Abstract
Protein function is a complicated interplay between structure and dynamics, which can be heavily influenced by environmental factors and conditions. This is particularly true in the case of membrane proteins, such as the visual receptor rhodopsin. It has been well documented that lipid headgroups, polyunsaturated tails, and the concentration of cholesterol in membranes all play a role in the function of rhodopsin. Recently, we used all-atom simulations to demonstrate that different lipid species have preferential interactions and possible binding sites on rhodopsin’s surface, consistent with experiment. However, the limited timescales of the simulations meant that the statistical uncertainty of these results was substantial. Accordingly, we present here 32 independent 1.6 µs coarse-grained simulations exploring lipids and cholesterols surrounding rhodopsin and opsin, in lipid bilayers mimicking those found naturally. Our results agree with those found experimentally and in previous simulations, but with far better statistical certainty. The results demonstrate the value of combining all-atom and coarse-grained models with experiment to provide a well-rounded view of lipid-protein interactions.
Keywords: rhodopsin, molecular dynamics, gpcr, coarse-grain
1. Lipid-Protein Interactions
Protein structure and dynamics play a major role in function. It is not surprising then that membrane protein function would be strongly influenced by interactions at the protein-lipid interface(1). There are many reasons that the lipid environment plays a major part in the structure and thus the function of integral membrane proteins. First, lipid composition of membranes is incredibly diverse, meaning that there is no single object that defines a membrane; rather, membranes from different cells (or different parts of the same cell) can differ wildly from each other(2). Moreover, membranes are intrinsically highly heterogeneous, with the membrane-water interface a “region of tumultuous chemical heterogeneity”(3), giving membrane proteins a highly diverse surrounding environment(4).
A number of interactions are at play in the membrane environment, including hydrophobic mismatch(5), lipid fluidity, membrane tension, hydrocarbon chain packing(6), bilayer free volume(7), the intrinsic curvature of lipids(8) and elastic strain resulting from bilayer curvature frustration(9). Furthermore, lipid-protein interactions can be subdivided into general interactions, or those that result from bilayer properties, and specific interactions, or those that involve an association with individual lipids(10). Specific interactions can be further subdivided into those with annular lipids, or the boundary lipids forming the first shell around a protein, and non-annular lipids, which can be described as “co-factor” lipids with unique binding sites (11,12). In some cases, lipids can even be structural elements of membrane proteins(12). A more complete survey of the suggested lipid-protein interaction models can be found in a number of thorough reviews(13,14).
1.1. Model Systems for Lipid-Protein Interactions
One class of proteins that senses mechanical forces within the bilayer are known as mechanosensitive channels. The function of these channels depends on the relationship between membrane properties, protein structure and flexibility, and protein function(15). One such channel, MscL (mechanosensitive channel of large conductance), responds to turgor pressure changes and hypotonic shock by opening a 30 Å pore to release osmolytes and solutes(16,17). Such a large pore shows no clear specificity, serving as an emergency release of cellular components in the event of hypotonic shock. The mechanism by which MscL “senses” these conditions is by responding structurally to tension changes in the lipid bilayer. Turgor pressure, as well as cellular processes like cell division, result in a stretching or compressing of the bilayer. Thorough reviews of this interplay between membrane structure and protein function for these channels exist elsewhere(15,18,19).
Ion channels are another class of membrane protein that can depend on environmental conditions, including general bilayer properties and the presence of specific lipids. The bacterial potassium channel, KcsA, is a prototypical example. It has been demonstrated that KcsA requires a lipid bilayer for proper folding, despite being stable and active in experiments using detergent micelles, and requires the binding of a single negatively charged lipid on each monomer for activation(10). Simulation studies confirmed a Arg64-Arg89 binding motif for acidic lipids that was discovered in the experimental studies(20).
Linear gramicidins are a family of bilayer-spanning antibacterial cation channels that increase the permeability of target membranes(21). The natural folding preference is to form intertwined (double-stranded) dimers(22). In the presence of lipid bilayers they refold into the functional end-to-end (single-stranded) dimers(23), with all four Trp residues in each subunit hydrogen bonding with the bilayer at the membrane surface(24), effectively anchoring the structure in a bilayer spanning configuration. The preference for this conformation in the presence of a lipid bilayer is driven by these Trp residues at the bilayer-solvent interface, which would create a penalty if they were buried in the hydrophobic core. These effects have been noted for other proteins that show a similar Trp anchoring pattern(25).
2. Rhodopsin
Rhodopsin, a G protein-coupled receptor (GPCR) responsible for dim-light vision, is a well-characterized transmembrane protein activated by the isomerization of 11-cis-retinal to the all-trans configuration via light absorption. The ligand’s isomerization initiates a cascade of thermal relaxations in the protein, ending with metarhodopsin I (MI). MI exists in equilibrium with metarhodopsin II (MII), the tranducin-binding (or “active”) form of the protein(26). The MI-MII transition features large structural motions that lead to the activation of the G protein and ultimately results in signal transduction.
Rhodopsin is found in large concentrations in the rod outer segment disks (ROS) of rod cells, making up the vast majority of the protein component of each disk’s membrane and occupying about a third of the total area(27,28). The disk membrane phospholipid distribution is about 44 % phosphatidylcholine (PC), 41 % phosphatidylethanolamine (PE), 13 % phosphatidylserine (PS), and 2 % phosphatidylinositol(29), with a high concentration polyunsaturated docosahexaenoyl fatty acid (DHA) tails(30). The concentration of cholesterol in new disks is high (30 %) and decreases as the disk ages(31). Given that rhodopsin is an integral membrane protein with a cascade of structural changes implicated in its function, it is not surprising that this unique membrane environment has been shown to affect the behavior of rhodopsin, particularly the equilibrium between the MI and MII states; recent reviews of these effects are available(32). Given the biomedical importance of GPCRs(33) and studies of polyunsaturated fatty acids in dietary intake(34), the implications for bilayer regulation of GPCRs to human diseases are clear. Here, we intend to not only highlight experimental and simulation work that explores these effects, but also present long time-scale coarse-grained simulations that provide near-atomic resolution into the possible general and specific mechanisms by which the lipid bilayer interacts with rhodopsin.
2.1. Rhodopsin-Lipid Interactions
A number of studies by the Brown lab, starting in the late 1980’s and early 90’s(35–37), focused on MII production as a result of the photoisomerization of rhodopsin reconstituted in membranes with a variety of phospholipid and fatty acid combinations. They showed that the population of MII depended on the lipid headgroup composition as well as the concentration of polyunsaturated acyl chains. Native ROS membranes, as expected, showed the greatest quantities of MII. In membranes of PC with short, saturated acyl chains, for instance di(14:0)PC, rhodopsin is essentially inactive. Using di(22:6)PC, they demonstrated that polyunsaturation increased the degree of activity, but not to native levels. The addition of PE lipids also increased activity, though this increase was minor. The presence of polyunsaturation or PE lipids alone does not recreate native activity. Instead, a mixture of phospholipids containing both polyunsaturated chains (22:6ω3) and PE head groups had the highest activity among non-native systems(35).
Exploring the role of chain length, and in turn the hydrophobic mismatch between the bilayer and rhodopsin, has shown that the MII population is maximized with chain lengths around 18 carbons, with the equilibrium shifting back towards MI with chain lengths above 20 carbons or below 16. This is coupled to local bilayer compression and stretching effects(38). This mismatch and the resulting bilayer deformations affect rhodopsin activation by altering the helical content of the protein(39). All of this suggests that mechanical and physical properties of the bilayer, including the bilayer thickness and area per lipid, likely modulate the MI-MII transition. However, these bulk effects do not exclude the possibility of localized lipid binding sites on the surface of the protein.
Some preliminary tests were included exploring the role of headgroups in rhodopsin activation, by comparing the effects of PE and PS headgroups(36). Later, the role of the membrane potential at the membrane-water interface due to lipid headgroups was explored more thoroughly. It was demonstrated that lipids with PS headgroups have two contradictory effects: they alter the bilayer’s properties in ways that oppose MII formation, but their net charge creates an electrostatic environment rich with H3O+ ions that promotes MII formation(40). This is in agreement with other results that showed that MII formation is enhanced in acidic conditions(41). Finally, they concluded that both PS headgroups and the combination of PE headgroups and DHA chains are needed to maximize the population of MII. The theory suggests that membranes with only PC and PS headgroups would favor MII based only on electrostatics, but this would be counteracted by the structural unfavorability of a charged bilayer surface. As a result, ROS membranes have high concentrations of lipids with highly negative spontaneous curvature (PE) and polyunsaturated chains to counteract this by providing curvature stress and thus promote MII(42). These results led Brown and coworkers to a general model, known as the flexible surface model (FSM), where composition of the lipid matrix actively regulates rhodopsin function(43).
