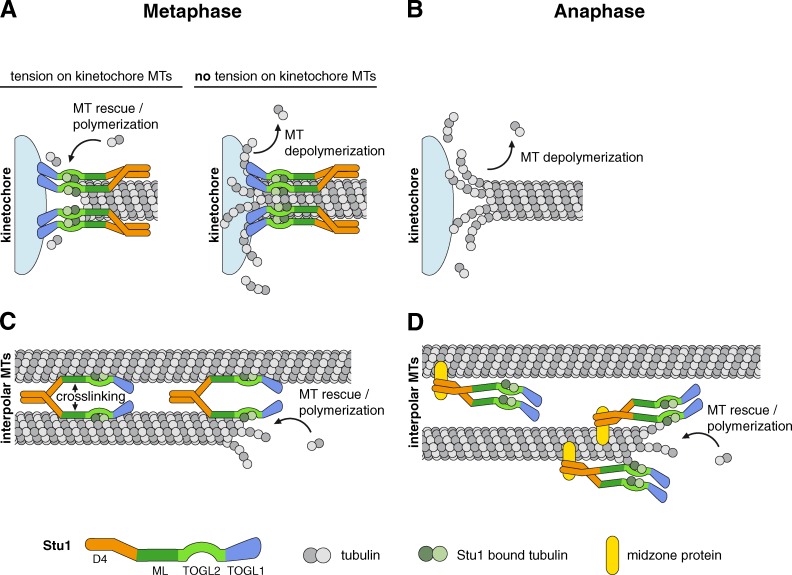
The two different TOGL domains of the budding yeast CLASP Stu1 are responsible for its distinct mitotic activities, and these activities are only partially mediated by tight microtubule binding.
Abstract
Cytoplasmic linker–associated proteins (CLASPs) are proposed to function in cell division based on their ability to bind tubulin via arrayed tumor overexpressed gene (TOG)–like (TOGL) domains. Structure predictions suggest that CLASPs have at least two TOGL domains. We show that only TOGL2 of Saccharomyces cerevisiae CLASP Stu1 binds to tubulin and is required for polymerization of spindle microtubules (MTs) in vivo. In contrast, TOGL1 recruits Stu1 to kinetochores (KTs), where it is essential for the stability and tension-dependent regulation of KT MTs. Stu1 is also recruited to spindle MTs by different mechanisms depending on the mitotic phase: in metaphase, Stu1 binds directly to the MT lattice, whereas in anaphase, it is localized indirectly to the spindle midzone. In both phases, the activity of TOGL2 is essential for interpolar MT stability, whereas TOGL1 is not involved. Thus, the two TOGL domains of yeast CLASP have different activities and execute distinct mitotic functions.
Introduction
The cytoplasmic linker–associated proteins (CLASPs), including the human CLASP1 and CLASP2, Drosophila melanogaster multiple asters (MAST)/orbit, Caenorhabditis elegans Cls2, Schizosaccharomyces pombe Cls1, and Saccharomyces cerevisiae Stu1, are microtubule (MT)-associated proteins that regulate the dynamic behavior of MTs. Most CLASPs contain two TOG or TOG-like (TOGL) domains at the N terminus followed by a basic serine-rich domain. In vitro, this domain suffices for high-affinity binding of Cls1 to the MT lattice (Al-Bassam et al., 2010), whereas Stu1 additionally requires part of the TOGL2 domain (Yin et al., 2002). TOG or TOGL domains that are found in CLASPs, and proteins of the XMAP215 protein family consist of six HEAT repeats that bind α/β-tubulin dimers via the intra-HEAT loops (Al-Bassam et al., 2007; Ayaz et al., 2012; Leano et al., 2013). Based on EM data, a model has been presented in which the four TOGL domains of a Cls1 dimer enclose one tubulin dimer (Al-Bassam et al., 2010). However, the recently obtained structure of the TOG1 domain from Stu2 (a member of the XMAP215 protein family) in a complex with a tubulin dimer indicated that one TOG per Stu2 monomer provides sufficient interface to bind a complete tubulin dimer (Ayaz et al., 2012). In vitro, Cls1 promotes MT rescue for which the basic serine-rich domain as well as a functional TOGL2 domain are essential (Al-Bassam et al., 2010). In vivo, CLASPs only regulate a subset of cellular MTs, most likely caused by specific CLASP localization (Al-Bassam and Chang, 2011). In mammalian cells, CLASP1 localizes to the centrosome and to the plus ends of MTs during interphase and mitosis (Maiato et al., 2003). The latter is facilitated by the interaction of the S-X-I-P motifs found in mammalian CLASPs with the plus end–binding protein EB1 (Mimori-Kiyosue et al., 2005). In preanaphase, CLASP1 localizes along and at the plus ends of spindle MTs. In anaphase, it localizes to the spindle midzone and in telophase to the midbody, that is, to sites of antiparallel MT overlap (Maiato et al., 2003). Similarly, in S. pombe, Cls1 localizes to the spindle midzone in anaphase (Bratman and Chang, 2007). Furthermore, Cls1 localizes to interphase antiparallel MT bundles. In both cases, the localizations depend on Ase1 (Bratman and Chang, 2007). Upon interfering with CLASP function during mitosis, spindle poles collapse into close proximity, resulting in monopolar spindles (Yin et al., 2002; Maiato et al., 2003; Inoue et al., 2004), a phenotype best explained by a failure to stabilize antiparallel MT overlaps. Metazoan CLASPs also localize to kinetochores (KTs; Maiato et al., 2003; Inoue et al., 2004), and this has been implicated in the regulation of KT MT (kMT) dynamics. In particular, the incorporation of tubulin dimers at the plus ends of kMTs (Maiato et al., 2005) and the positive or negative regulation of kMT stability by CLASP1 in a complex with astrin or Kif2b, respectively (Manning et al., 2010), have been described as CLASP-dependent events. However, in neither case have the dependence on KT localization and the required domains been demonstrated. Furthermore, phosphorylation of CLASP2 has been described to regulate KT–MT attachment (Maia et al., 2012).
Like other CLASPs, Stu1 localizes to the spindle midzone in anaphase (Yin et al., 2002), and as mentioned for Cls1, this localization depends on Ase1 (Khmelinskii et al., 2007). Furthermore, Stu1 localizes along spindle MTs in metaphase (Yin et al., 2002; Ortiz et al., 2009). However, it is not known how this interaction is achieved and whether it depends on Ase1. KT localization of Stu1 is very robust at KTs that are not attached to MTs (Ortiz et al., 2009). In fact, Stu1 is so efficiently sequestered at unattached KTs (uaKTs) that this competes with the localization at MTs or attached KTs (Ortiz et al., 2009). Consequently uaKTs interfere with the formation of a normal spindle. After a uaKT is captured and transported to a spindle pole, most of the bound Stu1 translocates to the spindle. Stu1 cannot be detected at KTs in anaphase (Ortiz et al., 2009). It is, however, unclear whether Stu1 localizes to metaphase KTs and, if so, what role it has there.
Here, we describe how different Stu1 domains contribute to Stu1 localization and function at KTs or MTs, and we suggest a model of how the spatial and temporal localization of Stu1 regulates mitosis. In particular, we show that the localization of Stu1 to uaKTs versus metaphase KTs and the localization to metaphase versus anaphase spindles are facilitated by the cooperation of distinct Stu1 domains. Furthermore, we provide evidence that the localization to metaphase KTs stabilizes kMTs and allows the regulation of kMT stability in correlation to the tension at the KT–MT interface.
Results
Putative Stu1 domains
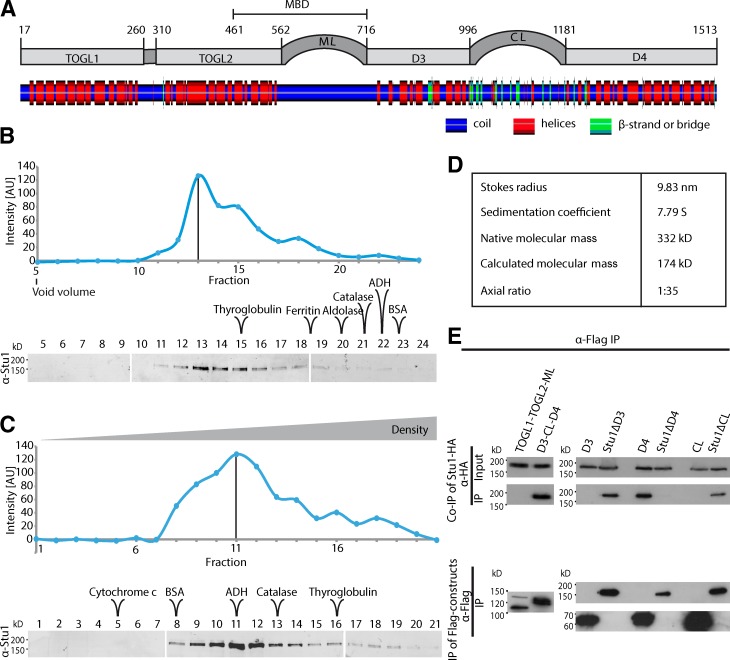
Based on Al-Bassam and Chang (2011) and secondary structure analysis (http://www.ibi.vu.nl/programs/sympredwww/), Stu1 may be organized into six domains as shown in Fig. 1 A: two N-terminal TOGL domains (TOGL1 and TOGL2), an unstructured region that includes a basic serine-rich sequence (middle loop [ML]), a domain that is predicted to consist of predominantly α helices (D3), a putatively largely unstructured region (C-terminal loop [CL]), and a C-terminal domain predicted to consist predominantly of α helices (D4). Part of TOGL2 and the ML contains the MT-binding domain (MBD) of Stu1 (Yin et al., 2002). To identify which domains are required for cell viability, we tested single domain deletions tagged with a SV40 NLS to ensure nuclear localization. TOGL2 was the only essential domain. TOGL1, ML, D3, CL, and D4 were not required for viability but enhanced the fidelity of chromosome segregation (Table 1). Particularly, the deletion of TOGL1, ML, and D4 resulted in high chromosome loss phenotypes (Table 1).
Figure 1.
Stu1 dimerizes via D4. (A) Putative domain organization of Stu1. Secondary structure prediction was performed using SYMPRED and visualized by POLYVIEW-2D (Porollo et al., 2004). (B–D) Stu1 is a dimer with an elongated rod shape. The data shown are from a single representative experiment out of two analyses. (B) Gel filtration analysis of FLAG-Stu1. (C) Sucrose gradient analysis of FLAG-Stu1. (B and C) The samples were run on three separate gels as shown. Fractions containing Stu1 were detected by Western analysis and quantified. Standard proteins as indicated were used to determine the Stokes radius or sedimentation coefficient of Stu1 (see also Fig. S1, A and B). White lines indicate that intervening lanes have been spliced out. The black lines indicate the peak fraction. AU, arbitrary unit. (D) Native molecular mass and axial ratio of Stu1 as calculated from the determined hydrodynamic properties as previously described (Schuyler and Pellman, 2002). (E) D4 is the dimerization domain. FLAG-Stu1-GFP constructs were coexpressed with Stu1-HA in S. cerevisiae, and FLAG-Stu1-GFP constructs were affinity purified.
Table 1.
