Abstract
Bacteriocins are a kind of ribosomal synthesized antimicrobial peptides produced by bacteria, which can kill or inhibit bacterial strains closely-related or non-related to produced bacteria, but will not harm the bacteria themselves by specific immunity proteins. Bacteriocins become one of the weapons against microorganisms due to the specific characteristics of large diversity of structure and function, natural resource, and being stable to heat. Many recent studies have purified and identified bacteriocins for application in food technology, which aims to extend food preservation time, treat pathogen disease and cancer therapy, and maintain human health. Therefore, bacteriocins may become a potential drug candidate for replacing antibiotics in order to treat multiple drugs resistance pathogens in the future. This review article summarizes different types of bacteriocins from bacteria. The latter half of this review focuses on the potential applications in food science and pharmaceutical industry.
Keywords: bacteriocin, protein, natural product, food, cancer treatment
Introduction
There are many antibacterial substances produced by animals, plants, insects, and bacteria, such as hydrogen peroxide, fatty acids, organic acids, ethanol, antibiotics, and bacteriocins. Antimicrobial peptides (AMPs) or proteins produced by bacteria are categorized as bacteriocins. Scant nutrients in the environment trigger microbial production of a variety of bacteriocins for competition of space and resources. Bacteriocins are abundant, have large diversity, and the genes encode ribosomally synthesized antimicrobial peptides or proteins, which kill other related (narrow spectrum) or non-related (broad spectrum) microbiotas as one of the inherent defense system weapons of bacteria (Figure 1) (Cotter et al., 2005). More than 99% of bacteria can produce at least one bacteriocin, most of which are not identified (Riley and Wertz, 2002). The killing ability of bacteriocins is considered a successful strategy for maintaining population and reducing the numbers of competitors to obtain more nutrients and living space in environments. Unlike most antibiotics, which are secondary metabolites, bacteriocins are ribosomally synthesized and sensitive to proteases while generally harmless to the human body and surrounding environment.
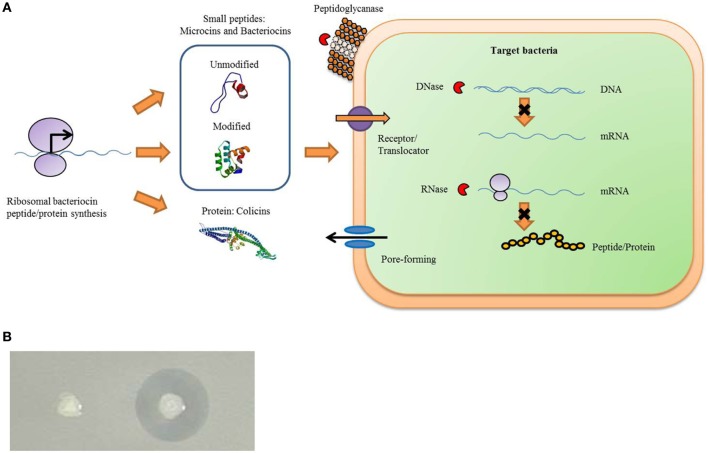
Figure 1.
Bacteriocins function as a natural bacterial immune weapon system. Gram-positive and Gram-negative can produce many kinds of bacteriocins that allow bacteriocin-producing bacteria to have the ability to inhibit the growth of sensitive bacteria. (A) General process of bacteriocins production and antibacterial functions. Bacteriocins are proteins or peptides synthesized by the ribosomal. When released by bacteriocin-producing bacteria, it can become combined with the corresponding receptor on the surface of the sensitive bacteria to kill the bacteria. The sensitive bactericidal mechanisms include the pore-forming type, a nuclease type with DNase and RNase function, and peptidoglycanase type etc. The structure of an unmodified bacteriocin peptide of small molecular weight is subtilisin A (PDB cord 1PXQ). The structure of the modified bacteriocin peptide of small molecular weight is bacteriocin AS-48 (PDB cord 1O83), and the colicin protein structure is colicin Ia (PDB cord 1CII). These structural diagrams are taken from the website of a protein data bank (PDB). (B) When the bacteriocin-producing strains are growing on sensitive bacteria LB soft agar, an inhibition zone will be produced around the bacteriocin-producing strains (right colony). However, there is no inhibition zone around bacteriocin non-producing strains (left colony).
Modern society is more conscious of the importance of food safety, as many of the chemical additives used in food may elicit toxic concern; therefore, it is beneficial to claim natural resources and health benefits of diets. The health benefits of natural foods without chemical additives have become more popular; however, most commercially available preservatives and antibiotics are produced by chemical synthesis, and long-term consumption of such products can affect human health as they reduce the counts of bacteria in the gut. Moreover, the use of antibiotics or residues in food is illegal. Unlike chemical preservatives and antibiotics, “generally recognized as safe” (GRAS) bacteriocins, such as nisin, promise safe use as a food preservative in vegetables, dairy, cheese, meats, and other food products, as they inhibit microorganisms contamination during the production process (Deegan et al., 2006; Settanni and Corsetti, 2008).
This review focuses on the classification of bacteriocins from Gram-negative and Gram-positive bacteria. The application of bacteriocin-producing bacteria and bacteriocins from natural resources for human life is also elucidated upon in the report.
Classification of bacteriocins
Bacteriocins from gram-negative bacteria
Colicins
Colicins are antibacterial proteins produced by bacteria, which can kill bacterial strains closely related to a produced species, in order to reduce environmental competitors for acquiring nutrients and living space. Colicins are organized in three specific domains, an amino-terminal translocation (T) domain, which is implicated in the transfer across the outer membrane via the translocator protein; a central receptor-binding (R) domain, which is bound with a bacterial outer membrane receptor; and a carboxy-terminal cytotoxic (C) domain, which has antibacterial activity (Cascales et al., 2007; Kleanthous, 2010). In order to avoid poisoning by self-produced colicins, specificity immunity proteins will be simultaneously produced to inactivate colicins (Kleanthous, 2010). When a bacterial outer membrane surface has the colicins recognition receptors protein and the translocators protein system, the colicins are transported into the bacteria, which kills it, and are known as sensitive strains. For a particular colicin, non-receptor protein bacteria are classified as resistant strains. Bacteria with a deficiency of translocator protein system are classified as tolerant strains, which produce immunity proteins are classified as immune strains. Resistant, tolerant, and immune strains of bacteria would not be killed by corresponding colicins. There are many colicins found in succession, most of which are encoded on plasmids, while few are located in chromosomes. A typical colicin gene cluster encodes the toxin protein, immunity protein, and lysis gene (Guasch et al., 1995; Kleanthous, 2010). The lysis protein, known as bacteriocin release protein (BRP), can induce the release of colicins from bacteria.
According to translocation across the outer membrane (translocator) system, colicins are categorized into two groups: groups A and B (Table 1). Group A colicins use the Tol protein system (Tol system) to penetrate the outer membrane of sensitive bacteria, for example: colicins E1 to E9, colicin A, K, N… Group B barteriocins use the Ton system (Ton system) to penetrate the outer membrane of sensitive bacteria, for example: colicin 5, 10, B, D, M, V, Ia, Ib… (Dimov et al., 2005; Kleanthous, 2010). In the general, group A colicins are encoded on small plasmids with a lysis gene and can be released out of the bacteria, while group B colicins are encoded on large plasmids without a lysis gene (Cascales et al., 2007).
Table 1.
