Significance
We have used a bioinformatics approach to discover a role for the homeodomain-interacting protein kinase 2 (Hipk2) gene in adipogenesis. Using gene correlation networks from skin and mammary gland from genetically heterogeneous mice, we predicted a function for the Hipk2 gene in fat development. In support of this hypothesis, silencing of Hipk2 potently suppressed adipocyte differentiation in vitro, and deletion of Hipk2 in mice led to reduced adiposity, increased insulin sensitivity, and partial resistance to high-fat diet–induced obesity. These data demonstrate the value of gene network approaches for analysis of gene function in vivo, and provide a biological framework for discovery of potential target genes, such as Hipk2, in metabolic and other diseases.
Abstract
Homeodomain-interacting protein kinase 2 (Hipk2) has previously been implicated in the control of several transcription factors involved in embryonic development, apoptosis, cell proliferation, and tumor development, but very little is understood about the exact mechanisms through which Hipk2 influences these processes. Analysis of gene expression in normal tissues from genetically heterogeneous mouse or human populations can reveal network motifs associated with the structural or functional components of the tissue, and may predict roles for genes of unknown function. Here we have applied this network strategy to uncover a role for the Hipk2 gene in the transcriptional system controlling adipogenesis. Both in vitro and in vivo models were used to show that knockdown or loss of Hipk2 specifically inhibits white adipose cell differentiation and tissue development. In addition, loss of Hipk2 leads to induction of pockets of multilocular brown fat-like cells in remaining white adipose depots, which express markers of brown and beige fat such as uncoupling protein 1 and transmembrane protein 26. These changes are accompanied by increased insulin sensitivity in Hipk2 knockout mice and reduced high-fat diet–induced weight gain, highlighting a potential role for this kinase in diseases such as diabetes and obesity. Our study underscores the versatility and power of a readily available tissue, such as skin, for network modeling of systemic transcriptional programs involved in multiple pathways, including lipid metabolism and adipogenesis.
The highly conserved serine/threonine nuclear kinase homeodomain-interacting protein kinase 2 (Hipk2), in common with many transcriptional coactivators, corepressors, and kinases, affects the expression of multiple genes involved in a broad spectrum of signaling pathways (1, 2). Among the known binding partners of Hipk2 are Trp53, C-terminal binding protein 1 (Ctbp1), c-Myb, p300, Hmga1, Zyxin, H2B, Pc2, β-catenin, Siah2, and MeCP2 (1, 2). Despite this plethora of pathways linked to Hipk2, the known consequences of Hipk2 deletion in the mouse germ line are relatively modest (3–5), including an expansion of trigeminal sensory neurons (4) and altered maturation of dopaminergic neurons (5). Hipk2 has also been implicated in cancer development, either as a suppressor of skin tumors and lymphoma or as an oncogene amplified in pilocytic astrocytomas, but the mechanisms that underlie these phenotypes are not known (1, 2, 6–8).
In this study, we now demonstrate that Hipk2 is required for white adipocyte differentiation and development. Hipk2 knockout mice have reduced white adipose tissue mass and augmented insulin sensitivity. Moreover, white adipose tissue in knockout mice displayed an induction of brown adipocyte-like cells, which expressed markers of brown and beige fat such as uncoupling protein 1 (Ucp1) and transmembrane protein 26 (Tmem26) (9), and thermogenic genes including peroxisome proliferative activated receptor gamma, coactivator 1 alpha (Ppargc1a) and cell death-inducing DNA fragmentation factor, alpha-subunit–like effector A (Cidea).
Results and Discussion
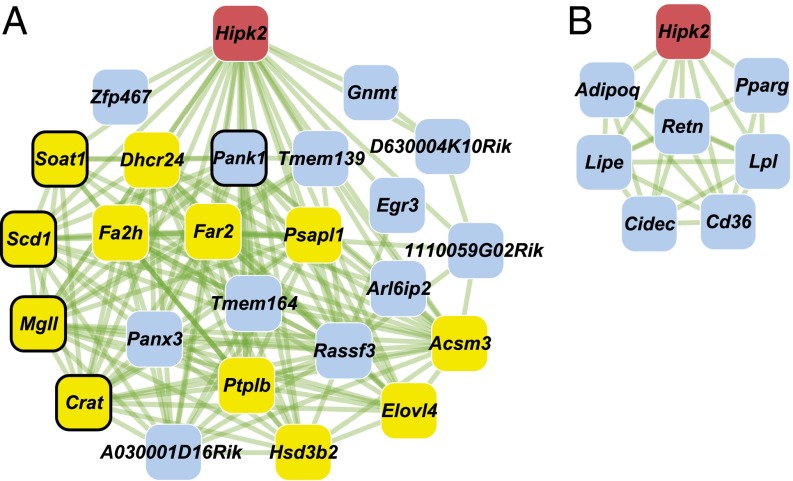
