The Cancer Genome Atlas and other large-scale cancer genome sequencing projects have identified an impressive number of significantly mutated genes that are likely drivers of cancer progression (1). These studies have definitively confirmed that the gene encoding the p53 tumor suppressor is the most frequently mutated gene in human cancers. A major component of the p53 anticancer response is its ability to arrest cell division at different stages of the cell cycle. In PNAS, a study by Shaltiel et al. provides additional insights into the mechanisms of reversal of the p53-mediated cell cycle arrest (2). Such insights may provide new cancer therapeutic opportunities.
In the normal cell, the p53 protein functions as a stress integrator that becomes activated in response to myriad dysfunctional states (e.g., damaged DNA, aberrant oncogenic signaling, and hypoxia). Once activated, p53 can initiate stress response programs to eliminate the dysfunctional state, such as repair of DNA damage. If the stressed cell is dividing, p53 can enact any one of several antiproliferative programs. Under low to moderate stress conditions, the dividing cell may be transiently arrested. Alternatively, under conditions of high stress, the cell may be permanently arrested (senescence) or eliminated completely in an ordered manner (apoptosis). In those cases where cell division is merely arrested, time is allowed for the damage or dysfunction to be repaired without being transmitted to progeny cells. Thus, p53 has been called the “guardian of the genome” (3). Given the importance of p53 as a cellular failsafe mechanism, it is not surprising that its inactivation is a highly selected event in cancer progression.
Activated p53 can halt cell division in both the G1 and G2 phases of the cell division cycle. G1 is the preparation phase of the cell before replication of its DNA and G2 prepares the cell for mitosis. Arrest and repair of cells before DNA replication or mitosis are likely to prevent damage-induced mutational events or abnormal chromosome segregation events, respectively. Once repair of damage is achieved, the cell uses pathways to relieve the p53-mediated cell cycle arrest and return to a normal dividing state.
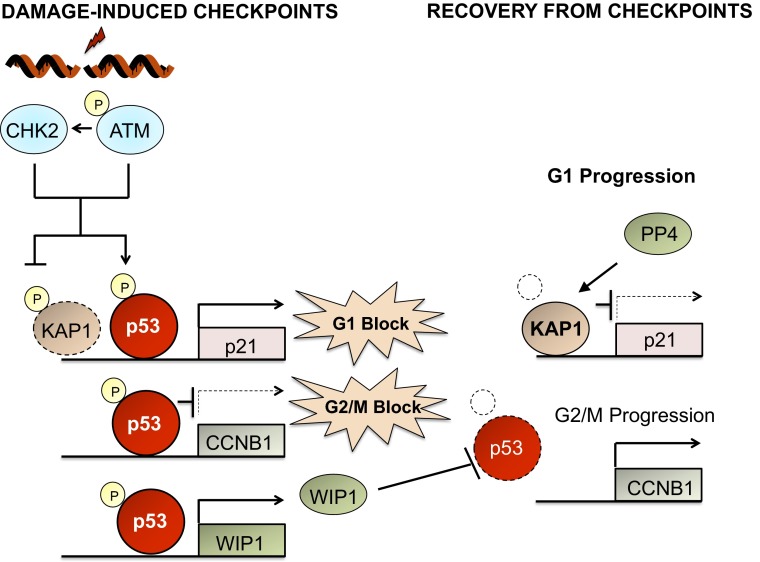
Much of the p53-mediated cell cycle arrest is effected by kinases that phosphorylate p53 (Fig. 1). Therefore, phosphatases acting on the p53 phosphorylation sites are natural candidates to reverse cell cycle arrest. Unfortunately, our knowledge of phosphatases that remove phosphate groups from p53 (about seven identified thus far) is less advanced than our knowledge of the kinases that phosphorylate it (at least 33 identified) (4). In 2005, my laboratory reported that the serine/threonine phosphatase WIP1 (PPM1D) could dephosphorylate p53 at serine 15, the same site phosphorylated by the p53-activating kinase ATM (5), and act to reverse the G2/M cell cycle arrest caused by DNA damage. The Medema laboratory later showed that WIP1 was able to relieve G2 arrest by interfering with the ability of p53 to repress expression of key G2/M progression factors such as cyclin B1 (6) (Fig. 1). Interestingly, WIP1 had earlier been discovered as a p53 transcriptional target gene (7). Thus, WIP1 is likely to operate as part of a negative feedback regulatory loop with p53 in which damage-activated p53 up-regulates WIP1 that accumulates and inactivates p53 once damage is repaired. The suppressor (WIP1) of a tumor suppressor (p53) is a strong candidate to have oncogenic properties. Indeed, the gene encoding WIP1, PPM1D, has been shown to be amplified and overexpressed in a number of human cancers (8).
