Abstract
As part of our ongoing small-molecule metabotropic glutamate (mGlu) receptor positive allosteric modulator (PAM) research, we performed structure–activity relationship (SAR) studies around a series of group II mGlu PAMs. Initial analogues exhibited weak activity as mGlu2 receptor PAMs and no activity at mGlu3. Compound optimization led to the identification of potent mGlu2/3 selective PAMs with no in vitro activity at mGlu1,4–8 or 45 other CNS receptors. In vitro pharmacological characterization of representative compound 44 indicated agonist-PAM activity toward mGlu2 and PAM activity at mGlu3. The most potent mGlu2/3 PAMs were characterized in assays predictive of ADME/T and pharmacokinetic (PK) properties, allowing the discovery of systemically active mGlu2/3 PAMs. On the basis of its overall profile, compound 74 was selected for behavioral studies and was shown to dose-dependently decrease cocaine self-administration in rats after intraperitoneal administration. These mGlu2/3 receptor PAMs have significant potential as small molecule tools for investigating group II mGlu pharmacology.
Introduction
Glutamate is the major excitatory neurotransmitter in the mammalian central nervous system (CNS), mediating fast synaptic transmission through ion channels, primarily the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and kainate ionotropic glutamate receptor subtypes.1 The metabotropic glutamate (mGlu) receptors are a family of eight G protein-coupled receptors that are activated by glutamate and perform a modulatory function in the nervous system.2−4 The group II mGlu receptors include the mGlu2 and mGlu3 receptor subtypes, which couple with Gi/o proteins to negatively regulate the activity of adenylyl cyclase.3,5 Localization studies suggest that mGlu2 receptors act predominantly as presynaptic autoreceptors to modulate the release of glutamate into the synaptic cleft.6 On the other hand, mGlu3 receptors exhibit a broad distribution in the brain and have been shown to be present on astrocytes.7 In addition, it has been shown that activation of mGlu3 receptors is required for the neuroprotective effects of mGlu2/3 agonists toward N-methyl-d-aspartate (NMDA) neurotoxicity in mixed cultures of astrocytes and neurons, whereas activation of mGlu2 receptors may be harmful.8
Various brain regions, including the cerebral cortex, hippocampus, striatum, amygdala, frontal cortex, and nucleus accumbens, display high levels of mGlu2 and mGlu3 receptor binding.9,10 This distribution pattern suggests a role for the mGlu2/3 receptor subtypes in the pathology of neuropsychiatric disorders such as anxiety,11 depression,12,13 schizophrenia,14,15 drug dependence,16−22 neuroprotection,23,24 Alzheimer’s disease,25 and sleep/wake architecture.26 Thus, there is significant potential for the development of selective group II mGlu receptor activators, including agonists and positive allosteric modulators (PAMs), for the treatment of CNS diseases caused by aberrant glutamatergic signaling.
Orthosteric (glutamate site) mGlu2/3 agonists such as LY37926827 are constrained amino acid analogues that are typically equipotent at both mGlu2 and mGlu3, presumably because of the high degree of sequence homology at the glutamate-binding site for these two receptors.3 Although LY541850, an orthosteric ligand with mixed mGlu2 agonist/mGlu3 antagonist activity, has been reported,28,29 currently there are no truly mGlu2 selective orthosteric ligands available. The systemically active mGlu2/3 receptor agonist LY379268 has been shown to reduce glutamate release from presynaptic terminals in many brain regions and thus to decrease glutamatergic neurotransmission.30,31 LY379268 is active in several different rodent models of CNS disorders including anxiety,32 schizophrenia,33 Huntington’s disease,34−36 and drug dependence, where it attenuates cocaine self-administration both in rats20,37 and in squirrel monkeys.18 However, LY379268 also inhibits responding for food and food-seeking behavior,20,21,37,38 suggesting that mGlu2/3 receptor agonists exhibit nonselective actions on responding for drug and nondrug reinforcers.
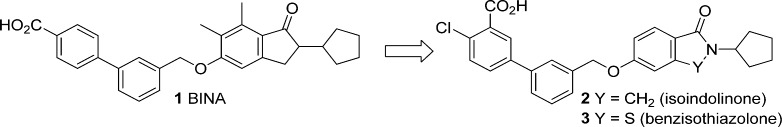
In addition to orthosteric agonists of Group II mGlu receptors, multiple reports have recently described the synthesis and characterization of selective mGlu2 receptor PAMs.5,39−46 These compounds invariably resulted from the optimization of hits obtained from high-throughput screening (HTS) of small molecule libraries. PAMs, through their interaction at allosteric sites on the mGlu receptor, positively modulate (i.e. potentiate) the effects of the endogenous orthosteric mGlu agonist glutamate. The advantages of PAMs compared with orthosteric agonists includes enhanced subtype selectivity, the potential for spatial and temporal modulation of receptor activation, and ease of optimization and fine-tuning of druglike properties.47 We recently reported the design, synthesis, and in vitro and in vivo characterization of a series of potent and selective mGlu2 receptor PAMs.48,49 The optimized compounds 2 and 3 (Figure 1), which were developed using the prototypical mGlu2 receptor PAM BINA (1) as a starting point, were employed to determine the effects of selectively activating mGlu2 receptors on cocaine or nicotine dependence.48,49 In these studies we showed that compound 3, unlike the mGlu2/3 orthosteric agonist LY379268, decreased cocaine self-administration in rats at doses that did not affect responding for food.48 We also showed that compound 2 dose-dependently decreased nicotine self-administration in rats following oral (po) administration.49 Taken together, our data suggest that mGlu2 receptor PAMs have the potential for therapeutic utility in the treatment of drug dependence.
Figure 1.
Structures of mGlu2 receptor PAMs developed from BINA (1).
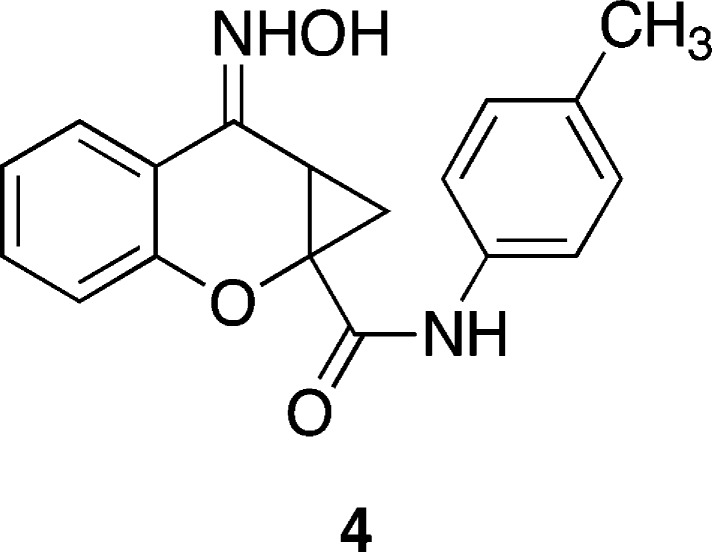
As noted above, there have been many accounts in the literature describing selective mGlu2 receptor PAMs, whereas very little has been reported on compounds that potentiate the effects of glutamate at mGlu3 receptors. This is somewhat surprising given the significant sequence homology (approximately 75%) within the transmembrane regions of mGlu2 and mGlu3 receptors. A single disclosure by Schann and co-workers described compound 4 (Figure 2) which was reported to be a mixed mGlu2 receptor negative allosteric modulator (NAM)/mGlu3 receptor PAM.50 We hypothesized that it might be possible to design and synthesize compounds that activate both mGlu2 and mGlu3 receptors through an allosteric mechanism. Considering the dearth of information on mixed mGlu2/3 receptor PAMs, the development of such compounds would provide valuable pharmacological tools. For example, a CNS penetrant mGlu2/3 receptor PAM could facilitate investigations into whether effects on food responding in rats are due to general activation of mGlu3 receptors or an effect specific to direct activation of the mGlu receptor by agonists that act at the mGlu orthosteric binding site.
Figure 2.
Structure of recently reported mGlu2 receptor NAM/mGlu3 receptor PAM 4.
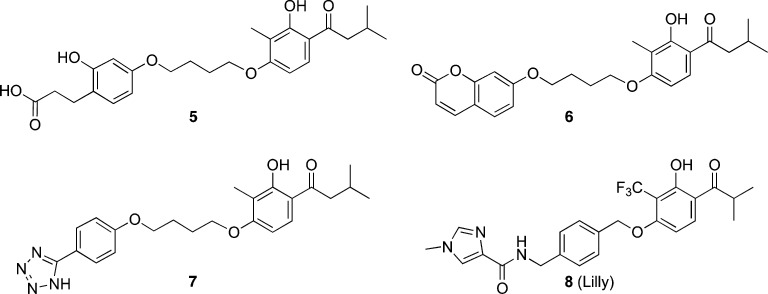
The strategy for the design and synthesis of mGlu2/3 receptor PAMs grew out of our general approach to the development of selective mGlu2 receptor PAMs. In addition to exploring the structure–activity relationship (SAR) around compound 1, which led to the series of selective mGlu2 receptor PAMs exemplified by 2 and 3,48,49 we also investigated compounds such as 5–7 (Figure 3). The mGlu2 receptor PAMs 5–7 are representative of a series developed by Pinkerton and co-workers at Merck.51 The mGlu2 receptor PAMs in this structural class were reported to exhibit varying degrees of in vitro potency and efficacy with little information provided regarding subtype selectivity. Compound 8, a selective mGlu2 receptor PAM that bears a resemblance to compounds 5–7, was recently reported by researchers at Eli Lilly.52 With the exception of the optimized compound 8, most of the compounds in this series were reported to be modestly potent at mGlu2 receptors in vitro and displayed suboptimal pharmacokinetic (PK) profiles and brain penetration in vivo. Our initial goal was to develop group II mGlu receptor PAMs having the potential for systemic activity, with a focus on maintaining or improving potency and efficacy at mGlu2 while in parallel investigating the PAM activity at mGlu3 receptors. Herein we describe the design, synthesis, and pharmacological characterization of a library of analogues from which a series of mixed mGlu2/3 receptor PAMs with unique pharmacology were discovered. Furthermore, because the selective mGlu2 receptor PAM BINA decreased cocaine- but not food-maintained responding,37 we investigated the in vivo efficacy of one of the synthesized mGlu2/3 receptor PAMs on cocaine- and food-maintained responding in rats.
Figure 3.
Structures of mGlu2 receptor PAMs in the acetophenone series.
Chemistry
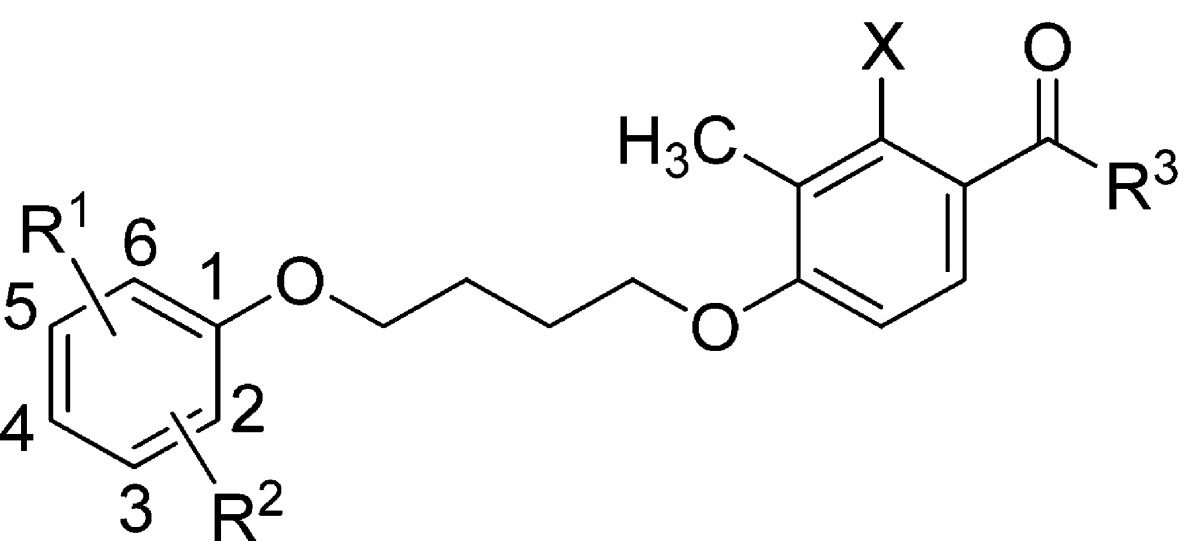
Examination of compounds 5–7 suggested the presence of common structural elements. Specifically, they consist of an aryl ether connected via a butyl ether linker to an acetophenone derivative. Thus, we envisioned a synthetic strategy wherein the key building blocks were synthesized and systematically assembled to generate a compound library rapidly and efficiently. This would allow the production of analogues with sufficient diversity to investigate the requirements for optimal potency, efficacy, and pharmacokinetic properties. The structural components were synthesized or purchased and incorporated into the new group II mGlu receptor PAMs as shown in Scheme 1. Briefly, commercially available carboxylic acids were converted to the corresponding acyl chloride derivatives (9) using oxalyl chloride in CH2Cl2. After removal of solvents, the acyl chlorides were employed in a Friedel–Crafts acylation of substituted phenols (10) using aluminum chloride to provide the key acetophenone derivatives 11. The phenol derivatives (12) were coupled with 1,4-dibromobutane by heating with potassium carbonate in acetonitrile to provide the corresponding bromobutoxybenzoate derivative (13). Finally, Finkelstein alkylation of intermediate 11 with 13 under microwave conditions delivered the ester derivatives (14–17 and 18′–75′), which were saponified with potassium hydroxide to provide the target carboxylic acid derivatives 18–75.
Scheme 1. General Synthesis of New Group II mGlu Receptor PAMs.
Results and Discussion
The initial in vitro evaluation of analogues was performed in thallium flux assays in human embryonic kidney 293 (HEK) cells expressing heteromeric G-protein-coupled inwardly rectifying potassium (GIRK) 1/2 channels and rat mGlu2 or rat mGlu3 receptors.41 The potency and efficacy of compounds were established by measuring the concentration–response relationship that potentiates the effect of an EC20 concentration of glutamate. After determination of the concentration–response relationship, the effect of a maximally effective concentration (% Max) of each compound was determined. For the allosteric potentiators, potency is expressed as the EC50 for potentiation of the glutamate EC20. Table 1 shows the results of testing the compounds against rat mGlu2 or mGlu3 receptors in vitro.
Table 1. In Vitro Potency and Efficacy Data at mGlu2 or mGlu3 Receptors for PAMsa.
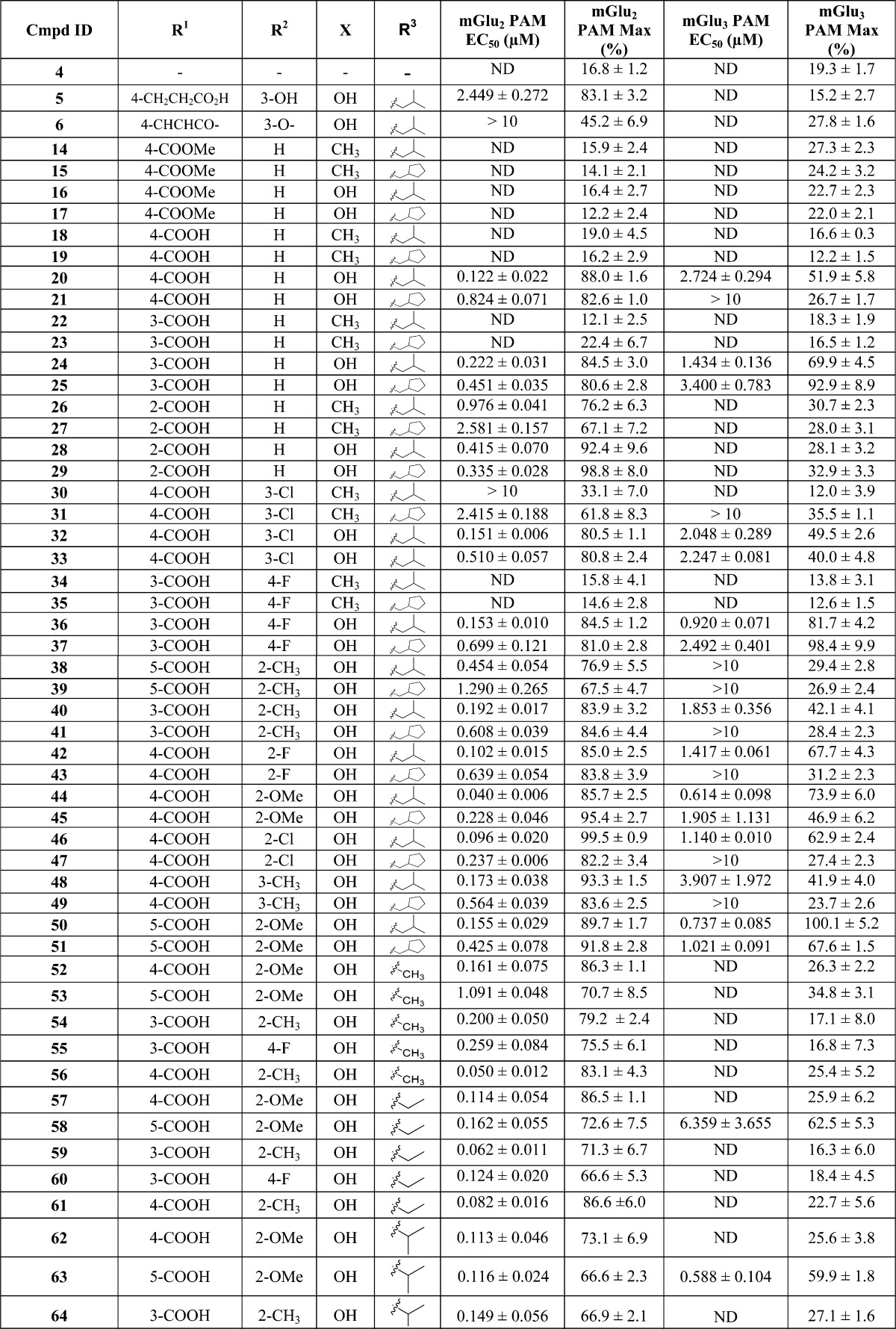
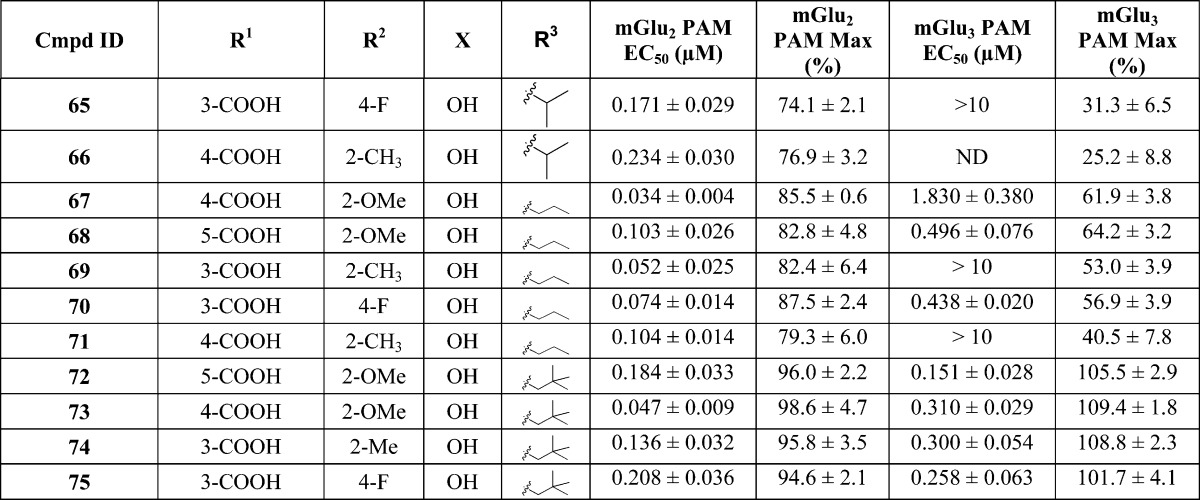
mGlu2 PAM EC50 (μM) data and glutamate Max (%) data represent the mean ± SEM for at least three independent experiments performed in triplicate. ND = not determined.