The normal tissue functions of Hipk2 in vivo, and how these pathways are disrupted during disease development, are major unanswered questions. To address this problem, we first investigated the network of genes that are correlated with Hipk2 in normal skin from a Mus spretus/Mus musculus backcross [(SPRET/Ei x FVB/N) x FVB/N; hereafter FVBBX] (10). The perturbations induced by polymorphisms inherited by individual mice in this backcross lead to changes in gene expression that can be used to create a network view of the genetic architecture of normal tissues (10, 11). This architecture can be used to suggest functions of genes based on their locations in motifs linked to specific cell compartments or signaling pathways (10). By using Hipk2 as a seed, we found that Hipk2 was correlated in expression with a group of genes associated with Pparg signaling (12, 13) and adipogenesis (Fig. 1A), suggesting that it may have functions within the adipocyte cell compartment of the skin. Several genes shown in Fig. 1A are targets of Pparg [e.g., Crat (14), Mgll (15), Pank1 (16), Scd1 (15), and Soat1 (17)], and/or have known functions in adipogenesis [e.g., Fa2h (18) and Zfp467 (19)]. Additional components of the adipocyte network were identified by using Pparg (20, 21) itself and the gluconeogenesis regulator phosphoenolpyruvate carboxykinase 1 (Pck1) (22) as seeds for correlation analysis. The network of significant correlations with Pparg revealed a conserved structure of Pparg-driven transcriptional programs that have been comprehensively characterized elsewhere (20, 21) (Fig. S1A). The energy storage regulators Cd36, Cidea, and adipose differentiation-related protein (Adfp; also known as Plin2) have been described as key targets of this nuclear receptor (20, 21). The Pck1-anchored network included several adipokines known to be secreted by adipocytes (23), such as adiponectin (Adipoq), retinol binding protein 4 (Rbp4), adipsin (Cfd), and resistin (Retn) (Fig. S1B). Although Hipk2 was not directly correlated with either Pparg or Pck1 motifs at a level significant after correction for multiple tests (Materials and Methods), further analysis of the same skin RNA samples using quantitative PCR (qPCR) showed significant connections between Hipk2, Pparg, and Pck1 (Table 1) as well as between several other representative genes from Fig. 1A and Fig. S1 A and B.
Fig. 1.
Skin gene networks suggest a role for Hipk2 in adipocyte transcriptional programs. (A) Hipk2 (red) correlation network in epidermis from FVBBX mice (n = 71). Network edges (green lines) denote coexpression (rho ≥ 0.64) links between genes (red, blue, and yellow). Nodes with black borders have been described as Pparg target genes, and yellow-colored nodes indicate genes annotated in the most significant Gene Ontology enrichment term for the gene network (lipid metabolic process; P = 4 × 10−8). (B) Hipk2 (red) correlation subnetwork (rho ≥ ±0.65) in normal mammary glands (n = 115) from an independent FVBBX mouse population.
Table 1.
Hipk2, Pparg, and Pck1 are positively correlated with genes involved in adipogenesis and lipid metabolism
| Gene correlations | Spearman’s rho | P value |
| Hipk2 correlations | ||
| Fa2h | 0.66 | 2.8 × 10−4 |
| Pank1 | 0.77 | 5.4 × 10−6 |
| Scd1 | 0.71 | 4.4 × 10−5 |
| Pparg | 0.91 | 2.0 × 10−10 |
| Pck1 | 0.72 | 5.6 × 10−5 |
| Pparg correlations | ||
| Adfp | 0.83 | 1.4 × 10−7 |
| CD36 | 0.87 | 6.9 × 10−9 |
| Cidea | 0.73 | 2.5 × 10−5 |
| Pck1 | 0.64 | 6.6 × 10−4 |
| Pck1 correlations | ||
| Adipoq | 0.93 | 9.4 × 10−12 |
| Cfd | 0.78 | 5.5 × 10−6 |
| Lipe | 0.66 | 3.9 × 10−4 |
| Rbp4 | 0.71 | 9.2 × 10−5 |
For validation of microarray results, qPCR analysis was performed using FVBBX skin samples (n = 26) and correlation coefficients (Spearman’s rho) with matching P values were calculated using Spearman’s rank correlation.
We further investigated a possible function for Hipk2 in adipogenesis by analysis of a completely independent gene expression microarray dataset derived from 115 normal FVBBX mammary glands. In contrast to skin, white fat is a major cell compartment of the mammary gland, and this analysis also showed a striking link between Hipk2 expression and lipid metabolism (Fig. 1B), particularly to Pparg itself (rho = 0.66) and several genes shown in Fig. S1 A and B (e.g., Retn, Lipe, Adipoq, Lpl, Cidec, and Cd36). We conclude that analysis of normal tissue gene expression networks reveals strong transcriptional cross-talk between many of the known genes and pathways controlling adipogenesis and lipid metabolism, unveiling a role for Hipk2 in these processes. We therefore carried out a series of in vitro and in vivo studies to explore possible functional links between Hipk2 and adipogenesis.