Fig. 1.
Phosphatases PP4 and WIP1 reverse G1 and G2/M checkpoints. Following DNA damage, ATM and CHK2 kinases phosphorylate p53 and KAP1. Phosphorylation of p53 stimulates its transcriptional activation of the p21 gene, resulting in a G1 cell cycle arrest. ATM and CHK2 phosphorylation of KAP1 prevents its normal repression of p21 RNA expression. In addition, p53 directly binds to genes that mediate G2/M progression such as cyclin B1 (CCNB1) and inhibits their expression, enhancing G2/M cell cycle arrest. The phosphatase WIP1 is up-regulated by p53 and participates in a negative feedback loop that relieves the G2/M block through dephosphorylation of p53 (facilitating degradation of p53). The G1 block is relieved by PP4 dephosphorylation of KAP1, resulting in its reactivation and suppression of p21 RNA transcription.
A second phosphatase shown to act on p53 was the dual-specificity phosphatase 26 (DUSP26). It is an inhibitor of p53-mediated functions through dephosphorylation of serine 20 and 37, reversing the effects of the damage-induced kinases CHK1/2 and ATR (9). DUSP26 was shown to be overexpressed in primary neuroblastomas and neuroblastoma cell lines, implicating it as a potential oncogene. Yet another phosphatase that inhibits p53 signaling is PP1, which has been shown to dephosphorylate serine 15 on p53 and repress p53 activity through dephosphorylation and stabilization of the p53 inhibitor MDM4 (10). The catalytic subunit of PP1, PP1CA, has also been demonstrated to be amplified and overexpressed in oral squamous cell carcinomas (11).
The key role of WIP1 in relieving G2 arrest raised the question of whether WIP1 also functions in regulating the p53-mediated G1 arrest. Shaltiel et al. address this question in an elegant set of experiments using immortalized retinal pigment epithelial cells expressing G1 and S phase-specific fluorescent fusion proteins in cells transfected with WIP1 and control siRNAs (2). When these cells were DNA damaged with 1 Gy of ionizing radiation during G2 phase, siRNA depletion of WIP1 reduced entry of cells into mitosis by half relative to control cells, consistent with a role for WIP1 in regulating the G2 cell cycle checkpoint. In contrast, WIP1 siRNA had no apparent effect on cells irradiated in G1 in their subsequent entry into S phase. Experiments in another cell type confirmed that WIP1 has no direct role in regulation of G1 cell cycle arrest.
Because the phosphorylation of p53 is so closely linked to its effect on the G1 checkpoint, the authors initiated a screen for novel phosphatases that might reverse the p53-mediated G1 arrest. They screened an siRNA library containing 224 phosphatases and phosphatase regulators for those phosphatases that affected the kinetics of S phase entry following ionizing radiation exposure of siRNA-treated cells. A number of candidate phosphatases were identified as potential regulators of G1 checkpoint recovery, and PP4, PTPRN2, PTPN6, and DUSP2 were confirmed with additional siRNA experiments. The authors chose to further investigate the serine/threonine phosphatase PP4. PP4 is a type 2A phosphatase previously implicated in reversing phosphorylation of proteins involved in ATM-driven DNA damage response pathways (12). When the authors depleted PP4 or its regulatory subunit PP4R2 from irradiated test cells, recovery from G1 arrest was dramatically reduced. However, depletion of both PP4 and PP4R2 had no effect on G2 arrest recovery, indicating a critical role for PP4 only in regulating the G1 checkpoint. Importantly, it was demonstrated that the PP4-mediated checkpoint regulation was p53 dependent, as cells codepleted for PP4 and p53 showed a complete checkpoint recovery.
PP4 regulation of the p53-mediated G1 checkpoint was shown to be related to the activities of p53 transcriptional target gene p21CIP1, a key inhibitor of the cyclin–CDK complexes that drive cell cycle progression. KRAB domain-associated protein 1 (KAP1) is a corepressor associated with the promoter of the p21 gene (13). In unstressed cells, it inhibits p21 expression. However, on DNA damage, ATM and CHK2 phosphorylate KAP1 at serines 824 and 473, respectively. KAP1 phosphorylation deactivates KAP1 suppressive function and p21 expression is increased (Fig. 1). The authors decided to examine whether PP4 might reactivate KAP1 activity through dephosphorylation of S824 and S473. Indeed, they found that PP4 or PP4R2 depletion produced an enhanced and prolonged phosphorylation of S473, associated with increased p21 expression. The authors concluded that PP4 is an essential component of the recovery from G1 arrest through its effects on p21 expression. That PP4 may also inhibit p53 signaling in a cancer context is suggested by findings that PP4 is overexpressed in breast, lung, and pancreatic cancers (14, 15).