Classification of colicins by different translocators system: Tol- and Ton-dependent in the E. coli.
| Colicins | Antibacterial activity | Receptor | Translocators | Molecular weight (Da) | Producing strain | References |
|---|---|---|---|---|---|---|
| GROUP A | ||||||
| A | Pore-forming | BtuB | OmpF, TolABQR | 62989 | Citrobacter freudii | Varenne et al., 1981; Morlon et al., 1983 |
| E1 | Pore-forming | BtuB | TolC, TolAQ | 57279 | Escherichia coli | Yamada et al., 1982 |
| K | Pore-forming | Tsx | OmpAF, TolABQR | 59611 | Escherichia coli | Pilsl and Braun, 1995a,b,c |
| N | Pore-forming | OmpF | OmpF, TolAQR | 41696 | Escherichia coli | Pugsley, 1987 |
| S4 | Pore-forming | OmpW | OmpF, TolABQR | 54085 | Escherichia coli | Pilsl et al., 1999 |
| U | Pore-forming | OmpA | OmpF, TolABQR | 66289 | Shigella boydii | Smajs et al., 1997 |
| 28b | Pore-forming | OmpA | OmpF, TolABQR | 47505 | Serratia marcescens | Guasch et al., 1995 |
| GenBank: CAA44310.1 | ||||||
| E2 | DNase | BtuB | OmpF, TolABQR | 61561 | Escherichia coli | Herschman and Helinski, 1967; Cursino et al., 2002 |
| Shigella sonnei | ||||||
| E7 | DNase | BtuB | OmpF, TolABQR | 61349 | Escherichia coli | Chak et al., 1991; Cursino et al., 2002 |
| E8 | DNase | BtuB | OmpF, TolABQR | ~70000 | Escherichia coli | Toba et al., 1988 |
| E9 | DNase | BtuB | OmpF, TolABQR | 61587 | Escherichia coli | Chak et al., 1991; Macdonald et al., 2004 |
| E3 | 16S rRNase | BtuB | OmpF, TolABQR | 57960 | Escherichia coli | Herschman and Helinski, 1967; Cursino et al., 2002 |
| E4 | 16S rRNase | BtuB | OmpF, TolABQR | ND | Escherichia coli | Males and Stocker, 1982 |
| E6 | 16S rRNase | BtuB | OmpF, TolABQR | 58011 | Escherichia coli | Akutsu et al., 1989; Cursino et al., 2002 |
| DF13 | 16S rRNase | IutA | OmpF, TolAQR | 59293 | Escherichia coli | van den Elzen et al., 1983 |
| E5 | tRNase | BtuB | OmpF, TolABQR | 58254 | Escherichia coli | Males and Stocker, 1982 |
| Shigella sonnei | GenBank: KF925332.1 | |||||
| GROUP B | ||||||
| B | Pore-forming | FepA | TonB-ExbBD | 54742 | Escherichia coli | Schramm et al., 1987 |
| Ia | Pore-forming | Cir | TonB-ExbBD | 69429 | Escherichia coli | Konisky and Richards, 1970 GenBank: AAA23182.2 |
| Ib | Pore-forming | Cir | TonB-ExbBD | 69923 | Escherichia coli | Konisky and Richards, 1970 |
| Shigella sonnei | GenBank: AAA23188.1 | |||||
| 5 | Pore-forming | Tsx | TolC, TonB-ExbBD | 53137 | Escherichia coli | Pilsl and Braun, 1995a |
| 10 | Pore-forming | Tsx | TolC, TonB-ExbBD | 53342 | Escherichia coli | Pilsl and Braun, 1995b |
| D | tRNase | FepA | TonB-ExbBD | 74683 | Escherichia coli | Roos et al., 1989 |
| M | Peptidoglycanase | FhuA | TonB-ExbBD | 29453 | Escherichia coli | Kock et al., 1987 |
ND, not determined.
When colicins enter the target cell, they can be divided into three categories based on bactericidal mechanisms: (1) Pore-forming type colicins: the formation of pores or channels in the inner-membrane cause leakage of cytoplasmic compounds, destruct electrochemical gradient, ion loss, and cell death. These include colicin A, B, E1, Ia, Ib, K, and N; (2) Nuclease type colicins: colicins containing DNase, 16S rRNase, and tRNase to non-specifically digest DNA and RNA of bacteria. These include colicin E2 to E9; (3) Peptidoglycanase type colicins: these proteins can digest the peptidoglycan precursor, leading to an inability to synthesize peptidoglycan and bacterial death (Cascales et al., 2007).
Microcins
Microcins are low molecular weight ribosomal synthesized hydrophobic antimicrobial peptides (<10 kDa), which is distinguished by 25–80 kDa high molecular weight colicins protein. Microcins are produced as precursor peptides, including N-terminal leader peptide and core peptides. Microcin precursor peptides may or may not undergo a post-translational modification process in the course of maturation to an active microcin. Microcins are predominantly produced by Enterobacteriaceae showing great tolerance to heat, extreme pH, and proteases (Rebuffat, 2012). The bactericidal mechanisms of microcins are diverse, including the pore-forming type, the nuclease type, such as DNase and RNase functions, and inhibitors of protein synthesis or DNA replication. No microcin genes have a corresponding lysis gene, and microcins are secreted outside the bacteria through the Type I ABC (ATP binding cassette) transporter secretion system, which is composed of a number of proteins (Duquesne et al., 2007).
Microcins are classified as two categories according to molecular masses, disulfide bonds in structure, and post-translational modifications (Table 2). Class I microcins, such as microcin B17, C7-C51, D93, and J25 are of low molecular weight (<5 kDa) post-translationally modified peptides. The molecular weight of class II microcins are larger (5–10 kDa) than that of class I microcins. Class II microcins can be further divided into two subclasses, including class IIa and IIb. Class IIa microcins, such as microcin L, V, and N require three different genes to synthesize and assemble functional peptides. Class IIb microcins, such as microcin E492, M, and H47, are linear peptides with or without post-translational modifications at the C-terminal (Severinov et al., 2007).
Table 2.
Classification scheme for gram-negative microcins.
| Classification | Characteristics | Microcins | Molecular weight (Da) | Producing strain | References |
|---|---|---|---|---|---|
| Class I | Low molecular weight peptides (<5 kDa), post-translationally modified | B17 | 3094 | Escherichia coli | Collin et al., 2013 |
| C7/C51 | 1177 | Escherichia coli | Severinov et al., 2007 | ||
| D93 | <1000 | Escherichia coli | Martinez and Perez-Diaz, 1986 | ||
| J25 | 2107 | Escherichia coli | Wilson et al., 2003 | ||
| Class II | Larger (5–10 kDa) peptides, with or without post-translational modifications | ||||
| class IIa | Required more than one genes to synthesize and assemble functional peptides | L | 8884 | Escherichia coli | Pons et al., 2004 |
| V | 8741 | Escherichia coli | Fath et al., 1994 | ||
| N/24 | 7274 | Escherichia coli | Corsini et al., 2010 | ||
| class IIb | Linear peptides with post-translational modifications or not at C-terminal | E492 | 7886 | Klebsiella pneumoniae | Pons et al., 2002 |
| M | 7284 | Escherichia coli | Vassiliadis et al., 2010 | ||
| H47 | 4865 | Escherichia coli | Vassiliadis et al., 2010 |
Bacteriocins from gram-positive bacteria
Unlike colicins from Gram-negative bacteria, which are plasmid or chromosome encoded 25–80 kDa proteins, the Gram-positive bacteria bacteriocins exert similar characteristics to microsins. These gene-encoded bacteriocins are low molecular weight antimicrobial peptides with less than 60 amino acids. In Gram-positive bacteria, lactic acid bacteria (LAB) are the typical bacteria producing a variety of bacteriocins of different sizes, structures, physicochemical properties, and inhibitory spectrum. Due to the large diversity of bacteriocins, some investigations show different ways to classify bacteriocins from Gram-positive bacteria (Dimov et al., 2005; Cotter et al., 2013). The Gram-positive bacteriocins are generally divided into class I (modified peptides, lantibiotics), class II (unmodified peptides, non-lanthionine), and class III (large proteins, heat unstable) (Table 3).
Table 3.