The effects of headgroups on the MI-MII equilibrium are not limited to electrostatics and membrane elasticity. In fact, in work intended to discern the energetic contribution of membrane elasticity to rhodopsin function, Gawrisch and coworkers noted that PE headgroups also induce a shift toward MII that correlates with their hydrogenbonding ability(44). Furthermore, saturation transfer NMR studies of rhodopsin in mixed PC/PE bilayers showed greater magnetization transfer to PE lipids when compared to PC lipids(45), suggesting that in addition to their effects on bulk bilayer properties, there may be a specific role for PE headgroups at the surface of rhodopsin.
2.2. Role of Cholesterol
A major component of the rod disk membranes, cholesterol has been shown to have an effect on rhodopsin activation as well. The presence of cholesterol in membranes drives the MI-MII equilibrium towards MI, reducing signaling(7). However, it is unclear from these experiments whether this is caused by cholesterol’s effects on bilayer properties, direct interactions between it and the protein, or some combination of these effects.
Cholesterol’s effects on membrane liquid crystalline structure are well documented. The presence of cholesterol causes tighter packing of lipid hydrocarbons(46) and increases the thickness of the bilayer, leading to changes in the lateral compressibility(47). These bilayer effects may create an environment that inhibits the conformational transition from MI to MII, though cholesterol also promotes negative curvature elastic stress, a property of PE headgroups that tends to promote MII formation(48).
The alternative means by which cholesterol may affect rhodopsin activation is through direct and possibly specific interactions. Studies of cholesterol, cholestatrienol and ergosterol in ROS membranes have suggested that there is at least one cholesterol binding site on rhodopsin(49). Sites of preferential cholesterol interaction have been identified via molecular simulation as well(50), although these and other simulations suggested that cholesterol is on the whole depleted at the protein surface(51).
2.3. Simulation Applied to Rhodopsin
In 2000, the first crystal structure of a GPCR was solved in the form of bovine rhodopsin at 2.8 Å resolution(52), opening the way for the use of molecular dynamics simulation to probe the atomic-level interactions between rhodopsin and its environment. An early simulation of rhodopsin, performed by Huber et al.(53), was compared to available NMR data to quantify membrane deformation in the presence of protein and compute cross-sectional protein areas. They found that rhodopsin imposed curvature in the bilayer, which could facilitate selection for polyunsaturated lipids at the surface of rhodopsin. This was seen as further support for the flexible surface model(54).
As the available computer power improved, simulations began to explore the interactions between rhodopsin and bilayer constituents via longer molecular dynamics trajectories of the protein embedded in a more realistic bilayer. These systems featured cholesterol, as well as a mixture of lipids with two different headgroups, each of which was linked to one polyunsaturated tail and one saturated tail. Early results showed a preference for the polyunsaturated tails at the surface of rhodopsin and little indication of specific binding sites for cholesterol(51,55)
Later, a series of 26 independent 100 ns simulations of rhodopsin in a realistic lipid composition was used to address these questions, with better statistical sampling(50). Polyunsaturated tails were again enriched at the surface of the protein. It was also now possible to identify residues that preferentially interacted with cholesterol and each of the lipid components. Although this work required a heroic effort at the time, the sampling was not sufficient to give high confidence in the predictions about specific residues, particularly for cholesterol, since any given interaction was only seen in a small fraction the trajectories(50). Nonetheless, the results demonstrated that computational methods could confirm the experimentally suggested trend of preferential interactions with polyunsaturated tails, allowing some to speculate on the roles these flexible lipids could play in the activation of rhodopsin(56). Preferential sites of interaction between rhodopsin and cholesterol were also identified(57).
More recently, the same data (supplemented by an additional 1.6 µs simulation) was reanalyzed to explore the interactions between the palmitoyl moieties attached to a pair of cysteines in the cytoplasmic helix H8 and the helical bundle of rhodopsin(58). There was a high degree of contact between both chains and the protein, but also a significant difference between the two, even though they are attached to consecutive residues. The high level of contact between the palmitate on Cys322 and the protein helices suggests that it may play an important role beyond that of a nonspecific lipid anchor.
Recently, some groups have begun using coarse-grained simulations to explore rhodopsin-lipid interactions as well. For example, Periole et al (59) explored the effects of varying the bilayer hydrophobic thickness on the oligomerization of rhodopsin, using the MARTINI force field(60,61). The results demonstrated that oligomerization is driven by frustration of lipid-protein interactions, something that had already been seen experimentally(38). Rhodopsin significantly altered the local membrane thickness, encouraging oligomerization as a way to reduce unfavorable protein-lipid interactions. While useful for exploring bilayer adaptations attributed to hydrophobic mismatch, each simulation featured homogeneous bilayers, so no new evidence was gleaned about the role for cholesterol and different lipid headgroups in rhodopsin activation.
In this work, we employed simulations featuring membranes with native-like compositions to more thoroughly explore protein-lipid interactions. Our focus in these systems was the role played by the saturation state of the lipids, the headgroups of the lipids, and the presence of cholesterol in modulating lipid-protein interactions.
3. Methods
3.1. Simulation Systems
The goal of this research was to explore lipid-protein interactions in a system with rhodopsin and a biologically relevant model membrane. We sought to extend previously published work that featured 26 separate 100 ns all-atom simulations of rhodopsin in a membrane(50). These systems featured a 2:2:1 ratio of 1-stearoyl-2-docosahexaenoyl-phosphatidylcholine (SDPC), 1-stearoyl-2-docosahexaenoyl-phosphatidylethanolamine (SDPE), and cholesterol, and were intended to mimic those membranes found biologically and used in experimental studies with model membranes(30,35,49). The systems described here mimic these all-atom systems in an attempt to bring together a great deal of experimental work that has been done investigating the role of various membrane properties on rhodopsin activation, taking advantage of the ability of molecular dynamics simulation to explore interactions on the molecular level.
While being a remarkable effort at the time, previous all-atom simulations had some limitations that faster computers and more efficient simulation methods have allowed us to overcome. By employing a coarse-grained model, we simulated larger systems for longer timescales, allowing for better sampling of long time-scale processes critical for bilayer-protein interactions, such as the lateral reorganization of the lipid bilayer. The work described here features systems nearly three times larger than the all-atom work, with timescales an order of magnitude longer. Furthermore, simulations of “dark” and “active” rhodopsin were done to make comparisons about the lipid and cholesterol binding surfaces in these differing structures. Overall, the new results agree with the earlier all-atom work, while providing more robust statistics and allowing us to make some conclusions about specific and general interactions between the lipid environment and rhodopsin.
3.2. Construction
Rhodopsin was modeled after the same crystal structure used in the previous work (PDB ID: 1U19(62)). For comparison, retinal-free opsin was also modeled (PDB ID: 3CAP(63)). The MARTINI coarse-grained force field was used(60) with the extension for proteins(61). There is recent precedent that the MARTINI forcefield is capable of reproducing the physicochemical properties of the surface of rhodopsin, at least for aggregation(64,65). To create the coarse-grained model for each protein, all molecules present other than rhodopsin/opsin were removed from the crystal structure. We then mapped the coarse amino acids onto this structure using the martinize.py script available online at the MARTINI website(66). The acyl chains on the cysteine residues at positions 322 and 323 were manually added and the system energy was minimized. Retinal was not explicitly represented in the rhodopsin model; in the MARTINI protein model, protein fluctuations are dominated by the network of restraints required to stabilize the tertiary structure, and since the retinal itself does not interact with the membrane environment in any significant way, we believe it is sufficient to model the retinal-stabilized dark-state rhodopsin structure of the protein in its absence.
For the rhodopsin system, we initially inserted the protein into an SDPC lipid bilayer using a bilayer expansion and compression technique(67). In this method, the lipids are translated in the plane of the bilayer by a large scaling factor, creating space for the protein insertion without clashes. Then, the lipids are scaled back to the ideal area-per-lipid using a number of cycles of translation and minimization. This reduces lipid-lipid and lipid-protein clashes.
Water and ions were added to solvate and neutralize the system; additional NaCl was added to bring the concentration to 100 mM. Afterwards, we held the protein position fixed and performed a 10 nanosecond simulation to allow the lipid and solvent environment to relax. To generate unique starting states for the bilayer, randomly selected SDPC lipids were converted to SDPE lipids by simply changing the head group beads, while others were swapped with cholesterol. The result was a set of 16 unique rhodopsin systems with 2:2:1 SDPC:SDPE:cholesterol bilayers. We minimized the resulting systems and performed another round of equilibration, again holding the protein fixed.
To make the equivalent opsin model, rhodopsin was replaced with opsin after backbone alignment for each of the rhodopsin systems. These were then equilibrated with a short simulation (100 picoseconds) to ensure that the starting configurations for the opsin systems and the rhodopsin systems were similar.
The final systems included 1 protein molecule (either rhodopsin or opsin), 180 SDPC, 180 SDPE, 90 cholesterol, about 19,000 water beads (each representing 4 water molecules), and about 140 each of Na+ and Cl−. This brings the system size to about 26,000 CG beads, which is roughly equivalent to an all-atom system with 105,000 atoms (double the total system size and more than triple the bilayer size of the previous work).
