Abstract
NMR spectroscopy in combination with 13C-labeled substrate infusion is a unique technique to obtain information about dynamic metabolic fluxes noninvasively in vivo. In many cases, the in vivo information content obtained during dynamic 13C studies in rodents can be enhanced by high-resolution 1H-[13C] NMR spectroscopy on brain extracts. Previously, it has been shown that 1H NMR spectra from rat brain extracts can be accurately quantified with a spectral fitting routine utilizing simulated basis sets using complete prior knowledge of chemical shifts and scalar couplings. The introduction of 13C label into the various metabolites presents complications that demand modifications of the spectral fitting routine. As different multiplets within a given molecule accumulate various amounts of 13C label, the fixed amplitude relationship between multiplets typical for 1H NMR spectra must be abandoned. In addition, 13C isotope effects lead to spectral multiplet patterns that become dependent on the amount of 13C label accumulation, thereby preventing the use of a common basis set. Here a modified spectral fitting routine is presented that accommodates variable 13C label accumulation and 13C isotope effects. Spectral fitting results are quantitatively compared to manual integration on column-separated samples in which spectral overlap is minimized.
The 1H NMR spectroscopy technique is a powerful method in the assignment and quantification of chemical compounds. In the field of metabolomics, 1H NMR has been developed as one of the two primary detection techniques, providing complementary information to the various forms of mass-spectrometry-based detection. 1H NMR is routinely able to detect and quantify a wide range of metabolites in a number of biofluids, including plasma, cerebrospinal fluid and urine. The noninvasive nature of NMR has led to the development of in vivo 1H NMR spectroscopy that can currently detect up to 18 unique metabolites in cerebral tissues.1
Information about dynamic metabolic fluxes can be obtained when NMR spectroscopy is combined with the intravenous infusion of 13C-labeled substrates, such as [1-13C]-glucose. As the substrate is metabolized, the 13C label is transferred to metabolic products downstream. A classic example is the metabolism of [1-13C]-glucose, in which the 13C label transfers, via glycolysis and the tricarboxylic acid (TCA) cycle, to [4-13C]-glutamate and [4-13C]-glutamine. Figure S1 (Supporting Information) shows a schematic illustration of the 13C labeling patterns expected with glycolysis and the TCA cycle when using [1-13C]-glucose for a substrate. Detection of the 13C label in downstream metabolic products allows one, in combination with a mathematical model of metabolism, to determine the dynamic fluxes through the various metabolic pathways. Reviews on the principles and applications of dynamic metabolic studies with NMR spectroscopy are readily available.2 The 13C label turnover can be detected by direct 13C NMR spectroscopy,3 or by indirect 1H-[13C] NMR4 in which the 1H NMR signal from protons attached to a 13C nucleus is detected. Although 1H NMR has a much smaller chemical shift dispersion, thus leading to increased spectral overlap, it also has the advantage of a greatly enhanced sensitivity and the ability to detect the total metabolic pool (i.e., detect both the protons attached to 12C as well as to 13C).
1H-[13C] NMR is ideally performed directly in vivo, as this allows one to obtain the complete 13C turnover curves in a single experiment. In vivo 1H-[13C] NMR has been performed on rat brains4b,5 and human brains4a,6 and allows the detection of total energy metabolism3,4b,6a and glutamatergic3,4b and GABAergic4b neurotransmission. However, for a variety of reasons, direct in vivo 1H-[13C] NMR spectroscopy is not always feasible. These reasons include (1) the detection of low concentration metabolites, like GABA, (2) the detection from very small or poorly positioned volumes, like the hypothalamus, (3) the detection from regions with low magnetic field homogeneity, (4) the study of metabolism in nonanesthetized, awake rodents or (5) the unavailability of an in vivo NMR system. Under these conditions, brain extracts can be used in combination with high-resolution NMR to obtain the required information. In addition, brain extracts are often used to validate and complement the in vivo 1H-[13C] NMR results.
High-resolution 1H NMR spectra from brain extracts are typically analyzed by integration to obtain the relative peak areas. Although this approach is perfectly valid for simple 1H NMR spectra from pure compounds, 1H NMR spectra from brain extracts contain many partially overlapping resonances. Under these conditions, integration is, in general, no longer suitable and data is often only partially analyzed in areas of limited spectral overlap. Partial analysis is generally an inefficient use of available information and may also lead to inaccurate results when the baseline cannot be accurately estimated.
Recently,7 it has been demonstrated that high-resolution liquid-state 1H NMR spectra from rat brain extracts can be reliably quantified with spectral fitting, provided that appropriate modifications were made. These modifications included the allowance of separate line widths and frequency shifts for various multiplets within a given molecule and the inclusion of a global line shape distortion due to imperfect magnetic field homogeneity.
Here the spectral fitting algorithm is extended to allow the quantification of 1H-[13C] NMR spectra. There are two primary modifications necessary to achieve this goal. First, the 13C isotope effect, leading to a frequency shift of a 13C-labeled compound relative to the unlabeled 12C compound, is readily observable in high-resolution 1H-[13C] NMR spectra and must be accounted for. Second, the amount of 13C label accumulation is, in general, not identical for different carbon positions within a given compound. As a result, the amplitudes in a 1H-[13C] difference spectrum can therefore also not be made equal. Here the modified spectral fitting algorithm is described and applied to quantify 1H-[13C] NMR spectra from rat brain at 11.7 T. Quantification results are validated by comparing 13C fractional enrichments obtained by spectral fitting with those obtained by spectral integration from column-separated samples in which spectral overlap is minimal.