Our initial strategy for the design of the butyl ether derived PAMs library was to systematically explore different substitution patterns on the left-hand aryl ether moiety combined with a set of four right-hand acetophenones. Specifically, acetophenone derivatives were synthesized in which X was either methyl (e.g., 14) or hydroxyl (e.g., 16) and where R3 was either isopropylmethylene (e.g., 14 and 16) or cyclopentylmethylene (e.g., 15 and 17). In the first phase of library synthesis, analogues in which R1 was a 4-carboxylate moiety (14–21) were prepared and tested in vitro. Many of the compounds were inactive or only very weakly active in the assays. Known compounds 5 and 6 were synthesized and tested as part of the library to provide a benchmark of activity for the series. Some important observations were made during this phase regarding the activity of the analogues tested. First, benchmark compounds 5 and 6 exhibited PAM activity at mGlu2 but not at mGlu3 receptors in the GIRK assay. Moreover, compound 6 was only very weakly active (EC50 > 10 μM, 45% efficacy) in the GIRK assay against mGlu2. Second, in general, compounds containing X = Me were less potent than compounds in which X = OH. Third, whereas the methyl 4-benzoate derivatives 16 and 17 were inactive, the corresponding free carboxylic acid derivatives 20 and 21 were active (20) or weakly active (21) as PAMs at both mGlu2 and mGlu3 receptors. Furthermore, compound 20 was at least an order of magnitude more potent as a PAM at mGlu2 than benchmark compound 5 and had an EC50 of 2.7 μM (52% efficacy) against mGlu3. Encouraged by the PAM activity found for compounds 20 and 21, analogues containing free carboxylic acid moieties at other positions on the aryl ether ring were synthesized and tested (compounds 22–29). In the 3-carboxyl series compounds 22 and 23 (X = Me) were inactive whereas compounds 24 and 25 (X = OH) exhibited PAM activity at both mGlu2 and mGlu3 receptors. Interestingly, in the 2-carboxyl series (compounds 26–29), all four compounds were active as mGlu2 receptor PAMs with no activity at mGlu3. These results prompted us to synthesize and test the next set of analogues (compounds 30–51) containing carboxylic acid moieties at the 3- or 4-position (R1) in addition to substituents at other positions around the aryl ether ring (i.e., R2 = F, Cl, Me, or OMe). In this series, the analogues containing X = Me (30, 31, 34, 35) continued the trend of very weak (31, EC50 > 10 μM, 36% efficacy) or no mGlu3 activity (30, 34, 35, 12%–14% efficacy), and therefore, all subsequent analogues were synthesized with X = OH. Furthermore, several of the analogues in this group (39, 41, 43, 47, and 49) were active as PAMs at mGlu2 but were essentially inactive at mGlu3 receptors (24%–31% efficacy). However, in addition to being submicromolar PAMs at mGlu2, compounds 32, 33, 37, 40, 42, 45, 46, 48, and 51 were active as PAMs at mGlu3 receptors in the micromolar range (EC50 ≈ 1–4 μM, 40%–98% efficacy). Most promising of all, compounds 36, 44, and 50 possessed submicromolar PAM activity at mGlu3 receptors as well as good potency at mGlu2. Especially noteworthy were compounds 44 and 50, with EC50 values in the 600–700 nM range at mGlu3 with excellent efficacy as PAMs (74% and 100%, respectively). These results suggested that our goal of producing PAMs with potent activity at both mGlu2 and mGlu3 receptors was attainable. Having identified favorable substitution patterns for the left-hand aryl ether moiety, we next designed a series of analogues (compounds 52–75) to investigate which ketone alkyl group (R3) would impart the best potency and efficacy at both mGlu2 and mGlu3 receptors. Thus, in this set of analogues, compounds were synthesized with R3 = methyl (52–56), ethyl (57–61), isopropyl (62–66), n-propyl (67–71), or tert-butylmethylene (72–75). Interestingly, none of the methyl ketone derivatives (52–56) had activity at mGlu3 receptors, while in the ethyl ketone series (57–61) only compound 58 (5-CO2H, 2-OMe) had activity at mGlu3 (EC50 = 6359 nM, 63% efficacy). In the isopropyl ketone series, only compound 63 (5-CO2H, 2-OMe) had activity at mGlu3 (EC50 = 588 nM, 60% efficacy). On the other hand, in the n-propyl series, all compounds demonstrated some level of activity. While 69 and 71 were weakly active at mGlu3 (EC50s > 10 μM, 53% and 41% efficacy, respectively), 67 (4-CO2H, 2-OMe), 68 (5-CO2H, 2-OMe), and 70 (3-CO2H, 4-F) demonstrated more potent mGlu3 activity (EC50 = 1830 nM, 62% efficacy, EC50 = 496 nM, 64% efficacy, EC50 = 438 nM, 57% efficacy, respectively). It is noteworthy that compound 67 (4-CO2H, 2-OMe) was the most potent PAM and selective for mGlu2 with good efficacy (EC50 = 34 nM, 86% efficacy). Finally, in the tert-butylmethylene series (compounds 72–75) all four analogues exhibited good potency and excellent efficacy for both mGlu2 and mGlu3 receptors. Compound 72 was the most potent against mGlu3 (EC50 = 151 nM, 106% efficacy) with similar potency against mGlu2, while compounds 73–75 fell into the 258–310 nM potency range (102%–110% efficacy) against mGlu3 receptors. Thus, a combination of favored left-hand aryl ether moieties with R3 = tert-butylmethylene provides compounds with unique PAM activity at both mGlu2 and mGlu3 receptors.
With these results in hand, we tested some of the most promising new PAMs to provide an estimation of their druglike properties in vitro using absorption, distribution, metabolism, and excretion (ADME) assays. The results of these studies are shown in Table 2. The permeability across artificial membranes was assessed via the parallel artificial membrane permeability assay (PAMPA) assay and showed a range of results, with most compounds showing some degree of permeability. Plasma and microsomal stability was likewise acceptable, with only four of the analogues showing less than 20% remaining after 1 h. In addition, an algorithm was used to provide an estimate of plasma protein binding for 11 of the analogues. The data, which are provided in the Supporting Information (Table S1), suggest that the compounds in this series are all >90% plasma protein bound.
Table 2. In Vitro ADME Data for Group II mGlu PAMs.
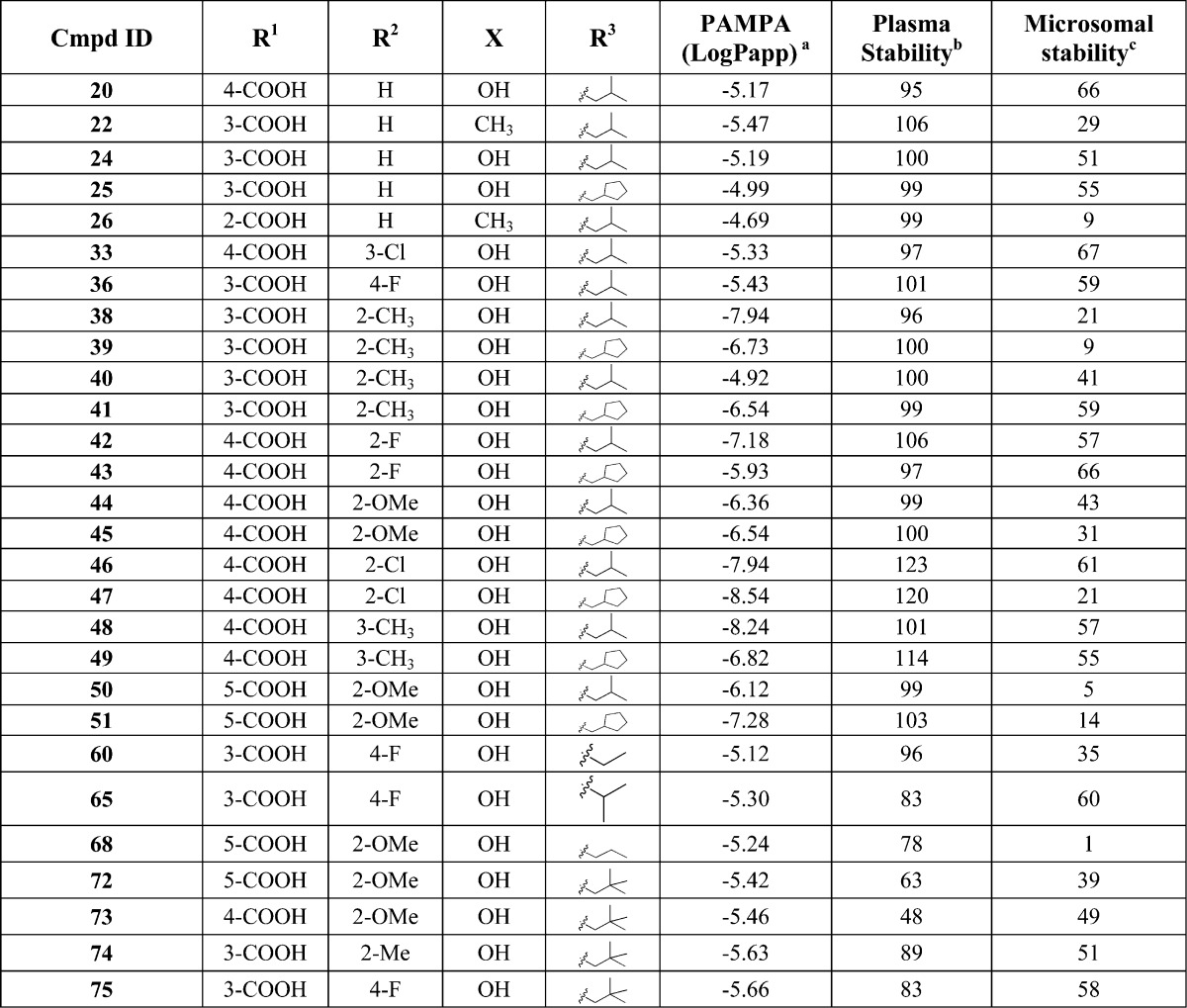
Permeability is monitored by measuring the amount of compound that can diffuse through a polar brain lipid membrane to predict BBB permeability.42
Percent remaining after incubation for 60 min at 37.5 °C.
Compounds 20, 25, 33, 44, 67, 72, and 73 were profiled against the remaining mGlu receptor subtypes to determine their selectivity relative to mGlu2 and mGlu3 (Table 3). With the exception of 67, which displayed weak antagonist/NAM activity (IC50 > 10 μM) at mGlu1 and mGlu4, and compound 73, which showed weak PAM activity at mGlu6 (EC50 > 10 μM), these compounds were found to be highly selective for mGlu2 and mGlu3. As a representative of this series, compound 72 was profiled against a representative panel of CNS receptors through the NIMH Psychoactive Drug Screening Program (PDSP; see http://pdsp.med.unc.edu/indexR.html for details). As shown in Table S2 (see Supporting Information), at 10 μM, no binding activity was detected for compound 72 at 45 CNS receptors, suggesting that the new mGlu2/3 PAMs have a low likelihood of off-target activity.
Table 3. mGlu Receptor Subtype Selectivitya.
| compound |
|||||||
|---|---|---|---|---|---|---|---|
| 20 | 25 | 33 | 44 | 67 | 72 | 73 | |
| mGlu1 | inactiveb | inactiveb | inactiveb | inactiveb | antagonist | inactiveb | inactiveb |
| FS = 0.3 | |||||||
| Emin = 3% | |||||||
| Emax = 79% | |||||||
| mGlu2 | Ago-PAM | Ago-PAM | Ago-PAM | Ago-PAM | Ago-PAM | Ago-PAM | Ago-PAM |
| FS = 3.2 | FS = 12.5 | FS = 13.3 | FS = ND | FS = ND | FS = ND | FS = ND | |
| Emin = 54% | Emin = 45% | Emin = 54% | Emin = 90% | Emin = 79% | Emin = 72% | Emin = 79% | |
| Emax = 77% | Emax = 71% | Emax = 77% | Emax = 95% | Emax = 92% | Emax = 95% | Emax = 92% | |
| mGlu3 | PAM | PAM | PAM | PAM | PAM | Ago-PAM | Ago-PAM |
| FS = 2.7 | FS = 4.5 | FS = 2.5 | FS = 3.9 | FS = 2.9 | FS = 7.1 | FS = 8.9 | |
| Emin = 9% | Emin = 6% | Emin = 10% | Emin = 4% | Emin = 1% | Emin = 54% | Emin = 30% | |
| Emax = 153% | Emax = 143% | Emax = 143% | Emax = 116% | Emax = 88% | Emax = 95% | Emax = 99% | |
| mGlu4 | inactiveb | inactiveb | inactiveb | inactiveb | antagonist | inactiveb | inactiveb |
| FS = 1.9 | |||||||
| Emin = 0% | |||||||
| Emax = 74% | |||||||
| mGlu5 | inactiveb | inactiveb | inactiveb | inactiveb | inactiveb | inactiveb | inactiveb |
| mGlu6 | inactiveb | inactiveb | inactiveb | inactiveb | inactiveb | inactiveb | PAM |
| FS = 2.0 | |||||||
| Emin = 8% | |||||||
| Emax = 93% | |||||||
| mGlu7 | inactiveb | inactiveb | inactiveb | inactiveb | inactiveb | inactiveb | inactiveb |
| mGlu8 | inactiveb | inactiveb | inactiveb | inactiveb | inactiveb | inactiveb | inactiveb |
In these selectivity experiments, for all receptors a full concentration–response of agonist was performed once in triplicate in the presence and absence of a 10 μM final concentration of each compound. This allows determination of positive allosteric modulator (PAM) (left shift of the agonist concentration response curve), antagonist (right shift in the agonist concentration response with a possible decrease in maximal agonist response), and agonist (increase in baseline response) activity in a single experiment. General activity for each compound at each mGlu is listed (PAM, antagonist, Ago-PAM, inactive) followed by the fold-shift (FS) of the agonist concentration–response obtained. Where tested compounds demonstrate activity toward an mGlu receptor subtype, the maximal (Emax) and minimal (Emin) responses of the concentration–response of agonist are indicated. Where 10 μM test compound induced greater than a 2-fold shift (FS) of the glutamate concentration–response curve (L-AP4 in the case of mGlu7), full compound concentration–response curves were performed in triplicate on 3 different days to assess compound potency. Compound 67 showed weak antagonist/NAM activity (IC50 > 10 μM) at mGlu1 and mGlu4, and compound 73 showed weak PAM activity at mGlu6 (EC50 > 10 μM).
Inactive compounds show no ability to left- or right-shift the agonist concentration response curve at 10 μM. ND = not determined.
At a relatively early stage of the project and on the basis of the overall in vitro profile of the PAMs (Tables 1 and 2), we selected compounds 20, 36, 44, and 50 for in vivo assessment of pharmacokinetic (PK) properties in rats. For this initial evaluation we determined the PK properties of the compounds by oral (po) and intravenous (iv) routes of administration as shown in Tables 4 and 5, respectively. The PAMs were found to be systemically bioavailable with half-life (t1/2) values of greater than 90 min when dosed po and demonstrate a range of maximal plasma levels from a low of 1.05 μM (50) to a high of 12.46 μM (36) (Table 4). All compounds had moderate volume of distribution at steady state (Vdss) and medium to high clearance (CL) values, indicating moderate metabolism with a primary distribution in plasma and extracellular fluids, suggesting that one or more of the PAMs might have promise as candidates for in vivo studies (Table 5). The four compounds exhibited low (20) to good oral bioavailability (50) (% F) albeit at the relatively high oral dose of 20 mg/kg; however, the brain levels of 20, 36, and 44 were low, resulting in low brain/plasma ratios. Although the brain/plasma ratios of these compounds are low, the total brain concentrations of 20 and 44 are 9-fold and 18-fold above the in vitro EC50 for mGlu2, respectively, and close to the in vitro EC50s for mGlu3.
Table 4. In Vivo PK Data for mGlu2/mGlu3 PAMs in Rats after po Administration (20 mg/kg)a.
| compd | Cmax (μM) | Tmax (min) | AUC(0→t) (μmol/L)·min | T1/2 (min) | F (%) | brain (μM) | plasma (μM) | brain/plasma |
|---|---|---|---|---|---|---|---|---|
| 20 | 2.57 ± 0.45 | 135 ± 15 | 544.8 ± 75.4 | 303 ± 55 | 23.5 | 1.12 ± 0.43 | 5.29 ± 1.98 | 0.20 ± 0.03 |
| 36 | 12.46 ± 6.42 | 30 ± 0 | 720.5 ± 283.4 | 89 ± 17 | 58.2 | 0.23 ± 0.06 | 4.23 ± 1.19 | 0.06 ± 0.03 |
| 44 | 2.75 ± 0.47 | 90 ± 17 | 1274.6 ± 246.3 | 471 ± 89 | 41.9 | 0.81 ± 0.17 | 16.38 ± 1.13 | 0.05 ± 0.01 |
| 50 | 1.05 ± 0.15 | 96 ± 40 | 466.2 ± 105.2 | 569 ± 199 | 60.4 | ND | ND | ND |
Cmax: maximum concentration of the compound detected in plasma. Tmax: time at Cmax. AUC: area under the curve. t1/2: terminal half-life. F: oral bioavailability. Brains and plasma were harvested at or near the Tmax. Compounds were dosed in a volume of 2 mL/kg po (n = 3–4) at 20 mg/kg in 0.6% Tween 80. ND = not determined.
Table 5. In Vivo PK Data for mGlu2/mGlu3 PAMs in Rats after iv Administration (2 mg/kg)a.
| compd | Cmax (μM) | CL (mL·min–1·kg–1) | Vdss (L·kg–1) | AUC(0→t) (μmol/L)·min | T1/2 (min) |
|---|---|---|---|---|---|
| 20 | 5.00 ± 0.60 | 21.09 ± 3.66 | 0.81 ± 0.14 | 231.7 ± 42.12 | 30 ± 2 |
| 36 | 3.52 ± 0.13 | 38.78 ± 2.55 | 0.89 ± 0.03 | 123.7 ± 7.7 | 18 ± 3 |
| 44 | 6.43 ± 0.39 | 14.58 ± 1.18 | 0.55 ± 0.04 | 304.6 ± 25.8 | 28 ± 0 |
| 50 | 2.15 ± 0.14 | 58.29 ± 7.98 | 2.87 ± 0.60 | 77.2 ± 8.0 | 57 ± 12 |
Cmax: maximum concentration of the compound detected in plasma. AUC: area under the curve. t1/2: terminal half-life. CL: clearance. Vdss: steady state volume of distribution. Compounds were injected in a volume of 1 mL/kg iv (n = 3–4) through an iv catheter at 2 mg/kg in 0.6% Tween 80 or in 1 M NaOH. pH was adjusted to ∼7.
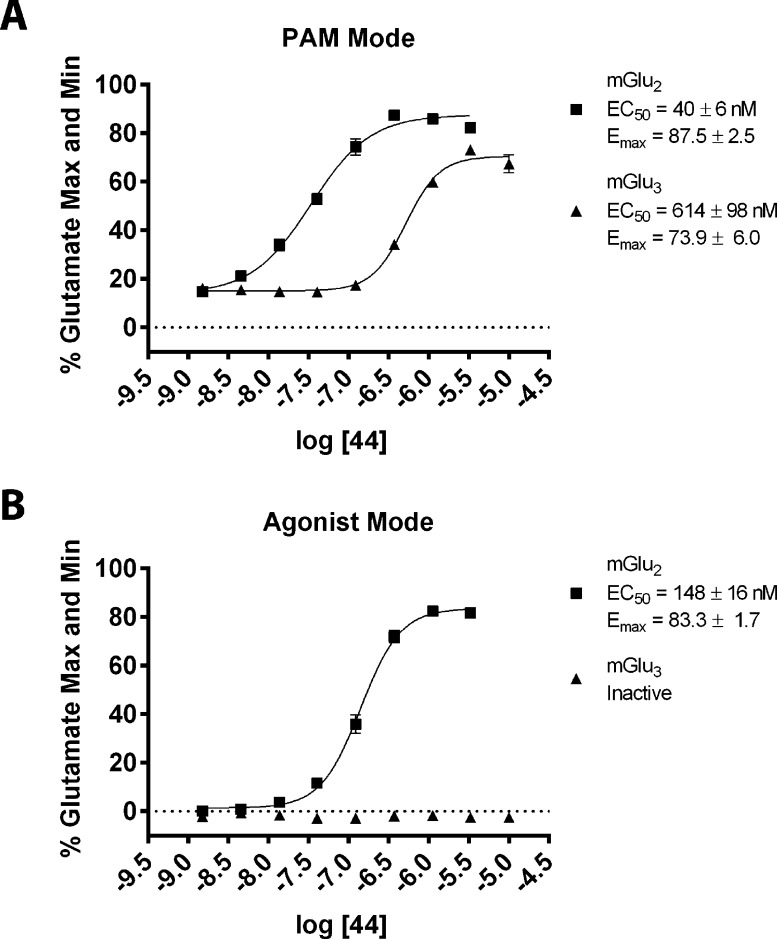
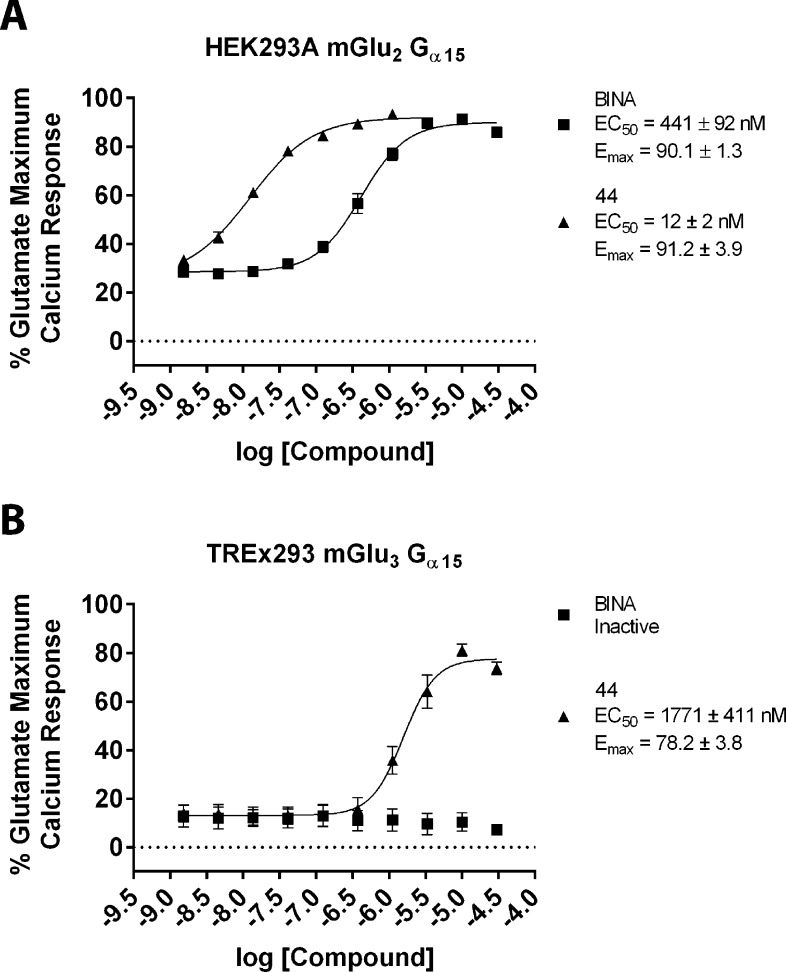
Since compound 44 had shown the best mix of in vitro and in vivo properties at this stage, including potent activity and efficacy at mGlu3 receptors, this PAM was selected for comprehensive in vitro pharmacological characterization. We began with a more detailed investigation of the activity of 44 in the mGlu2 and mGlu3 GIRK thallium flux assays. The nature of the GIRK assay requires that each compound is screened for a single mode of pharmacology at a time, since activity is only detected through the GIRK channel when thallium is added to the assay. The data presented in Table 1 represent compounds screened for activity in “PAM mode”, where a test compound is added, followed 2.5 min later by an EC20 concentration of glutamate in the presence of thallium. We had noted that the response of 44 in “PAM mode” toward mGlu2 decreased slightly at higher concentrations of test compound (Figure 4A). This decrease could be caused by either receptor desensitization or intrinsic agonist activity of 44 that was not detected because of the mode in which the functional assay was performed. To investigate this further, we carried out the same experiments in the absence of an EC20 concentration of agonist (agonist mode). For these experiments, test compounds were added in the presence of thallium and GIRK activity was immediately monitored. We found that 44 displayed intrinsic agonist activity toward mGlu2 but not mGlu3 in the GIRK assay (Figure 4B). Thus, this compound is best characterized as having mGlu2 agonist-PAM activity and mGlu3 PAM activity in the GIRK thallium flux assays.