Classification scheme for gram-positive bacteriocins.
| Classification/features | Bacteriocins | Molecular weight (Da) | Producing strain | References |
|---|---|---|---|---|
| CLASS I | ||||
| The bacteriocins are post-translationally modified, linear or globular peptides containing lanthionine, β-methyl lanthionine and dehydrated amino acids | Nisin A | 3352 | Lactococcus lactis subsp. lactic | Field et al., 2012 |
| Nisin U | 3029 | Streptococcus uberis | Wirawan et al., 2006 | |
| Nisin Z | 3493 | Lactococcus lactis subsp. lactic | Mulders et al., 1991 | |
| Mersacidin | 1824 | Bacillus sp.Y85,54728 | Chatterjee et al., 1992 | |
| Labyrinthopeptin A2 | 1922 | Actinomadura sp. | Meindl et al., 2010 | |
| subtilosin A | 3399 | Bacillus subtilis 168 | Babasaki et al., 1985 | |
| CLASS II | ||||
| Heat stable, unmodified, non-lanthionine-containing bacteriocins, heterogeneous class of small peptides | ||||
| Class IIa (pediocin PA-1like bacteriocins) | pediocin PA-1 | 4629 | Pediococcus acidilactici PAC-1.0 | Henderson et al., 1992 |
| carnobacteriocin X | 3602 | Carnobacterium maltaromaticum C2 | Tulini et al., 2014 | |
| Class IIb (composed of two peptides) | lactacin F | 4755 | Lactobacillus spp. | Fremaux et al., 1993 |
| ABP-118 | 4096 | Lactobacillus salivarius subsp. salivarius UCC118 | Flynn et al., 2002 | |
| Class IIc (circular peptide) | carnocyclin A | 5862 | Carnobacterium maltaromaticum UAL307 | Martin-Visscher et al., 2008 |
| enterocin AS-48 | 7149 | Enterococcus faecalis | Samyn et al., 1994 | |
| Class IId (linear, non-pediocin like, single-peptide) | epidermicin NI01 | 6074 | Staphylococcus epidermidis | Sandiford and Upton, 2012 |
| lactococcin A | 5778 | Lactococcus lactis subsp. Cremoris | Holo et al., 1991 | |
| CLASS III | ||||
| Large, heat unstable proteins | Caseicin 80 | ~42000 | Lactobacillus casei B80 | Muller and Radler, 1993 |
| Enterolisin A | 34501 | Enterococcus faecalis LMG 2333 | Nilsen et al., 2003 | |
| Helveticin J | 37511 | Lactobacillus helveticus 481 | Joerger and Klaenhammer, 1990 | |
Class I peptides are post-translationally modified bacteriocins or lantibiotics with less than 28 amino acids small membrane-active peptides (<5 kDa), linear or globular peptides which contain lanthionine, β-methyl lanthionine, and dehydrated amino acids. Linear structure peptides are membrane disrupting mode of action, and globular structure peptides with cellular enzymatic reaction. Class I bacteriocins are further subdivided into lantibiotics, such as linear peptide nisin and globular peptide mersacidin (Chatterjee et al., 1992), labyrinthopeptins, such as globular peptide labyrinthopeptin A2 (Meindl et al., 2010), and sactibiotics, such as globular peptide subtilosin A (Kawulka et al., 2004).
Class II bacteriocins are 30–60 amino acids (<10 kDa), which always exhibit the unique properties of heat tolerance, unmodified non-lanthionine, and positive charge. The class II bacteriocins are subdivided into five sub-classes by Cotter et al. (2013). Class IIa bacteriocins are Listeria-active peptides with a consensus amino acid sequence of YGNGVXaaC in the N-terminal, and include pediocin PA-1 (Henderson et al., 1992) and carnobacteriocin X (Tulini et al., 2014). Class IIb bacteriocins require two different unmodified peptides for forming a fully active poration complex, such as lactacin F (Allison et al., 1994) and ABP-118 (Flynn et al., 2002). Class IIc bacteriocins are circular peptide bacteriocins, such as carnocyclin A (Gong et al., 2009; Martin-Visscher et al., 2009), and enterocin AS-48 (Martínez-Bueno et al., 1994). Class IId bacteriocins are linear, non-pediocin-like, single-peptide bacteriocins, including epidermicin NI01 (Sandiford and Upton, 2012) and lactococcin A (Holo et al., 1991). Class IIe bacteriocins are non-ribosomal siderophore-type post-translation modification at the serine-rich carboxy-terminal region, such as microcin E492 (de Lorenzo and Pugsley, 1985). As microcin E492 was isolated from Klebsiella pneumonia, which is not a Gram-positive bacteria, the class IIe bacteriocins should be categorized to microcins of Gram-negative bacteria.
Class III bacteriocins are large molecular weight (>30 kDa), heat unstable proteins. Class III can be further subdivided into two distinct groups. Group A bacteriocins are the bacteriolytic enzymes which killing the sensitive strains by lysis of the cell well, such as Enterolisin A (Nilsen et al., 2003). Group B bacteriocins are non-lytic proteins such as Caseicin 80 (Müller and Radler, 1993) and Helveticin J (Joerger and Klaenhammer, 1986).
Potential applications of bacteriocins in food science, pharmaceutics, and clinical medicine
Bacteriocins are now widely used in food science to extend food preservation duration (Ghrairi et al., 2012), which inhibit pathogen infection of animal diseases (van Heel et al., 2011), and pharmaceutical industry and medical society to treatment for malignant cancers (Figure 2) (Lancaster et al., 2007).
Figure 2.
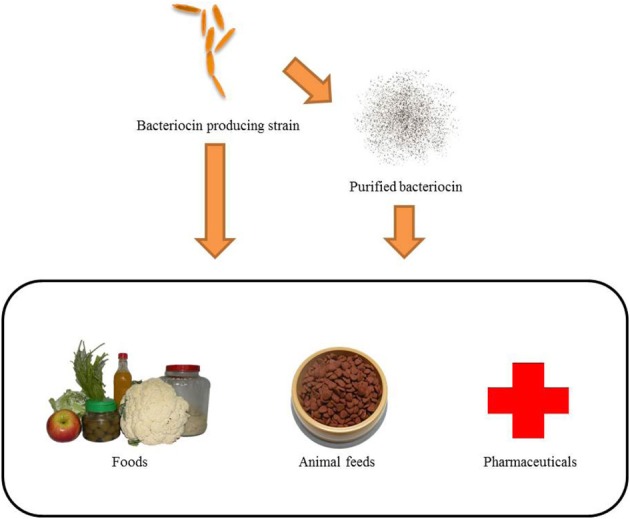
Bacteriocin-producing strains and purified bacteriocins can be applied to food, animals, and medicine. If bacteriocin-producing strains are applied through start culture or co-culture in food, it can extend the preservation of food. Probiotics produced by bacteriocins can balance the bacteria in the digestive tract to reduce gastrointestinal diseases. Purified bacteriocins can be added directly to foods as a natural preservative. Bacteriocins can be added to animal feed as an anti-pathogen additive to protect livestock against pathogen damage. The bacteriocins used in medicine can improve the quality of human life. Bacteriocins have the potential to replace antibiotics as an antibacterial drug, and are a novel anti-cancer drug.
The first bacteriocin was discovered by Gratia (1925), and in recent years, many bacteriocins are successively identified by scientists. Bacteriocins are considered as a natural product because they are the peptides or proteins produced by bacteria present in many fermented or non-fermented foods since ancient times. Many microorganisms, such as bacteriocin producing LAB, are used to start cultures or co-cultures in food production processes for increasing flavor and prolonging shelf-life. In addition, many bacteriocin producing bacteria are isolated from foods and their raw materials (Settanni and Corsetti, 2008).
Probiotics
The term “probiotic” is derived from the Greek pro bios, meaning “for life” or “in support of life,” which was first used by Lilly and Stillwell (1965). The probiotic is generally considered to promote the balance of intestinal microbiota and increase health benefits. The World Health Organization (WHO) defines probiotics as, “Live microorganisms which, when administrated in adequate amounts, confer a health benefit on the host” (Dobson et al., 2012). The characteristics of probiotics should include: a group of strains beneficial to the host animal that can stably survive and have metabolic activity in the intestinal environment, and being non-pathogenic and non-toxic, remain stable and viable for long periods of storage and harsh conditions (Fuller, 1989). There are many ways for probiotics to control intestinal pathogens. Probiotics demonstrate the capabilities of antimicrobial substance production, competitive exclusion of pathogen binding, competition for nutrients, and modulation of the immune system (FAO/WHO, 2001). Many antibacterial substances, such as bacteriocins, short chain fatty acids, and hydrogen peroxide, are produced by probiotics for inhibiting gastrointestinal microorganisms or pathogens. Dobson et al. (2012) considered that bacteriocins are one of the traits of probiotics. Currently many probiotics are used in daily life, including LAB, non-pathogenic E. coli, bacilli, and yeasts.