Methods
NMR Spectroscopy
All experiments were performed on a Bruker Avance spectrometer (Bruker Instruments, Billerica, MA) operating at 500.13 MHz for 1H and equipped with a 5 mm triple resonance probe incorporating triple-axis gradient coils. The magnetic field homogeneity on each sample was optimized with an automated 3D field mapping algorithm capable of adjusting up to fifth order spherical harmonics.
1H-[13C] NMR spectra were acquired with a spin-echo method (TR/TE = 15 000/8 ms) executed with adiabatic excitation and refocusing BIR-4 pulses (tanh/tan modulation,8 pulse length, T = 1.0 ms, maximum frequency sweep, Δνmax = 100 kHz, maximum RF amplitude, B1max = 10 kHz). On alternate scans, an adiabatic full passage inversion pulse (HS8 modulation,9T = 1.0 ms, Δνmax = 20 kHz, B2max = 10 kHz) was executed on the carbon-13 channel (125.76 MHz) at the same time as the proton refocusing pulse. Broadband adiabatic 13C decoupling was applied during the total acquisition time. The decoupling sequence was executed with 2.0 ms AFP pulses [HS8 modulation,9 Δνmax = 10 kHz, B2max = 2.5 kHz, center frequency = 34.2 ppm) incorporated in a 20-step supercycle.10 Water suppression was achieved with a six-pulse SWAMP (solvent suppression with adiabatic-modulated pulses11) sequence executed with 20 ms adiabatic full passage pulses (Lorentzian modulation,9 Δνmax = 1.0 kHz, offset = +1.0 kHz, B1max = 2.0 kHz).
Single-transient free induction decays (FIDs) were acquired as 8192 complex points over 1.36 s and the 256 repetitions separately stored. 1H NMR spectra were obtained through zero-filling to 65 536 points, fast Fourier transformation (no apodization) and zero-order phase correction. Following frequency alignment of all spectra, the even and odd-numbered scans were summed to give NMR spectra obtained in the absence and presence of a 13C inversion pulse, respectively. The difference between these two data sets provides the 1H-[13C] NMR spectrum displaying the 13C-bound proton signals, whereas the data set without the 13C inversion pulse shows the total proton signal (i.e., protons bound to 12C or 13C).
Substrate Infusion, Brain Extraction and Column Separation of Amino Acids
All procedures on animals were performed under approved protocols by the Yale Animal Care and Use Committee in accordance with American Veterinary Medical Association (AVMA) guidelines on euthanasia.
Five nonfasted, male Sprague-Dawley rats (192 ± 8 g, mean ± SD) received an intravenous infusion of [1-13C]-glucose through the tail vein. Briefly, animals received an initial 250 μL intravenous bolus of 0.75 M [1-13C]-glucose (per 200 g body weight) followed by an intravenous infusion of 0.75 M [1-13C]-glucose. The infusion rate was decreased every 30 s according to a decreasing exponential function during the first 8 min and was constant at 13.7 μL/min for the remainder of the experiment. Animals were awake during the infusion and were allowed free movement in their cages. Animals were infused for 15 (n = 1), 30 (n = 2) or 60 (n = 2) minutes.
Animals were euthanized by focused-beam microwave irradiation (4.5 kW for 0.9 s, Muromachi Microwave Fixation System, Stoelting Co, Wood Dale, IL, USA), halting enzyme activity and cerebral metabolism rapidly. Following microwave irradiation, the medial prefrontal cortex (MPFC) was manually dissected (60–90 mg) and homogenized with 0.1 M HCl/methanol (2:1 vol/wt) followed by extraction with ethanol. The supernatant was clarified by centrifugation after which heavy metals were removed on Chelex-100 columns. Samples were lyophilized for long-term storage at −80 °C or resuspended in 600 μL of phosphate-buffered (50 mM, pH 7.0) D2O/H2O (85/15%) solution containing 0.5 mM 2,2-dimethyl-2-silapentane-5-sulfonate-D6 (DSS-D6).
After the samples were measured by 1H-[13C] NMR spectroscopy, the samples were passed through an AG 1-X8 anion exchange column (200 to 400 mesh, 1.4 mL bed volume; Bio-Rad Laboratories, Hercules, CA, USA) in order to separate the amino acids. Glutamine and GABA were eluted with deionized water and glutamate was eluted with 2 M acetic acid. The pH of both fractions was reduced to ca. 3 before lyophilization to enhance the volatility of acetic acid. Both fractions were lyophilized, and resuspended in a phosphate-buffered (pH 7.0) D2O/H2O (85/15%) solution containing 0.5 mM DSS-D6. The new samples underwent the same NMR protocol as described earlier, except that because of the lack of spectral overlap, spectral processing could now be limited to baseline correction and manual integration.