Viability and chromosome loss of Stu1 deletion constructs
| Strain | Viability | Chromosome loss per 1,000 cells |
| WT | + | 2.4 |
| Δmad2 | + | 27.5 |
| stu1ΔTOGL1 | + | 364.5 |
| stu1ΔTOGL2 | − | ND |
| stu1ΔML | + | 955.6 |
| stu1ΔD3 | + | 9.4 |
| stu1ΔCL | + | 3.2 |
| stu1ΔD4 | + | 967.7 |
| stu1ΔD4-ZIP | + | 59.4 |
Stu1 is a homodimer in solution, and D4 is the dimerization domain
Stu1 has been shown to interact with itself (Yin et al., 2002). However, the exact oligomeric state of Stu1 is unclear. We therefore determined the hydrodynamic properties of FLAG-tagged Stu1 purified from S. cerevisiae (Schuyler and Pellman, 2002). The gel filtration analysis revealed a Stokes radius of 9.83 nm (Figs. 1, B and D; and S1 A), and a sedimentation coefficient of 7.79 S was determined by sucrose gradient sedimentation analysis (Figs. 1, C and D; and S1 B). Based on this, we calculated the molecular mass of Stu1 as 332 kD (Schuyler and Pellman, 2002). We thus conclude that Stu1 is a dimer (theoretical molecular mass = 348 kD) in solution. Modeling the Stu1 dimer as a prolate ellipsoid, the axial ratio of this molecule, as deduced from the Stokes radius and the native molecular mass, is 1:35. This predicts that the Stu1 dimer has an extended rod shape.
To determine the dimerization domain of Stu1, we checked for copurification of wild-type (WT) Stu1 with Stu1 deletion constructs purified from S. cerevisiae (Fig. 1 E). All constructs that contained the D4 domain, including the C-terminal half of Stu1 (D3-CL-D4) and the D4 domain alone, were able to copurify WT Stu1, whereas the N-terminal half of Stu1 (TOGL1-TOGL2-ML) and Stu1 constructs that lacked only D4 were not. We therefore conclude that D4 is the dimerization domain.
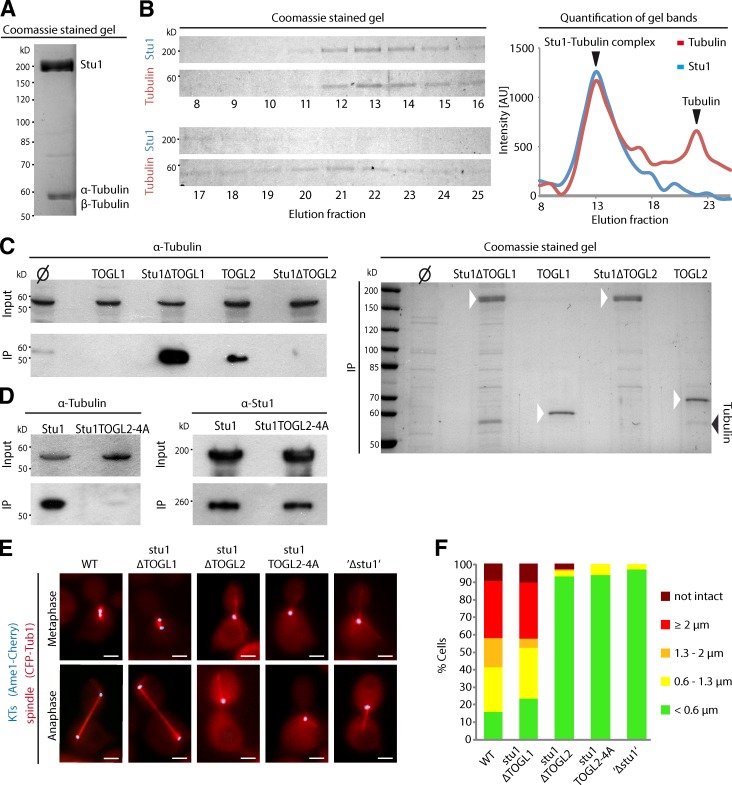
TOGL2, but not TOGL1, interacts with tubulin and is required for spindle stability
TOG domains have been shown to bind tubulin dimers and function as an MT polymerase or rescue factor (Al-Bassam et al., 2006, 2010; Brouhard et al., 2008; Ayaz et al., 2012). When FLAG-tagged Stu1 was purified from S. cerevisiae, we found that α- and β-tubulin copurified (Fig. 2 A) and that this interaction was maintained during gel filtration (Fig. 2 B).
Figure 2.
TOGL2, but not TOGL1, binds tubulin and is essential for spindle formation. (A) Tubulin copurifies with Stu1. FLAG-Stu1-GFP was affinity purified (see Materials and methods). Copurified tubulin was identified by mass spectrometry. (B) Stu1 and tubulin coelute as a complex during gel filtration. Fractions were analyzed as indicated and quantified. The data shown are from a single representative experiment out of two analyses. AU, arbitrary unit. (C) TOGL2, but not TOGL1, mediates tubulin binding. The constructs were expressed and purified as in A. Similar amounts of the IPs were loaded and subjected to Western analyses (α-tubulin) or PAGE/Coomassie. Ø indicates that no FLAG-tagged Stu1 construct was expressed. White arrowheads indicate the Stu1 constructs. (D) Mutations in the intra-HEAT repeat loops of TOGL2 abolish the Stu1–tubulin interaction. Constructs were expressed and purified as in A. (E) Cells were released from G1 arrest and visualized in meta- and anaphase. To analyze stu1ΔTOGL2, stu1TOGL2-4A, and ‘Δstu1’ cells, WT Stu1 expressed in the background was depleted. Bars, 2 µm. (F) Spindle phenotypes were quantified as indicated 2 or 2.5 h (Fig. S2) after G1 release. n > 100.
Purifying deletion constructs of Stu1 revealed that the tubulin binding activity of Stu1 was completely abolished when these constructs lacked the TOGL2 domain (Fig. 2 C) or if Stu1 harbored four point mutations in two of the predicted intra-HEAT repeat loops of TOGL2 (TOGL2-4A: W339A, R342A, K428A, and K429A; Fig. 2 D). Corresponding loops in Cls1 are important for tubulin interaction (Al-Bassam et al., 2010). Surprisingly, deletion of TOGL1 did not disrupt tubulin binding. Accordingly, tubulin copurified with an isolated TOGL2 but not with an isolated TOGL1 domain (Fig. 2 C). We thus conclude that TOGL2 of Stu1 has tubulin binding activity, but TOGL1 does not. This is in agreement with the fact that the intra-HEAT repeat loops of TOGL1 lack strong homology to those TOG domains that have been shown to interact with tubulin (Al-Bassam and Chang, 2011). In particular, conserved residues essential for tubulin binding are absent in TOGL1 of Stu1, that is, tryptophan of loop T1, the lysine of loop T4, and the arginine in the helix after loop T5 (Al-Bassam et al., 2007, 2010; Ayaz et al., 2012). Stu1 dimerization is not absolutely required for tubulin binding because TOGL2 alone cannot form a homodimer (see above). It may, however, increase the affinity or avidity of the Stu1–tubulin interaction because the tubulin amount copurified with WT Stu1 or Stu1ΔTOGL1 was consistently higher than that copurified with TOGL2 alone (Fig. 2, C and D). We were asking how the absence of either TOGL1 or TOGL2 and how compromised tubulin binding of TOGL2 affected the formation of prometa- or metaphase spindles (0.6–2 µm) as well as anaphase spindles (≥2 µm). 2 h after the release from G1, most WT cells had separated the spindle pole bodies (SPBs) beyond 0.6 µm and formed prometa-, meta-, and anaphase spindles (Fig. 2, E and F). About 10% of cells had disassembled spindles most likely because they had entered telophase. As described for the stu1-5 mutant (Yin et al., 2002) and similar to cells depleted of Stu1 (‘Δstu1’), stu1ΔTOGL2 and stu1-TOGL2-4A cells predominantly failed to separate their spindle poles beyond 0.6 µm (Fig. 2, E and F). Part of TOGL2 (amino acid 461 and higher) contributes to the MBD (Yin et al., 2002). However, the mutations of Stu1TOGL2-4A do not reside in the MBD but selectively interfere with tubulin binding (see above). This indicates that TOGL2 provides an additional function for spindle stabilization besides MT interaction. In contrast to TOGL2, the absence of TOGL1 did not affect the formation of metaphase and anaphase spindles severely (Fig. 2, E and F). In summary, these results indicate that TOGL2 may function, similar to other tubulin-binding TOG domains, as an MT polymerase or MT rescue factor and that TOGL1 has a different function.
Efficient interaction of Stu1 with uaKTs requires TOGL1, the CL, and Stu1 dimerization
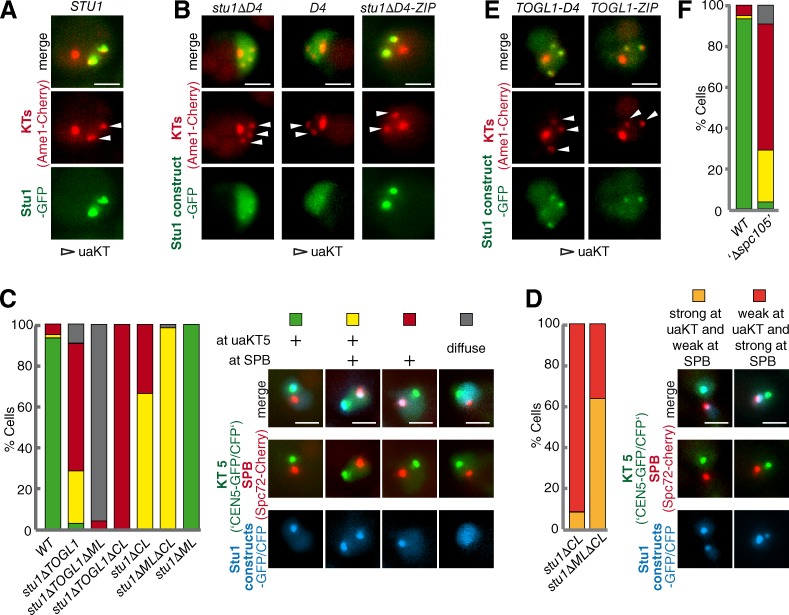
As shown previously (Ortiz et al., 2009), Stu1 localizes very efficiently to uaKTs (Fig. 3 A), and this localization depends on D4 (Fig. 3 B). The finding that D4 is required for Stu1 dimerization raised the question whether D4 confers Stu1–KT interaction directly or whether Stu1 dimerization is required. We therefore replaced the D4 domain with the dimerization domain of Gcn4 (ZIP [Zipper]; Trybus et al., 1997) and found that the hybrid protein (Stu1ΔD4-ZIP) strongly associated with uaKTs (Fig. 3 B). Also, the D4 domain alone was unable to localize to uaKTs (Fig. 3 B). We thus conclude that D4 does not directly interact with uaKTs. Rather, the dimerization of Stu1, as mediated by D4 or the Gcn4 dimerization domain, is required for efficient Stu1–KT interaction.
Figure 3.
Stu1 localization to uaKTs depends on TOGL1, CL, dimerization, and Spc105. (A–F) Cells were analyzed 3 h after release from G1 into medium containing Nz. Bars, 2 µm. (A) Stu1 selectively associates with uaKTs. (B) Dimerization is required for Stu1 localization to uaKTs. To analyze D4-GFP cells, WT Stu1 expressed in the background was depleted. (C and D) TOGL1 and CL are essential for Stu1 localization to uaKTs. (C) Phenotypes were quantified as indicated in cells with a uaKT5. n > 70. (D) More detailed analyses of the Stu1 signal intensity at SPBs and uaKTs. n > 70. (E) TOGL1 interacts with uaKTs. WT Stu1 expressed in the background was depleted. (F) uaKT localization depends on Spc105. Depletion of Spc105 (‘Δspc105’) was as described in the Materials and methods section. Phenotypes were quantified as in C. n = 72. The result obtained for WT cells is shown as a comparison.