Figure 4.
Compound 44 displays Ago-PAM activity toward mGlu2 and PAM activity toward mGlu3 in GIRK thallium-flux assays. A concentration–response of 44 was performed in the presence (A) and absence (B) of an EC20 of glutamate in either the mGlu2 GIRK assay (squares) or mGlu3 GIRK assay (triangles). In the mGlu2 assay, 44 displays both agonist and PAM activity and is characterized as an Ago-PAM. In the mGlu3 assay, only PAM activity is detected. Data were analyzed using nonlinear regression, providing EC50 values for each curve. Data were obtained from three separate experiments performed in triplicate, normalized to the response to 100 μM glutamate in each experiment, and are expressed as the mean ± SEM.
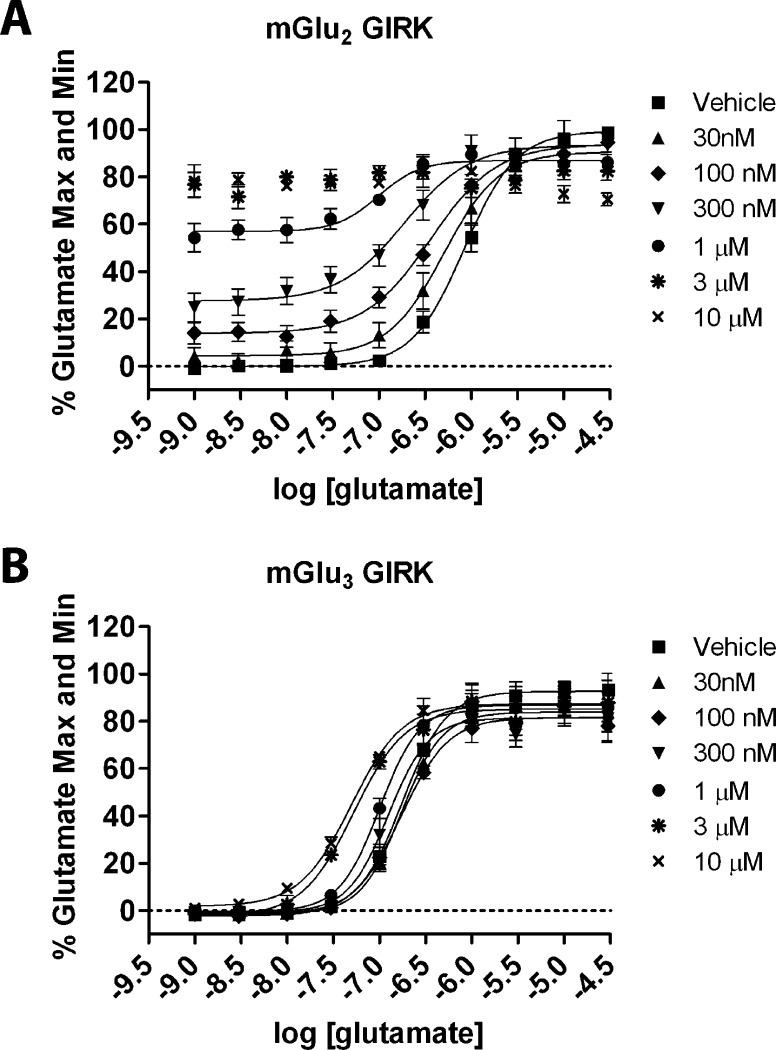
We next evaluated 44 in a fold-shift assay, another measure of the potentiating activity of a PAM toward the orthosteric ligand glutamate (Figure 5). Fold-shift values are calculated by determining the ratio of the potency of the orthosteric agonist glutamate in the presence and absence of increasing concentrations of an allosteric modulator. Increasing fixed concentrations of 44 dose-dependently shifted the glutamate concentration–response of mGlu2 (Figure 5A) and mGlu3 (Figure 5B) to the left, consistent with an enhancement of glutamate responses. For these assays, the Ago-PAM activity toward mGlu2 is readily apparent as the increase in baseline at low concentrations of glutamate (Figure 5A). These data are in contrast with the results for mGlu3 (Figure 5B), which does not show a change in baseline of the glutamate dose–response.
Figure 5.
Compound 44 dose-dependently induces a leftward shift in the glutamate concentration response at (A) mGlu2 and (B) mGlu3 in GIRK thallium flux assays. The increase in baseline in the mGlu2 GIRK assay at higher concentrations of 44 is due to the Ago-PAM activity of this compound. The leftward shifts induced by 44 indicate a potentiation of the response of mGlu2 and mGlu3 to glutamate. The maximal fold-shift at mGlu2 is 4.50 ± 0.96 and was derived from the test concentration of 44 (300 nM) due to Ago-PAM activity. The maximal fold-shift at mGlu3 is 5.48 ± 0.27 for the 10 μM test concentration. Concentration–response relationships were generated by adding a fixed concentration of 44 to cells as indicated, followed by increasing concentrations of glutamate. Data were analyzed using nonlinear regression, providing EC50 values for each curve. Data were obtained from three separate experiments performed in duplicate, normalized to the response to 100 μM glutamate in each experiment, and are expressed as the mean ± SEM.
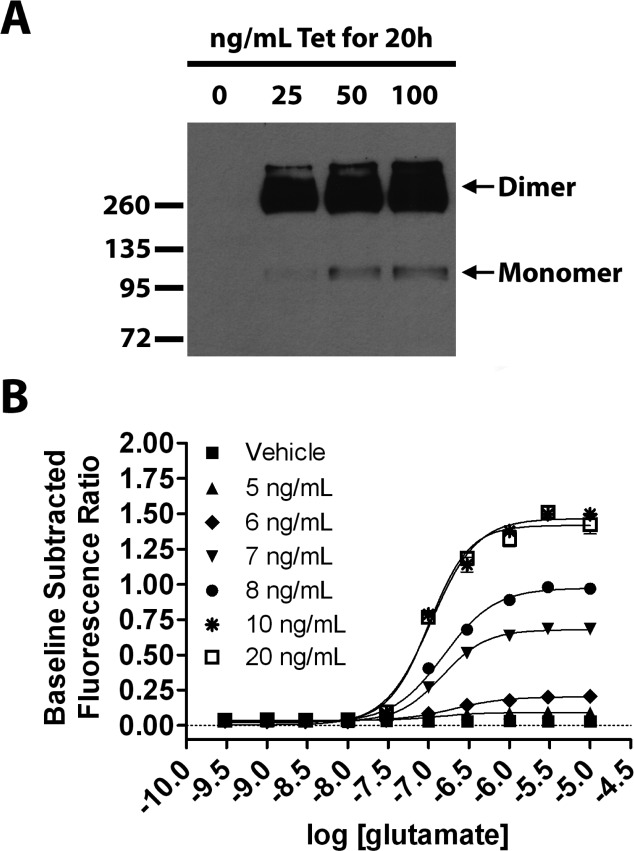
In the final set of pharmacological characterization experiments, we evaluated the potent PAM 44 in an orthogonal assay of mGlu3 and mGlu2 activity. For mGlu3 we utilized the TREx tetracycline-inducible system (Invitrogen). We developed a cell line in which the expression of mGlu3 is dose-dependently induced by tetracycline (Tet) and functionally coupled to calcium mobilization by the promiscuous G protein Gα15 (Figure 6). In the absence of Tet, no measurable expression of mGlu3 is detected either by Western blot (Figure 6A) or by functional response to calcium mobilization (Figure 6B). The optimal calcium mobilization response for this cell line was achieved at 20 ng/mL Tet for 20 h prior to assay. This concentration of Tet was then utilized for further characterization of 44 in the TREx293 mGlu3 Gα15 calcium assay (Figure 7B), which shows that 44 demonstrates mGlu3 PAM activity in this orthogonal assay whereas the mGlu2 selective PAM BINA remains inactive. These compounds were also evaluated in calcium assays utilizing HEK293A mGlu2 Gα15 cells as shown in Figure 7A. Unlike the mGlu2 GIRK assay where it displays Ago-PAM activity, compound 44 behaves as a PAM in the calcium assay. In future experiments, it will be essential to determine the potency and selectivity of 44 in native tissue preparations to determine whether the observed profile in vitro is replicated in vivo.
Figure 6.
Development of a cell line with inducible mGlu3 expression coupled to calcium mobilization via the promiscuous G protein Gα15. (A) TREx293 mGlu3 Gα15 cells were stimulated with the indicated concentrations of tetracycline (Tet) for 20 h. Protein lysates were prepared. Equivalent amounts of protein were loaded for all lanes, and mGlu2/3 expression was detected by Western blot. Both monomeric and dimeric mGlu3 were detected as indicated. (B) TREx293 mGlu3 Gα15 cells were stimulated with the indicated concentrations of Tet for 20 h, and a calcium mobilization assay was performed. Tet dose-dependently induced a glutamate-simulated calcium response that was maximal at 20 ng/mL Tet. Data were analyzed using nonlinear regression. Data were obtained from three separate experiments performed in triplicate, normalized to the response to 100 μM glutamate in each experiment, and are expressed as the mean ± SEM.
Figure 7.
Compound 44 displays PAM activity toward mGlu2 and mGlu3 in calcium assays utilizing the promiscuous G protein Gα15. A concentration–response of 44 (triangles) and the control mGlu2 selective PAM BINA (squares) was performed in the presence of an EC20 of glutamate in either the (A) HEK293A mGlu2 Gα15 calcium assay or (B) TREx293 mGlu3 Gα15 calcium assay. In both assays, 44 displays PAM activity. BINA displays PAM activity in the mGlu2 calcium assay but is inactive in the mGlu3 calcium assay. For this assay, mGlu3 expression was induced with 20 ng/mL Tet for 20 h prior to assay. Data were analyzed using nonlinear regression, providing EC50 values for each curve. Data were obtained from three separate experiments performed in triplicate, normalized to the response to 100 μM glutamate in each experiment, and are expressed as the mean ± SEM.
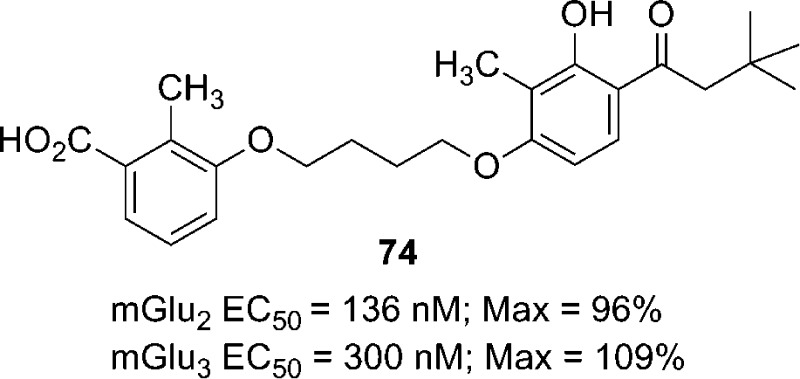
Following the in vitro pharmacological characterization of 44 as a representative analogue of this class of mGlu2/3 PAMs, we wished to evaluate a member of this series in efficacy studies in rats. Given the low brain levels achieved by po dosing for compounds 20, 36, 44, and 50 (Table 4), we evaluated nine compounds by intraperitoneal (ip) dosing in order to avoid first pass metabolism and to cast a wider net for a compound suitable for rat efficacy studies (Table 6). All compound plasma levels were determined, but only those with the highest plasma concentrations (i.e., 44, 73, 74, and 75) were evaluated for brain levels. On the basis of its combination of potency, selectivity, and PK properties, compound 74 was selected for efficacy studies in rats.
Table 6. In Vivo PK for mGlu2/3 PAMs in Rats after ip Administration (10 mg/kg)a.
| compd | plasma (μM)a | plasma t1/2 (min) | brain (μM)a | brain/plasma |
|---|---|---|---|---|
| 44 | 10.57 ± 2.52 | 43 | 0.23 ± 0.10 | 0.03 ± 0.01 |
| 50 | 3.29 ± 1.27 | 20 | ND | ND |
| 60 | 4.49 ± 0.49 | 22 | ND | ND |
| 65 | 5.44 ± 2.92 | 30 | ND | ND |
| 68 | 2.18 ± 0.58 | 28 | ND | ND |
| 72 | 2.44 ± 2.19 | 27 | ND | ND |
| 73 | 6.81 ± 0.84 | 132 | 0.47 ± 0.35 | 0.02 ± 0.01 |
| 74 | 17.05 ± 0.19 | 106 | 0.56 ± 0.10 | 0.03 ± 0.01 |
| 75 | 6.52 ± 0.54 | 80 | 0.22 ± 0.04 | 0.01 ± 0.01 |
Maximum concentration of the compound detected in plasma or brain. t1/2: terminal half-life. Compounds were dosed ip (n = 3) at 10 mg/kg in 10% EtOH/1% Tween 80. pH was adjusted to ∼7. Brains and plasma were harvested at the Tmax (30 min for all tested). ND = not determined.
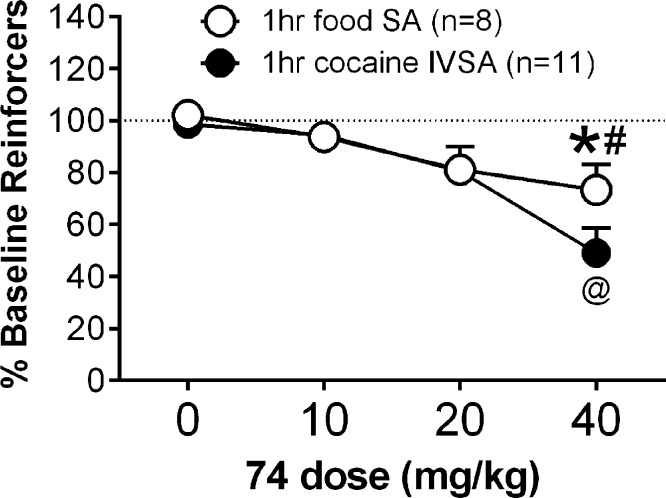
Taking all the relevant data into account, we determined that the mGlu2/3 receptor PAM 74 would be a good candidate for evaluation in a rat model of cocaine dependence. When assessed in vivo, compound 74 dose-dependently decreased cocaine- and food-maintained responding [compound 74 dose main effect: F3,51 = 14.55; p < 0.0001] (Figure 8). Interestingly, however, cocaine-maintained responding was decreased to a greater extent than food-maintained responding at the highest dose tested (40 mg/kg; p < 0.05). Because of the within-subjects design of the dose response (i.e., each rat received each dose of compound 74 using a Latin-square design), it was not possible to collect brain samples to determine brain concentrations of compound 74 at 40 mg/kg during behavioral testing. It is unlikely that brain concentrations of compound 74 differed between cocaine- and food-maintained rats at this dose, suggesting that the observed differences in behavior were not a function of group differences in brain pharmacokinetic properties of compound 74, although this should be confirmed in future studies. We have previously shown that the mGlu2/3 receptor agonist LY379268 similarly decreased both cocaine- and food-maintained responding.37,53 Moreover, the selective mGlu2 receptor PAM BINA decreased only cocaine-maintained responding while having no effect on food-maintained responding.37 Consistent with our previous results, Morishima et al. (2005) have demonstrated that mGlu2 receptor knockout mice exhibited increased conditioned place preference for cocaine.54 While these prior studies focused only on the role of mGlu2 receptors in responding for drug and natural rewards, our present findings begin to delineate the individual roles of mGlu2 and mGlu3 receptors in reward processing. These patterns of results possibly suggest that activation of mGlu2 receptors selectively modulates drug-reinforced behavior, whereas activation of mGlu3 receptors either selectively modulates responding for natural rewards or nonselectively modulates responding for both drug and natural rewards. Use of an mGlu2/3 receptor PAM, as reported here, provided an initial tool by which to indirectly test these hypotheses. However, future development and use of selective mGlu3 receptor PAMs are necessary to test these hypotheses directly. Moreover, increasing mGlu2/3 receptor activity using a PAM compared to a receptor agonist may affect behaviors reinforced by natural rewards to a lesser extent relative to drug-reinforced behaviors. Thus, targeting mGlu2 receptors with a PAM may be an effective strategy for treating drug dependence without affecting other motivated behaviors.
Figure 8.
The mGlu2/3 PAM, 74, decreased cocaine-maintained responding, and to a lesser extent food-maintained responding, in rats. At the highest dose tested (40 mg/kg), cocaine-maintained responding was significantly lower than food-maintained responding. The 40 mg/kg dose decreased responding for food compared to vehicle only, whereas the same dose decreased responding for cocaine compared to all other doses tested. (∗) Responding was significantly different from 0 mg/kg. (#) Responding was significantly different from cocaine. (@) Responding was significantly different from 0, 10, and 20 mg/kg. Data are expressed as the mean ± SEM of responding at baseline.
In conclusion, a ligand–based rational design approach utilizing certain previously reported selective mGlu2 receptor PAMs as starting points led to a series of new mGlu2/3 PAMs. A library of more than 60 analogues was synthesized and tested, providing compounds with excellent potency and efficacy at mGlu2 and mGlu3 receptors in vitro. The most promising mGlu2/3 PAMs were profiled in in vitro ADME assays, and on the basis of these data, several compounds were selected for PK studies in rats. The lead structures were found to be essentially devoid of activity at other mGlu receptor subtypes and 45 additional CNS receptors. Representative PAM 44 was characterized extensively in vitro and was found to demonstrate ago-PAM activity at mGlu2 and PAM activity at mGlu3 receptors. Finally, the systemically active mGlu2/3 PAM 74 dose-dependently decreased cocaine self-administration in rats following a single intraperitoneal dose. Future studies will focus on additional characterization of the new mGlu2/3 PAMs and further optimization of compounds with enhanced PAM activity at mGlu3 receptors.
Experimental Section
General Chemistry
All reactions were performed in oven-dried glassware under an atmosphere of argon with magnetic stirring. All solvents and chemicals used were purchased from Sigma-Aldrich or Acros, and were used as received without further purification. Purity of compounds was established by liquid chromatography–mass spectroscopy (HPLC–MS) and was >95% for all tested compounds. Silica gel column chromatography was carried out using prepacked silica cartridges from RediSep (ISCO Ltd.) and eluted using an Isco Companion system. 1H and 13C NMR spectra were obtained on a Jeol 400 spectrometer at 400 and 100 MHz, respectively. Chemical shifts are reported in δ (ppm) relative to residual solvent peaks or TMS as internal standards. Coupling constants are reported in Hz. Melting points were obtained using a capillary melting point apparatus (MEL-TEMP) and are uncorrected. High-resolution ESI-TOF mass spectra were acquired from the Mass Spectrometry Core at The Sanford-Burnham Medical Research Institute (Orlando, FL). HPLC–MS analyses were performed on a Shimadzu 2010EV LCMS instrument using the following conditions: Kromisil C18 column (reverse phase, 4.6 mm × 50 mm); a linear gradient from 10% acetonitrile and 90% water to 95% acetonitrile and 5% water over 4.5 min; flow rate of 1 mL/min; UV photodiode array detection from 200 to 300 nm.
General Methods for the Synthesis of mGlu2/3 Receptor PAMs
General Method A
To a stirred solution of methyl 4-hydroxybenzoate (1 mmol, 1 equiv) and 1,4-dibromobutane (3 mmol, 3 equiv) in ACN was added potassium carbonate (2 mmol, 2 equiv). The reaction mixture was heated at reflux for 6 h, at which time it was cooled to room temperature. The crude reaction mixture was diluted with CH2Cl2 and washed twice with 5% aqueous HCl (200 mL). The organic layers were collected and washed twice with saturated NaHCO3 solution (200 mL). The organic layers were collected, dried over Na2SO4, and evaporated to dryness. To a stirred solution of AlCl3 (0.039 mol, 1 equiv) in CH2Cl2 at 0 °C under nitrogen, the acyl chloride (0.039 mol, 1 equiv) was dissolved in CH2Cl2 and added dropwise to the stirred solution. The phenol (0.039 mol, 1 equiv) was added to the reaction mixture, and the mixture was allowed to warm to room temperature over 12 h. The reaction was quenched with HCl (5% aqueous), and CH2Cl2 was added (50 mL). The organic layer was separated and washed with saturated NaHCO3 solution, then brine and dried over Na2SO4. The solvents were removed by rotary evaporation, and the products were isolated by flash chromatography [SiO2, hexanes/EtOAc (4:1)] and concentrated in vacuo. To a crimp top microwave vial were added the phenol (1 mmol, 1 equiv), bromobutoxy benzoate (1 mmol, 1 equiv), potassium carbonate (2 mmol, 2 equiv), and potassium iodide (0.1 mmol, 0.1 equiv), all dissolved in ACN (0.2 M). The reaction mixture was heated in the microwave at 160 °C for 15 min. Following filtration and evaporation of solvents, the products were isolated by flash chromatography or reverse phase HPLC and lyophilized to provide the final compounds which were determined to be >95% pure by HPLC–UV, HPLC–MS, and 1H NMR.