Spectral Fitting
The general principle behind the spectral fitting routine is that a measured 1H NMR spectrum can, in principle, be approximated as a linear sum of the individual constituent metabolite 1H NMR spectra. This principle also forms the basis for the LCModel spectral fitting program.12 The excellent spectral resolution available in 1H NMR spectra of brain extracts demands accurate knowledge of the chemical shifts and scalar couplings of metabolites. To maintain full flexibility in the adjustment of prior knowledge the fitting basis set was simulated using home-written Matlab (Matlab 8.0, The Mathworks, Natick, MA, USA) software employing the density matrix formalism.
The spectral basis set used in the current study was composed of 29 metabolites and included acetate, acetone, alanine, ascorbate, aspartate, choline, creatine, γ-amino butyric acid (GABA), glucose, glutamate, glutamine, glycine, glycerophosphocholine (GPC), glutathione (oxidized form, GSSG), lactate, myo-inositol, N-acetyl aspartate (NAA), N-acetylaspartylglutamate (NAAG), phosphocholine (PC), phosphocreatine (PCr), phosphoethanolamine (PE), pyroglutamate, serine, scyllo-inositol, succinate, taurine and threonine. In addition to the cerebral metabolites, the extraction procedure contaminants ethanol and methanol were also included. For acetate, alanine, aspartate, GABA, glucose, glutamate, glutamine, lactate and pyroglutamate, a second, separate 13C labeled basis set was created. The number of metabolites and multiplets used during the actual spectral fitting could be significantly lower depending on the spectral window used for fitting.
The general model function used to approximate the experimental 1H-[13C] NMR difference data is given by
| 1 |
with
| 2 |
and
| 3 |
where S13C(ν) is the 1H-[13C] NMR difference spectrum showing only the 13C-bound proton signals. fmn(t) represents the simulated time domain signal of multiplet n from metabolite m. M and N are the total number of metabolites and the total number of independent multiplets per metabolite, respectively. For example, glutamate has five spins in an AMNPQ spin system. The A spin is weakly coupled to the M and N spins, and its spectral pattern can be shifted independently. The M and N (ΔνMN/JMN = 2.48) and P and Q (ΔνPQ/JPQ = 0.57) spins are strongly coupled, and the corresponding spectral patterns can, therefore, not be shifted relative to each other. The spectral patterns of the H3/H3′ and H4/H4′ protons are, thus, combined, making the number of independent multiplets, N, equal to 3 for glutamate. Δνmn and T2,mn represent the frequency shift and T2 relaxation time constant, which is inversely proportional to the line broadening for each resonance, respectively. For all resonances, these parameters were restricted to Δνmn ∈ [−1.0, 1.0] Hz and 1/(πT2,mn) ∈ [0.6, 1.2] Hz. Whereas eq 1 depicts Lorentzian line shapes, the line broadening is readily modified to Gaussian by changing the exponent from −t/T2,mn to −(t/T2,mn).2 The final term in eq 1 represents the spectral baseline modeled as a finite polynomial expansion with amplitudes Bp. For all data sets, a low-order polynomial (P = 2) was sufficient. Although the spectral baseline can be accommodated by an infinite number of functions, the low-order polynomial function in eq 1 provided adequate results with only three independent parameters (for P = 2). D(t) represents the spectral line shape distortion common among all resonances included in the spectral fit. Line shape distortions are modeled as a sum of frequency-shifted, amplitude-scaled complex exponentials and can account for deviations from the ideal Lorentzian or Gaussian line shape due to imperfect magnetic field homogeneity (i.e., “shimming”). Line shape variations due to differences in T2 relaxation are accommodated by the T2,mn terms in eq 1. However, multiexponential T2 relaxation and differences therein among the various resonances are currently not accommodated by the spectral model function. For all data sets, an optimal balance between line shape approximation and number of additional parameters (=3K) was obtained for K = 5. The amplitudes Ak were initialized as 1 whereas the frequencies Δνk and phases ϕk were initialized as 0. The amplitudes Ak were constrained to be positive, the phases ϕk were not constrained and the frequencies, Δνk, were constrained to the range [−1.0 to +1.0] Hz. The term DSB(t), as described in eq 3, represents signal modulation due to decoupling sidebands. Decoupling sidebands appear at symmetrical frequency positions surrounding a main decoupled signal and arise from small modulations in scalar coupling evolution with the application of the multiple inversion pulses during broadband decoupling. For adiabatic decoupling the decoupling sidebands are typically limited to a few signals at frequencies that are a multiple of the reciprocal of the inversion pulse length. The decoupling sidebands frequency Δνl is thus typically known, whereas the amplitude Al is typically limited to 1–2% of the main decoupled signal. The phase ϕl of the decoupling sidebands is normally not known and must be fitted unconstrained. In the current study, only the signals that were far off-resonance on the 13C channel ([2-13C]-glutamate and [2-13C]-glutamine) required decoupling sideband modulation with L = 1.
The spectral model described by eq 1 is different from that used for regular 1H NMR spectra in that the amplitudes A13C,mn for individual multiplets n do not have to be the same, because different carbon positions can incorporate 13C label at different rates. Equation 1 accommodates both natural abundance 13C-bound proton signals, as well as 13C-bound proton signals formed by net metabolic turnover of 13C label. For natural abundance (NA) signals, A13C,mn are equal for all n, whereas for 13C-bound proton signals formed by metabolic turnover, A13C,mn can vary for different n.