Subsequently, the remaining Stu1 deletion constructs were tested for uaKT localization by releasing cells from a G1 arrest into nocodazole (Nz; Ortiz et al., 2009). The deletion of TOGL2, ML, or D3 did not result in observable localization defects. Like WT Stu1 (Fig. 3 A), these proteins localized exclusively to uaKTs. The deletion of TOGL1 or CL caused a severe localization defect with reduced amounts or no detectable Stu1 at uaKTs and increased Stu1 localization in the vicinity of SPBs (most likely bound to short MTs that resisted Nz treatment; Fig. 3 C). In general, the deletion of TOGL1 was more severe than the deletion of CL. Deletion of both domains abolished uaKT localization completely. Interestingly, the concomitant deletion of CL and ML considerably restored the localization of Stu1 to uaKTs and diminished the localization close to the SPB (Fig. 3, C and D). ML is part of the MBD as defined by an MT sedimentation assay. Furthermore, we will show below that ML, similar to the basic serine-rich sequences of Cls1 (Al-Bassam et al., 2010), confers lateral MT interaction. Thus, the results obtained upon ML deletion indicate that uaKT localization of Stu1ΔCL can be partially rescued by interfering with the Stu1–MT interaction. This indicates that CL is required to shift the competition between uaKT and MT interaction toward uaKT interaction. In contrast, the residual localization of Stu1ΔTOGL1 to uaKTs was completely abolished by interfering with the Stu1–MT interaction (Fig. 3 C). This suggests that the residual apparent localization at uaKTs observed for Stu1ΔTOGL1 reflects interaction with short, Nz-resistant KT-derived MTs (Kitamura et al., 2010) and not KT interaction. Furthermore, TOGL1 fused to a dimerization domain (D4 or ZIP) was sufficient to associate with uaKTs although less efficient than full-length Stu1 (Fig. 3 E). In summary, we thus conclude that the TOGL1s of a Stu1 dimer confer uaKT–Stu1 interaction directly and that CL shifts the competition between uaKT and MT interaction toward uaKT interaction. See Discussion.
Localization to uaKTs depends on Spc105
The sequestering of Stu1 at uaKTs depends on Ndc80 (Ortiz et al., 2009). We now asked whether Stu1, like the spindle assembly checkpoint proteins (Gillett et al., 2004; Pagliuca et al., 2009), also requires Spc105 beside Ndc80. We were unable to detect Stu1 at uaKTs in Nz-treated cells after Spc105 depletion by microscopy (Fig. 3 F). Because the KT localizations of the Ndc80 complex and the Spc105 complex do not depend on each other (Pagliuca et al., 2009), we conclude that Stu1 binding to uaKTs depends on Ndc80 and Spc105 independently.
Stu1 localizes to metaphase KTs, and this is dependent on TOGL1, ML, and dimerization
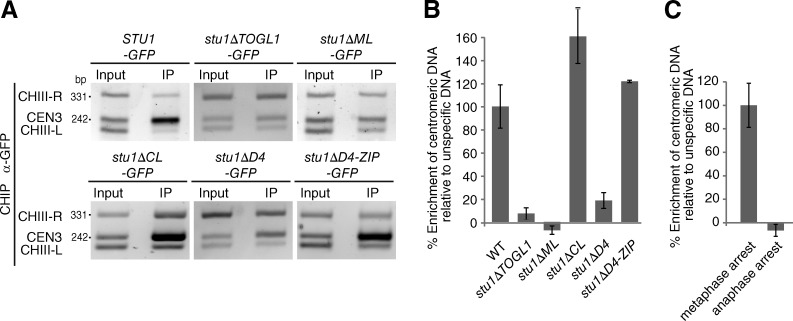
In metaphase, Stu1 localizes along spindle MTs. It is therefore difficult to clarify whether Stu1 localizes to KTs by microscopy. We thus analyzed KT localization of cells arrested in metaphase by chromatin immunoprecipitation (IP; ChIP). This revealed that Stu1 clearly localizes to metaphase KTs (Fig. 4, A and B) but not to anaphase KTs (Fig. 4 C). When we tested the aforementioned Stu1 deletion constructs, we found that KT localization depended on TOGL1 and dimerization (compare Stu1ΔD4 and Stu1ΔD4-ZIP cells; Fig. 4, A and B), similar to what we found for uaKTs. However, the metaphase KT localization was dependent on ML but not CL (Fig. 4 A, B), indicating that lateral MT interaction (see below) was required in this case.
Figure 4.
TOGL1 and ML localize the Stu1 dimer to metaphase KTs but not to anaphase KTs. (A and B) The Stu1–KT interaction in metaphase requires TOGL1, ML, and dimerization. Cells were arrested in metaphase by Cdc20 depletion for 3 h and analyzed by ChIP. (A) The presence of CEN3 DNA and two flanking DNA regions (CHIII-R and CHIII-L) was detected by triplex PCR. The input was 0.1% of the immunoprecipitation (IP). (B) Quantitative ChIP. The mean (3.5-fold) enrichment of CEN3 DNA over a control DNA (PHO5) observed for the WT was set to 100%. Error bars represent the SDs of three PCR experiments. (C) Stu1 is absent from anaphase KTs. cdc15-1 cells were arrested in anaphase by incubation at 37°C and analyzed by quantitative ChIP as in B. The result obtained for cells arrested in metaphase (set to 100%) is shown as a comparison.
TOGL1 regulates kMT length by localizing Stu1 to KTs
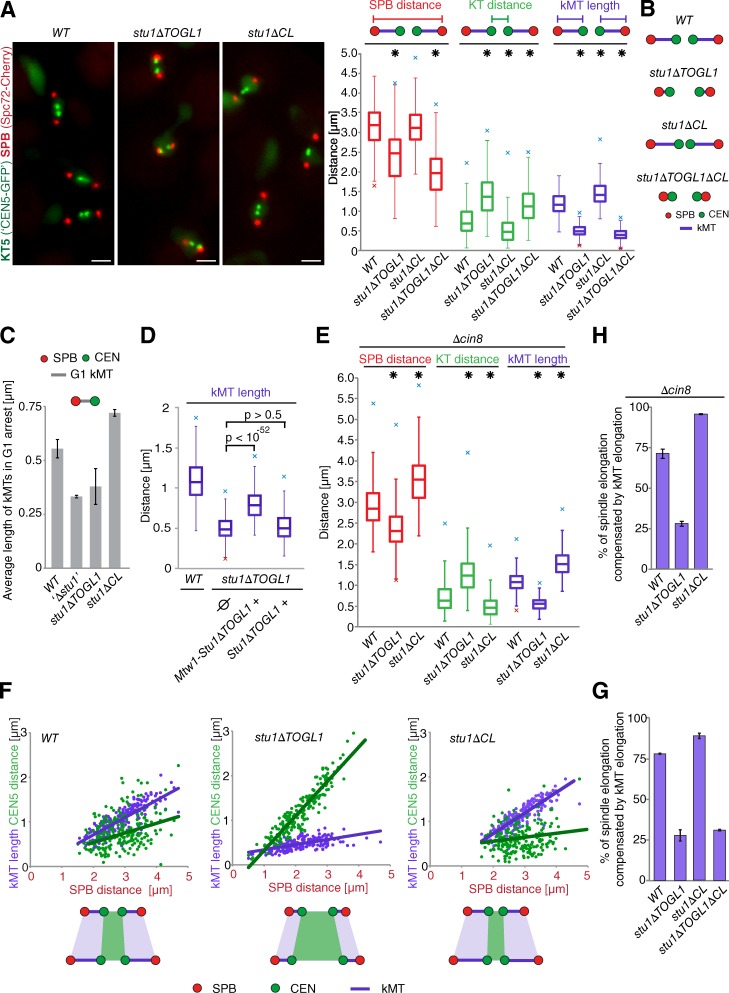
Because TOGL1 confers Stu1 binding to metaphase KTs, we asked whether TOGL1 affects kMTs. We determined kMT length as the distance between an individual CEN5-labeled KT and its corresponding SPB in metaphase. At this point, sister KTs are pulled apart along the spindle axis, whereas sister chromatid cohesion is maintained (Goshima and Yanagida, 2000). Consequently, the KT and the KT–MT interface are experiencing tension. Comparing WT and stu1ΔTOGL1 cells revealed that the mean kMT length was dramatically shorter in stu1ΔTOGL1 cells, and in agreement, the intersister KT distance was significantly increased (Fig. 5, A and B). As a consequence of the severely shortened kMTs, SPBs were pulled closer together in stu1ΔTOGL1 cells, resulting in a shorter spindle length. In principle, shorter spindles may also be caused by compromised interpolar MT (ipMT) function. This is expected to result in reduced tension on sister KTs and consequently a shorter inter-KT distance (Gardner et al., 2005). However, this distance was significantly larger in stu1ΔTOGL1 cells, and several observations indicate that the tension was increased. First, the fluorescent signal linked to tetracycline operators (tetOs) integrated in the vicinity of CEN5 appeared consistently more stretched in metaphase-arrested stu1ΔTOGL1 cells (Fig. S4 A). Second, they could cope less well with subnormal levels of the cohesion Scc1 compared with WT cells (Fig. S4 B). Collectively, we therefore conclude that deletion of TOGL1 results in destabilization of kMTs and that cells respond to the short kMTs by decreasing the SPB distance to keep the tension at the KTs at a reasonable level. Also, we analyzed the effect of TOGL1 deletion on kMT length in G1, when tension at the KT is not an issue. Again, the kMT length was shorter in stu1ΔTOGL1 cells; in fact, it was similar to Stu1 depletion (Fig. 5 C). In contrast, deletion of TOGL1 had, if anything, a stabilizing effect on non-kMTs (Fig. S4 C). Thus, the G1 data support a model in which a deletion of TOGL1 specifically destabilizes kMTs.
Figure 5.
Stu1 at metaphase KTs allows adaption of kMT length in correlation to tension at the KT–MT interface. (A–C) The kMT length is decreased in TOGL1 and increased in CL deletion mutants. (A) Cells were arrested in metaphase by Cdc20 depletion for 5 h. Bars, 2 µm. n > 200. Asterisks indicate significant differences to WT with P < 0.0001. (B) Schemes illustrating the results described in A. (C) Cells were arrested in G1 for 3 h. n > 100. Error bars represent the SDs for two experiments. (D) KT-linked Stu1ΔTOGL1 partially rescues kMT lengths in metaphase. stu1ΔTOGL1 cells additionally expressing either an Mtw1-Stu1ΔTOGL1 fusion protein or Stu1ΔTOGL1 were analyzed as in A. n > 100. Ø indicates that no additional Stu1 construct was expressed. (E) CIN8 deletion intensifies the ΔCL but fails to rescue the ΔTOGL1 phenotype. Cells with a Δcin8 background were analyzed as in A. n > 100. Asterisks indicate significant differences to WT with P < 0.0001. (A, D, and E) Boxes cover the middle 50% of the data, with a horizontal line at the median. Whiskers show the range of data (maximal 1.5× interquartile range). Maximal outliers are shown as crosses. (F) Cells were arrested in metaphase by Cdc20 depletion. (F and G) The percentage of spindle elongation that is compensated by kMT elongation is strongly decreased in the absence of TOGL1 but increased in the absence of CL in comparison with WT cells. After 2 and 5 h, inter-KT distances and kMT lengths of individual cells were determined and blotted against the corresponding inter-SPB distance. The models illustrate how kMTs and inter-KT distances react to spindle elongation as revealed by the presented data. n > 100. Lines indicate the linear regression. (G) Percentage of spindle elongation that is compensated by kMT elongation. The values represent the twofold slopes of the kMT graphs (purple) as shown in F. Error bars represent the SDs for two experiments. (H) Δcin8 strains with the indicated Stu1 constructs were analyzed as described in F and G. n > 100. Error bars represent the SDs for two experiments.