General Method B
To a stirred solution of the product from “General Method A” (1 mmol, 1 equiv) in dioxane at room temperature was added KOH (6 mmol, 6 equiv) in water (0.5 mL). The mixture was stirred continuously for an additional 12 h. The reaction was quenched with HCl (5% aqueous), and CH2Cl2 (50 mL) was added. The organic layer was separated and dried over Na2SO4. The solvents were removed by rotary evaporation, and the products were isolated by flash chromatography [SiO2, hexanes/EtOAc (1:1)] or reverse phase HPLC and lyophilized to provide the final compounds which were determined to be >95% pure by HPLC–UV, HPLC–MS, and 1H NMR. Compounds 4, 5, and 6 were synthesized according to the published procedures.50,51 The following compounds were prepared using the general procedures A and B from the appropriate starting materials.
Methyl 4-(4-(2,3-Dimethyl-4-(3-methylbutanoyl)phenoxy)butoxy)benzoate (14)
14 was prepared according to general procedure A. Colorless solid (0.070 g, 17%); mp 55–57 °C. 1H NMR (CDCl3): δ 7.86 (d, J = 7.8 Hz, 2H), 7.17–6.94 (m, 4H), 4.05 (t, J = 5.9 Hz, 2H), 3.88 (s, 3H), 3.80 (t, J = 6.0 Hz, 2H), 2.81 (d, J = 6.9 Hz, 2H), 2.29 (s, 3H), 2.20 (s, 3H), 2.10–2.01 (m, 1H), 1.88–1.84 (m, 4H), 0.93 (d, J = 6.4 Hz, 6H). 13C NMR (CDCl3): δ 203.3, 166.8, 162.7, 155.5, 142.3, 132.2, 131.5, 130.6, 125.7, 125.4, 122.5, 114.0, 75.1, 67.7, 51.8, 51.7, 26.9, 25.9, 25.1, 22.63, 20.0, 12.3. ESI-MS m/z 413 [M + H]+. HRMS m/z calcd for C25H32O5 [M + H]+: 413.2323. Found: 413.2256.
Methyl 4-(4-(4-(2-Cyclopentylacetyl)-2,3-dimethylphenoxy)butoxy)benzoate (15)
15 was prepared according to general procedure A. Colorless solid (0.039 g, 9%). 1H NMR (CDCl3): δ 7.87 (d, J = 9.2 Hz, 2H), 7.17 (d, J = 7.8 Hz, 1H), 7.00–6.97 (m, 3H), 4.05 (t, J = 5.9 Hz, 2H), 3.88 (s, 3H), 3.80 (t, J = 5.9 Hz, 2H), 2.95 (d, J = 6.9 Hz, 2H), 2.29 (s, 3H), 2.20 (s, 3H), 2.21–2.10 (m, 1H), 1.99–1.89 (m, 4H), 1.80–1.76 (m, 2H), 1.59–1.46 (m, 4H), 1.14–1.01 (m, 2H). 13C NMR (CDCl3): δ 203.3, 166.8, 162.7, 155.5, 142.3, 132.2, 131.5, 130.6, 126.9, 126.2, 114.0, 74.7, 67.7,51.8, 49.0, 36.1, 32.7, 26.9, 25.9, 24.9, 20.0, 12.3. ESI-MS m/z 439 [M + H]+. HRMS m/z calcd for C27H34O5 [M + H]+: 439.2479. Found: 439.2419.
Methyl 4-(4-(3-Hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy)butoxy)benzoate (16)
16 was prepared according to general procedure A. Colorless solid (0.112 g, 27%); mp 72–74 °C. 1H NMR (CDCl3): δ 7.98–7.96 (m, 2H), 7.58 (d, J = 8.7 Hz, 1H), 6.90–6.88 (m, 2H), 6.41(d, J = 9.2 Hz, 1H), 4.09–4.03 (m, 4H), 3.86 (s, 3H), 2.76 (d, J = 7.3 Hz, 2H), 2.20–2.15 (m, 1H), 2.09 (s, 3H), 1.96–1.94 (m, 4H), 0.97 (d, J = 6.9 Hz, 6H). 13C NMR (CDCl3): δ 205.1, 166.8, 162.6, 162.5, 162.3, 131.5, 129.3, 122.5, 114.0, 113.7, 102.4, 67.7, 67.5, 51.8, 46.8, 26.9, 26.8, 25.9, 22.7, 7.6. ESI-MS m/z 415 [M + H]+. HRMS m/z calcd for C24H30O6 [M + H]+: 415.2115. Found: 415.2137.
Methyl 4-(4-(4-(2-Cyclopentylacetyl)-3-hydroxy-2-methylphenoxy)butoxy)benzoate (17)
17 was prepared according to general procedure A. Colorless solid (0.277 g, 63%). 1H NMR (CDCl3): δ 7.86 (d, J = 8.7 Hz, 2H), 7.79 (d, J = 8.7 Hz, 1H), 6.90 (d, J = 9.2 Hz, 2H), 6.61 (d, J = 9.2 Hz, 1H), 4.11–4.09 (m, 4H), 3.87 (s, 3H), 2.90 (d, J = 7.3 Hz, 2H), 2.35–2.20 (m, 1H), 1.96 (s, 3H), 1.94–1.92 (m, 4H), 1.85–1.76 (m, 2H), 1.59–1.47 (m, 4H), 1.87–1.19 (m, 2H). 13C NMR (CDCl3): δ 205.3, 162.6, 162.5, 162.3, 131.6, 129.2, 122.5, 114.0, 113.8, 113.7, 102.4, 67.7, 67.5, 51.8, 44.0, 36.8, 32.6, 25.9, 24.8, 7.6. ESI-MS m/z 441 [M + H]+. HRMS m/z calcd for C26H32O6 [M + H]+: 441.2272. Found: 441.2278.
4-(4-(2,3-Dimethyl-4-(3-methylbutanoyl)phenoxy)butoxy)benzoic Acid (18)
Colorless solid (0.159 g, 40%). 1H NMR (DMSO): δ 7.84 (d, J = 8.7 Hz, 2H), 7.20 (d, J = 8.2 Hz, 1H), 6.99 (d, J = 8.7 Hz, 3H), 4.11 (t, J = 5.5 Hz, 2H), 3.75 (t, J = 5.5 Hz, 2H), 2.78 (d, J = 6.9 Hz, 2H), 2.24 (s, 3H), 2.15 (s, 3H), 2.02–1.98 (m, 1H), 1.87–1.80 (m, 4H), 0.86 (d, J = 6.9 Hz, 6H). 13C NMR (DMSO): δ 203.6, 167.0, 162.1, 155.1, 142.1, 131.7, 131.3, 130.4, 125.9, 125.2, 123.1, 114.2, 74.5, 67.5, 51.1, 27.2, 26.3, 25.3, 24.5, 22.3, 20.0, 12.0. ESI-MS m/z 399 [M + H]+. HRMS m/z calcd for C24H30O5 [M + H]+: 399.2166. Found: 399.2191.
4-(4-(4-(2-Cyclopentylacetyl)-2,3-dimethylphenoxy)butoxy)benzoic Acid (19)
Colorless solid (0.093 g, 22%). 1H NMR (DMSO): δ 7.83 (d, J = 8.7 Hz, 2H), 7.21 (d, J = 7.8 Hz, 1H), 6.98 (d, J = 8.7 Hz, 3H), 4.12 (t, J = Hz, 2H), 3.75 (t, J = Hz, 2H), 2.90 (d, J = 6.9 Hz, 2H), 2.25 (s, 3H), 2.14 (s, 3H), 2.09–2.07 (m, 1H), 1.86–1.84 (m, 4H), 1.69–1.62 (m, 2H), 1.51–1.37 (m, 4H), 1.04–0.98 (m, 2H). 13C NMR (DMSO): δ 207.8, 167.0, 162.3, 154.2, 142.1, 131.4, 130.4, 125.8, 125.6, 122.9, 114.3, 74.1, 67.9, 49.3, 35.6, 32.1, 25.6, 25.1, 24.5, 20.2, 12.1. ESI-MS m/z 425 [M + H]+. HRMS m/z calcd for C26H32O5 [M + H]+: 425.2323. Found: 425.2366.
4-(4-(3-Hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy)butoxy)benzoic Acid (20)
Colorless solid (0.208 g, 52%); mp 153–155 °C. 1H NMR (DMSO): δ 7.84 (d, J = 8.7 Hz, 2H), 7.76 (d, J = 8.7 Hz, 1H), 6.96 (d, J = 9.2 Hz, 2H), 6.60 (d, J = 9.2 Hz, 1H), 4.11–4.06 (m, 4H), 2.79 (d, J = 6.9 Hz, 2H), 2.10–2.08 (m, 1H), 1.93 (s, 3H), 1.89–1.86 (m, 4H), 0.89 (d, J = 6.9 Hz, 6H). 13C NMR (DMSO): δ 206.2, 167.0, 162.5, 162.3, 161.4, 131.4, 130.4, 122.9, 114.3, 113.5, 112.0, 103.4, 67.7, 67.5, 51.8, 44.0, 36.8, 32.6, 25.4, 22.4, 7.6. ESI-MS m/z 401 [M + H]+. HRMS m/z calcd for C23H28O6 [M + H]+: 401.1956. Found: 401.1966.
4-(4-(4-(2-Cyclopentylacetyl)-3-hydroxy-2-methylphenoxy)butoxy)benzoic Acid (21)
Colorless solid (0.192 g, 45%); mp 158–160 °C. 1H NMR (DMSO): δ 7.83–7.81 (m, 3H), 6.69 (d, J = 8.7 Hz, 2H), 6.59 (d, J = 8.7 Hz, 1H), 4.11–4.08 (m, 4H), 2.97 (d, J = 6.9 Hz, 2H), 2.22–2.01 (m, 1H), 1.96 (s, 3H), 1.89–1.86 (m, 4H), 1.72–1.71 (m, 2H), 1.59–1.55 (m, 2H), 1.45–1.53 (m, 2H), 1.15–1.12 (m, 2H). 13C NMR (DMSO): δ 207.8, 167.0, 162.5, 162.3, 161.4, 131.6, 131.3, 122.9, 114.2, 112.8, 111.9, 103.8, 69.0, 68.9, 43.8, 36.2, 31.1, 24.5, 22.4, 8.9. ESI-MS m/z 427 [M + H]+. HRMS m/z calcd for C25H30O6 [M + H]+: 427.2115. Found: 427.2120.
3-(4-(2,3-Dimethyl-4-(3-methylbutanoyl)phenoxy)butoxy)benzoic Acid (22)
Colorless solid (0.167 g, 42%). 1H NMR (DMSO): δ 7.49–7.46 (m, 2H), 7.41–7.39 (m, 2H), 7.16–7.14 (m, 1H), 6.73–6.69 (m, 1H), 4.04–4.03 (m, 4H), 2.74 (d, J = 6.9 Hz, 2H), 2.11 (s, 3H), 2.03 (s, 3H), 1.90–1.87 (m, 1H), 1.86–1.84 (m, 4H), 0.82 (d, J = 6.9 Hz, 6H). 13C NMR (DMSO): δ 205.6, 167.2, 162.9, 161.4, 158.6, 132.7, 131.4, 129.7, 124.7, 121.8, 119.8, 114.2, 109.8, 74.2, 67.6, 51.7, 25.6, 25.3, 22.4, 24.1, 19.9, 19.8, 12.0, 11.9. ESI-MS m/z 399 [M + H]+. HRMS m/z calcd for C24H30O5 [M + H]+: 399.2166. Found: 399.2239.
3-(4-(4-(2-Cyclopentylacetyl)-2,3-dimethylphenoxy)butoxy)benzoic Acid (23)
Colorless solid (0.114 g, 27%). 1H NMR (DMSO): δ 7.47–7.40 (m, 3H), 7.19–7.15 (m, 2H), 6.97 (d, J = 7.8 Hz, 1H), 4.04 (t, J = 5.6 Hz, 2H), 3.73 (t, J = 5.9 Hz, 2H), 2.89 (d, J = 6.9 Hz, 2H), 2.21 (s, 3H), 2.11–2.04 (ovelapping singlet and multiplets, 4H), 1.85–1.83 (m, 4H), 1.66–1.64 (m, 2H), 1.48–1.39 (m, 4H), 1.04–1.01 (m, 2H). 13C NMR (DMSO): δ 205.6, 167.2, 167.2, 158.2, 155.2, 141.9, 132.7, 131.4, 130.2, 129.7, 125.9, 125.2, 121.8, 119.3, 114.5, 74.4, 67.6, 48.4, 35.5, 32.0, 26.3, 25.4, 24.5, 20.0, 11.9. ESI-MS m/z 425 [M + H]+. HRMS m/z calcd for C26H32O5 [M + H]+: 425.2323. Found: 425.2383.
3-(4-(3-Hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy)butoxy)benzoic Acid (24)
Colorless solid (0.148 g, 37%); mp 118–120 °C. 1H NMR (DMSO): δ 7.81 (d, J = 9.2 Hz, 1H), 7.46 (d, J = 7.8 Hz, 1H), 7.39–7.34 (m, 2H), 7.14–7.13 (m, 1H), 6.58 (d, J = 9.2 Hz, 1H), 4.13 (t, J = 5.9 Hz, 2H), 4.07 (t, J = 5.9 Hz, 2H), 2.81 (d, J = 6.9 Hz, 2H), 2.11–2.08 (m, 1H), 1.95 (s, 3H), 1.88–1.86 (m, 4H), 0.91 (d, J = 6.9 Hz, 6H). 13C NMR (DMSO): δ 205.6, 167.2, 162.5, 161.4, 158.6, 132.3, 130.4, 129.7, 121.6, 119.4, 114.5, 113.5, 112.0, 103.4, 67.9, 67.3, 46.0, 25.6, 25.4, 22.5, 7.5. ESI-MS m/z 401 [M + H]+. HRMS m/z calcd for C23H28O6 [M + H]+: 401.1959. Found: 401.1967.
3-(4-(4-(2-Cyclopentylacetyl)-3-hydroxy-2-methylphenoxy)butoxy)benzoic Acid (25)
Colorless solid (0.230 g, 54%); mp 125–127 °C. 1H NMR (DMSO): δ 7.76 (d, J = 9.2 Hz, 1H), 7.46 (d, J = 7.8 Hz, 1H), 7.41–7.33 (m, 2H), 7.13–7.12 (m, 1H), 6.58 (d, J = 9.2 Hz, 1H), 4.08–4.03 (m, 4H), 2.94 (d, J = 7.3 Hz, 2H), 2.17–2.16 (m, 1H), 1.95 (s, 3H), 1.87–1.85 (m, 4H), 1.71–1.68 (m, 2H), 1.55–1.42 (m, 4H), 1.12–1.09 (m, 2H). 13C NMR (DMSO): δ 205.8, 162.5, 161.3, 158.6, 132.3, 130.3, 129.7, 121.6, 119.4, 114.6, 113.3, 112.0, 103.3, 67.9, 67.4, 43.6, 36.3, 32.1, 25.4, 24.5, 7.5. 167.2, ESI-MS m/z 427 [M + H]+. HRMS m/z calcd for C25H30O6 [M + H]+: 427.2115. Found: 427.2122.
2-(4-(2,3-Dimethyl-4-(3-methylbutanoyl)phenoxy)butoxy)benzoic Acid (26)
Colorless solid (0.207 g, 52%); mp 48–50 °C. 1H NMR (DMSO): δ 7.58 (d, J = 7.8 Hz, 1H), 7.45 (t, J = 7.8 Hz, 1H), 7.11–6.99 (m, 3H), 6.73–6.68 (m, 1H), 4.09–4.04 (m, 4H), 2.77 (d, J = 7.3 Hz, 2H), 2.24 (s, 3H), 2.18 (s, 3H), 2.01–2.00 (m, 1H), 1.89–1.86 (m, 4H), 0.86 (d, J = 6.8 Hz, 6H). 13C NMR (DMSO): δ 205.8, 167.4, 157.5, 156.5, 137.2, 133.0, 130.7, 125.9, 121.9, 120.1, 113.5, 109.3, 67.9, 67.4, 51.2, 48.7, 25.6, 24.6, 22.4, 19.8, 12.1, 11.5. ESI-MS m/z 399 [M + H]+. HRMS m/z calcd for C24H30O5 [M + H]+: 399.2166. Found: 399.2284.
2-(4-(4-(2-Cyclopentylacetyl)-2,3-dimethylphenoxy)butoxy)benzoic Acid (27)
Colorless solid (0.199 g, 47%); mp 90–92 °C. 1H NMR (DMSO): δ 7.59 (d, J = 7.9 Hz, 1H), 7.46–7.44 (m, 2H), 7.06 (d, J = 8.2 Hz, 1H), 6.92 (t, J = 7.8 Hz, 1H), 6.81 (d, J = 8.7 Hz, 1H), 4.05–4.04 (m, 4H), 2.79 (d, J = 6.9 Hz, 2H), 2.24 (s, 3H), 2.19 (s, 3H), 2.11–2.04 (m, 1H), 1.89–1.86 (m, 4H), 1.66–1.64 (m, 2H), 1.51–1.40 (m, 4H), 1.05–1.02 (m, 2H). 13C NMR (DMSO): δ 204.1, 167.5, 158.1, 157.4, 136.7, 132.8, 132.2, 130.6, 127.4, 125.5, 121.7, 120.0, 113.4, 107.9, 67.8, 67.5, 47.6, 36.1, 32.0, 25.5, 24.5, 16.5, 11.5. ESI-MS m/z 425 [M + H]+. HRMS m/z calcd for C26H32O5 [M + H]+: 425.2323. Found: 425.2357.
2-(4-(3-Hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy)butoxy)benzoic Acid (28)
Colorless solid (0.164 g, 41%); mp 101–103 °C. 1H NMR (DMSO): δ 7.78 (d, J = 8.7 Hz, 1H), 7.59 (d, J = 9.6 Hz, 1H), 7.42–7.41 (m, 1H), 7.06 (d, J = 8.3 Hz, 1H), 6.95 (t, J = 7.4 Hz, 1H), 6.60 (d, J = 9.2 Hz, 1H), 4.11–4.07 (m, 4H), 2.79 (d, J = 6.9 Hz, 2H), 2.09–2.02 (m, 1H), 1.95 (s, 3H), 1.89–1.86 (m, 4H), 0.90 (d, J = 6.4 Hz, 6H). 13C NMR (CDCl3): δ 205.6, 167.4, 162.5, 161.3, 157.4, 132.9, 130.6, 130.3, 121.6, 120.0, 113.4, 111.9, 103.3, 67.9, 67.8, 45.8, 36.3, 25.4, 22.4, 7.5. ESI-MS m/z 401 [M + H]+. HRMS m/z calcd for C23H28O6 [M + H]+: 401.1959. Found: 401.1990.
2-(4-(4-(2-Cyclopentylacetyl)-3-hydroxy-2-methylphenoxy)butoxy)benzoic Acid (29)
Colorless solid (0.158 g, 37%); mp 93–95 °C. 1H NMR (DMSO): δ 7.78 (d, J = 8.7 Hz, 1H), 7.62 (d, J = 9.6 Hz, 1H), 7.45 (t, J = 7.6 Hz, 1H), 7.10 (d, J = 8.2 Hz, 1H), 6.69 (t, J = 7.3 Hz, 1H), 6.63 (d, J = 9.2 Hz, 1H), 4.14–4.11 (m, 4H), 2.98 (d, J = 6.8 Hz, 2H), 2.23–2.18 (m, 1H), 1.97 (s, 3H), 1.92–1.87 (m, 4H), 1.75–1.71 (m, 2H), 1.58–1.47 (m, 4H), 1.15–1.11 (m, 2H). 13C NMR (DMSO): δ 205.8, 167.4, 162.5, 161.3, 157.4, 132.9, 130.6, 130.3, 121.6, 120.0, 113.4, 113.2, 103.3, 111.9, 67.8, 67.9, 43.9, 36.3, 32.0, 25.6, 24.4, 7.5. ESI-MS m/z 427 [M + H]+. HRMS m/z calcd for C25H30O6 [M + H]+: 427.2115. Found: 427.2235.
2-Chloro-4-(4-(2,3-dimethyl-4-(3-methylbutanoyl)phenoxy)butoxy)benzoic Acid (30)
Colorless solid (0.078 g, 18%); mp 77–79 °C. 1H NMR (DMSO): δ 7.72–7.68 (m, 1H), 7.09 (d, J = 7.8 Hz, 1H), 6.91–4–6.91(m, 3H), 4.06–4.03 (m, 4H), 2.62 (d, J = 6.9 Hz, 2H), 2.11 (s, 3H), 2.01 (s, 3H), 1.88–1.86 (m, 4H), 1.74–1.72 (m, 1H), 0.78 (d, J = 6.4 Hz, 6H). 13C NMR (DMSO): δ 204.1, 167.5, 162.0, 155.6, 143.3, 134.1, 133.9, 131.8, 131.7, 126.0, 125.9, 122.3, 117.1, 113.8, 75.8, 69.9, 51.7, 26.1, 25.8, 25.2, 22.8, 20.6, 12.5. ESI-MS m/z 433 [M + H]+. HRMS m/z calcd for C24H29ClO5 [M + H]+: 433.1776. Found: 433.1799.
2-Chloro-4-(4-(4-(2-cyclopentylacetyl)-2,3-dimethylphenoxy)butoxy)benzoic Acid (31)
Colorless solid (0.115 g, 25%); mp 97–99 °C. 1H NMR (DMSO): δ 7.68 (d, J = 8.2 Hz, 1H), 7.38 (d, J = 8.7 Hz, 1H), 6.88 (s, 1H), 6.88 (d, J = 8.7 Hz, 1H), 6.74 (d, J = 8.7 Hz, 1H), 4.07–4.03 (m, 4H), 2.71 (d, J = 6.9 Hz, 2H), 2.10 (s, 3H), 1.98–1.94 (m, 1H), 1.95 (s, 3H), 1.79–1.77 (m, 4H), 1.66–1.64 (m, 2H), 1.51–1.40 (m, 4H), 1.05–1.02 (m, 2H). 13C NMR (DMSO): δ 205.8, 166.0, 162.5, 161.4, 133.2, 130.4, 122.3, 116.6, 113.5, 111.7, 103.4, 68.0, 67.8, 46.0, 26.1, 25.6, 25.3, 25.1, 22.5, 7.5. ESI-MS m/z 459 [M + H]+. HRMS m/z calcd for C26H31ClO5 [M + H]+: 459.1933. Found: 459.1942.