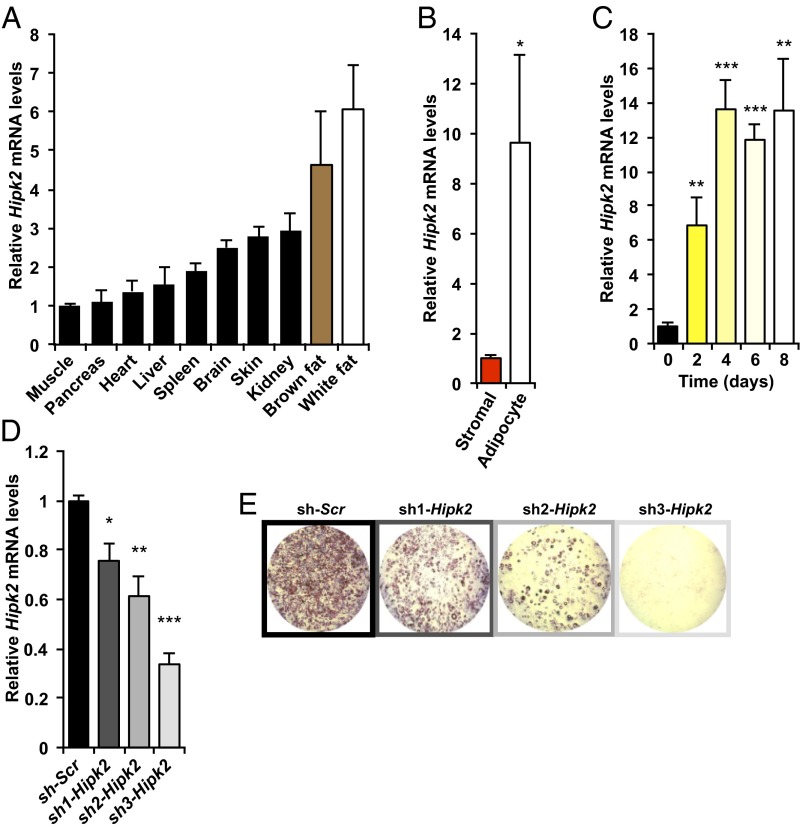
Mammals have two major functionally distinct types of adipose tissue, white and brown. White adipose tissue (WAT) stores energy primarily in the form of triglycerides (obesity arises from excessive WAT deposition of lipids). In contrast to WAT, brown adipose tissue (BAT) dissipates energy through thermogenesis (20, 21, 24). We analyzed Hipk2 expression in a wide array of mouse tissues, revealing the highest expression in BAT and WAT (Fig. 2A). In the latter, Hipk2 was associated with the adipocyte fraction of adipose tissue, whereas the stromal vascular fraction showed markedly lower levels (Fig. 2B). We next assessed the expression of Hipk2 during adipocyte differentiation of 3T3-L1 cells, a well-characterized in vitro model of adipocyte differentiation. Expression of Hipk2 in undifferentiated 3T3-L1 cells was low (Fig. 2C), but was significantly increased as early as 2 d after treatment with an adipogenic mixture and was induced up to 10-fold after 8 d of differentiation. These findings support the prediction from network analysis that Hipk2 is linked to adipocyte-associated transcriptional programs and therefore is positively associated with adipose tissue development. To more directly address the function of Hipk2 during adipocyte differentiation, we expressed short hairpin RNAs (shRNAs) targeting Hipk2 in 3T3-L1 cells. Knockdown of Hipk2 markedly reduced 3T3-L1 differentiation into fully mature adipocytes (Fig. 2 D and E).
Fig. 2.
Hipk2 expression is enriched in white fat in vivo and is positively associated with white adipocyte differentiation in vitro. (A) Hipk2 mRNA tissue distribution in 8-wk-old female 129/SvJ:C57BL/6J mice (n = 3). Data are presented as mean ± SEM. (B) mRNA levels of Hipk2 in the adipocyte and stromal vascular fractions of ovarian WAT from female 129/SvJ:C57BL/6J FVB mice (n = 4). *P < 0.05 for stromal vascular fraction versus adipocyte fraction. Data are presented as mean ± SEM. (C) Hipk2 mRNA levels in undifferentiated (day 0) and differentiation-induced 3T3-L1 cells (n = 3–4). (D) Hipk2 expression levels in 3T3-L1 cells stably expressing control shRNA (sh-Scr) or shRNAs against Hipk2 (sh1-Hipk2, sh2-Hipk2, and sh3-Hipk2) (n = 4). *P < 0.05, **P < 0.01, and ***P < 0.001 versus day 0 or sh-Scr in C and D, respectively. Data are presented as mean ± SEM. (E) Oil red O staining of control (sh-Scr) and Hipk2 knockdown (sh1-Hipk2, sh2-Hipk2, and sh3-Hipk2) 3T3-L1 cells that were induced to differentiate into adipocytes for 10 d.