Because TOGL1 is essential for KT localization in metaphase, it is reasonable to assume that Stu1 has to localize to KTs to stabilize kMTs. To support this, we permanently linked Stu1ΔTOGL1 to the C terminus of the KT protein Mtw1 in cells that also expressed unlinked Stu1ΔTOGL1. We found that the kMT stability was significantly rescued in these cells, in contrast to cells that expressed two copies of unlinked Stu1ΔTOGL1 (Fig. 5 D). Thus, a major role of TOGL1 in kMT stabilization stems clearly from KT localization of Stu1.
In G1, kMTs are shorter than non-kMTs in WT as well as stu1ΔTOGL1 cells. This suggests that MT-destabilizing factors work at KTs. Cin8 has been proposed to be such a factor (Gardner et al., 2008). We thus wondered whether Stu1’s function at KTs is to counteract Cin8 and whether deletion of CIN8 would rescue kMT stability. However, we found the mean kMT length to be as short in stu1ΔTOGL1 Δcin8 cells as in stu1ΔTOGL1 cells (compare Fig. 5, A and E). Thus, there is probably additional MT-destabilizing activity at KTs. Moreover, the data support a model that Cin8 activity increases with kMT length (Gardner et al., 2008): because Cin8 is predestined to correct too long kMTs, deletion of CIN8 provides little help to rescue kMTs that are too short.
Finally, we asked whether Stu1 might assist a tension-dependent regulation of kMT dynamics. To investigate KT–MT interfaces under different tension, we took advantage of the fact that in metaphase-arrested cells, the SPB distance increases with increased arrest time and also varies from cell to cell. Consequently, the tension at the KT–MT interface varies till changes in kMT length compensate the changes in SPB distance. We therefore gathered data for cells arrested in metaphase for 3 and 5 h and plotted the kMT length and intersister KT distance of individual cells against the corresponding inter-SPB distances (Fig. 5 F).
For WT cells, kMT length increased with SPB length. Notably, the increase in kMT length only compensated 78% of the increase in inter-SPB distance (Fig. 5, F and G), causing an increase of 22% of the inter-KT distance and consequently a higher tension at the KT–MT interface. These observations are in agreement with a model that assumes that the stability of kMTs increases with the tension at the KT–MT interface (Gardner et al., 2005). Consequently, an increase in the inter-SPB distance would promote an increase in kMT length to a point when the kMT length fits the appropriate tension at the KT–MT interface. In stu1ΔTOGL1, the percentage of spindle elongation that was compensated by kMT elongation was severely reduced (Fig. 5 F). In fact, it compensated only 28% of the increase in inter-SPB distance (Fig. 5 G). Consequently, the inter-KT distance changed drastically with the inter-SPB distance. Again, deletion of CIN8 did not rescue this phenotype (Fig. 5 H). In conclusion, these data suggest that localizing Stu1 to metaphase KTs via TOGL1 is important to allow kMT polymerization in response to increased tension.
As reported earlier in this paper, Stu1 clearly dissociates from KTs during anaphase (Fig. 4 C). Because KT-bound Stu1 is required to stabilize kMTs in metaphase, the dissociation of Stu1 in anaphase is likely to cause shortening of kMTs and thus may regulate the initiation of anaphase A.
In stu1ΔCL cells, the kMT length is increased
When we analyzed kMT length in stu1ΔCL cells (as described for stu1ΔTOGL1 cells), we found that the mean kMT length in metaphase was significantly increased compared with WT cells (Fig. 5, A and B). Because the mean inter-SPB distance remained unchanged, the inter-KT distance was reduced for stu1ΔCL cells. In G1 or upon CIN8 deletion, kMT length was considerably longer in stu1ΔCL cells than in WT cells (Fig. 5, C and E). Thus, CL apparently attenuates Stu1’s ability to stabilize kMTs. The observation that this effect is intensified in a Δcin8 background is in agreement with a model that kMT destabilization by Cin8 increases with kMT length (see previous section; Gardner et al., 2008). stu1ΔCL cells increased kMT length more upon the increase of inter-SPB distance than WT cells (Fig. 5, F and G). Again, this effect was intensified in a Δcin8 background (Fig. 5 H). Consequently, these cells kept the inter-KT distance and thus kept the tension at the KT–MT interface nearly constant irrespective of the inter-SPB distance. Thus, stu1ΔCL Δcin8 cells elongate kMTs until a minimum standard tension is reached, and consequently, in these cells, increased kMT length does not correlate with higher tension at KTs. Irrespective of the exact function of CL in this respect, it depends on KT localization of Stu1 because the additional deletion of CL in stu1ΔTOGL1 cells did not elongate kMT nor increase the tension-dependent kMT elongation (Fig. 5, A, B, and G).
ML is required for the interaction of Stu1 with the MT lattice and for metaphase spindle localization
TOGL1 directs TOGL2 to KTs. However, Stu1’s spindle localization was largely independent of TOGL1. In cells released from G1, as well as metaphase-arrested cells and in vitro, the spindle localization of Stu1ΔTOGL1 was very similar to WT Stu1 (Fig. 6, A, B, G, and H). In agreement with this and as mentioned above, stu1ΔTOGL1 cells exhibited no severe spindle defects (Fig. 6, B and F).
Figure 6.
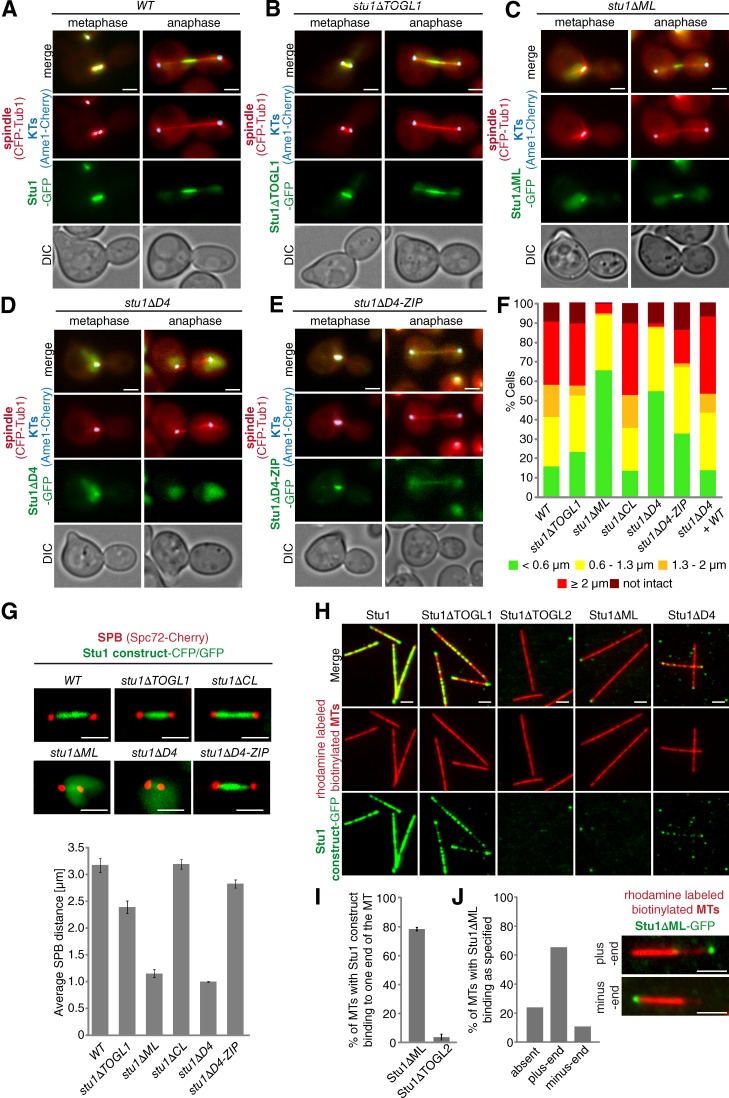
ML and TOGL2 confer interaction of Stu1 with the MT lattice. (A–G) Localization to metaphase spindles depends on ML and Stu1 dimerization. (A–E) Cells were released from G1 arrest and visualized in metaphase and anaphase. DIC, differential interference contrast. (F) Spindle phenotypes were quantified as indicated 2 or 2.5 h (Fig. S2) after G1 release. n > 100. (G) Cells were arrested in metaphase by Cdc20 depletion for 5 h. Spindle length was measured as the distance between SPB signals for n > 100. Error bars represent SDs for two experiments. (H) ML and TOGL2 confer MT lattice binding in vitro. Stabilized and immobilized MTs were incubated with purified Stu1 constructs (Fig. S5 A) and visualized as indicated. (I and J) TOGL2 confers MT plus-end binding in vitro. (I) Quantification of MTs with Stu1 at one of the ends. Experiment performed as in H. Error bars represent the SDs of two experiments. n > 150. (J) Quantification of MTs with Stu1 at the plus end. The experiment was performed as in H with the exception that polarity-marked MTs were used. The fraction of MTs with Stu1ΔML bound to plus or minus ends was determined. n = 174 . Bars, 2 µm.
Thus, we were wondering which Stu1 domains govern the localization of TOGL2 to MTs. ML and TOGL2 have previously been found to contribute to the MBD in an MT sedimentation assay (Yin et al., 2002). Our in vivo analyses showed that Stu1ΔML indeed localized only very weakly along prometa- and metaphase spindles in cells released from G1 (Fig. 6 C). In agreement, stu1ΔML cells failed to separate their SPBs (65%) or formed only short prometaphase spindles (29%; Fig. 6, C and F). In metaphase-arrested cells, their spindle length was on average ∼3× shorter than in WT cells (Fig. 6 G). In vitro, Stu1ΔML completely failed to interact with the MT lattice (Fig. 6 H), but it still associated with MT ends, predominantly plus ends (Fig. 6, I and J). The effect of TOGL2 on MT interaction was difficult to analyze in vivo because stu1ΔTOGL2 cells completely failed to form spindles (Fig. 2 E and F). The in vitro analysis showed that Stu1ΔTOGL2 completely failed to interact with the MT lattice as well as with the MT plus ends (Fig. 6, H and I).
Collectively, these results show that ML and TOGL2 strongly contribute to the interaction of Stu1 with the MT lattice and that TOGL2 additionally mediates (plus) end binding in vitro. Thus, ML mediates the interaction with the MT lattice as well as the interaction of Stu1 with metaphase spindles. This strongly indicates that Stu1 interacts with the MT lattice in metaphase.
