of tubulin-polymerization, which is a common phenomenon of apoptosis-related structural protein change [66]. We also performed a comparable study with another inducer of apoptosis, CPT, to investigate the specificity of Vpr mediated proteomic alterations (data not shown). CPT can cause similar proteomic alterations in C8166 cells to endogenously expressed Vpr. However, the two inducers may differentially impact the regulation of cell cycle associated protein, 14-3-3. These data provide a clearer depiction of the Vpr-induced apoptosis signaling network.
Indeed, other than inducing apoptosis, Vpr itself has anti-inflammatory activity through down-regulating NF-κB and the production of pro-inflammatory cytokines such as TNF-α, and IL-12 [67]. This further avoids the inflammation response risks and makes Vpr better qualified as a candidate for ATL gene therapy. Thioredoxin, which is a redox protein modulating TNF-α signaling and NF-κB activation [68], was found in this study to be up-regulated. This implies the possible anti-inflammatory functions of endogenously expressed Vpr in ATL cells.
Serum Vpr is a very important parameter in determining AIDS progression. It has been suggested that the breakdown or lysis of infected cells and virus particles is likely to release free Vpr into the sera and the CSF [10]. Thus, the risk of free Vpr release and bystander effects of killing healthy cells is a potential drawback if introducing the vpr gene into peripheral blood. However, this risk could be mitigated by several factors: 1) apoptosis related phagocytotic clearance of Vpr containing apoptotic bodies, 2) progression on the development of ATL specific vector systems, which reduces unnecessary Vpr expression and 3) Vpr inactivation via surface processing [69] or chaperone protein binding [70].
In conclusion, the HIV-1 vpr gene shows promising characteristics and may be a well-qualified candidate for ATL gene therapy. Endogenously expressed HIV-1 Vpr can mediate apoptosis of ATL cells, as confirmed by multiple approaches of cellular biology and proteomics. The cell cycle G2/M phase arrest, mitochondria membrane potential loss, nucleic acid condensation and caspases 3&7 activation are the major phenotypes in apoptotic HTLV-1 transformed C8166 cells induced by endogenously expressed Vpr. The proteomic confirmation and endpoint discovery involves multiple structural, regulatory and redox proteins. Our findings suggest that Vpr can kill HTLV-1 transformed C8166 cells effectively via apoptosis, while avoiding inflammation responses.
Acknowledgments
The authors would like to thank Hailan Tang, Xinqiang Lai, Core Facility of Experimental Technology, Jinan University, for their great technical assistance in laser confocal and flow cytometry; Xin Zhang, Bing Song and Xiaoyu Lu, Jninan University, for fluorescence microscopy, and other technical supports. We thank Gregory Dann, Jazel Dolores and Charlotte Pan, University of Washington School of Medicine, for their critical reading of this manuscript. This study is a part of the thesis of Fang He, supervised by both Drs. Yaoying Zeng and Tong Wang, submitted in fulfillment of the requirements for the degree of Doctor of Medicine at the Jinan University, Guangzhou, China. This work was supported by the Natural Science Foundation of Guangdong Province, China (Grant No. 5300413, TW), New Investigator Award, USA (UW/FHCRC CFAR, NIH grant P30 AI 027757, TW), the Major State Basic Research Program (973), China (Grant No. 2006CB504200, YZ), and the Key Subject of Biochemistry and Molecular Biology of Guangdong Province, China (TW and XH).