3.3. Simulation Protocol
Simulations were performed with version 4.5.4 of the GROMACS molecular dynamics package(68,69) on a Linux cluster. We used a time step of 10 fs, as suggested for accurate integration(70,71), with the neighbor list updated every 5 steps. We held the temperature at 300 K using Nosé-Hoover temperature coupling(72,73) and treated the pressure semi-isotropically with a reference of 1 bar using the Parrinello-Rahman barostat(74). A shift function was employed for electrostatics with a coulomb cutoff of 12 Å. The Lennard-Jones potential was shifted between of 9 Å and 12 Å.
It is well established that external restraints are required when simulating native proteins using MARTINI. Initially, we performed our simulations with only the parameters necessary to maintain the secondary structure. However, under these conditions the rhodopsin structure moved rapidly away from the crystal structure, reaching a transmembrane alpha carbon RMSD as high as 6 Å (data not shown). This problem in the MARTINI force field with maintaining protein tertiary structure has been noted before and protocols have been developed for overcoming these limitations utilizing distance-based restraints(75). To maintain the integrity of our proteins, we included a similar network model, restraining the distances between backbone beads between 2 and 10 Å apart. We tested multiple force constants using short trajectories to try to match the amplitude of fluctuations to previous all-atom simulations; with a force constant of 800 kJ/mol·nm2, the rhodopsin RMSD fluctuated between 2.0 and 2.5 Å, consistent with previous all-atom results(76), and the opsin RMSD fluctuated between 2.5 and 3.0 Å.
We ran 32 independent simulations, 16 each for rhodopsin and opsin. Each simulation was 1.6 µs, for a total of 51.2 µs of simulation time (effective time of about 205 µs if we apply a 4× scaling to the time, as suggested by previous authors to account for the enhanced kinetics of the coarse-grained model(60)). All times we report here are the actual simulation times, without the 4× scaling.
3.4. Analysis
All simulation analyses were performed using tools developed using the LOOS library. LOOS is an object-oriented library implemented in C++ and Boost for rapidly creating new tools for analyzing molecular dynamics simulations(77,78). All analysis was performed on trajectories subsampled to 1 ns resolution.
3.4.1. Lateral Radial Distribution Function
We computed the 2D radial distribution function (RDF) of various bilayer species relative to the center of mass of the protein structure in the membrane plane using available LOOS tools. Each molecule was treated as a single unit, located at its centroid.
3.4.2. Density Maps
We created 2D density maps to show average density of each lipid component in the plane of the bilayer. We aligned the protein structure of each frame of the simulation to the initial structure. Then we binned the centroid of each component in a grid on the plane parallel to the membrane, with 1 Å2 bins. The resulting density histogram is displayed as a heat map.
To probe the 3-dimensional distribution of lipid components about the membrane, we first aligned our trajectories using transmembrane Cα’s. Then we used a 1 Å3 grid superimposed over the protein’s bounding box, padded by 30 Å. Each atom is then placed in the nearest bin and the resulting histogram is convolved with a gaussian for a smoother visualization.
3.4.3. Residue-Based Binding Scans
To highlight interactions between lipid components and individual residues, we computed a residue binding score for each residue of the protein to each lipid component. The residue score R for residue n and lipid component m can be expressed as:
| (1) |
Where N is the number of atoms in residue n, M is the sum of all atoms for all molecules of lipid component m in the system, rij is the distance between atoms i and j. The normalized residue score is then simply the residue score divided by the average residue score for all transmembrane alpha helix residues:
| (2) |
3.4.4. Statistical significance
Because we have multiple independently constructed trajectories of each system, we have attempted to assess the statistical significance of our results. This was done using a standard T-test, treating the average result (e.g. the radial distribution density at a particular distance) from each trajectory as a single data point. We typically plot the p-values on the same axes as the results themselves. To compute these p-values, we used statistical tools available in SciPy(79).
4. Results and Discussion
4.1. RDFs Demonstrate Surface Preferences for DHA
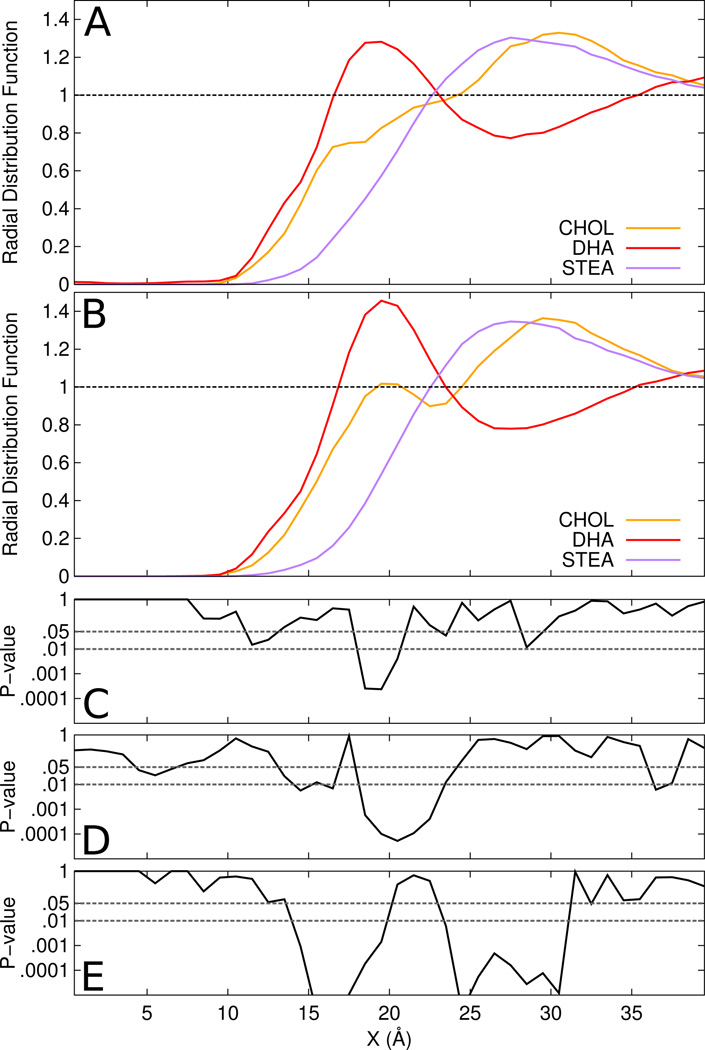
To begin our analysis, we used a two-dimensional radial distribution function (RDF) in the plane of the membrane, as was done in previous work(55), to assess the packing of the different members of the bilayer against rhodopsin. In Figure 1, we show RDFs for the two lipid tail types and cholesterol. Here we can see a drastic enrichment of DHA between 15 and 20 Å from the center of the protein, with the peaks for cholesterol and stearoyl beyond 20 Å. Interestingly, while cholesterol is not enriched at the surface, it has significant density deeper into the protein than stearoyl, and nearly as deep as DHA. Given its smaller size, this may be indicative of regions accessible only to cholesterol.
Figure 1.
Lateral radial distribution functions of each lipid tails and cholesterol for the (A) rhodopsin and (B) opsin systems. Comparison of means tests between rhodopsin and opsin for (C) cholesterol, (D) DHA, and (E) stearoyl.
To accurately assess the significance of the difference between RDF curves for opsin and rhodopsin, we calculated p-values for each point. Given that each point is the mean of a set of 16 independent samples, we have a fairly large set of data from which to do this assessment (unusual, if not unique, in the simulation community). The resulting p-values are plotted below the RDF curves with the same x-axis, with confidence levels of 0.01 and 0.05 shown for reference. Panels C, D and E show these p-value plots for cholesterol, DHA and stearoyl, comparing the means between rhodopsin and opsin. Statistically significant (P < 0.01) differences appear between all analyzed bilayer constituents, but some for only brief stretches of the RDF curves.
The short region of cholesterol significance coincides with a peak in the opsin curve that is not present in rhodopsin, indicating a region of bulk density at the very surface of opsin where the DHA density is the highest. The significant regions are much more substantial for the lipids tails. For stearoyl, large stretches of the curve show very significant differences (P < 0.001). Visual inspection of the RDFs shows greater penetration of the lipid tails between 10 and 20 Å. This can be explained to some degree by the greater flexibility and more “open” structure for the opsin system; there is a greater area accessible to the lipid tails between the helices and in the protein interior.
4.2. Density Maps Show DHA Preference
The above results are consistent with previous simulation and experimental results. However, simple lateral radial distribution functions contain limited information, because they treat rhodopsin as a featureless cylinder, integrating out the distinctions between different portions of the protein surface. Moreover, in these plots, both leaflets were treated together, again averaging away potentially valuable information. Accordingly, we instead project our results along 2 dimensions, using lateral density heat maps.