2-Chloro-4-(4-(3-hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy)butoxy)benzoic Acid (32)
Colorless solid (0.191 g, 44%); mp 113–115 °C. 1H NMR (DMSO): δ 7.78 (d, J = 8.7 Hz, 2H), 7.00 (s, 1H), 6.94 (d, J = 8.7 Hz, 1H), 6.60 (d, J = 9.2 Hz, 1H), 4.10–4.08 (m, 4H), 2.80 (d, J = 6.9 Hz, 2H), 2.13–2.01 (m, 1H), 1.92 (s, 3H), 1.89–1.86 (m, 4H), 0.90 (d, J = 6.4 Hz, 6H). 13C NMR (DMSO): δ 205.8, 166.0, 162.5, 161.4, 133.2, 130.4, 122.3, 116.6, 113.5, 111.7, 103.4, 68.0, 67.8, 46.0, 26.1, 25.6, 25.3, 25.1, 22.5, 7.5. ESI-MS m/z 435 [M + H]+. HRMS m/z calcd for C23H27ClO6 [M + H]+: 435.1569. Found: 435.1563.
2-Chloro-4-(4-(4-(2-cyclopentylacetyl)-3-hydroxy-2-methylphenoxy)butoxy)benzoic Acid (33)
Colorless solid (0.168 g, 30%); mp 130–132 °C. 1H NMR (DMSO): δ 7.78 (d, J = 8.7 Hz, 2H), 7.03 (s, 1H), 7.00 (d, J = 9.2 Hz, 1H), 6.58 (d, J = 9.2 Hz, 1H), 4.10–4.08 (m, 4H), 2.95 (d, J = 7.3 Hz, 2H), 2.21–2.19 (m, 1H), 1.94 (s, 3H), 1.89–1.86 (m, 4H), 1.77–1.71 (m, 2H), 1.56–1.45 (m, 4H), 1.13.1.10 (m, 2H). 13C NMR (DMSO): δ 205.8, 166.0, 162.5, 161.4, 133.2, 130.4, 122.3, 116.6, 113.5, 111.7, 103.4, 68.0, 67.8, 46.0, 26.1, 25.6, 25.3, 25.1, 22.5, 7.5. ESI-MS m/z 461 [M + H]+. HRMS m/z calcd for C25H29ClO6 [M + H]+: 461.1725. Found: 461.1721.
5-(4-(2,3-Dimethyl-4-(3-methylbutanoyl)phenoxy)butoxy)-2-fluorobenzoic Acid (34)
Colorless solid (0.133 g, 32%). 1H NMR (DMSO): δ 7.27–7.15 (m, 4H), 6.94 (d, J = 7.8 Hz, 1H), 3.98–3.68 (m, 4H), 2.71 (d, J = 6.9 Hz, 2H), 2.19 (s, 3H), 2.09 (s, 3H), 2.05–1.95 (m, 1H), 1.89–1.81 (m, 4H), 0.80 (d, J = 6.4 Hz, 6H). 13C NMR (DMSO): δ 202.9, 155.2, 154.4, 142.2, 131.8, 130.5, 126.0, 125.3, 120.6, 118.0, 117.7, 116.1, 74.5, 68.1, 51.1, 26.3, 25.5, 24.6, 22.4, 20.1, 12.0. ESI-MS m/z 417 [M + H]+. HRMS m/z calcd for C25H29FO5 [M + H]+: 417.2072. Found: 417.2116.
5-(4-(4-(2-Cyclopentylacetyl)-2,3-dimethylphenoxy)butoxy)-2-fluorobenzoic Acid (35)
Colorless solid (0.177 g, 40%); mp 118–120 °C. 1H NMR (DMSO): δ 7.44 (d, J = 8.2 Hz, 1H), 7.27–7.11 (m, 3H), 6.77 (d, J = 8.7 Hz, 1H), 3.98–3.70 (m, 4H), 2.77 (d, J = 7.3 Hz, 2H), 2.18 (s, 3H), 2.10–2.01 (m, 1H), 2.02 (s, 3H), 1.89–1.83 (m, 4H), 1.67–1.64 (m, 2H), 1.48–1.38 (m, 4H), 1.05–1.03 (m, 2H). 13C NMR (DMSO): δ 204.0, 165.0, 158.1, 154.3, 136.8, 132.2, 127.3, 125.6, 117.8, 117.6, 116.0, 107.9, 67.9, 67.4, 47.6, 36.1, 32.0, 25.6, 24.5, 16.5, 11.4. ESI-MS m/z 443 [M + H]+. HRMS m/z calcd for C26H31FO5 [M + H]+: 443.2228. Found: 443.2214.
2-Fluoro-5-(4-(3-hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy)butoxy)benzoic Acid (36)
Colorless solid (0.188 g, 45%); mp 99–101 °C. 1H NMR (DMSO): δ 7.76 (d, J = 9.2 Hz, 1H), 7.28–7.15 (m, 3H), 6.58 (d, J = 9.2 Hz, 1H), 4.10–4.03 (m, 4H), 2.78 (d, J = 7.3 Hz, 2H), 2.10–2.01 (m, 1H), 1.94 (s, 3H), 1.87–1.84 (m, 4H), 0.89 (d, J = 6.9 Hz, 6H). 13C NMR (DMSO): δ 205.7, 165.0, 162.5, 161.4, 154.4, 130.4, 121.3, 117.8, 116.0, 113.5, 112.0, 103.4, 67.9, 67.8, 46.0, 25.6, 25.5, 22.5, 7.5. ESI-MS m/z 419 [M + H]+. HRMS m/z calcd for C23H27FO6 [M + H]+: 419.1864. Found: 419.1863.
5-(4-(4-(2-Cyclopentylacetyl)-3-hydroxy-2-methylphenoxy)butoxy)-2-fluorobenzoic Acid (37)
Colorless solid (0.208 g, 47%); mp 110–112 °C. 1H NMR (DMSO): δ 7.76 (d, J = 9.2 Hz, 1H), 7.28 (d, J = 8.7 Hz, 1H), 7.15–7.13 (m, 2H), 6.59 (d, J = 9.2 Hz, 1H), 4.10–4.03 (m, 4H), 2.94 (d, J = 7.3 Hz, 2H), 2.20–2.10 (m, 1H), 1.94 (s, 3H), 1.87–1.84 (m, 4H), 1.72–1.70 (m, 2H), 1.56–1.44 (m, 4H), 1.12–1.01 (m, 2H). 13C NMR (DMSO): δ 206.5, 165.0, 162.5, 161.3, 154.4, 130.3, 121.3, 118.0, 117.8, 116.0, 113.3, 112.0, 103.3, 67.9, 67.8, 44.0, 36.0, 32.1, 25.5, 25.4, 24.5, 7.5. ESI-MS m/z 445 [M + H]+. HRMS m/z calcd for C25H29FO6 [M + H]+: 445.2021. Found: 445.2027.
3-{4-[3-Hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy]butoxy}-4-methylbenzoic Acid (38)
Colorless solid (0.277 g, 67%); mp 151–153 °C. 1H NMR (DMSO): δ 7.79 (d, J = 9.2 Hz, 1H), 7.43–7.41 (m, 2H), 7.20 (d, J = 7.8 Hz, 1H), 6.60 (d, J = 8.7 Hz, 1H), 4.14–4.10 (m, 4H), 2.81 (d, J = 6.9 Hz, 2H), 2.15 (s, 3H), 2.13–2.12 (m, 1H), 1.97 (s, 3H), 1.91–1.89 (m, 4H), 0.90 (d, J = 6.9 Hz, 6H). 13C NMR (DMSO): δ 205.8, 167.4, 162.5, 161.3, 157.4, 132.9, 130.6, 130.3, 121.6, 120.0, 113.4, 113.2, 111.9, 103.3, 67.9, 67.8, 43.9, 36.3, 32.0, 25.6, 24.4, 7.5. ESI-MS m/z 415 [M + H]+. HRMS m/z calcd for C24H30O6 [M + H]+: 415.2115. Found: 415.2009.
3-{4-[4-(2-Cyclopentylacetyl)-3-hydroxy-2-methylphenoxy]butoxy}-4-methylbenzoic Acid (39)
Colorless solid (0.277 g, 63%). 1H NMR (DMSO): δ 7.79 (d, J = 9.2 Hz, 1H), 7.42–7.40 (m, 2H), 7.20 (d, J = 7.3 Hz, 1H), 6.59 (d, J = 9.2 Hz, 1H), 4.16–4.08 (m, 4H), 2.95 (d, J = 7.3 Hz, 2H), 2.23–2.21 (m, 1H), 2.15 (s, 3H), 1.96 (s, 3H), 1.94–1.90 (m, 4H), 1.73–1.71 (m, 2H), 1.57–1.45 (m, 4H), 1.17–1.14 (m, 2H). 13C NMR (DMSO): δ 205.8, 167.3, 162.4, 161.2, 156.4, 131.3, 130.3, 129.6, 121.4, 113.2, 111.9, 111.2, 103.3, 67.8, 67.2, 43.2, 36.3, 32.0, 25.4, 24.4, 16.1, 7.5. ESI-MS m/z 441 [M + H]+. HRMS m/z calcd for C26H32O6 [M + H]+: 441.2272. Found: 441.2245.
4-{4-[3-Hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy]butoxy}-3-methylbenzoic Acid (40)
Colorless solid (0.282 g, 68%); mp 125–127 °C. 1H NMR (DMSO): δ 7.78 (d, J = 9.2 Hz, 1H), 7.73–7.71 (m, 1H), 7.67 (d, J = 1.4 Hz, 1H), 6.96 (d, J = 8.7 Hz, 1H), 6.59 (d, J = 9.2 Hz, 1H), 4.13–4.10 (m, 4H), 2.80 (d, J = 6.9 Hz, 2H), 2.11 (s, 3H), 2.10–2.07 (m, 1H), 1.93 (s, 3H), 1.92–1.89 (m, 4H), 0.89 (d, J = 6.4 Hz, 6H). 13C NMR (DMSO): δ 205.7, 167.2, 162.4, 161.3, 160.2, 131.5, 130.3, 129.2, 125.7, 122.3, 113.4, 111.9, 110.6, 103.3, 67.8, 67.4, 45.9, 25.4, 22.4, 15.8, 7.5. ESI-MS m/z 415 [M + H]+. HRMS m/z calcd for C24H30O6 [M + H]+: 415.2115. Found: 415.2084.
4-{4-[4-(2-Cyclopentylacetyl)-3-hydroxy-2-methylphenoxy]butoxy}-3-methylbenzoic Acid (41)
Colorless solid (0.273 g, 62%). 1H NMR (DMSO): δ 7.78 (d, J = 8.7 Hz, 1H), 7.73 (d, J = 8.7 Hz, 1H), 7.68 (s, 1H), 6.95 (d, J = 8.7 Hz, 1H), 6.58 (d, J = 8.7 Hz, 1H), 4.12–4.08 (m, 4H), 2.94 (d, J = 6.9 Hz, 2H), 2.20–2.01 (m, 1H), 2.12 (s, 3H), 1.93 (s, 3H), 1.90–1.89 (m, 4H), 1.75–1.73 (m, 2H), 1.55–1.44 (m, 4H), 1.13–1.01 (m, 2H). 13C NMR (DMSO): δ 205.8, 167.2, 162.4, 161.2, 160.2, 131.5, 130.3, 129.2, 125.7, 122.3, 113.2, 111.9, 110.6, 103.3, 67.8, 67.4, 43.2, 36.3, 32.0, 25.4, 24.4, 15.8, 7.4. ESI-MS m/z 441 [M + H]+. HRMS m/z calcd for C26H32O6 [M + H]+: 441.2272. Found: 441.2270.
3-Fluoro-4-{4-[3-hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy]butoxy}benzoic Acid (42)
Colorless solid (0.188 g, 45%). 1H NMR (DMSO): δ 7.80 (d, J = 8.7 Hz, 1H), 7.70 (d, J = 9.2 Hz, 1H), 7.61–7.59 (m, 1H), 7.23 (t, J = 8.5 Hz, 1H), 6.60 (d, J = 9.2 Hz, 1H), 4.18–4.13 (m, 4H), 2.81 (d, J = 6.9 Hz, 2H), 2.20–2.10 (m, 1H), 1.93 (s, 3H), 1.89–1.87 (m, 4H), 0.90 (d, J = 6.4 Hz, 6H). 13C NMR (DMSO): δ 206.3, 166.7, 163.0, 161.8, 150.8, 130.9, 127.2, 117.1, 114.7, 114.0, 112.5, 103.9, 69.0, 68.2, 46.5, 26.0, 25.7, 22.9, 8.0. ESI-MS m/z 419 [M + H]+. HRMS m/z calcd for C23H27FO6 [M + H]+: 419.1864. Found: 419.1848.
4-{4-[4-(2-Cyclopentylacetyl)-3-hydroxy-2-methylphenoxy]butoxy}-3-fluorobenzoic Acid (43)
Colorless solid (0.213 g, 48%); mp 160–162 °C. 1H NMR (DMSO): δ 7.79 (d, J = 9.2 Hz, 1H), 7.70–7.76 (m, 1H), 7.60–7.58 (m, 1H), 7.22 (t, J = 8.7 Hz, 1H), 6.58 (d, J = 9.2 Hz, 1H), 4.17–4.12 (m, 4H), 2.94 (d, J = 7.3 Hz, 2H), 2.20–2.18 (m, 1H), 1.92 (s, 3H), 1.89–1.87 (m, 4H), 1.72–1.69 (m, 2H), 1.55–1.44 (m, 4H), 1.12–1.10 (m, 2H). 13C NMR (DMSO): δ 206.3, 166.7, 162.9, 161.8, 150.8, 130.8, 127.2, 123.8, 117.0, 114.7, 113.8, 112.5, 103.8, 69.0, 68.2, 43.8, 36.8, 32.6, 25.7, 25.0, 8.0. ESI-MS m/z 445 [M + H]+. HRMS m/z calcd for C25H29FO6 [M + H]+: 445.2021. Found:.445.2014.
4-{4-[3-Hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy]butoxy}-3-methoxybenzoic Acid (44)
Colorless solid (0.301 g, 70%); mp 133–135 °C. 1H NMR (DMSO): δ 7.78 (d, J = 8.7 Hz, 1H), 7.53–7.51 (m, 1H), 7.40 (d, J = 1.8 Hz, 1H), 7.00 (d, J = 8.2 Hz, 1H), 6.59 (d, J = 9.2 Hz, 1H), 4.13–4.07 (m, 4H), 3.76 (s, 3H), 2.83 (d, J = 6.9 Hz, 2H), 2.12–2.10 (m, 1H), 1.94 (s, 3H), 1.90–1.88 (m, 4H), 0.89 (d, J = 6.9 Hz, 6H). 13C NMR (DMSO): δ 205.7, 167.1, 162.5, 161.3, 151.9, 148.4, 130.3, 122.9, 113.4, 111.9, 103.3, 67.9, 67.8, 55.4, 45.9, 25.4, 22.4, 7.5. ESI-MS m/z 431 [M + H]+. HRMS m/z calcd for C24H30O7 [M + H]+: 431.2064. Found: 431.2061.
4-{4-[4-(2-Cyclopentylacetyl)-3-hydroxy-2-methylphenoxy]butoxy}-methoxybenzoic Acid (45)
Colorless solid (0.292 g, 64%). 1H NMR (DMSO): δ 7.80 (d, J = 9.2 Hz, 1H), 7.50–7.49 (m, 1H), 7.39 (d, J = 1.8 Hz, 1H), 7.00 (d, J = 8.7 Hz, 1H), 6.60 (d, J = 9.2 Hz, 1H), 4.14–4.07 (m, 4H), 3.75 (s, 3H), 2.95 (d, J = 6.9 Hz, 2H), 2.21–2.19 (m, 1H), 1.94 (s, 3H), 1.89–1.87 (m, 4H), 1.74–1.72 (m, 2H), 1.56–1.45 (m, 4H), 1.14–1.12 (m, 2H). 13C NMR (DMSO): δ 205.8, 167.1, 162.4, 161.2, 151.8, 148.4, 130.3, 123.1, 113.2, 112.1, 111.9, 103.3, 67.9, 67.8, 55.5, 43.2, 36.3, 32.0, 25.5, 25.2, 24.4, 7.5. ESI-MS m/z 457 [M + H]+. HRMS m/z calcd for C26H32O7 [M + H]+: 457.2221. Found: 457.2218.
3-Chloro-4-{4-[3-hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy]butoxy}benzoic Acid (46)
Colorless solid (0.252 g, 58%); mp 147–149 °C. 1H NMR (DMSO): δ 7.86–7.84 (m, 3H), 7.21 (d, J = 8.7 Hz, 1H), 6.61 (d, J = 9.2 Hz, 1H), 4.20–4.15 (m, 4H), 2.81 (d, J = 6.9 Hz, 2H), 2.21–2.10 (m, 1H), 1.93 (s, 3H), 1.92–1.89 (m, 4H), 0.90 (d, J = 6.4 Hz, 6H). 13C NMR (DMSO): δ 206.0, 166.6, 163.0, 161.8, 150.7, 130.9, 127.2, 117.1, 114.9, 114.0, 112.5, 103.8, 69.0, 68.1, 46.3, 26.0, 25.8, 22.4, 8.0. ESI-MS m/z 435 [M + H]+. HRMS m/z calcd for C23H27ClO6 [M + H]+: 435.1569. Found: 435.1557.
3-Chloro-4-{4-[4-(2-cyclopentylacetyl)-3-hydroxy-2-methylphenoxy]butoxy}benzoic Acid (47)
Colorless solid (0.253 g, 55%). 1H NMR (DMSO): δ 7.87–7.82 (m, 3H)), 7.23 (d, J = 8.2 Hz, 1H), 6.62 (d, J = 9.2 Hz, 1H), 4.22–4.17 (m, 4H), 2.98 (d, J = 6.9 Hz, 2H), 2.23–2.21 (m, 1H), 1.95 (s, 3H), 1.94–1.91 (m, 4H), 1.76–1.74 (m, 2H), 1.56–1.44 (m, 4H), 1.18–1.15 (m, 2H). 13C NMR (DMSO): δ 206.0, 166.7, 162.9, 161.8, 151.0, 130.8, 127.2, 123.7, 117.0, 114.6, 113.9, 112.5, 103.8, 69.1, 68.2, 43.7, 36.8, 32.7, 25.7, 25.1, 8.0. ESI-MS m/z 461 [M + H]+. HRMS m/z calcd for C25H29ClO6 [M + H]+: 461.1725. Found: 461.1720.
4-{4-[3-Hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy]butoxy}-2-methylbenzoic Acid (48)
Colorless solid (0.269 g, 65%): mp 136–138 °C. 1H NMR (DMSO): δ 7.79 (d, J = 9.6 Hz, 2H), 6.78–6.76 (m, 2H), 6.59 (d, J = 9.2 Hz, 1H), 4.11–4.06 (m, 4H), 2.80 (d, J = 7.3 Hz, 2H), 2.45 (s, 3H), 2.10–2.01 (m, 1H), 1.94 (s, 3H), 1.86–1.84 (m, 4H), 0.89 (d, J = 6.9 Hz, 6H). 13C NMR (DMSO): δ 205.7, 168.0, 162.4, 161.3, 161.1, 142.1, 132.8, 130.3, 122.0, 117.2, 113.4, 111.9, 111.5, 103.3, 67.7, 67.2, 45.9, 25.3, 22.4, 21.8, 7.5. ESI-MS m/z 415 [M + H]+. HRMS m/z calcd for C24H30O6 [M + H]+: 415.2115. Found: 415.2071.
4-{4-[4-(2-Cyclopentylacetyl)-3-hydroxy-2-methylphenoxy]butoxy}-2-methylbenzoic Acid (49)
Colorless solid (0.273 g, 62%). 1H NMR (DMSO): δ 7.78 (d, J = 9.2 Hz, 2H), 6.77–6.76 (m, 2H), 6.58 (d, J = 8.7 Hz, 1H), 4.11–4.05 (m, 4H), 2.94 (d, J = 7.3 Hz, 2H), 2.46 (s, 3H), 2.20–2.01 (m, 1H), 1.94 (s, 3H), 1.86–1.84 (m, 4H), 1.74–1.71 (m, 2H), 1.56–1.45 (m, 4H), 1.13–1.01 (m, 2H). 13C NMR (DMSO): δ 205.8, 168.0, 162.4, 161.1, 142.1, 132.8, 130.3, 129.6, 122.0, 117.2, 113.2, 111.9, 111.5, 103.3, 67.7, 67.2, 43.2, 36.3, 32.0, 25.2, 24.4, 21.8, 7.5. ESI-MS m/z 441 [M + H]+. HRMS m/z calcd for C26H32O6 [M + H]+: 441.2272. Found: 441.2248.
3-{4-[3-Hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy]butoxy}-4-methoxybenzoic Acid (50)
Colorless solid (0.254 g, 59%); mp 143–145 °C. 1H NMR (DMSO): δ 7.80 (d, J = 8.7 Hz, 1H), 7.51 (d, J = 8.2 Hz, 1H), 7.41 (d, J = 2.3 Hz, 1H), 7.00 (d, J = 8.7 Hz, 1H), 6.60 (d, J = 9.2 Hz, 1H), 4.14–4.04 (m, 4H), 3.77 (s, 3H), 2.81 (d, J = 6.9 Hz, 2H), 2.10–2.01 (m, 1H), 1.95 (s, 3H), 1.88–1.84 (m, 4H), 0.90 (d, J = 6.9 Hz, 6H). 13C NMR (DMSO): δ 205.7, 167.1, 162.5, 161.3, 152.7, 147.5, 130.3, 123.0, 113.4, 111.9, 111.1, 103.3, 67.9, 55.7, 45.9, 25.5, 22.4, 7.5. ESI-MS m/z 431 [M + H]+. HRMS m/z calcd for C24H30O7 [M + H]+: 431.2064. Found: 431.2060.