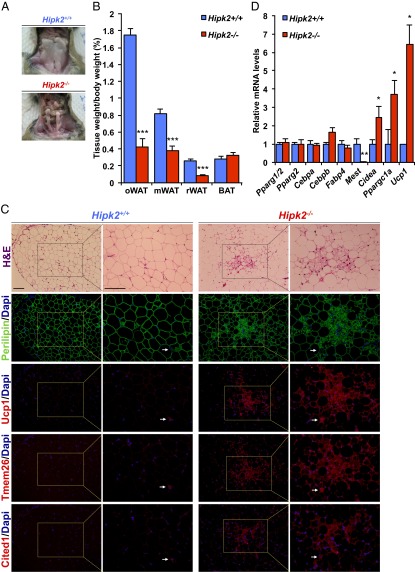
Consistent with these in vitro observations, analysis of tissues from Hipk2 knockout mice revealed a striking and selective depletion of white fat masses (Fig. 3 A and B and Fig. S2A). This depletion was accompanied by a reduction in adipocyte size and the presence of groups of smaller multiloculated cells, suggesting that loss of Hipk2 impinges on adipocyte differentiation (Fig. 3C and Fig. S2B). This appears to be a WAT-specific effect, as there was no evidence that ablation of Hipk2 affected other tissues investigated (Fig. S3). In support of the evident morphological browning of the white fat depots, these pockets showed robust immunostaining for the mitochondrial protein and BAT marker Ucp1 (21, 24) (Fig. 3C). Recently, a novel type of fat cell, known as beige fat (9) [also known as “brite” (25)], was identified within WAT that is functionally very similar to brown fat. To test the possibility that these Ucp1-positive cells are indeed adaptive beige adipocytes, we investigated the specific expression of beige fat markers in Hipk2−/− mice by immunohistochemistry. This analysis revealed that the beige adipocyte markers Tmem26 (9) and Cited1 (26) were indeed expressed specifically in the morphologically altered cells in the white fat depots of Hipk2−/− mice (Fig. 3C). Additional analysis of gene expression in WAT from Hipk2−/− mice furthermore demonstrated changes consistent with the appearance of brown-like adipocytes in the WAT depots. TaqMan analysis of gene expression in white fat from control and Hipk2−/− mice using a range of probes for adipocyte genes showed that BAT-selective genes such as Ucp1, Ppargc1a, and Cidea were induced whereas general adipogenic differentiation markers (Pparg1/2, Pparg2, Cebpa, Cebpb, and Fabp4) remained unchanged in WAT of Hipk2 knockout mice (Fig. 3D). Mesoderm-specific transcript (Mest), a gene known to regulate adipocyte size (27), was significantly down-regulated in WAT of Hipk2−/− mice (Fig. 3D). Pparg forms heterodimers with the retinoid X receptor alpha (Rxra) and modulates gene transcription by binding to Ppar response elements (PPREs) located in the promoter regions of target genes (20, 21). Consistent with the unchanged mRNA levels of Pparg between the genotypes in Fig. 3D, Hipk2 had no effect on a PPRE-luciferase reporter (Fig. S4A), further indicating that the role of Hipk2 in adipogenesis is not likely mediated through a Pparg-dependent mechanism. PR domain containing 16 (Prdm16) and Cebpb form a transcriptional complex that induces the expression of brown/beige fat genes such as Ppargc1a and Ucp1 (24). Hipk2 WT and kinase-dead Hipk2 (Hipk2 KD) repressed a Ppargc1a-luciferase reporter in the presence of Prdm16 and Cebpb, indicating that Hipk2 negatively regulates this brown fat fate regulator in a kinase-independent manner (Fig. S4B). Altogether, these results demonstrate that Hipk2 is required for normal differentiation of white adipocytes and that Hipk2 knockout mice display a pronounced browning phenotype.
Fig. 3.
Hipk2 ablation in vivo is associated with impaired white adipose tissue development and browning/beigeing of WAT depots. (A) Representative abdominal images of 8-wk-old female Hipk2+/+ and Hipk2−/− mice. (B) Weight of adipose depots (o, ovarian; m, mesenteric; r, retroperitoneal) from 8- to 10-wk-old female Hipk2+/+ (n = 8) and Hipk2−/− (n = 8) mice after normalization to body weight. (C) Consecutive sections of ovarian WAT from Hipk2+/+ and Hipk2−/− 8-wk-old female mice stained with H&E or with antibodies against the general adipocyte marker perilipin (green), the brown adipocyte marker Ucp1 (red), and the beige adipocyte markers Tmem26 (red) and Cited1 (red). Nuclei were counterstained with DAPI (blue). White arrows denote blood vessels in serial sections. (Scale bars, 100 μm.) (D) mRNA levels of Pparg1/2, Pparg2, Cebpa, Cebpb, Fabp4, Mest, Cidea, Ppargc1a, and Ucp1 in ovarian WAT of 8- to 10-wk-old female Hipk2+/+ (n = 4) and Hipk2−/− (n = 5) mice. *P < 0.05, **P < 0.01, and ***P < 0.001 for Hipk2+/+ versus Hipk2−/− mice. Data are presented as mean ± SEM.