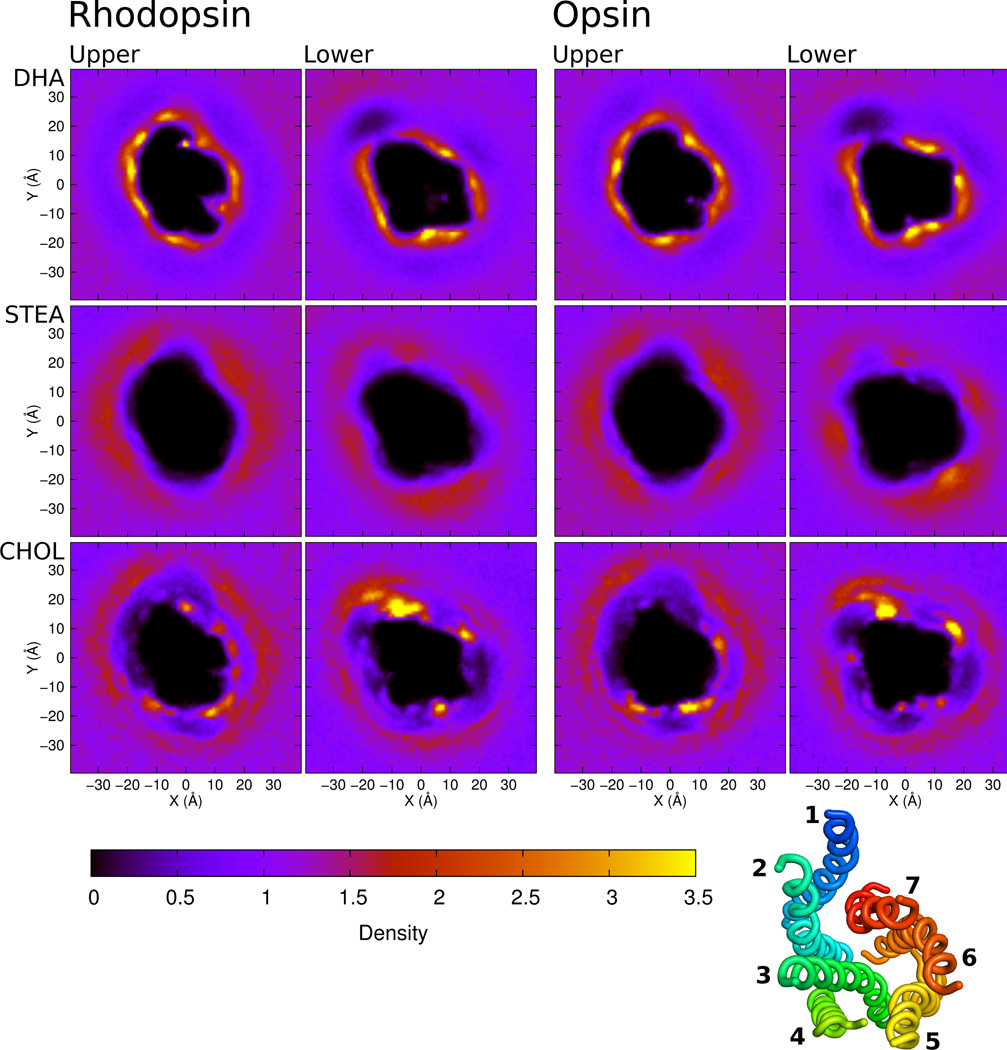
Figure 2 shows density maps for the different lipid components for both rhodopsin and opsin, in both the upper and lower leaflets. The dark region in the center of each frame represents the excluded volume of the helix bundle, as we look down from extracellular side. Opsin and rhodopsin were aligned by their backbones so that they are oriented the same way in all of the heat maps for comparison. These images represent the average of all 16 simulations for each system.
Figure 2.
Density maps of each bilayer component, for each leaflet, in each system (rhodopsin or opsin). Density is reported as lipid components per Å2. All images are viewed from the extracellular side of the protein. The upper leaflet refers to the leaflet on the extracellular side. For every map, rhodopsin or opsin was centered at the origin and aligned against a reference structure so that the maps can be directly compared. The orientation of the helices is shown in the small panel in the bottom right corner, also viewed from the extracellular side of the protein.
In the plots of DHA density, we see a bright, thin ring tracing the protein space. In contrast with the low densities for stearoyl and cholesterol, this indicates that DHA is preferentially packed against the surface of the protein, with the exception of a bright cholesterol spot next to helices H1 and H7. The corresponding stearoyl densities show rings as well, immediately outside the DHA ring. The stearoyl rings are dimmer and, in general, more diffuse.
The lateral radial distribution functions, coupled with the density heat maps, suggest a strong preference for DHA at the surface of both opsin and rhodopsin, in agreement with previous experimental and computational results. Previous work suggests that this preference is entropically driven(80). It has been demonstrated that DHA is extremely flexible(81) and rapidly interconverts between conformations(82), making it ideal for packing against the relatively rigid but uneven surface of the protein.
The region just beyond the first shell of DHA chains is enriched in stearate. This result is not surprising, because the lipids used in these simulations each have one DHA and one stearoyl. For every lipid with a DHA tail packed against rhodopsin, there is also a stearoyl facing away from the protein, accounting for the inner DHA ring and the outer stearoyl ring. This outer ring is not as bright in the heat maps as the DHA ring because the accessible surface area in this ring is far greater, so the motion of these tails is more diffuse.
4.3. SDPE is Preferred at Protein Surface
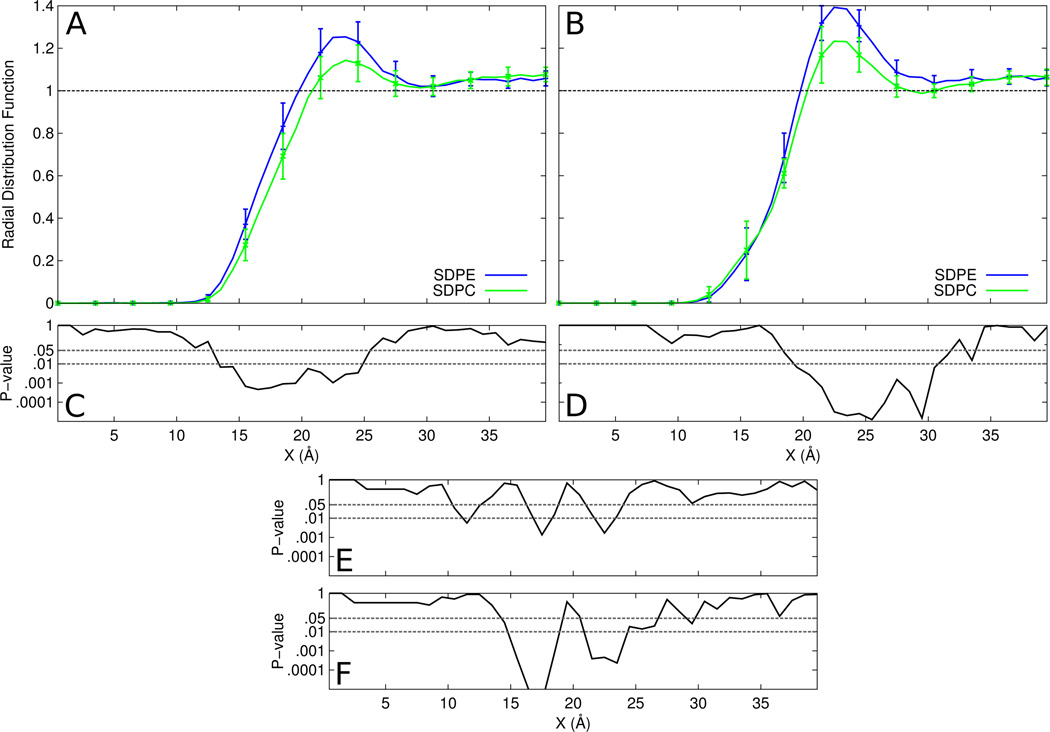
In Figure 3, we compare preferences between SDPE and SDPC within each system to explore a possible preference for one headgroup over another. Visual inspection suggests a slight preference for SDPE over SDPC at the surface of the protein, which is confirmed to be statistically significant; the lower panels show the p-value for the difference between SDPC and SDPE and demonstrates significance at the 0.01 level out to for the entire first “solvation” shell of rhodopsin, and most of that region for opsin. Panel E shows that the differences in the SDPC RDF between rhodopsin and opsin are marginally significant at best, but the SDPE RDF (Panel F) does show a significant difference in the location of this initial rise at the protein surface.
Figure 3.
Lateral radial distribution functions of SDPE and SDPC for the (A) rhodopsin and (B) opsin systems. Comparison of means tests were performed comparing SDPE to SDPC within the (C) rhodopsin and (D) opsin systems. Comparisons were also made to test differences in mean between rhodopsin to opsin for (E) SDPC and (F) SDPE.