3-{4-[4-(2-Cyclopentylacetyl)-3-hydroxy-2-methylphenoxy]butoxy}-4-methoxybenzoic Acid (51)
Colorless solid (0.273 g, 60%). 1H NMR (DMSO): δ 7.80 (d, J = 9.2 Hz, 1H), 7.51 (d, J = 8.2 Hz, 1H), 7.41 (d, J = 1.8 Hz, 1H), 6.99 (d, J = 8.7 Hz, 1H), 6.60 (d, J = 9.2 Hz, 1H), 4.14–4.04 (m, 4H), 3.77 (s, 3H), 2.95 (d, J = 6.9 Hz, 2H), 2.21–2.10 (m, 1H), 1.94 (s, 3H), 1.88–1.84 (m, 4H), 1.72–1.69 (m, 2H), 1.56–1.45 (m, 4H), 1.14–1.12 (m, 2H). 13C NMR (DMSO): δ 205.8, 167.1, 162.4, 161.2, 152.7, 147.5, 130.3,123.1, 113.2, 111.9, 111.1, 103.3, 67.9, 55.7, 43.2, 36.3, 32.0, 25.4, 24.4, 7.5. ESI-MS m/z 457 [M + H]+. HRMS m/z calcd for C26H32O7 [M + H]+: 457.2221. Found: 457.2224.
4-(4-(4-Acetyl-3-hydroxy-2-methylphenoxy)butoxy)-3-methoxybenzoic Acid (52)
Colorless solid (0.255 g, 58%); mp 138–140 °C. 1H NMR (DMSO): δ 7.75 (d, J = 9.2 Hz, 1H), 7.48 (d, J = 8.2 Hz, 1H), 7.39 (s, 1H), 7.02 (d, J = 8.2 Hz, 1H), 6.62 (d, J = 8.7 Hz, 1H), 4.13–4.07 (m, 4H), 3.75 (s, 3H), 2.53 (s, 3H), 1.93 (s, 3H), 1.89–1.87 (m, 4H). 13C NMR (DMSO): δ 204.4, 167.7, 163.1, 161.5, 152.3, 148.9, 131.4, 123.6, 11.0, 112.6, 112.4, 112.3, 103.9, 68.5, 68.3, 55.9, 26.9, 26.0, 25.7, 8.0. LC–MS (ESI) calcd for C21H24O7 [M + H]+: 389.15. Found: 389.05. HRMS (ESI) calcd for C21H24O7 [M + H]+: 389.1595. Found: 389.1587.
3-(4-(4-Acetyl-3-hydroxy-2-methylphenoxy)butoxy)-4-methoxybenzoic Acid (53)
Colorless solid (0.264 g, 68%); mp 135–136 °C. 1H NMR (DMSO): δ 7.76 (d, J = 9.2 Hz, 1H), 7.50 (d, J = 10.0 Hz, 1H), 7.42 (s, 1H), 6.99 (d, J = 8.7 Hz, 1H), 6.60 (d, J = 9.2 Hz, 1H), 4.13–4.03 (m, 4H), 3.77 (s, 3H), 2.53 (s, 3H), 1.94 (s, 3H), 1.92–1.88 (m, 4H). 13C NMR (DMSO): δ 204.3, 167.6, 163.1, 161.5, 153.3, 148.0, 131.4, 123.7, 123.5, 114.0, 113.7, 112.3, 111.7, 103.9, 68.4, 56.2, 26.8, 25.9, 8.0. LC–MS (ESI) calcd for C21H24O7 [M + H]+: 389.15. Found: 389.00. HRMS (ESI) calcd for C21H24O7 [M + H]+: 389.1595. Found: 389.1585.
3-(4-(4-Acetyl-3-hydroxy-2-methylphenoxy)butoxy)-2-methylbenzoic Acid (54)
Colorless solid (0.204 g, 55%); mp 140–142 °C. 1H NMR (DMSO): δ 7.73 (d, J = 9.2 Hz, 1H), 7.27 (d, J = 8.7 Hz, 1H), 7.16 (t, J = 7.8 Hz, 1H), 7.16 (d, J = 7.8 Hz, 1H), 6.58 (d, J = 8.7 Hz, 1H), 4.11–4.02 (m, 4H), 2.52 (s, 3H), 2.28 (s, 3H), 1.93 (s, 3H), 1.89–1.87 (m, 4H). 13C NMR (DMSO): δ 204.3, 169.7, 163.0, 161.5, 157.4, 133.2, 131.4, 127.1, 126.7, 121.9, 114.8, 114.0, 112.3, 103.8, 68.3, 68.1, 26.8, 25.9, 13.1, 8.0. LC–MS (ESI) calcd for C21H24FO6 [M + H]+: 373.16. Found: 373.00. HRMS (ESI) calcd for C21H24O6 [M + Na]+: 395.1465. Found: 395.1464.
5-(4-(4-Acetyl-3-hydroxy-2-methylphenoxy)butoxy)-2-fluorobenzoic Acid (55)
Colorless solid (0.229 g, 0.253 g, 61%); mp 102–104 °C. 1H NMR (DMSO): δ 7.75 (d, J = 8.7 Hz, 1H), 7.26–7.13 (m, 3H), 6.59 (d, J = 9.2 Hz, 1H), 4.11–4.03 (m, 4H), 2.52 (s, 3H), 1.94 (s, 3H), 1.92–1.86 (m, 4H). 13C NMR (DMSO): δ 204.3, 165.4, 163.0, 161.5, 154.8, 154.6, 131.4, 118.4, 118.2, 116.5, 114.0, 112.3, 103.8, 68.3, 26.9, 25.8, 8.0. LC–MS (ESI) calcd for C20H21FO6 [M + H]+: 377.13. Found: 377.00. HRMS (ESI) calcd for C20H21FO6 [M + H]+: 377.1395. Found: 377.1395.
4-(4-(4-Acetyl-3-hydroxy-2-methylphenoxy)butoxy)-3-methylbenzoic Acid (56)
Colorless solid (0.201 g, 54%); mp 188–190 °C. 1H NMR (DMSO): δ 7.70–7.65 (m, 2H), 7.64 (s, 1H), 6.93 (d, J = 8.7 Hz, 1H), 6.57 (d, J = 9.2 Hz, 1H), 4.09–4.04 (m, 4H), 2.49 (s, 3H), 2.08 (s, 3H), 1.89 (s, 3H), 1.88–1.86 (m, 4H). 13C NMR (DMSO): δ 204.4, 167.7, 163.0, 161.5, 160.8, 132.0, 131.4, 129.7, 126.2, 122.8, 114.0, 112.3, 112.2, 108.8, 68.3, 67.9, 26.8, 25.9, 25.8, 16.4, 8.0. LC–MS (ESI) calcd for C21H24O6 [M + H]+: 373.16. Found: 373.00. HRMS (ESI) calcd for C21H24O6 [M + Na]+: 395.1465. Found: 395.1463.
4-(4-(3-Hydroxy-2-methyl-4-propionylphenoxy)butoxy)-3-methoxybenzoic Acid (57)
Colorless solid (0.253 g, 63%); mp 128–130 °C. 1H NMR (DMSO): δ 7.80 (d, J = 9.2 Hz, 1H), 7.49 (d, J = 8.2 Hz, 1H), 7.40 (s, 1H), 7.01 (d, J = 8.7 Hz, 1H), 6.60 (d, J = 9.2 Hz, 1H), 4.13–4.03 (m, 4H), 3.74 (s, 3H), 2.99 (q, J = 7.5 Hz, 2H), 1.96 (s, 3H), 1.90–1.88 (m, 4H), 1.07 (t, J = 7.3 Hz, 3H). 13C NMR (DMSO): δ 206.8, 162.9, 161.5, 152.3, 148.9, 130.5, 123.6, 113.5, 112.6, 112.4, 103.9, 98.5, 68.5, 66.9, 66.2, 60.6, 60.0, 31.1, 26.0, 25.7, 8.9, 8.0. LC–MS (ESI) calcd for C22H26O7 [M + H]+: 403.17. Found: 403.00. HRMS (ESI) calcd for C22H26O7 [M + Na]+: 425.1571. Found: 425.1568.
3-(4-(3-Hydroxy-2-methyl-4-propionylphenoxy)butoxy)-4-methoxybenzoic Acid (58)
Colorless solid (0.293 g, 73%); mp 120–122 °C. 1H NMR (DMSO): δ 7.77 (d, J = 8.6 Hz, 1H), 7.50 (d, J = 10.5 Hz, 1H), 7.42 (s, 1H), 6.99 (d, J = 8.7 Hz, 1H), 6.60 (d, J = 9.1 Hz, 1H), 4.13–4.03 (m, 4H), 3.77 (s, 3H), 2.98 (q, J = 7.5 Hz, 2H), 1.95 (s, 3H), 1.90–1.88 (m, 4H), 1.05 (t, J = 7.3 Hz, 3H). 13C NMR (DMSO): δ 206.8, 167.7, 162.9, 161.5, 153.3, 148.0, 130.4, 123.7, 113.7, 113.4, 112.4, 111.6, 103.8, 68.5, 68.4, 56.2, 30.6, 25.5, 25.4, 8.9, 8.0. LC–MS (ESI) calcd for C22H26O7 [M + H]+: 403.17. Found: 403.35. HRMS (ESI) calcd for C22H26O7 [M + Na]+: 425.1571. Found: 425.1569.
3-(4-(3-Hydroxy-2-methyl-4-propionylphenoxy)butoxy)-2-methylbenzoic Acid (59)
Colorless solid (0.228 g, 59%); 103–105 °C. 1H NMR (DMSO): δ 7.77 (d, J = 9.2 Hz, 1H), 7.24 (d, J = 9.2 Hz, 1H), 7.16 (t, J = 7.8 Hz, 1H), 7.08 (d, J = 8.2 Hz, 1H), 6.59 (d, J = 9.2 Hz, 1H), 4.12–4.02 (m, 4H), 3.00 (q, J = 7.3 Hz, 2H), 2.28 (s, 3H), 1.94 (s, 3H), 1.90–1.89 (m, 4H), 1.05 (t, J = 7.4 Hz, 3H). 13C NMR (DMSO): δ 206.8, 169.7, 162.8, 161.5, 157.4, 133.2, 130.4, 127.1, 126.8, 121.9, 114.8, 113.5, 112.4, 103.8, 63.3, 31.8, 24.6, 13.1, 8.9, 8.0. LC–MS (ESI) calcd for C22H26O6 [M + H]+: 387.17. Found: 387.05. HRMS (ESI) calcd for C22H26O6 [M + H]+: 387.1802. Found: 387.1802.
2-Fluoro-5-(4-(3-hydroxy-2-methyl-4-propionylphenoxy)butoxy)benzoic Acid (60)
Colorless solid (0.254 g, 65%); mp 123–125 °C. 1H NMR (DMSO): δ 7.76 (d, J = 8.7 Hz, 1H), 7.26–7.13 (m, 3H), 6.59 (d, J = 9.2 Hz, 1H), 4.11–4.02 (m, 4H), 2.98 (q, J = 7.3 Hz, 2H), 1.93 (s, 3H), 1.90–1.86 (m, 4H), 1.05 (t, J = 7.3 Hz, 3H). 13C NMR (DMSO): δ 206.8, 165.5, 162.8, 161.5, 154.8, 154.6, 130.4, 118.4, 116.5, 113.5, 112.4, 103.8, 68.3, 31.1, 25.8, 8.9, 8.0. LC–MS (ESI) calcd for C21H23FO6 [M + H]+: 391.15. Found: 391.00. HRMS (ESI) calcd for C21H23FO6 [M + Na]+: 413.1371. Found: 413.1366.
4-(4-(3-Hydroxy-2-methyl-4-propionylphenoxy)butoxy)-3-methylbenzoic Acid (61)
Colorless solid (0.205 g, 53%); mp 178–180 °C. 1H NMR (DMSO): δ 7.80–7.78 (m, 2H), 7.70 (s, 1H), 6.99 (d, J = 8.7 Hz, 1H), 6.63 (d, J = 9.2 Hz, 1H), 4.15–4.10 (m, 4H), 3.02 (q, J = 7.3 Hz, 2H), 2.24 (s, 3H), 1.96 (s, 3H), 1.95–1.92 (m, 4H), 1.08 (t, J = 7.3 Hz, 3H). 13C NMR (DMSO): δ 206.8, 167.7, 162.8, 161.5, 160.8, 132.0, 130.5, 129.7, 126.2, 122.8, 113.5, 112.4, 111.2, 103.8, 68.3, 67.9, 31.1, 25.9, 25.8, 16.4, 8.9, 8.0. LC–MS (ESI) calcd for C22H26O6 [M + H]+: 387.17. Found: 387.05. HRMS (ESI) calcd for C22H26O6 [M + Na]+: 409.1622. Found: 409.1625.
4-(4-(3-Hydroxy-4-isobutyryl-2-methylphenoxy)butoxy)-3-methoxybenzoic Acid (62)
Colorless solid (0.283 g, 68%); mp 118–120 °C. 1H NMR (DMSO): δ 7.84 (d, J = 8.7 Hz, 1H), 7.49 (d, J = 10.5 Hz, 1H), 7.39 (s, 1H), 7.00 (d, J = 8.2 Hz, 1H), 6.61 (d, J = 9.2 Hz, 1H), 4.14–4.07 (m, 4H), 3.74 (s, 3H), 3.74–3.63 (m, 1H), 1.94 (s, 3H), 1.90–1.88 (m, 4H), 1.09 (d, J = 6.9 Hz, 6H). 13C NMR (DMSO): δ 210.3, 167.6, 163.0, 162.3, 152.4, 148.9, 130.5, 123.6, 123.5, 112.7, 112.6, 112.4, 112.2, 103.9, 68.5, 56.0, 34.8, 26.0, 25.7, 20.0, 8.1. LC–MS (ESI) calcd for C23H28O7 [M + H]+: 417.18. Found: 417.05. HRMS (ESI) calcd for C23H28O7 [M + Na]+: 439.1727. Found: 439.1725.
3-(4-(3-Hydroxy-4-isobutyryl-2-methylphenoxy)butoxy)-4-methoxybenzoic Acid (63)
Colorless solid (0.279 g, 67%); 82–84 °C. 1H NMR (DMSO): δ 7.84 (d, J = 9.2 Hz, 1H), 7.50 (d, J = 10.5 Hz, 1H), 7.42 (s, 1H), 6.99 (d, J = 8.7 Hz, 1H), 6.61 (d, J = 9.2 Hz, 1H), 4.14 (brs, 2H), 4.07 (brs, 2H), 3.77 (s, 3H), 3.74–3.63 (m, 1H), 1.95 (s, 3H), 1.88 (brs, 4H), 1.08 (d, J = 6.4 Hz, 6H). 13C NMR (DMSO): δ 210.2, 167.6, 163.0, 162.3, 153.3, 148.1, 130.4, 123.7, 113.7, 112.7, 112.2, 111.7, 103.9, 68.4, 56.2, 26.0, 19.9, 8.1. LC–MS (ESI) calcd for C23H28O7 [M + H]+: 417.18. Found: 417.05. HRMS (ESI) calcd for C23H28O7 [M + H]+: 417.1908. Found: 417.1895.
3-(4-(3-Hydroxy-4-isobutyryl-2-methylphenoxy)butoxy)-2-methylbenzoic Acid (64)
Colorless solid (0.264 g, 66%); mp 115–117 °C. 1H NMR (DMSO): δ 7.85 (d, J = 9.2 Hz, 1H), 7.28 (d, J = 6.8 Hz, 1H), 7.18 (t, J = 7.8 Hz, 1H), 7.10 (d, J = 7.3 Hz, 1H), 6.64 (d, J = 9.2 Hz, 1H), 4.16–4.05 (m, 4H), 3.66–3.54 (m, 1H), 2.03 (s, 3H), 1.93 (s, 3H), 1.91–1.88 (m, 4H), 1.10 (d, J = 6.8 Hz, 6H). 13C NMR (DMSO): δ 210.2, 169.7, 162.9, 157.4, 133.2, 130.4, 127.1, 126.8, 121.9, 114.8, 112.7, 112.2, 103.9, 68.3, 68.1, 34.3, 26.0, 19.9, 13.1, 8.0. LC–MS (ESI) calcd for C23H28O6[M + H]+: 401.19. Found: 401.05. HRMS (ESI) calcd for C23H28O6 [M + H]+: 401.1959. Found: 401.1948.
2-Fluoro-5-(4-(3-hydroxy-4-isobutyryl-2-methylphenoxy)butoxy)benzoic Acid (65)
Colorless solid (0.234 g, 58%); mp 122–124 °C. 1H NMR (DMSO): δ 7.82 (d, J = 9.2 Hz, 1H), 7.27–7.13 (m, 3H), 6.61 (d, J = 8.7 Hz, 1H), 4.12–4.02 (m, 4H), 3.64–3.46 (m, 1H), 1.94 (s, 3H), 1.90–1.86 (m, 4H), 1.08 (d, J = 6.9 Hz, 6H). 13C NMR (DMSO): δ 210.2, 165.5, 162.9, 162.3, 154.8, 154.6, 130.5, 118.2, 116.4, 112.7, 112.2, 103.9, 68.3, 33.6, 25.8, 19.9, 8.0. LC–MS (ESI) calcd for C22H25FO6 [M + H]+: 405.16. Found: 405.00. HRMS (ESI) calcd for C22H25FO6 [M + H]+: 405.1708. Found: 405.1705.
4-(4-(3-Hydroxy-4-isobutyryl-2-methylphenoxy)butoxy)-3-methylbenzoic Acid (66)
Colorless solid (0.224 g, 56%); mp 160–162 °C. 1H NMR (DMSO): δ 7.84 (d, J = 9.2 Hz, 1H), 7.75–7.71 (m, 1H), 7.76 (s, 1H), 6.98 (d, J = 8.7 Hz, 1H), 6.61 (d, J = 9.2 Hz, 1H), 4.14–4.09 (m, 4H), 3.71–3.61 (m, 1H), 2.12 (s, 3H), 1.94 (s, 3H), 1.94–1.90 (m, 4H), 1.08 (d, J = 5.5 Hz, 6H). 13C NMR (DMSO): δ 210.2, 167.7, 162.9, 162.3, 160.8, 132.0, 130.5, 129.7, 126.2, 122.8, 1127, 112.3, 111.2, 103.9, 68.3, 67.9, 34.6, 25.9, 19.9, 16.4, 8.0. LC–MS (ESI) calcd for C23H28O6 [M + H]+: 401.19. Found: 401.05. HRMS (ESI) calcd for C23H28O6 [M + Na]+: 423.1778. Found: 423.1778.
(4-(4-Butyryl-3-hydroxy-2-methylphenoxy)butoxy)-3-methoxybenzoic Acid (67)
Colorless solid (0.270 g, 65%); mp 148–150 °C. 1H NMR (DMSO): δ 7.80 (d, J = 9.2 Hz, 1H), 7.48 (d, J = 10.5 Hz, 1H), 7.39 (s, 1H), 7.00 (d, J = 8.2 Hz, 1H), 6.60 (d, J = 8.7 Hz, 1H), 4.13–4.07 (m, 4H), 3.75 (s, 3H), 2.93 (t, J = 7.5 Hz, 2H), 1.94 (s, 3H), 1.91–1.88 (m, 4H), 1.61–1.57 (m, 2H), 0.90 (t, J = 7.3 Hz, 3H). 13C NMR (DMSO): δ 206.4, 167.4, 162.9, 161.7, 152.4, 148.9, 130.7, 123.6, 123.5, 112.5, 112.4, 103.8, 68.5, 68.3, 55.9, 26.0, 25.7, 18.4, 14.1, 8.0. LC–MS (ESI) calcd for C23H28O7 [M + H]+: 417.18. Found: 417.00. HRMS (ESI) calcd for C23H28O7 [M + H]+: 417.1908. Found: 417.1903.
3-(4-(4-Butyryl-3-hydroxy-2-methylphenoxy)butoxy)-4-methoxybenzoic Acid (68)
Colorless solid (0.279 g, 67%); mp 129–131 °C. 1H NMR (DMSO): δ 7.79 (d, J = 9.1 Hz, 1H), 7.43 (d, J = 10.0 Hz, 1H), 7.43 (s, 1H), 7.02 (d, J = 8.7 Hz, 1H), 6.63 (d, J = 9.2 Hz, 1H), 4.13–4.07 (m, 4H), 3.79 (s, 3H), 2.99 (t, J = 7.3 Hz, 2H), 1.96 (s, 3H), 1.89–1.86 (m, 4H), 1.63–1.61 (m, 2H), 0.92 (d, J = 7.6 Hz, 3H). 13C NMR (DMSO): δ 206.4, 167.6, 162.9, 161.7, 153.3, 148.0, 130.6, 123.7, 123.4, 113.6, 112.4, 111.6, 103.8, 68.44, 68.41, 56.2, 26.0, 25.6, 18.4, 14.1, 8.0. LC–MS (ESI) calcd for C23H28O7 [M + H]+: 417.18. Found: 417.00. HRMS (ESI) calcd for C23H28O7 [M + H]+: 417.1908. Found: 417.1896.