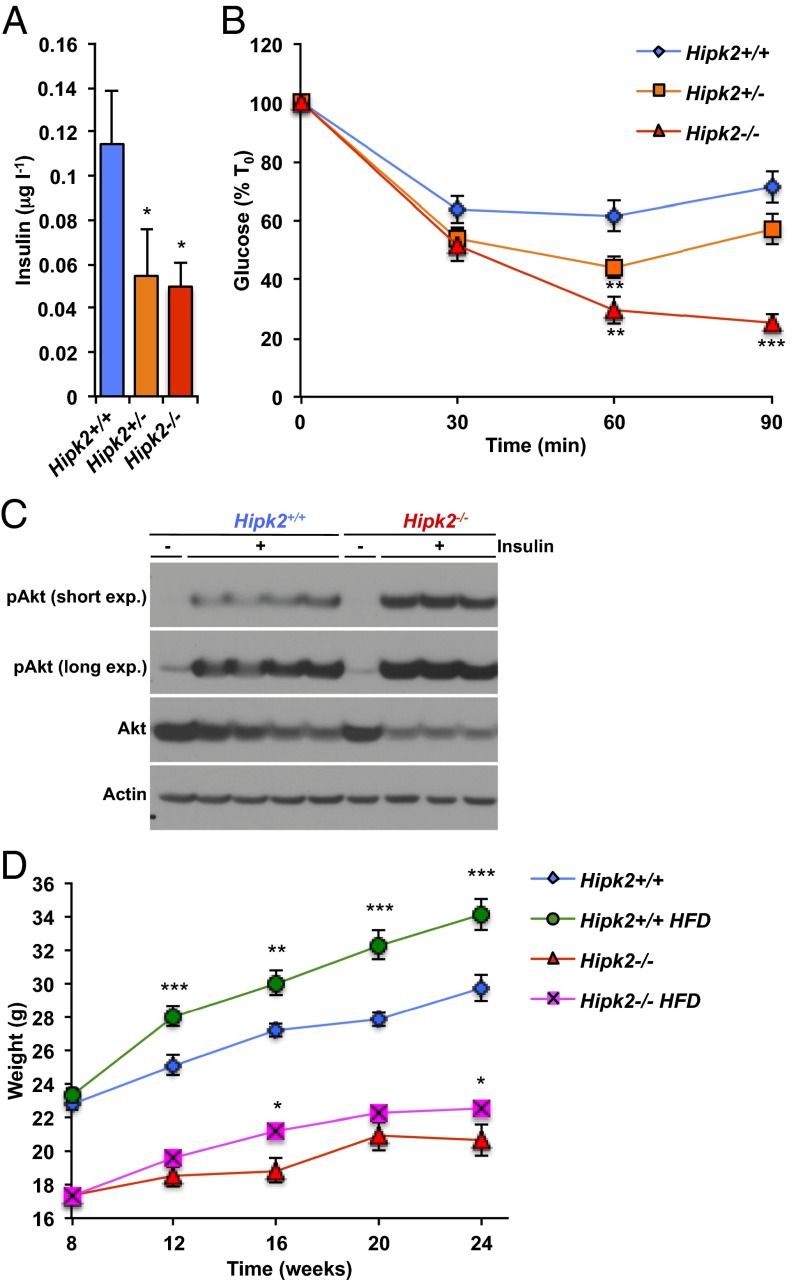
The dramatic effects of Hipk2 loss on adipose tissue development prompted us to evaluate the metabolic effects of targeted deletion of this kinase in mice. Whereas free fatty acid levels (Fig. S5A), glucose levels (Fig. S5 B and C), and glucose tolerance (Fig. S5 D and E) remained unchanged in Hipk2 knockout mice, basal insulin levels were significantly reduced (Fig. 4A and Fig. S6A) and insulin sensitivity was increased, as evidenced by a considerable lowering of plasma glucose levels in response to insulin administration (Fig. 4B and Fig. S6B). Augmented insulin-induced phosphorylation of Akt on Ser473 in skeletal muscle confirmed the enhanced insulin sensitivity of Hipk2-null mice relative to wild-type mice (Fig. 4C). Interestingly, female mice were apparently more sensitive than males to Hipk2 loss, as higher insulin sensitivity was seen in heterozygous female mice (Fig. 4B) but not in heterozygous male mice (Fig. S6B). A similar sex effect was seen in overall weight between males and females, as the latter showed significant weight loss even in the heterozygous state (Fig. S7 A and B). In an attempt to investigate the reasons for the sex-specific consequences of Hipk2 deletion, we carried out further characterization of the transcriptional signature observed in Hipk2-centric correlation analysis, segregating samples according to sex. To our surprise, the correlations to the adipocyte-associated genes were strongly decreased in male samples (Table S1) but were substantially more prominent in female mice (Table S1). We propose that sex-specific wiring of Hipk2 transcription networks, by mechanisms that remain to be found, underlies the stronger phenotypes seen in female Hipk2 heterozygous mice compared with their male counterparts. Finally, paucity of adipose tissue development in Hipk2-null mice led us to evaluate whether mice defective for this gene are resistant to high-fat diet–induced obesity. Indeed, a 16-wk period of high-fat diet (45% kcal from fat) feeding revealed that Hipk2 knockout mice are partially resistant to diet-induced weight gain (Fig. 4D). This difference in response to high-fat diet was not influenced by changes in food consumption, which was the same for the two genotypes (Fig. S8A). Serum leptin levels were significantly lower in Hipk2−/− mice than in wild-type mice when fed a high-fat diet whereas no significant differences in leptin levels were observed on normal chow (Fig. S8B), indicating that leptin levels only correlate with the adipose tissue mass in high-fat diet–fed mice.
Fig. 4.
Loss of Hipk2 increases insulin sensitivity and reduces high-fat diet–induced weight gain. (A) Plasma insulin levels of 8- to 10-wk-old female Hipk2+/+, Hipk2+/−, and Hipk2−/− mice (n = 14, Hipk2+/+; n = 16, Hipk2+/−; n = 10, Hipk2−/−). (B) Insulin tolerance tests on 8- to 10-wk-old female Hipk2+/+, Hipk2+/−, and Hipk2−/− mice (n = 14, Hipk2+/+; n = 17, Hipk2+/−; n = 8, Hipk2−/−). T0, time zero (start of the experiment, i.e., the basal level when insulin was injected). (C) Western blot analysis of Ser273-phosphorylated Akt (pAkt), Akt, and actin of skeletal muscle lysates from control (-) and insulin-treated (+) female Hipk2+/+ and Hipk2−/− mice. (D) Body weights of male Hipk2+/+ (n = 12–20) and Hipk2−/− (n = 26–28) mice on a standard diet or high-fat diet (HFD). In A and B, *P < 0.05, **P < 0.01, and ***P < 0.001 for Hipk2+/+ versus Hipk2+/− or Hipk2−/− mice, and in D, for standard diet versus HFD within each genotype. Data are presented as mean ± SEM.