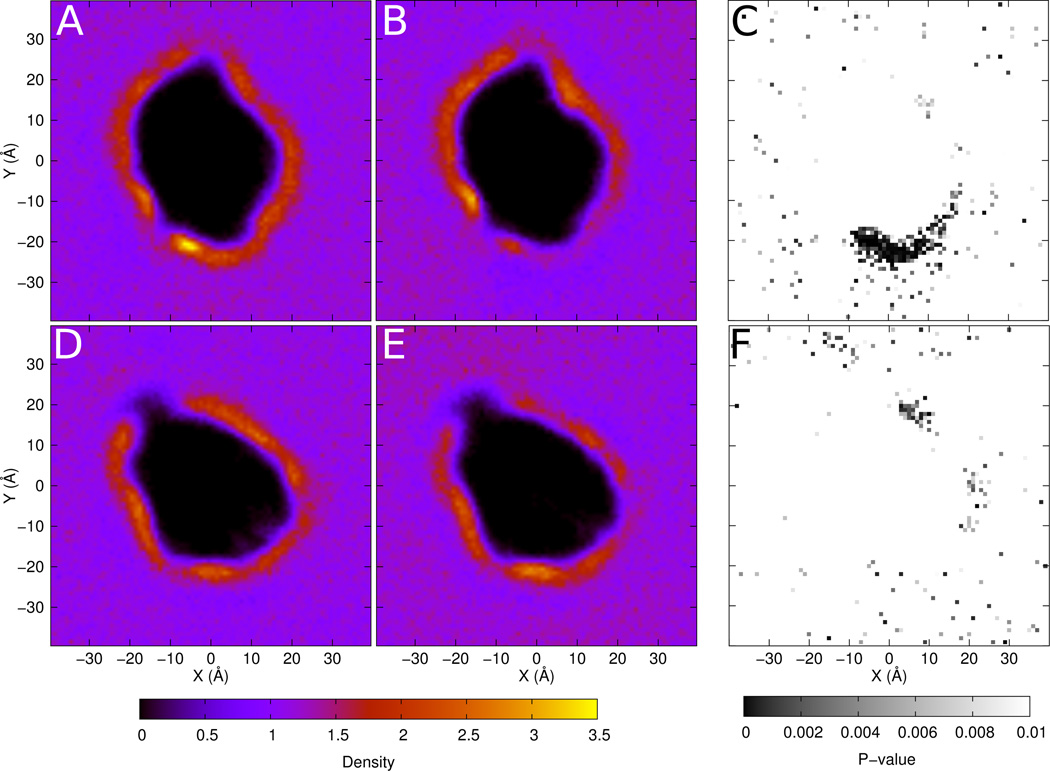
This is in agreement with experimental results that suggest that PE is a preferred partner for rhodopsin(44,45). To explore the possibility of specific PE interaction sites, we generated density maps of SDPE and SDPC for rhodopsin, found in Figure 4. For quantitative comparison, p-values were computed for every point in the maps and plotted. Panel C shows the p-value plot comparing SDPE (panel A) and SDPC (panel B) for the upper leaflet. There is a large region of statistical significance that indicates differences between the densities of the two headgroups along helices H3, H4 and H5 on the extracellular side of the protein. No such regions of interest are seen in the other leaflet, and it is less pronounced for the opsin system (data not shown).
Figure 4.
Density maps of (A) SDPE in the upper leaflet of rhodopsin, (B) SDPC in the upper leaflet rhodopsin, (D) SDPE in the lower leaflet of rhodopsin and (E) SDPC in the lower leaflet of rhodopsin. Density is reported as lipid components per Å2. Maps with p-values comparing SDPE and SDPC for (C) the upper leaflet and (F) the lower leaflet are also shown.
4.4. Mapping Density to Structure Probes Cholesterol Binding Sites
The present simulations, by accessing the microsecond timescale, are much more effective at allowing for sampling of the lateral motion of cholesterol and lipids in the bilayer than previous work. Considering the enhanced rate of lipid diffusion in the MARTINI forcefield, each of our simulations arguably samples nearly 6.5 µs of lateral reorganization. With this in mind, we have the ability to probe for the appearance of cholesterol binding sites in all of our simulations and hopefully converge on representative sites, whereas previous work could only note sites where some fraction of the simulations had seen strong contacts.
In the lateral radial distribution plots, there is evidence that cholesterol can pack inside the helical bundle (deeper than either lipid tail), albeit not with high abundance. Indeed, integrating the RDF suggests the presence of roughly 1 cholesterol immediately at the protein surface (data not shown). The heat maps clarify this result; visual examination shows one site of clear cholesterol preference, next to helices H1 and H7, approximately where we would find the palmitoyls attached to helix H8. DHA lipids are excluded from this location, as seen by a patch of low density. Whether this site represents a true competitive binding site, where binding to cholesterol is preferred over other bilayer constituents, or simply a deep protein pocket or crevice that is only accessible to cholesterol, is unclear.
In order to better understand the details of the cholesterol binding, we further expanded the data to look at the full 3-dimensional distribution. Figure 5 shows contours of regions with high average cholesterol density superimposed onto the structures of rhodopsin and opsin. The brightest cholesterol spot in the 2D maps, which as mentioned excludes other lipids, corresponds with the region of high density beside H8, the intracellular helix lying parallel to the bilayer, and packed against H1 and H7 of the protein. The density surface packs neatly behind the pair of post-translational palmitates on Cys322 and Cys323. In these simulations, H8 is embedded in the bilayer interface, creating a pocket in the hydrophobic core that is too small for lipids to comfortably diffuse, leading us to conclude that in this case, cholesterol is able to pack where other lipids cannot penetrate efficiently. This cholesterol “hot spot” is found in both the opsin and rhodopsin simulations, as is a second region found on the opposite side of the protein, between the cytoplasmic end of helix H3 and helices H4 and H5. This corresponds with a cholesterol interaction site predicted from a pair of 800 ns simulations of the adenosine A2A receptor(83).
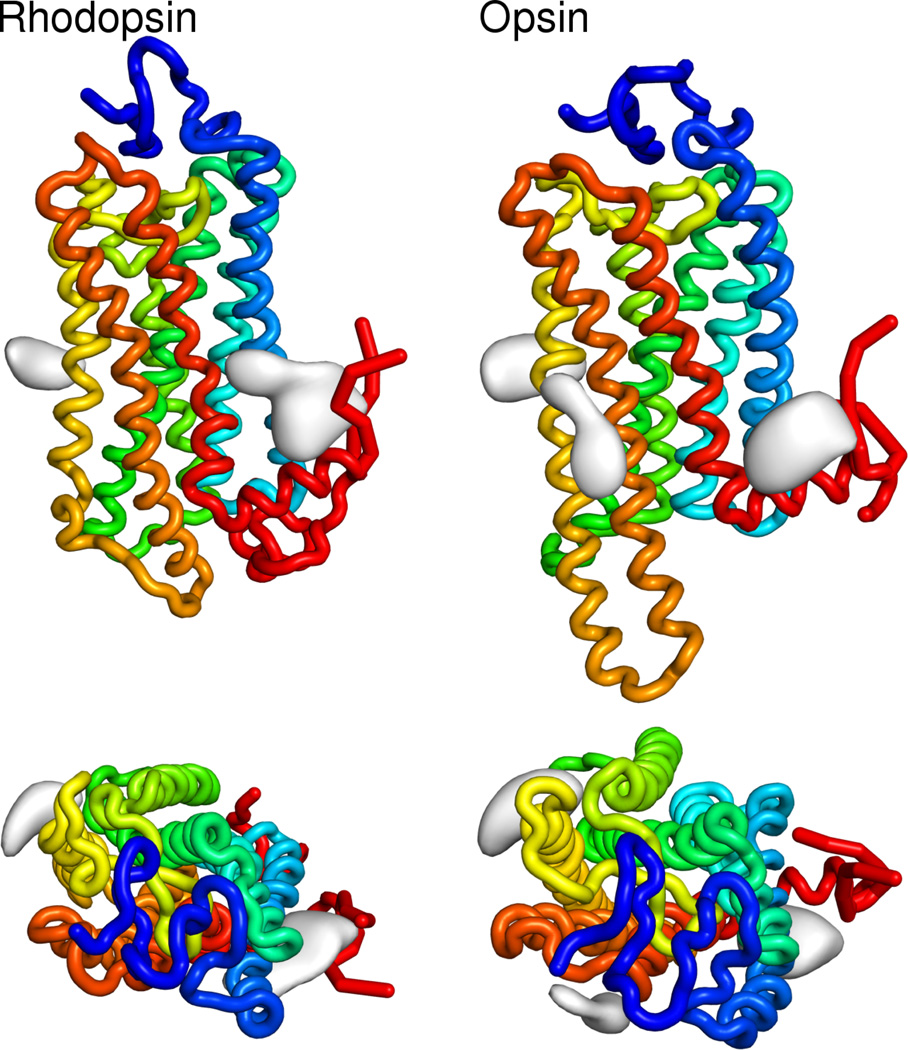
Figure 5.
3D images of regions of high cholesterol density (gray) for both rhodopsin and opsin. Bottom panels are top down views, as seen from the extracellular side. Rhodopsin and opsin are colored with a spectrum from the N-terminus (blue) to the C-terminus (red).