Dimerization is required for efficient interaction of Stu1 with the MT lattice and for metaphase spindle localization
Most stu1ΔD4 cells either failed to separate their SPBs (55%; 2 h after G1) or formed short prometaphase spindles (32%; 2 h after G1; Fig. 6, D and F). When arrested in metaphase, stu1ΔD4 cells formed spindles that were on average three times shorter than in WT cells (Fig. 6 G). Furthermore, Stu1ΔD4 localized only weakly along prometa- or metaphase spindles (Fig. 6, D and G). D4 represents the dimerization domain (see above). Reconstituting Stu1ΔD4 dimerization in stu1ΔD4-ZIP cells resulted in a nearly complete rescue of prometa- or metaphase spindles and Stu1ΔD4-ZIP localized efficiently to these spindles (Figs. 6, E–G; and S3). Also, in vitro Stu1ΔD4 interacted with the MT lattice only weakly (Fig. 6 H). Collectively, these results demonstrate that efficient lateral MT interaction requires dimerization of Stu1. The fact that dimerization is also important for the localization to ipMT in metaphase confirms that lateral MT–Stu1 interaction is essential in metaphase. Our observation that the interaction of Stu1ΔD4 with the MT lattice is severely compromised in vivo and in vitro somewhat contradicts the sedimentation experiments that defined MBD (Yin et al., 2002). We thus tested the Stu1 deletion constructs in an MT sedimentation assay (Fig. S5 B). The results mirrored the microscopy experiments. WT Stu1 interacted with the MTs, whereas Stu1ΔML and Stu1ΔTOGL2 did not. Stu1ΔD4 interacted very weakly. Possible reasons for the discrepancy in respect to the MT interaction of Stu1ΔD4 might be that in our case the proteins were purified from S. cerevisiae and that we had to work at concentrations several fold below the dissociation constant (Yin et al., 2002).
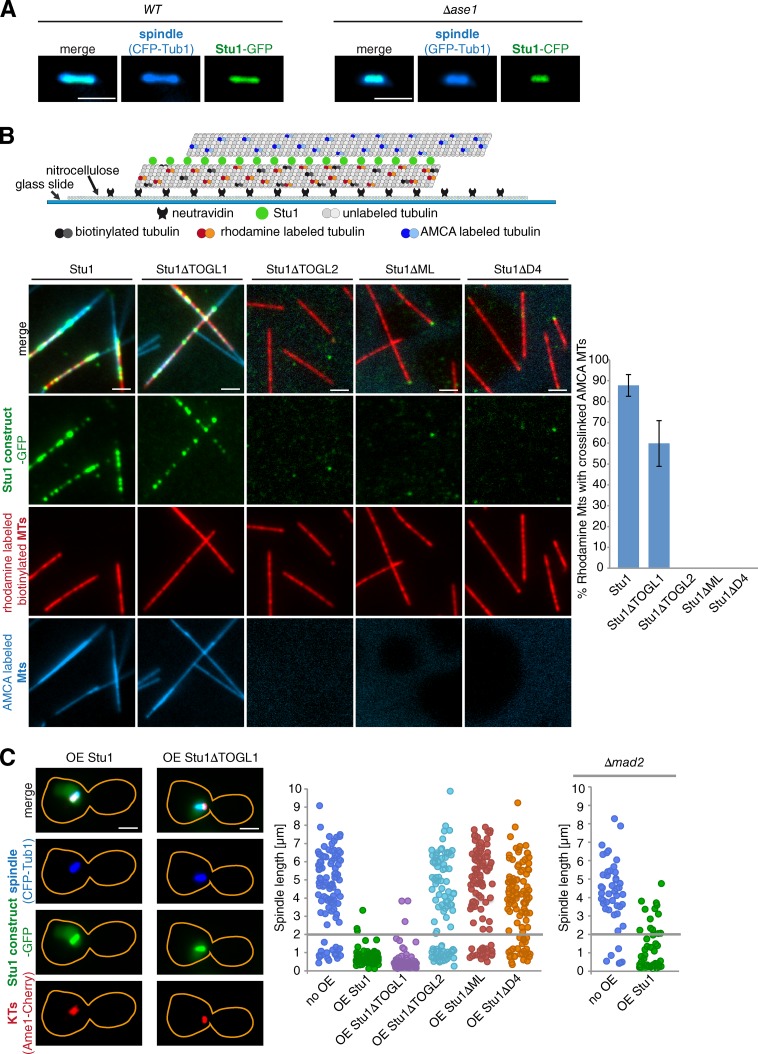
Laterally bound Stu1 may cross-link ipMTs in prometa- or metaphase independent of Ase1
The localization of Stu1 to overlapping ipMTs in anaphase (midzone) depends on Ase1 (Khmelinskii et al., 2007). The finding that Stu1 interacts with the MT lattice in metaphase suggested that the spindle localization of Stu1 is independent of Ase1 at this time. Indeed, the localization of Stu1 to metaphase spindles in Δase1 cells was indistinguishable from that of WT cells (Fig. 7 A). Ase1 is thought to cross-link ipMTs (Schuyler et al., 2003). However, Ase1, in contrast to Stu1, is not essential, and short metaphase spindles form in the absence of Ase1 (Fig. 7 A). We therefore asked whether Stu1 is involved in the cross-linking of ipMTs in metaphase. In vitro, Stu1 clearly was able to cross-link mobile MTs to MTs immobilized on a glass slide, and this depended on the interaction of Stu1 with the MT lattice (Fig. 7 B). This suggests that Stu1 might also bundle ipMT in vivo when it is bound to the MT lattice in prometa- or metaphase. Consistently, overexpression of WT Stu1 resulted in large budded cells with short, prometaphase-like spindles (Fig. 7 C). Notably, this phenotype was only observed for Stu1 constructs that interacted with the MT lattice and showed MT cross-linking activity in vitro (Fig. 7 C). It was not dependent on a functional spindle assembly checkpoint because this arrest did also occur in Δmad2 cells (Fig. 7 C). Collectively, this suggests that overexpression of Stu1 cross-links ipMTs to an extent that stalls spindle elongation. Thus, we propose that laterally bound Stu1 forms direct cross-links between ipMTs in prometa- and metaphase.
Figure 7.
Laterally bound Stu1 cross-links MTs. (A) Stu1 spindle localization in metaphase is independent of Ase1. Cells were arrested in metaphase by Cdc20 depletion for 2 h. (B) Stu1 cross-links MTs in vitro. As illustrated, stabilized MTs were immobilized, incubated with the indicated Stu1 constructs, and subsequently incubated with mobile MTs. Colocalization of immobile and mobile MTs was quantified. Error bars represent the SDs of two experiments. n > 100. (C) Overexpression (OE) of Stu1 arrests cells with metaphase-like spindles independent of the spindle assembly checkpoint. Yellow lines indicate the outline of the cells. Overexpression of the indicated Stu1 constructs was started after G1 release. The distribution of spindle lengths for cells with large buds (>2/3 of mother) was determined. Lines indicate the cutoff for metaphase spindles. n > 100 for WT, and n > 45 for Δmad2 cells. AMCA, aminomethylcoumarin acetate. Bars, 2 µm.
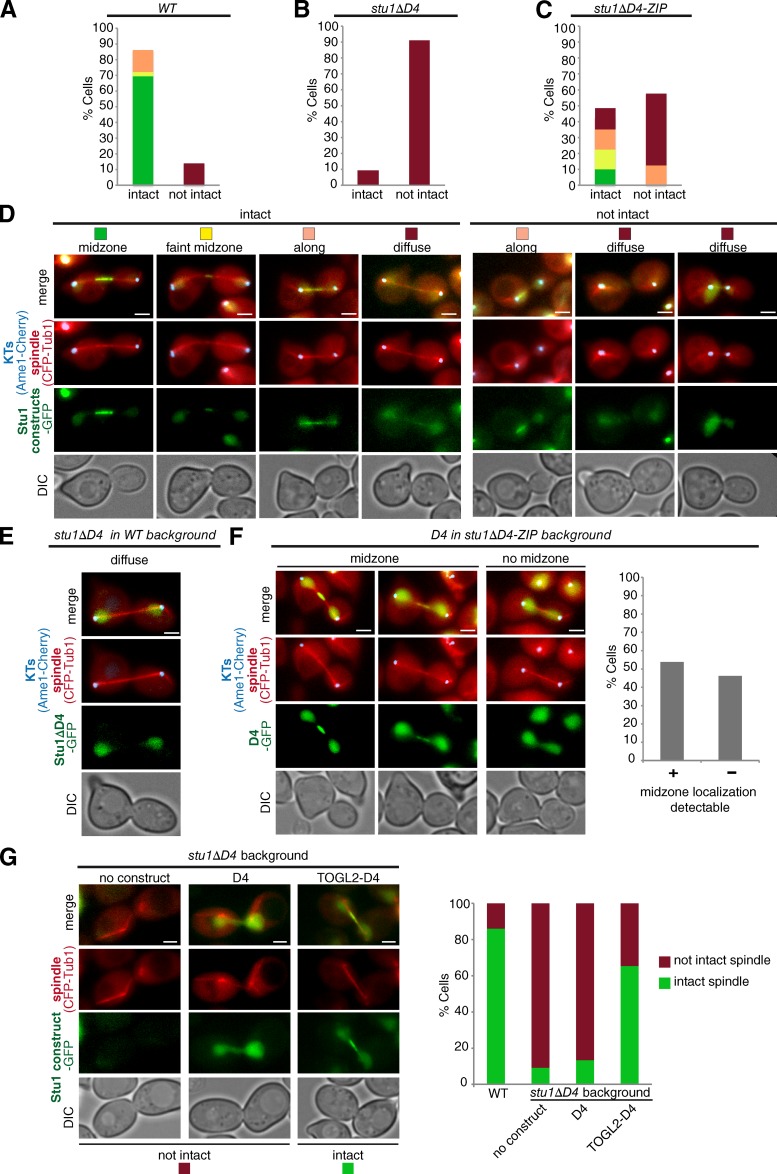
Midzone localization of Stu1 in anaphase depends on D4 but not on lateral MT interaction
As described above, Stu1ΔML fails to associate with ipMTs, and stu1ΔML cells exhibit severely compromised spindles in metaphase. Despite this, 18% of these cells had formed intact anaphase spindles when analyzed 2.5 h after the release from G1 and Stu1ΔML localized efficiently to the midzone of these spindles (Figs. 6, C and F; and S3). This indicates that in anaphase (in contrast to metaphase) Stu1 is not required to interact with the MT lattice to form stable spindles and to localize to the spindle midzone.
Also, Stu1ΔD4 failed to bind to ipMTs efficiently and produced very compromised spindles in metaphase (see above). In contrast to stu1ΔML cells, however, stu1ΔD4 cells produced predominantly defective anaphase spindles that failed to interdigitate (Fig. 8, B and D), and StuΔD4 localization was diffuse (Fig. 8, B and D). To address whether Stu1ΔD4 could localize to the spindle midzone (if present), we expressed Stu1ΔD4 in a WT STU1 background to rescue anaphase spindle and midzone formation. However, Stu1ΔD4 also failed to localize to the midzone in these cells (Fig. 8 E). Furthermore, reconstituting Stu1 dimerization (Stu1ΔD4-ZIP) rescued spindle localization of Stu1 and spindle formation in prometa- or metaphase (see above) but only moderately improved the formation of intact anaphase spindles in comparison to stu1ΔD4 cells (Fig. 8, A–D). Also, in comparison to WT Stu1, Stu1ΔD4-ZIP localized very inefficiently to midzones of intact anaphase spindles (Fig. 8 A, C, and D). This indicates that dimerization is not sufficient to localize Stu1 efficiently to the spindle midzone and that a compromised midzone localization of Stu1 causes defective spindles. Furthermore, it indicates that D4 may play a direct role in midzone localization. This was supported by the fact that, when expressed in stu1ΔD4-ZIP cells, D4 localized to the midzone of 54% of the cells that had formed intact anaphase spindles (Fig. 8 F). Thus, D4 has a direct role in midzone localization beyond dimerization, for example, by binding to other midzone proteins.
Figure 8.