Besides knowledge of the 13C-bound proton signals, it is also important to obtain a quantitative estimate of the total (12C + 13C) or the 12C-bound proton signals of metabolites. For in vivo 1H-[13C] NMR studies, the total proton signals of metabolites are typically obtained by fitting the 1H NMR spectrum without the 13C inversion pulse. For high-resolution, in vitro 1H NMR, this is not an option due to the 13C isotope effect, which causes a frequency shift for 13C-labeled compounds relative to the native 12C compound that is larger than the typical spectral line width. As a result, the line shape of a given multiplet (e.g., alanine-H3) becomes a function of the 13C isotope shift and the fractional enrichment, which is different for different compounds and multiplets. To circumvent complications arising from the 13C isotope effect, the 13C fractional enrichments are obtained by quantification of the 1H-[12C] NMR spectrum rather than the total 1H-[12C + 13C] NMR spectrum. The NMR spectrum containing the 12C-bound proton signals, S12C(ν) can be calculated as
| 4 |
where Stotal(ν) is the NMR spectrum containing all (12C and 13C-bound) proton signals obtained in the absence of a carbon-13 inversion pulse and S13C(ν) is given by eq 1, i.e., the spectral fit to the 1H-[13C] edited NMR spectrum. Note that shifted signals due to the 13C isotope effect are completely removed during the subtraction shown in eq 4. However, S12C(ν) is different from a regular 1H NMR spectrum in that the amplitudes A12C,mn do not have to be equal because different amounts of 13C-bound proton signal were subtracted. The spectrum S12C(ν) can thus be modeled as
| 5 |
where Am is the amplitude of metabolite m, which adheres to the relation Am = A12C,mn + A13C,mn for every multiplet n. Note that, with the exception of A13C,mn, none of the variables and functions in eq 5 have to be the same as in eq 1. For example, the small frequency shifts Δνmn are different for S12C(ν) and S13C(ν) due to the 13C isotope effect. T2,mn can be slightly different as imperfect decoupling can lead to slight line broadening for S13C(ν), which does not play a role for S12C(ν). The simulated time domain signals f12C,mn(t) and f13C,mn(t) are typically different as the presence of a 13C nucleus can significantly change the scalar coupling modulation during the echo time TE. These time domain signals therefore have to be simulated separately with complete prior knowledge on the carbon-13 chemical shifts and 1H–13C scalar coupling constants. All density matrix calculations were performed in Matlab (Matlab 8.0, The Mathworks, Natick, MA, USA) using the exact pulse sequence details, but ignoring relaxation. Unlike a previous implementation,7 the current density matrix calculations were performed with Spinach,13 a novel software package that can handle up to several hundred spins.
Results
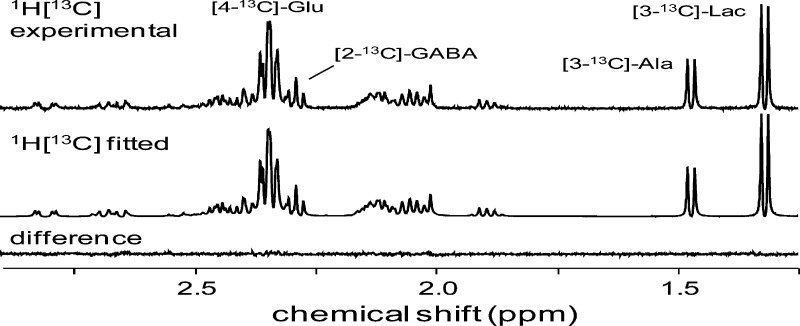
Figure 1A shows a typical result obtained for the 1H-[13C] NMR spectrum containing the 13C-bound proton signals. Following 30 min of [1-13C]-glucose infusion, significant 13C label accumulation is evident in [4-13C]-glutamate and [2-13C]-GABA, as well as a number of other compounds and multiplets. The 1H-[13C] NMR spectrum is readily fitted, as shown in Figure 1B. The quality of the spectral fit can be judged from the difference/residual between the experimental and fitted NMR spectra (Figure 1C). All 1H-[13C] NMR spectra were processed with the same basis set, f13C,mn(t), consisting for the spectral fitting window [1.25–2.9 ppm] of M = 14 metabolites and N = 21 multiplets, of which 6 metabolites and 9 multiplets were considered at natural abundance (acetone, GSSG, NAA, NAAG, succinate, threonine), whereas the rest were assumed to have significant 13C label accumulation (acetate, alanine, aspartate, GABA, glutamate, glutamine, lactate, pyroglutamate). For longer [1-13C]-glucose infusions, NAA may exhibit some 13C label accumulation and should then switch groups so that the multiplet amplitudes in the 1H-[13C] NMR spectrum are no longer equal.