This study by Shaltiel et al. represents a major advance in our understanding of p53-mediated checkpoint functions and the importance of multiple phosphatases in promoting recovery from the checkpoint. They identified a phosphatase (PP4) involved in p53 checkpoint recovery and mechanistically showed how PP4 and WIP1 act in checkpoint relief. WIP1 acts primarily in G2 by preventing p53 suppression of key G2/M drivers, whereas PP4 acts in G1 by promoting down-regulation of p21CIP1 (Fig. 1). Naturally, a study such as this raises additional questions. Are there additional mechanisms of p53 checkpoint relief? Are there undiscovered phosphatases that act on p53 to relieve the checkpoint? As of this study, four phosphatases, WIP1, DUSP26, PP1, and PP4, have been definitively shown to dephosphorylate p53 or its effectors in the regulation of p53 checkpoint functions. Additional screening approaches should provide new candidates and new mechanisms of checkpoint relief in the coming years.
Of the four phosphatases that have been shown to inhibit p53 checkpoint functions, all four have been shown to be overexpressed or amplified in some tumor types, implicating them as oncogenic cancer drivers (8, 9, 11, 14, 15). A number of laboratories and pharmaceutical companies have developed small molecule inhibitors to WIP1 that have shown significant efficacy in preclinical studies in cancer cells with intact p53 (16, 17). With the discovery of PP4 as a p53 G1 checkpoint regulator, it may be worth considering combinatorial approaches with PP4 and WIP1 inhibitors in which both p53-mediated G1 and G2 checkpoints can be simultaneously enhanced, perhaps leading to more robust antiproliferative or apoptotic responses in cancer cells. At the very least, this exciting study should lead to increased interest in phosphatases as key regulators of stress responses and cancer signaling pathways.
Supplementary Material
Footnotes
The author declares no conflict of interest.
See companion article on page 7313.
References
- 1.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaltiel IA, et al. Distinct phosphatases antagonize the p53 response in different phases of the cell cycle. Proc Natl Acad Sci USA. 2014;111:7313–7318. doi: 10.1073/pnas.1322021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 4.Meek DW, Anderson CW. Posttranslational modification of p53: Cooperative integrators of function. Cold Spring Harb Perspect Biol. 2009;1(6):a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19(10):1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindqvist A, et al. Wip1 confers G2 checkpoint recovery competence by counteracting p53-dependent transcriptional repression. EMBO J. 2009;28(20):3196–3206. doi: 10.1038/emboj.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiscella M, et al. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci USA. 1997;94(12):6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulavin DV, et al. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet. 2002;31(2):210–215. doi: 10.1038/ng894. [DOI] [PubMed] [Google Scholar]
- 9.Shang X, et al. Dual-specificity phosphatase 26 is a novel p53 phosphatase and inhibits p53 tumor suppressor functions in human neuroblastoma. Oncogene. 2010;29(35):4938–4946. doi: 10.1038/onc.2010.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Z, Wan G, Guo H, Zhang X, Lu X. Protein phosphatase 1 inhibits p53 signaling by dephosphorylating and stabilizing Mdmx. Cell Signal. 2013;25(4):796–804. doi: 10.1016/j.cellsig.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu LC, Huang X, Seasholtz S, Potter DM, Gollin SM. Gene amplification and overexpression of protein phosphatase 1alpha in oral squamous cell carcinoma cell lines. Oncogene. 2006;25(40):5517–5526. doi: 10.1038/sj.onc.1209563. [DOI] [PubMed] [Google Scholar]
- 12.Lee DH, et al. Phosphoproteomic analysis reveals that PP4 dephosphorylates KAP-1 impacting the DNA damage response. EMBO J. 2012;31(10):2403–2415. doi: 10.1038/emboj.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, et al. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282(50):36177–36189. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- 14.Wang B, et al. Protein phosphatase PP4 is overexpressed in human breast and lung tumors. Cell Res. 2008;18(9):974–977. doi: 10.1038/cr.2008.274. [DOI] [PubMed] [Google Scholar]
- 15.Weng S, et al. Overexpression of protein phosphatase 4 correlates with poor prognosis in patients with stage II pancreatic ductal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2012;21(8):1336–1343. doi: 10.1158/1055-9965.EPI-12-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rayter S, et al. A chemical inhibitor of PPM1D that selectively kills cells overexpressing PPM1D. Oncogene. 2008;27(8):1036–1044. doi: 10.1038/sj.onc.1210729. [DOI] [PubMed] [Google Scholar]
- 17.Gilmartin AG, et al. Allosteric Wip1 phosphatase inhibition through flap-subdomain interaction. Nat Chem Biol. 2014;10(3):181–187. doi: 10.1038/nchembio.1427. [DOI] [PubMed] [Google Scholar]