Lastly, at the density contour level chosen, a third high density cholesterol region is present in the opsin system packed against helices H5 and H6. At lower contour thresholds, this region appears in the rhodopsin systems as well, but its presence here indicates a possible difference in cholesterol packing interfaces between the two structures. Given that the greatest structural changes between the chosen opsin and rhodopsin structures are the orientations of helices H5 and H6, as well as the elongation of helix H5 in opsin, these changes in cholesterol packing suggest a role for helix-helix interactions and arrangement in cholesterol preferences. The presence of the extra cholesterol binding spot is consistent with Figure 1, which shows greater cholesterol binding at the surface of the opsin system. This is indicative of a general trend that the opsin structure is more amenable to cholesterol binding, again either because the surface has greater preference for cholesterol or the more “open” opsin structure provides a greater number of cholesterol accessible pockets.
The above visualizations are convenient, but do not in themselves tell us precisely which protein residues are involved in the “binding sites.” Accordingly, we decided to track specific lipid-residue contacts, using a variant of the packing score applied in previous work(50), as discussed in the methods section. Unlike that work, here we account for the size of the residues, and report the ratio of the packing score to that of the average score of all residues in the transmembrane region (Equation 1). Figure 6 shows the protein structures colored by residue score. The residues clustered into 3 well-defined groups based on physical location; Table 1 lists the residues with values at least 3 times greater than the average. These groupings correspond nicely with the three high density regions for cholesterol as seen previously for the opsin structure. Overall, the high-scoring clusters bear a striking resemblance to the groups of residues identified in the previous all-atom work(50).
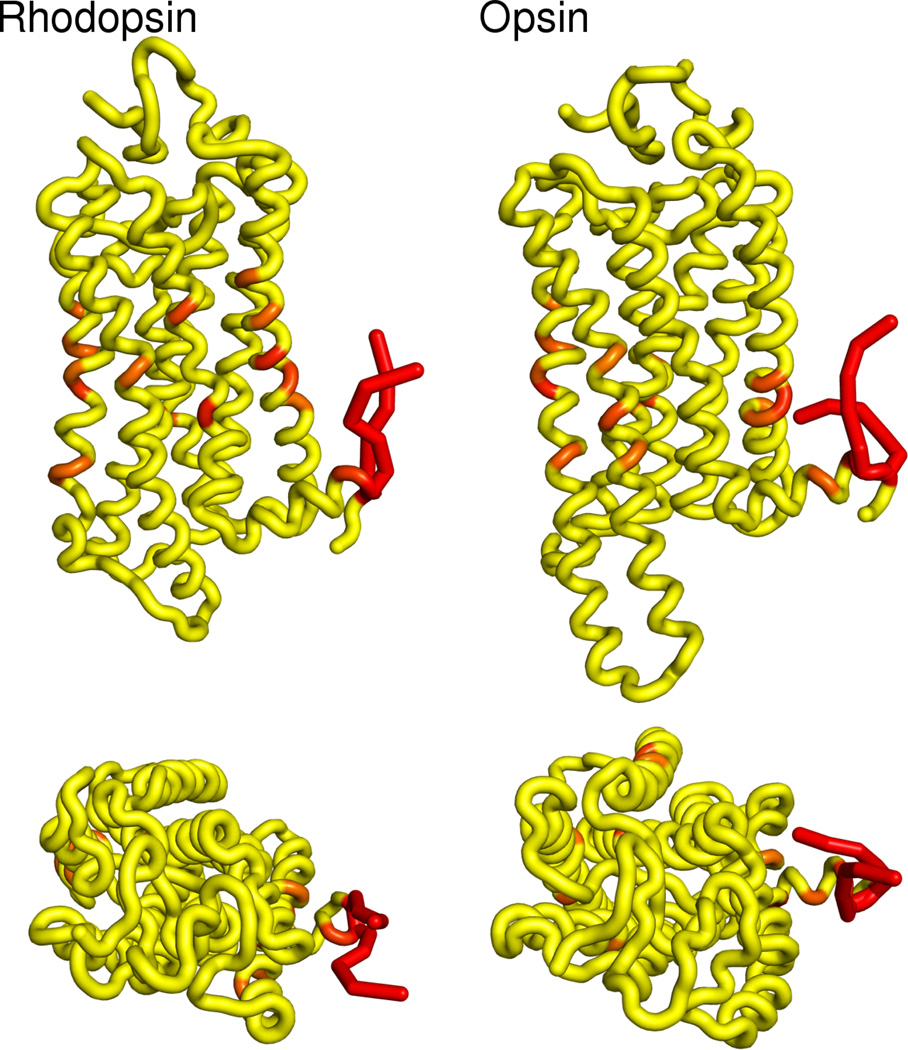
Figure 6.
3D images of rhodopsin and cholesterol interactions, color coded from yellow (low contact) to red (high contact).
Table 1.
Clusters of key cholesterol-binding residues identified via binding scan.
| Structure | Group | Residues | Average Score |
|---|---|---|---|
| Rhodopsin | 1 | 43, 46, 50, 53, 56, 294, 301, 321, 322, 323 | 6.0683 |
| 2 | 126, 159, 206, 209, 210, 213, 214, 220, 221 | 3.6042 | |
| 3 | 263 | 3.3597 | |
| Opsin | 1 | 50, 53, 54, 56, 318, 321, 322, 323 | 6.7907 |
| 2 | 126, 159, 162, 206, 209, 210, 213, 214, 220 | 3.9135 | |
| 3 | 256, 259, 263 | 3.4568 | |
The most dominant cluster is a collection of residues that pack between helices H1 and H8, with the greatest contribution to the cluster score coming from the palmitoyl chains attached to H8. In recent β2-adrenergic receptor crystal structures, cholesterol and palmitic acids have been resolved in the H1/H8 interface, close to where the palmitoyl post-translational modifications in rhodopsin are located(84). However, it has been suggested that these cholesterols may exist as an artifact of crystal packing or protein dimerization. Our simulations suggest a real effect exists, showing preferential interactions between cholesterol and rhodopsin at helix H8, despite the presence of only one rhodopsin molecule.
Despite success correlating our results with previous simulation and crystallographic data, we have been unable to detect significant contacts between cholesterol and the groove on the intracellular ends of helices H1, H2, H3 and H4, which had been detected in previous all-atom simulations(57) and in structures of the β2-adrenergic GPCR(84,85). We were also unable to locate cholesterol binding sites that correspond with three sites identified in a recent high-resolution structure of the adenosine A2A receptor(86). One of these sites, between the intracellular ends of helices H1 and H2, was noted in all-atom simulations(87) and crystal structures of the β2-adrenergic structure(85), and absent in other simulations of A2A (83).
5. Conclusions
The membrane environment around rhodopsin contains a diverse set of constituents that impact receptor activation, from general bilayer structural properties to specific binding interactions. Utilizing coarse-grained simulation and a combination of radial distribution functions, density representations and quantitative binding scans, we explored and identified a number of bilayer-rhodopsin interactions.
DHA chains were found in higher concentrations at the protein surface, with stearoyl chains excluded to a second solvation shell in the bilayer. There was enrichment of PE headgroups over PC headgroups at the surface of the protein. A region of significant difference was discovered, suggesting a possible specific binding site for lipids with PE headgroups. Possible cholesterol-binding sites were also identified, with the predominant one at the helix H1 and helix H8 interface, behind the palmitoyl chains attached to the protein.
We also found differences between the rhodopsin and opsin systems for these lipid constituents. In the opsin system, the concentrations of cholesterol, stearoyl and DHA reach bulk levels deeper in the protein and the stearoyl and DHA peaks were much higher at the surface of the protein, suggesting a more open structure with greater available surface area.
The use of restraints to maintain protein stability limited the motions available to rhodopsin and opsin, likely preventing any major structural changes that would result from bilayer-protein interactions. We do not feel that this is an issue for our particular simulations, as we are probing preferential interaction sites along the surface of the protein, not the effects of these interactions on protein structure. Our chosen rhodopsin and opsin structures represent distant endpoints along the activation path of rhodopsin, allowing us to explore the effect of these major structural changes on the surface available for interaction.
Coarse-grained models limit the ability of the simulations to capture specific binding interactions. For example, coarse-grained representations of cholesterol maintain the molecule’s hydrophobicity, but cannot capture the chemically distinct faces. Interaction sites that are uniquely suited to interactions with cholesterol are then unable to interact. The use of an all-atom model would overcome these limitation to some degree, but the loss of sufficient sampling remains a barrier to utilizing these models to explore a process as slow as bilayer lateral reorganization.
In the future, a number of other variables could be explored. First, it would be worth exploring variations in concentration of lipid components in the bilayer. Cholesterol concentrations vary with the age of the disc membrane and can result in a drastic change in rhodopsin activity. Also, an improved MARTINI model that accurately maintains tertiary structure would remove the need for restraints and allow us to explore the effect of the lipid bilayer and specific lipid species on the structure of rhodopsin.