D4 mediates Stu1 midzone association. (A–G) Cells were analyzed 2 h after G1 release. (A–D) D4 promotes Stu1 midzone positioning. Stu1 localization and anaphase spindle phenotypes were quantified as indicated. n > 40. (E) Stu1ΔD4 fails to localize to the midzone of intact anaphase spindles. Stu1ΔD4 was expressed in the WT background. n > 40. (F) D4 associates with the spindle midzone. Midzone localization of D4 was quantified in stu1ΔD4-ZIP cells with intact anaphase spindles as indicated. n > 100. (G) Localizing TOGL2 to the spindle midzone via D4 rescues the stu1ΔD4 spindle defect. Anaphase spindle phenotypes were quantified in stu1ΔD4 cells expressing the indicated constructs. n > 30. DIC, differential interference contrast. Bars, 2 µm.
Localization of TOGL2 to the midzone is required for anaphase spindle stability
D4 localizes Stu1, and consequently TOGL2, to the midzone in anaphase. Clearly, deletion of TOGL2 leads to cells that cannot form anaphase spindles (see above). However, because these cells also completely fail to form metaphase spindles, it is not clear whether the absence of anaphase spindles is a result of an inability to enter anaphase or because TOGL2 cannot localize to the spindle midzone. To clarify whether the formation of stable anaphase spindles requires TOGL2 at the spindle midzone, we expressed a TOGL2-D4 construct in a stu1ΔD4 background. stu1ΔD4 cells form a very high number of defective anaphase spindles (Fig. 8, B and D), but the expression of TOGL2-D4 saved spindle formation to a large extent (Fig. 8 G). In contrast, the expression of D4 in stu1ΔD4 cells had no effect (Fig. 8 G). Thus, the localization of TOGL2 to the spindle midzone via D4 is essential for the formation of stable anaphase spindles.
Discussion
TOGL1 is an unusual TOG domain
The finding that TOGL1 does not bind to tubulin but confers KT interaction in budding yeast is surprising in two ways. First, it was shown for human CLASP1 that the very C terminus is required and also sufficient to bind to KTs in the WT background (Maiato et al., 2003). Similarly, the very C terminus of Stu1 harboring D4 is required for KT binding. However, it does not confer KT binding itself. Instead, its function in dimerization is required. Thus, for Stu1, it is TOGL1 that mediates KT binding and not the very C terminus. Second, it has been assumed that an array of TOGL domains is required for their activity in MT polymerization and/or stabilization (Widlund et al., 2011). However, in agreement with our finding that TOGL1 does not interact with tubulin, sequence alignments of the predicted intra-HEAT repeat loops indicate that known tubulin determinants are missing in the TOGL1 but were found in the TOGL2 domain of Stu1 (Fig. S2). This raises the question whether TOGL1s in other CLASPs also fail to bind tubulin dimers. In general, the TOGL1s of higher eukaryotes exhibit stronger homologies to bona fide TOG domains than TOGL1 of Stu1 (Fig. S2). This includes residues (the lysine in T4 or the arginine in T5) that are important for tubulin interaction. On the other hand, like TOGL1 of Stu1, they lack the conserved tryptophan residue of T1 that is essential for tubulin binding in TOG1 of Stu2 and TOGL2 of Cls1 and the function of TOG2 in MAST (Al-Bassam et al., 2007, 2010; Ayaz et al., 2012). Although further investigation is required, this may support the view that TOGL1s of all CLASPs function as nonclassical TOG domains. In support of this, a recent publication showed that the TOGL1 domain of MAST/Orbit failed to bind tubulin in vitro (De la Mora-Rey et al., 2013).
TOGL1 confers binding and CL drives sequestration of Stu1 at uaKTs
In the presence of a uaKT, most nuclear Stu1 is sequestered at that uaKT (Ortiz et al., 2009). As a consequence, Stu1 is not available to interact with spindle MTs, and SPBs remain in close proximity and attached KTs stay close to a SPB. Both of these effects are likely to facilitate efficient biorientation of sister KTs. Domain analyses revealed that the interaction of Stu1 with uaKTs is mediated by TOGL1, and the sequestration is driven by the activity of CL because in the absence of CL, Stu1 binds to uaKTs as well as prometaphase KTs and MTs. There are several possibilities how CL could accomplish this. CL may interfere with lateral MT interaction via a direct interaction with ML. Electrostatic forces could drive this interaction because ML has a strong positive and CL has a strong negative net charge. Furthermore, interfering with lateral MT interaction could also explain why Stu1 binds preferably to uaKTs and not to attached KTs because the interaction of Stu1 with the latter depends on lateral MT interaction. Alternatively, CL may strengthen the interaction with uaKTs by providing a second (weak) KT binding site or facilitate Stu1 conformations that position the two TOGL1 domains of the dimer optimally for uaKT interaction. Finally, CL may support oligomerization of Stu1 at uaKTs (Ortiz et al., 2009).
Stu1 association with metaphase KTs requires TOGL1 and the interaction with the MT lattice via ML
When all KTs are attached to spindle MTs, Stu1 is no longer sequestered by CL and thus available to associate with MTs and attached KTs. In contrast to uaKTs, Stu1 localization to metaphase KTs also requires ML as well as TOGL1 and dimerization. This indicates that the interaction of TOGL1 with uaKTs has a different quality than with attached KTs. One possibility is that the dependency for MT interaction reflects that Stu1 reaches metaphase KTs via a kMT-facilitated transport. However, the in vivo localization of Stu1 to kMTs was barely detectable in our hands. We therefore favor the model that Stu1 requires a concomitant interaction with the KT via TOGL1 and with its kMT via ML (Fig. 9 A). In agreement with a position at the KT–MT interface, Stu1 was found to be essential for kMT stability and for the tension-dependent adaptation of kMT dynamics. Assuming Stu1 to be the effector of the tension signal, a possible mechanism is proposed in Fig. 9 A: With TOGL1 bound to the KT and TOGL2 plus ML to the kMT, at low tension, the TOGL2 domain may be displaced from the MT plus end and consequently cannot promote MT polymerization and/or stability. Upon tension at the KT caused by MT depolymerization, TOGL2 may be placed at the kMT plus end to execute its role. Interestingly, Stu1 cannot be detected at anaphase KTs (Ortiz et al., 2009). How the dissociation of Stu1 from KTs is regulated at the meta- to anaphase transition requires further investigation. However, the dissociation of Stu1 from KTs at this point clearly is a straightforward way to initiate the shortening of kMTs in anaphase A (Fig. 9 B).
Figure 9.
Model: How individual Stu1 domains direct Stu1 localization and function to KTs and MTs in metaphase versus anaphase. (A–D) See Discussion for details.
Different strategies of Stu1 localization are used to serve the individual needs of pro/metaphase and anaphase spindles
In prometa- and metaphase, Stu1 interacts directly (independent of Ase1) with the lattice of ipMTs via ML and probably TOGL2 (Fig. 9 C). We have shown that both of these domains are important for lateral MT interaction in vitro. Furthermore, we have shown that laterally bound Stu1 is able to cross-link MTs in vitro. Thus, we suggest that localizing a Stu1 dimer laterally to ipMTs in pro/metaphase not only stabilizes them via TOGL2 activity but also cross-links them. The latter may serve to form spindles sufficiently stable to withstand the forces that occur when sister KTs are kept under tension. But it may also allow only limited antiparallel gliding of ipMTs. In contrast, during anaphase, ipMTs must glide to allow spindle elongation, and Stu1 cross-linking activity would be a hindrance for this. Consistent with this, the Stu1 association with the ipMTs in anaphase is realized in a different manner. It is independent of the interaction with the MT lattice via ML but dependent on the D4 domain that localizes Stu1 to the spindle midzone (Fig. 9 D) and on Ase1 (Khmelinskii et al., 2007). In agreement with the idea that lateral MT interaction of Stu1 needs to be efficiently attenuated upon anaphase entry to allow ipMT gliding, overexpression of WT Stu1 (but not Stu1ΔML) arrested cells with metaphase-like spindles. Nevertheless, it is clear that also anaphase spindle formation requires TOGL2 activity at MTs, and the midzone localization of Stu1 is a way to verify this. Accordingly, we have shown that the spindle defect of stu1ΔD4 cells can be rescued by localizing TOGL2 to the midzone. Collectively, we propose that Stu1 interaction with the MT lattice is attenuated during the metaphase to anaphase transition, but TOGL2 activity at ipMTs is maintained to allow gliding and stabilization at the same time.
Materials and methods
Strain and plasmid construction
Yeast strains and plasmids are described in Tables S1 and S2. All used yeast strains are derivatives of YPH 499 (Sikorski and Hieter, 1989). Tagging of endogenous genes and promoter replacements were performed by PCR-mediated integration (Janke et al., 2004). To produce strains harboring STU1 deletions, either the construct was released from a corresponding plasmid and integrated into the endogenous STU1 locus, or a plasmid carrying the deletion construct was integrated into the LYS2 locus. To disrupt CIN8 and MAD2, a disruption cassette (pBK1503 and pJO607) was used. STU1 and ASE1 were disrupted with the HIS3 marker by PCR-mediated integration.
Microscopy and image processing
Live cell imaging was performed on a life science imaging station (Cell^R; Olympus) with a 100× Plan Apochromat, NA 1.4, objective and a charge-coupled device camera (ORCA-ER; Hamamatsu Photonics) at 22°C as previously described (Kemmler et al., 2009). For image acquisition, the xcellence rt software (Olympus) was used. Images had 15.625 pixels/µm and a bit depth of 16 bit. Both features were not altered by image processing. CFP, GFP, and mCherry were used as fluorochromes. For live cell imaging, cells were resuspended in nonfluorescent medium (0.9 g/liter KH2PO4, 0.23 g/liter K2HPO4, 0.5 g/liter MgSO4, 3.5 g/liter (NH4)2SO4, 0.79 g/liter Complete Supplement Mixture + all [MP Biomedicals], 0.5 mg/liter β-alanine, 0.2 mg/liter thiamin HCl, 3 mg/liter Ca-pantothenat, 2 mg/liter inositol, and 0.4 mg/liter biotin), and seven to nine z stacks with a distance of 0.25–0.35 µm were acquired. Imaging of in vitro assays was performed in buffer H (BH)/0.10 (25 mM Hepes, pH 8.0, 2 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 0.1% NP-40, 150 mM KCl, and 10% glycerol) supplemented with 2 mM taxol, 2 mM DTT, 40 mM glucose, 40 mg/ml glucose oxidase, and 16 mg/ml catalase, and one z stack was acquired. Image processing as z projection and adjustments of contrast and brightness were performed using the Fiji software (National Institutes of Health). Color channels were adjusted individually. The lookup table was linear and covered the full range of data. Length and distance measurements were performed using the measure feature of Fiji.
Statistical analyses
P-values were calculated using a two-tailed unpaired t test. Box–whisker plots were constructed as follows. The box covers the middle 50% of the data, with a horizontal line at the median. Whiskers show the range of data (maximal 1.5× interquartile range). Maximal outliers are shown as crosses.