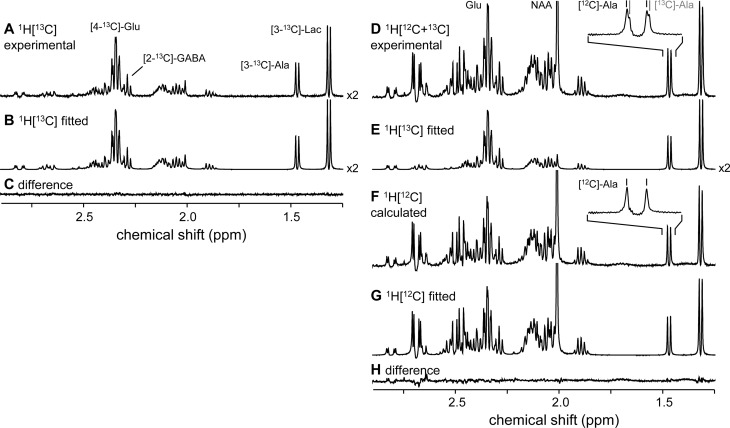
Figure 1.
Experimental (A) and fitted (B) 1H-[13C] NMR spectra from rat brain extract. (C) Difference (A–B) spectrum. (D) Experimental, total 1H-[12C+13C] NMR spectrum minus the fitted 1H-[13C] NMR spectrum (B/E) gives a calculated 1H-[12C] NMR spectrum (F), which can be fitted without a 13C isotope shift (G). (H) Difference (F–G) spectrum. The 13C isotope shift is visualized for alanine in panel D, but can be detected for all metabolites. The inset in panel F shows that the doublet signal from [3-13C]-alanine is effectively eliminated during the subtraction (D–E), leaving only the doublet signal from [3-12C]-alanine.
Figure 1D shows the total 1H-[12C+13C] NMR spectrum obtained without a 13C inversion pulse (but with broadband 13C decoupling). The 13C isotope effect can be observed on many signals, for example on the alanine-H3 doublet at 1.47 ppm. The 13C-labeled alanine-H3 signal appears slightly upfield as a shoulder on the unlabeled alanine-H3 signal. Even though the 13C isotope effect is small (−1.98 ± 0.11 ppb for [3-13C]-alanine), it is readily observable due to the narrow line widths obtained in vitro. The result of the 13C isotope effect is that the combined [12C + 13C] alanine-H3 signal can no longer be modeled with Lorentzian lines. The line shape distortion factor, D(t), could, in principle, account for the observed distortion, but only for a single multiplet. As multiplets in various compounds have different 13C isotope shifts and 13C fractional enrichments, the line shape distortion varies across the spectrum and can therefore not be accommodated by a global line shape distortion factor. Table 1 summarizes the 13C isotope shifts observed for the commonly observed 13C-labeled compounds. It should be noted that although 1H NMR is sensitive to 13C isotope shifts, it does not allow detailed isotopomer analysis, as done with direct 13C NMR, because the 13C isotope shifts for different isotopomers are indistinguishable.
Table 1. 1H NMR Detected 13C Isotope Shifts (in ppb).
| metabolite | multiplet | isotope shift | errora |
|---|---|---|---|
| alanine | H2 | –2.16 | 0.09 |
| H3 | –1.98 | 0.11 | |
| aspartate | H2 | –2.13 | 0.12 |
| H3 | –1.75 | 0.10 | |
| GABA | H2 | –1.67 | 0.15 |
| H3 | –1.90 | 0.10 | |
| H4 | –2.44 | 0.09 | |
| glucose | αH1 | –2.55 | 0.06 |
| glutamate | H2 | –2.11 | 0.10 |
| H3 | –1.86 | 0.11 | |
| H4 | –1.71 | 0.12 | |
| glutamine | H2 | –2.11 | 0.10 |
| H3 | –1.86 | 0.11 | |
| H4 | –1.68 | 0.07 | |
| glycine | H2 | –2.11 | 0.10 |
| lactate | H2 | –2.25 | 0.07 |
| H3 | –2.00 | 0.07 |
Error represents the standard deviation (in ppb) over five 1H NMR spectra from rat brain extracts following 1 h of [1-13C]-glucose infusion.
The 13C isotope effect is effectively eliminated by calculating the 1H-[12C] NMR spectrum (Figure 1F) as the difference between the total 1H-[12C+13C] NMR spectrum (Figure 1D) and the fitted 1H-[13C] NMR spectrum (Figure 1B/E). As all signals from 13C-labeled compounds have been subtracted, the 1H-[12C] NMR spectrum only contains unshifted 1H NMR resonances, which can be modeled like a regular 1H NMR spectrum with the exception that the amplitudes of the multiplets must be corrected according to eq 4 for the amount of 13C signal subtracted. Figure 1G shows the spectral fit of the 1H-[12C] NMR spectrum (Figure 1F) according to eq 4. Figure 1H shows the difference between the calculated and fitted 1H-[12C] NMR spectra.