Purified bacteriocins, or bacteriocin producing probiotics, can reduce the number of pathogens or change the composition of intestinal microbiota in animal models, such as mice, chickens, and pigs. Bernbom et al. (2006) report the ability of pure nisin, nisin-producing Lactococcus lactis strain CHCC5826 and non-nisin-producing Lactococcus lactis strain CHCH2862, could affect the composition of intestinal microbiota in human flora-associated rats. They found that the number of Bifidobacterium in the feces of rats fed with nisin-producing and non-nisin-producing Lactococcus lactis for 8 days significantly increased, while the number of enterococci/streptococci in duodenum, ileum, cecum, and the colon were reduced. However, the above effect was not found after feeding of purified nisin. Lactococcus lactis may affect the intestinal microbiota through competition of nutrients or adhesion sites. Therefore, the effect of changes in the composition of intestinal microbiota may be not related to the presence of nisin in Lactococcus lactis. Cursino et al. (2006) indicated that colicin Ib, E1, and microcin C7, as derived from E. coli strain H22, possess the ability to inhibit the growth of pathogenic or non-pathogenic bacteria, including Enterobacter, Escherichia, Klebsiella, Morganella, Salmonella, Shigella, and Yersinia. In a germ-free mouse model, E. coli strain H22 showed the ability to reduce the population of Shigella flexneri 4 to undetectable levels in feces after a 6-day oral inoculation (Cursino et al., 2006). The results suggested that E. coli strain H22, and other multiple bacteriocin producing strains, had potential to be employed as a probiotic for livestock and humans. Corr et al. (2007) demonstrated that the human origin probiotic strains Lactobacillus salivarius UCC118 produced bacteriocin Abp118 for inhibiting infection with foodborne pathogen Listeria monocytogenes in mice. The bacteriocin Abp118 mutant strain UCC118Δabp118 failed to protect mice against infection by Listeria monocytogenes. Riboulet-Bisson et al. (2012) also demonstrated that bacteriocins of UCC118 were one of the points in changing the composition of microbiota in the mice and pig models. Millette et al. (2008) isolated two bacteriocin-producing LAB, Lactococcus lactis MM19 and Pediococcus acidilactici MM33, from human feces, and found the ability of LAB to reduce the numbers of vancomycin-resistant Enterococci (VRE) in the C57BL/6 mice model. Nisin Z and pediocin PA-1/AcH were produced by Lactococcus lactis MM19 and Pediococcus acidilactici MM33, respectively, which showed strong activity against clinical VRE isolate. The results revealed that Lactococcus lactis and non-pediocin PA-1/AcH producing mutant Pediococcus acidilactici MM33A had the ability to increase the total LAB and anaerobes populations, while Pediococcus acidilactici MM33 decreased the Enterobacteriaceae populations in healthy mice feces. Moreover, after feeding MM19 or MM33 for 3 days, the VRE populations in feces were respectively reduced to 2.50 and 1.85 log10 CFU/g than PBS control. A similar study (Dabour et al., 2009) demonstrated that pediocin PA-1 producing probiotic Pediococcus acidilactici UL5 had the ability to inhibit Listeria monocytogenes in vitro. However, Pediococcus acidilactici UL5 did not reduce the population of Listeria monocytogenes in the mouse intestinal, and was not detectable in fecal samples. Pediococcus acidilactici UL5 did not protect from infection of Listeria monocytogenes, which was due to the failure to compete with other intestinal microbes or inhibit the production of pediocin PA-1 by other microbial in the intestine. There is another ideal situation for artificially establishing probiotic, by transforming a bacteriocin gene into an E. coli strain. The benefits to humans by the more powerful probiotics can be produced using genetic engineering techniques. Gillor et al. (2009) utilized six different bacteriocin-encoding plasmids, including colicin A, E1, E2, E7, K, and N, in order to transform into the E. coli strain BZB1011. Four-week-old CD-1 mice were inoculated with the BZB1011 control strain, or one of the six colicinogenic BZB1011 strains. The fecal bacteria were monitored for 112 days. After 112 days incubation, the density of colicinogenicity BZB1011 in the feces was significantly higher than that of control BZB1011. The results suggest that colicin expression is helpful to increase E. coli colonization in the mouse gastrointestinal tract.
Food technology
In order to extend shelf-life, antibiotics or food preservative are incorporated (e.g., nitrite and sulfur dioxide) into foods to delay microbial growth and possible corruption. However, most commercial preservatives are developed via chemical synthesis, and long-term consumption of the synthetic preservatives may have an adverse impact on the human body. Moreover, it is illegal to use antibiotics in food products. The bacteriocins produced by Gram-positive or Gram-negative bacteria are gene encoded peptides or proteins, which are suitable as natural preservatives in food products. Due to the sensitivity of bacteriocins to some proteases, harmless bacteriocins are possibly digested, thus, un-functional small peptides and amino acids are bacteriocin-loaded foods digested in the gastrointestinal tract (Cleveland et al., 2001; Bernbom et al., 2006). Thus, bacteriocins are considered as basically safe food additives after intake by the gastrointestinal system.
Bacteriocins are natural food additives due to the bacteriocin producing bacteria presence in many types of foods since ancient times, such as cheeses, yogurts, and Portuguese fermented meat (Yang et al., 2012; Todorov et al., 2014). In food technology, nisin is produced by Lactococcus lactis and was the first antibacterial peptide found in LAB (Rogers, 1928). It is also a commercial bacteriocin used as a food preservative against contamination by microorganism, which is marketed as Nisaplin®. It is the only bacteriocin approved for utilization as a preservative in many foods by the U.S. Food and Drug Administration (USFDA), and licensed as a food additive in over 45 countries (Settanni and Corsetti, 2008). Another commercially available bacteriocin is pediocin PA-1, marketed as Alta® 2341, which inhibits the growth of Listeria monocytogenes in meat products (Settanni and Corsetti, 2008). In many food products, such as traditional European cheeses, the milk used in the manufacturing process is easily contaminated with animal excrement. The bacteriocinogenic Enterococci as starter cultures or co-cultures can be used for reducing microbiota contamination (Foulquié Moreno et al., 2006). Settanni and Corsetti (2008) reviewed bacteriocinogenic LAB strains as a co-culture, protective, or starter cultures in fermented and non-fermented vegetables, such as olives, sourdough, miso, sauerkrauts, refrigerated pickles, and mungbean sprouts. Moreover, they introduced bacteriocins as foods additives, such as nisin, which is used in kimchi, mashed potatoes, and fresh-cut products. Enterocin AS-48 is used in cider, fruit and vegetable juices, and canned vegetables for contamination inhibition. Enterocin CCM4231 and EJ97 are used in soy milk and zucchini purée for suppression of contamination, respectively (Settanni and Corsetti, 2008).
Treatment of pathogen-associated diseases
Since the first antibiotic penicillin was discovered in 1928 by Alexander Fleming, many discovered antibiotics are applied to treat pathogens. Antibiotics were first approved by USFDA in 1951, and used in animal feed, which significantly reduced the number of deaths due to bacterial infection cases. However, the problem with multiple drug resistance pathogens has become increasingly serious, due to concerns regarding the abuse of antibiotics (Joerger, 2003). Bacteriocins are reported to inhibit important animal and plant pathogens, such as Shiga toxin-producing E. coli (STEC), enterotoxigenic E. coli (ETEC), methicillin-resistant Staphylococcus aureus (MRSA), VRE, Agrobacterium, and Brenneria spp. (Grinter et al., 2012; Cotter et al., 2013). The bactericidal mechanism of bacteriocins are mainly located in the receptor-binding of bacteria surfaces, and then through the membrane, which causes bacteria cytotoxicity. In addition, bacteriocins are low-toxic peptides or proteins sensitive to proteases, such as trypsin and pepsin (Cleveland et al., 2001).