Western blot analyses
SDS-PAGE and Western blotting were performed as previously described (Scharfenberger et al., 2003). The primary antibodies as indicated in the respective figures were rat α-HA purchased from Roche, mouse α-FLAG purchased from Sigma-Aldrich, mouse α-Tub1 purchased from Sigma-Aldrich, and rabbit α-Stu1 raised against the complete Stu1 protein. Secondary antibodies were mouse α-rat-AP purchased from Sigma-Aldrich (Fig. 1 E, α-HA), goat α-mouse-HRP purchased from Sigma-Aldrich (Fig. 1 E, α-FLAG), goat α-mouse-AP purchased from Sigma-Aldrich (Fig. 2, C and D, α-tubulin), goat α-rabbit-HRP purchased from Sigma-Aldrich (Fig. 2 D, α-Stu1), and Alexa Fluor 680 goat α-rabbit purchased from Invitrogen (Fig. 1, B and C). Fluorescence of the Alexa Fluor 680 goat α-rabbit antibody was detected by the LI-COR Biosciences system. Signal quantification was performed using the Fiji software.
Quantification of Western blots and Coomassie-stained gels
Quantifications were performed using the Fiji software. A rectangular area was chosen to quantify the intensity of each band, and the respective background intensity (above or below) was subtracted.
Mass spectrometry
Coomassie-stained proteins were cut out from gels, and in-gel digestion was performed as follows. Gel slices were washed with 150 µl of water (10 min at RT) and shrunk by incubation with 175 µl acetonitrile (15 min at RT). Acetonitrile was removed, and gel slices were dried in the vacuum. Thiol groups were reduced by incubation with 10 mM DTT in 100 mM ammonium bicarbonate (30 min at 56°C). Gel slices were shrunk, and thiol groups were alkylated by incubation in 55 mM iodoacetamide in the dark (20 min at RT). Gel slices were washed with 100 mM ammonium bicarbonate (15 min at RT), shrunk as above, and rehydrated with 20–24 µl of 20-ng/µl trypsin (Trypsin Gold; Promega) in 40 mM ammonium bicarbonate (40 min at 0°C). With the exception of 12 µl, the liquid was removed, and samples were incubated at 37°C overnight. Subsequently, the tryptic peptides present in the supernatant were purified with C18 ZipTips (EMD Millipore) according to the manufacturer’s instructions and eluted with 5 mg/ml α-cyano-4-hydroxy cinnamic acid in 50% acetonitrile and 0.1% trifluoroacetic acid onto the target plate. Analysis was performed on a mass spectrometer instrument (Reflex III MALDI-TOF; Bruker) in the reflector mode, and proteins were identified using the Mascot software (Matrix Science).
Chromosome loss assay
Cells carrying a chromosomal fragment (Shero et al., 1991) were grown in Synthetic Dextrose Complete medium lacking uracil, shifted to YPD (yeast, peptone, dextrose) medium for 3 h, diluted to 10−3 OD/ml, and then plated on YPD plates containing 4% glucose. After 3 d at 30°C, loss of the chromosomal fragment was quantified by counting colonies with red sectors that were at least half the size of the colony. More than 5,000 colonies were quantified per strain.
Protein purification and IP
For purification of proteins from S. cerevisiae, Stu1 constructs were expressed from their native promoter (Fig. 1 E) or from a pGal promoter at 2% galactose for 8–9 h (all other experiments). For protein purification, a protocol applied to isolate KTs (Akiyoshi et al., 2010) was adapted. Yeast cells were lysed for 30–40 min at 2,000 rpm with glass beads in BH/0.10 containing protease inhibitors, phosphatase inhibitors, and 2 mM DTT as described. For clarification, the cell lysate was centrifuged at 43,000 g for 30 min at 4°C. The supernatant was incubated for 4 h under constant rotation with anti-FLAG beads (anti-FLAG M2 Affinity Gel [Sigma-Aldrich]; Figs. 1 and 2, C and D) or Dynabeads (Invitrogen) coupled to α-FLAG (Sigma-Aldrich; Figs. 2 A and S5 A). Subsequently, the beads were washed six times with BH/0.10 buffer containing 200 mM PMSF and 2 mM DTT. Elution was performed by incubating the beads under gentle agitation at 25°C for 25 min with 0.5 mg/ml 3×FLAG peptide (Sigma-Aldrich) in BH/0.10 containing protease inhibitors and 2 mM DTT.
Sucrose gradient analyses
For the sucrose gradient analyses shown in Fig. 1, 80 µl of sample containing FLAG-tagged Stu1 with some associated tubulin purified from S. cerevisiae as described above was layered on a 10–40% sucrose gradient in BH/0.10 buffer and centrifuged 10 h at 55,000 rpm at 4°C in an ultracentrifuge (Optima L-90K; Beckman Coulter) using a rotor (SW 60; Beckman Coulter). 200-µl fractions were sampled. Note that the Stu1–tubulin complex dissociated during the analyses, and both proteins were obtained in different density fractions. The following standards (sedimentation coefficients are indicated in brackets) were used for determination of the Stu1 sedimentation coefficient: thyroglobulin (19.4 S), catalase (11.3 S), yeast alcohol dehydrogenase (7.4 S), bovine serum albumin (4.3 S), and cytochrome c (1.9 S).
Analytical size exclusion chromatography
Analytical size exclusion chromatography was performed on a Superose 6 10/300 column (GE Healthcare) using an ÄKTApurifier 100 system (GE Healthcare). The sample used for the experiment shown in Fig. 1 contained the FLAG-tagged Stu1 from the peak fractions of the sucrose gradient analysis. It was thus tubulin free. Chromatography was performed at 4°C and a flow rate of 0.2 ml. 0.5-ml fractions were collected. The following standards (corresponding Stokes radii are indicated in brackets) were used for the determination of the Stu1 Stokes radius: thyroglobulin (8.5 nm), ferritin (6.1 nm), catalase (5.2 nm), aldolase (4.8 nm), yeast alcohol dehydrogenase (4.6 nm), bovine serum albumin (4.6 nm), and cytochrome c (1 nm).
In vitro MT binding and cross-linking assay
Fluorescently labeled MTs were polymerized using labeled and unlabeled tubulin (bovine unlabeled tubulin, bovine X-rhodamine–labeled tubulin, porcine biotinylated tubulin, and porcine aminomethylcoumarin acetate–labeled tubulin; Cytoskeleton, Inc.) at a ratio of 1:6 at 35°C for 20 min in BRB80 (80 mM Pipes, pH 6.9, 1 mM MgCl2, and 1 mM EGTA, pH 6.9). Polarity marked MTs were first polymerized using labeled and unlabeled tubulin at a ratio of 1:3 for 10 min, and subsequently, the ratio was reduced to 1:6 for another 10-min incubation time. After polymerization, MTs were diluted 1:20 in prewarmed BRB80 containing 20 µM taxol (AppliChem) and pelleted through a BH/0.40 cushion (25 mM Hepes, pH 8.0, 2 mM MgCl2, 0.1 mM EDTA 0.5 mM EGTA 0.1% NP-40, 150 mM KCl, and 40% glycerol) supplemented with 20 µM taxol at 20,000 g for 50 min at 23°C. The pellet was resuspended in BH/0.10 buffer containing 20 µM taxol and 2 mM DTT. Slides and coverslips used for perfusion chambers were covered with a film of nitrocellulose (Collodion Solution; Sigma-Aldrich) before they were assembled into 10-µl perfusion chambers. Flow channels were incubated with 0.2 mg/ml NeutrAvidin (Thermo Fisher Scientific) in PBS for 5 min. After a wash with 30 µl BH/0.10 containing 20 µM taxol and 2 mM DTT, rhodamine-labeled, biotinylated MTs were infused into the channel. After 15 min, the channel was washed with 30 µl BH/0.10 containing 20 µM taxol and 2 mM DTT. Subsequently, Stu1 constructs supplemented with 20 µM taxol were infused into the channel at a concentration of 125 nM. For MT binding assays, Stu1 constructs were incubated for 15 min before the channel was washed with 40 µl BH/0.10 containing 20 µM taxol, 2 mM DTT, 40 mM glucose, 40 mg/ml glucose oxidase, and 16 mg/ml catalase. For cross-linking assays, Stu1 constructs were incubated for 8 min before aminomethylcoumarin acetate–labeled (but not biotinylated) MTs were infused into the flow cell. After 18 min, the channel was washed with 30 µl BH/0.10 supplemented with 20 µM taxol, 2 mM DTT, 40 mM glucose, 40 mg/ml glucose oxidase, and 16 mg/ml catalase. Fluorescence microscopy and image processing were performed as described previously in this paper.
MT sedimentation assay
We used the MT Binding Protein Spin-Down Assay kit (Cytoskeleton, Inc.) with the following exceptions. To prepare MTs, 10-µl aliquots of 5 mg/ml tubulin were polymerized according to the Cytoskeleton, Inc. protocol. To remove free tubulin, MTs were centrifuged (20,000 g for 50 min at 23°C) through a 40% glycerol cushion made in BH buffer (25 mM Hepes, pH 8.0, 2 mM MgCl2, 150 mM KCl, 0.1 mM EDTA, pH 8.0, 0.5 mM EGTA, pH 8.0, and 0.1% NP-40) supplemented with 20 µM taxol. The MT pellet was resuspended in BH buffer supplemented with 20 µM taxol, 2 mM DTT, and 10% glycerol. To remove aggregates, the Stu1 constructs used for the binding assay were centrifuged in the presence of 10 mM DTT (120,000 g for 30 min at 4°C). For the binding assay, taxol-stabilized MTs (310 nM polymerized tubulin) were incubated with 100 nM Stu1 constructs in BH buffer supplemented with 10% glycerol, 20 µM taxol, and 2 mM DTT in a total reaction volume of 25 µl at RT for 40 min. The samples were centrifuged through a 40% glycerol cushion in BH buffer supplemented with 20 µM taxol and 2 mM DTT (120,000 g for 40 min at 23°C). 20 µl of 25-µl supernatant was removed and mixed with 5 µl of 5× Laemmli sample buffer without glycerol. The pellet was resuspended in Laemmli sample buffer.
ChIP
50 OD578 yeast cells were resuspended in 45 ml of 1% formaldehyde and incubated (RT for 45 min). Cells were washed two times in cold PBS, resuspended in 200 µl of lysis buffer (50 mM Hepes/KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] sodium desoxycholate, 2 mM PMSF, and 20 µl Protease-Inhibitor Mix FY [Serva]), and lysed mechanically with glass beads. The DNA of the lysates was sheared to a size of 500–800 bp by sonication and clarified by centrifugation (10,000 g for 30 min at 4°C). 20 µl (input) was removed from the clarified lysate. The remaining lysate was incubated with 10 µl protein A–Sepharose CL-4B (Pharmacia Biotech) and 10 µl rabbit anti-GFP (Molecular Probes) antibody (overnight on a rotating wheel at 4°C). Sepharose beads were washed three times with lysis buffer, two times with wash buffer 1 (50 mM Hepes/KOH, pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% [vol/vol] Triton X-100, and 0.1% [wt/vol] sodium deoxycholate), two times with wash buffer 2 (10 mM Tris-HCl, pH 8, 0.25 M LiCl, 0.5% NP-40, 0.5% [wt/vol] sodium deoxycholate, and 1 mM EDTA), and one time with TE (10 mM Tris-HCl, pH 8, and 1 mM EDTA, pH 8) and eluted in 130 µl TE and 1% SDS (65°C; 10 min). The cross-linking in the input (supplemented with 110 µl TE and 1% SDS) and IP was reversed by incubation at 65°C for 6 h. Subsequently, the samples were incubated with 1.5 µg/µl proteinase K (2 h at 37°C), extracted with phenol, and ethanol precipitated. The nucleic acid pellets were resuspended in 50 or 20 µl TE for the input or IP, respectively. 5 µl of input (diluted 1:40) or IP (diluted 1:5) was used for the PCR. Three sets of primers specific for the CEN3 DNA (CEN3-12, 5′-GATCAGCGCCAAACAATATGG-3′; CEN3-13, 5′-AACTTCCACCAGTAAACGTTTC-3′) and the two flanking sequences CEN3-L (ChrIII-1, 5′-ACTTTGGCTTTCCGCTCGTG-3′; ChrIII-2, 5′-GAAAGTCTTCTAGAGTTACAGG-3′) and ChrIII-R (ChrIII-3, 5′-GACCAGCATGTAGGAAGGTG-3′; ChrIII-4, 5′-ACATTGATAAATTGCTCTCACCA-3′) were used for triplex PCR.