Abbreviations
- HTLV-1
Human T-cell leukemia virus-1
- MFI
Mean fluorescence intensity
- Pfu
Plaque-forming unit
- Vpr
Viral protein R
- 2DE
Two-dimensional electrophoresis
References
- 1.Lilienbaum A, Paulin D. Activation of the human vimentin gene by the Tax human T-cell leukemia virus. I. Mechanisms of regulation by the NF-kappa B transcription factor. J Biol Chem. 1993;268:2180–2188. [PubMed] [Google Scholar]
- 2.Miyake K, Inokuchi K, Miyake N, Dan K, Shimada T. HIV vector-mediated targeted suicide gene therapy for adult T-cell leukemia. Gene Ther. 2007;14:1662–1667. doi: 10.1038/sj.gt.3303024. [DOI] [PubMed] [Google Scholar]
- 3.Tsukasaki K, Tanosaki S, DeVos S, et al. Identifying progression-associated genes in adult T-cell leukemia/lymphoma by using oligonucleotide microarrays. Int J Cancer. 2004;109:875–881. doi: 10.1002/ijc.20028. [DOI] [PubMed] [Google Scholar]
- 4.Kami M, Hamaki T, Miyakoshi S, et al. Allogeneic haematopoietic stem cell transplantation for the treatment of adult T-cell leukaemia/lymphoma. Br J Haematol. 2003;120:304–309. doi: 10.1046/j.1365-2141.2003.04054.x. [DOI] [PubMed] [Google Scholar]
- 5.Michelfelder S, Lee MK, deLima-Hahn E, et al. Vectors selected from adeno-associated viral display peptide libraries for leukemia cell-targeted cytotoxic gene therapy. Exp Hematol. 2007;35:1766–1776. doi: 10.1016/j.exphem.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 6.Alimonti JB, Ball TB, Fowke KR. Mechanisms of CD4+ T lymphocyte cell death in human immunodeficiency virus infection and AIDS. J Gen Virol. 2003;84:1649–1661. doi: 10.1099/vir.0.19110-0. [DOI] [PubMed] [Google Scholar]
- 7.Borgne-Sanchez A, Dupont S, Langonne A, et al. Targeted Vpr-derived peptides reach mitochondria to induce apoptosis of alphaVbeta3-expressing endothelial cells. Cell Death Differ. 2007;14:422–435. doi: 10.1038/sj.cdd.4402018. [DOI] [PubMed] [Google Scholar]
- 8.Patel CA, Mukhtar M, Pomerantz RJ. Human immunodeficiency virus type 1 Vpr induces apoptosis in human neuronal cells. J Virol. 2000;74:9717–9726. doi: 10.1128/jvi.74.20.9717-9726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersen JL, Planelles V. The role of Vpr in HIV-1 pathogenesis. Curr HIV Res. 2005;3:43–51. doi: 10.2174/1570162052772988. [DOI] [PubMed] [Google Scholar]
- 10.Tungaturthi PK, Sawaya BE, Singh SP, et al. Role of HIV-1 Vpr in AIDS pathogenesis: relevance and implications of intravirion, intracellular and free Vpr. Biomed Pharmacother. 2003;57:20–24. doi: 10.1016/s0753-3322(02)00328-1. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Li HS, Pauza CD, Bukrinsky M, Zhao RY. Roles of HIV-1 auxiliary proteins in viral pathogenesis and host-pathogen interactions. Cell Res. 2005;15:923–934. doi: 10.1038/sj.cr.7290370. [DOI] [PubMed] [Google Scholar]
- 12.Toborek M, Lee YW, Flora G, et al. Mechanisms of the blood-brain barrier disruption in HIV-1 infection. Cell Mol Neurobiol. 2005;25:181–199. doi: 10.1007/s10571-004-1383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones GJ, Barsby NL, Cohen EA, et al. HIV-1 Vpr causes neuronal apoptosis and in vivo neurodegeneration. J Neurosci. 2007;27:3703–3711. doi: 10.1523/JNEUROSCI.5522-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahalingam S, MacDonald B, Ugen KE, et al. In vitro and in vivo tumor growth suppression by HIV-1 Vpr. DNA Cell Biol. 1997;16:137–143. doi: 10.1089/dna.1997.16.137. [DOI] [PubMed] [Google Scholar]