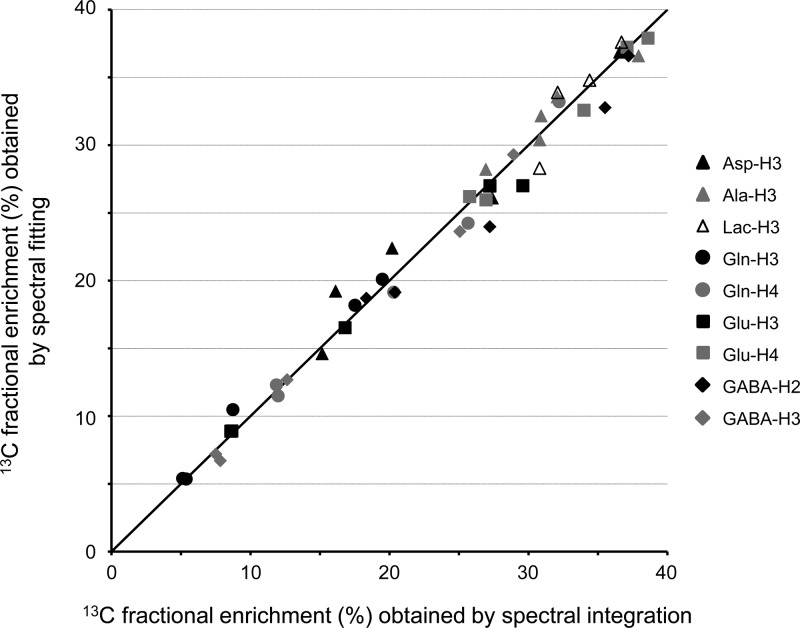
Figure 2 shows the relationship between the 13C fractional enrichments as obtained by spectral fitting and manual integration of 1H NMR spectra of column-separated samples. The correlation is near unity with the best linear fit given by FEfitting = 0.975FEintegration + 0.437 (R = 0.992) with FE representing the 13C fractional enrichment (in %) obtained by spectral fitting or integration.
Figure 2.
Correlation between the 13C fractional enrichments obtained by spectral integration on column-separated samples and spectral fitting on nonseparated samples. Nine multiplets from five samples are displayed. The samples were obtained from rat brain extracts following 15 (n = 1), 30 (n = 2) and 60 (n = 2) minutes of [1-13C]-glucose infusion. The solid line represents the identity line. The best linear fit is characterized by a slope of 0.975 and an intercept of 0.437% (R = 0.992). The exact 13C fractional enrichments can be found in Table S1 (Supporting Information).
Discussion
Here it has been demonstrated that high-resolution 1H-[13C] NMR spectra of rat brain extracts can be reliably quantified through the use of a spectral fitting algorithm. The flexibility of a recently introduced spectral fitting algorithm7 for high-resolution 1H NMR spectra provided a good starting point for the quantification of 1H-[13C] NMR spectra, requiring essentially two modifications specific for 1H-[13C] NMR. First, complications introduced by 13C isotope shifts prevent the direct quantification of the total 1H-[12C + 13C] NMR spectrum. This complication was circumvented by first quantifying the 1H-[13C] NMR spectrum, which could then be used to calculate a 1H-[12C] NMR spectrum. As the 13C isotope-shifted multiplets are cleanly subtracted, the complications associated with the 13C isotope effect are effectively eliminated. Second, in a 1H NMR spectrum the multiplet signals from a given molecule are linked in amplitude. As different chemical groups within a given molecule can accumulate different amounts of 13C label during a 13C-labeled substrate infusion, the multiplet signals in a 1H-[13C] NMR spectrum are no longer linked in amplitude. The presented spectral fitting algorithm incorporated both of these modifications to allow the robust quantification of 1H-[13C] NMR spectra.
The most important complication to the quantification of 1H or 1H-[13C] NMR spectra is the use of incorrect prior knowledge on the chemical shifts and scalar couplings. Even though all lyophilized brain extracts were dissolved in the same phosphate buffer and measured at 298 K, it was occasionally observed that small variations in spectral patterns could not be accommodated by the spectral fitting routine. Small variations in chemical shifts, even between multiplets within the same molecule, are accommodated by allowing each multiplet to frequency shift over a limited [−1 to +1] Hz range. However, variations in scalar coupling constants cannot be accommodated in the current version of the algorithm and would lead to an inability to properly model an experimental multiplet pattern. Variations in chemical shifts and scalar couplings may be caused by a number of effects. The use of a phosphate buffer minimizes pH effects but does not completely eliminate them as the buffer capacity is limited. Increasing the buffer capacity by increasing the phosphate concentration is possible but would also lead to increased coil loading and a lower NMR sensitivity. Even though heavy metals ions were removed with a Chelex-100 column, it is still possible that certain ions like Ca2+, Mg2+ and Zn2+ remain present. Binding of the ions to negatively charged functional groups could lead to changes in chemical shift and scalar couplings. Studies are currently in progress to quantify chemical shifts and scalar couplings as a function of pH, temperature, ionic strength and the presence of specific cations in order to better understand the factors affecting the observed multiplet patterns. In future studies a pH sensitive compound, e.g., imidazole, could be added during the extraction procedure to accurately determine the pH of each sample. When the pH-sensitivity of all multiplets is known, the correct prior knowledge could be used on every sample.