For quantification of ChIPs, the enrichment of a DNA sequence close to the centromeric region of chromosome III over an unspecific DNA region (DNA close to the PHO5 gene) was determined using RT-PCR. The following primers were used: Pho5-foward, 5′-GAATAGGCAATCTCTAAATGAA-3′; Pho5-reverse, 5′-GAAAACAGGGACCAGAATCATAAATT-3′; Pho5-probe, 5′-ACCTTGGCACTCACACGTGGGACTAGC-3′; Cen3-foward, 5′-CCTTCCGCTTATAGTACAGTACCTA-3′; Cen3-reverse, 5′-TTCAATGAATAGCTTTCTGTGGA-3′; and Cen3-probe, 5′-CGATCAGCGCCAAACAATATGGAA-3′ (Thermo Fisher Scientific). PCR reactions were performed in a volume of 20 µl in 96-well plates and contained TaqMan Gene Expression Master Mix (Applied Biosystems), 0.9 µM forward primer, 0.9 µM reverse primer, 0.25 µM TaqMan probe, and 4 µl of the probe to be analyzed. Inputs were diluted 1:50 (analyzed as controls), and co-IP probes were 1:10. The PCR was performed in the LightCycler 480 (Roche). After an initial step at 50°C for 2 min and 95°C for 10 min, 40 cycles were performed. The amplification time was 1 min with a temperature of 57°C for the DNA close to the centromeric region and 60°C for the DNA close to the PHO5 gene. Denaturation was at 95°C for 15 s. Crossing point values were derived from the absolute quantification analyses tool of the LightCycler 480 software. A standard curve was generated for both primer sets using 0.625-, 1.25-, 2.5-, 5-, and 10-attomol/µl concentrations of yeast genomic DNA and used to determine the amounts of the DNA close to the centromeric region and close to the PHO5 gene present in the probe. The x-fold enrichment of the centromeric DNA over the PHO5 gene was calculated.
Growth conditions of yeast cells
Yeast cells were grown in YP medium (yeast extract, peptone) or synthetic complete medium (yeast nitrogen base and synthetic complete dropout mixture) containing 2% glucose or 2% raffinose. To synchronize cells in G1, logarithmically growing Δsst1 cells were incubated with 250 ng/ml α factor for 2–3 h. For G1 release, cells were washed with H2O and suspended in fresh medium. To arrest cells in metaphase, logarithmically growing pMET-CDC20 cells were shifted to YPD medium containing 2 mM methionine to suppress CDC20 expression. To enrich for uaKTs and arrest cells in prometaphase, Nz was added after G1 release at 15 µg/ml. To deplete WT Stu1, cells with Ubiquitin-R-STU1 under the control of a galactose promoter were grown at YPRG (yeast extract, peptone, raffinose, galactose medium; 0.8%), shifted to YPRG (0.1%) overnight, and released into YPD 3 h before addition of α factor. To deplete Spc105, cells with Ubiquitin-R-SPC105 under the control of a galactose promoter were grown at YPRG (0.8%), shifted to YPRG (0.1%) overnight, and released into YPD 2 h before the addition of α factor.
Online supplemental material
Fig. S1 shows the calibration curves of the gel filtrations and sucrose gradient analyses used to determine the hydrodynamic properties of Stu1. Fig. S2 shows the alignment of intra-HEAT repeat loops of TOG domains from the XMAP215 and CLASP family. Fig. S3 depicts the spindle phenotype of various Stu1 mutants 2.5 h after G1 release (related to Fig. 5 G, which shows the spindle phenotype after 2 h). Fig. S4 shows data affirming that deletion of TOGL1 compromises specifically kMTs. Fig. S5 shows Stu1 constructs used for in vitro MT binding experiments and MT sedimentation assay. Table S1 shows the yeast strains used in this study. Table S2 shows plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201310018/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201310018.dv.
Supplementary Material
Acknowledgments
We thank Maria Knapp and Britta Klem for excellent technical assistance as well as Petra Ihrig and Jürgen Reichert for the mass spectrometry analyses.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft. C. Funk was additionally supported by the MD/PhD program of the medical faculty Heidelberg and Hartmut Hoffmann-Berling International Graduate School.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BH
- buffer H
- ChIP
- chromatin IP
- CL
- C-terminal loop
- CLASP
- cytoplasmic linker–associated protein
- IP
- immunoprecipitation
- ipMT
- interpolar MT
- kMT
- KT MT
- KT
- kinetochore
- MAST
- multiple asters
- MBD
- MT-binding domain
- ML
- middle loop
- MT
- microtubule
- Nz
- nocodazole
- SPB
- spindle pole body
- tetO
- tetracycline operator
- TOG
- tumor overexpressed gene
- TOGL
- TOG-like
- uaKT
- unattached KT
- WT
- wild type
References
- Akiyoshi B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., Biggins S. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 468:576–579 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., Chang F. 2011. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 21:604–614 10.1016/j.tcb.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., van Breugel M., Harrison S.C., Hyman A. 2006. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J. Cell Biol. 172:1009–1022 10.1083/jcb.200511010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., Larsen N.A., Hyman A.A., Harrison S.C. 2007. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 15:355–362 10.1016/j.str.2007.01.012 [DOI] [PubMed] [Google Scholar]
- Al-Bassam J., Kim H., Brouhard G., van Oijen A., Harrison S.C., Chang F. 2010. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev. Cell. 19:245–258 10.1016/j.devcel.2010.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P., Ye X., Huddleston P., Brautigam C.A., Rice L.M. 2012. A TOG:αβ-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 337:857–860 10.1126/science.1221698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratman S.V., Chang F. 2007. Stabilization of overlapping microtubules by fission yeast CLASP. Dev. Cell. 13:812–827 10.1016/j.devcel.2007.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., Hyman A.A. 2008. XMAP215 is a processive microtubule polymerase. Cell. 132:79–88 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Mora-Rey T., Guenther B.D., Finzel B.C. 2013. The structure of the TOG-like domain of Drosophila melanogaster Mast/Orbit. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69:723–729 10.1107/S1744309113015182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.K., Pearson C.G., Sprague B.L., Zarzar T.R., Bloom K., Salmon E.D., Odde D.J. 2005. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol. Biol. Cell. 16:3764–3775 10.1091/mbc.E05-04-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.K., Bouck D.C., Paliulis L.V., Meehl J.B., O’Toole E.T., Haase J., Soubry A., Joglekar A.P., Winey M., Salmon E.D., et al. 2008. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 135:894–906 10.1016/j.cell.2008.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett E.S., Espelin C.W., Sorger P.K. 2004. Spindle checkpoint proteins and chromosome–microtubule attachment in budding yeast. J. Cell Biol. 164:535–546 10.1083/jcb.200308100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Yanagida M. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 100:619–633 10.1016/S0092-8674(00)80699-6 [DOI] [PubMed] [Google Scholar]
- Inoue Y.H., Savoian M.S., Suzuki T., Máthé E., Yamamoto M.T., Glover D.M. 2004. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J. Cell Biol. 166:49–60 10.1083/jcb.200402052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Kemmler S., Stach M., Knapp M., Ortiz J., Pfannstiel J., Ruppert T., Lechner J. 2009. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 28:1099–1110 10.1038/emboj.2009.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Lawrence C., Roostalu J., Schiebel E. 2007. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 177:981–993 10.1083/jcb.200702145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E., Tanaka K., Komoto S., Kitamura Y., Antony C., Tanaka T.U. 2010. Kinetochores generate microtubules with distal plus ends: their roles and limited lifetime in mitosis. Dev. Cell. 18:248–259 10.1016/j.devcel.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leano J.B., Rogers S.L., Slep K.C. 2013. A cryptic TOG domain with a distinct architecture underlies CLASP-dependent bipolar spindle formation. Structure. 21:939–950 10.1016/j.str.2013.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia A.R., Garcia Z., Kabeche L., Barisic M., Maffini S., Macedo-Ribeiro S., Cheeseman I.M., Compton D.A., Kaverina I., Maiato H. 2012. Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore–microtubule attachments. J. Cell Biol. 199:285–301 10.1083/jcb.201203091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Fairley E.A., Rieder C.L., Swedlow J.R., Sunkel C.E., Earnshaw W.C. 2003. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 113:891–904 10.1016/S0092-8674(03)00465-3 [DOI] [PubMed] [Google Scholar]
- Maiato H., Khodjakov A., Rieder C.L. 2005. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat. Cell Biol. 7:42–47 10.1038/ncb1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A.L., Bakhoum S.F., Maffini S., Correia-Melo C., Maiato H., Compton D.A. 2010. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 29:3531–3543 10.1038/emboj.2010.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., Akhmanova A. 2005. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168:141–153 10.1083/jcb.200405094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J., Funk C., Schäfer A., Lechner J. 2009. Stu1 inversely regulates kinetochore capture and spindle stability. Genes Dev. 23:2778–2791 10.1101/gad.541309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca C., Draviam V.M., Marco E., Sorger P.K., De Wulf P. 2009. Roles for the conserved spc105p/kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS ONE. 4:e7640 10.1371/journal.pone.0007640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porollo A.A., Adamczak R., Meller J. 2004. POLYVIEW: a flexible visualization tool for structural and functional annotations of proteins. Bioinformatics. 20:2460–2462 10.1093/bioinformatics/bth248 [DOI] [PubMed] [Google Scholar]
- Scharfenberger M., Ortiz J., Grau N., Janke C., Schiebel E., Lechner J. 2003. Nsl1p is essential for the establishment of bipolarity and the localization of the Dam-Duo complex. EMBO J. 22:6584–6597 10.1093/emboj/cdg636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler S.C., Pellman D. 2002. Analysis of the size and shape of protein complexes from yeast. Methods Enzymol. 351:150–168 10.1016/S0076-6879(02)51845-0 [DOI] [PubMed] [Google Scholar]
- Schuyler S.C., Liu J.Y., Pellman D. 2003. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. J. Cell Biol. 160:517–528 10.1083/jcb.200210021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shero J.H., Koval M., Spencer F., Palmer R.E., Hieter P., Koshland D. 1991. Analysis of chromosome segregation in Saccharomyces cerevisiae. Methods Enzymol. 194:749–773 10.1016/0076-6879(91)94057-J [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus K.M., Freyzon Y., Faust L.Z., Sweeney H.L. 1997. Spare the rod, spoil the regulation: necessity for a myosin rod. Proc. Natl. Acad. Sci. USA. 94:48–52 10.1073/pnas.94.1.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund P.O., Stear J.H., Pozniakovsky A., Zanic M., Reber S., Brouhard G.J., Hyman A.A., Howard J. 2011. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc. Natl. Acad. Sci. USA. 108:2741–2746 10.1073/pnas.1016498108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., You L., Pasqualone D., Kopski K.M., Huffaker T.C. 2002. Stu1p is physically associated with beta-tubulin and is required for structural integrity of the mitotic spindle. Mol. Biol. Cell. 13:1881–1892 10.1091/mbc.01-09-0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.