- 15.Muthumani K, Choo AY, Hwang DS, Ugen KE, Weiner DB. HIV-1 Vpr: enhancing sensitivity of tumors to apoptosis. Curr Drug Deliv. 2004;1:335–344. doi: 10.2174/1567201043334614. [DOI] [PubMed] [Google Scholar]
- 16.Elder RT, Benko Z, Zhao Y. HIV-1 VPR modulates cell cycle G2/M transition through an alternative cellular mechanism other than the classic mitotic checkpoints. Front Biosci. 2002;7:d349–d357. doi: 10.2741/elder. [DOI] [PubMed] [Google Scholar]
- 17.Blazquez MV, Macho A, Ortiz C, et al. Extracellular HIV type 1 Tat protein induces CD69 expression through NF-kappaB activation: possible correlation with cell surface Tat-binding proteins. AIDS Res Hum Retroviruses. 1999;15:1209–1218. doi: 10.1089/088922299310304. [DOI] [PubMed] [Google Scholar]
- 18.Boirivant M, Viora M, Giordani L, et al. HIV-1 gp120 accelerates Fas-mediated activation-induced human lamina propria T cell apoptosis. J Clin Immunol. 1998;18:39–47. doi: 10.1023/a:1023235803948. [DOI] [PubMed] [Google Scholar]
- 19.Goh WC, Rogel ME, Kinsey CM, et al. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 20.Bolton DL, Lenardo MJ. Vpr cytopathicity independent of G2/M cell cycle arrest in human immunodeficiency virus type 1-infected CD4+ T cells. J Virol. 2007;81:8878–8890. doi: 10.1128/JVI.00122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chowdhury IH, Wang XF, Landau NR, et al. HIV-1 Vpr activates cell cycle inhibitor p21/Waf1/Cip1: a potential mechanism of G2/M cell cycle arrest. Virology. 2003;305:371–377. doi: 10.1006/viro.2002.1777. [DOI] [PubMed] [Google Scholar]
- 22.Ayyavoo V, Mahalingam S, Rafaeli Y, et al. HIV-1 viral protein R (Vpr) regulates viral replication and cellular proliferation in T cells and monocytoid cells in vitro. J Leukoc Biol. 1997;62:93–99. doi: 10.1002/jlb.62.1.93. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Hasui K, Utsunomiya A, Jia X, Matsuyama T, Murata F. Association of high proliferation in adult T-cell leukemia cells with apoptosis, and expression of p53 protein in acute type ATL. J Clin Exp Hematop. 2008;48:1–10. doi: 10.3960/jslrt.48.1. [DOI] [PubMed] [Google Scholar]
- 24.Takemoto S, Mulloy JC, Cereseto A, et al. Proliferation of adult T cell leukemia/lymphoma cells is associated with the constitutive activation of JAK/STAT proteins. Proc Natl Acad Sci U S A. 1997;94:13897–13902. doi: 10.1073/pnas.94.25.13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, Gong N, Liu J, et al. HIV-1-infected astrocytes and the microglial proteome. J Neuroimmune Pharmacol. 2008;3:173–186. doi: 10.1007/s11481-008-9110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T, Gong N, Liu J, et al. Proteomic modeling for HIV-1 infected microglia-astrocyte crosstalk. PLoS ONE. 2008;3:e2507. doi: 10.1371/journal.pone.0002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu S, Xie Y, Ramachandran P, et al. Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics. 2005;5:1714–1728. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- 29.Paul R, Haydon RC, Cheng H, et al. Potential use of Sox9 gene therapy for intervertebral degenerative disc disease. Spine. 2003;28:755–763. [PMC free article] [PubMed] [Google Scholar]