Jordi et al. (2001) found that 20 kinds of E. coli could express colicin, which inhibited five kinds of Shiga toxin-producing E. coli (O26, O111, O128, O145, and O157: H7). These E. coli can cause diarrhea and hemolytic uremic syndrome in humans. In a simulated cattle rumen environment, colicin E1, E4, E8-J, K, and S4 producing E. coli can significantly inhibit the growth of STEC. Stahl et al. (2004) utilized purified colicin E1 and colicin N for effective activity against enterotoxigenic E. coli pathogens F4 (K88) and F18 in vitro, which caused post-weaning diarrhea in piglets. Furthermore, purified coilicin E1 proteins were mixed with the dietary intake of young pigs. The results showed a reduction of the incidence of post-weaning diarrhea by F-18 positive E. coli (Cutler et al., 2007). The growth of the piglets was thus ameliorated. Józefiak et al. (2013) used the nisin-supplemented bird diet to feed broiler chickens, and found a reduced number of Bacteroides and Enterobacteriacae in ileal digesta of nisin-supplemented chickens. The action of nisin was similar to that of salinomycin. After a 35-day growth, the average body weight gain of nisin-supplemented (2700 IU nisin/g) chickens is 1918 g/bird, which was higher than the 1729 g of non-nisin supplemented or the 1763 g of salinomycin-supplemented chickens. Stern et al. (2006) reported that class II low molecule mass bacteriocin OR-7 was purified from the Lactobacillus salivarius strain NRRL B-30514. This bacteriocin exhibited an ability against human gastroenteritis pathogen Campylobacter jejuni. OR-7 was stable when treated with lysozyme, lipase, and heat to 90°C, or at pH ranges from 3.0 to 9.1. The purified OR-7 was encapsulated in polyvinylpyrrolidone for chicken feed. The populations of C. jejuni were reduced at least one million fold over that of non-OR-7 supplemented chickens in cecal material of OR-7-treated chickens. These results suggest that nisin, OR-7, and other bacteriocins, illustrated potential when applied to replace antibiotics in poultry and other animal feeds.
Cancer therapy
Over the past half century, cancer has become a serious problem, and a threat to human health. According to new information from the WHO website, there were 8.2 million people died from cancer and 14.1 million new cancer cases worldwide in 2012, with 60% of world's total new annual cases occurring in Africa, Asia, and Central and South America. Of the world's top 10 leading causes of death, lung cancer (2.7%, including trachea and bronchus cancer) was 7th and caused 1.5 million (2.7%) deaths in 2011, higher than 1.2 million (2.2%, 9th) deaths in 2000. In the United States, there were 1,660,290 new cancer cases, with 580,350 cancer deaths projected to occur in 2013, pedestalling cancer as the second leading cause of death, exceeded only by heart diseases (Center, 2013; Siegel et al., 2013).
In cancer therapy, some researches indicate that bacteriocins show activity against tumor cells. Considering that bacteriocins are naturally and legally added in foods, bacteriocins may be suitable as a potential anti-tumor drug candidate. Some bacteriocins, such as pore-forming colicin A and E1 inhibited the growth of one human standard fibroblast line MRC5 and 11 human tumor-cell lines (Chumchalová and Smarda, 2003). Contrarily, pore-forming colicin U and RNAase activity colicin E3 did not display this growth inhibition capability. Colicin D, E2, E3, and pore-forming colicin A could inhibit the viability of murine leukaemia cells P388, while pore-forming colicin E1 and colicin E3 suppressed v-myb-transformed chicken monoblasts (Fuska et al., 1979; Lancaster et al., 2007). Bures et al. (1986) isolated E. coli from the feces of 77 colorectal carcinoma patients, where 32 patients (41.6%) had barteriocins-producing E. coli. In the feces of 160 healthy people, 102 people (63.8%) had barteriocins-producing E. coli, which also showed that colicins from bacteria in the intestine may be one of the factors in reducing human colorectal carcinoma. Colicins could act as an anti-cancer drug of moderate potential. Supplements of bacteriocin-producing probiotics may be another way to prevent cancer occurrence. In a recent study, Joo et al. (2012) found that nisin had capabilities to prevent cancer cell growth. Three head and neck squamous cell carcinoma (HNSCC) 17B, HSC, and 14A were treated by nisin at concentrations of 40 and 80 μg/ml. After 24 h nisin increased DNA fragmentation or apoptosis, arrested cell cycle and reduced cell proliferation of HNSCC occurred. The floor-of-mouth oral cancer xenograft mouse model was used to test the anti- HNSCC function of nisin. A 150-mg/kg dose of nisin was administered orally every day for 3 weeks, and tumor volumes were significantly reduced in nisin-treated mice, as compared with the control mice, which received only water. These results indicated that nisin provides a safe and novel therapy for treating HNSCC.
Conclusions and future perspectives
In general, the genes and immune genes of bacteriocins are encoded on the same plasmid or in adjacent regions of a chromosome. The bactericins genes can enter into other bacterial cells via conjugation (Ito et al., 2009; Burton et al., 2013). Accordingly, by conjugation or transposon for insertion, the bacterial pathogen can obtain some immune genes of bacteriocins. In this way, in environment or food pathogens prevention strategies, the use of purified bacterial peptide or protein is superior to the use of bacteria to produce bacteriocins. In terms of disease control, many papers report only the ability to fight pathogens of the probiotic produced by a single or specific bacteriocin, and the ability of the probiotic generated by a single bacteriocin to balance the bacteria strains in the intestinal tract. However, there are many different kinds of pathogens and mutants in nature. The specific use of a particular bacteriocin cannot eliminate all bacterial pathogens. Therefore, we can mix a variety of bacteriocin proteins to make cocktail drugs for application to the prevention of certain human or animal pathogens to cause death or reduce its diffusion rate. In disease control, bacteriocin can solve some of the most challenging problems of multi-drug resistant pathogens.
In recent years, the increased number of multi-drug resistant pathogens has become a serious problem, and finding or developing a new generation of antimicrobial agents is becoming increasingly important. Many new antibacterial substances have also been found by scientists to replace the old antibiotics; however, finding and identifying new antimicrobial substances is a difficult task, and the use of biotechnology to fuse two known bacteriocins into a new bacteriocins may be a quick fix method. By using recombinant PCR techniques, Acuña et al. (2012) integrated enterocin CRL35 and microcin V genes to obtain a bacteriocins called Ent35-MccV. Ent35-MccV has bactericidal capacity against clinically isolated enterohemorrhagic E. coli and Listeria monocytogenes, as well as some Gram-positive and Gram-negative bacteria, which are not clinically isolated. Such a technique can crease new or multi-functional bacteriocins, which are more powerful in functionality and germicidal range. As a result, they can be widely used in food, animal husbandry, and medicine.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the financial support from Chang Gung University (grant numbers: EMRPD1D901 and CMRPD1B0332).