3-(4-(4-Butyryl-3-hydroxy-2-methylphenoxy)butoxy)-2-methylbenzoic Acid (69)
Colorless solid (0.244 g, 61%); mp 97–99 °C. 1H NMR (DMSO): δ 7.78 (d, J = 8.7 Hz, 1H), 7.27 (d, J = 7.3 Hz, 1H), 7.17 (t, J = 8.2 Hz, 1H), 7.08 (d, J = 7.8 8 Hz, 1H), 6.59 (d, J = 9.2 Hz, 1H), 4.13–4.02 (m, 4H), 2.92 (t, J = 7.3 Hz, 2H), 2.28 (s, 3H), 1.94 (s, 3H), 1.89–1.86 (m, 4H), 1.61–1.57 (m, 2H), 0.89 (t, J = 7.8 Hz, 3H). 13C NMR (DMSO): δ 206.4, 169.6, 162.9, 161.7, 157.4, 133.1, 130.6, 127.1, 126.8, 121.8, 114.8, 113.7, 112.5, 103.8, 68.3, 68.2, 26.0, 18.4, 14.1, 13.1, 8.0. LC–MS (ESI) calcd for C23H28O6 [M + H]+: 401.19. Found: 401.05. HRMS (ESI) calcd for C23H28O6 [M + Na]+: 439.1727. Found: 439.1727.
5-(4-(4-Butyryl-3-hydroxy-2-methylphenoxy)butoxy)-2-fluorobenzoic Acid (70)
Colorless solid (0.255 g, 63%); mp 88–90 °C. 1H NMR (DMSO): δ 7.78 (d, J = 8.7 Hz, 1H), 7.27–7.13 (m, 3H), 6.59 (d, J = 9.2 Hz, 1H), 4.11–4.02 (m, 4H), 2.92 (t, J = 7.3 Hz, 2H), 1.94 (s, 3H), 1.89–1.87 (m, 4H), 1.61–1.59 (m, 2H), 0.89 (t, J = 7.3 Hz, 3H). 13C NMR (DMSO): δ 206.4, 165.4, 162.9, 161.7, 154.8, 154.6, 130.6, 118.2, 116.4, 113.6, 112.4, 103.8, 68.3, 25.8, 18.3, 14.1, 8.0. LC–MS (ESI) calcd for C22H25FO6 [M + H]+: 405.43. Found: 405.05. HRMS (ESI) calcd for C22H25FO6 [M + Na]+: 427.1527. Found: 427.1523.
4-(4-(4-Butyryl-3-hydroxy- 2-methylphenoxy)butoxy)-3-methylbenzoic Acid (71)
Colorless solid (0.232 g, 58%); mp 161–163 °C. 1H NMR (DMSO): δ 7.79 (d, J = 9.2 Hz, 1H), 7.71 (d, J = 8.7 Hz, 1H), 7.68 (s, 1H), 6.97 (d, J = 8.7 Hz, 1H), 6.59 (d, J = 8.7 Hz, 1H), 4.13–4.08 (m, 4H), 2.92 (t, J = 7.3 Hz, 2H), 2.12 (s, 3H), 1.94 (s, 3H), 1.90–1.88 (m, 4H), 1.63–1.61 (m, 2H), 0.89 (t, J = 7.3 Hz, 3H). 13C NMR (DMSO): δ 206.4, 167.7, 162.9, 161.7, 160.8, 132.0, 130.7, 129.7, 126.3, 122.8, 113.7, 112.5, 111.2, 103.9, 68.3, 67.9, 26.1, 25.9, 18.4, 16.3, 14.1, 8.0. LC–MS (ESI) calcd for C23H28O6 [M + H]+: 401.19. Found: 401.05. HRMS (ESI) calcd for C23H28O6 [M + Na]+: 423.1778. Found: 423.1778.
3-(4-(4-(3,3-Dimethylbutanoyl)-3-hydroxy-2-methylphenoxy)butoxy)-4-methoxybenzoic Acid (72)
Colorless solid (0.270 g, 61%); mp 140–142 °C. 1H NMR (DMSO): δ 7.84 (d, J = 9.2 Hz, 1H), 7.50 (d, J = 10.0 Hz, 1H), 7.49 (s, 1H), 7.00 (d, J = 8.7 Hz, 1H), 6.58 (d, J = 9.2 Hz, 1H), 4.13–4.03 (m, 4H), 3.79 (s, 3H), 2.80 (s, 2H), 1.94 (s, 3H), 1.89–1.87 (m, 4H), 0.96 (s, 9H). 13C NMR (DMSO): δ 206.4, 167.6, 163.1, 162.1, 153.3, 148.1, 131.8, 123.7, 123.5, 114.9, 113.6, 111.6, 103.6, 68.4, 56.2, 49.1, 32.1, 30.5, 26.0, 25.8, 8.0. LC–MS (ESI) calcd for C25H32O7 [M + H]+: 445.21. Found: 445.00. HRMS (ESI) calcd for C25H32O7 [M + Na]+: 467.2040. Found: 467.2041.
4-(4-(4-(3,3-Dimethylbutanoyl)-3-hydroxy-2-methylphenoxy)butoxy)-5-methoxycyclohexa-1,3-dienecarboxylic Acid (73)
Colorless solid (0.258 g, 58%); mp 133–135 °C. 1H NMR (DMSO): δ 7.86 (d, J = 9.2 Hz, 1H), 7.48 (d, J = 10.1 Hz, 1H), 7.39 (s, 1H), 7.01 (d, J = 8.7 Hz, 1H), 6.58 (d, J = 9.2 Hz, 1H), 4.13–4.07 (m, 4H), 3.77 (s, 3H), 2.80 (s, 2H), 1.94 (s, 3H), 1.89–1.87 (m, 4H), 0.96 (s, 9H). 13C NMR (DMSO): δ 206.4, 167.6, 163.1, 162.1, 152.4, 148.9, 131.8, 123.6, 114.9, 112.6, 112.4, 103.6, 68.4, 55.9, 49.1, 32.1, 30.5, 26.0, 25.8, 8.0. LC–MS (ESI) calcd for C25H32O7 [M + H]+: 445.21. Found: 445.00. HRMS (ESI) calcd for C25H32O7 [M + Na]+: 467.2040. Found: 467.2045.
3-(4-(4-(3,3-Dimethylbutanoyl)-3-hydroxy-2-methylphenoxy)butoxy)-2-methylbenzoic Acid (74)
Colorless solid (0.278 g, 65%); mp 123–125 °C. 1H NMR (DMSO): δ 7.84 (d, J = 8.7 Hz, 1H), 7.26 (d, J = 7.8 Hz, 1H), 7.16 (t, J = 8.2 Hz, 1H), 7.08 (d, J = 7.3 Hz, 1H), 6.58 (d, J = 9.2 Hz, 1H), 4.13–4.02 (m, 4H), 2.80 (s, 2H), 2.28 (s, 3H), 1.94 (s, 3H), 1.90–1.87 (m, 4H), 0.96 (s, 9H). 13C NMR (DMSO): δ 206.4, 169.7, 163.0, 162.1, 157.4, 131.1, 131.8, 127.1, 126.8, 121.9, 114.9, 114.8, 112.4, 103.6, 68.3, 66.8, 49.1, 32.1, 30.4, 13.1, 8.0. LC–MS (ESI) calcd for C25H32O7 [M + H]+: 429.22. Found: 429.00. HRMS (ESI) calcd for C25H32O6 [M + Na]+: 451.2091. Found: 451.2093.
5-(4-(4-(3,3-Dimethylbutanoyl)-3-hydroxy-2-methylphenoxy)butoxy)-2-fluorobenzoic Acid (75)
Colorless solid (0.258 g, 60%); mp 130–132 °C. 1H NMR (DMSO): δ 7.83 (d, J = 9.2 Hz, 1H), 7.27–7.13 (m, 3H), 6.57 (d, J = 9.2 Hz, 1H), 4.11–4.02 (m, 4H), 2.79 (s, 2H), 1.94 (s, 3H), 1.86–1.85 (m, 4H), 0.96 (s, 9H). 13C NMR (DMSO): δ 206.4, 165.5, 163.0, 162.1, 154.8, 154.6, 131.7, 121.2, 118.5, 116.4, 114.9, 112.4, 103.6, 68.4, 66.8, 49.1, 32.2, 30.4, 8.0. LC–MS (ESI) calcd for C24H29FO6 [M + H]+: 433.19. Found: 433.00. HRMS (ESI) calcd for C24H29FO6 [M + Na]+: 455.1840. Found: 455.1840.
mGlu receptor in Vitro Assays
Human embryonic kidney (HEK-293) cell lines coexpressing rat mGlu receptors 2, 3, 4, 6, 7, or 8 and G protein-coupled inwardly rectifying potassium (GIRK) channels55 were grown in growth medium containing 45% DMEM, 45% F-12, 10% FBS, 20 mM HEPES, 2 mM l-glutamine, antibiotic/antimycotic, nonessential amino acids, 700 μg/mL G418, and 0.6 μg/mL puromycin at 37 °C in the presence of 5% CO2. Cells expressing rat mGlu1 and mGlu5 receptor were cultured as described in Hemstapat et al.56 All cell culture reagents were purchased from Invitrogen Corp. (Carlsbad, CA) unless otherwise noted. Calcium assays were used to assess activity of compounds at mGlu1 and mGlu5, as previously described by Engers et al.57 Calcium assays at mGlu3 were performed as described for mGlu5 with the exception that TREx293 mGlu3 Gα15 cells were treated with tetracycline at 20 ng/mL for 20 h prior to assay.
Compound activity at group II (mGlu2 and mGlu3) and group III (mGlu4, mGlu6, mGlu7, and mGlu8) was assessed using thallium flux through GIRK channels, a method that has been described in detail.55 Briefly, cells were plated into 384-well, black-walled, clear-bottomed poly-d-lysine-coated plates at a density of (15 000 cells/20 μL)/well in DMEM containing 10% dialyzed FBS, 20 mM HEPES, and 100 units/mL penicillin/streptomycin (assay medium). Plated cells were incubated overnight at 37 °C in the presence of 5% CO2. The following day, the medium was exchanged from the cells to assay buffer [Hanks’ balanced salt solution (Invitrogen) containing 20 mM HEPES, pH 7.3] using an ELX405 microplate washer (BioTek), leaving 20 μL/well, followed by the addition of 20 μL/well FluoZin2-AM (330 nM final concentration) indicator dye (Invitrogen, prepared as a stock in DMSO and mixed in a 1:1 ratio with Pluronic acid F-127) in assay buffer. Cells were incubated for 1 h at room temperature, and the dye was exchanged to assay buffer using an ELX405, leaving 20 μL/well. Test compounds were diluted to 2 times their final desired concentration in assay buffer (0.3% DMSO final concentration). Agonists were diluted in thallium buffer [125 mM sodium bicarbonate (added fresh on the morning of the experiment), 1 mM magnesium sulfate, 1.8 mM calcium sulfate, 5 mM glucose, 12 mM thallium sulfate, and 10 mM HEPES, pH 7.3] at 5 times the final concentration to be assayed. Cell plates and compound plates were loaded onto a kinetic imaging plate reader (FDSS 6000 or 7000; Hamamatsu Corporation, Bridgewater, NJ). Appropriate baseline readings were taken (10 images at 1 Hz; excitation, 470 ± 20 nm; emission, 540 ± 30 nm), and test compounds were added in a 20 μL volume and incubated for approximately 2.5 min before the addition of 10 μL of thallium buffer with or without agonist. After the addition of agonist, data were collected for approximately an additional 2.5 min. Data were analyzed using Excel (Microsoft Corp, Redmond, WA). The slope of the fluorescence increase beginning 5 s after thallium/agonist addition and ending 15 s after thallium/agonist addition was calculated, corrected to vehicle and maximal agonist control slope values, and plotted using either XLfit (ID Business Solutions Ltd.) or Prism software (GraphPad Software, San Diego, CA) to generate concentration–response curves. Potencies were calculated from fits using a four-point parameter logistic equation. For concentration–response curve experiments, compounds were serially diluted 1:3 into 10 point concentration–response curves and were transferred to daughter plates using an Echo acoustic plate reformatter (Labcyte, Sunnyvale, CA). Test compounds were applied and followed by EC20 concentrations of glutamate. For selectivity experiments, full concentration–response curves of glutamate or L-AP4 (for mGlu7) were obtained in the presence of a 10 μM concentration of compound, and compounds that affected the concentration–response by less than 2-fold in terms of potency or efficacy were designated as inactive.
TREx293 mGlu3 Gα15 Cell Line Creation
In order to generate a tetracycline (Tet) inducible rat mGlu3 stable cell line to be used for a calcium mobilizaion assay, TREx293 cells (Invitrogen) were transfected with mouse Gα15-pCMV6 plasmid (Origene) using Fugene6 (Promega). The cells were selected for Gα15 expression with 1 mg/mL G418 in the presence of 10 μg/mL blasticidins to maintain Tet repressor expression. Two weeks after the selection, polyclonal TREx293 Gα15 cells were obtained. The entire coding sequence of rat mGlu3 was amplified by polymerase chain reaction (PCR) and cloned into the Tet-inducible expression plasmid pcDNA5/TO (Invitrogen). Rat mGlu3-pcDNA5/TO was transfected into TREx293 Gα15 cells and selected for mGlu3 expression with 200 μg/mL hygromycin in the presence of G418 and blasticidins. The resulting polyclonal TREx293 mGlu3 Gα15 cells were plated for monoclonal selection, and positive monoclones were identified in the calcium mobilization assay. Cells were maintained in growth medium containing DMEM, 10% Tet-tested FBS (Atlanta Biogicals), 20 mM HEPES, 2 mM l-glutamine, antibiotic/antimycotic, nonessential amino acids, 500 μg/mL G418, 100 μg/mL hygromycin, and 5 μg/mL blasticidin S at 37 °C in the presence of 5% CO2.
HEK293A mGlu2 Gα15 Cell Line Creation
In order to generate a rat mGlu2 stable cell line to be used for a calcium mobilization assay, HEK293A cells (ATCC) were transfected with mouse Gα15-pCMV6 plasmid (Origene) using Fugene6 (Promega). The cells were selected for Gα15 expression with 1 mg/mL G418. Two weeks after the selection, polyclonal HEK293A Gα15 cells were obtained. The entire coding sequence of rat mGlu2 was amplified by PCR and cloned into the expression plasmid pIRESpuro3 (Invitrogen). Rat mGlu2-pIRESpuro3 was transfected into HEK293A Gα15 cells and selected for mGlu2 expression with 0.6 μg/mL puromycin in the presence of G418. The resulting polyclonal HEK293A mGlu2 Gα15 cells were then utilized for calcium mobilization assays. Cells were maintained in growth medium containing DMEM, 10% FBS, 20 mM HEPES, 2 mM l-glutamine, antibiotic/antimycotic, nonessential amino acids, 700 μg/mL G418, and 0.6 μg/mL puromycin at 37 °C in the presence of 5% CO2.
Western Blotting
Western blotting was performed as detailed previously58 utilizing 10% polyacrylamide gels and a rabbit polyclonal mGlu2/3 antibody (Millipore, catalog no. 06-676) for detection of mGlu3.
Microsomal Stability in Vitro Assay
Pooled rat liver microsomes (BD Biosciences, no. 452701) were preincubated with test compounds at 37.5 °C for 5 min in the absence of NADPH. The reaction was initiated by addition of NADPH, and the mixture was incubated under the same conditions. The final incubation concentrations were 4 μM test compound, 2 mM NADPH, and 1 mg/mL (total protein) liver microsomes in phosphate buffered saline (PBS) at pH 7.4. One aliquot (100 μL) of the incubation mixture was withdrawn at 15 min time points and combined immediately with 100 μL of ACN/MeOH. After mixing, the sample was centrifuged at approximately 13 000 rpm for 12 min. The supernatant was filtered and transferred into an autosampler vial, and the amount of test compound was quantified using a Shimadzu LCMS 2010EV mass spectrometer. The change of the AUC (area under the curve) of the parent compound as a function of time was used as a measure of microsomal stability. Test compounds were run in duplicate with a positive control.
Plasma Stability in Vitro Assay
A 20 μL aliquot of a 10 mM solution in DMSO of the test compound was added to 2.0 mL of heparinized rat plasma (Lampire, P1-150N) to obtain a 100 μM final solution. The mixture was incubated for 1 h at 37.5 °C. Aliquots of 100 μL were taken at 15 min intervals and diluted with 100 μL of MeOH/ACN. After mixing, the sample was centrifuged at approximately 13 000 rpm for 12 min. The supernatant was filtered and transferred into an autosampler vial, and the amount of test compound was quantified using the Shimadzu LCMS-2010EV system. The change of the AUC of the parent compound as a function of time was used as a measure of plasma stability.
Parallel Artificial Membrane Permeation Assay (PAMPA)
A 96-well microtiter plate (Millipore, MSSACCEPTOR) was filled with 300 μL of aqueous buffer solution (in general, phosphate pH 7.2 buffer was used) and covered with a microtiter filterplate (Millipore, MAIPNTR10) to create a sort of sandwich construction. The hydrophobic filter material was impregnated with a 10% solution of polar brain lipid extract in chloroform (Avanti) as the artificial membrane, and the organic solvent was allowed to completely evaporate. Permeation studies were started by the transfer of 150 μL of a 100 μM test compound solution on top of the filter plate. The maximum DMSO content of the stock solutions was <1.5%. In parallel, an equilibrium solution lacking a membrane was prepared using the exact concentrations and specifications but lacking the membrane. The concentrations of the acceptor and equilibrium solutions were determined using the Shimadzu LCMS-2010EV and AUC methods. The acceptor plate and equilibrium plate concentrations were used to calculate the permeability rate (log Pe) of the compounds. The log Pe values were calculated using the following equations:
In this equation, VD (cm3) is the donor volume (0.150 cm3), VA (cm3) is the acceptor volume (0.300 cm3), area (cm2) is the accessible filter area (0.168 cm2), and time (s) is the incubation time. Variables [drug]acceptor and [drug]equilibruim are concentrations of the test drug for the sample (acceptor) and reference (equilibrium) solutions in the acceptor compartment.
Behavioral Assessments
Subjects
Male Wistar rats (Charles River Laboratories, Raleigh, NC) weighing 300–350 g at the beginning of each experiment were housed in pairs in standard rat Plexiglas cages with food and water available ad libitum except during food training and the food self-administration experiment (see below). Rats were maintained in a climate-controlled room at 21 °C on a 12 h reverse light/dark cycle, and all experiments were conducted during the dark (i.e., active) phase (7:00 to 19:00 h) of the cycle under dim red lighting. All procedures were conducted in accordance with the guidelines from the National Institutes of Health and the Association for the Assessment and Accreditation of Laboratory Animal Care and were approved by the Institutional Animal Care and Use Committee.
Drugs
Cocaine hydrochloride (National Institute on Drug Abuse, Bethesda, MD) was dissolved in sterile physiological saline and filtered through a 0.22 μm syringe filter (Fisher Scientific, Pittsburgh, PA) for sterilization purposes. 74 was mixed into a 10% EtOH, 1% Tween 80 solution.
Food Training
Details regarding the experimental procedures have been described previously.37 All rats were placed under food restriction (20 g of food/day) and trained during daily 1 h sessions to lever-press for 45 mg food pellets (Research Diets, New Brunswick, NJ) under a fixed ratio 1 reinforcement schedule with a 1 s time-out period (FR1 TO1s). Successful responses were followed by illumination of a cue light for the duration of the time-out period, when lever presses had no consequence. Successful acquisition of food responding, defined as earning 100 pellets during each session, resulted in progression of the training program to FR1 TO10s and FR1 TO20s. Training lasted approximately 5 days.
Cocaine Self-Administration Experiment
After successful acquisition of food training, rats (n = 11) were fed ad libitum, surgically prepared with intravenous catheters inserted into the right jugular vein under isoflurane anesthesia (1–1.5% isoflurane/oxygen mixture), and allowed 7 days to recover (see Jin et al. (2010) for details). Rats were then trained to self-administer cocaine under a FR1 TO20s reinforcement schedule during daily 1 h sessions. Each response at the active lever resulted in an intravenous infusion of cocaine (0.5 mg/kg/infusion) over a 2 s period in a volume of 0.05 μL. Rats were trained for approximately 10 days until responding for cocaine stabilized (i.e., >10 infusions/session; <20% variability in number of infusions over three consecutive sessions). After stabilization of responding, rats were administered 74 (0, 10, 20, 40 mg/kg, ip; 3 mL/kg volume; 60 min pretreatment time) according to a within-subjects Latin-square design. At least 4 days elapsed between drug/vehicle injections to re-establish stable self-administration behavior (<20% variability over three consecutive sessions).
Food Self-Administration Experiment
To assess nonspecific actions of 74, after successful acquisition of food training and stabilization of responding (<20% variability over three consecutive sessions), rats (n = 8) were administered 74 (0, 10, 20, 40 mg/kg, ip; 3 mL/kg volume; 60 min pretreatment time) according to a within-subjects Latin-square design. All test parameters, including the FR1 TO20s reinforcement schedule, were identical to the parameters under which cocaine was self-administered.
Statistical Analyses
The number of cocaine infusions/food pellets earned during test sessions with 74 was calculated as a percentage of the average number of infusions/pellets earned during the prior three baseline sessions. Data were then analyzed with a mixed design analysis of variance (ANOVA) with 74 dose (within-subjects) and self-administration (i.e., cocaine vs food; between-subjects) as factors. Significant effects were further analyzed with Tukey post hoc tests. The level of significance was set at α = 0.05.
Acknowledgments
This work was supported by National Institutes of Health Grant R01 DA023926 to N.D.P.C. and NIDA grant DA011946 to A.M. D.J.S. is a recipient of a National Alliance for Research on Schizophrenia and Depression (NARSAD), Dylan Tauber Young Investigator Award. The binding profile for compound 72 was generously provided by the National Institute of Mental Health’s Psychoactive Drug Screening Program, Contract HHSN-271-2008-00025-C (NIMH PDSP).