- 30.Hu ZB, Wu CT, Wang H, et al. A simplified system for generating oncolytic adenovirus vector carrying one or two transgenes. Cancer Gene Ther. 2008;15:173–182. doi: 10.1038/sj.cgt.7701105. [DOI] [PubMed] [Google Scholar]
- 31.Cossarizza A, Baccarani-Contri M, Kalashnikova G, Franceschi C. A new method for the cytofluorimetric analysis of mitochondrial membrane potential using the J-aggregate forming lipophilic cation 5,5',6,6'-tetrachloro-1,1',3,3'-tetraethylbenzimidazolcarbocyanine iodide (JC-1) Biochem Biophys Res Commun. 1993;197:40–45. doi: 10.1006/bbrc.1993.2438. [DOI] [PubMed] [Google Scholar]
- 32.Smiley ST, Reers M, Mottola-Hartshorn C, et al. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc Natl Acad Sci U S A. 1991;88:3671–3675. doi: 10.1073/pnas.88.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds JE, Li J, Eastman A. Detection of apoptosis by flow cytometry of cells simultaneously stained for intracellular pH (carboxy SNARF-1) and membrane permeability (Hoechst 33342) Cytometry. 1996;25:349–357. doi: 10.1002/(SICI)1097-0320(19961201)25:4<349::AID-CYTO6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 34.El-Aneed A. An overview of current delivery systems in cancer gene therapy. J Control Release. 2004;94:1–14. doi: 10.1016/j.jconrel.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Goncalves MA, de Vries AA. Adenovirus: from foe to friend. Rev Med Virol. 2006;16:167–186. doi: 10.1002/rmv.494. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Ma H, Zhang J, Liu S, Liu Y, Zheng D. AAV-mediated TRAIL gene expression driven by hTERT promoter suppressed human hepatocellular carcinoma growth in mice. Life Sci. 2008;82:1154–1161. doi: 10.1016/j.lfs.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 37.Suh W, Chung JK, Park SH, Kim SW. Anti-JL1 antibody-conjugated poly (L-lysine) for targeted gene delivery to leukemia T cells. J Control Release. 2001;72:171–178. doi: 10.1016/s0168-3659(01)00273-5. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X, Li H, Lee RJ. Targeted drug delivery via folate receptors. Expert Opin Drug Deliv. 2008;5:309–319. doi: 10.1517/17425247.5.3.309. [DOI] [PubMed] [Google Scholar]
- 39.Metzl C, Mischek D, Salmons B, Gunzburg WH, Renner M, Portsmouth D. Tissue- and tumor-specific targeting of murine leukemia virus-based replication-competent retroviral vectors. J Virol. 2006;80:7070–7078. doi: 10.1128/JVI.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blumenthal M, Skelton D, Pepper KA, Jahn T, Methangkool E, Kohn DB. Effective suicide gene therapy for leukemia in a model of insertional oncogenesis in mice. Mol Ther. 2007;15:183–192. doi: 10.1038/sj.mt.6300015. [DOI] [PubMed] [Google Scholar]
- 41.Staal FJ, Pike-Overzet K, Ng YY, van Dongen JJ. Sola dosis facit venenum. Leukemia in gene therapy trials: a question of vectors, inserts and dosage? Leukemia. 2008;22:1849–1852. doi: 10.1038/leu.2008.219. [DOI] [PubMed] [Google Scholar]
- 42.Choi J, Walker J, Talbert-Slagle K, Wright P, Pober JS, Alexander L. Endothelial cells promote human immunodeficiency virus replication in nondividing memory T cells via Nef-, Vpr-, and T-cell receptor-dependent activation of NFAT. J Virol. 2005;79:11194–11204. doi: 10.1128/JVI.79.17.11194-11204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng X, Mukhtar M, Acheampong EA, et al. HIV-1 Vpr potently induces programmed cell death in the CNS in vivo. DNA Cell Biol. 2007;26:116–131. doi: 10.1089/dna.2006.0541. [DOI] [PubMed] [Google Scholar]