Acknowledgments
We would like to thank Nick Leioatts, Dejun Lin and Tod Romo for critical reviews of this manuscript. We would also like to gratefully acknowledge financial support from the U.S. National Institutes of Health (1R01GM095496). We also thank the University of Rochester’s Center for Integrated Research Computing for the computing resources necessary to support this work.
References
- 1.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. Protein Modulation of Membrane Structure. [DOI] [PubMed] [Google Scholar]
- 2.Brügger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nanoelectrospray ionization tandem mass spectrometry. Proceedings of the National Academy of Sciences. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiener MC, White SH. Structure of a fluid dioloeoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data: III. complete structure. Biophys. J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 5.Mouritsen OG, Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys. J. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fattal DR, Ben-Shaul A. A molecular model for lipid-protein interactions in membranes: the role of hydrophobic mismatch. Biophys. J. 1993;65:1795–1809. doi: 10.1016/S0006-3495(93)81249-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell DC, Straume M, Miller JL, Litman BJ. Modulation of metarhodopsin formation by cholesterol-induced ordering of bilayer lipids. Biochemistry. 1990;29:9143–9149. doi: 10.1021/bi00491a007. [DOI] [PubMed] [Google Scholar]
- 8.Gruner SM. Intrinsic curvature hypothesis for biomembrane lipid composition: a role for nonbilayer lipids. Proceedings of the National Academy of Sciences. 1985;82:3665–3669. doi: 10.1073/pnas.82.11.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee AG. How lipids affect the activities of integral membrane proteins. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. Lipid-Protein Interactions. [DOI] [PubMed] [Google Scholar]
- 10.Valiyaveetil FI, Zhou Y, MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. doi: 10.1021/bi026215y. PMID:12196015. [DOI] [PubMed] [Google Scholar]
- 11.Simmonds A, East J, Jones O, Rooney E, McWhirter J, Lee A. Annular and non-annular binding sites on the (Ca2+ + Mg2+)-ATPase. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1982;693:398–406. doi: 10.1016/0005-2736(82)90447-3. [DOI] [PubMed] [Google Scholar]
- 12.Lee A. Lipid-protein interactions in biological membranes: a structural perspective. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 13.Mouritsen OG, Bloom M. Models of lipid-protein interactions in membranes. Ann. Rev. Biophys. Biomol. Struct. 1993;22:145–171. doi: 10.1146/annurev.bb.22.060193.001045. [DOI] [PubMed] [Google Scholar]
- 14.Andersen OS, Koeppe RE. Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 15.Haswell E, Phillips R, Rees D. Mechanosensitive channels: What can they do and how do they do it? Structure. 2011;19:1356–1369. doi: 10.1016/j.str.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinac B, Buechner M, Delcour AH, Adler J, Kung C. Pressure-sensitive ion channel in Escherichia coli. Proceedings of the National Academy of Sciences. 1987;84:2297–2301. doi: 10.1073/pnas.84.8.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cruickshank C, Minchin R, Dain AL, Martinac B. Estimation of the pore size of the large-conductance mechanosensitive ion channel of Escherichia coli. Biophysical Journal. 1997;73:1925–1931. doi: 10.1016/S0006-3495(97)78223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kung C, Martinac B, Sukharev S. Mechanosensitive channels in microbes. Annual Review of Microbiology. 2010;64:313–329. doi: 10.1146/annurev.micro.112408.134106. [DOI] [PubMed] [Google Scholar]
- 19.Martinac B. Bacterial mechanosensitive channels as a paradigm for mechanosensory transduction. Cell Physiol Biochem. 2011;28:1051–1060. doi: 10.1159/000335842. [DOI] [PubMed] [Google Scholar]
- 20.Sansom MS, Bond PJ, Deol SS, Grottesi A, Haider S, Sands ZA. Molecular simulations and lipid-protein interactions: potassium channels and other membrane proteins. Biochem Soc Trans. 2005;33:916–920. doi: 10.1042/BST20050916. [DOI] [PubMed] [Google Scholar]
- 21.Harold FM, Baarda JR. Gramicidin, valinomycin, and cation permeability of Streptococcus faecalis. Journal of Bacteriology. 1967;94:53–60. doi: 10.1128/jb.94.1.53-60.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burkhart BM, Li N, Langs DA, Pangborn WA, Duax WL. The conducting form of gramicidin a is a right-handed double-stranded double helix. Proceedings of the National Academy of Sciences. 1998;95:12950–12955. doi: 10.1073/pnas.95.22.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen O, Apell H-J, Bamberg E, Busath D, Koeppe R, Sigworth F, Szabo G, Urry D, Woolley A. Gramicidin channel controversy – the structure in a lipid environment. Nat Struct Mol Biol. 1999;6:609–609. doi: 10.1038/10648. [DOI] [PubMed] [Google Scholar]
- 24.O’Connell A, Koeppe R, Andersen O. Kinetics of gramicidin channel formation in lipid bilayers: transmembrane monomer association. Science. 1990;250:1256–1259. doi: 10.1126/science.1700867. [DOI] [PubMed] [Google Scholar]
- 25.Killian J, von Heijne G. How proteins adapt to a membrane-water interface. Trends in Biochemical Sciences. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 26.Liebman PA, Parker KR, Dratz EA. The molecular mechanism of visual excitation and its relation to the structure and composition of the rod outer segment. Annual Review of Physiology. 1987;49:765–791. doi: 10.1146/annurev.ph.49.030187.004001. [DOI] [PubMed] [Google Scholar]
- 27.Molday RS. Photoreceptor membrane proteins, phototransduction, and retinal degenerative diseases. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1998;39:2491–2513. [PubMed] [Google Scholar]
- 28.Buzhynskyy N, Salesse C, Scheuring S. Rhodopsin is spatially heterogeneously distributed in rod outer segment disk membranes. Journal of Molecular Recognition. 2011;24:483–489. doi: 10.1002/jmr.1086. [DOI] [PubMed] [Google Scholar]
- 29.Boesze-Battaglia K, Albert AD. Phospholipid distribution among bovine rod outer segment plasma membrane and disk membranes. Exp Eye Res. 1992;54:821–823. doi: 10.1016/0014-4835(92)90040-y. [DOI] [PubMed] [Google Scholar]
- 30.Boesze-Battaglia K, Albert AD. Fatty acid composition of bovine rod outer segment plasma membrane. Exp Eye Res. 1989;49:699–701. doi: 10.1016/s0014-4835(89)80064-8. [DOI] [PubMed] [Google Scholar]
- 31.Boesze-Battaglia K, Hennessey T, Albert AD. Cholesterol heterogeneity in bovine rod outer segment disk membranes. J. Biol. Chem. 1989;264:8151–8155. [PMC free article] [PubMed] [Google Scholar]
- 32.Soubias O, Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim Biophys Acta. 2012;1818:234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drews J. Drug discovery: a historical perspective. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 34.Neuringer M. Infant vision and retinal function in studies of dietary long-chain polyunsaturated fatty acids: methods, results, and implications. Am J Clin Nutr. 2000;71:256S–267S. doi: 10.1093/ajcn/71.1.256S. [DOI] [PubMed] [Google Scholar]
- 35.Wiedmann TS, Pates RD, Beach JM, Salmon A, Brown MF. Lipid-protein interactions mediate the photochemical function of rhodopsin. Biochemistry. 1988;27:6469–6474. doi: 10.1021/bi00417a041. [DOI] [PubMed] [Google Scholar]
- 36.Gibson NJ, Brown MF. Lipid headgroup and acyl chain composition modulate the MI-MII equilibrium of rhodopsin in recombinant membranes. Biochemistry. 1993;32:2438–2454. doi: 10.1021/bi00060a040. [DOI] [PubMed] [Google Scholar]
- 37.Brown MF. Modulation of rhodopsin function by properties of the membrane bilayer. Chemistry And Physics Of Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 38.Botelho AV, Huber T, Sakmar TP, Brown MF. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys. J. 2006;91:4464–4477. doi: 10.1529/biophysj.106.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soubias O, Niu S-L, Mitchell DC, Gawrisch K. Lipid-rhodopsin hydrophobic mismatch alters rhodopsin helical content. J Am Chem Soc. 2008;130:12465–12471. doi: 10.1021/ja803599x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown MF. Influence of non-lamellar-forming lipids on rhodopsin. Curr. Top. Membr. 1997;44:285–356. [Google Scholar]
- 41.Delange F, Merkx M, Bovee-Geurts PHM, Pistorius AMA, Degrip WJ. Modulation of the Metarhodopsin I/Metarhodopsin II equilibrium of bovine rhodopsin by ionic strength. European Journal of Biochemistry. 1997;243:174–180. doi: 10.1111/j.1432-1033.1997.0174a.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Botelho AV, Martinez GV, Brown MF. Electrostatic properties of membrane lipids coupled to metarhodopsin II formation in visual transduction. J Am Chem Soc. 2002;124:7690–7701. doi: 10.1021/ja0200488. [DOI] [PubMed] [Google Scholar]