Glossary
Abbreviations Used
- mGlu2
metabotropic glutamate receptor subtype 2
- PAM
positive allosteric modulator
- BINA
3′-((2-cyclopentyl-6,7-dimethyl-1-oxo-2,3-dihydro-1H-inden-5-yloxy)methyl)biphenyl-4-carboxylic acid
- HTS
high-throughput screening
- PK
pharmacokinetic
- POC
proof-of-concept
- ADME
absorption, distribution, metabolism, excretion
- AUC
area under curve
- LOQ
limit of quantitation
- HEK
human embryonic kidney
- GIRK
G protein-coupled inwardly rectifying potassium
- PBS
phosphate buffered saline
- DMEM
Dulbecco’s modified Eagle medium
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- FBS
fetal bovine serum
Supporting Information Available
Computationally determined plasma–protein binding for 11 compounds and off-target profiling data against a panel of CNS receptors for compound 72. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
# R.-P.D. and D.J.S. contributed equally.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Marmiroli P.; Cavaletti G. The glutamatergic neurotransmission in the central nervous system. Curr. Med. Chem. 2012, 19, 1269–1276. [DOI] [PubMed] [Google Scholar]
- Sheffler D. J.; Gregory K. J.; Rook J. M.; Conn P. J. Allosteric modulation of metabotropic glutamate receptors. Adv. Pharmacol. 2011, 62, 37–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn P. J.; Pin J. P. Pharmacology and functions of metabotropic glutamate receptors. Annu. Rev. Pharmacol. Toxicol. 1997, 37, 205–237. [DOI] [PubMed] [Google Scholar]
- Enz R. Metabotropic glutamate receptors and interacting proteins: evolving drug targets. Curr. Drug Targets 2012, 13, 145–156. [DOI] [PubMed] [Google Scholar]
- Sheffler D. J.; Pinkerton A. B.; Dahl R.; Markou A.; Cosford N. D. Recent progress in the synthesis and characterization of group II metabotropic glutamate receptor allosteric modulators. ACS Chem. Neurosci. 2011, 2, 382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartmell J.; Schoepp D. D. Regulation of neurotransmitter release by metabotropic glutamate receptors. J. Neurochem. 2000, 75, 889–907. [DOI] [PubMed] [Google Scholar]
- Durand D.; Carniglia L.; Caruso C.; Lasaga M. mGlu3 receptor and astrocytes: partners in neuroprotection. Neuropharmacology 2013, 66, 1–11. [DOI] [PubMed] [Google Scholar]
- Corti C.; Battaglia G.; Molinaro G.; Riozzi B.; Pittaluga A.; Corsi M.; Mugnaini M.; Nicoletti F.; Bruno V. The use of knock-out mice unravels distinct roles for mGlu2 and mGlu3 metabotropic glutamate receptors in mechanisms of neurodegeneration/neuroprotection. J. Neurosci. 2007, 27, 8297–8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi H.; Neki A.; Mizuno N. Distribution of a metabotropic glutamate receptor, mGluR2, in the central nervous system of the rat and mouse: an immunohistochemical study with a monoclonal antibody. Neurosci. Res. 1998, 30, 65–82. [DOI] [PubMed] [Google Scholar]
- Wright R. A.; Arnold M. B.; Wheeler W. J.; Ornstein P. L.; Schoepp D. D. [3H]LY341495 binding to group II metabotropic glutamate receptors in rat brain. J. Pharmacol. Exp. Ther. 2001, 298, 453–460. [PubMed] [Google Scholar]
- Swanson C. J.; Bures M.; Johnson M. P.; Linden A. M.; Monn J. A.; Schoepp D. D. Metabotropic glutamate receptors as novel targets for anxiety and stress disorders. Nat. Rev. Drug Discovery 2005, 4, 131–144. [DOI] [PubMed] [Google Scholar]
- Krystal J. H.; Mathew S. J.; D’Souza D. C.; Garakani A.; Gunduz-Bruce H.; Charney D. S. Potential psychiatric applications of metabotropic glutamate receptor agonists and antagonists. CNS Drugs 2010, 24, 669–693. [DOI] [PubMed] [Google Scholar]
- Chaki S.; Ago Y.; Palucha-Paniewiera A.; Matrisciano F.; Pilc A. mGlu2/3 and mGlu5 receptors: potential targets for novel antidepressants. Neuropharmacology 2013, 66, 40–52. [DOI] [PubMed] [Google Scholar]
- Conn P. J.; Lindsley C. W.; Jones C. K. Activation of metabotropic glutamate receptors as a novel approach for the treatment of schizophrenia. Trends Pharmacol. Sci. 2009, 30, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffler D. J.; Conn P. J.. Activation of Group II Metabotropic Glutamate Receptors (mGluR2 and mGluR3) as a Novel Approach for Treatment of Schizophrenia. In Glutamate-Based Therapies for Psychiatric Disorders; Skolnick P., Ed.; Birkhäuser: Basel, Switzerland, 2010; pp 101–116. [Google Scholar]
- Kenny P. J.; Markou A. The ups and downs of addiction: role of metabotropic glutamate receptors. Trends Pharmacol. Sci. 2004, 25, 265–272. [DOI] [PubMed] [Google Scholar]
- Xi Z. X.; Ramamoorthy S.; Baker D. A.; Shen H.; Samuvel D. J.; Kalivas P. W. Modulation of group II metabotropic glutamate receptor signaling by chronic cocaine. J. Pharmacol. Exp. Ther. 2002, 303, 608–615. [DOI] [PubMed] [Google Scholar]
- Adewale A. S.; Platt D. M.; Spealman R. D. Pharmacological stimulation of group II metabotropic glutamate receptors reduces cocaine self-administration and cocaine-induced reinstatement of drug seeking in squirrel monkeys. J. Pharmacol. Exp. Ther. 2006, 318, 922–931. [DOI] [PubMed] [Google Scholar]
- Aujla H.; Martin-Fardon R.; Weiss F. Rats with extended access to cocaine exhibit increased stress reactivity and sensitivity to the anxiolytic-like effects of the mGluR 2/3 agonist LY379268 during abstinence. Neuropsychopharmacology 2008, 33, 1818–1826. [DOI] [PubMed] [Google Scholar]
- Baptista M. A.; Martin-Fardon R.; Weiss F. Preferential effects of the metabotropic glutamate 2/3 receptor agonist LY379268 on conditioned reinstatement versus primary reinforcement: comparison between cocaine and a potent conventional reinforcer. J. Neurosci. 2004, 24, 4723–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.; Kalivas P. W. The group II metabotropic glutamate receptor agonist, LY379268, inhibits both cocaine- and food-seeking behavior in rats. Psychopharmacology (Berlin, Ger.) 2006, 186, 143–149. [DOI] [PubMed] [Google Scholar]
- Xi Z. X.; Baker D. A.; Shen H.; Carson D. S.; Kalivas P. W. Group II metabotropic glutamate receptors modulate extracellular glutamate in the nucleus accumbens. J. Pharmacol. Exp. Ther. 2002, 300, 162–171. [DOI] [PubMed] [Google Scholar]
- Flor P. J.; Battaglia G.; Nicoletti F.; Gasparini F.; Bruno V. Neuroprotective activity of metabotropic glutamate receptor ligands. Adv. Exp. Med. Biol. 2002, 513, 197–223. [DOI] [PubMed] [Google Scholar]
- Allen J. W.; Ivanova S. A.; Fan L.; Espey M. G.; Basile A. S.; Faden A. I. Group II metabotropic glutamate receptor activation attenuates traumatic neuronal injury and improves neurological recovery after traumatic brain injury. J. Pharmacol. Exp. Ther. 1999, 290, 112–120. [PubMed] [Google Scholar]
- Richards G.; Messer J.; Faull R. L.; Stadler H.; Wichmann J.; Huguenin P.; Bohrmann B.; Mutel V. Altered distribution of mGlu2 receptors in beta-amyloid-affected brain regions of Alzheimer cases and aged PS2APP mice. Brain Res. 2010, 1363, 180–190. [DOI] [PubMed] [Google Scholar]
- Ahnaou A.; Dautzenberg F. M.; Geys H.; Imogai H.; Gibelin A.; Moechars D.; Steckler T.; Drinkenburg W. H. Modulation of group II metabotropic glutamate receptor (mGlu2) elicits common changes in rat and mice sleep–wake architecture. Eur. J. Pharmacol. 2009, 603, 62–72. [DOI] [PubMed] [Google Scholar]
- Monn J. A.; Valli M. J.; Massey S. M.; Hansen M. M.; Kress T. J.; Wepsiec J. P.; Harkness A. R.; Grutsch J. L. Jr.; Wright R. A.; Johnson B. G.; Andis S. L.; Kingston A.; Tomlinson R.; Lewis R.; Griffey K. R.; Tizzano J. P.; Schoepp D. D. Synthesis, pharmacological characterization, and molecular modeling of heterobicyclic amino acids related to (+)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid (LY354740): identification of two new potent, selective, and systemically active agonists for group II metabotropic glutamate receptors. J. Med. Chem. 1999, 42, 1027–1040. [DOI] [PubMed] [Google Scholar]
- Dominguez C.; Prieto L.; Valli M. J.; Massey S. M.; Bures M.; Wright R. A.; Johnson B. G.; Andis S. L.; Kingston A.; Schoepp D. D.; Monn J. A. Methyl substitution of 2-aminobicyclo[3.1.0]hexane 2,6-dicarboxylate (LY354740) determines functional activity at metabotropic glutamate receptors: identification of a subtype selective mGlu2 receptor agonist. J. Med. Chem. 2005, 48, 3605–3612. [DOI] [PubMed] [Google Scholar]
- Hanna L.; Ceolin L.; Lucas S.; Monn J.; Johnson B.; Collingridge G.; Bortolotto Z.; Lodge D. Differentiating the roles of mGlu2 and mGlu3 receptors using LY541850, an mGlu2 agonist/mGlu3 antagonist. Neuropharmacology 2013, 66, 114–121. [DOI] [PubMed] [Google Scholar]
- Anwyl R. Metabotropic glutamate receptors: electrophysiological properties and role in plasticity. Brain Res. Rev. 1999, 29, 83–120. [DOI] [PubMed] [Google Scholar]
- Cartmell J.; Monn J. A.; Schoepp D. D. The metabotropic glutamate 2/3 receptor agonists LY354740 and LY379268 selectively attenuate phencyclidine versus d-amphetamine motor behaviors in rats. J. Pharmacol. Exp. Ther. 1999, 291, 161–170. [PubMed] [Google Scholar]
- Wieronska J. M.; Stachowicz K.; Branski P.; Palucha-Poniewiera A.; Pilc A. On the mechanism of anti-hyperthermic effects of LY379268 and LY487379, group II mGlu receptors activators, in the stress-induced hyperthermia in singly housed mice. Neuropharmacology 2012, 62, 322–331. [DOI] [PubMed] [Google Scholar]
- Galici R.; Jones C. K.; Hemstapat K.; Nong Y.; Echemendia N. G.; Williams L. C.; de Paulis T.; Conn P. J. Biphenyl-indanone A, a positive allosteric modulator of the metabotropic glutamate receptor subtype 2, has antipsychotic- and anxiolytic-like effects in mice. J. Pharmacol. Exp. Ther. 2006, 318, 173–185. [DOI] [PubMed] [Google Scholar]
- Reiner A.; Lafferty D. C.; Wang H. B.; Del Mar N.; Deng Y. P. The group 2 metabotropic glutamate receptor agonist LY379268 rescues neuronal, neurochemical and motor abnormalities in R6/2 Huntington’s disease mice. Neurobiol. Dis. 2012, 47, 75–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A.; Wang H. B.; Del Mar N.; Sakata K.; Yoo W.; Deng Y. P. BDNF may play a differential role in the protective effect of the mGluR2/3 agonist LY379268 on striatal projection neurons in R6/2 Huntington’s disease mice. Brain Res. 2012, 1473, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefer J.; Sprunken A.; Puls C.; Luesse H. G.; Milkereit A.; Milkereit E.; Johann V.; Kosinski C. M. The metabotropic glutamate receptor 5 antagonist MPEP and the mGluR2 agonist LY379268 modify disease progression in a transgenic mouse model of Huntington’s disease. Brain Res. 2004, 1019, 246–254. [DOI] [PubMed] [Google Scholar]
- Jin X.; Semenova S.; Yang L.; Ardecky R.; Sheffler D. J.; Dahl R.; Conn P. J.; Cosford N. D.; Markou A. The mGluR2 positive allosteric modulator BINA decreases cocaine self-administration and cue-induced cocaine-seeking and counteracts cocaine-induced enhancement of brain reward function in rats. Neuropsychopharmacology 2010, 35, 2021–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert J. M.; Poles G. C.; Sheffler-Collins S. I.; Ghitza U. E. The mGluR2/3 agonist LY379268 attenuates context- and discrete cue-induced reinstatement of sucrose seeking but not sucrose self-administration in rats. Behav. Brain Res. 2006, 173, 148–152. [DOI] [PubMed] [Google Scholar]
- Cacciola J.; Empfield J.; Folmer J.; Hunter A. M.; Throner S.. Metabotropic glutamate receptor isoxazole ligands and their use as potentiators 286. U.S. Patent 7,790,760, September 7, 2010.
- Pinkerton A. B.; Cube R. V.; Hutchinson J. H.; Rowe B. A.; Schaffhauser H.; Zhao X.; Daggett L. P.; Vernier J. M. Allosteric potentiators of the metabotropic glutamate receptor 2 (mGlu2). Part 1: Identification and synthesis of phenyl-tetrazolyl acetophenones. Bioorg. Med. Chem. Lett. 2004, 14, 5329–5332. [DOI] [PubMed] [Google Scholar]
- Pinkerton A. B.; Cube R. V.; Hutchinson J. H.; James J. K.; Gardner M. F.; Rowe B. A.; Schaffhauser H.; Rodriguez D. E.; Campbell U. C.; Daggett L. P.; Vernier J. M. Allosteric potentiators of the metabotropic glutamate receptor 2 (mGlu2). Part 3: Identification and biological activity of indanone containing mGlu2 receptor potentiators. Bioorg. Med. Chem. Lett. 2005, 15, 1565–1571. [DOI] [PubMed] [Google Scholar]
- Johnson M. P.; Baez M.; Jagdmann G. E. Jr.; Britton T. C.; Large T. H.; Callagaro D. O.; Tizzano J. P.; Monn J. A.; Schoepp D. D. Discovery of allosteric potentiators for the metabotropic glutamate 2 receptor: synthesis and subtype selectivity of N-(4-(2-methoxyphenoxy)phenyl)-N-(2,2,2- trifluoroethylsulfonyl)pyrid-3-ylmethylamine. J. Med. Chem. 2003, 46, 3189–3192. [DOI] [PubMed] [Google Scholar]
- Pinkerton A. B.; Vernier J. M.; Schaffhauser H.; Rowe B. A.; Campbell U. C.; Rodriguez D. E.; Lorrain D. S.; Baccei C. S.; Daggett L. P.; Bristow L. J. Phenyl-tetrazolyl acetophenones: discovery of positive allosteric potentiatiors for the metabotropic glutamate 2 receptor. J. Med. Chem. 2004, 47, 4595–4599. [DOI] [PubMed] [Google Scholar]
- Govek S. P.; Bonnefous C.; Hutchinson J. H.; Kamenecka T.; McQuiston J.; Pracitto R.; Zhao L. X.; Gardner M. F.; James J. K.; Daggett L. P.; Rowe B. A.; Schaffhauser H.; Bristow L. J.; Campbell U. C.; Rodriguez D. E.; Vernier J. M. Benzazoles as allosteric potentiators of metabotropic glutamate receptor 2 (mGluR2): efficacy in an animal model for schizophrenia. Bioorg. Med. Chem. Lett. 2005, 15, 4068–4072. [DOI] [PubMed] [Google Scholar]
- Duplantier A. J.; Efremov I.; Candler J.; Doran A. C.; Ganong A. H.; Haas J. A.; Hanks A. N.; Kraus K. G.; Lazzaro J. T. Jr.; Lu J.; Maklad N.; McCarthy S. A.; O’Sullivan T. J.; Rogers B. N.; Siuciak J. A.; Spracklin D. K.; Zhang L. 3-Benzyl-1,3-oxazolidin-2-ones as mGluR2 positive allosteric modulators: hit-to lead and lead optimization. Bioorg. Med. Chem. Lett. 2009, 19, 2524–2529. [DOI] [PubMed] [Google Scholar]
- Tresadern G.; Cid J. M.; Macdonald G. J.; Vega J. A.; de Lucas A. I.; Garcia A.; Matesanz E.; Linares M. L.; Oehlrich D.; Lavreysen H.; Biesmans I.; Trabanco A. A. Scaffold hopping from pyridones to imidazo[1,2-a]pyridines. New positive allosteric modulators of metabotropic glutamate 2 receptor. Bioorg. Med. Chem. Lett. 2010, 20, 175–179. [DOI] [PubMed] [Google Scholar]
- Melancon B. J.; Hopkins C. R.; Wood M. R.; Emmitte K. A.; Niswender C. M.; Christopoulos A.; Conn P. J.; Lindsley C. W. Allosteric modulation of seven transmembrane spanning receptors: theory, practice, and opportunities for central nervous system drug discovery. J. Med. Chem. 2012, 55, 1445–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanya R. P.; Sidique S.; Sheffler D. J.; Nickols H. H.; Herath A.; Yang L.; Dahl R.; Ardecky R.; Semenova S.; Markou A.; Conn P. J.; Cosford N. D. Design and synthesis of an orally active metabotropic glutamate receptor subtype-2 (mGluR2) positive allosteric modulator (PAM) that decreases cocaine self-administration in rats. J. Med. Chem. 2011, 54, 342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidique S.; Dhanya R. P.; Sheffler D. J.; Nickols H. H.; Yang L.; Dahl R.; Mangravita-Novo A.; Smith L. H.; D’Souza M. S.; Semenova S.; Conn P. J.; Markou A.; Cosford N. D. Orally active metabotropic glutamate subtype 2 receptor positive allosteric modulators: structure–activity relationships and assessment in a rat model of nicotine dependence. J. Med. Chem. 2012, 55, 9434–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schann S.; Mayer S.; Franchet C.; Frauli M.; Steinberg E.; Thomas M.; Baron L.; Neuville P. Chemical switch of a metabotropic glutamate receptor 2 silent allosteric modulator into dual metabotropic glutamate receptor 2/3 negative/positive allosteric modulators. J. Med. Chem. 2010, 53, 8775–8779. [DOI] [PubMed] [Google Scholar]
- Cube R. V.; Vernier J. M.; Hutchinson J. H.; Gardner M. F.; James J. K.; Rowe B. A.; Schaffhauser H.; Daggett L.; Pinkerton A. B. 3-(2-Ethoxy-4-{4-[3-hydroxy-2-methyl-4-(3-methylbutanoyl)phenoxy]butoxy}phenyl)propanoic acid: a brain penetrant allosteric potentiator at the metabotropic glutamate receptor 2 (mGluR2). Bioorg. Med. Chem. Lett. 2005, 15, 2389–2393. [DOI] [PubMed] [Google Scholar]
- Fell M. J.; Witkin J. M.; Falcone J. F.; Katner J. S.; Perry K. W.; Hart J.; Rorick-Kehn L.; Overshiner C. D.; Rasmussen K.; Chaney S. F.; Benvenga M. J.; Li X.; Marlow D. L.; Thompson L. K.; Luecke S. K.; Wafford K. A.; Seidel W. F.; Edgar D. M.; Quets A. T.; Felder C. C.; Wang X.; Heinz B. A.; Nikolayev A.; Kuo M. S.; Mayhugh D.; Khilevich A.; Zhang D.; Ebert P. J.; Eckstein J. A.; Ackermann B. L.; Swanson S. P.; Catlow J. T.; Dean R. A.; Jackson K.; Tauscher-Wisniewski S.; Marek G. J.; Schkeryantz J. M.; Svensson K. A. N-(4-((2-(Trifluoromethyl)-3-hydroxy-4-(isobutyryl)phenoxy)methyl)benzyl)-1-methy l-1H-imidazole-4-carboxamide (THIIC), a novel metabotropic glutamate 2 potentiator with potential anxiolytic/antidepressant properties: in vivo profiling suggests a link between behavioral and central nervous system neurochemical changes. J. Pharmacol. Exp. Ther. 2011, 336, 165–177. [DOI] [PubMed] [Google Scholar]
- Liechti M. E.; Lhuillier L.; Kaupmann K.; Markou A. Metabotropic glutamate 2/3 receptors in the ventral tegmental area and the nucleus accumbens shell are involved in behaviors relating to nicotine dependence. J. Neurosci. 2007, 27, 9077–9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima Y.; Miyakawa T.; Furuyashiki T.; Tanaka Y.; Mizuma H.; Nakanishi S. Enhanced cocaine responsiveness and impaired motor coordination in metabotropic glutamate receptor subtype 2 knockout mice. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 4170–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender C. M.; Johnson K. A.; Luo Q.; Ayala J. E.; Kim C.; Conn P. J.; Weaver C. D. A novel assay of Gi/o-linked G protein-coupled receptor coupling to potassium channels provides new insights into the pharmacology of the group III metabotropic glutamate receptors. Mol. Pharmacol. 2008, 73, 1213–1224. [DOI] [PubMed] [Google Scholar]
- Hemstapat K.; de Paulis T.; Chen Y.; Brady A. E.; Grover V. K.; Alagille D.; Tamagnan G. D.; Conn P. J. A novel class of positive allosteric modulators of metabotropic glutamate receptor subtype 1 interact with a site distinct from that of negative allosteric modulators. Mol. Pharmacol. 2006, 70, 616–626. [DOI] [PubMed] [Google Scholar]
- Engers D. W.; Niswender C. M.; Weaver C. D.; Jadhav S.; Menon U. N.; Zamorano R.; Conn P. J.; Lindsley C. W.; Hopkins C. R. Synthesis and evaluation of a series of heterobiarylamides that are centrally penetrant metabotropic glutamate receptor 4 (mGluR4) positive allosteric modulators (PAMs). J. Med. Chem. 2009, 52, 4115–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffler D. J.; Conn P. J.. Activation of Group II Metabotropic Glutamate Receptors (mGluR2 and mGluR3) as a Novel Approach for Treatment of Schizophrenia. In Glutamate-Based Therapies for Psychiatric Disorders; Skolnick P., Ed.; Birkhäuser: Basel, Switzerland, 2010; pp 101–116. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.