- 44.Wheeler ED, Achim CL, Ayyavoo V. Immunodetection of human immunodeficiency virus type 1 (HIV-1) Vpr in brain tissue of HIV-1 encephalitic patients. J Neurovirol. 2006;12:200–210. doi: 10.1080/13550280600827377. [DOI] [PubMed] [Google Scholar]
- 45.Azuma A, Matsuo A, Suzuki T, Kurosawa T, Zhang X, Aida Y. Human immunodeficiency virus type 1 Vpr induces cell cycle arrest at the G(1) phase and apoptosis via disruption of mitochondrial function in rodent cells. Microbes Infect. 2006;8:670–679. doi: 10.1016/j.micinf.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Planelles V, Jowett JB, Li QX, Xie Y, Hahn B, Chen IS. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J Virol. 1996;70:2516–2524. doi: 10.1128/jvi.70.4.2516-2524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolton DL, Barnitz RA, Sakai K, Lenardo MJ. 14-3-3 theta binding to cell cycle regulatory factors is enhanced by HIV-1 Vpr. Biol Direct. 2008;3:17. doi: 10.1186/1745-6150-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Yu BW, Rahman KM, Ahmad F, Sarkar FH. Induction of growth arrest and apoptosis in human breast cancer cells by 3,3-diindolylmethane is associated with induction and nuclear localization of p27kip. Mol Cancer Ther. 2008;7:341–349. doi: 10.1158/1535-7163.MCT-07-0476. [DOI] [PubMed] [Google Scholar]
- 49.Chang JH, Kwon HY. Expression of 14-3-3delta, cdc2 and cyclin B proteins related to exotoxin A-induced apoptosis in HeLa S3 cells. Int Immunopharmacol. 2007;7:1185–1191. doi: 10.1016/j.intimp.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 50.Chen JS, Lin SY, Tso WL, et al. Checkpoint kinase 1-mediated phosphorylation of Cdc25C and bad proteins are involved in antitumor effects of loratadine-induced G2/M phase cell-cycle arrest and apoptosis. Mol Carcinog. 2006;45:461–478. doi: 10.1002/mc.20165. [DOI] [PubMed] [Google Scholar]
- 51.Gate L, Lunk A, Tew KD. Resistance to phorbol 12-myristate 13-acetate-induced cell growth arrest in an HL60 cell line chronically exposed to a glutathione S-transferase pi inhibitor. Biochem Pharmacol. 2003;65:1611–1622. doi: 10.1016/s0006-2952(03)00152-7. [DOI] [PubMed] [Google Scholar]
- 52.Fischer C, Sanchez-Ruderisch H, Welzel M, et al. Galectin-1 interacts with the {alpha}5{beta}1 fibronectin receptor to restrict carcinoma cell growth via induction of p21 and p27. J Biol Chem. 2005;280:37266–37277. doi: 10.1074/jbc.M411580200. [DOI] [PubMed] [Google Scholar]
- 53.Marshall JC, Watson RW. Programmed cell death (apoptosis) and the resolution of systemic inflammation. Can J Surg. 1997;40:169–174. [PMC free article] [PubMed] [Google Scholar]
- 54.Williams GT. Programmed cell death: apoptosis and oncogenesis. Cell. 1991;65:1097–1098. doi: 10.1016/0092-8674(91)90002-g. [DOI] [PubMed] [Google Scholar]
- 55.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 56.Szabadkai G, Duchen MR. Mitochondria: the hub of cellular Ca2+ signaling. Physiology (Bethesda) 2008;23:84–94. doi: 10.1152/physiol.00046.2007. [DOI] [PubMed] [Google Scholar]
- 57.Kroemer G, Dallaporta B, Resche-Rigon M. The mitochondrial death/life regulator in apoptosis and necrosis. Annu Rev Physiol. 1998;60:619–642. doi: 10.1146/annurev.physiol.60.1.619. [DOI] [PubMed] [Google Scholar]