References
- Acuña L., Picariello G., Sesma F., Morero R. D., Bellomio A. (2012). A new hybrid bacteriocin, Ent35-MccV, displays antimicrobial activity against pathogenic Gram-positive and Gram-negative bacteria. FEBS Open Bio 2, 12–19 10.1016/j.fob.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akutsu A., Masaki H., Ohta T. (1989). Molecular structure and immunity specificity of colicin E6, an evolutionary intermediate between E-group colicins and cloacin DF13. J. Bacteriol. 171, 6430–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison G. E., Fremaux C., Klaenhammer T. R. (1994). Expansion of bacteriocin activity and host range upon complementation of two peptides encoded within the lactacin F operon. J. Bacteriol. 176, 2235–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babasaki K., Takao T., Shimonishi Y., Kurahashi K. (1985). Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J. Bacteriol. 98, 585–603 [DOI] [PubMed] [Google Scholar]
- Bernbom N., Licht T. R., Brogren C. H., Jelle B., Johansen A. H., Badiola I., et al. (2006). Effects of Lactococcus lactis on composition of intestinal microbiota: role of nisin. Appl. Environ. Microbiol. 72, 239–244 10.1128/AEM.72.1.239-244.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bures J., Horák V., Fixa B., Komárková O., Zaydlar K., Lonskı V., et al. (1986). Colicinogeny in colorectal cancer. Neoplasma 33, 233–237 [PubMed] [Google Scholar]
- Burton J. P., Wescombe P. A., Macklaim J. M., Chai M. H., Macdonald K., Hale J. D., et al. (2013). Persistence of the oral probiotic Streptococcus salivarius M18 is dose dependent and megaplasmid transfer can augment their bacteriocin production and adhesion characteristics. PLoS ONE 8:e65991 10.1371/journal.pone.0065991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E., Buchanan S. K., Duche D., Kleanthous C., Lloubès R., Postle K., et al. (2007). Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229 10.1128/MMBR.00036-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center N. (2013). Cancer Facts and Figures 2013. Goyang: Director of National Cancer Center Minister of Health and Welfare [Google Scholar]
- Chak K. F., Kuo W. S., Lu F. M., James R. (1991). Cloning and characterization of the ColE7 plasmid. J. Gen. Microbiol. 137, 91–100 10.1099/00221287-137-1-91 [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Chatterjee S., Lad S. J., Phansalkar M. S., Rupp R. H., Ganguli B. N., et al. (1992). Mersacidin, a new antibiotic from Bacillus. Fermentation, isolation, purification and chemical characterization. J. Antibiot. (Tokyo) 45, 832–838 10.7164/antibiotics.45.832 [DOI] [PubMed] [Google Scholar]
- Chumchalová J., Smarda J. (2003). Human tumor cells are selectively inhibited by colicins. Folia Microbiol. (Praha) 48, 111–115 10.1007/BF02931286 [DOI] [PubMed] [Google Scholar]
- Cleveland J., Montville T. J., Nes I. F., Chikindas M. L. (2001). Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71, 1–20 10.1016/S0168-1605(01)00560-8 [DOI] [PubMed] [Google Scholar]
- Collin F., Thompson R. E., Jolliffe K. A., Payne R. J., Maxwell A. (2013). Fragments of the bacterial toxin microcin B17 as gyrase poisons. PLoS ONE 8:e61459 10.1371/journal.pone.0061459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corr S. C., Li Y., Riedel C. U., O'Toole P. W., Hill C., Gahan C. G. (2007). Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U.S.A. 104, 7617–7621 10.1073/pnas.0700440104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini G., Karahanian E., Tello M., Fernandez K., Rivero D., Saavedra J. M., et al. (2010). Purification and characterization of the antimicrobial peptide microcin N. FEMS Microbiol. Lett. 312, 119–125 10.1111/j.1574-6968.2010.02106.x [DOI] [PubMed] [Google Scholar]
- Cotter P. D., Hill C., Ross R. P. (2005). Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788 10.1038/nrmicro1273 [DOI] [PubMed] [Google Scholar]
- Cotter P. D., Ross R. P., Hill C. (2013). Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105 10.1038/nrmicro2937 [DOI] [PubMed] [Google Scholar]
- Cursino L., Chartone-Souza E., Nascimento A. (2002). Recent updated aspects of colicins of Enterobacteriaceae. Braz. J. Microbiol. 33, 185–195 10.1590/S1517-83822002000300001 [DOI] [Google Scholar]
- Cursino L., Smajs D., Smarda J., Nardi R. M., Nicoli J. R., Chartone-Souza E., et al. (2006). Exoproducts of the Escherichia coli strain H22 inhibiting some enteric pathogens both in vitro and in vivo. J. Appl. Microbiol. 100, 821–829 10.1111/j.1365-2672.2006.02834.x [DOI] [PubMed] [Google Scholar]
- Cutler S. A., Lonergan S. M., Cornick N., Johnson A. K., Stahl C. H. (2007). Dietary inclusion of colicin e1 is effective in preventing postweaning diarrhea caused by F18-positive Escherichia coli in pigs. Antimicrob. Agents Chemother. 51, 3830–3835 10.1128/AAC.00360-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabour N., Zihler A., Kheadr E., Lacroix C., Fliss I. (2009). In vivo study on the effectiveness of pediocin PA-1 and Pediococcus acidilactici UL5 at inhibiting Listeria monocytogenes. Int. J. Food Microbiol. 133, 225–233 10.1016/j.ijfoodmicro.2009.05.005 [DOI] [PubMed] [Google Scholar]
- Deegan L. H., Cotter P. D., Hill C., Ross P. (2006). Bacteriocins: biological tools for bio-preservation and shelf-life extension. Int. Dairy J. 16, 1058–1071 10.1016/j.idairyj.2005.10.026 [DOI] [Google Scholar]
- de Lorenzo V., Pugsley A. P. (1985). Microcin E492, a low-molecular-weight peptide antibiotic which causes depolarization of the Escherichia coli cytoplasmic membrane. Antimicrob. Agents Chemother. 27, 666–669 10.1128/AAC.27.4.666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimov S., Ivanova P., Harizanova N., Ivanova I. (2005). Bioactive peptides used by bacteria in the concurrence for the ecological niche: eneral classification and mode of action (overview). Biotechnol. Biotechnol. Eq. 3, 3–22 10.1080/13102818.2005.10817185 [DOI] [Google Scholar]
- Dobson A., Cotter P. D., Ross R. P., Hill C. (2012). Bacteriocin production: a probiotic trait? Appl. Environ. Microbiol. 78, 1–6 10.1128/AEM.05576-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquesne S., Petit V., Peduzzi J., Rebuffat S. (2007). Structural and functional diversity of microcins, gene-encoded antibacterial peptides from enterobacteria. J. Mol. Microbiol. Biotechnol. 13, 200–209 10.1159/000104748 [DOI] [PubMed] [Google Scholar]
- FAO/WHO. (2001). Report on Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk With Live Lactic Acid Bacteria. Córdoba: FAO/WHO [Google Scholar]
- Fath M. J., Zhang L. H., Rush J., Kolter R. (1994). Purification and characterization of colicin V from Escherichia coli culture supernatants. Biochemistry 33, 6911–6917 10.1021/bi00188a021 [DOI] [PubMed] [Google Scholar]
- Field D., Begley M., O'Connor P. M., Daly K. M., Hugenholtz F., Cotter P. D., et al. (2012). Bioengineered nisin A derivatives with enhanced activity against both Gram positive and Gram negative pathogens. PLoS ONE 7:e46884 10.1371/journal.pone.0046884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn S., van Sinderen D., Thornton G. M., Holo H., Nes I. F., Collins J. K. (2002). Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148, 973–984 [DOI] [PubMed] [Google Scholar]
- Foulquié Moreno M. R., Sarantinopoulos P., Tsakalidou E., De Vuyst L. (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106, 1–24 10.1016/j.ijfoodmicro.2005.06.026 [DOI] [PubMed] [Google Scholar]
- Fremaux C., Ahn C., Klaenhammer T. R. (1993). Molecular analysis of the lactacin F operon. Appl. Environ. Microbiol. 59, 3906–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. (1989). Probiotics in man and animals. Experientia 35, 406–407 [PubMed] [Google Scholar]
- Fuska J., Fuskova A., Smarda J., Mach J. (1979). Effect of colicin E3 on leukemia cells P388 in vitro. Experientia 35, 406–407 10.1007/BF01964380 [DOI] [PubMed] [Google Scholar]