The available data suggest that Hipk2 may regulate adipocyte differentiation and thermogenesis through its known complex with Ctbp1 and other corepressor proteins (2, 7, 28, 29). Alternative binding partners for Ctbp1, for example Rip140, Ppargc1a, and Prdm16, have been shown to play important roles in control of fat differentiation (24, 30–32). Hipk2 has been shown to bind Ctbp1 through the C-terminal YH domain (7), resulting in repression of transcription of target genes. Although deletion of the Hipk2 kinase domain does not affect the recruitment of Ctbp1 (7), the kinase activity may have other regulatory roles in adipogenesis, as it is known to phosphorylate Ctbp1 in kinase assays (29). Identification of these regulatory mechanisms may provide a novel route to manipulation of this pathway to influence energy balance and identify drug targets for obesity or other metabolic diseases. The latter possibility is supported by studies identifying an extensively replicated obesity linkage peak containing HIPK2 and a genome-wide association study that has identified a significant association between a SNP (rs10954654) located close to HIPK2 on human chromosome 7q32–q34 and type 2 diabetes (33–36).
Materials and Methods
Animals.
The generation of M. spretus/M. musculus backcross mice (FVBBX mice) and gene expression analysis of FVBBX mouse tail skin have previously been described (10). RNA extraction and gene expression analysis of FVBBX mammary glands were essentially done as described for tail skin (10). Microarray results have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) and are accessible under accession nos. GSE12248 and GSE46077. Hipk2−/− mice were maintained on a mixed 129/SvJ and C57BL/6J background and have been described previously (4). All animals were maintained in a temperature-controlled colony room with a 12-h light/dark cycle and kept on a standard diet (13% kcal from fat; LabDiet; PicoLab) with free access to food and water. For high-fat diet experiments, 8-wk-old male mice were started on a diet containing 45% kcal from fat (Harlan; Teklad) for 16 wk. Food intake normalized to body weight was measured during 4 consecutive weeks by the age of 10–14 wk. Experimental protocols were approved by the Laboratory Animal Resource Center at the University of California, San Francisco.
Isolation of Adipocytes and Stromal Vascular Fraction.
Freshly isolated adipose tissue was minced with scissors and incubated with 1.5 mg ml−1 collagenase (Sigma-Aldrich) at 37 °C for 45 min. Samples were then filtered through a nylon filter (100-μm). The resulting suspension was centrifuged at 1,000 × g for 5 min and the floating fraction (adipocytes) and pellet (stromal vascular fraction) were recovered.
Cell Culture.
3T3-L1 cells were obtained from the American Type Culture Collection and maintained in DMEM [University of California, San Francisco (UCSF) Cell Culture Core Facility] supplemented with 10% (vol/vol) FCS (UCSF Cell Culture Core Facility). For differentiation assays, 2-d postconfluent cells (defined as day 0) were exposed to DMEM containing 10% FCS, 5 μg ml−1 insulin, 1 μM dexamethasone, and 0.5 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich) until day 2. Induction medium was then replaced with DMEM supplemented with 5 μg ml−1 insulin, which was replaced every second day until cells were harvested or stained at the indicated time points. Adipocyte lipid content of cells was detected by oil red O staining (Sigma-Aldrich) using a standard protocol.
Short Hairpin RNA Construction.
Double-stranded DNA sequences encoding Hipk2 were cloned into the pSUPER vector (Oligoengine). The following nucleotide sequences were used for cloning into the retroviral vector: sh1-Hipk2 5′-GATCCCCGCTCCATGACCAACACCTATGTTCAAGAGACATAGGTGTTGGTCATGGAGCTTTTTGGAAA-3′; sh2-Hipk2 5′-GATCCCCGCATGCTCATGGAAGTCATTATTCAAGAGATAATGACTTCCATGAGCATGCTTTTTGGAAA-3′; sh3-Hipk2 5′-GATCCCCGGTGAAGCGAGTTATTAATTGTTCAAGAGACAATTAATAACTCGCTTCACCTTTTTGGAAA-3′. All shRNA constructs were confirmed by DNA sequencing. Vectors were transfected into HEK293 cells, and supernatants were collected 48 h after transfection. 3T3-L1 cells were transduced with 8 μg ml−1 polybrene (Millipore) and stable infected cells were selected by 1 μg ml−1 puromycin.
Luciferase Reporter Assays.
3T3-L1 cells were transiently transfected with PPRE ×3-TK-luc, Ppargc1a-luc, Pparg, Rxra, Prdm16, Cebpb, Hipk2 WT, and/or Hipk2 KD constructs by using Lipofectamine LTX (Invitrogen). phRL-TK Renilla expression vector (Promega) was used as a control for transfection efficiency. Cells were lysed and assayed for luciferase and Renilla activities using the Dual-Luciferase Reporter Assay System (Promega). The Hipk2 expression vectors were kindly provided by Yongsok Kim (National Institutes of Health, Bethesda) and Qinghong Zhang (University of Colorado, Denver).
Histology and Immunofluorescence.