- 43.Botelho AV, Gibson NJ, Thurmond RJ, Wang Y, Brown MF. Conformational energetics of rhodopsin modulated by nonlamellar-forming lipids. Biochemistry. 2002;41:6354–6368. doi: 10.1021/bi011995g. [DOI] [PubMed] [Google Scholar]
- 44.Soubias O, Teague WE, Hines KG, Mitchell DC, Gawrisch K. Contribution of membrane elastic energy to rhodopsin function. Biophys J. 2010;99:817–824. doi: 10.1016/j.bpj.2010.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soubias O, Teague WE, Gawrisch K. Evidence for specificity in lipid-rhodopsin interactions. J Biol Chem. 2006;281:33233–33241. doi: 10.1074/jbc.M603059200. [DOI] [PubMed] [Google Scholar]
- 46.Niu SL, Mitchell DC, Litman BJ. Manipulation of cholesterol levels in rod disk membranes by methyl-β-cyclodextrin. J. Bio. Chem. 2002;277:20139–20145. doi: 10.1074/jbc.M200594200. [DOI] [PubMed] [Google Scholar]
- 47.Needham D, McIntosh TJ, Evans E. Thermomechanical and transition properties of dimyristoylphosphatidylcholine/cholesterol bilayers. Biochemistry. 1988;27:4668–4673. doi: 10.1021/bi00413a013. [DOI] [PubMed] [Google Scholar]
- 48.Chen Z, Rand R. The influence of cholesterol on phospholipid membrane curvature and bending elasticity. Biophysical Journal. 1997;73:267–276. doi: 10.1016/S0006-3495(97)78067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Albert AD, Young JE, Yeagle P. Rhodopsin-cholesterol interactions in bovine rod out segment disk membranes. Biochim. Biophys. Acta. 1996;1285:47–55. doi: 10.1016/s0005-2736(96)00145-9. [DOI] [PubMed] [Google Scholar]
- 50.Grossfield A, Feller SE, Pitman MC. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proc Natl Acad Sci U S A. 2006;103:4888–4893. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pitman MC, Grossfield A, Suits F, Feller SE. Role of cholesterol and polyunsaturated chains in lipid-protein interactions: molecular dynamics simulation of rhodopsin in a realistic membrane environment. J Am Chem Soc. 2005;127:4576–4577. doi: 10.1021/ja042715y. [DOI] [PubMed] [Google Scholar]
- 52.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BJ, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 53.Huber T, Rajamoorthi K, Kurze VF, Beyer K, Brown MF. Structure of docosahexaenoic acid-containing phospholipid bilayers as studied by 2H NMR and molecular dynamics simulation. J. Am. Chem. Soc. 2002;124:298–309. doi: 10.1021/ja011383j. [DOI] [PubMed] [Google Scholar]
- 54.Huber T, Botelho AV, Beyer K, Brown MF. Membrane model for the G-protein-coupled receptor rhodopsin: hydrophobic interface and dynamical structure. Biophys. J. 2004;86:2078–2100. doi: 10.1016/S0006-3495(04)74268-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feller SE, Gawrisch K, Woolf TB. Rhodopsin exhibits a preference for solvation by polyunsaturated docosohexaenoic acid. J Am Chem Soc. 2003;125:4434–4435. doi: 10.1021/ja0345874. [DOI] [PubMed] [Google Scholar]
- 56.Feller SE, Gawrisch K. Properties of docosahexaenoic acid-containing lipids and their influence on the function of the GPCR rhodopsin. Curr. Opin. Struct. Bio. 2005;15:416–422. doi: 10.1016/j.sbi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 57.Khelashvili G, Grossfield A, Feller SE, Pitman MC, Weinstein H. Structural and dynamic effects of cholesterol at preferred sites of interaction with rhodopsin identified from microsecond length molecular dynamics simulations. Proteins. 2009;76:403–417. doi: 10.1002/prot.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olausson BES, Grossfield A, Pitman MC, Brown MF, Feller SE, Vogel A. Molecular dynamics simulations reveal specific interactions of post-translational palmitoyl modifications with rhodopsin in membranes. J Am Chem Soc. 2012;134:4324–4331. doi: 10.1021/ja2108382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Periole X, Huber T, Marrink S-J, Sakmar TP. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. Journal of the American Chemical Society. 2007;129:10126–10132. doi: 10.1021/ja0706246. [DOI] [PubMed] [Google Scholar]
- 60.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 61.Monticelli L, Kandasamy S, Periole X, Larson R, Tieleman D, Marrink S. The MARTINI coarse grained forcefield: extension to proteins. J. Chem. Theo. Comp. 2008;4:819–839. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 62.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 angstrom crystal structure. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 63.Park JH, Scheerer P, Hofmann KP, Choe H-W, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 64.Periole X, Knepp AM, Sakmar TP, Marrink SJ, Huber T. Structural determinants of the supramolecular organization of g protein-coupled receptors in bilayers. J Am Chem Soc. 2012;134:10959–10965. doi: 10.1021/ja303286e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knepp AM, Periole X, Marrink S-J, Sakmar TP, Huber T. Rhodopsin forms a dimer with cytoplasmic helix 8 contacts in native membranes. Biochemistry. 2012;51:1819–1821. doi: 10.1021/bi3001598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MARTINI. 2011 http://md.chem.rug.nl/cgmartini/ [Google Scholar]
- 67.Kandt C, Ash WL, Tieleman DP. Setting up and running molecular dynamics simulations of membrane proteins. Methods. 2007;41:475–488. doi: 10.1016/j.ymeth.2006.08.006. Structural Biology of Membrane Proteins. [DOI] [PubMed] [Google Scholar]
- 68.van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 69.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 70.Winger M, Trzesniak D, Baron R, van Gunsteren WF. On using a too large integration time step in molecular dynamics simulations of coarse-grained molecular models. Phys Chem Chem Phys. 2009;11:1934–1941. doi: 10.1039/b818713d. [DOI] [PubMed] [Google Scholar]
- 71.Marrink SJ, Periole X, Tieleman DP, de Vries AH. Winger M, Trzesniak D, Baron R, van Gunsteren WF, editors. Comment on “on using a too large integration time step in molecular dynamics simulations of coarse-grained molecular models”. Phys. Chem. Chem. Phys. 2009;11:1934. doi: 10.1039/b818713d. Phys Chem Chem Phys 12 (2010) 2254–6; author reply 2257–8. [DOI] [PubMed] [Google Scholar]
- 72.Nosé S, Klein ML. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 73.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 74.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics. 1981;52:7182–7190. [Google Scholar]
- 75.Periole X, Cavalli M, Marrink S-J, Ceruso MA. Combining an elastic network with a coarse-grained molecular force field: Structure, dynamics, and intermolecular recognition. Journal of Chemical Theory and Computation. 2009;5:2531–2543. doi: 10.1021/ct9002114. [DOI] [PubMed] [Google Scholar]
- 76.Grossfield A, Pitman MC, Feller SE, Soubias O, Gawrisch K. Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. J Mol Biol. 2008;381:478–486. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Romo TD, Grossfield A. LOOS: an extensible platform for the structural analysis of simulations. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:2332–2335. doi: 10.1109/IEMBS.2009.5335065. [DOI] [PubMed] [Google Scholar]
- 78.Romo TD, Grossfield A. LOOS: A lightweight object-oriented software library. 2010 http://loos.sourceforge.net. [Google Scholar]
- 79.Jones E, Oliphant T, Peterson P, et al. SciPy: Open source scientific tools for Python. 2001 [Google Scholar]
- 80.Grossfield A, Feller SE, Pitman MC. Contribution of omega-3 fatty acids to the thermodynamics of membrane protein solvation. J Phys Chem B. 2006;110:8907–8909. doi: 10.1021/jp060405r. [DOI] [PubMed] [Google Scholar]
- 81.Feller SE, Gawrisch K, MacKerell AD., Jr Polyunsaturated fatty acids in lipid bilayers: Intrinsic and environmental contributions to their unique physical properties. Journal of the American Chemical Society. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 82.Soubias O, Gawrisch K. Docosahexaenoyl chains isomerize on the sub-nanosecond time scale. Journal of the American Chemical Society. 2007;129:6678–6679. doi: 10.1021/ja068856c. [DOI] [PubMed] [Google Scholar]
- 83.Lee JY, Lyman E. Predictions for cholesterol interaction sites on the A(2A) adenosine receptor. Journal of the American Chemical Society. 2012;134:16512–16515. doi: 10.1021/ja307532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Kuhn P, Weis WI, Kobilka BK, Stevens RC. Highresolution crystal structure of an engineered human beta2-adrenergic G proteincoupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola V-P, Chien EY, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 angstrom structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, IJzerman AP, Cherezov V, Stevens RC. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lyman E, Higgs C, Kim B, Lupyan D, Shelley JC, Farid R, Voth GA. A role for a specific cholesterol interaction in stabilizing the apo configuration of the human A(2A) adenosine receptor. Structure. 2009;17:1660–1668. doi: 10.1016/j.str.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]