- Ghrairi T., Chaftar N., Hani K. (2012). Bacteriocins: recent advances and opportunities in Progress in Food Preservation, Chapter 23. eds Bhat R., Karim Alias A., Paliyath G. (Oxford: Wiley-Blackwell; ), 485–511 [Google Scholar]
- Gillor O., Giladi I., Riley M. A. (2009). Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol. 9:165 10.1186/1471-2180-9-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X., Martin-Visscher L. A., Nahirney D., Vederas J. C., Duszyk M. (2009). The circular bacteriocin, carnocyclin A, forms anion-selective channels in lipid bilayers. Biochim. Biophys. Acta 1788, 1797–1803 10.1016/j.bbamem.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Gratia A. (1925). Sur un remarquable example d'antagonisme entre deux souches de colibacille. Comput. Rend. Soc. Biol. 93, 1040–1042 [Google Scholar]
- Grinter R., Milner J., Walker D. (2012). Bacteriocins active against plant pathogenic bacteria. Biochem. Soc. Trans. 40, 1498–1502 10.1042/BST20120206 [DOI] [PubMed] [Google Scholar]
- Guasch J. F., Enfedaque J., Ferrer S., Gargallo D., Regue M. (1995). Bacteriocin 28b, a chromosomally encoded bacteriocin produced by most Serratia marcescens biotypes. Res. Microbiol. 146, 477–483 10.1016/0923-2508(96)80293-2 [DOI] [PubMed] [Google Scholar]
- Henderson J. T., Chopko A. L., van Wassenaar P. D. (1992). Purification and primary structure of pediocin PA-1 produced by Pediococcus acidilactici PAC-1.0. Arch. Biochem. Biophys. 295, 5–12 10.1016/0003-9861(92)90480-K [DOI] [PubMed] [Google Scholar]
- Herschman H. R., Helinski D. R. (1967). Purification and characterization of colicin E2 and colicin E3. J. Biol. Chem. 242, 5360–5368 [PubMed] [Google Scholar]
- Holo H., Nilssen O., Nes I. F. (1991). Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J. Bacteriol. 173, 3879–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Kawai Y., Arakawa K., Honme Y., Sasaki T., Saito T. (2009). Conjugative plasmid from Lactobacillus gasseri LA39 that carries genes for production of and immunity to the circular bacteriocin gassericin A. Appl. Environ. Microbiol. 75, 6340–6351 10.1128/AEM.00195-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger M. C., Klaenhammer T. R. (1986). Characterization and purification of helveticin J and evidence for a chromosomally determined bacteriocin produced by Lactobacillus helveticus 481. J. Bacteriol. 167, 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger M. C., Klaenhammer T. R. (1990). Cloning, expression, and nucleotide sequence of the Lactobacillus helveticus 481 gene encoding the bacteriocin helveticin J. J. Bacteriol. 172, 6339–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger R. D. (2003). Alternatives to antibiotics: bacteriocins, antimicrobial peptides and bacteriophages. Poult. Sci. 82, 640–647 10.1093/ps/82.4.640 [DOI] [PubMed] [Google Scholar]
- Joo N. E., Ritchie K., Kamarajan P., Miao D., Kapila Y. L. (2012). Nisin, an apoptogenic bacteriocin and food preservative, attenuates HNSCC tumorigenesis via CHAC1. Cancer Med. 1, 295–305 10.1002/cam4.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordi B. J., Boutaga K., van Heeswijk C. M., van Knapen F., Lipman L. J. (2001). Sensitivity of Shiga toxin-producing Escherichia coli (STEC) strains for colicins under different experimental conditions. FEMS. Microbiol. Lett. 204, 329–334 10.1111/j.1574-6968.2001.tb10906.x [DOI] [PubMed] [Google Scholar]
- Józefiak D., Kieroñczyk B., Juœkiewicz J., Zduñczyk Z., Rawski M., Długosz J., et al. (2013). Dietary nisin modulates the gastrointestinal microbial ecology and enhances growth performance of the broiler chickens. PLoS ONE 8:e85347 10.1371/journal.pone.0085347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawulka K. E., Sprules T., Diaper C. M., Whittal R. M., McKay R. T., Mercier P., et al. (2004). Structure of subtilosin A, a cyclic antimicrobial peptide from Bacillus subtilis with unusual sulfur to alpha-carbon cross-links: formation and reduction of alpha-thio-alpha-amino acid derivatives. Biochemistry 43, 3385–3395 10.1021/bi0359527 [DOI] [PubMed] [Google Scholar]
- Kleanthous C. (2010). Swimming against the tide: progress and challenges in our understanding of colicin translocation. Nat. Rev. Microbiol. 8, 843–848 10.1038/nrmicro2454 [DOI] [PubMed] [Google Scholar]
- Kock J., Olschlager T., Kamp R. M., Braun V. (1987). Primary structure of colicin M, an inhibitor of murein biosynthesis. J. Bacteriol. 169, 3358–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konisky J., Richards F. M. (1970). Characterization of Colicin Ia and Colicin Ib purification and some physical properties. J. Biol. Chem. 245, 2972–2978 [PubMed] [Google Scholar]
- Lancaster L. E., Wintermeyer W., Rodnina M. V. (2007). Colicins and their potential in cancer treatment. Blood Cells Mol. Dis. 38, 15–18 10.1016/j.bcmd.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Lilly D. M., Stillwell R. H. (1965). Porbiotics: growth-promoting factors produced by microorganisms. Science 147, 747–748 10.1126/science.147.3659.747 [DOI] [PubMed] [Google Scholar]
- Macdonald C. J., Tozawa K., Collins E. S., Penfold C. N., James R., Kleanthous C., et al. (2004). Characterisation of a mobile protein-binding epitope in the translocation domain of colicin E9. J. Biomol. NMR 30, 81–96 10.1023/B:JNMR.0000042963.71790.19 [DOI] [PubMed] [Google Scholar]
- Males B. M., Stocker B. A. (1982). Colicins E4, E5, E6 and A and properties of btuB+ colicinogenic transconjugants. J. Gen. Microbiol. 128, 95–106 [DOI] [PubMed] [Google Scholar]
- Martinez J. L., Perez-Diaz J. C. (1986). Isolation, characterization, and mode of action on Escherichia coli strains of microcin D93. Antimicrob. Agents Chemother. 29, 456–460 10.1128/AAC.29.3.456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Bueno M., Maqueda M., Gálvez A., Samyn B., Van Beeumen J., Coyette J., et al. (1994). Determination of the gene sequence and the molecular structure of the enterococcal peptide antibiotic AS-48. J. Bacteriol. 176, 6334–6339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Visscher L. A., Gong X., Duszyk M., Vederas J. C. (2009). The three-dimensional structure of carnocyclin A reveals that many circular bacteriocins share a common structural motif. J. Biol. Chem. 284, 28674–28681 10.1074/jbc.M109.036459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Visscher L. A., van Belkum M. J., Garneau-Tsodikova S., Whittal R. M., Zheng J., McMullen L. M., et al. (2008). Isolation and characterization of carnocyclin a, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 74, 4756–4763 10.1128/AEM.00817-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl K., Schmiederer T., Schneider K., Reicke A., Butz D., Keller S., et al. (2010). Labyrinthopeptins: a new class of carbacyclic lantibiotics. Angew. Chem. Int. Ed. Engl. 49, 1151–1154 10.1002/anie.200905773 [DOI] [PubMed] [Google Scholar]
- Millette M., Cornut G., Dupont C., Shareck F., Archambault D., Lacroix M. (2008). Capacity of human nisin- and pediocin-producing lactic Acid bacteria to reduce intestinal colonization by vancomycin-resistant enterococci. Appl. Environ. Microbiol. 74, 1997–2003 10.1128/AEM.02150-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlon J., Lloubes R., Varenne S., Chartier M., Lazdunski C. (1983). Complete nucleotide sequence of the structural gene for colicin A, a gene translated at non-uniform rate. J. Mol. Biol. 170, 271–285 10.1016/S0022-2836(83)80148-X [DOI] [PubMed] [Google Scholar]
- Mulders J. W., Boerrigter I. J., Rollema H. S., Siezen R. J., de Vos W. M. (1991). Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur. J. Biochem. 201, 581–584 10.1111/j.1432-1033.1991.tb16317.x [DOI] [PubMed] [Google Scholar]