WAT, BAT, liver, pancreas, and skeletal muscle were fixed in 10% formalin, embedded in paraffin, and sectioned at 5 μm. Slide tissues were stained with hematoxylin and eosin (H&E). For immunofluorescence of ovarian WAT, the paraffin-embedded specimens were deparaffinized and microwave-treated according to standard procedures with citrate buffer (pH 6) as antigen retrieval solution. After incubation in blocking solution [10% donkey serum in PBS (1% BSA)] for 1 h, slides were incubated overnight at 4 °C with rabbit anti-perilipin (Cell Signaling Technology), rabbit anti-Cited1 (Abcam), rabbit anti-Ucp1 (Abcam), and rabbit anti-Tmem26 (Sigma). The sections were then washed and incubated for 1 h at room temperature with fluorochrome-conjugated secondary antibodies [Alexa Fluor 488 anti-rabbit and Alexa Fluor 555 anti-rabbit (Molecular Probes)] diluted in PBS-T [PBS with 0.1% Tween 20 (Sigma-Aldrich)]. Slides were counterstained with DAPI. Tissue sections were analyzed using an Olympus BX60 microscope and images were captured with an Olympus DP71 camera. Cell size was analyzed using ImageJ software (National Institutes of Health). We analyzed at least 200 cells per sample from six random nonoverlapping fields.
Quantitative Real-Time PCR Analyses.
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Purified RNA was reverse-transcribed into cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative PCR was performed using mouse-specific TaqMan probes (Applied Biosystems) and TaqMan Universal PCR Master Mix (Applied Biosystems). Primers used for qPCR are shown in Table S2. Samples were run in triplicate on the Applied Biosystems PRISM 7900HT System. Relative mRNA levels were calculated using the comparative Ct method (37) and normalized to three endogenous reference genes (Sdha, Ywhaz, and Actin). The normalized quantities were rescaled relative to the sample with lowest or highest relative quantity.
Free Fatty Acid, Glucose, Insulin, and Leptin Levels.
Free fatty acid levels were measured with a commercially available kit (Wako Diagnostics) in mice that were fasted for 6 h. For glucose, insulin, and leptin measurements, mice were fasted overnight (16 h) and blood samples were collected at euthanasia. Serum glucose levels were assessed with a glucometer (Abbott Laboratories) and serum insulin levels were measured using an insulin ELISA system (Millipore). Plasma leptin levels were determined by ELISA kit (R&D Systems) according to the manufacturer’s instructions.
Glucose and Insulin Tolerance Tests.
For glucose tolerance tests, an i.p. dose of d-glucose (2 g kg−1 body weight) was administered to overnight-fasted (16 h) mice. For insulin tolerance tests, animals were fasted for 6 h before an i.p. injection of insulin (0.3 U kg−1 body weight). Tail blood glucose levels were monitored immediately before and at the indicated time points after injection using a glucometer (Abbott Laboratories).
Western Blot Analysis.
Mice were injected with insulin (0.3 U kg−1 body weight) 5 min before skeletal muscles were harvested for analysis. Total protein extracts were prepared from mouse skeletal muscle (gastrocnemius) with STEN lysis buffer (50 mM Tris, pH 7.4, 2 mM EDTA, 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, and 0.5% Triton X-100) containing Complete Protease Inhibitor mixture (Roche). Lysates were separated by SDS/PAGE and blotted onto Immobilon-P (Millipore) membranes. The membranes were incubated with rabbit anti-Akt (Cell Signaling Technology), rabbit anti-Ser473 pAkt (Cell Signaling Technology), and rabbit anti-actin (Sigma-Aldrich). HRP-conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories. Proteins were detected by enhanced chemiluminescence (Pierce).
Statistical Analysis.
Statistical significance was evaluated using a two-tailed, unpaired Student t test or ANOVA. Differences were considered significant at P values of less than 0.05. Microarrays were background-corrected and normalized by RMA using the oligo package (38) in the R statistical environment (39). A 5% genome-wide significance cutoff for correlation was assessed using the permutation approach as described (40). Correlation networks were plotted using Cytoscape (41).
Supplementary Material
Acknowledgments
We are grateful to Jian-Hua Mao for useful discussions at an early stage of this work and to other members of the A.B. laboratory for technical assistance and comments on the manuscript. This work was supported by National Institutes of Health/National Cancer Institute (NCI) Grants U01 CA84244 and U01 CA141455 (NCI Mouse Models of Human Cancer Consortium) and The Barbara Bass Bakar Professorship of Cancer Genetics (to A.B.). J.S. was supported by a postdoctoral fellowship from the Swedish Research Council and the Tegger Foundation. F.G.P. was supported by a postdoctoral fellowship from the Pew Latin American Fellows Program.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The microarray results reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE12248 and GSE46077).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322275111/-/DCSupplemental.