The use of spectral fitting lends itself well for a high degree of automation. In the current implementation of the software, all steps from loading of the raw FID data to storing of the final spectral fitting results can be performed in a fully automated version. It is therefore possible to process many data sets in an overnight run. The main consideration is the time it takes to complete the spectral fit of one spectrum. When a large spectral range (0.5–5.5 ppm) is included, a spectral fit can take up to 30 min to complete (2.6 GHz, 32-bit processor with 4 GB RAM). It is therefore important to include only the spectral range of interest, which can also include multiple ranges separated from each other. In addition, when the entire spectral range is included, the large number of fitting parameters (>200 in some cases) greatly increases the chance of the spectral fit ending in a local minimum, rather than a global minimum. To decrease the chance of this possibility, the software has been extended with a prescan option that fits the desired spectral range as multiple smaller, independent spectral ranges (e.g., the 0.5–5.5 ppm range is fitted as ten 0.5 ppm wide ranges). The spectral fit over the small ranges is typically very fast (<1 min/range). The spectral fitting results of the small spectral ranges are then combined into one overview file that is then used as a starting point for the spectral fit of the entire spectral range. Even though some time (<10 min) is lost to perform the prescan, the improved starting values ensure (1) that the spectral fit converges much faster (thereby saving time) and (2) greatly increases the likelihood that the final spectral fitting result is close to the global minimum.
Despite the successful implementation of spectral fitting for both 1H and 1H-[13C] NMR spectra, the process is undeniably complex, requiring complete prior knowledge of all metabolites, a means to obtain the simulated basis sets and a critical assessment of all parameters potentially affecting the fitting quality (e.g., B0 magnetic field inhomogeneity, incorrect prior knowledge, temporal magnetic field drifts). As such, the simplicity of integration together with its robustness under the correct conditions should not be dismissed too quickly. When dealing with (parts of) spectra with minimal spectral crowding and overlap, manual integration is a valid, fast and perhaps even desirable method to quantify 1H and 1H-[13C] NMR spectra. Under these conditions, integration can even be fully automated when used in combination with robust chemical shift referencing and baseline correction. However, the utility of integration quickly diminishes when significant spectral overlap is present, either in the 1H or 1H-[13C] NMR spectra, or both. Examples can be found in [3-13C]-glutamate and [3-13C]-glutamine overlap in 1H-[13C] NMR spectra and glutamate, glutamine, GSSG and NAA overlap in 1H NMR spectra.7 In these cases, the use of spectral fitting is recommended and can provide robust results as demonstrated here for 1H-[13C] NMR.
Although the spectral fitting algorithm was extended to accommodate decoupling sideband modulations, the option was required only for two multiplets, [2-13C]-glutamate and [2-13C]-glutamine, with 13C resonances relatively far from the 34.2 ppm on-resonance condition. The decoupling sideband modulation option was successful in adequately modeling the observed sidebands (data not shown) at the expense of introducing four additional fitting parameters (two amplitudes and two phases). A better option is to eliminate the decoupling sidebands experimentally by varying the phase of the decoupling sidebands in subsequent acquisitions.14 This is now the default in our laboratory, thereby making the decoupling sideband option in eq 1 effectively obsolete.
It is generally assumed that the inversion efficiency of the 13C editing pulse is perfect. The use of an adiabatic inversion pulse is recommended to guarantee this assumption. On [2-13C]-acetate, it was experimentally verified that the inversion efficiency is >99% over the entire relevant spectral range, thereby validating this assumption. However, when working with nonadiabatic RF pulses and with very large spectral ranges, this assumption should always be verified experimentally. Although the inversion efficiency is perfect, the resonances from 13C-labeled compounds do not necessarily invert perfectly. An exact 180° phase accrual is only achieved when the echo-time TE is equal to 1/1JCH. Since 1JCH varies among different multiplets and compounds, the inversion efficiency therefore also varies. This is not necessarily a problem for spectral fitting as prior knowledge of 1JCH is automatically used during the creation of the spectral basis sets. However, for manual integration this effect will lead to a direct underestimation of the 13C fractional enrichments unless correction factors are established using phantoms or by density matrix simulations.
1H-[13C] NMR is not the only option for analyzing the 13C label content of brain extracts. Direct 13C NMR enhanced by polarization transfer, nuclear Overhauser enhancement or cryoprobes can be a valuable alternative, especially when 13C–13C isotopomers15 or carbon positions without a directly attached proton are the focus of study. In addition, the large chemical shift dispersion minimizes spectral overlap, even for similar compounds like glutamate and glutamine. However, despite the aforementioned sensitivity enhancement methods, the sensitivity of 13C observation is generally lower than that of 1H detection. This can become a critical consideration for mass-limited samples (e.g., mouse brain extracts) or samples with low 13C label accumulation (e.g., following [2-13C]-glucose infusion). In addition, whereas the 13C fractional enrichment is readily obtained from 1H-[13C] NMR data, direct 13C observation is limited to the detection of the 13C fraction. Nevertheless, the choice of detection method is ultimately dictated by the application and it has been demonstrated here that 1H-[13C] NMR is a robust method that allows automated spectral quantification provided that all spectral details are taken into account.