- Müller E., Radler F. (1993). Caseicin, a bacteriocin from Lactobacillus casei. Folia Microbiol. (Praha) 38, 441–446 10.1007/BF02814392 [DOI] [PubMed] [Google Scholar]
- Nilsen T., Nes I. F., Holo H. (2003). Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 69, 2975–2984 10.1128/AEM.69.5.2975-2984.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsl H., Braun V. (1995a). Evidence that the immunity protein inactivates colicin 5 immediately prior to the formation of the transmembrane channel. J. Bacteriol. 177, 6966–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsl H., Braun V. (1995b). Novel colicin 10: assignment of four domains to TonB-and TolC dependent uptake via the Tsx receptor and to pore formation. Mol. Microbiol. 16, 57–67 10.1111/j.1365-2958.1995.tb02391.x [DOI] [PubMed] [Google Scholar]
- Pilsl H., Braun V. (1995c). Strong function-related homology between the pore-forming colicins K and 5. J. Bacteriol. 177, 6973–6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsl H., Smajs D., Braun V. (1999). Characterization of colicin S4 and its receptor, OmpW, a minor protein of the Escherichia coli outer membrane. J. Bacteriol. 181, 3578–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons A. M., Delalande F., Duarte M., Benoit S., Lanneluc I., Sable S., et al. (2004). Genetic analysis and complete primary structure of microcin L. Antimicrob. Agents Chemother. 48, 505–513 10.1128/AAC.48.2.505-513.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons A. M., Zorn N., Vignon D., Delalande F., Van Dorsselaer A., Cottenceau G. (2002). Microcin E492 is an unmodified peptide related in structure to colicin V. Antimicrob. Agents Chemother. 46, 229–230 10.1128/AAC.46.1.229-230.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A. P. (1987). Nucleotide sequencing of the structural gene for colicin N reveals homology between the catalytic, C-terminal domains of colicins A and N. Mol. Microbiol. 1, 317–325 10.1111/j.1365-2958.1987.tb01938.x [DOI] [PubMed] [Google Scholar]
- Rebuffat S. (2012). Microcins in action: amazing defence strategies of Enterobacteria. Biochem. Soc. Trans. 40, 1456–1462 10.1042/BST20120183 [DOI] [PubMed] [Google Scholar]
- Riboulet-Bisson E., Sturme M. H., Jeffery I. B., O'Donnell M. M., Neville B. A., Forde B. M., et al. (2012). Effect of Lactobacillus salivarius bacteriocin Abp118 on the mouse and pig intestinal microbiota. PLoS ONE 7:e31113 10.1371/journal.pone.0031113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M. A., Wertz J. E. (2002). Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56, 117–137 10.1146/annurev.micro.56.012302.161024 [DOI] [PubMed] [Google Scholar]
- Rogers L. A. (1928). The inhibiting effect of Streptococcus lactis on Lactobacillus bulgaricus. J. Bacteriol. 16, 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos U., Harkness R. E., Braun V. (1989). Assembly of colicin genes from a few DNA fragments. Nucleotide sequence of colicin D. Mol. Microbiol. 3, 891–902 10.1111/j.1365-2958.1989.tb00238.x [DOI] [PubMed] [Google Scholar]
- Samyn B., Martinez-Bueno M., Devreese B., Maqueda M., Galvez A., Valdivia E., et al. (1994). The cyclic structure of the enterococcal peptide antibiotic AS-48. FEBS Lett. 352, 87–90 10.1016/0014-5793(94)00925-2 [DOI] [PubMed] [Google Scholar]
- Sandiford S., Upton M. (2012). Identification, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against Staphylococci. Antimicrob. Agents Chemother. 56, 1539–1547 10.1128/AAC.05397-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm E., Mende J., Braun V., Kamp R. M. (1987). Nucleotide sequence of the colicin B activity gene cba: consensus pentapeptide among TonB-dependent colicins and receptors. J. Bacteriol. 169, 3350–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settanni L., Corsetti A. (2008). Application of bacteriocins in vegetable food biopreservation. Int. J. Food Microbiol. 121, 123–138 10.1016/j.ijfoodmicro.2007.09.001 [DOI] [PubMed] [Google Scholar]
- Severinov K., Semenova E., Kazakov A., Kazakov T., Gelfand M. S. (2007). Low-molecular-weight post-translationally modified microcins. Mol. Microbiol. 65, 1380–1394 10.1111/j.1365-2958.2007.05874.x [DOI] [PubMed] [Google Scholar]
- Siegel R., Naishadham D., Jemal A. (2013). Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- Smajs D., Pilsl H., Braun V. (1997). Colicin U, a novel colicin produced by Shigella boydii. J. Bacteriol. 179, 4919–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl C. H., Callaway T. R., Lincoln L. M., Lonergan S. M., Genovese K. J. (2004). Inhibitory activities of colicins against Escherichia coli strains responsible for postweaning diarrhea and edema disease in swine. Antimicrob. Agents Chemother. 48, 3119–3121 10.1128/AAC.48.8.3119-3121.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern N. J., Svetoch E. A., Eruslanov B. V., Perelygin V. V., Mitsevich E. V., Mitsevich I. P., et al. (2006). Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob. Agents Chemother. 50, 3111–3116 10.1128/AAC.00259-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba M., Masaki H., Ohta T. (1988). Colicin E8, a DNase which indicates an evolutionary relationship between colicins E2 and E3. J. Bacteriol. 170, 3237–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov S. D., Franco B. D., Wiid I. J. (2014). In vitro study of beneficial properties and safety of lactic acid bacteria isolated from Portuguese fermented meat products. Benef. Microbes. 24, 1–16 10.3920/BM2013.0030 [DOI] [PubMed] [Google Scholar]
- Tulini F. L., Lohans C. T., Bordon K. C., Zheng J., Arantes E. C., Vederas J. C., et al. (2014). Purification and characterization of antimicrobial peptides from fish isolate Carnobacterium maltaromaticum C2: Carnobacteriocin X and carnolysins A1 and A2. Int. J. Food Microbiol. 173, 81–88 10.1016/j.ijfoodmicro.2013.12.019 [DOI] [PubMed] [Google Scholar]
- van den Elzen P. J., Walters H. H., Veltkamp E., Nijkamp H. J. J. (1983). Molecular structure and function of the bacteriocin gene and bacteriocin protein of plasmid Clo DF13. Nucleic Acids Res. 11, 2465–2477 10.1093/nar/11.8.2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel A. J., Montalban-Lopez M., Kuipers O. P. (2011). Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert Opin. Drug Metab. Toxicol. 7, 675–680 10.1517/17425255.2011.573478 [DOI] [PubMed] [Google Scholar]
- Varenne S., Cavard D., Lazdunski C. (1981). Biosynthesis and export of colicin A in Citrobacter freundii CA31. Eur. J. Biochem. 116, 615–620 10.1111/j.1432-1033.1981.tb05380.x [DOI] [PubMed] [Google Scholar]
- Vassiliadis G., Destoumieux-Garzon D., Lombard C., Rebuffat S., Peduzzi J. (2010). Isolation and characterization of two members of the siderophore-microcin family, microcins M and H47. Antimicrob. Agents Chemother. 54, 288–297 10.1128/AAC.00744-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. A., Kalkum M., Ottesen J., Yuzenkova J., Chait B. T., Landick R., et al. (2003). Structure of microcin J25, a peptide inhibitor of bacterial RNA polymerase, is a lassoed tail. J. Am. Chem. Soc. 125, 12475–12483 10.1021/ja036756q [DOI] [PubMed] [Google Scholar]
- Wirawan R. E., Klesse N. A., Jack R. W., Tagg J. R. (2006). Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl. Environ. Microbiol. 72, 1148–1156 10.1128/AEM.72.2.1148-1156.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Ebina Y., Miyata T., Nakazawa T., Nakazawa A. (1982). Nucleotide sequence of the structural gene for colicin E1 and predicted structure of the protein. Proc. Natl. Acad. Sci. U.S.A. 79, 2827–2831 10.1073/pnas.79.9.2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E., Fan L., Jiang Y., Doucette C., Fillmore S. (2012). Antimicrobial activity of bacteriocin-producing lactic acid bacteria isolated from cheeses and yogurts. AMB Exp. 2, 48 10.1186/2191-0855-2-48 [DOI] [PMC free article] [PubMed] [Google Scholar]