References
- 1.Hofmann TG, Glas C, Bitomsky N. HIPK2: A tumour suppressor that controls DNA damage-induced cell fate and cytokinesis. Bioessays. 2013;35(1):55–64. doi: 10.1002/bies.201200060. [DOI] [PubMed] [Google Scholar]
- 2.D’Orazi G, Rinaldo C, Soddu S. Updates on HIPK2: A resourceful oncosuppressor for clearing cancer. J Exp Clin Cancer Res. 2012;31:63. doi: 10.1186/1756-9966-31-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trapasso F, et al. Targeted disruption of the murine homeodomain-interacting protein kinase-2 causes growth deficiency in vivo and cell cycle arrest in vitro. DNA Cell Biol. 2009;28(4):161–167. doi: 10.1089/dna.2008.0778. [DOI] [PubMed] [Google Scholar]
- 4.Wiggins AK, et al. Interaction of Brn3a and HIPK2 mediates transcriptional repression of sensory neuron survival. J Cell Biol. 2004;167(2):257–267. doi: 10.1083/jcb.200406131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, et al. Essential function of HIPK2 in TGFbeta-dependent survival of midbrain dopamine neurons. Nat Neurosci. 2007;10(1):77–86. doi: 10.1038/nn1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao J-H, et al. Hipk2 cooperates with p53 to suppress γ-ray radiation-induced mouse thymic lymphoma. Oncogene. 2012;31(9):1176–1180. doi: 10.1038/onc.2011.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei G, et al. HIPK2 represses beta-catenin-mediated transcription, epidermal stem cell expansion, and skin tumorigenesis. Proc Natl Acad Sci USA. 2007;104(32):13040–13045. doi: 10.1073/pnas.0703213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshmukh H, et al. High-resolution, dual-platform aCGH analysis reveals frequent HIPK2 amplification and increased expression in pilocytic astrocytomas. Oncogene. 2008;27(34):4745–4751. doi: 10.1038/onc.2008.110. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quigley DA, et al. Genetic architecture of mouse skin inflammation and tumour susceptibility. Nature. 2009;458(7237):505–508. doi: 10.1038/nature07683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet. 2012;44(8):841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kliewer SA, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci USA. 1994;91(15):7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barak Y, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4(4):585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 14.Sharma S, et al. PPAR-γ regulates carnitine homeostasis and mitochondrial function in a lamb model of increased pulmonary blood flow. PLoS ONE. 2012;7(9):e41555. doi: 10.1371/journal.pone.0041555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen R, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22(21):2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siersbæk MS, et al. Genome-wide profiling of peroxisome proliferator-activated receptor γ in primary epididymal, inguinal, and brown adipocytes reveals depot-selective binding correlated with gene expression. Mol Cell Biol. 2012;32(17):3452–3463. doi: 10.1128/MCB.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bugge A, Grøntved L, Aagaard MM, Borup R, Mandrup S. The PPARgamma2 A/B-domain plays a gene-specific role in transactivation and cofactor recruitment. Mol Endocrinol. 2009;23(6):794–808. doi: 10.1210/me.2008-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L, Zhou D, Pryse KM, Okunade AL, Su X. Fatty acid 2-hydroxylase mediates diffusional mobility of raft-associated lipids, GLUT4 level, and lipogenesis in 3T3-L1 adipocytes. J Biol Chem. 2010;285(33):25438–25447. doi: 10.1074/jbc.M110.119933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quach JM, et al. Zinc finger protein 467 is a novel regulator of osteoblast and adipocyte commitment. J Biol Chem. 2011;286(6):4186–4198. doi: 10.1074/jbc.M110.178251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anghel SI, Wahli W. Fat poetry: A kingdom for PPAR gamma. Cell Res. 2007;17(6):486–511. doi: 10.1038/cr.2007.48. [DOI] [PubMed] [Google Scholar]
- 21.Tontonoz P, Spiegelman BM. Fat and beyond: The diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 22.Beale EG, Harvey BJ, Forest C. PCK1 and PCK2 as candidate diabetes and obesity genes. Cell Biochem Biophys. 2007;48(2-3):89–95. doi: 10.1007/s12013-007-0025-6. [DOI] [PubMed] [Google Scholar]
- 23.Ouchi N, Parker JL, Lugus JJ, Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol. 2011;11(2):85–97. doi: 10.1038/nri2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seale P, Kajimura S, Spiegelman BM. Transcriptional control of brown adipocyte development and physiological function—Of mice and men. Genes Dev. 2009;23(7):788–797. doi: 10.1101/gad.1779209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrovic N, et al. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285(10):7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharp LZ, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS ONE. 2012;7(11):e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi M, Kamei Y, Ezaki O. Mest/Peg1 imprinted gene enlarges adipocytes and is a marker of adipocyte size. Am J Physiol Endocrinol Metab. 2005;288(1):E117–E124. doi: 10.1152/ajpendo.00244.2004. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115(2):177–186. doi: 10.1016/s0092-8674(03)00802-x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q, Nottke A, Goodman RH. Homeodomain-interacting protein kinase-2 mediates CtBP phosphorylation and degradation in UV-triggered apoptosis. Proc Natl Acad Sci USA. 2005;102(8):2802–2807. doi: 10.1073/pnas.0409373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kajimura S, et al. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22(10):1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: Is beige the new brown? Genes Dev. 2013;27(3):234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jack BHA, Pearson RC, Crossley M. C-terminal binding protein: A metabolic sensor implicated in regulating adipogenesis. Int J Biochem Cell Biol. 2011;43(5):693–696. doi: 10.1016/j.biocel.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 33.Feitosa MF, et al. Quantitative-trait loci influencing body-mass index reside on chromosomes 7 and 13: The National Heart, Lung, and Blood Institute Family Heart Study. Am J Hum Genet. 2002;70(1):72–82. doi: 10.1086/338144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W-D, et al. Linkage and linkage disequilibrium mapping of genes influencing human obesity in chromosome region 7q22.1–7q35. Diabetes. 2003;52(6):1557–1561. doi: 10.2337/diabetes.52.6.1557. [DOI] [PubMed] [Google Scholar]
- 36.Platte P, et al. A study of linkage and association of body mass index in the Old Order Amish. Am J Med Genet C Semin Med Genet. 2003;121C(1):71–80. doi: 10.1002/ajmg.c.20005. [DOI] [PubMed] [Google Scholar]
- 37.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Development Core Team 2011. R: A Language and Environment for Statistical Computing (R Found Stat Comput, Vienna). Available at www.r-project.org. Accessed April 2, 2014.
- 40.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138(3):963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.