The spectral fitting algorithm demonstrated here for high-resolution 1H-[13C] NMR spectra is equally applicable to analysis of in vivo 1H-[13C] NMR data. The workflow for in vivo 1H-[13C] NMR spectral analysis is similar to that outlined here, with possible modifications to the line width and frequency shift ranges. To obtain the 13C fractional enrichment, the 1H-[12C] NMR spectrum could be calculated and quantified as shown here. However, as the 13C isotope effect represents a much smaller fraction of the in vivo spectral line widths, the total 1H-[12C+13C] NMR spectrum could also be quantified directly without noticeable effects.
The spectral fitting routines may be downloaded from http://mrrc.yale.edu or can be obtained by contacting the corresponding author.
Acknowledgments
The authors thank Xiaoxian Ma for his assistance with animal preparation. This research was funded, in part, by NIH grants NIMH R01-MH095104 and NINDS P30-NS052519.
Supporting Information Available
Figures of the metabolic model of glycolysis and the tricarboxylic acid (TCA) cycle and 13C fractional enrichments obtained with integration and spectral fitting. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- a de Graaf R. A.In Vivo NMR Spectroscopy. Principles and Techniques, 2 ed.; John Wiley: Chichester, U.K, 2007. [Google Scholar]; b Pfeuffer J.; Tkac I.; Provencher S. W.; Gruetter R. J. Magn. Reson. 1999, 141, 104–120. [DOI] [PubMed] [Google Scholar]; c Mlynarik V.; Cudalbu C.; Xin L.; Gruetter R. J. Magn. Reson. 2008, 194, 163–168. [DOI] [PubMed] [Google Scholar]
- a Rothman D. L.; De Feyter H. M.; de Graaf R. A.; Mason G. F.; Behar K. L. NMR Biomed. 2011, 24, 943–957. [DOI] [PMC free article] [PubMed] [Google Scholar]; b de Graaf R. A.; Rothman D. L.; Behar K. L. NMR Biomed. 2011, 24, 958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Shen J.; Petersen K. F.; Behar K. L.; Brown P.; Nixon T. W.; Mason G. F.; Petroff O. A.; Shulman G. I.; Shulman R. G.; Rothman D. L. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 8235–8240. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Gruetter R.; Seaquist E. R.; Ugurbil K. Am. J. Physiol. Endocrinol. Metab. 2001, 281, E100–E112. [DOI] [PubMed] [Google Scholar]
- a Rothman D. L.; Novotny E. J.; Shulman G. I.; Howseman A. M.; Petroff O. A.; Mason G.; Nixon T.; Hanstock C. C.; Prichard J. W.; Shulman R. G. Proc. Natl. Acad. Sci. U. S. A. 1992, 89, 9603–9606. [DOI] [PMC free article] [PubMed] [Google Scholar]; b van Eijsden P.; Behar K. L.; Mason G. F.; Braun K. P.; de Graaf R. A. J. Neurochem. 2010, 112, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Pfeuffer J.; Tkac I.; Choi I. Y.; Merkle H.; Ugurbil K.; Garwood M.; Gruetter R. Magn. Reson. Med. 1999, 41, 1077–1083. [DOI] [PubMed] [Google Scholar]; b de Graaf R. A.; Mason G. F.; Patel A. B.; Rothman D. L.; Behar K. L. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 12700–12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Chen W.; Zhu X. H.; Gruetter R.; Seaquist E. R.; Adriany G.; Ugurbil K. Magn. Reson. Med. 2001, 45, 349–355. [DOI] [PubMed] [Google Scholar]; b Pan J. W.; Stein D. T.; Telang F.; Lee J. H.; Shen J.; Brown P.; Cline G.; Mason G. F.; Shulman G. I.; Rothman D. L.; Hetherington H. P. Magn. Reson. Med. 2000, 445673–9. [DOI] [PubMed] [Google Scholar]
- de Graaf R. A.; Chowdhury G. M.; Behar K. L. Anal. Chem. 2011, 83, 216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood M.; Ke Y. J. Magn. Reson. 1991, 94, 511–525. [Google Scholar]
- Tannus A.; Garwood M. J. Magn. Reson., Ser. A 1996, 120, 133–137. [Google Scholar]
- Fujiwara T.; Nagayama K. J. Magn. Reson. 1988, 77, 53–63. [Google Scholar]
- de Graaf R. A.; Nicolay K. Magn. Reson. Med. 1998, 40, 690–696. [DOI] [PubMed] [Google Scholar]
- Provencher S. W. Magn. Reson. Med. 1993, 30, 672–679. [DOI] [PubMed] [Google Scholar]
- Hogben H. J.; Krzystyniak M.; Charnock G. T.; Hore P. J.; Kuprov I. J. Magn. Reson. 2011, 208, 179–194. [DOI] [PubMed] [Google Scholar]
- a Kupce E.; Freeman R.; Wider G.; Wuthrich K. J. Magn. Reson., Ser. A 1996, 122, 81–84. [Google Scholar]; b Zhang S.; Gorenstein D. G. J. Magn. Reson. 2000, 144, 316–321. [DOI] [PubMed] [Google Scholar]
- a Malloy C. R.; Sherry A. D.; Jeffrey F. M. H. Am. J. Physiol. 1990, 259, H987–H995. [DOI] [PubMed] [Google Scholar]; b Henry P. G.; Oz G.; Provencher S.; Gruetter R. NMR Biomed. 2003, 16, 400–412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.