- 58.Zhang MH, Lee JS, Kim HJ, et al. HSP90 protects apoptotic cleavage of vimentin in geldanamycin-induced apoptosis. Mol Cell Biochem. 2006;281:111–121. doi: 10.1007/s11010-006-0638-x. [DOI] [PubMed] [Google Scholar]
- 59.Krieser RJ, Eastman A. Cleavage and nuclear translocation of the caspase 3 substrate Rho GDP-dissociation inhibitor, D4-GDI, during apoptosis. Cell Death Differ. 1999;6:412–419. doi: 10.1038/sj.cdd.4400515. [DOI] [PubMed] [Google Scholar]
- 60.Choi MR, Groot M, Drexler HC. Functional implications of caspase-mediated RhoGDI2 processing during apoptosis of HL60 and K562 leukemia cells. Apoptosis. 2007;12:2025–2035. doi: 10.1007/s10495-007-0121-5. [DOI] [PubMed] [Google Scholar]
- 61.Ikonen E, Fiedler K, Parton RG, Simons K. Prohibitin, an antiproliferative protein, is localized to mitochondria. FEBS Lett. 1995;358:273–277. doi: 10.1016/0014-5793(94)01444-6. [DOI] [PubMed] [Google Scholar]
- 62.Fassl S, Leisser C, Huettenbrenner S, et al. Transferrin ensures survival of ovarian carcinoma cells when apoptosis is induced by TNFalpha, FasL, TRAIL, or Myc. Oncogene. 2003;22:8343–8355. doi: 10.1038/sj.onc.1207047. [DOI] [PubMed] [Google Scholar]
- 63.Lanteri M, Giordanengo V, Hiraoka N, et al. Altered T cell surface glycosylation in HIV-1 infection results in increased susceptibility to galectin-1-induced cell death. Glycobiology. 2003;13:909–918. doi: 10.1093/glycob/cwg110. [DOI] [PubMed] [Google Scholar]
- 64.De Santis ML, Hammamieh R, Das R, Jett M. Adipocyte-fatty acid binding protein induces apoptosis in DU145 prostate cancer cells. J Exp Ther Oncol. 2004;4:91–100. [PubMed] [Google Scholar]
- 65.Vavrova J, Janovska S, Rezacova M, et al. Proteomic analysis of MOLT-4 cells treated by valproic acid. Mol Cell Biochem. 2007;303:53–61. doi: 10.1007/s11010-007-9455-0. [DOI] [PubMed] [Google Scholar]
- 66.Iguchi T, Yachide-Noguchi T, Hashimoto Y, et al. Novel tubulin-polymerization inhibitor derived from thalidomide directly induces apoptosis in human multiple myeloma cells: possible anti-myeloma mechanism of thalidomide. Int J Mol Med. 2008;21:163–168. [PubMed] [Google Scholar]
- 67.Muthumani K, Desai BM, Hwang DS, et al. HIV-1 Vpr and anti-inflammatory activity. DNA Cell Biol. 2004;23:239–247. doi: 10.1089/104454904773819824. [DOI] [PubMed] [Google Scholar]
- 68.Lemarechal H, Anract P, Beaudeux JL, Bonnefont-Rousselot D, Ekindjian OG, Borderie D. Expression and extracellular release of Trx80, the truncated form of thioredoxin, by TNF-alpha- and IL-1beta-stimulated human synoviocytes from patients with rheumatoid arthritis. Clin Sci. 2007;113:149–155. doi: 10.1042/CS20070067. [DOI] [PubMed] [Google Scholar]
- 69.Xiao Y, Chen G, Richard J, et al. Cell-surface processing of extracellular human immunodeficiency virus type 1 Vpr by proprotein convertases. Virology. 2008;372:384–397. doi: 10.1016/j.virol.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iordanskiy S, Zhao Y, Dubrovsky L, et al. Heat shock protein 70 protects cells from cell cycle arrest and apoptosis induced by human immunodeficiency virus type 1 viral protein R. J Virol. 2004;78:9697–9704. doi: 10.1128/JVI.78.18.9697-9704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]